
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Earth Sci., 27 January 2022
Sec. Paleontology
Volume 9 - 2021 | https://doi.org/10.3389/feart.2021.805271
This article is part of the Research TopicTechnological Frontiers in Dinosaur Science Mark a New Age of Opportunity for Early Career ResearchersView all 9 articles
Recently, deep learning has reached significant advancements in various image-related tasks, particularly in medical sciences. Deep neural networks have been used to facilitate diagnosing medical images generated from various observation techniques including CT (computed tomography) scans. As a non-destructive 3D imaging technique, CT scan has also been widely used in paleontological research, which provides the solid foundation for taxon identification, comparative anatomy, functional morphology, etc. However, the labeling and segmentation of CT images are often laborious, prone to error, and subject to researchers own judgements. It is essential to set a benchmark in CT imaging processing of fossils and reduce the time cost from manual processing. Since fossils from the same localities usually share similar sedimentary environments, we constructed a dataset comprising CT slices of protoceratopsian dinosaurs from the Gobi Desert, Mongolia. Here we tested the fossil segmentation performances of U-net, a classic deep neural network for image segmentation, and constructed a modified DeepLab v3+ network, which included MobileNet v1 as feature extractor and practiced an atrous convolutional method that can capture features from various scales. The results show that deep neural network can efficiently segment protoceratopsian dinosaur fossils, which can save significant time from current manual segmentation. But further test on a dataset generated by other vertebrate fossils, even from similar localities, is largely limited.
Vertebrate paleontology is based on fossils that are remains of ancient organisms. Because fossils usually do not preserve molecular or behavior information, paleontologists mainly focus on their morphology, which not only includes exterior features but also interior structures like brain endocasts and inner ears. Traditionally, researchers used destructive thin sectioning to reveal interior structures, which totally destroys the fossils. With the application of non-destructive 3D imaging techniques, like CT (computed tomography) scan and synchrotron radiation scanning, paleontologists can observe and interact with previously hidden structures without making damages. CT scan and other non-destructive imaging techniques have greatly facilitated the development of vertebrate paleontology not only in revealing hidden structures but also providing 3D models for teaching and exhibition.
Recently, CT scans have been successfully applied to various branches in vertebrate paleontology (Figures 1A–E), for example the studies of archosaur brain endocasts (Watanabe et al., 2019), mammalian inner ears (Luo and Ketten 1991), theropod dinosaur body mass estimation (Allen et al., 2013), finite element analysis of the Allosaurus skull (Rayfield et al., 2001), and horned dinosaur tooth replacement patterns (He et al., 2018). Figure 1F illustrates a general workflow in fossil CT scan processing from original fossil to unsegmented volumes, then to individually segmented bones. For fragile specimens that cannot be mechanically prepared, CT scan (or other non-invasive scanning techniques) is the only way to make detailed observations. Such scanners are now common elements in many paleontological laboratories. However, processing the data generated by CT and other observational equipment is both arduous and time-consuming.

FIGURE 1. Application of CT scans in vertebrate paleontology. (A) mammal inner ear structure reconstruction, from Luo and Ketten (1991); (B) archosaur brain evolution, from Watanabe et al. (2019); (C) Allosaurus skull mechanic analysis, from Rayfield et al. (2001); (D) Tooth replacement in Liaoceratops, from He et al. (2018); (E) dinosaur body mass estimation, from Allen et al. (2013); (F) generalized workflow of CT data processing in vertebrate paleontology, from above: original fossils, raw 3D rendering, and segmented bone volumes.
Current CT data processing is laborious, especially for morphologically complex objects like human bodies and vertebrate fossils. CT scanners differentiate volumes by their absorptions of X-ray radiation, which is primarily controlled by their densities. However, fossils and their surrounding matrices are often similar or even identical in densities (Figures 2B,C), making the differentiation between matrix and specimen difficult. Many researchers have to rely on manual segmentation, sometimes slice by slice, according to their own judgment and understanding of anatomy. One of the major flaws in current CT processing is the lack of reproducibility. Most reconstruction have to be based on anatomical knowledge and professional experience.
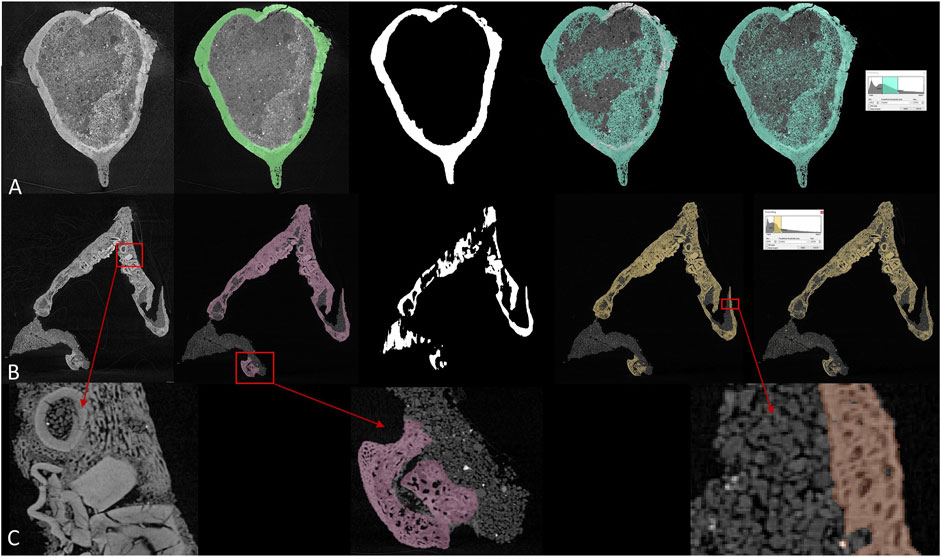
FIGURE 2. Comparison between different segmentation methods in (A) brain endocast of Protoceratops and (B) toothed lower jaw of Protoceratops, from left to right: CT slice, manual segmentation, U-net, region growing, and thresholding. (C) detailed structure in toothed area in dentary and fossil-rock boundaries.
Most paleontological CT data are processed in commercial software such as VGstudio by Volume Graphics and Mimics by Materialise. There is also a handful of opensource software (e.g., ImageJ developed by the National Institute of Health, United States) but most of them provide users with less functionality comparing to commercial ones. A lot of 3D image processing software offers thresholding and region growing functions for more efficient segmentation. Thresholding differentiates voxels according to their grey values, therefore it is functionless when ROI (region of interest) and surrounding areas are similar in density. Region growing requires a seed voxel to grow from in given directions following a grey value range, which means all connected voxels within a given grey value range will be labeled. Thresholding and region growing work best when there is significant contrast between ROI and surrounding areas, and their boundaries can be clearly defined. Although current software can process multiple slices at the same time based on linear interpolation, it is often insufficient for more rapid and accurate segmentation, especially of intricately preserved fossils. In CT data processing, if there are significant grey value differences (which represent densities) between target elements (bony fossils, stained soft tissue, and cavity structures) and undesired elements (i.e., rock matrices or surrounding tissues), it is easy to segment. But due to the complex taphonomy, fragmentary preservation, similarity between rocks and embedded fossils, CT data processing is usually exhausting. Figures 2A,B show two example slices of fossil protoceratopsians segmented by manual labelling, U-net, region growing, and thresholding. While manual labelling outperforms all other methods, it costs much more time and labor especially facing high-resolution scans, and either thresholding or region growing cannot properly segment the desired regions therefore requires subsequent manual processing. Details of segmentation maps in Figure 2C indicates the complexity (e.g. tooth surrounded by dentary bone matrix) in fossil CT scans. In conclusion, there exist three major drawbacks in current 3D imaging processing:
1. Manual processing is necessary when ROI and surrounding areas are similar in densities or their boundaries are ambiguous.
2. Only linear interpolation (i.e., thresholding and region growing) are applied to track density and morphology changes.
3. There lacks a consistent standard and enough validation for annotation.
Over the last decade, deep learning has shown incredible potential when applied in complicated tasks including image processing. Since 2013, automated processing (classification, object detection, and segmentation) of medical images has utilized by deep learning (Ker et al., 2017; Litjens et al., 2017; Shen et al., 2017; Cardoso et al., 2017; Roth et al., 2018; Ting et al., 2018). Many image-related tasks in medical sciences can be effectively accomplished by deep learning algorithms such as brain lesion segmentation (Kamnitsas et al., 2017), nodule classification (Ciompi et al., 2017), skin cancer detection (Esteva et al., 2017), etc. Considering the similarity between medical and vertebrate paleontological CT images, it is natural to apply deep learning to fossil data. However, there are three major differences between fossil and medical CT scans:
1. To reduce the harm to patients, medical scans are usually restricted to relatively low energy and short exposure time. Therefore, they cannot produce as much contrast as the higher energy beams used for fossils. Also, paleontological scan data usually have higher resolutions, thus requires greater computational capability.
2. Most fossils are deformed and fragmentary. Many image-related tasks in medical sciences focus on morphologically alike structures, for example hearts, livers, and lungs.
3. Paleontological scans generally have much small sample sizes in comparison to medical scans because of the uniqueness of fossil specimens.
In this study, we generate a dataset from the CT-scans of three well-preserved protoceratopsian skulls (Ornithischia, Dinosauria) from Gobi Desert, Mongolia, and manually annotated bone structures in each slice for subsequent analysis. We test the performance of classic U-net (Ronneberger et al., 2015) and DeepLab v3+ (Chen et al., 2016) with various modifications (Figure 3).
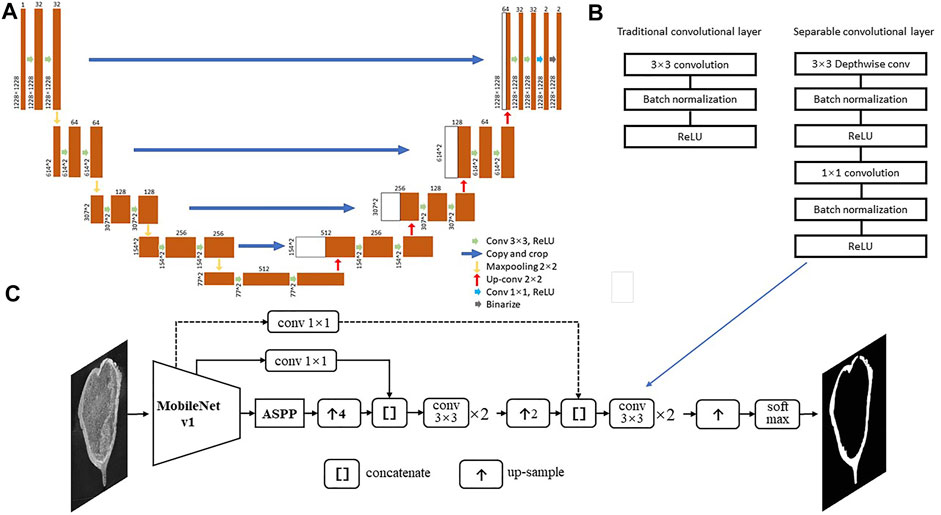
FIGURE 3. Deep neural network structures used in this study. (A) U-net from Ronneberger et al. (2015). (B) Separable convolutional layers; (C) Deeplab v3+ with MobileNet v1 as feature extractor, dash line indicates the connection may be skipped in certain models.
Three protoceratopsian skulls, in which two (IGM 100/3654 and IGM 100/3655) were collected during the 1992 AMNH-Mongolian Academy of Sciences expeditions in Tugrugeen Shireh, Mongolia and IGM 100/1021 was collected in Ukhaa Tolgod, Mongolia, were CT scanned at the Department of Earth & Planetary Sciences, Yale University, United States. Specimen IGM 100/3654 and IGM 100/3655 are two nearly complete and undeformed skulls identified as embryonic elements due to their smallest sizes (less than 4 cm in lateral length) among all discovered protoceratopsian dinosaurs, ossification status, and morphology differences from later developmental stages. They are smaller and less ossified than IGM 100/1021 (a known embryo), which is a dorsal-ventrally deformed skull but preserves most facial elements.
Scanning detail of three protoceratopsian specimens are shown in Table 1. These specimens were manually segmented and cross validated in all three directions (axial, sagittal, and coronal) by authors in Mimics 19.0 (Materialise, Belgium). The details of datasets are shown in Table 1. Then, manually annotated masks as well as associated raw slices were exported correspondingly. Before preparing the dataset, slices containing too little information (e.g., slices close to the boundaries) were excluded. In total 7986 images are prepared as the training dataset and 3329 images as the testing dataset.
In this study, we first test the performance of classic U-net for image segmentation (Ronneberger et al., 2015, Figure 3A), and then modified DeepLab v3+ (Chen et al., 2016). We use MobileNet v1 (Howard et al., 2017) or ResNet v2 (He et al., 2016) in the encoder, which introduces separable convolutional layers with depthwise and pointwise layers followed by BatchNorm and ReLU layer, respectively (Figure 3B&C), thus to reduce the computational cost and number of parameters but keep most of the performance. The skip connections between Astrous Spatial Pyramid Pooling (ASPP) and decoders allow the recognition of features in different scale, thus to better capture patterns that distinguishing fossils and rock matrices. The Deeplab v3+ model structures are shown in Figure 3C and Table 2.
Part of the segmentation results are shown in Figure 4. Classic U-net model reached a high accuracy of 98.09 and 97.44% in test and validation dataset, however, the cross-entropy loss for both datasets are relatively considerable of 4.94 and 9.92%, respectively. Segmentation results from varieties of modified DeepLab v3+ reaches mean dice between 0.738 and 0.894, and mean IOU (intersection over union) between 0.612 and 0.817. The Sørensen–Dice coefficient and IOU scores are calculated as following:
Therefore,
where TP represents true positive, FP false positive, and FN false negative, respectively. The DeepLab v3+ model with MobileNet v1 as feature extractor and two skip connections outperformed other models. The prediction maps are shown in Figure 4B.
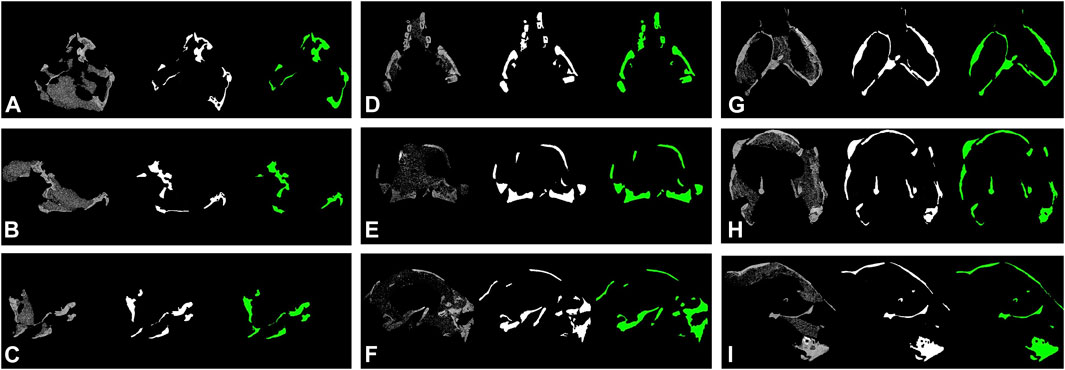
FIGURE 4. Segmentation results of protoceratopsian dinosaurs by DeepLab v3+ with MobileNet v1 as feature extractor. In each subfigure, from left to right: original CT slice, groundtruth, and prediction by models. (A–C). IGM 100/1021; (D–F). IGM 100/3654; (G–I), IGM 100/3655.
The fast advancements in techniques not only enable unprecedented resolution in observation of fossil material, but also increase the cost in data processing. Currently paleontologists are spending days to weeks in segmenting fossil scans, the introduction of deep learning can reduce that time to minutes. Although the 3D renderings from automated segmented slices are not as meticulous as manual results (Figure 5), the comparison between raw reconstruction and deep learning segmented volumes has shown that deep learning can at least reduce processing time in differentiating fossils and rock matrices.

FIGURE 5. Comparison of different 3D renderings. From left: raw reconstruction (after thresholding), manual segmentation, and deep learning segmentation (by Deeplab v3+MobileNet v1) (A) IGM 100/1021 (B) IGM 100/3654 (C) IGM 100/3655.
To further test the generalization performance of deep neural network, we use the trained best-performed Deeplab v3+ network to segment other vertebrate fossils from the Gobi Desert, Mongolia with similar sedimentary environments, which should reduce the bias from sampling to the minimum. The new dataset (see supplementary material) including CT-scanned fossil slices of ornithischian dinosaur Haya (IGM 100-3178, IGM 100-3181) and Pinacosaurus (IGM 100-3186). However, these non-protoceratopsian fossil images are extremely poorly segmented (Figure 6). Although both U-net and modified DeepLab v3+ shows close to manual segmentation performance in at least part of the protoceratopsian dataset, the capability of extrapolation on other datasets seems to be largely limited. If we are applying deep learning in future paleontological studies (e.g., facilitate CT segmentation), generalization performance is a must. The Gobi Desert in Mongolia has yielded a wide range of both vertebrate and invertebrate fossils, it cannot be expected to have a training dataset comprising fossil data from all known taxa. On the other hand, since our training dataset is solely derived from protoceratopsian dinosaurs, more specifically from specimens in early developmental stages, it is likely to have intrinsic bias in the dataset due to the poor ossification status of bones and other possible factors. Such overfitting problem is a major concern in deep learning studies. Comparing to medical applications that focus on a particular structure (e.g., brain lesion Kamnitsas et al., 2017) mentioned before, fossil segmentation tasks in paleontological studies seem to be more non-specific, therefore, the models constructed in this study may improve by feeding more generalized training data, possibly covering more taxa and developmental stages. It should be also noted that in this study, we only performed semantic segmentation on fossil bone without distinguishing different structures, but the grey value patterns are obviously varied across different tissues like teeth and dentary bones (Figure 2C).
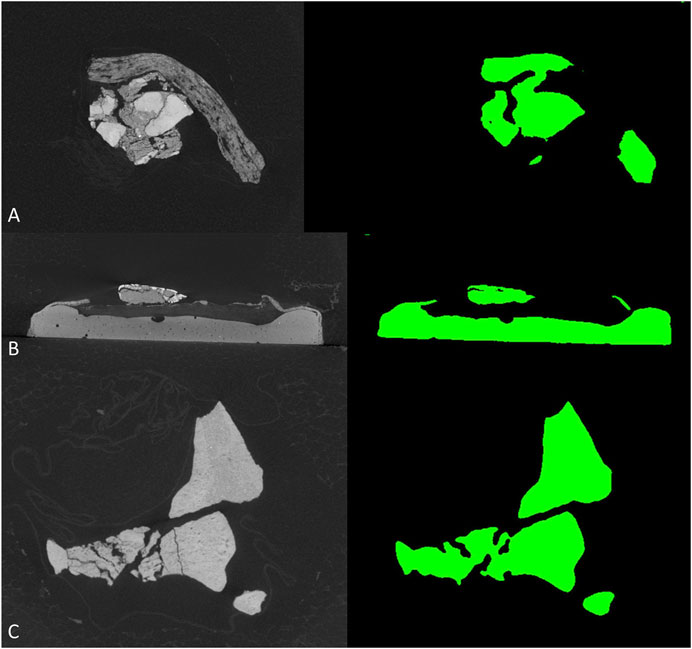
FIGURE 6. Segmentation results of non-protoceratopsian dinosaurs by DeepLab v3+ with MobileNet v1 as feature extractor. (A). Haya, IGM 100/3178; (B). Haya, IGM 100/3181; (C). Pinacosaurus, IGM 100/3186.
Deep learning has already shown its power in many research branches such as Go game playing, medical imaging processing, and earth science studies (Silver et al., 2016; Esteva et al., 2017; Litjens et al., 2017; Reichstein et al., 2019; Silver et al., 2017; Bergen et al., 2019). The recent advancements in observation techniques and increasing available fossil specimens facilitate paleontological data growing in an exponential manner, which enable large-scale data-driven paleontological and earth science project, for example the Deep-time Digital Earth (DDE, Wang et al., 2021). There have been only few studies introducing deep learning in the field of paleontology until recently, for example the segmentation of fish fossil bones (Hou et al., 2020; Hou et al., 2021), planktonic foraminiferal species identification (Hsiang et al., 2019), and automated pollen recognition (Bourel et al., 2020). The studied materials cover plants, invertebrates, and vertebrates distributed in different ages, indicating the potential to apply deep learning in various branches of paleontological studies. Including this study, most of paleontological deep learning studies focus on faster and automated imaging classification and segmentation, other potential application scenarios in paleontology are less investigated, for example digitally reconstruction of incomplete fossils based on GAN (generative adversarial network) and data mining from published literatures to reconstruct macro evolutionary patterns. With more available paleontological and maturation of algorithms, we shall expect wider applications of deep learning in paleontological studies soon.
The datasets presented in this study can be found in online repositories at https://doi.org/10.5061/dryad.k6djh9w7w.
CY design the project and prepare the datasets, CY, FQ, YL and ZQ proceeded the analysis. Everyone put efforts in preparing the manuscript.
CY and MN are funded by the Newt and Calista Gingrich Endowment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the 1992 Mongolia-American Museum of Natural History Expedition crews for collecting those Protoceratops fossils and Bhart-Anjan Bhullar in Yale University for providing CT-scans. We thank two reviewers for their comments and suggestions on improving this study.
Allen, V., Bates, K. T., Li, Z., and Hutchinson, J. R. (2013). Linking the Evolution of Body Shape and Locomotor Biomechanics in Bird-Line Archosaurs. Nature 497, 104–107. doi:10.1038/nature12059
Bergen, K. J., Johnson, P. A., de Hoop, M. V., and Beroza, G. C. (2019). Machine Learning for Data-Driven Discovery in Solid Earth Geoscience. Science 363. doi:10.1126/science.aau0323
Bourel, B., Marchant, R., de Garidel-Thoron, T., Tetard, M., Barboni, D., Gally, Y., et al. (2020). Automated Recognition by Multiple Convolutional Neural Networks of Modern, Fossil, Intact and Damaged Pollen Grains. Comput. Geosciences 140, 104498. doi:10.1016/j.cageo.2020.104498
Cardoso, M. J., Arbel, T., Carneiro, G., Syeda-Mahmood, T., Tavares, J. M. R. S., Moradi, M., et al. (2017). “Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support,” in Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, September 14, Proceedings (Québec City, QC: Springer), 10553.
Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K., and Yuille, A. L. (2016). Semantic Image Segmentation with Deep Convolutional Nets and Fully Connected CRFs. arXiv:1412.7062v4 [cs].
Ciompi, F., Geessink, O., Bejnordi, B. E., de Souza, G. S., Baidoshvili, A., Litjens, G., et al. (2017). “The Importance of Stain Normalization in Colorectal Tissue Classification with Convolutional Networks,” in Proceeding of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI), Melbourne, VIC, Australia, 18-21 April 2017 (IEEE), 160–163. doi:10.1109/isbi.2017.7950492
Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M., Blau, H. M., et al. (2017). Dermatologist-level Classification of Skin Cancer with Deep Neural Networks. Nature 542, 115–118. doi:10.1038/nature21056
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Identity Mappings in Deep Residual Networks. Lecture Notes in Computer Science, Editor B. Leibe, J. Matas, N. Sebe, and M. Welling 9908 (Amsterdam, The Netherlands: Springer, Cham) arXiv:1603.05027 [cs]. doi:10.1007/978-3-319-46493-0_38
He, Y., Makovicky, P. J., Xu, X., and You, H. (2018). High-resolution Computed Tomographic Analysis of Tooth Replacement Pattern of the Basal Neoceratopsian Liaoceratops Yanzigouensis Informs Ceratopsian Dental Evolution. Sci. Rep. 8, 1–15. doi:10.1038/s41598-018-24283-5
Hou, Y., Canul-Ku, M., Cui, X., Hasimoto-Beltran, R., and Zhu, M. (2021). Semantic Segmentation of Vertebrate Microfossils from Computed Tomography Data Using a Deep Learning Approach. J. Micropalaeontol. 40, 163–173. doi:10.5194/jm-40-163-2021
Hou, Y., Cui, X., Canul-Ku, M., Jin, S., Hasimoto-Beltran, R., Guo, Q., et al. (2020). ADMorph: A 3D Digital Microfossil Morphology Dataset for Deep Learning. IEEE Access 8, 148744–148756. doi:10.1109/access.2020.3016267
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D., Wang, W., Weyand, T., et al. (2017). MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv:1704.04861 [cs].
Hsiang, A. Y., Brombacher, A., Rillo, M. C., Mleneck‐Vautravers, M. J., Conn, S., Lordsmith, S., et al. (2019). Endless Forams: >34,000 Modern Planktonic Foraminiferal Images for Taxonomic Training and Automated Species Recognition Using Convolutional Neural Networks. Paleoceanography and Paleoclimatology 34, 1157–1177. doi:10.1029/2019pa003612
Kamnitsas, K., Baumgartner, C., Ledig, C., Newcombe, V., Simpson, J., Kane, A., et al. (2017). “Unsupervised Domain Adaptation in Brain Lesion Segmentation with Adversarial Networks,” in International Conference on Information Processing in Medical Imaging (Boone, USA: Springer), 597–609. doi:10.1007/978-3-319-59050-9_47
Ker, J., Wang, L., Rao, J., and Lim, T. (2017). Deep Learning Applications in Medical Image Analysis. Ieee Access 6, 9375–9389. doi:10.1109/ACCESS.2017.2788044
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M., et al. (2017). A Survey on Deep Learning in Medical Image Analysis. Med. image Anal. 42, 60–88. doi:10.1016/j.media.2017.07.005
Luo, Z., and Ketten, D. (1991). CT Scanning and Computerized Reconstructions of the Inner Ear of Multituberculate Mammals. J. Vertebr. Paleontol. 11, 220–228. doi:10.1080/02724634.1991.10011389
Rayfield, E. J., Norman, D. B., Horner, C. C., Horner, J. R., Smith, P. M., Thomason, J. J., et al. (2001). Cranial Design and Function in a Large Theropod dinosaur. Nature 409, 1033–1037. doi:10.1038/35059070
Reichstein, M., Camps-Valls, G., Stevens, B., Jung, M., Denzler, J., Carvalhais, N., et al. (2019). Deep Learning and Process Understanding for Data-Driven Earth System Science. Nature 566, 195–204. doi:10.1038/s41586-019-0912-1
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-net: Convolutional Networks for Biomedical Image Segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Munich, Germany: Springer), 234–241. doi:10.1007/978-3-319-24574-4_28
Roth, H. R., Shen, C., Oda, H., Oda, M., Hayashi, Y., Misawa, K., et al. (2018). Deep Learning and its Application to Medical Image Segmentation. Med. Imaging Technology 36, 63–71. doi:10.11409/mit.36.63
Shen, D., Wu, G., and Suk, H.-I. (2017). Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 19, 221–248. doi:10.1146/annurev-bioeng-071516-044442
Silver, D., Huang, A., Maddison, C. J., Guez, A., Sifre, L., van den Driessche, G., et al. (2016). Mastering the Game of Go with Deep Neural Networks and Tree Search. Nature 529, 484–489. doi:10.1038/nature16961
Silver, D., Schrittwieser, J., Simonyan, K., Antonoglou, I., Huang, A., Guez, A., et al. (2017). Mastering the Game of Go without Human Knowledge. Nature 550, 354–359. doi:10.1038/nature24270
Ting, D. S. W., Liu, Y., Burlina, P., Xu, X., Bressler, N. M., and Wong, T. Y. (2018). AI for Medical Imaging Goes Deep. Nat. Med. 24, 539–540. doi:10.1038/s41591-018-0029-3
Wang, C., Hazen, R. M., Cheng, Q., Stephenson, M. H., Zhou, C., Fox, P., et al. (2021). The Deep-Time Digital Earth Program: Data-Driven Discovery in Geosciences. Natl. Sci. Rev. 8 (9), nwab027. doi:10.1093/nsr/nwab027
Keywords: deep learning, CT, segmentation, fossil, dinosaur
Citation: Yu C, Qin F, Li Y, Qin Z and Norell M (2022) CT Segmentation of Dinosaur Fossils by Deep Learning. Front. Earth Sci. 9:805271. doi: 10.3389/feart.2021.805271
Received: 30 October 2021; Accepted: 29 December 2021;
Published: 27 January 2022.
Edited by:
Matteo Belvedere, Università degli Studi di Firenze, ItalyCopyright © 2022 Yu, Qin, Li, Qin and Norell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Congyu Yu, Y3l1QGFtbmgub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.