- 1Instituto de Recursos Naturales y Agrobiologia, IRNAS-CSIC, Sevilla, Spain
- 2Laboratorio HERCULES, Universidade de Evora, Evora, Portugal
- 3Department of Biological, Geological and Environmental Sciences, Italian Institute of Speleology, Bologna University, Bologna, Italy
- 4Gruppo Grotte Milano CAI-SEM, Federazione Speleologica Lombarda, Milano, Italy
Morgana Cave is located in Val di Scerscen, Central Italian Alps. The cave opens at an altitude of 2,600 m a.s.l. close to the retreating glacier Vedretto di Scerscen, and its entrance was discovered 30 years ago hidden underneath the glacier. A characteristic of this cave is the occurrence of vermiculation deposits on the walls and ceiling. In general, the composition of the microbial communities in cave vermiculations is relatively unknown and rarely investigated. Here we present the data of a geomicrobiological study of vermiculations from an Alpine cave subjected to extreme climate conditions. The microbial communities were dominated by 13 main phyla of Bacteria, and contained a negligible percentage (<1%) of Archaea. The two major bacterial classes were Gammaproteobacteria and Betaproteobacteria, whose metabolic traits were mainly associated with the nitrogen cycle. In addition, psychrophilic and methanotrophic bacterial groups were identified. The occurrence of a large number of uncultured members, at the lowest taxonomic ranks, indicated the presence of still unexplored microbial taxa in the vermiculations.
Introduction
Global warming is causing a rapid glacial retreat. The total glacier-covered surface area in the Alps decreased from an estimated 4,500 km2 around 1850 to around 2,200 km2 in 2001, 1,800 km2 in 2010 (Haeberli et al., 2013) and 1,650 km2 in 2015 (Sommer et al., 2020).
The glacier Vedretta di Scerscen is located in Val di Scerscen, Central Italian Alps (Northern Lombardy), in the scenic setting of Valmalenco (Figures 1A,G). The glacier retreat has uncovered a few caves, the most important of which, opening at about 2,600 m a.s.l., are: Veronica Cave (638 m long), Morgana Cave (770 m), and Tana dei Marsooi (77 m). These caves represent very interesting systems from both pure geological-geomorphological and microbiological standpoints (De Waele et al., 2014).

FIGURE 1. Morgana Cave. (A) Panoramic view of the caves location; (B) Entrance partially covered by snow; (C) Phreatic morphologies; (D) Phreatic and vadose morphologies (sampling site M1); (E) Glacial sediments; (F) Phreatic and condensation-corrosion features; (G) Planimetry and cross-section of the cave. The six sampling sites from the innermost M1 to the outermost zone M5-M6 have been indicated on the map. Photos (A) and (B) by A. Granati, photos (C–F) by M. Inglese. Planimetry from Gruppo Grotte Milano.
Morgana Cave was hidden underneath the rapidly retreating glacier until the end of the last century (Figure 1B). In the Morgana Cave area, snow and ice remain until the beginning of June. At the sampling time (July 17th, 2017) the entrance of the cave was open, but there was still snow around and above the cave. The cave had very low temperatures, slightly above 0°C, with freezing occurring close to the entrance, and rather stable conditions occurring deeper into the cave (reaching 2°C 50 m inward).
Morgana Cave is characterized by phreatic morphologies (Figures 1C,D) and contains glacial sediments (Figure 1E). The cave probably formed before the last ice age. The entrance area of Morgana cave and many smaller nearby caves were all apparently cut both by valley slope retreat and ice scouring. The caves are characterized by shallow vadose entrenchments (Figure 1D) which record a later phase of free-running meltwaters in the cave passages. Air circulation triggered by the temperature difference between the cave interior and exterior induces condensation-corrosion processes, which are actually shaping the cave wall and ceiling morphologies (Figure 1F), with smooth reliefs, notches and cupola (D’Angeli et al., 2018).
Vermiculation deposits are among the most characteristic features of Morgana Cave. Vermiculation patterns developing on wall surfaces have been found in many caves around the world (Hose and Macalady, 2006; Merino et al., 2014; Bojar et al., 2015; Faucher and Lauriol, 2016; D’Angeli et al., 2017, 2019a, 2019b; Addesso et al., 2020).
Cave vermiculations are thin, irregular and discontinuous deposits of incoherent particles. Generally, they exhibit several kinds of morphologies (i.e. dots, dendritic, hieroglyphic), colors (red, brown, grey or white) and dimensions (De Waele et al., 2014; Addesso et al., 2020). According to Bini et al. (1978) vermiculations are mainly composed of clays and are the consequence of climatic and environmental conditions, including condensation-evaporation processes. However, other authors reported a different type of vermiculations, the so-called biovermiculations with complex geometric forms resembling clay vermiculations, but lacking significant clay content. These vermiculations are typical of sulfuric acid cave systems in the United States, Mexico, and Italy (Hose et al., 2000; Hose and Macalady, 2006; D’Angeli et al., 2017; D’Angeli et al., 2019a).
According to Bojar et al. (2015) the formation of vermiculation patterns is driven by evaporation and water film rupture of particle aggregates. The rheology of the clayey sediments of vermiculations and the chemistry of the fluids soaking the deposits appear to be important drivers in the formation of the patterns (Freydier et al., 2019). Boston (2016) recognized biovermiculation patterns, observed in limestone caves, lava tubes, mines, etc., as universal signatures of extant and extinct life in which the mathematical similarity of the patterns are the morphological expression of behavior in response to ecological drivers.
Three papers dealing with the microbiological characterization of vermiculations are worth mentioning. In the first one, Jones et al. (2008) investigated the biovermiculations from the Frasassi Cave system, Italy, and reported that the community was diverse with at least 15 major bacterial lineages, mainly Betaproteobacteria, Gammaproteobacteria, Acidobacteria, Nitrospirae, and Planctomycetes. These authors identified potential sulfur- and nitrite-oxidizing bacteria, in addition to auto- and heterotrophic microorganisms in the biovermiculations.
D’Angeli et al. (2019b) carried out an extensive geomicrobiological study on the vermiculations from Fetida Cave, Santa Cesarea Terme, Italy, an active sulfuric acid cave influenced by seawater. These vermiculations contained 18 phyla, including autotrophic taxa associated with sulfur and nitrogen cycles and biomineralization processes. The bacterial community was composed of Proteobacteria (including Alpha-, Beta-, Gamma-, Delta-, Epsilonproteobacteria), Planctomycetes, Acidobacteria, Chloroflexi, Bacteroidetes, Actinobacteria, and Nitrospirae in decreasing order of abundance. Archaea constituted <4.3% of the total community and were dominated by members of the phylum Thaumarchaeota.
Adesso et al. (2020) found that the most abundant phylum in vermiculations from the Italian Pertosa-Auletta Cave was Proteobacteria, followed by Acidobacteria, Actinobacteria, Nitrospirae, Firmicutes, Planctomycetes, Chloroflexi, Gemmatimonadetes, Bacteroidetes and Latescibacteria. Less-represented taxonomic groups (<1%), as well as unclassified ones, were also detected.
Recently uncovered caves located close to retreating glaciers are ideal places to study microbial life in cold ecosystems. The geomicrobiology of Alpine caves and their vermiculations remains largely understudied due to the remoteness and difficult access to the caves, as most of them are far away from mountain paths. In addition, sampling in high mountain caves is not an easy task due to severe working conditions. Opportunities to obtain samples are rare, and the samples are small and hard to collect due to their mineral composition. The small sample size sometimes makes it difficult to perform a complete microbiological analysis.
Vermiculations, being aggregates of weathered minerals and clays, represent an interesting and suitable niche for microorganisms. In addition, organic compounds are largely adsorbed onto the clays. Clays also increase water retention with respect to the host rock, facilitating colonization and thriving of microorganisms. Therefore, vermiculation deposits are ideal sites for studying the diversity of cold-adapted microbial communities in Morgana Cave.
Here we report the microbial community of different vermiculation deposits (Figure 2) collected on the walls of Morgana Cave, in an attempt to expand the understanding on the geomicrobiology of Alpine caves.
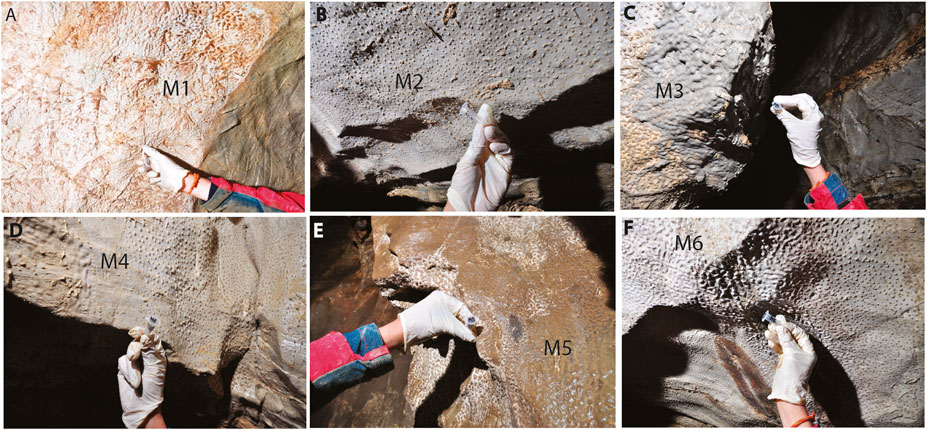
FIGURE 2. (A-F) Vermiculation deposits (from M1 to M6) collected from the walls of Morgana Cave. They present different shapes and colors such as whitish, brown, and greyish (photos by M. Inglese).
Materials and Methods
Sampling
Morgana Cave (44° 08′ 46.6″ N, 8° 09′ 31.7″ E, WGS84), was visited on the July 17–18th 2017. Six samples of vermiculations were collected in different passages of the cave (from the innermost part M1 towards the outermost zone of the cave in M5-6, as shown in Figure 1), using sterilized scalpels.
Sample M1 was composed of whitish-grey vermiculations close to a flowing water body, M2 of whitish vermiculations (small dots), M3 of dense grey vermiculations, M4 of whitish vermiculations (dots), M5 of dense brown vermiculations and M6 of white vermiculations (elongated) (Figure 2).
The samples, stored in sterile containers, were maintained at 5°C until laboratory arrival and those used for field emission scanning electron microscopy (FESEM) were fixed in situ. The samples for molecular biology were preserved in RNAlater and then conserved at −80°C in the laboratory.
Vermiculations showed different colors (whitish, greyish, brownish); the whitish are dominated by dolomite clasts, whereas clay minerals have been solely observed in the greyish and brownish deposits.
Field Emission Scanning Electron Microscopy
The morphology of the different vermiculations was studied by field emission scanning electron microscopy (FESEM) combined with energy dispersive X-ray spectroscopy (EDS). Previously, samples were dried at 50°C for 24 h and directly mounted on sample stubs, sputter coated with gold, and subsequently examined in a FEI Teneo FESEM equipped with an EDS detector. FESEM examinations were operated using the secondary electron (SE) detection mode (Everhart-Thomley) with an acceleration voltage of 5 kV for ultra-high resolution images and 10 kV for elemental microanalysis.
Nucleic Acid Extraction
Genomic DNA and RNA were extracted from 300 mg of samples using the DNeasy PowerBiofilm DNA and RNA isolation kits according to manufacturer’s protocols (Qiagen) and quantified using a Qubit 2.0 fluorometer (Invitrogen). DNA and RNA were stored at −80°C until their use.
Genomic DNA and RNA were extracted from all samples. Only DNA from samples M2, M3, M4, M5, and M6 reached sufficient concentration (>10 ng/μl) for next generation sequencing. RNA concentrations were very low, <0.2 ng/μl, insufficient for sequencing.
Next-Generation Sequencing and Data Analysis
The extracted DNA of all samples (except M1) was analyzed by next generation sequencing (NGS). We investigated the bacterial V3 and V4 regions of the 16S SSU rRNA gene using Illumina MiSeq and 2 × 250 paired end sequencing, according to the Illumina 16S metagenomic library preparation protocol used by Macrogen (Seoul, Korea).
The primer sequences used in this study were 341F (CCTACGGGNGGCWGCAG) and 805R (GACTACHVGGGTATCTAATCC).Takahashi et al. (2014) showed that this prokaryotic universal primer for simultaneous analysis had a coverage rate of 98% for Bacteria and 94.6% for Archaea. Raw data were processed in QIIME 1.9.1 (Caporaso et al., 2010). Quality control and trimming were performed using FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Trimmomatic (0.36 version), respectively. Paired end reads were assembled using PEAR (Zhang et al., 2014). Primers removal was carried out using a PERL script developed by the Environmental Microbiology group (IRNAS-CSIC). Operational Taxonomic Units (OTUs) were clustered at 97% cutoff using UCLUST (Edgar, 2010). The SILVA database (version 132) was used to assign the taxonomic classification of each 16S rRNA gene sequence with a threshold of 80% (Quast et al., 2013). The alpha_diversity.py command was employed for alpha diversity measurements.
Amplification of Functional Genes and Clone Library Construction
Amplification of functional genes from genomic DNA, nifH (as a measure of N-fixing bacteria) and amoA (as a measure of ammonia-oxidizing archaea and bacteria) were previously described by Mao et al. (2011). We used primers PolF (5′-TGC GAY CCS AAR GCB GAC TC-3′) and PolR (5′-ATS GCC ATC ATY TCR CCG GA -3′) for detecting nifH gene (Poly et al., 2001); Arch-amoAF (5′-STA ATG GTC TGG CTT AGA CG-3′) and Arch-amoAR (5′-GCG GCC ATC CAT CTG TAT CT-3′) for archaeal amoA gene (Francis et al., 2005); and amoA-1F (5′-GGG GTT TCT ACT GGT GGT-3′) and amoA-2R (5′-CCC CTC KGS AAA GCC TTC TTC-3′) for bacterial amoA gene (Rotthauwe et al., 1997). Clone libraries protocol was detailed by Gonzalez-Pimentel et al. (2018). Plasmids were sequenced by Secugen Sequencing Services (CSIC, Madrid, Spain). Sequences were checked for chimera by chimera.slayer as implemented in the software package mothur (Schloss et al., 2009). Sequences were aligned using mothur. After this analysis, all sequences were compared to the nonredundant database of sequences deposited at the National Center for Biotechnology (NCBI; http://www.ncbi.nlm.nih.org/blast/) using BLASTN algorithm. Statistical data (Chao1, Shannon and Simpson indices) were calculated as described by Riquelme et al. (2017).
Phylogenetic trees were constructed using the maximum-likelihood (Felsenstein, 1981) method in MEGA X (Kumar et al., 2018). A bootstrap analysis of 1000 re-samplings was used to evaluate the trees’ robustness.
Deposit of Sequences
Raw sequences were deposited in the NCBI SRA database under accession numbers ERS3506978-ERS3506982. Clone sequences (≈360 bp for nifH gene, 650 bp form archaeal amoA gene and 490 bp for bacterial amoA gene) were deposited at GenBank under the accession numbers LR596555-LR5965594, LR596553, and LR778119-LR778132.
Results
Field Emission Scanning Electron Microscopy
The study of the samples by field emission scanning electron microscopy (FESEM) revealed the presence of filamentous microorganisms in the vermiculations of Morgana Cave (Figure 3). Most of the microbial features observed in samples M1, M3, M4, M5, and M6 resemble the enigmatic reticulated filaments reported in caves worldwide (Melim et al., 2008; Miller et al., 2012), which are long tubular filaments with peculiar surface ornamentation (Miller et al., 2012; Melim et al., 2015).
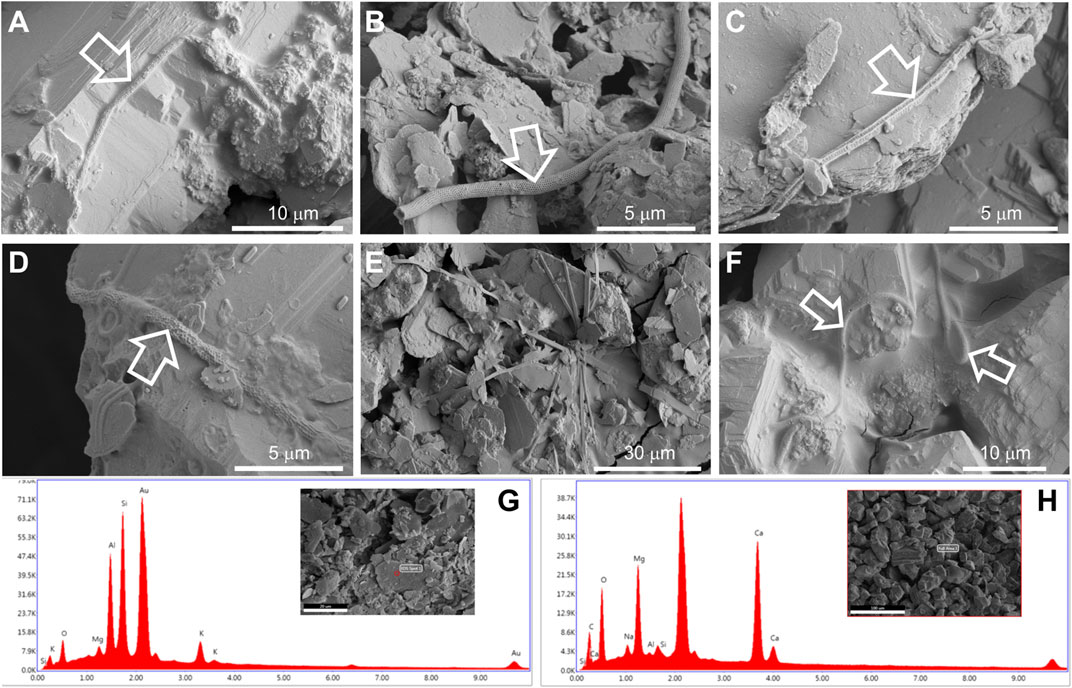
FIGURE 3. FESEM images from samples M1 (A), M3 (B,C), M4 (D), M5 (E), and M6 (F) depicting microbial structures associated with vermiculations. FESEM with corresponding EDS spectra from sample M5 (G) denotes the presence of clay minerals, due to the abundance of Al and Si, and sample M6 (H), rich in Ca and Mg, of dolomite.
In general, these long filaments (<100 µm) were found embedded in a slimy matrix of extracellular polymeric substances (Figures 3A,D,F). These filaments depict mineralized empty sheaths (Figure 3B), which can be attributed to the lysis of the cells and disappearance of living microorganisms.
The reticulated quadrangular-shaped filament depicted in Figure 3C is another type of intriguing structure, which, to our knowledge, has not been observed in other caves. In addition, flat filaments were found, which are probably mineral based on their morphology (Figure 3E).
The analysis of all the samples with energy dispersive X-ray spectroscopy (EDS) revealed that the bulk samples M1, M2, M4, and M6 contain high amounts of Ca and Mg (Supplementary Figure S1), suggesting the presence of dolomite derived from the host rock of Morgana Cave. Samples M3 and M5 showed the presence of Al and Si, indicating clay minerals (Supplementary Figure S1), consistent with the morphology of the mineral grains. EDS microanalysis of the mineral grains from samples M5 (clay minerals) and M6 (dolomite) are depicted in Figures 3G,H.
Microbial Diversity
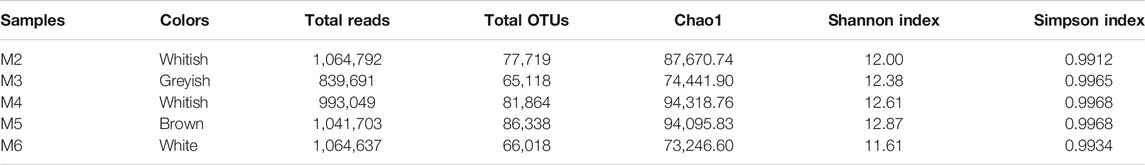
The amplicons for Illumina MiSeq analysis ranged from 839,691 to1,064,792 reads across the five vermiculation deposits, corresponding to 65,118–86,338 OTUs (Table 1).
Chao1, Shannon and Simpson indices were calculated to estimate alpha diversity (Table 1). Regardless of variation in community composition, Shannon and Simpson indices were similar across samples. The species richness and evenness in M4 and M5 were higher than in M2, M3, and M6.
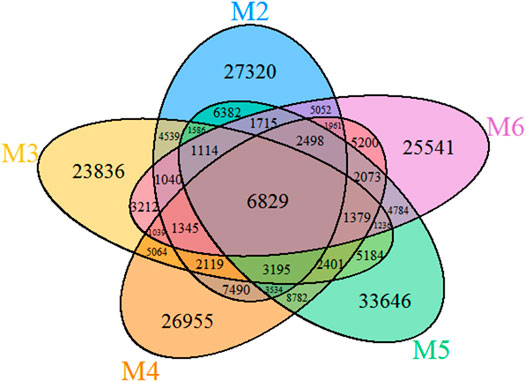
The VENN diagram of the OTUs distribution in the five vermiculations (Figure 4) revealed the presence of 137,298 OTUs that are unique to vermiculation deposits. Most of these unique taxa were found in sample M5 (33,646), followed by M3 (23,836) and M2 (27,320). The common microbial core comprised 6,829 distinct OTUs. The largest OTUs number (30,691) was shared between M4 and M5 with the highest bacterial diversity, whilst the lowest common prokaryotic OTUs were found between M2 and M3 (21,767) and M5 and M6 (21,628).
Table 2 shows that the microbial communities of the samples from Morgana Cave were almost totally composed of Bacteria, ranging from 99.33 (in M2) up to 99.79% (in M5). Archaea exhibited a low percentage ranging from 0.65 (in M2) to 0.19% (in M5). Unclassified members at domain level ranged from 0.05 to 0.02%. This distribution of taxa is similar to those reported by Itcus et al. (2018) from a perennial ice cave, where the range was 94.90–99.03% for Bacteria and 0–1.03% for Archaea.
Archaeal Phyla
Four Archaea phyla were found in the vermiculations of Morgana Cave, Diapherotrites, Euryarchaeota, Nanoarchaeota and Thaumarchaeota (Figure 5), although with very low relative abundances of OTUs. The class Thermoplasmata (Euryarchaeota phylum) ranged between 0.17 and 0.04%, Woesearchaeia (Nanoarchaeota phylum) between 0.37 and 0.08%, and Nitrososphaeria (Thaumarchaeota phylum) between 0.16 and 0.06%. The class Thermoplasmata was mainly present in M2, M3, and M6, and includes the order Methanomassiliicoccales with methanogenic archaea which accounted for 9.7% of all the Archaea present in the vermiculations. In addition, we observed an uncultured order of Thermoplasmata accounting for 17.3%.
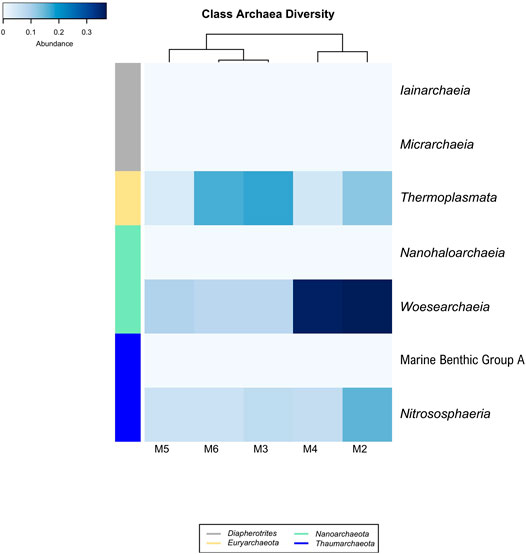
FIGURE 5. Heat-map analysis of Morgana Cave archaeal community relative abundance at class level. The identified groups are presented in the right column and their phylum level indicated in left-colors column.
The class Woesearchaeia was the most abundant (50%), considering all Archaea classes, particularly in M2 and M4. The class Nitrososphaeria was mainly present in M2, with the order Nitrosopumilales, accounting for 17.4% of all the Archaea. This last order includes Nitrosopumilus maritimus, an ammonia-oxidizing archaea (Pester et al., 2011).
Bacterial Phyla
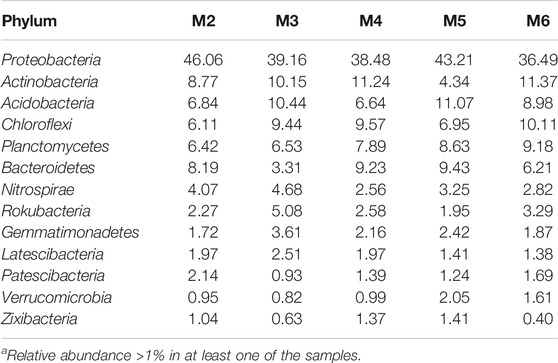
Thirteen major phyla (>1% relative abundance) were found in the vermiculations from Morgana Cave, but predominant phyla were Proteobacteria, Actinobacteria, Acidobacteria, Chloroflexi, Planctomycetes, and Bacteroidetes (Table 3). Less abundant phyla were Nitrospirae, Rokubacteria, Gemmatimonadetes, Latescibacteria, Patescibacteria, Verrucomicrobia, and Zixibacteria. In addition to the 13 major phyla, another 35 phyla were retrieved with relative abundances <1%. They encompass Armatimonadetes, Chlamydiae, Dependentiae, GAL15, and Omnitrophicaeota with abundances between 0.8 and 0.5%; and Cyanobacteria, Elusimicrobia and WS2 with abundances of 0.1%. The rest of phyla: Acetothermia, Atribacteria, Calditrichaeota, Coprothermobacteraeota, Dadabacteria, Entotheonellaeota, Epsilonbacteraeota, Fibrobacteres, Firmicutes, Hydrogenedentes, Kiritimatiellaeota, Lentisphaerae, Margulisbacteria, Nitrospinae, Poribacteria, Spirochaetes, Synergistetes, Tenericutes, Anck6, BRC1, FBP, FCPU426, LCP-89, WPS-2, TA06, and PAUC34f were present in abundances below 0.1%.
Figure 6 displays the bacterial classes with a relative abundance >1% in at least one of the samples retrieved from Morgana Cave vermiculations. The most abundant classes were largely represented by Gamma- and Betaproteobacteria, followed by Planctomycetacia, Acidimicrobiia, Rubrobacteria, Ignavibacteria, NC10 from the phylum Rokubacteria, and Blastocatellia, with percentages above 5% in at least one of the vermiculations.
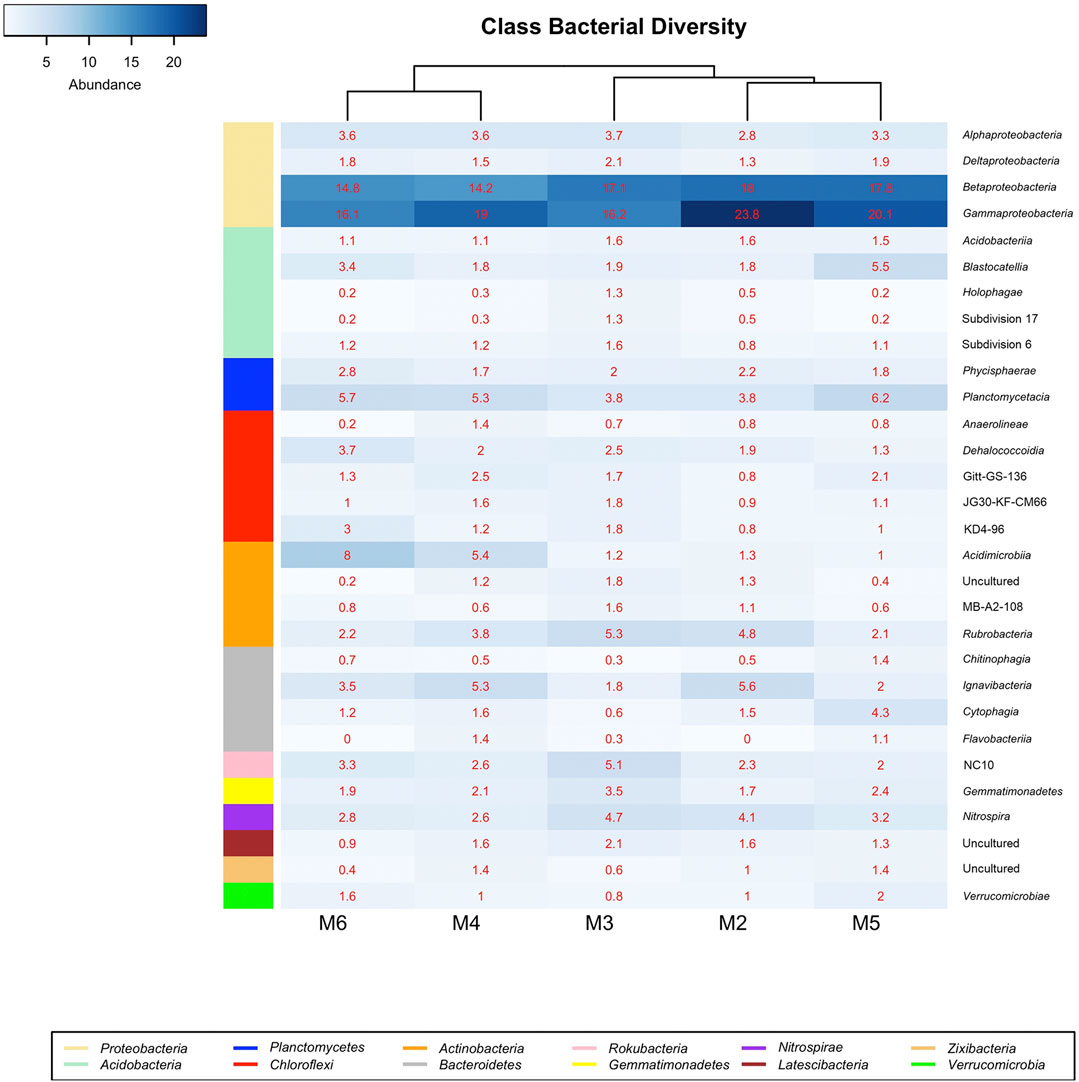
FIGURE 6. Heat-map analysis of Morgana Cave bacterial community relative abundance at class level. The identified groups are presented in the right column and their phylum level indicated in left-colors column.
Gammaproteobacteria were represented by the orders Methylococcales, wb1-P19, and PLTA13 (Figure 7; Table 4; Supplementary Table S1). The most abundant uncultured lineage was wb1-P19, seconded by PLTA13. The third and least abundant order was represented by the methanotrophic Methylococcales.
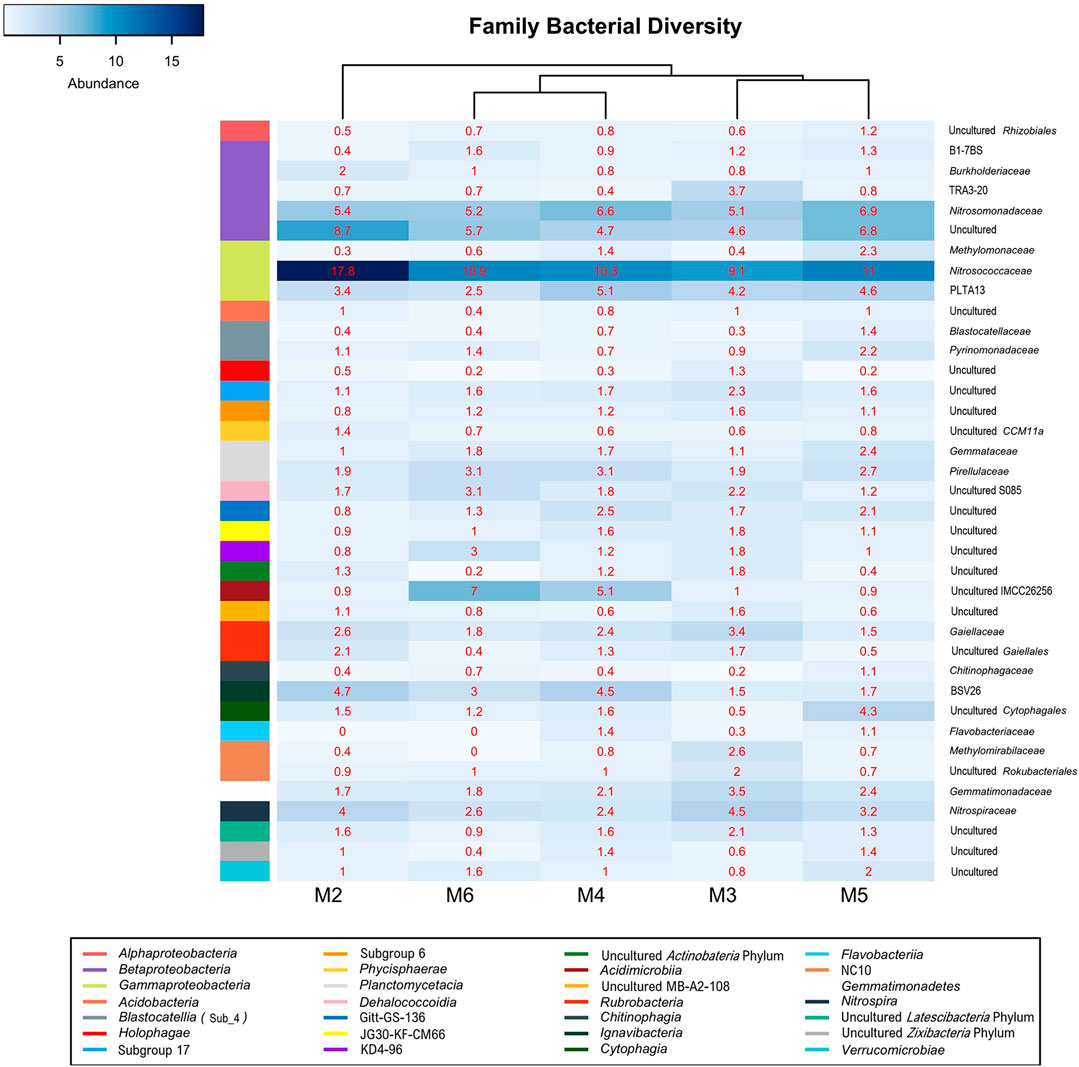
FIGURE 7. Heat-map analysis of Morgana Cave bacterial community relative abundance at family level. The identified groups are presented in the right column and their class level indicated in left-colors column.
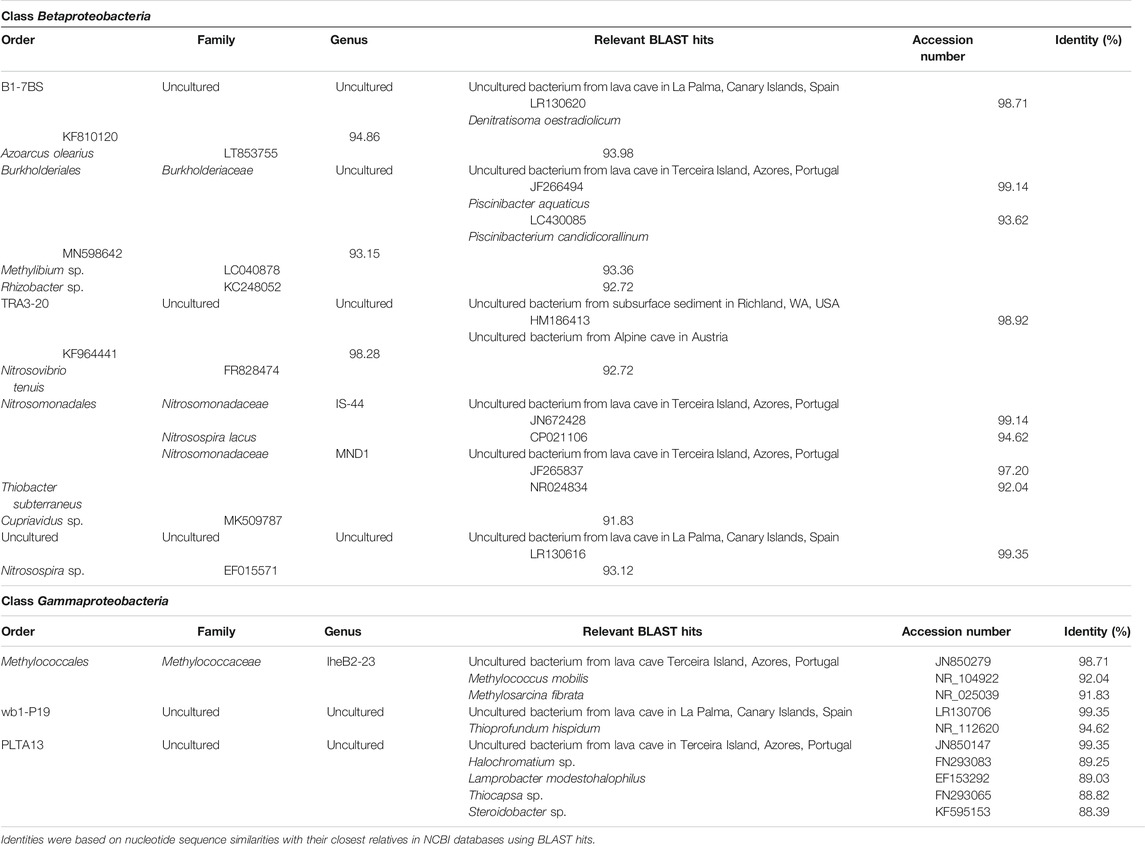
TABLE 4. Taxonomic affiliation of the sequences of Betaproteobacteria and Gammaproteobacteria, based on 16S rRNA amplicon sequence reads.
The class Betaproteobacteria was composed of sequences of uncultured members of families from which only Nitrosomonadaceae and Burkholderiaceae were identified (Figure 7; Table 4; Supplementary Table S1).
In Morgana Cave Planctomycetacia members were affiliated with the family Gemmataceae and other uncultured families and genera, with relatively low relative abundances (Supplementary Table S1).
Actinobacteria were mainly composed by the classes Acidimicrobiia and Rubrobacteria (Figure 6). In the class Acidimicrobiia we observed sequences of the uncultured actinobacterium IMCC26256 (Figure 7), clearly separated from other Acidimicrobiia species (Hu et al., 2018). The class Rubrobacteria with the genus Gaiella is well represented in the different vermiculations (Supplementary Table S1).
Ignavibacteria is a recently established class of Bacteroidetes and was denoted by clone BVS26 in the vermiculations of Morgana Cave (mainly in M2 and M4).
The class NC10 of the Rokubacteria phylum was most abundant in M3 (Figure 6), with a slightly higher relative abundance of the order MIZ17 over Rokubacteriales, whereas in other vermiculations (M2, M4, M5, and M6) Rokubacteriales dominated over MIZ17 (Supplementary Table S1).
Of the 26 existing subdivisions in the phylum Acidobacteria only five were found in Morgana Cave (Figure 6): Acidobacteriia, Blastocatellia, Holophagae and subdivisions 17 and 6. Within the Blastocatellales the families Blastocatellaceae and Pyrinomonadaceae contained the uncultured genera JGI and RB41, respectively (Supplementary Table S1). In general, the relative abundances of the different subdivisions were low, and in most cases rarely exceeded 1%.
Regarding the distribution in classes in the different vermiculations (Figure 6; Supplementary Table S1) vermiculation M2 presented the highest relative abundances in Beta- and Gammaproteobacteria. M3 showed a high diversity with the majority of relative abundances in 12 different classes, corresponding to eight phyla. In this vermiculation, Beta- and Gammaproteobacteria were also highly significant.
Vermiculation M4 revealed high relative abundances with respect to other vermiculations only in two classes of Chloroflexi and one of Bacteroidetes. M5 showed high relative abundances in 2 classes of Bacteroidetes, and one each of Acidobacteria, Planctomycetes, Zixibacteria and Verrucomicrobia. M6 presented high relative abundances in two classes of Chloroflexi, and one of Planctomycetes and Actinobacteria.
Functional Genes
To determine the involvement of the vermiculation communities in the nitrogen cycle, we surveyed the presence of functional genes involved in nitrogen fixation (nifH) and ammonia oxidation to nitrite (amoA). We observed nifH presence in the vermiculations M1, M2, M5, and M6. The phylogenetic analysis placed the sequences among nitrogen-fixing species (Supplementary Figure S2).
In addition, we detected the presence of functional genes involved in ammonia oxidation (amoA) in M1, M2, and M6 corresponding to Archaea. Regarding the bacteria, we identified genes related to Nitrosospira in M2 and M3. Phylogenetic analyses (Supplementary Figure S3A) indicated that the closest relatives of bacterial amoA gene sequences was the genus Nitrosospira, while the genus Nitrosopumilus was the closest to archaeal sequences (Supplementary Figure S3B).
Discussion
Microbial communities typically show a few dominant phyla and simultaneously a high number of phyla with a quite low abundance, in most cases <0.1%. Sogin et al. (2006) referred to this as the “rare biosphere”, microorganisms that cannot be identified without deep-sequencing NGS approaches. Only one third of the 35 less abundant phyla retrieved from the vermiculations of Morgana Cave have been also found in other caves (Ortiz et al., 2013; De Mandal et al., 2016; Hershey and Barton, 2018). For Pascoal et al. (2020) the rare biosphere constitutes an important “genomic reservoir” or “pool of diversity” and likely plays fundamental roles in ecosystem functioning.
Vermiculations form an ecological niche suitable for microorganisms due to the presence of different mineral particles and water retention. Samples from vermiculations M3 and M5 were mainly composed of clay minerals (Figure 3; Supplementary Figure S1), and M5 showed greater diversity than dolomite-rich vermiculations. Organic compounds and bacteria are generally adsorbed onto clay minerals (Bingham and Cotrufo, 2015; Cuadros, 2017), which might explain the abundance of taxa in the clay-containing vermiculations with respect to dolomite-rich vermiculations. In both types of vermiculations, EPS and reticulated filaments were evident. These enigmatic filaments have been found in microbial mats from caves worldwide, such as limestone caves and basaltic lava tubes (Melim et al., 2008; Jones, 2009, 2010, 2011; Miller et al., 2014; Riquelme et al., 2015a), as well as in a granite spring water tunnel in Portugal (Miller et al., 2012). They were characterized as long microbial filaments with hexagonal and diamond-shaped chambers resembling honeycombed structures, but attempts to decipher their taxonomic affiliation have failed thus far.
Melim et al. (2008) reported long reticulated filaments (up to 75 μm long and 0.5 μm in diameter) in calcite speleothems from limestone caves in New Mexico, and in lava tubes from Cape Verde Islands. (Jones 2009, 2010, 2011) described the occurrence of reticulated filaments in caves from the British West Indies, and suggested they are calcified filaments. Contrasting with those filaments described by Melim et al. (2008) and (Jones 2009, 2010), rich in carbon and calcium, respectively, Miller et al. (2012) found reticulated filaments rich in Mn oxides. These filaments were composed of an inner cellular structure coated by an exterior sheath of manganese oxides (Miller et al., 2012). Melim et al. (2015) reported living reticulated filaments associated with clay minerals in a limestone cave in Germany. They were rich in carbon, without mineralized sheaths. Most of these filaments were coated with clays, obscuring wall ornamentation, as also observed in samples from Morgana Cave. In fact, this association with clays could explain their presence in the vermiculations M3 and M5. Shedding light on the origin and nature of these intricate reticulated filaments is a still a challenge for geomicrobiologists.
Archaea are ubiquitous in terrestrial and marine ecosystems (Eme et al., 2017), but the richness of this domain in the vermiculations of Morgana Cave was scarce. Tebo et al. (2015) did not find any archaeal rRNA gene sequences in the volcanic ice caves of Mt. Erebus, Antarctica. Itcus et al. (2018) reported the lack of Archaea in surface ice blocks from a perennial ice cave and negligible percentages in ice cores. In addition, Icelandic and Antarctic lakes were dominated by Bacteria with no detectable or few archaeal members (Achberger et al., 2017).
In Morgana Cave a few groups of microorganisms could be involved in the nitrogen cycle. Ward et al. (2019) recognized four clades of ammonia oxidizers, which included members of the Thaumarchaeota (Archaea), the Nitrococcaceae family (Gammaproteobacteria), the Nitrosomonadaceae family (Betaproteobacteria) and some members of Nitrospirae.
The order Nitrosopumilales (Thaumarchaeota) contains the families Nitrosopumilaceae and Candidatus Nitrosotenuaceae, which includes Nitrosopumilus maritimus and Candidatus Nitrosotenuis cloacae, respectively (Qin et al., 2017). Ammonia-oxidizing archaea were detected in the vermiculations through the identification of the gene encoding monooxygenae subunit A (amoA), however the relative abundance is very low as shown in Table 2. Among the bacteria, amoA genes related to Nitrosospira (Betaproteobacteria) were also found.
The high proportion of bacteria over archaea found in the vermiculations is consistent with previous reports indicating that in terrestrial ecosystems nitrate stimulates the activity of ammonia-oxidizing bacteria, but not that of archaea (Meinhardt et al., 2018). In addition, Zhang et al. (2019a) reported that bacterial amoA gene abundance was significantly higher than that of archaea due to the activity of cold adapted Nitrosospira in rivers from the Qinghai-Tibet Plateau. In this context, Zhang et al. (2019b) showed that soil microbial communities were adapted to high nitrogen levels, strengthening the function of ammonia oxidation in order to deplete the excess of nitrogen.
Among Bacteria, the phylum Proteobacteria and the classes Gammaproteobacteria and Betaproteobacteria were the majority in the vermiculations, with percentages in all the samples exceeding 14%.
The gammaproteobacterial order wb1-P19 was the most abundant group in the vermiculations. Holmes et al. (2001) proposed that wb1-P19 clustered phylogenetically with sulfur or nitrite-oxidizing autotrophic bacteria. Unfortunately, records on this lineage were very scarce, with only three references in the literature (Holmes et al., 2001; Schabereiter-Gurtner et al., 2002; Zhu et al., 2019). These three papers reported this order in caves. Remarkably, Zhu et al. (2019) found that in Chinese caves, in addition to wb1-P19, uncultured Rokubacteriales and Gaiellales were also present, similar to our results in the vermiculations.
Table 4 shows the taxonomic affiliation of the representative sequences of OTUs classified as Gammaproteobacteria. In this case, the closest related sequences corresponded to uncultured bacteria from volcanic caves in the Terceira and La Palma islands, located in the Macaronesia region (Riquelme et al., 2015b; Gonzalez-Pimentel et al., 2018). This is striking due to the different environmental and climate (subtropical) conditions of the region and the mineral composition of the volcanic rocks (basalts) (Morgana is an Alpine cave developed on dolomitic marble).
Unfortunately, little information on metabolic traits has been reported for these uncultured volcanic cave bacteria. Gonzalez-Pimentel et al. (2018) stated that sulfur and nitrogen were associated with the chemical composition of basaltic rocks in lava tubes, and sulfur and nitrate oxidizers or reducers, such as bacteria from the Proteobacteria phylum, were very abundant in La Palma cave. Also, this phylum has similar abundances in Terceira cave (Riquelme et al., 2015b). Interestingly, Hathaway et al. (2014a) studied the diversity of ammonia oxidation (amoA) and nitrogen fixation (nifH) genes in lava caves from the Azores. They found that Nitrosospira-like sequences dominated the ammonia-oxidizing bacteria community, and the nitrogen fixation community was dominated by Klebsiella pneumoniae-like sequences. Note that Nitrosospira sequences were also present in vermiculations from Morgana Cave.
Apart from the close relationship between analyzed representative sequences and uncultured bacteria from the lava caves, secondary affiliations are indicated in Table 4. Some sequences from Morgana Cave were closely related to the gammaproteobacterial genera Methylococcus, Methylosarcina, Thioprofundum, Halochromatium, Lamprobacter, Thiocapsa, and Steroidobacter, with identity percentages ranging between 88.39 and 94.62%. According to Yarza et al. (2014), who established taxonomic boundaries for higher taxonomic levels for taxa of bacteria, the Morgana Cave uncultured bacterium close to Methylococcus and Methylosarcina must be included in the family Methylococcaceae. This family comprises methane-oxidizing bacteria that carry out the first step of nitrification, the oxidation of ammonia to nitrite, through the intermediate hydroxylamine (Hazeu et al., 1980; Wise et al., 2001; Sutka et al., 2003; Poret-Peterson et al., 2008).
The uncultured bacterium close to Thioprofundum must be placed within this genus. Thioprofundum hispidum is an obligately chemolithoautotrophic sulfur-oxidizing bacterium that bases its metabolism on sulfur oxidation and carbon dioxide fixation (Mori and Suzuki, 2014).
The uncultured bacteria related to Halochromatium, Lamprobacter, and Thiocapsa should be included in the Chromatiaceae, a family of phototrophic purple sulfur bacteria growing under anoxic conditions using sulfide as photosynthetic electron donor, which is oxidized to sulfate via intermediate accumulation of globules of elemental sulfur (Imhoff, 2014).
The uncultured bacterium related to Steroidobacter, a genus classified under incertae sedis, is close to the family Solimonadaceae. Steroidobacter anaerobically metabolizes steroids with nitrate reduction to dinitrogen monoxide, and further to dinitrogen (Fahrbach et al., 2008) A Steroidobacter OTU was also found in Californian lava caves (Lavoie et al., 2017).
The class Betaproteobacteria was placed with lower relative abundance, represented by Nitrosomonadales and some uncultured members. The order Nitrosomonadales is composed of the genera IS-44 and MND1 of the family Nitrosomonadaceae. All the cultivated representatives of this family are lithoautotrophic ammonia oxidizers (Prosser et al., 2014). IS-44 is rarely cited in the literature and, among the scarce reports, was found to be a member of the microbial community of composts (Neher et al., 2013). MND1 was found in ferromanganese nodules, soils and caves and may be diverse in functions in a wide range of environmental conditions (Jones et al., 2008; Spain et al., 2009).
Table 4 shows the complete taxonomic affiliation of the representative sequences of OTUs classified as Betaproteobacteria. In this class, also OTUs from Morgana Cave were closely related (97–99%) with uncultured bacteria from the volcanic caves. In addition, OTUs affiliated to the order TRA3-20 were closely related to uncultured bacteria from a United States subsurface basaltic site (Lin et al., 2012) and from an Alpine cave in Austria (Reitschuler et al., 2015). However, other more distant affiliations, within the family Nitrosomonadaceae, were Nitrosospira lacus, a psychrotolerant ammonia-oxidizing bacterium (Urakawa et al., 2015). The finding of bacterial ammonia oxidizer communities at elevated altitude on Mt. Everest suggests that this group is sufficiently adapted to survive in Alpine areas (Zhang et al., 2009). Nitrosospira sp. and Nitrosovibrio tenuis are also ammonia-oxidizing bacteria (Prosser et al., 2014).
Other uncultured bacteria included in the betaproteobacterial order B1-7BS are related to the denitrifying Denitratisoma oestradiolicum (Fahrbach et al., 2008) and the nitrogen-fixing Azoarcus olearius (Chen et al., 2013).
An uncultured bacterium within the family Burkholderiaceae is related to sequences of Piscinibacter aquaticus, Piscinibacterium candidicorallinum, Rhizobacter sp. and Methylibium sp., genera of uncertain position placed as unclassified Burkholderiales. Most strains from these genera are able to perform nitrate reduction (Sheu et al., 2016).
Within the genus MND1, family Nitrosomonadaceae, an uncultured bacterium closely related to another from Terceira lava cave, and more distant from Thiobacter subterraneus, a thermophilic, sulfur-oxidizing bacterium (Hirayama et al., 2005) and Cupriavidus sp., a genus including nitrate reducing strains (Vandamme and Coenye, 2004) was included.
Therefore, the scope of the metabolic diversity within the two main Proteobacteria classes, Gamma- and Betaproteobacteria, in the Morgana Cave vermiculations remains largely unknown, although there is great evidence of possible participations in the carbon, nitrogen and sulfur cycles.
In a lower level of abundance, two orders were found in the vermiculations: the actinobacterial Gaiellales, and Nitrospirales. Gaiellales comprises the genus Gaiella with only one known species: G. occulta, isolated from a deep aquifer with poorly mineralized waters. Distinctive characteristics are the presence of internally branched iso-fatty acids found in no other bacterium and the utilization of organic compounds (Albuquerque et al., 2011). The genome was studied by Severino et al. (2019) identifying genes of nitrate reduction. This genus was also found in volcanic caves (Riquelme et al., 2015a; Gonzalez-Pimentel et al., 2018).
The genus Nitrospira comprises ammonia-oxidizing bacteria which are relatively common in caves (Saiz-Jimenez, 2015). Its metabolic versatility, including the participation in different nitrogen cycling processes, is a key factor for the ubiquity and the high diversity of Nitrospira in caves (Tomczyk-Żak and Zielenkiewicz, 2016). Nitrospira was previously found in volcanic caves from the Azores, New Mexico, Hawai’i, and California (Northup et al., 2011; Riquelme et al., 2015b; Lavoie et al., 2017).
The phyla Rokubacteria, Gemmatimonadetes, Latescibacteria, Patescibacteria, Verrucomicrobia, and Zixibacteria had the lowest abundances in the vermiculations. They are commonly represented by uncultured bacteria that can only be identified in deep-sequencing NGS (Sogin et al., 2006). Most of these phyla are part of a rare cave biosphere formed by uncultured bacteria present in low numbers (Hershey and Barton, 2018).
Nitrogen fixation (nifH) genes have been found in Azorean and Brazilian caves (Hathaway et al., 2014a; Marques et al., 2018), suggesting this metabolic trait is widespread in the subsurface environment. Nearly all known diazotrophs contain a minimum of six conserved genes: nifH, nifD, nifK, nifE, nifN, and nifB, but for practical reasons in environmental studies only the nifH gene is commonly used as an indicator of nitrogen fixation activity. Although Dos Santos et al. (2012) stated that false positives can be obtained if only nifH is used, the data here reported and the phylogenetic tree of Supplementary Figure S2 place the sequences retrieved from the vermiculations among well-known nitrogen-fixing species, thus suggesting that potential nitrogen-fixing bacteria are present in the cave.
In addition, Table 4 shows that some sequences from the vermiculations were related to the betaproteobacterial nitrogen-fixing Azoarcus, and the gammaproteobacterial Thiocapsa. Furthermore, the sequence similarities of nifH genes with the genera Thiocystis, Chlorobium, and Aquaspirillum support the belief that uncultured bacteria have metabolic traits related to nitrogen-fixation in the vermiculations. Uncultured members of the Rhizobiales, Bulkholderiales, Methylococcales, Dehalococcoidia, and Euryarchaeota were also observed in the vermiculations and many species of these groups were reported as diazotrophs by Dos Santos et al. (2012).
In Alpine caves it is important to investigate how microbes acquire energy and obtain organic carbon. In this context, one of the major impacts on oligotrophic caves is organic carbon, which enriches the ecosystem, brought into the cave from outside.
Atmospheric transport and deposition of carbon and nitrogen is an important dispersal process on the earth’s surface. Several papers have reported the presence of organic carbon in the ice and snow of the Alps (Gröllert et al., 1997; Legrand et al., 2007; Preunkert and Legrand, 2013). This organic carbon derives from anthropogenic and biogenic sources, and is mainly composed of mono- and dicarboxylic acids, aliphatic alcohols, monoterpenes, phenols and humic-like substances (Guilhermet et al., 2013; Legrand et al., 2013; Müller-Tautges et al., 2016; Materic et al., 2019). These compounds constitute suitable carbon sources for microorganisms.
In addition, the Alpine region is characterized by nitrogen deposition with a rate of 10.5 kg ha−1 yr−1 at elevations around 1,200 m a.s.l. (Balestrini et al., 2013), and 0.5–3 kg N ha−1 yr−1 at elevations above 2,000 m a.s.l. (Filippa et al., 2010). Colombo et al. (2019) studied the nitrogen forms in meltwater samples collected in the period 2012–2015 from the Indren Glacier (ca. 4,000 m a.s.l., Mt. Rosa, NW Italian Alps), 150 km west of Val di Scerscen. Among the nitrogen forms, they found abundant NO3−, NH4+, and organic nitrogen. Similar results for nitrogen and sulfur deposition in the Alps were reported by Rogora et al. (2006), Balestrini et al. (2013), and Muri (2013).
The presence of organic carbon, nitrogen and sulfur in the Alps suggests that its content in the meltwaters from the glacier Vedretto di Scerscen has likely enhanced the activity of cave microorganisms involved in the geochemical cycles, and particularly of the most copiotrophic groups, such as Betaproteobacteria and Gammaproteobacteria involved in the nitrogen cycle (Figure 6; Table 4).
Chemolithoautotrophic bacteria provide a way to evaluate whether microbes are capable of producing their own energy and organic carbon to sustain an ecosystem. A possible metabolic pathway in vermiculations, in addition to nitrogen and sulfur, is methanotrophy. This is supported by the presence of the Rokubacteria class NC10, able of anaerobic oxidation of methane coupled to denitrification, as well as members of the Methanomassiliicoccales order which includes methanogenic archaea (Borrel et al., 2014). Uncultured members of the Methylococcales were present in the vermiculations suggesting that gammaproteobacterial methanotrophs may occur in this cave. Rokubacteria abundance ranged from 2 to 5%, Methylococcales from 0.3 to 2.3%, while Methanomassiliicoccales abundance was lower than 0.1% in all the vermiculations. These data indicate that bacteria metabolizing organic matter brought into the cave from external sources probably contribute more to the ecosystem than methanotrophic bacteria.
On the other hand, Alpine ecosystems are covered by snow and ice for most of the year, and ice and glacier caves are characterized by low temperatures below 0°C and inhabited by extremophiles. In these ecosystems, cave microorganisms have to cope with severe environmental constraints.
Bowman (2017) studied the genomes of 257 bacterial and archaeal genera related to cold ecosystems, from which 59 genera could be considered highly cold adapted and 137 genera included psychrotolerant species. From the list only Flavobacterium was present in the vermiculations analyzed in this work. Members of the Flavobacteria group colonized cold springs in the Qinghai-Tibetan Plateau (Li et al., 2012) and represented 12% of the isolates from high-altitude ice cores in the Tibetan Plateau (Liu et al., 2019).
Flavobacterium, Cytophaga species and Ignavibacteria contain antifreeze DUF3494-type proteins, which are present only in cold-adapted taxa (Bowman, 2017). The finding in the vermiculations of uncultured members of the classes Cytophagia and Ignavibacteria, in addition to the genus Terrimonas, from which novel species were isolated from Arctic environments (Kim et al., 2017), suggests that psychrophilic bacteria may be present in the vermiculations. Psychrophilic ammonia-oxidizing bacteria were also reported in cold environments (Zhang et al., 2009; Urakawa et al., 2015).
In spite of the extensive investigations on the microbiology of several glacial environments, very little is known on the diversity, activity and environmental role of microorganisms colonizing Alpine caves. In previous studies of ice caves, the presence of unclassified bacteria was common (Itcus et al., 2018; Purcarea, 2018). This also occurs in the vermiculations of Morgana Cave, where most of the retrieved phyla contain a large number of uncultured representatives, with unknown metabolic traits. As a matter of fact, many taxa common in subsurface soils are represented by high proportions of novel and undescribed microbial lineages, less amenable to currently known cultivation techniques (Brewer et al., 2019).
It is noteworthy that some of the Beta- and Gammaproteobacteria OTUs from the Alpine Morgana Cave (Table 4) were closely related to sequences from two subtropical volcanic caves in the Macaronesia region (Azores and Canary islands). Hathaway et al. (2014b) suggested that the distribution of lava cave organisms is not cosmopolitan, as they found substantial differences in the microbial diversity of lava caves from the Azores and Hawai’i. In fact, at the OTU level, there was only 5% overlap between the two archipelagoes. Nevertheless, the similarities of OTUs between our high Alpine and two subtropical volcanic caves are remarkable. Porca et al. (2012) studied microbial communities from geographically distinct limestone caves and reported the existence of a common core of microorganisms that were morphologically similar and related in phylogeny. These authors suggested that, in nutrient-limited environments, in order to efficiently exploit nutrients, cave microorganisms could develop complex cooperative and mutualistic associations which could explain the broad distribution of bacterial phyla in caves.
Conversely, it was clear that the comparison of the microbial communities from Morgana Cave vermiculations with those of the Frasassi and Fetida sulfuric acid caves revealed striking differences, due to the extremely different environments (alkaline in Morgana and highly acid in sulfuric caves). In fact, Fetida Cave vermiculations showed high abundance of unclassified Betaproteobacteria and sulfur-oxidizing Hydrogenophilales (including Sulfuriferula), Acidiferrobacterales (including Sulfurifustis), sulfur-reducing Desulfurellales, and ammonia-oxidizing Planctomycetes and Nitrospirae (D’Angeli et al., 2019b).
The Frasassi Cave vermiculation community was diverse, including as major lineages Betaproteobacteria (composed of unclassified bacteria, some related to sulfur-oxidizing bacteria), Gammaproteobacteria (composed of relatives of Piscirickettsiaceae, Nevskiaceae, Acidothiobacillaceae, and unclassified bacteria), Acidobacteria, Nitrospirae, and Planctomycetes (Jones et al., 2008). The data from Frasassi and Fetida sulfuric acid caves suggests that pH and sulfur are important drivers in the composition of their vermiculation microbial communities. On the contrary, vermiculations from the carbonate Morgana Cave have different communities, indicating that the microbial composition of vermiculations largely depends on environmental conditions and other factors. To summarize, the microbial groups identified in the vermiculations of this Alpine cave differed significantly from those retrieved from sulfuric acid caves, suggesting that mineral composition and pH play a major role in the colonization and establishment of communities.
This study provides new insights into vermiculation microbial communities from Morgana Cave which are composed of cosmopolitan bacteria in addition to a few niche-specific members. It is remarkable that this Alpine cave, supposed to be oligotrophic in nature due to the altitude and its remoteness, revealed that the 2 major bacterial classes Gammaproteobacteria and Betaproteobacteria, are composed of copiotrophic members whose metabolic traits are mainly associated with the nitrogen cycle. Evidences of the presence of methane- and sulfur-oxidizing bacteria are also found. This highlights the importance of atmospheric transport and deposition of anthropogenic pollutants which enhance the colonization and activity of microorganisms in pristine environments.
Conclusion
The microbial communities from Morgana Cave vermiculations were composed of different major evolutionary lineages. The vermiculations were dominated by the phyla Proteobacteria, Actinobacteria, Acidobacteria, Chloroflexi, Planctomycetes, and Bacteroidetes. The most abundant classes were Gamma- and Betaproteobacteria, followed by Planctomycetacia, Acidimicrobiia, Rubrobacteria, Ignavibacteria, NC10 from the phylum Rokubacteria, and Blastocatellia. The microorganisms linked to the nitrogen cycle are noteworthy, and their abundance must be related to atmospheric deposition of different forms of nitrogen coming from glacier meltwaters and rain/snow melt infiltrating into the cave. In addition, psychrophilic and methanotrophic bacterial groups were identified. The high number of phyla and candidate phyla (48) found in this oligotrophic high mountain cave is remarkable. The high percentage of uncultured bacteria present in the vermiculations, particularly at the lowest taxonomic ranks (only four genera were identified) indicates that the microbial diversity of Alpine caves deserves further research.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
VJ performed the molecular biology, VJ, BH, and JG-P contributed to bioinformatics and statistical data analyses and AM to FESEM. ID’A and PT participated to cave sampling and described the sampling locations and the caves. JDW and CS-J led the research. JDW participated in discussions and corrected the manuscript. CS-J wrote the manuscript. All of the authors reviewed the manuscript and approved the final version.
Funding
Funding was provided by the Spanish project MINECO CGL2016-75590-P with ERDF funds. The Rotary Club in Sondrio (Italy) also provided funds for sample analysis thanks to C. Mazza, M. Boccardi, and P. Nana, with the Municipality of Lanzada (S. Bardea) and the Istituto Mineralogico Valtellinese (I. Foianini) supporting the cave research project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI). Thanks to Mauro Inglese and Angelo Granati for the pictures in Figures 1, 2. We gratefully acknowledge the valuable comments of Cristina Purcarea and Diana E. Northup that helped to improve the initial submission of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feart.2020.586248/full#supplementary-material.
References
Achberger, A. M., Michaud, A. B., Vick-Majors, T. J., Christner, B. C., Skidmore, M. L., Priscu, J. C., et al. (2017). “Microbiology of subglacial environments,” in Psychrophiles: from biodiversity to biotechnology. Editor R. Margesin (Cham: Springer Nature Switzerland), 83–110.
Addesso, R., Gonzalez-Pimentel, J. L., D’Angeli, I. M., De Waele, J., Saiz-Jimenez, C., Jurado, V., et al. (2020). Microbial community characterizing vermiculations from karst caves and its role in their formation. Microb. Ecol. 2020. doi:10.1007/s00248-020-01623-5
Albuquerque, L., França, L., Rainey, F. A., Schumann, P., Nobre, M. F., and Da Costa, M. S. (2011). Gaiella occulta gen. nov., sp. nov., a novel representative of a deep branching phylogenetic lineage within the class Actinobacteria and proposal of Gaiellaceae fam. nov. and Gaiellales ord. nov. Syst. Appl. Microbiol. 34, 595–599. doi:10.1016/j.syapm.2011.07.001
Balestrini, R., Arese, C., Freppaz, M., and Buffagni, A. (2013). Catchment features controlling nitrogen dynamics in running waters above the tree line (central Italian Alps). Hydrol. Earth Syst. Sci. 17, 989–1001. doi:10.5194/hess-17-989-2013
Bingham, A. H., and Cotrufo, M. F. (2015). Organic nitrogen storage in mineral soil: implications for policy and management. Soil Discuss. 2, 587–618. doi:10.1016/j.scitotenv.2016.02.020
Bini, A., Cavalli Gori, M., and Gori, S. (1978). A critical review of hypotheses on the origin of vermiculations. Int. J. Speleol. 10, 11–33. doi:10.5038/1827-806X.10.1.2
Bojar, A-V., Guja, O., and Stefanescu, D. (2015). Vermiculation patterns in Coiba Mare Cave, Bihor Mountains, Romania. Quater. Int. 357, 212–219. doi:10.1016/j.quaint.2014.07.028
Borrel, G., Parisot, N., Harris, H. M. B., Peyretaillade, E., Gaci, N., Tottey, W., et al. (2014). Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genom. 15, 679. doi:10.1186/1471-2164-15-679
Boston, P. J. (2016). Biovermiculation biopatterns as universal signatures of extant and extinct life. Geol. Soc. America Abstr. Prog. 48, 7, Paper No. 109-8. doi:10.1130/abs/2016AM-286766
Bowman, J. P. (2017). “Genomics of psychrophilic bacteria and archaea,” in Psychrophiles: from biodiversity to biotechnology. Editor R. Margesin (Cham: Springer Nature Switzerland), 345–387.
Brewer, T. E., Aronson, E. L., Arogyaswamy, K., Billings, S. A., Botthoff, J. K., Campbell, A. N., et al. (2019). Ecological and genomic attributes of novel bacterial taxa that thrive in subsurface soil horizons. mBio 10, e01318-19. doi:10.1128/mBio.01318-19
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi:10.1038/nmeth.f.303
Chen, M.-H., Sheu, S.-Y., James, E. K., Young, C.-C., and Chen, W.-M. (2013). Azoarcus olearius sp. nov., a nitrogen-fixing bacterium isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 63, 3755–3761. doi:10.1099/ijs.0.050609-0
Colombo, N., Bocchiola, D., Martin, M., Confortola, G., Salerno, F., Godone, D., et al. (2019). High export of nitrogen and dissolved organic carbon from an Alpine glacier (Indren Glacier, NW Italian Alps). Aquat. Sci. 81, 74. doi:10.1007/s00027-019-0670-z
Cuadros, J. (2017). Clay minerals interaction with microorganisms: a review. Clay Miner. 52, 235–261. doi:10.1180/claymin.2017.052.2.05
D’Angeli, I. M., De Waele, J., Ieva, M. G., Leuko, S., Cappelletti, M., Parise, M., et al. (2017). Next-generation sequencing for microbial characterization of biovermiculation from a sulfuric acid cave in Apulia (Italy),” in Proceedings 17th international congress of speleology. Editors K. Moore, and S. White (Sydney: Australian Speleological Federation), 377–380.
D’Angeli, I. M., Parise, M., Vattano, M., Madonia, G., Galdenzi, S., and De Waele, J. (2019a). Sulfuric acid caves of Italy: a review. Geomorphology 333, 105–122. doi:10.1016/j.geomorph.2019.02.025
D’Angeli, I. M., Santagata, T., Tognini, P., Foianini, I., Audra, P., Cahiol, D. A., et al. (2018). Late stage condensation-corrosion in high mountain marble cave (Val di Scerscen, Bernina Massif, Valtellina, Italy). Geophys. Res. Abstr. 20, EGU2018-8296. doi:10.1007/978-3-642-12486-0
D’Angeli, I. M., Ghezzi, D., Leuko, S., Firrincieli, A., Parise, M., Fiorucci, A., et al. (2019b). Geomicrobiology of a seawater-influenced active sulfuric acid cave. PLoS One 14, e0220706. doi:10.1371/journal.pone.0220706
De Mandal, S., Zothansanga, , Panda, A. K., Bisht, S. S., and Kumar, N. S. (2016). MiSeq HV4 16S rRNA gene analysis of bacterial community composition among the cave sediments of Indo-Burma biodiversity hotspot. Environ. Sci. Pollut. Res. 23, 12216–12226. doi:10.1007/s11356-016-6423-9
De Waele, J., Galdenzi, S., Madonia, G., Menichetti, M., Parise, M., Piccini, L., et al. (2014). “A review on hypogene caves in Italy,” in Hypogene cave morphologies. Editors A. Klimchouk, I. D. Sasowsky, J. Mylroie, S. A. Engel, and A. S. Engel (Leesburg: Karst Waters Institute Special Publication),Vol. 18, 28–30.
Dos Santos, P. C., Fang, Z., Mason, S. W., Setubal, J. C., and Dixon, R. (2012). Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genom. 13, 162. doi:10.1186/1471-2164-13-162
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi:10.1093/bioinformatics/btq461
Eme, L., Spang, A., Lombard, J., Stairs, C. W., and Ettema, T. J. G. (2017). Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 15, 711–723. doi:10.1038/nrmicro.2017.133
Fahrbach, M., Kuever, J., Remesch, M., Huber, B. E., Kämpfer, P., Dott, W., et al. (2008). Steroidobacter denitrificans gen. nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 58, 2215–2223. doi:10.1099/ijs.0.65342-0
Faucher, B., and Lauriol, B. (2016). Les vermiculations de la grotte Wilson (Lac la Pêche, Québec, Canada). Contexte morphoclimatique, analyses sédimentologiques et distribution spatiale. Géomorphologie 22, 95–103. doi:10.4000/geomorphologie.11280
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi:10.1007/BF01734359
Filippa, G., Freppaz, M., Williams, M. W., and Zanini, E. (2010). Major element chemistry in inner alpine snowpacks (Aosta Valley Region, NW Italy). Cold Reg. Sci. Technol. 64, 158–166. doi:10.1016/j.coldregions.2010.07.005
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A. 102, 14683–14688. doi:10.1073/pnas.0506625102
Freydier, P., Martin, J., Guerrier, B., Jeannin, P. Y., and Doumenc, F. (2019). Rheology of cave sediments: application to vermiculation. Rheol. Acta 58, 675–685. doi:10.1007/s00397-019-01167-7
Gonzalez-Pimentel, J. L., Miller, A. Z., Jurado, V., Laiz, L., Pereira, M. F. C., and Saiz-Jimenez, C. (2018). Yellow colored mats from lava tube of La Palma (Canary Islands, Spain) are dominated by metabolically active Actinobacteria. Sci. Rep. 8, 1944. doi:10.1038/s41598-018-20393-2
Gröllert, C., Kasper, A., and Puxbaum, H. (1997). Organic compounds in high alpine snow. Int. J. Environ. Anal. Chem. 67, 213–222. doi:10.1080/03067319708031405
Guilhermet, J., Preunkert, S., Voisin, D., Baduel, C., and Legrand, M. (2013). Major 20th century changes of water-soluble humic-like substances (HULISWS) aerosol over Europe inferred from Alpine ice cores. J. Geophys. Res. Atmos. 11, 3869–3878. doi:10.1002/jgrd.50201
Haeberli, W., Paul, F., and Zemp, M. (2013). “Vanishing glaciers in the European Alps,” in Fate of mountain glaciers in the anthropocene. Editors P. J. Crutzen, L. Bengtsson, and V. Ramanathan (Vatican city: Pontifical Academy of Sciences, Scripta Varia), Vol. 118.
Hathaway, J. J. M., Garcia, M. G., Moya Balasch, M., Spilde, M. N., Stone, F. D., Dapkevicius, M. L. N. E., et al. (2014b). Comparison of bacterial diversity in Azorean and Hawai’ian lava cave microbial mats. Geomicrobiol. J. 31, 205–220. doi:10.1080/01490451.2013.777491
Hathaway, J. J. M., Sinsabaugh, R. L., Dapkevicius, M. L. N. E., and Northup, D. E. (2014a). Diversity of ammonia oxidation (amoA) and nitrogen fixation (nifH) genes in lava caves of Terceira, Azores, Portugal. Geomicrobiol. J. 31, 221–235. doi:10.1080/01490451.2012.752424
Hazeu, W., Batenburg-van der Vegte, W. H., and de Bruyn, J. C. (1980). Some characteristics of Methylococcus mobilis sp. nov. Arch. Microbiol. 124, 211–220. doi:10.1007/BF00427729
Hershey, O. S., and Barton, H. A. (2018). “The microbial diversity of caves,” in Cave ecology. Editors O. T. Moldovan, L. Kovác, and S. Halse (Cham: Springer Nature Switzerland), 69–90.
Hirayama, H., Takai, K., Inagaki, F., Nealson, K. H., and Horikoshi, K. (2005). Thiobacter subterraneus gen. nov., sp. nov., an obligately chemolithoautotrophic, thermophilic, sulfur-oxidizing bacterium from a subsurface hot aquifer. Int. J. Syst. Evol. Microbiol. 55, 467–472. doi:10.1099/ijs.0.63389-0
Holmes, A. J., Tujula, N. A., Holley, M., Contos, A., James, J. M., Rogers, P., et al. (2001). Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ. Microbiol. 3, 256–264. doi:10.1046/j.1462-2920.2001.00187.x
Hose, L. D., and Macalady, J. L. (2006). “Observations from active sulfuric karst systems: is the present the key to understanding Guadalupe Mountain speleogenesis?,” in Caves and karst of Southeastern New Mexico. Editors L. Land, L. V. W. Lueth, W. Raatz, P. Boston, and D. L. Love (Socorro: New Mexico Geological Society), 185–194.
Hose, L. D., Palmer, A. N., Palmer, M. V., Northup, D. E., Boston, P. J., and DuChene, H. R. (2000). Microbiology and geochemistry in a hydrogen-sulphide-rich karst environment. Chem. Geol. 169, 399–423. doi:10.1016/50009-2541(00)00217-5
Hu, D., Cha, G., and Gao, B. (2018). A phylogenomic and molecular markers based analysis of the class Acidimicrobiia. Front. Microbiol. 9, 987. doi:10.3389/fmicb.2018.00987
Imhoff, J. F. (2014). “The family Chromatiaceae” in The prokaryotes. Gammaproteobacteria Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer-Verlag), 151–178.
Itcus, C., Pascu, M. D., Lavin, P., Persoiu, A., Iancu, L., and Purcarea, C. (2018). Bacterial and archaeal community structures in perennial cave ice. Sci. Rep. 8, 15671. doi:10.1038/s41598-018-34106-2
Jones, B. (2009). Cave Pearls—the integrated product of abiogenic and biogenic processes. J. Sediment. Res. 79, 689–710. doi:10.2110/jsr.2009.071
Jones, B. (2010). Speleothems in a wave-cut notch, Cayman Brac, British West Indies: the integrated product of subaerial precipitation, dissolution, and microbes. Sediment. Geol. 232, 15–34. doi:10.1016/j.sedgeo.2010.09.003
Jones, B. (2011). Biogenicity of terrestrial oncoids formed in soil pockets, Cayman Brac, British West Indies. Sediment. Geol. 236, 95–108. doi:10.1016/j.sedgeo.2010.12.009
Jones, D. S., Lyon, E. H., and Macalady, J. L. (2008). Geomicrobiology of biovermiculations from the Frasassi cave system. J. Cave Karst Stud. 70, 78–93. https://caves.org/pub/journal/PDF/v70/cave-70-02-78.pdf.
Kim, M. C., Kang, O. C., Kim, C. M., Zhang, Y., Liu, Z., Wei, Z., et al. (2017). Terrimonas crocea sp. nov., isolated from the till of a high Arctic glacier. Int. J. Syst. Evol. Microbiol. 67, 868–874. doi:10.1099/ijsem.0.001689
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Lavoie, K. H., Winter, A. S., Read, K. J. H., Hughes, E. M., Spilde, M. N., and Northup, D. E. (2017). Comparison of bacterial communities from lava cave microbial mats to overlying surface soils from Lava Beds National Monument, USA. PloS One 12, e0169339. doi:10.1371/journal.pone.0169339
Legrand, M., Preunkert, S., Jourdain, B., Guilhermet, J., Faïn, X., Alekhina, I., et al. (2013). Water-soluble organic carbon in snow and ice deposited at Alpine, Greenland, and Antarctic sites: a critical review of available data and their atmospheric relevance. Clim. Past 9, 2195–2211. doi:10.5194/cp-9-2195-2013
Legrand, M., Preunkert, S., Schock, M., Cerqueira, M., Kasper-Giebl, A., Afonso, J., et al. (2007). Major 20th century changes of carbonaceous aerosol components (EC, WinOC, DOC, HULIS, carboxylic acids, and cellulose) derived from Alpine ice cores, J. Geophys. Res. 112, D23S11. doi:10.1029/2006JD008080
Li, G., Jiang, H., Hou, W., Wang, S., Huang, L., Ren, H., et al. (2012). Microbial diversity in two cold springs on the Qinghai-Tibetan Plateau. Geosci. Front. 3, 317–325. doi:10.1016/j.gsf.2011.12.004
Lin, X., Kennedy, D., Fredrickson, J., Bjornstad, B., and Konopka, A. (2012). Vertical stratification of subsurface microbial community composition across geological formations at the Hanford Site. Environ. Microbiol. 14, 414–425. doi:10.1111/j.1462-2920.2011.02659.x
Liu, Y., Priscu, J. C., Yao, T., Vick-Majors, T. J., Michaud, A. B., and Sheng, L. (2019). Culturable bacteria isolated from seven high-altitude ice cores on the Tibetan Plateau. J. Glaciol. 65, 29–38. doi:10.1017/jog.2018.86
Mao, Y., Yannarell, A. C., and Mackie, R. I. (2011). Changes in N-transforming archaea and bacteria in soil during the establishment of bioenergy crops. PloS One 6, e24750. doi:10.1371/journal.pone.0024750
Marques, E. L. S., Gross, E., Dias, J. C. T., Pirovani, C. P., and Rezende, R. P. (2018). Ammonia oxidation (amoA) and nitrogen fixation (nifH) genes along metasandstone and limestone caves of Brazil. Geomicrobiol. J. 35, 869–878. doi:10.1080/01490451.2018.1482386
Materic, D., Ludewig, E., Xu, K., Röckmann, T., and Holzinger, R. (2019). Brief communication: analysis of organic matter in surface snow by PTR-MS- implications for dry deposition dynamics in the Alps. Cryosphere 13, 297–307. doi:10.5194/tc-13-297-2019
Meinhardt, K. A., Stopnisek, N., Pannu, M. W., Strand, S. E., Fransen, S. C., Casciotti, K. L., et al. (2018). Ammonia-oxidizing bacteria are the primary N2O producers in an ammonia-oxidizing archaea dominated alkaline agricultural soil. Environ. Microbiol. 20, 2195–2206. doi:10.1111/1462-2920.14246
Melim, L. A., Northup, D. E., Spilde, M. N., Jones, B., Boston, P. J., and Bixby, R. J. (2008). Reticulated filaments in cave pool speleothems: microbe or mineral? J. Cave Karst Stud. 70, 135–141. doi:10.7939/R3J38KZ7V
Melim, L. A., Northup, D. E., Spilde, M. N., and Boston, P. J. (2015). Update: living reticulated filaments from Herbslabyrinth-Adventhöhle Cave system, Germany. J. Cave Karst Stud. 77, 87–90. doi:10.4311/2015MB0112
Merino, A., Ginés, J., Tuccimei, P., Soligo, M., and Fornós, J. J. (2014). Speleothems in Cova des Pas de Vallgornera: their distribution and characteristics within an extensive coastal cave from the eogenetic karst of southern Mallorca (Western Mediterranean). Int. J. Speleol. 43, 125–142. doi:10.5038/1827-806X.43.2.3
Miller, A. Z., Hernández-Mariné, M., Jurado, V., Dionísio, A., Barquinha, P., Fortunato, E., et al. (2012). Enigmatic reticulated filaments in subsurface granite. Environ. Microbiol. Rep. 4, 596–603. doi:10.1111/j.1758-2229.2012.00375.x
Miller, A. Z., Pereira, M. F. C., Calaforra, J. M., Forti, P., Dionísio, A., and Saiz-Jimenez, C. (2014). Siliceous speleothems and associated microbe- mineral interactions from Ana Heva Lava Tube in Easter Island (Chile). Geomicrobiol. J. 31, 236–245. doi:10.1080/01490451.2013.827762
Mori, K., and Suzuki, K. -I. (2014). “The family Thioalkalispiraceae”, in The Prokaryotes. Gammaproteobacteria. Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer-Verlag), 653–658.
Müller-Tautges, C., Eichler, A., Schwikowski, M., Pezzatti, G. B., Conedera, M., and Hoffmann, T. (2016). Historic records of organic compounds from a high Alpine glacier: influences of biomass burning, anthropogenic emissions, and dust transport. Atmos. Chem. Phys. 16, 1029–1043. doi:10.5194/acp-16-1029-2016
Muri, G. (2013). Atmospheric deposition chemistry in a subalpine area of the Julian Alps, North-West Slovenia. J. Limnol. 72, 291–300. doi:10.4081/jlimnol.2013.e23
Neher, D. A., Weicht, T. R., Bates, S. T., Leff, J. W., and Fierer, N. (2013). Changes in bacterial and fungal communities across compost recipes, preparation methods, and composting times. PloS One 8, e79512. doi:10.1371/journal.pone.0079512
Northup, D. E., Melim, L. A., Spilde, M. N., Hathaway, J. J. M., Garcia, M. G., Moya, M., et al. (2011). Lava cave microbial communities within mats and secondary mineral deposits: implications for life detection on other planets. Astrobiology 11, 601–618. doi:10.1089/ast.2010.0562
Ortiz, M., Neilson, J. W., Nelson, W. M., Legatzki, A., Byrne, A., Yu, Y., et al. (2013). Profiling bacterial diversity and taxonomic composition on speleothem surfaces in Kartchner Caverns, AZ. Microb. Ecol. 65, 371–383. doi:10.1007/s00248-012-0143-6
Pascoal, F., Magalhães, C., and Costa, R. (2020). The link between the ecology of the prokaryotic rare biosphere and its biotechnological potential. Front. Microbiol. 11, 231. doi:10.3389/fmicb.2020.00231
Pester, M., Schleper, C., and Wagner, M. (2011). The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr. Op. Microbiol. 14, 300–306. doi:10.1016/j.mib.2011.04.007
Poly, F., Ranjard, L., Nazaret, S., Gourbiere, F., and Monrozier, L. J. (2001). Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. App. Environ. Microbiol. 67, 2255–2262. doi:10.1128/AEM.67.5.2255-2262.2001
Porca, E., Jurado, V., Zgur-Bertok, D., Saiz-Jimenez, C., and Pasic, L. (2012). Comparative analysis of yellow microbial communities growing on the walls of geographically distinct caves indicates a common core of microorganisms involved in their formation. FEMS Microbiol. Ecol. 81, 255–266. doi:10.1111/j.1574-6941.2012.01383.x
Poret-Peterson, A. T., Graham, J. E., Gulledge, J., and Klotz, M. G. (2008). Transcription of nitrification genes by the methane-oxidizing bacterium, Methylococcus capsulatus strain Bath. ISME J. 2, 1213–1220. doi:10.1038/ismej.2008.71
Preunkert, S., and Legrand, M. (2013). Towards a quasi-complete reconstruction of past atmospheric aerosol load and composition (organic and inorganic) over Europe since 1920 inferred from Alpine ice cores. Clim. Past 9, 1403–1416. doi:10.5194/cp-9-1403-2013
Prosser, J. I., Head, I. M., and Stein, L. Y. (2014). “The family Nitrosomonadaceae,” in The Prokaryotes. Alphaproteobacteria and Betaproteobacteria. Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer-Verlag), 901–918.
Purcarea, C. (2018). “Microbial life in ice caves,” in Ice caves. Editors A. Persoiu, and A. S. E. Lauritzen (Amsterdam: Elsevier), 173–187.
Qin, W., Heal, K. R., Ramdasi, R., Kobelt, J. N., Martens-Habbena, W., Bertagnolli, A. D., et al. (2017). Nitrosopumilus maritimus gen. nov., sp. nov., Nitrosopumilus cobalaminigenes sp. nov., Nitrosopumilus oxyclinae sp. nov., and Nitrosopumilus ureiphilus sp. nov., four marine ammonia-oxidizing archaea of the phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 67, 5067–5079. doi:10.1099/ijsem.0.002416
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acid. Res. 41, D590–D596. doi:10.1093/nar/gks1219
Reitschuler, C., Lins, P., Schwarzenauer, T., Spötl, C., Wagner, A. O., and Illmer, P. (2015). New undescribed lineages of non-extremophilic Archaea form a constant and dominant element within alpine moonmilk microbiomes. Geomicrobiol. J. 32, 890–902. doi:10.1080/01490451.2015.1025317
Riquelme, C., Hathaway, J. J. M., Dapkevicius, M. L. N. E., Miller, A. Z., Kooser, A., Northup, D. E., et al. (2015a). Actinobacterial diversity in volcanic caves and associated geomicrobiological interactions. Front. Microbiol. 6, 1342. doi:10.3389/fmicb.2015.01342
Riquelme, C., Rigal, F., Hathaway, J. J. M., Northup, D. E., Spilde, M. N., Borges, P. A. V., et al. (2015b). Cave microbial community composition in oceanic islands: disentangling the effect of different colored mats in diversity patterns of Azorean lava caves. FEMS Microbiol. Ecol. 91, fiv141. doi:10.1093/femsec/fiv141
Riquelme, C., Dapkevicius, M. L. E., Miller, A. Z., Charlop-Powers, Z., Brady, S., Mason, C., et al. (2017). Biotechnological potential of Actinobacteria from Canadian and Azorean volcanic caves. Appl. Microbiol. Biotechnol. 101, 843–857. doi:10.1007/s00253-016-7932-7
Rogora, M., Mosello, R., Arisci, S., Brizzio, M. C., Barbieri, A., Balestrini, R., et al. (2006). An overview of atmospheric deposition chemistry over the Alps: present status and long-term trends. Hydrobiologia 562, 17–40. doi:10.1007/s10750-005-1803-z
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712. doi:10.1128/AEM.63.12.4704-4712.1997
Saiz-Jimenez, C. (2015). “The microbiology of show caves, mines tunnels and tombs: implications for management and conservation,” in Microbial life of cave systems. Editor A. S. Engel (Berlin: DeGruiter), 231–261.
Schabereiter-Gurtner, C., Saiz-Jimenez, C., Piñar, G., Lubitz, W., and Rölleke, S. (2002). Phylogenetic 16S rRNA analysis reveals the presence of complex and partly unknown bacterial communities in Tito Bustillo cave, Spain, and on its Paleolithic paintings. Environ. Microbiol. 4, 392–400. doi:10.1046/j.1462-2920.2002.00303.x
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describirng and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi:10.1128/AEM.01541-09
Severino, R., Froufe, H. J. C., Barroso, C., Albuquerque, L., Lobo-da-Cunha, A., da Costa, M. S., et al. (2019). High-quality draft genome sequence of Gaiella occulta isolated from a 150 meter deep mineral water borehole and comparison with the genome sequences of other deep-branching lineages of the phylum Actinobacteria. Microbiol. Open 8, e00840. doi:10.1002/mbo3.8407
Sheu, S.-Y., Li, Y.-S., and Chen, W.-M. (2016). Piscinibacterium candidicorallinum gen. nov., sp. nov., a member of the order Burkholderiales isolated from a fish pond. Int. J. Syst. Evol. Microbiol. 66, 5260–5267. doi:10.1099/ijsem.0.001505
Sogin, M. L., Morrison, H. G., Huber, J. A., Welch, D. M., Huse, S. M., Neal, P. R., et al. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120. doi:10.1073/pnas.0605127103
Sommer, C., Malz, P., Seehaus, T. C., Lippl, S., Zemp, M., and Braun, M. H. (2020). Rapid glacier retreat and downwasting throughout the European Alps in the early 21st century. Nat. Comm. 11, 3209. doi:10.1038/s41467-020-16818-0
Spain, A. M., Krumholz, L. R., and Elshahed, M. S. (2009). Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 3, 992–1000. doi:10.1038/ismej.2009.43
Sutka, R. L., Ostrom, N. E., Ostrom, P. H., Gandhi, H., and Breznak, J. A. (2003). Nitrogen isotopomer site preference of N2O produced by Nitrosomonas europaea and Methylococcus capsulatus Bath. Rapid Commun. Mass. Spectrom. 17, 738–745. doi:10.1002/rcm.968
Takahashi, S., Tomita, J., Nishioka, K., Hisada, T., and Nishijima, M. (2014). Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PloS One 9, e105592. doi:10.1371/journal.pone.0105592
Tebo, B. M., Davis, R. E., Anitori, R. P., Connell, L. B., Schiffman, P., and Staudigel, H. (2015). Microbial communities in dark oligotrophic volcanic ice cave ecosystems of Mt. Erebus, Antarctica. Front. Microbiol. 6, 179–192. doi:10.3389/fmicb.2015.00179
Tomczyk-Żak, K., and Zielenkiewicz, U. (2016). Microbial diversity in caves. Geomicrobiol. J. 33, 20–38. doi:10.1080/01490451.2014.1003341
Urakawa, H., Garcia, J. C., Nielsen, J. L., Le, V. Q., Kozlowski, J. A., Stein, L. Y., et al. (2015). Nitrosospira lacus sp. nov., a psychrotolerant, ammonia-oxidizing bacterium from sandy lake sediment. Int. J. Syst. Evol. Microbiol. 65, 242–250. doi:10.1099/ijs.0.070789-0
Vandamme, P., and Coenye, T. (2004). Taxonomy of the genus Cupriavidus: a tale of lost and found. Int. J. Syst. Evol. Microbiol. 54, 2285–2289. doi:10.1099/ijs.0.63247-0
Ward, L. M., Johnston, D. T., and Shih, P. M. (2019). Phanerozoic radiation of ammonia oxidizing bacteria. BioRxiv [preprint]. doi:10.1101/655399
Wise, M. G., McArthur, J. V., and Shimkets, L. J. (2001). Methylosarcina fibrata gen. nov., sp. nov. and Methylosarcina quisquiliarum sp. nov., novel type I methanotrophs. Int. J. Syst. Evol. Microbiol. 51, 611–621. doi:10.1099/00207713-51-2-611
Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F. O., Ludwig, W., Schleifer, K. -H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. doi:10.1038/nrmicro3330
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A. (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620. doi:10.1093/bioinformatics/btt593
Zhang, L-M., Wang, M., Prosser, J. I., Zheng, Y. -M., and He, J. -Z. (2009). Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest. FEMS Microbiol. Ecol. 70, 208–217. doi:10.1111/j.1574-6941.2009.00775.x
Zhang, S., Xia, X., Li, S., Zhang, L., Wang, G., Li, M., et al. (2019a). Ammonia oxidizers in high-elevation rivers of the Qinghai-Tibet Plateau display distinctive distribution patterns. Appl. Environ. Microbiol. 85, e01701-19. doi:10.1128/AEM.01701-19
Zhang, X., Johnston, E. R., Wang, Y., Yu, Q., Tian, D., Wang, Z., et al. (2019b). Distinct drivers of core and accessory components of soil microbial community functional diversity under environmental changes. mSystems 4, e00374-19. doi:10.1128/mSystems.00374-19
Keywords: vermiculation deposits, bacteria, archaea, nitrogen cycle, alpine cave
Citation: Jurado V, Gonzalez-Pimentel JL, Miller AZ, Hermosin B, D’Angeli IM, Tognini P, De Waele J and Saiz-Jimenez C (2020) Microbial Communities in Vermiculation Deposits from an Alpine Cave. Front. Earth Sci. 8:586248. doi: 10.3389/feart.2020.586248
Received: 22 July 2020; Accepted: 19 November 2020;
Published: 16 December 2020.
Edited by:
Aurel Perşoiu, Romanian Academy, RomaniaReviewed by:
Cristina Purcarea, Institute of Biology Bucharest of the Romanian Academy, RomaniaDiana Eleanor Northup, University of New Mexico, United States
Copyright © 2020 Jurado, Gonzalez-Pimentel, Miller, Hermosin, D'Angeli, Tognini, De Waele and Saiz-Jimenez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cesareo Saiz-Jimenez, c2FpekBpcm5hc2UuY3NpYy5lcw==
 Valme Jurado
Valme Jurado Jose Luis Gonzalez-Pimentel
Jose Luis Gonzalez-Pimentel Ana Zelia Miller
Ana Zelia Miller Bernardo Hermosin1
Bernardo Hermosin1 Jo De Waele
Jo De Waele Cesareo Saiz-Jimenez
Cesareo Saiz-Jimenez