- 1The Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD, United States
- 2Office of Surveillance and Epidemiology, Center for Drug Evaluation and Research, Food and Drug Administration, Silver Spring, MD, United States
The US Food and Drug Administration (FDA) receives millions of postmarket adverse event reports for drug and therapeutic biologic products every year. One of the most salient issues with these submissions is report duplication, where an adverse event experienced by one patient is reported multiple times to the FDA. Duplication has important negative implications for data analysis. We improved and optimized an existing deduplication algorithm that used both structured and free-text data, developed a web-based application to support data processing, and conducted a 6-month dedicated evaluation to assess the potential operationalization of the deduplication process in the FDA. Comparing algorithm predictions with reviewer determinations of duplicates for twenty-seven files for case series reviews (with a median size of 281 reports), the average pairwise recall and precision were equal to 0.71 (SD ± 0.32) and 0.67 (SD ± 0.34). Overall, reviewers felt confident about the algorithm and expressed their interest in using it. These findings support the operationalization of the deduplication process for case series review as a supplement to human review.
1 Introduction
At the US Food and Drug Administration (FDA), monitoring of drug and therapeutic biologic products for potential adverse events (AE) after marketing includes evaluating individual case safety reports (ICSRs) submitted to the FDA Adverse Event Reporting System (FAERS). A long-standing challenge with the evaluation of FAERS, and other pharmacovigilance (PV) databases, is the presence of “duplicate” reports, where an AE experienced by one patient is reported multiple times to the FAERS database. Duplicates have been defined as “separate and unlinked records that refer to one and the same case of a suspected adverse drug reaction” (Tregunno et al., 2014). A recent study of VigiBase reports from the United Kingdom and Spain showed that duplicate reports come from different sources including: 1) different report origin (e.g., patient and health care professional report the same AE); 2) unlinked follow-up (follow-up submitted with a new report identification number); 3) multiple companies submit the same report referencing their drug because of regulatory requirements; and 4) transmission errors (e.g., change of report identification number) (Tregunno et al., 2014). It is entirely possible for dozens of FAERS reports to describe the same patient case (Hauben et al., 2007; Khaleel et al., 2022).
The presence of duplicates in the FAERS database can potentially lead to false-positive signals or masking of signals, especially when using disproportionate reporting of a drug-event combination for signal identification (Hauben et al., 2007). Probabilistic models were used for duplicate identification in similar adverse event reporting systems before, notably by Norén et al. in the World Health Organization-Uppsala Monitoring Centre database (Norén et al., 2007). These models demonstrated average performance with confirmed duplicates in three data sources (33, 64, and 86%) (Tregunno et al., 2014). Identifying duplicates typically requires human expert review and is resource-intensive because some of the most salient information for duplicate detection is present in the report narrative.
We previously developed a deduplication algorithm that included clinical information extracted from the report narratives using natural language processing (NLP) (Kreimeyer et al., 2017). We analyzed its ability to support the identification of potential duplicates in the Vaccine Adverse Event Reporting System (VAERS) and FAERS (Kreimeyer et al., 2017). This algorithm compares two reports and decides how similar they are by examining information from the reports’ structured fields and free-text narratives. The inclusion of narrative-extracted clinical information in our algorithm and the enrichment of report details was a novel development with the potential to improve duplicate detection, according to Norén (Norén, 2017).
We are preparing the algorithm for implementation at the FDA’s Center for Drug Evaluation and Research (CDER) by assessing the algorithm’s utility in the PV workflow for case series reviews in a dedicated evaluation described in the current paper.
2 Methods
2.1 The Significance of Deduplication in the Pharmacovigilance Workflow
Safety reviewers are responsible for the surveillance of all marketed drugs and routinely review the corresponding postmarket FAERS reports. Reviews are also conducted when necessary according to CDER’s policies and procedures for the collaborative identification, evaluation and resolution of Newly Identified Safety Signals (NISS) associated with marketed drugs (FDA, 2020).
Duplication of FAERS reports may negatively impact the accuracy and validity of the findings and conclusions of these reviews unless adequately addressed. Reviewers allocate time and resources to review the structured data fields and the narrative of each FAERS report, as shown in Figure 1. Reviewers use standard data exploration techniques, such as sorting and text matching using an electronic spreadsheet to find duplicates in a manual process. On the other hand, our deduplication algorithm automates this search for duplicates using some of the same structured data fields and elements from the narrative. The overlap of the two processes is illustrated in Figure 1.
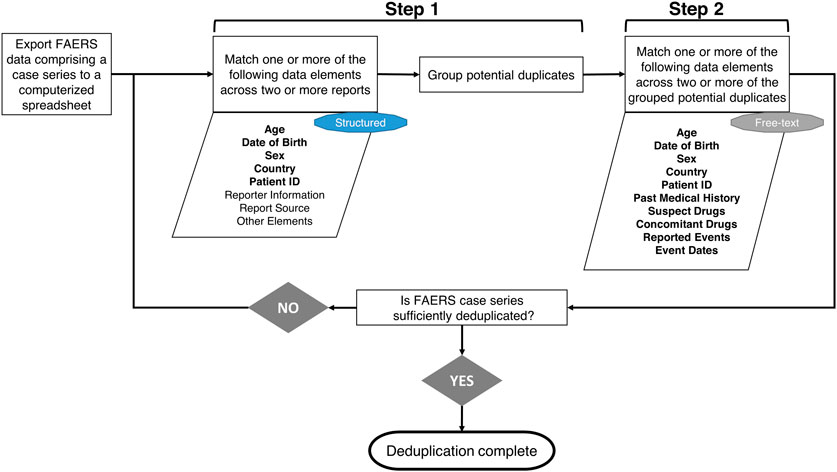
Figure 1. Flowchart of the manual deduplication procedure followed by the Safety Reviewers at the Center for Drug Evaluation and Research. In the first step, Safety Reviewers go over a line listing of FAERS reports in a spreadsheet, evaluate one or more of the structured data elements (included in the parallelogram shape) for potential matches across two or more reports, and group reports with matches together. The groups of potential duplicate reports are further examined in step 2 by reviewing the FAERS narratives and checking for potential matches across two or more reports using the free-text data elements shown in the second parallelogram shape. If case series is not sufficiently deduplicated after these two steps, Safety Reviewers will repeat the process by examining additional elements. It should be noted that some of the data elements manually evaluated by the reviewers are also used by the deduplication algorithm (shown in boldface) by retrieving them either from the FAERS structured data fields or the FAERS narratives using ETHER (described further in Supplementary Appendix A). “Family History” and “Patient or Report Number in Narrative” are the other two data elements contributing to the automated process that are not necessarily used in the manual deduplication and, therefore, are not shown in the flowchart.
To implement the deduplication algorithm at CDER, we performed a few changes to the originally published version of the algorithm. First, we changed the output presentation to display suggested groups of duplicates containing two or more reports. We also modified the part of the algorithm that compares the reported ages in the reports to first compare the structured fields for date of birth before comparing age information from the free-text narratives and structured fields. Detailed information about the algorithm can be found in Supplementary Appendix A and the original publication (Kreimeyer et al., 2017).
2.2 The Deduplication Evaluation
2.2.1 Web-Based Tool and Algorithm Optimization
We conducted a 6-month evaluation between 16 October 2020, and 15 April 2021, and assessed how well the deduplication algorithm could support reviewers’ routine work at CDER as described earlier. For this evaluation, a new standalone web-based program was deployed on FDA’s servers based on requirements provided by a working group of six reviewers. CDER’s reviewers were actively involved in the preliminary requirements-gathering and enhancement steps and provided additional feedback in an informal evaluation on four completed postmarket reviews containing 1735 reports prior to the evaluation described below. The core of the algorithm was also enhanced with two major speed optimizations. First, we added parallelization, making the process run simultaneously for several pairs using multiple CPU cores. Second, we combined all the narrative-based comparisons in one instead of two or three passes. Table 1 shows the reduction of the execution time for several datasets.

Table 1. The median execution time for running the deduplication algorithm on several sample data sets before and after optimization. The median execution time for each data set is reported because each one was run several different times to account for any external factors related to changes to operating system software.
2.2.2 Structure of the Evaluation
The evaluation intended to apply the algorithm to as many real-life PV case series use cases as possible and gather quantitative performance data on the algorithm and, primarily, qualitative feedback from reviewers about their experiences using the algorithm’s output. We invited all reviewers (approximately 60, including the six working group members) in CDER to use the tool and voluntarily provide feedback. It was published on an internal FDA URL and provided functionality to upload and download files. Feedback was provided voluntarily by reviewers in the context of actual day-to-day work. Each resulting file contained all the original uploaded data with two new columns, one for the algorithm’s suggested duplicate groupings and one for the true duplicate feedback, which are the reviewers’ true duplicate designations, as well as a separate page for qualitative feedback. The instructions provided for the evaluation are included in Supplementary Appendix B. We requested duplicate determinations on the entire dataset, not just on the reports suggested to be duplicates by the algorithm. For performance, we measured the recall and precision of the suggested pairs compared to the reviewers’ determinations. Following the pilot, one of the authors (OD), familiar with the case review process, performed a qualitative error analysis of the false positive pairs of the algorithm in the datasets where the pairwise performance was low (precision or F1-measure below 0.5).
3 Results
The deduplication evaluation began on 16 October 2020, and the first file was submitted the same day. New files were uploaded throughout the entire 6-month period, with an increasing rate over time. We also regularly received completed feedback files from reviewers. The largest file submitted contained 5700 reports, and the smallest contained 11 reports. Of the fifty-eight submissions from twenty unique reviewers, we received feedback for twenty-seven submissions. Twenty-three feedback files included labeled true duplicates and qualitative feedback, three files included true duplicates only (#16, #22, and #27), and one feedback file (#18) contained qualitative comments only. Reviewers often used the tool to do a quick check rather than a thorough review of large lists of reports. The five largest files (all >1900 reports) did not have any feedback returned.
The twenty-six feedback files with labeled true duplicates as determined by the reviewers allowed for a quantitative performance evaluation (Figure 2). The average pairwise recall and precision were equal to 0.71 (SD ± 0.32) and 0.67 (SD ± 0.34), respectively. The number of true duplicate pairs found within each data set varied considerably. Two data sets (#2 and #8) had zero true duplicate pairs. Data sets #9, #12, #15, #19, #22, and #23 had very high numbers of true duplicate pairs (1367, 4329, 2183, 1159, 2660, and 1501 pairs, respectively). Interestingly, two datasets (#16 and #17) had a (close to) perfect F-measure (0.99 and 1.00, respectively), and an additional three (#4, #13, and #25) had F-measure over 0.9. The size of the datasets did not necessarily affect pairwise recall, e.g., dataset #15 included 1060 reports and had a recall of 0.93. On the other hand, pairwise precision significantly dropped in large datasets, including over 800 reports, however, it varied in all other datasets, including less that 700 reports.
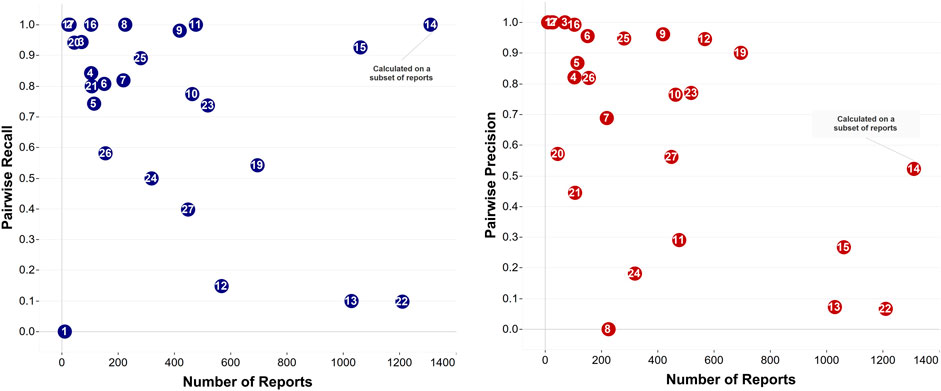
Figure 2. The two plots show the performance of the deduplication algorithm in naming pairs of reports as suggested duplicates for files where true duplicates were determined by reviewers. Datasets #1, #2, and #17 (with 11 reports, 29, and 25 reports, respectively) appear very close together on the precision plot. Datasets #1 and #17 as well as datasets #3 and #20 (with 71 and 46 reports, respectively) appear very close in the recall plot. Note: In data set #14, the reviewer considered evaluating only a subset of them for true duplicates served the purposes of that review (1820 reports) and the performance on the full data set is not known.
The qualitative feedback, based on twenty-four files, is summarized in Figure 3. Overall, reviewers expressed a fair amount of confidence about the algorithm and stated they were likely to use it in the future. Their responses varied widely in terms of time savings and detection of duplicates they may have otherwise missed. However, it should be noted that submissions represented different types of reviews, which may have had different characteristics. In particular, 13 of the qualitative feedback files we received were for reviews that required a detailed analysis of duplicates. The remaining reviews did not require a complete identification of duplicates because they were preliminary in nature.
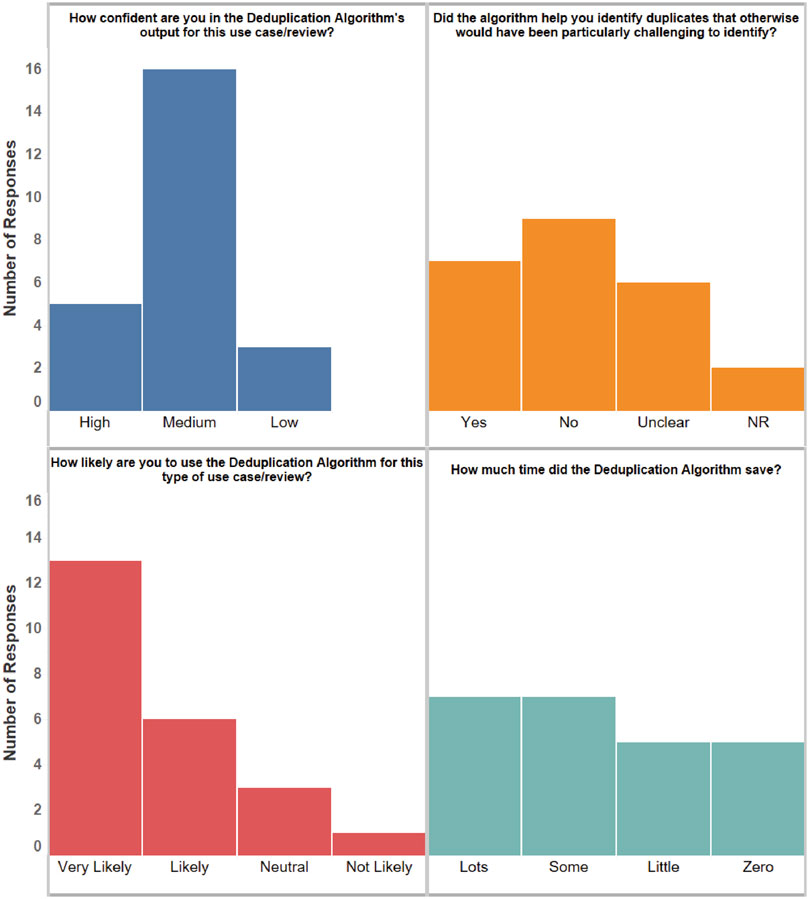
Figure 3. A summary of the qualitative feedback from the returned feedback files. The bars show the number of responses in each category. NR: No Response.
The qualitative error analysis of false positives was performed across eight datasets that demonstrated low performance, and a total of 331 reports in all false positive pairs (in the datasets where the pairwise performance was low) were checked. Three primary categories of errors were determined after reviewing these incorrect groupings: Mismatched Data Elements (MM) where at least one element was significantly not matching; Data Not Reported in FAERS Reports (NR) where there were important data elements missing and the non-missing data was unconvincing that the reports were duplicates; and reports with Similar Narratives for a Group of Patients (SN) where the same report text was used for multiple patients in a patient group, with the only difference being the number assigned to each individual patient.
The SN error group was the smallest, with only 13 reports. There were 62 reports in the NR category and 290 in the MM category. Both categories were also broken down by the type(s) of data that were not reported or mismatching, respectively. For the NR category, the most common data elements missing were details about concomitant medications (N = 58) and events (N = 52). Events included clinical events, other than the primary adverse event, that occurred during patients’ clinical course, medical history, or medical conditions. For the MM category, the most common data elements mismatching were information about the events (N = 173), with additional mismatches appearing in patient sex (N = 88), product and event dates (N = 73), and patient age (N = 72). Note that individual reports could be labeled as missing or mismatching multiple data elements; however, the labeling was not exhaustive for every data element in every report.
An assessability algorithm (Kreimeyer et al., 2021) designed to predict whether there was sufficient information in a report to assess causality was further applied to the 62 cases in the NR category to test the hypothesis that the relative importance of a particular feature used for deduplication is contingent on the total amount of information available in a report. Around a third of the cases (20 out of 62) were classified as unassessable suggesting that a complex interdependent relationship exists between missing and existing data elements in a report which could affect reviewers’ asssesment of duplicate reports.
4 Discussion
The deduplication algorithm was evaluated on multiple data sets and demonstrated promising performance, with recall and precision above 90% for some of the datasets. The qualitative error analysis has shown that reviewers place a high premium on mismatches found in most data elements (for example, they have requested firmer rules to never show potential duplicates with differing dates of birth or countries of occurrence to cut down on the number of false positive suggestions). These findings suggest that, while the algorithm may automate the process of duplicate detection, a human reviewer must still examine and confirm the output in most situations. Despite this limitation, the human reviewers still found the algorithm improved their efficiency at duplicate detection and gave them more confidence in the adequacy of their search for duplicates.
Overall, the deduplication algorithm still has two of the same limitations described in the original publication (Kreimeyer et al., 2017). The algorithm’s probabilistic approach assumes that fields are independent, which is unlikely to hold for many reports. Also, its narrative information relies on an NLP system that, like any other NLP tool, cannot perfectly identify every piece of textual information. While improving algorithm performance is an ongoing goal, it was evaluated in the real-life setting and reviewers think highly of its performance and potential routine use. A recent review highlighted that the utility of clinical decision support systems is generally not demonstrated (Ostropolets et al., 2020) and confirmed previous findings on the topic (Bright et al., 2012; Lobach et al., 2012). Although postmarket case series review differs from clinical decision making in many aspects, strict processes guide them both, and any support methods must meet specific requirements on performance and efficiency. Engaging end-users in the entire process helped us not only identify the algorithm’s weaknesses and make the appropriate improvements but primarily deliver an efficient solution tailored to their needs. We have to continue to work closely with reviewers, address any concerns about difficulties in specific use cases, and deliver a high-quality deduplication solution in the production environment.
Data Availability Statement
The datasets presented in this article are not readily available because this is FDA’s internal data that cannot be shared with the public. Requests to access the datasets should be directed to FDA’s Division of Drug Information (DDI) at druginfo@fda.hhs.gov.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was funded by the US Food and Drug Administration (grant #75F40121C00185 and #75F40119C10084).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdsfr.2022.918897/full#supplementary-material
References
Botsis, T., Jankosky, C., Arya, D., Kreimeyer, K., Foster, M., Pandey, A., et al. (2016). Decision Support Environment for Medical Product Safety Surveillance. J. Biomed. Inf. 64, 354–362. doi:10.1016/j.jbi.2016.07.023
Bright, T. J., Wong, A., Dhurjati, R., Bristow, E., Bastian, L., Coeytaux, R. R., et al. (2012). Effect of Clinical Decision-Support Systems. Ann. Intern Med. 157 (1), 29–43. [Published Online First: Epub Date]. doi:10.7326/0003-4819-157-1-201207030-00450
FDA (2020). “Collaborative Identification, Evaluation, and Resolution of a Newly Identified Safety Signal (NISS),” in CDER (MD: Silver Spring).
Hauben, M., Reich, L., DeMicco, J., and Kim, K. (2007). ‘Extreme Duplication’ in the US FDA Adverse Events Reporting System Database. Drug Saf. 30 (6), 551–554. [Published Online First: Epub Date]. doi:10.2165/00002018-200730060-00009
Khaleel, M. A., Khan, A. H., Ghadzi, S. M. S., Adnan, A. S., and Abdallah, Q. M. (2022). A Standardized Dataset of a Spontaneous Adverse Event Reporting System. Healthcare 10 (3), 420. [Published Online First: Epub Date]. doi:10.3390/healthcare10030420
Kreimeyer, K., Dang, O., Spiker, J., Muñoz, M. A., Rosner, G., Ball, R., et al. (2021). Feature Engineering and Machine Learning for Causality Assessment in Pharmacovigilance: Lessons Learned from Application to the FDA Adverse Event Reporting System. Comput. Biol. Med. 135, 104517. [Published Online First: Epub Date]. doi:10.1016/j.compbiomed.2021.104517
Kreimeyer, K., Menschik, D., Winiecki, S., Paul, W., Barash, F., Woo, E. J., et al. (2017). Using Probabilistic Record Linkage of Structured and Unstructured Data to Identify Duplicate Cases in Spontaneous Adverse Event Reporting Systems. Drug Saf. 40 (7), 571–582. [Published Online First: Epub Date]. doi:10.1007/s40264-017-0523-4
Lobach, D., Sanders, G. D., Bright, T. J., Wong, A., Dhurjati, R., Bristow, E., et al. (2012). Enabling Health Care Decisionmaking Through Clinical Decision Support and Knowledge Management. Evid. Rep. Technol. Assess. (Full Rep. 2012 (203), 1–784.
Norén, G. N., Orre, R., Bate, A., and Edwards, I. R. (2007). Duplicate Detection in Adverse Drug Reaction Surveillance. Data Min. Knowl. Disc. 14 (3), 305–328. [Published Online First: Epub Date]. doi:10.1007/s10618-006-0052-8
Norén, G. N. (2017). The Power of the Case Narrative - Can it Be Brought to Bear on Duplicate Detection? Drug Saf. 40 (7), 543–546. [Published Online First: Epub Date]. doi:10.1007/s40264-017-0548-8
Ostropolets, A., Zhang, L., and Hripcsak, G. (2020). A Scoping Review of Clinical Decision Support Tools that Generate New Knowledge to Support Decision Making in Real Time. J. Am. Med. Inf. Assoc. 27 (12), 1968–1976. [Published Online First: Epub Date]. doi:10.1093/jamia/ocaa200
Keywords: pharmacovigilance, deduplication, decision support, natural language processing, safety surveillance
Citation: Kreimeyer K, Dang O, Spiker J, Gish P, Weintraub J, Wu E, Ball R and Botsis T (2022) Increased Confidence in Deduplication of Drug Safety Reports with Natural Language Processing of Narratives at the US Food and Drug Administration. Front. Drug Saf. Regul. 2:918897. doi: 10.3389/fdsfr.2022.918897
Received: 12 April 2022; Accepted: 09 May 2022;
Published: 15 June 2022.
Edited by:
Juan M. Banda, Georgia State University, United StatesReviewed by:
Pantelis Natsiavas, Centre for Research and Technology Hellas (CERTH), GreeceEileen Navarro Almario, Paraclete Professionals, LLC, United States
Copyright © 2022 Kreimeyer, Dang, Spiker, Gish, Weintraub, Wu, Ball and Botsis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taxiarchis Botsis, dGJvdHNpczFAamhtaS5lZHU=
 Kory Kreimeyer1
Kory Kreimeyer1 Oanh Dang
Oanh Dang Jonathan Spiker
Jonathan Spiker Robert Ball
Robert Ball Taxiarchis Botsis
Taxiarchis Botsis