- 1Department of Life Sciences, University of Trieste, Trieste, Italy
- 2National Research Council (CNR), Institute of Crystallography, Trieste, Italy
- 3CIRPeB, Research Centre on Bioactive Peptides “Carlo Pedone”, University of Naples “Federico II”, Napoli, Italy
Cathelicidins are a ubiquitous family of host defence antimicrobial peptides in vertebrate animals. Unlike other antimicrobial peptide families, it is defined by a large and relatively well conserved proregion rather than by the mature bioactive peptides themselves, which are highly diverse and conform to at least five different structural types, resulting in distinct modes of action. Cathelicidin-derived host defence peptides have a pleiotropic role in immunity, displaying both a direct antimicrobial activity and the ability to boost other host responses to infection and injury. The presence of a relatively well conserved proregion attached to a vast repertoire of structurally and functionally diverse peptides allows mining the increasing number of vertebrate genomes for lead sequences to potentially useful new anti-infective and/or immunomodulatory agents. This should increase the number of cathelicidin-based peptides entering clinical trials, which has been limited to date, despite considerable efforts in the last 2 decades.
1 Introduction
Cathelicidins are vertebrate host defence proteins characterised by a large and relatively well conserved proregion associated with a highly variable antimicrobial peptide that becomes active upon proteolytic release (Zanetti et al., 1990; 1995; Scocchi et al., 1992; Mookherjee et al., 2013; Tossi et al., 2017). They form one of the most important and widespread vertebrate host defence peptide (HDP) families and are a prime example of the molecular diversity of antimicrobial peptides. Since their discovery by Romeo and co-workers in the early 1990s (Zanetti et al., 1995), cathelicidin-derived peptides have demonstrated a remarkably broad functional repertoire, with direct antibiotic activities reported against bacterial, fungal, viral and parasitic microorganisms, accompanied by the ability to orchestrate other aspects of the immune response to infection and modulate inflammation (Agier et al., 2015; Hancock et al., 2016; van Harten et al., 2018; Alford et al., 2020).
Strictly speaking, cathelicidin refers to the proforms (Zanetti et al., 1995), while the HDPs are referred to either by their origin (e.g., CRAMP for Cathelin-Related AMP) or origin and size (e.g., BMAP-28 for Bovine Myeloid Antimicrobial Peptide of 28 residues), or structural features (e.g., LL-37 from the first two sequence residues and size) (Tomasinsig and Zanetti, 2005). However, it has become customary to refer also to the active HDPs as cathelicidins. Over the last 3 decades, these have been intensely studied both for their role in vertebrate host defence and for their potential to develop new anti-infective agents for biomedical or veterinary purposes (Mahlapuu et al., 2016; Alford et al., 2020; Valdez-Miramontes et al., 2021; Dlozi et al., 2022; Zhu et al., 2022).
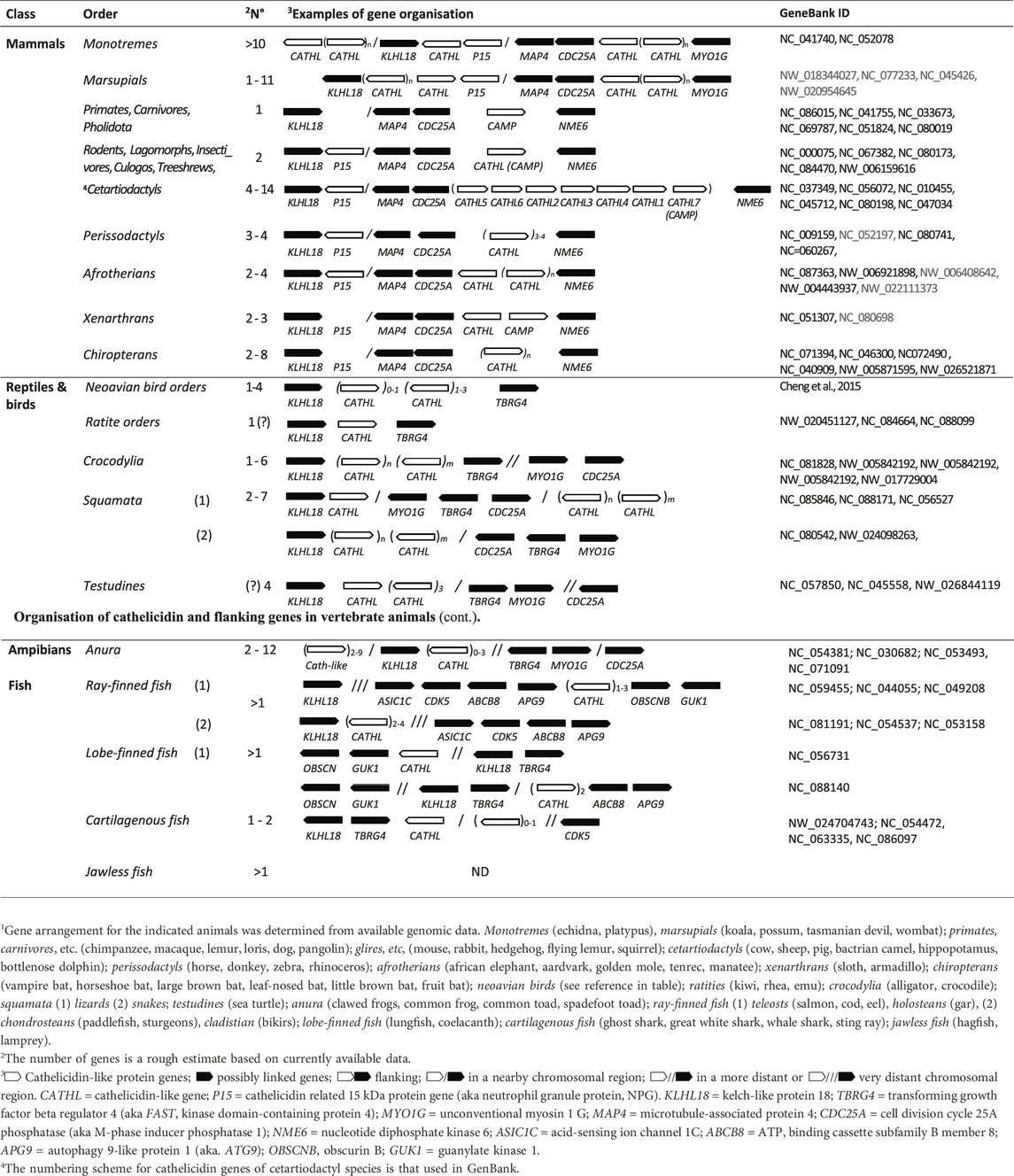
?A3B2 tlsb -.1pt?>The cathelicidins were discovered when Romeo’s group found that several structurally distinct and seemingly unrelated bovine HDPs all appeared to be synthesized in myeloid cells as larger precursors, from which they are proteolytically released. Cloning studies showed they all share a homologous proregion (Zanetti et al., 1993) with significant sequence identity to the porcine protein cathelin (Ritonja et al., 1989), hence the name cathelicidin (Zanetti et al., 1995). This and other research groups shortly added small disulphide-bridged peptides (Romeo et al., 1988; Kokryakov et al., 1993), and linear Trp-rich (Selsted et al., 1992), Pro-rich (Frank et al., 1990) and helical peptides (Tossi et al., 1994; Agerberth et al., 1995) from cow, pig, rabbit, human and other mammals (Zanetti, 2004). Some mammals (especially artiodactyls) were found to have several different cathelicidins, while others (e.g., primates, glires, rodents or carnivores) only one, orthologous to the only human cathelicidin HDP LL-37 (Xhindoli et al., 2016). Cathelicidins were then found to be present in all analysed vertebrates (see Figure 1), and many of their HDPs have been characterized with respect to their structure and antimicrobial and other roles within host defence. They differ significantly in size, sequence, structure, physico-chemical properties and biological functions, so that their relationship is essentially due to the presence of the relatively well conserved cathelin-like domain (CLD) in their proforms.
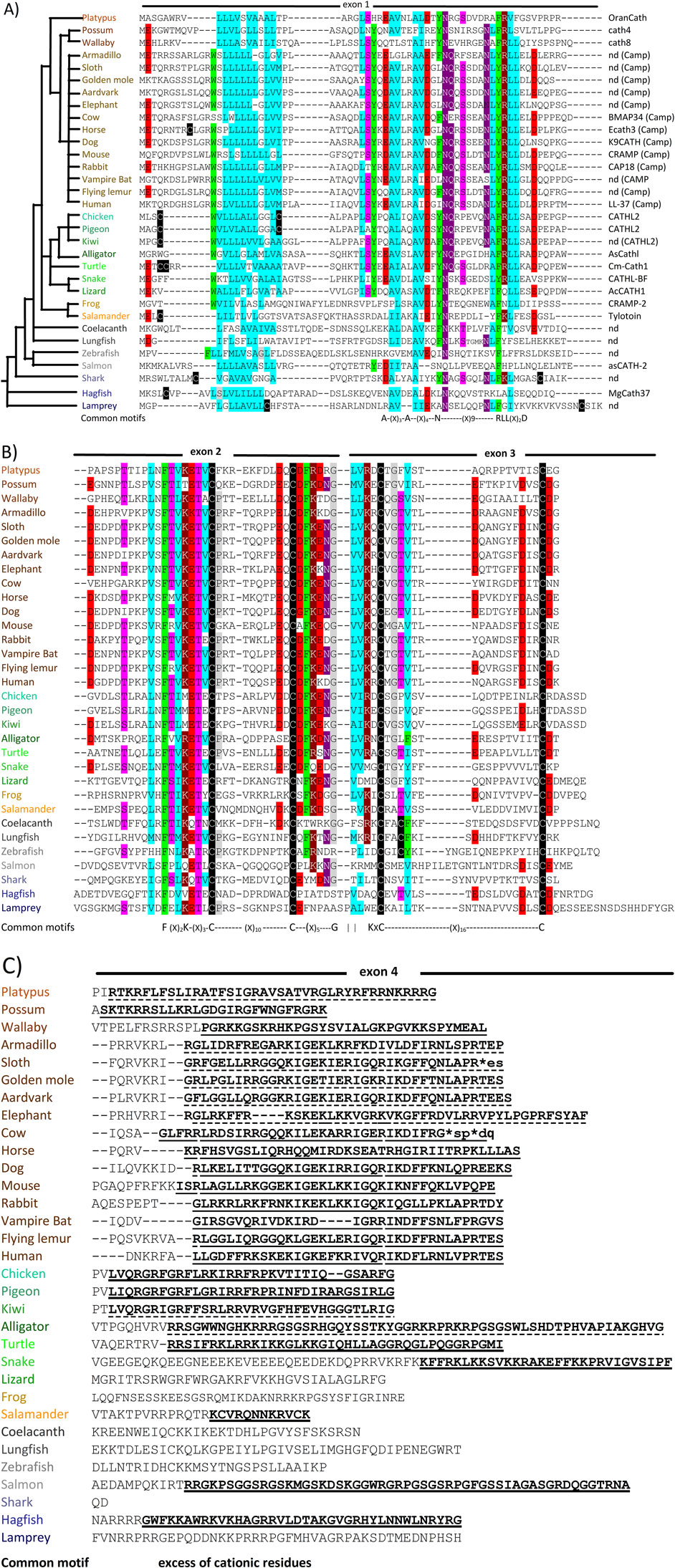
Figure 1. Examples of cathelicidin sequences selected from throughout the phylogenetic tree (shown top left of panel A) of vertebrate animals. The sequences are divided according to panel A) the encoding exon 1, panel B) exons 2 and 3 and panel C) exon 4 encoding the HDP. Gaps suggested by Clustall Ω are introduced to optimize alignment. Particularly conserved residues are highlighted in colours reflecting their physico-chemical characteristics (







This review provides a brief overview of the structural and functional features of cathelicidin proforms and HDPs, focusing on conserved aspects of the proregion and of genomic organisation that can facilitate mining of new HDPs with potentially interesting antimicrobial and/or immunomodulating functions. It provides a brief review of these functions, as well as considerations on their capacity to affect host cells, leading to beneficial or cytotoxic effects, and of their potential for the development of therapeutic agents. Many of these aspects have been the subject of several recent comprehensive reviews (Mookherjee et al., 2013; Hancock et al., 2016; Tossi et al., 2017; van Harten et al., 2018; Young-Speirs et al., 2018; Alford et al., 2020).
2 Distribution, expression and structural and functional characteristics of cathelicidins
2.1 Distribution and features of the proregion
Cathelicidins are ancient and widespread components of vertebrate innate immunity (see Figure 1), having been identified in basal vertebrate species (hagfish and lampreys) (Uzzell et al., 2003), all other types of fish (Masso-Silva and Diamond, 2014), amphibians (Hao et al., 2012), reptiles (van Hoek, 2014; van Hoek et al., 2019), birds (Wang et al., 2020; van Dijk et al., 2023) and all mammals (Mookherjee et al., 2013; Tossi et al., 2017; van Dijk et al., 2023). No cathelicidins are as yet reported in invertebrate animals. This implies that the cathelicidin gene family is at least 560 million years old, the estimated age of the latest common ancestor between vertebrates and lamprey/hagfish, according to TimeTree (Kumar et al., 2017).
Comparison of the HDP domains suggests that an ortholog of the human cathelicidin gene (CAMP, coding for the cathelicidin hCAP-18 and the HDP LL-37, see Figure 1) is present in all placental mammals, and often the only one present (primates, glires, carnivores and several other orders). For other orders (e.g., bats and perissodactyls) multiple cathelicidins are present and derive from duplication and diversification of the CAMP gene, while cetartiodactyls (ruminants, suids, camelids, hypopotamids and cetaceans) have a repertoir of structurally quite diverse HDPs, suggesting a more complex evolution (Xhindoli et al., 2016; Tossi et al., 2017; Zhu and Gao, 2017).
Cathelicidin genes consist of 4 exons, the first three encoding the preproregion and the fourth the HDP domain preceded by a variable number of proregion residues and the HDP cleavage site (see Figure 2). Despite indications otherwise in recent reviews, the HDP almost never begins in the 3rd exon. Mammalian cathelicidin genes cluster in syntenic chromosomal regions (Zhu, 2008a), and the presence of conserved flanking genes suggests this applies also to other vertebrates (see Table 1 in Section 3). Structural and sequence similarities place the proregion in the same superfamily as cystatins and kininogens, and it has been suggested that they derive from a common precursor (Zhu, 2008a), the cathelicidin gene having gained an additional 4th exon corresponding to the C-terminal HDP domain. The HDPs conform to several quite distinct structural types (Figures 2, 3).
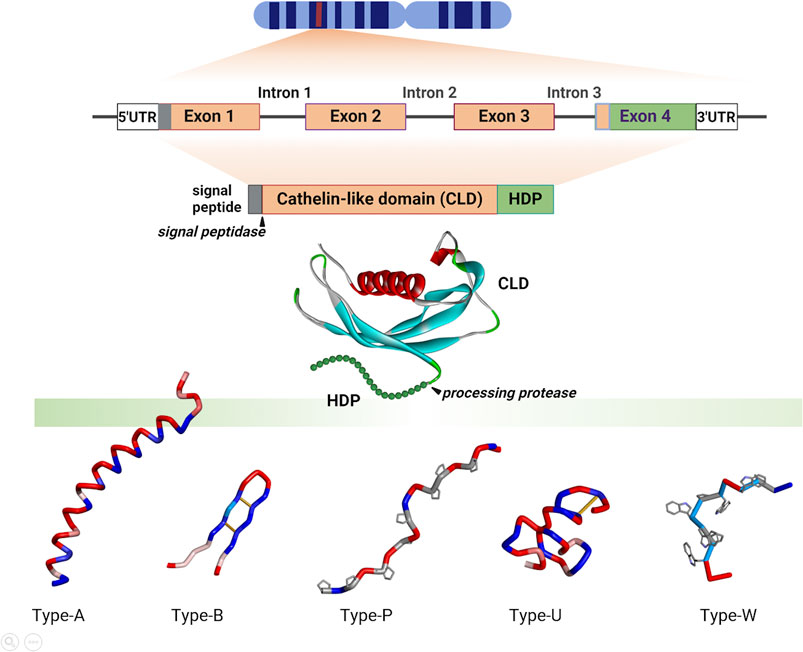
Figure 2. Schematic representation of cathelicidin gene, proform and HDP organisation and structures. The gene(s) encoding cathelicidin(s) are located on syntenic chromosomal regions with conserved flanking genes (see below Tables 1, 2). They are organized as four exons and three phase 0 introns, processed to the proform, with removal of the signal sequence by a signal peptidase before storage in granulocytes or secretion by epithelial cells. Mature HDPs of different structural types are released by extracellular or granule proteases (that differ depending on the organism and/or district). Some mammalian species express only one cathelicidin, which is invariably helical, other mammals and most non-mammalian vertebrates express multiple peptides. These can be α-helical (Type-A), disulfide stapled β-harpins (Type-B), Pro-rich peptides with extended conformations (Type-P) or Trp-rich peptides with wedge-shaped conformations (Type-W), or can be intrinsically unstructured (Type-U) as is the case of Gly/Ser-rich peptides found in some fish and reptile species. Structures were adapted from PDB coordinates using Biovia Discovery Studio 2021 [CLD, human cathelicidin pro-region (PDB 4EYC); Type-A, human LL-37 (PDB 2K6O); Type B, porcine protegrin (2MZ6); Type P, bovine Bac7 fragment 1-19 (5HAU); Type U, frog cathelicidin PY (2LR7); Type W, bovine Indolicidin (1G89)]. Created with the assistance of BioRender.com.
Figure 3. Selected cathelicidin HDP structures. These are divided into: Type-A, which display amphipathic, α-helical structures for at least part of their sequence [Homo sapiens LL-37, 2K6O; Bos taurus (cattle) BMAP27, 2KET; Oryctolagus cuniculus (rabbit) Cap18, 1LYP; Crotalus durissus terrificus (snake) Crotalicidin, 2MWT; Microhyla heymonsi (frog), CathMH; Gallus gallus (chicken) Fowlicidins 1, 2, and 3, (2AMN, 2GDL and 2HFR)]; Type B, with β-hairpin structures stapled by one or two disulphide bridges [Sus scrofa (pig) Protegrin (2MZ6); Physeter cathodon (whale) tandem dodecapeptide PcDode (7OSC)]; Type-P, with proline and arginine-rich extended structures [Bos taurus Bac7 (fragments 1-19, 5HAU, bound to bacterial ribosome, 15-21, 4JWD, bound to bacterial DnaK); Ovis aries (sheep) Bac7 (fragments 7-15, 4JWE and 35-43, 4JWI, bound to bacterial DnaK); Sus scrofa PR39 (fragment 5-12, 4EZO, bound to bacterial DnaK)]; Type-U, which are intrinsically unstructured and include some frog peptides and glycine- and serine-rich peptides from fish and reptiles [Aquarana catesbeiana Cathelicidin-PY (2LR7)]; Type-W [Bos taurus Indolicidin (1G89) and Sus scrofa Tritrpticin (1D6X)]. Structures were prepared using Biovia Discovery Studio 2021.
The original cathelicidin gene may have carried a helical peptide, as these are the most common and widespread, and was then duplicated, as most non-mammalian vertebrates, marsupials and monotremes have multiple cathelicidins. Some placental mammalian orders still have only one gene, carrying a helical peptide. Others have multiple helical genes. In cetartiodactyl species diversification appears to be driven by the insertion of new and very diverse HDP sequences after duplication (Zhu and Gao, 2009) so that they present a repertoire of different structural types (see Figure 2), which on the basis of characteristic conformations or representative amino acid residues are indicated as Type-A, -B, -P, -U and - W. The HDP domain in any case appears to be positively selected for variation (Zhu and Gao, 2017), while molecular mechanisms such as gene conversion may instead have acted to maintain a low variation in the proregion (Tomasinsig and Zanetti, 2005; Zanetti, 2005).
The structures of porcine and human CLDs are similar (PDB IDs: 1KWI, 1PFP, 4EYC, see Figure 2) and closely resemble that of cystatin (3GAX), consistent with the conservation of key sequence motifs (Sanchez et al., 2002; Yang et al., 2003; Kolodziejczyk et al., 2010; Pazgier et al., 2013). A long helical segment at the N-terminus nestles into a concave β-sheet platform, stabilized by two conserved disulphide bridges, to which the HDP is attached. The structures of the HDPs were determined separately (see Figure 3), and how these relate to the CLD in the proform has only been inferred by modelling (Sanchez et al., 2002). Although the CLD has a cystatin-like fold, it lacks key sequence elements required for cysteine protease inhibition (Pazgier et al., 2013).
The function(s) of the proregion remains controversial. It was initially hypothesised that it serves to keep the antimicrobial domain inactive until its release into the phagosome or extracellular medium (Scocchi et al., 1992; Sørensen et al., 1997; Zanetti et al., 2002), but it is questionable whether this alone justifies the conservation of the CLD. Proposals that the CLD has complementary antimicrobial activity to the HDP, or that it acts as a cathepsin inhibitor, are weakened by conflicting observations (Zaiou et al., 2003; Zhu, 2008a; Pazgier et al., 2013). In contrast, the hypothesis that the CLD serves as a pH-sensitive platform for the controlled proteolytic release of HDP at the right time and place (Sanchez et al., 2002) fits with the observations 1) that a substantial portion of the secreted human cathelicidin proform hCAP18 (see Figure 1) remains intact and bound to the surface of granulocytes or extracellular vesicles (Andersson et al., 2002; Stie et al., 2007), accompanying them to the site of infection and conferring a spatial specificity to HDP activation which concentrates the antimicrobial effect and minimizes cytotoxic effects, and 2) that relatively well conserved anionic residues form a strip on the surface of the CLD that would allow relevant interactions with the cationic HDP domain (Xhindoli et al., 2016). In addition, it would prevent active HDPs from being sequestered by plasma lipoproteins, which would occur if they were released too early (Wang et al., 1998; Sørensen et al., 1999).
2.2 Expression and processing
Cathelicidins are expressed in and secreted by various circulating immune cells or epithelial cells that respectively play an active role in host defence or form barriers against infection (Alford et al., 2020; Valdez-Miramontes et al., 2021). The expression pattern is varied, complex and regulated differently in different cell types and can be stimulated by both exogenous microbial components and endogenous factors, such as vitamin D in primates (Gombart et al., 2005; Lai and Gallo, 2009; Vandamme et al., 2012; van der Does et al., 2012). A similar expression pattern in leukocytes and epithelial cells is observed for bovine cathelicidins, but in this case the presence of several different genes allows differential expression at different sites (Tomasinsig et al., 2010; Kościuczuk et al., 2014; Whelehan et al., 2014). Avian and reptilian cathelicidins also generally derive from heterophils or epithelial cells (Alibardi, 2014; Chen et al., 2017; van Hoek et al., 2019; Wang et al., 2020), and are abundant in snake venom (de Barros et al., 2019). Cathelicidins are widely expressed in amphibian and fish tissues, both constitutively and upregulated by bacterial components during infection (Maier et al., 2008; Hao et al., 2012; Masso-Silva and Diamond, 2014). Manipulating this expression may help reduce the risk of infection in aquaculture environments, due to their direct antimicrobial and immunostimulatory capacities (D’Este et al., 2016).
In human and other mammals, cathelicidin gene products are channelled to storage granules, or secreted as proforms and the active HDP is released by serine proteases acting at appropriate cleavage sites. Elastase has been identified as the operational protease in several mammals (Scocchi et al., 1992), while proteinase-3 and kallikrein act in humans (Murakami et al., 2004; Zanetti, 2005; Yamasaki et al., 2006). The putative cleavage sites of avian, reptilian, amphibian and fish cathelicidins suggest that elastase-like proteases are involved (Maier et al., 2008; Gao et al., 2015; Sun et al., 2015; Furlan et al., 2018; van Hoek et al., 2019), but the operational proteases are largely unknown. Processing can be quite complex, and vary in different tissues; human LL-37, when secreted from eccrine glands or keratinocytes, can be further processed to shorter fragments in a manner that modulates its biological activities (Murakami et al., 2004). Furthermore, some cathelicidin HDPs are C-terminally amidated due to the presence of a Gly residue at the C-terminus or in a C-terminal sequences such as Gly-Lys-Arg or Gly-Arg-Arg, for example, see: (Agerberth et al., 1991; Selsted et al., 1992; Kokryakov et al., 1993; Zanetti et al., 1993; Skerlavaj et al., 1996).
2.3 Structural diversity of cathelicidin HDPs
Figure 3 shows some examples of cathelicidin HDP structures. The most common group is Type-A, with amphipathic helical structures, as found in hagfish, reptiles, amphibians, birds and mammals (see also Figure 1), suggesting that this is the ancestral type (Zhu, 2008a; Zhu and Gao, 2009). This conformation is also commonly found in AMPs unrelated to cathelicidins, and leads to a membranolytic antimicrobial mechanism (Tossi et al., 2000), suggesting convergent evolution of cathelicidin HDPs to this common function.
Cetartiodactyl cathelicidin HDPs show the greatest diversity of structural types (Tomasinsig and Zanetti, 2005; Scocchi et al., 2011). In addition to various Type-A peptides, there are long, Pro- and Arg-rich peptides with extended structures (Type-P), and small, wedge-shaped, Trp-rich peptides (type W) (see Figure 2) (Schibli et al., 1999; Rozek et al., 2000; Tomasinsig and Zanetti, 2005). Another type of cathelicidin HDP peculiar to this mammalian order are small peptides with β-hairpin conformations stapled by one or two disulphide bonds (Type-B). These include the bovid and cetacean dodecapeptides and the porcine protegrin, which exists in several allelic forms (PG-1 to −5) (Choi et al., 2014). Some fish and amphibian cathelicidins also show paired cysteine motifs, but in conjunction with other structural types (e.g., Type-U, see below).
While the majority of cathelicidins from ray-finned fish have sequences similar to that shown for zebrafish in Figure 1, they sometimes bear long, linear peptides that are particularly rich in Gly and Ser residues (Scocchi et al., 2009b). These are found especially in salmonids, and can be divided into two subgroups, with the presence of two cysteine residues in the N-terminal part of the HDP region in Cath1 type peptides, which is absent in Cath2 type peptides (Maier et al., 2008). The GS-rich stretch likely remains unstructured even in membrane-like environments, suggesting they are Type-U low complexity, intrinsically disordered sequences (D’Este et al., 2016). GS-rich cathelicidin peptides are also found in some amphibian species. In some cases, cathelicidin HDPs have structures conforming to several structural types. This is the case of the frog peptide Cathelicidin-PY (see Figure 3), which has a very short helical stretch and disulphide bridge within an essentially Type-U structure (Wei et al., 2013).
Cathelicidin HDPs are being identified in an increasing number of non-mammalian species, and it is not always facile to predict the structural type (see Table 1). In some cases, a low cationicity is apparent, which may be an indication that a direct antimicrobial action may not be the principal function.
2.4 Structure-dependent mode of action of cathelicidins
As described above, cathelicidin HDPs essentially conform to the five structural types shown in Figure 3, although some may have features of more than one type. A common aspect is a pronounced cationicity that favours their interaction with bacterial membranes upon proteolytic release from the proform, and many indeed proved to be membrane active, but for some the antimicrobial effect is not primarily based on disruption of the bacterial membrane. The mode of action is summarized schematically in Figure 4, where release and approach to the membrane are shown as step (1).
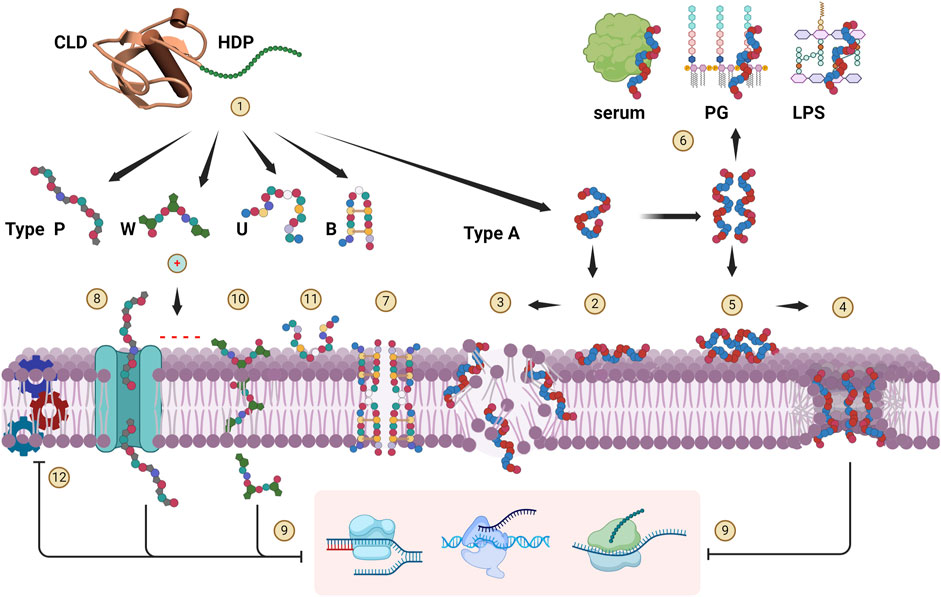
Figure 4. Cathelicidin HDP modes of action. Upon release from the CLD (1), cationic HDPs are attracted to the anionic surface of bacterial membranes. Type-A peptides tend to be disordered, and adopt an amphipathic, helical conformation at the membrane surface and insert into it (2). When a critical concentration is reached, they can breach the membrane bilayer in various ways. They can act in a detergent-like manner (carpet mechanism) (3), or form discrete toroidal pores (4). Helical peptides such as human LL-37 can adopt a helical structure already in bulk solution, which promotes aggregation so that they approach the membrane as oligomeric bundles (5), favouring pore formation. They are however more susceptible to sequestration interactions with serum or medium components and bacterial peptidoglycan (PG) components (e.g., LTA) or lipopolysaccharide (LPS) (6), so their antimicrobial activity is quite salt- and medium-sensitive. Type B peptides are reported to penetrate the membrane, oligomerize and form multimeric pores (7). Type P peptides can internalize into some bacteria via specific transport proteins (8) and inhibit cytoplasmic targets (9) (specifically ribosomal subunits) but can also act by membrane lysis at higher concentrations (mechanism 3). Type W peptides are reported to penetrate into the bacterial cytoplasm without compromising the membrane (10), where they then act by selective inhibition of DNA transcription. Inhibition of replication and/or transcription (9) has also been proposed for some Type A peptides once they enter the cell via mechanisms (3) or (4). Intrinsically unstructured type U peptides act on the bacterial surface (11), and although their mechanism of action is not yet known, the antimicrobial activity is quite salt-sensitive and not primarily membranolytic. All HDP types likely saturate the bacterial membrane surface at active concentrations, and inhibition of the membrane protein machinery (12) has been hypothesised to be an important aspect of their antimicrobial action. Created with BioRender.com.
Type Α cathelicidin HDPs are considered primarily membranolytic. Their cationic and amphipathic structure enables efficient interaction with, and insertion into the microbial membrane. The active conformation may form only upon contact with the membrane, where the peptides undergo a transition from an unstructured globule to a helical conformation partly inserted into the lipid bilayer [Figure 4 (2)], whereupon either a detergent-like disruption occurs when a critical concentration is reached (carpet model) (3), and/or cooperative formation of discrete cavities occurs (toroidal pore mechanism) (4) (Tossi et al., 2000). In some cases, peptides can adopt the amphipathic helical conformation in bulk solution, under physiological conditions, leading to oligomerization, so that they approach the membrane as helical bundles (5). These mechanisms may both contribute to bacterial killing, but peptides may favour one mechanism over the other. For example, rhesus RL-37 does not oligomerize and favours the carpet type mechanism (3), whereas the closely related human LL-37 oligomerizes and favours pore formation (4) (Morgera et al., 2009; Xhindoli et al., 2014; 2016). Another consequence of oligomerization is that the preformed helices are susceptible to sequestration by interacting with bacterial membrane or serum components (6), significantly affecting antimicrobial potency and sensitivity to the environment (Tomasinsig et al., 2009; Xhindoli et al., 2014).
With regard to Type-B peptides, the interaction of porcine protegrin with membranes has been extensively studied, and they are reported to act via a membranolytic mechanism (Bolintineanu et al., 2012; Lazaridis et al., 2013; Usachev et al., 2016). They interact with the membrane as monomers and then oligomerise to form an octameric pore [Figure 4 (7)] (Lazaridis et al., 2013; Usachev et al., 2016). Bovine dodecapeptide may also form S-S-stabilized β-hairpins in solution and dimerize at the bacterial surface (Raj et al., 2000), acting in a similar manner. However, the mode of action is made more complex by the possibility that it forms covalent dimers involving intermolecular disulfide bridge formation (Storici et al., 1996). Interestingly, in this case a parallel or antiparallel covalent dimeric arrangement does not seem to have a dramatic effect on the antimicrobial activity with respect to that of the β-hairpin monomer (Lee et al., 2008).
Type-P cathelicidin HDPs, which belong to the structural group of proline and arginine rich AMPs known as PrAMPs, have a distinct mode of action that relies less on membrane disruption (Scocchi et al., 2011; 2016; Li et al., 2014). Their extended structures do not change significanty upon contact with bacterial membrane surfaces, where they accumulate and then internalize into the cytoplasm also using specific membrane transporters, where they inactivate internal targets [Figure 4 (8) and (9)]. This mechanism has been shown to apply also to unrelated proline-rich peptides from arthropods, another example of convergent evolution of common strutural and functional features (Scocchi et al., 2011; Krizsan et al., 2014). This mechanism is selective with respect to the target bacteria, as only those expressing the transport system are strongly affected (e.g., Gram-negative bacteria such as E. coli, S. Typhimurium and A. baumanii, but not P. aeruginosa or any Gram-positive bacteria), and ii) stereoselective with respect to the peptide itself (Runti et al., 2013; Guida et al., 2015; Scocchi et al., 2016). In contrast to membranolytic peptides, in which the all-D enantiomer works just as well, for Type-P peptides it loses activity. This is likely due to stereoselective requirements for transport and/or for inactivation of the cytoplasmic target (Guida et al., 2015), the bacterial ribosome (Mardirossian et al., 2014; 2018a; Gagnon et al., 2016; Graf et al., 2017). Another bacterial protein that Pro-rich peptides in general bind to, including cathelicidin HDPs, is the chaperone DnaK (Cudic and Otvos Jr, 2002; Scocchi et al., 2009a).
Type-W peptides are so far limited to bovid indolicidins and porcine tritrpticin (see Figure 2) and act by another distinct mechanism. Due to the presence of Trp residues, they have a strong tendency to interact with bacterial membranes which they cross to hit internal targets [Figure 4 (9) and (10)]. They have a wedge-shaped conformation and after partitioning near the membrane-water interface then appear to enter the bacterial cell without significantly compromising membrane integrity, to then selectively inhibit DNA synthesis (Hsu et al., 2005; Chan et al., 2006; Ghosh et al., 2014; Shagaghi et al., 2016; Batista Araujo et al., 2022). It should be considered that indolicidin has 3 Pro residues so may have characteristics of P-type, and that cetacian Type-P peptides are also quite rich in Trp residues, so they may represent a cross between the two structural types and allow a more efficient penetration even in the absence of a suitable transporter (Mardirossian et al., 2018b; Sola et al., 2020).
Type-U peptides are mainly found in non-mammalian vertebrates and have the least well defined mechanism of action, which again appears to be distinct from the others, emphasising that cathelicidin HDPs cover a very broad structural and functional space. They contain Gly and Ser rich sequences (see Figure 1 for examples), which in salmonid fish are usually quite long and sometimes heterogeneous, with cysteine-bridged motifs or other types of flanking domains (Scocchi et al., 2009b). As a result, the mode of action has been studied for rationally selected GS-rich fragments rather than the whole peptide, and it appears that their intrinsically unstructured extended conformations are not strongly affected by membrane interaction (Broekman et al., 2011; D’Este et al., 2016). Moreover, they have a relatively low proportion of hydrophobic residues, so they probably only interact with the surface of membranes and do not insert into them [Figure 4 (11)]. This is consistent with their killing mechanism, which is quite salt sensitive, although the microbicidal mode of action is still unclear. Little is known about the mode of action of anuran GS-rich HDPs, but they probably have similar properties (Hao et al., 2012).
Regardless of the mode of action, all types initially interact with the membrane, and since their active concentrations are in the micromolar range, they completely saturate the bacterial surface (Loffredo et al., 2021), so they are likely to come into contact with vital protein machineries located in the bacterial membranes. This would affect bioenergetics, transport and maintenance of the cell wall. This is known as the sand-in-the-gearbox effect (Pag et al., 2008; Vaezi et al., 2014), and could be a relevant component of their killing mechanism [Figure 4 (12)].
2.5 Pleiotropic roles of cathelicidins in host defence
Most cathelicidin HDPs are first tested for their direct antimicrobial activity in vitro, even though this is not necessarily their main role in host defence. They often also show a significant ability to influence other aspects of immunity and healing, such as binding and sequestering bacterial components (e.g., LPS or LTA), recruiting or modulating the activities of cellular components of innate and adaptive immunity, and stimulating cell growth in wound healing (Lai and Gallo, 2009; Linde et al., 2013; Hancock et al., 2016; van Harten et al., 2018; Alford et al., 2020). The literature on direct antibiotic and immunomodulatory activities is extensive, partly due to the countless variants that have been developed over the years to investigate structure/function relationships or in an attempt to optimise activity for potential therapeutic applications.
In summary, the direct antibiotic activities of Type-Α and -Β cathelicidin HDPs, which act via membranolytic mechanisms, tend to be more potent and broad-spectrum in vitro. However, outside their physiological context, this is accompanied by appreciable toxicity for eukaryotic cells at their active concentrations, often measured in terms of their haemolytic activity. For this reason, when peptides of these types have been investigated as therapeutic agents, they exhibit appreciable toxicity close to their active concentrations. This also seems to be the case for Type-W peptides. Type-P peptides tend to be significantly less cytotoxic, but have a narrower activity range (Scocchi et al., 2016). Type U peptides are the least well characterized.
It is interesting to note that a certain ability to modulate host cell activities has been found for all structural types. Helical peptides, and in particular LL-37 and mouse CRAMP, have a wealth of reported activities, including attracting immune cells to the site of infection, modulating inflammatory responses via release of cytokines, binding to and inactivating endotoxins, promoting wound healing, etc (Kahlenberg and Kaplan, 2013; Fabisiak et al., 2016; Xhindoli et al., 2016; Krepel and Wang, 2019). Protegrin analogues (Type-B), indolicidin (Type-W) and Pro-rich (Type-P) cathelicidins also show analogous activities, despite their significant structural diversity (Bowdish et al., 2005; Djanani et al., 2006; Tomasinsig et al., 2006; Kin et al., 2011; Veldhuizen et al., 2014; Zughaier et al., 2014; Gupta et al., 2015; Nakagawa and Gallo, 2015). In general, immunomodulatory activities on host cells are thought to be due to receptor activation, but how cathelicidin HDPs exerts their action is not well understood. It could be that some of them act in a non-canonical manner by accumulating in the membrane surrounding various receptors, possibly favouring the cholesterol and sphingomyelin-rich lipid rafts where receptors often reside, and affecting their transmembrane domains rather than interacting with specific ligand binding sites. This is consistent with a frequently observed promiscuous and generally low-affinity activity, and is supported by the fact that at least for LL-37, activation of some receptors by the all-d enantiomer is as effective as the native enantiomer (Xhindoli et al., 2016). Different stereochemistry would not allow a similar interaction with a binding site, whereas the ability for helical structuring, oligomerization and membrane interaction is analogous.
3 Genomic organization and mining for cathelicidins
3.1 Bioinformatic strategies for the identification of novel cathelicidins
The Vertebrate Genome Project explicitly aims to generate the reference genomes of all nearly 70,000 extant vertebrate species (Rhie et al., 2021), providing new opportunities for data mining approaches that facilitate the identification of novel cathelicidins while bypassing traditional, labour-intensive isolation techniques. However, the primary sequence diversity among cathelicidin precursors and high variation of the HDP region make this task far from trivial, as with other bioactive peptides (Coelho et al., 2024). Despite conserved aspects of the proregion (see Figure 1), cathelicidins within the same vertebrate class can have up to 80% dissimilarity at the amino acid level. Therefore, although BLAST based methods have proven to be reliable for identifying new members when applied to phylogenetically closely related species (Zhu, 2008b; Ishige et al., 2017; Kim et al., 2017; Lastra et al., 2018; Choi et al., 2022; Kannoth et al., 2023), they may be inadequate in understudied or highly divergent animal groups. Although the reliability of these approaches can be improved by the use of strict orthology inference methods (Castellanos et al., 2023), they are not robust for vertebrate clades in which cathelicidin sequences are as yet poorly represented. The relatively limited number of cathelicidin sequences documented in fish, although coding genes are widely distributed, likely stems from these technical constraints.
In this respect, methods based on Hidden Markov Model (HMM) prove more efficient, as they allow to recognize common structural attributes inherent to all members of a given protein family, which facilitates an appropriate weighting of the evidence provided by conserved sites associated with specific positions, such as those defining the CLD. A benchmark HMM profile for cathelicidins is provided by Pfam entry PF00666, which belongs to the PepSY clan that also includes structurally similar molecules such as cystatins (Kordiš and Turk, 2009). Nonetheless, the cathelicidin HMM profile is based on the alignment of a limited data set of sequences that suffers from a taxonomic bias skewed heavily towards mammals. Consequently, while it has proven to be robust in identifying novel cathelicidins within mammalian and avian species (Cheng et al., 2015; Zhang et al., 2019; Xiao et al., 2020), it was less effective when applied to other vertebrate classes. Indeed, many authentic cathelicidin sequences from fish would escape unequivocal identification using this method, either due to overlap with the HMM associated with cystatins (PF00031), or failure to meet the standard detection threshold for significance. More advanced approaches can iteratively build a new HMM profile from scratch, a process that relies on the initial identification of reliable seed sequences through BLAST searches and may involve the use of the jackhammer tool from the HMMER package (Finn et al., 2011). This sophisticated strategy has recently demonstrated its efficacy and enabled the detection of numerous previously undiscovered cathelicidin genes in frogs (Tang et al., 2024).
Regardless of the chosen strategy, the ability to detect novel sequences strongly depends on the quality of annotation of the selected genomes, which varies greatly depending on the bioinformatics pipeline used and available supporting data. In practice, annotation of cathelicidin loci in many vertebrates lacks precision, with predictions being absent, incomplete and occasionally incorrect. For example, several genuine cathelicidin genes in non-mammalian species are mislabelled either as “kininogen” or as “secreted phosphoprotein 24,” which have cathelin-like regions, severely hampering data mining approaches based on keyword searches (Hu et al., 1995; Zhou et al., 2009; Pérez de la Lastra et al., 2021). Furthermore, a correctly identified proregion (exons 1–3) can be linked to an incorrect 4th exon encoding the HDP, as this is the most variable region.
From this perspective, the analysis of de novo assembled transcriptomes can be useful. First, it allows the identification of the complete sequence of the protein precursor without uncertainties regarding the correct identification of splicing acceptor and donor sites. Second, it is a concrete indication of the likely biological relevance of the encoded protein, as it distinguishes functional genes from cathelicidin-like pseudogenes that are often reported (Whelehan et al., 2014; Cheng et al., 2015; Zhang et al., 2019; Peel et al., 2021; van Dijk et al., 2023). This approach was initially successfully used to screen Expressed Sequence Tags (EST) datasets (Xiao et al., 2006), and has recently allowed the identification of novel cathelicidins in several animal species (Helbing et al., 2019; Zhong et al., 2020; Kannoth et al., 2023). On the other hand, the sole availability of transcriptome data is limiting, as it depends on the genes being expressed in the tissue that was selected for sequencing, and if similar paralogous genes are present the risk of overlooking some of them due to chimeric assemblies increases.
For genomics studies, information on synteny helps identifying cathelicidin gene clusters in newly released genomes. While significant structural genomic rearrangements are not uncommon in nature, there are indications that the genes flanking the cathelicidin gene clusters are generally well conserved (see below), at least within the same order, and often across higher taxonomic ranks as well. Therefore, locating the genomic region between these molecular markers, even in the absence of available gene annotations, can help identify novel cathelicidin genes (Cheng et al., 2015; van Hoek et al., 2019), by also exploiting the conserved architecture of all cathelicidin genes which comprises four exons and three phase 0 introns. This approach can leverage a combination of methods based on sequence homology detection and alignment with RNA-seq data to delineate putative exons. However, manual curation remains a crucial aspect for validating these predictive methods and correcting any errors.
3.2 Genomic organization of cathelicidins
The success of bioinformatic methods such as BLASTing of genome or transcriptome databases with known sequences or HMM profiles depends on the availability of a sufficiently representative number of known or inferred sequences. In this respect, InterPro entry PF00666 contains almost 1100 such sequences, which may include homologs from closely related species to the ones being searched. Otherwise, only some or no cathelicidin genes may be found, especially in vertebrate groups where cathelicidins are poorly represented. In these cases, it may be useful to rely on conserved flanking genes to facilitate identification. For example, we have found that in placental mammals the major cathelicidin gene cluster is located between NME6 and CDC25A and/or MAP4 (see Table 2), which is also confirmed in the literature (Ahn et al., 2022). In birds and reptiles, the indicated flanking genes are TBRG4 (a.k.a. FASTK) and KLHL18, respectively (Cheng et al., 2015; van Hoek et al., 2019). Based on this information, it was possible to scan well-annotated vertebrate genomes from different clades to build a picture of syntenic clusters and identify several other linked genes that either surround or are in proximity to cathelicidin gene clusters (Tossi et al., unpublished results), as shown in Table 2.
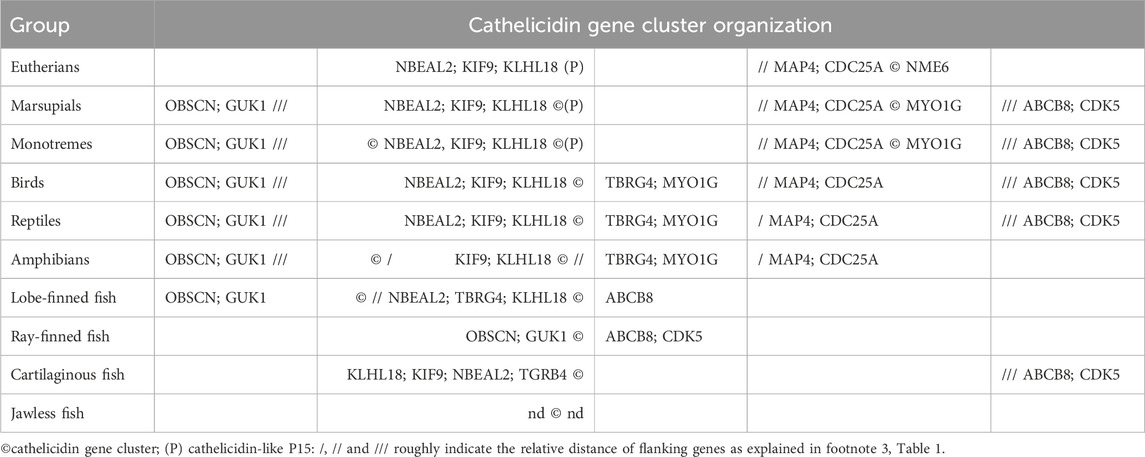
Table 2. Organization of cathelicidin gene clusters with respect to that of genes that are apparently linked to them.
Among mammals, monotremes exhibit the most diverse organization of cathelicidin-like gene clusters, with two closely spaced clusters flanked on one side by KLHL18, suggesting a link to syntenic clusters in non-mammalian vertebrates, and on the other side by MYO1G, with MAP4 and CDC25A in between, suggesting a link to syntenic clustering in placental mammals. A third cluster is located on the other side of KLHL18. The latter appears to be absent in marsupials, but otherwise the cathelicidin gene organization is similar. In placental mammals, two clusters are present in most orders, the principal one being flanked by MAP4/CDC25A on one side and NME6 on the other. The second cluster contains only the cathelicidin-like protein P15, also known as neutrophil granule protein, which was described some time ago as a divergent member of the cathelicidin family in rabbits. It does not release an antimicrobial HDP but rather synergizes with other immune proteins and binds LPS (Levy, 1996). P15 cathelicidin genes appear to be present in most placental mammals except for primates, carnivores and pangolins (see Supplementary Figure S1) that appear to have one only cathelicidin that is orthologous to the human CAMP gene.
Birds appear to have a single cathelicidin cluster flanked by KLHL18 and TBRG4, with the number of genes (1–4) varying according to order and species (Cheng et al., 2015). Reptiles have a more complex arrangement of cathelicidin genes - in general, crocodilians, snakes and turtles have a cathelicidin gene cluster flanked by KLHL18 on one side, and with MYO1G, TBRG4 and CDC25A at a short distance on the other side. For lizards, this cluster is reduced to one or two genes and a second larger cathelicidin gene cluster is located on the other side of CDC25A. Amphibians have a similar cathelicidin gene arrangement, but with the second large cluster on the other side of KLHL18.
The cathelicidin gene clusters of fish may be flanked by different genes than in other vertebrates, and determining the arrangement is complicated by the lack of annotations. In Teleostean and Holostean orders of ray-finned fish the cathelicidin gene cluster is located near KLHL18, whereas in Chondostrean and Cladistian orders it is located quite far from it and flanked by genes such as ASIC1C, CDK5, ABCB8, APG9 on one side and OBSCN and GUK1 on the other. In lungfish the cathelicidin gene cluster is flanked by OBSCN and GUK1 on one side and far from KLHL18 and TBRG4 on the other. With coelacanth there is a cathelicidin gene cluster between KLHL18 and TBRG4 on one side and ABCB8 and APG9 on the other. Cartilaginous fish generally have one or two cathelicidin genes. One type of gene that is always present and close to KLHL18 and TBRG4 encodes a rather odd cathelicidin in which the fourth exon consists of only one or two residues and therefore has no HDP domain. This is the only gene present in many sharks, whereas in rays there is a second gene in which the fourth exon encodes a small peptide. It is more distant from KLHL18 and TBRG4 and has CDK5 at some distance on the other side. For jawless fish, there are not enough well-annotated genomes for a comprehensive analysis, but the cathelicidin gene cluster does not seem to be linked to any of the genes found for other vertebrates.
Although the arrangement of cathelicidin genes shown in Table 1 differs somewhat between vertebrates from different orders it suggests a degree of syntenic conservation. Cathelicidin genes are largely located in clusters within a single chromosome, and the same types of linked genes generally occur on the same chromosome. The overall arrangement can be better appreciated in Table 2, which considers the approximate positioning of the cathelicidin gene clusters and possibly linked genes.
4 Therapeutic potential of cathelicidins
Since their discovery, the potential of antimicrobial peptides for therapeutic purposes has been recognised and widely discussed in the literature. After encouraging pre-clinical testing in in vitro, ex vivo and animal model studies, several AMPs or their synthetic derivatives have been investigated in human clinical trials, including cathelicidin peptides and analogues (Koo and Seo, 2019; Browne et al., 2020; Dijksteel et al., 2021; Moretta et al., 2021). Focusing on trials involving cathelicidin HDPs, despite significant efforts that have been made over the past 2 decades, none have as yet reached clinical use. In fact, some trials are still ongoing while others have been discontinued due to lack of efficacy, unfavourable pharmacokinetic profiles, adverse effects or failure to show improved efficacy with respect to conventional treatments.
Among the first cathelicidins to be developed and tested for potential clinical use were the Type-B peptide iseganan, based on protegrin, and Type-W peptide omiganan based on indolicidin (Toney, 2002; Isaacson, 2003) (see Supplementary Table S1). Both HDPs underwent a complex optimization processes to make them useful for therapeutic purposes, which involved a series of modifications such as residue replacements to increase cationicity or to modulate the hydrophobicity, introduction of non-proteogenic residues or cyclization and C-terminal capping or inverting and/or enantiomerizing the sequence to improve both the stability and activity (Chen et al., 2000; Staubitz et al., 2001; Rozek et al., 2003; Ryge et al., 2004; Ando et al., 2010).
As membrane active molecules, these peptides are therapeutically oriented towards topical use, and iseganan was initially investigated in a clinical trial for preventing oral mucositis in cancer patients undergoing radiation therapy for head and neck cancer, but failed in Phase III due to an apparent lack of efficacy (Giles et al., 2003; Trotti et al., 2004). However, a later study on patients undergoing chemotherapy found it significantly reduces the total oral aerobic bacterial and fungal load in patients undergoing stomatotoxic chemotherapy, showing a clear potential as an oral antimicrobial agent, and should be re-evaluated in the context of increasing resistance to conventional anti-infective agents (Dijksteel et al., 2021; Liang and Sonis, 2024).
Omiganan pentahydrochloride is a 12-mer derivative of indolicidin that demonstrated in vitro activity against a significant number of infective clinical isolates, including most ESKAPE pathogens and Candida spp. (Sader et al., 2004; Żyrek et al., 2021). It underwent clinical trials for different topical applications including the prevention of catheter-related infections, and treatment of acne, rosacea, atopic dermatitis and papillomavirus-induced genital lesions (Fritsche et al., 2008; Zouboulis et al., 2017; Niemeyer-van der Kolk et al., 2020) and is considered also for vulvovaginal candidiasis and other fungal infections (Rubinchik et al., 2009; Czechowicz et al., 2021; Żyrek et al., 2021). The results from these trials are mostly not yet disclosed or under review so it is not possible at the moment to assess their success.
With regard to Type-A peptides, a lot of interest has been placed on the human cathelicidin HDP LL-37, both as a potential therapeutic agent or as a marker for health or pathological conditions. For example, its levels have been correlated with conditions such as psoriasis, atopic dermatitis and periodontal disease (Kahlenberg and Kaplan, 2013; Lande et al., 2014; Hancock et al., 2016; Turkoglu et al., 2017; Antal et al., 2022; Bhattacharjya et al., 2024). This has prompted numerous clinical studies that are not only evaluating it as a drug for the treatment of pathologic conditions but also investigating its potential role as a biomarker (see Supplementary Table S1). Currently, LL37 is under clinical investigation for the treatment of hard-to-heal venous leg ulcers (HTH VLUs) in the Swedish study LL-37001B (EudraCT: 2012-002100-41). These are amongst the most prevalent type of chronic wounds and affect approximately 1%–3% of the older population in Western countries (Franks et al., 2016). The investigation was prompted by the observation that endogenous LL-37 is present in large quantities in acute wounds, whereas it is absent in chronic wounds, suggesting a critical role of this peptide in the healing process (Heilborn et al., 2003; Fabisiak et al., 2016). Moreover, it was observed that skin-targeted administration of LL-37 in ex and in vivo models of human acute and chronic wounds improved reepithelization and closure (Carretero et al., 2008; Steinstraesser et al., 2014). Results from phase I/II trials showed that local application of LL-37 twice a week significantly enhanced the healing rate without causing any systemic safety or local tolerability concerns (Grönberg et al., 2014). Subsequently, a phase II multicentric prospective trial (EudraCT: 2018-000536-10) indicated improved healing in relatively large wounds with a negative prognostic factor for healing (Mahlapuu et al., 2021). A recent clinical controlled trial in the USA (NCT04098562) was conducted to assess the effectiveness of a stable, LL-37-containing cream in diabetic foot ulcers (DFUs) and was found to improve the granulation index and healing rate of wounds (Wu et al., 2018; Miranda et al., 2023). These clinical studies support the notion that LL-37 topically-used is safe and well tolerated.
An improvement in the management of wounds may arise from the encapsulation of the HDP in a three-dimensional hydrogel system that allows effective delivery, prolonged stability and efficient release of the bioactive molecule. The use of dressings based on different types of self-healing multifunctional hydrogel system showed a significantly improved angiogenesis and reduced wound closure times in animal wound models (Hao et al., 2023; Jelodari et al., 2023).
As reported in section 2.2, expression of the single cathelicidin gene (CAMP) present in humans and other primates, is upregulated by 1,25-dihydroxyvitamin D3, in addition to its increased expression in response to pathogen exposure and inflammation. In fact, it has been determined that vitamin D response element (VDRE) is localized upstream to the promoter region of the CAMP gene, which binds the vitamin D-activated nuclear receptor (VDR), inducing gene expression (Wang et al., 2004; Gombart et al., 2005; 2009; White, 2010). Several clinical studies have suggested a link between vitamin D deficiency, inflammatory pathological conditions (such as urinary tract infections, ulcerative colitis and Crohn’s disease) and decreased expression of LL-37 (White, 2018; Deng et al., 2019; Gubatan et al., 2020). These and many other observations suggest that treatment with vitamin D could be a way to increase LL-37 production in contexts in which its levels are insufficient to counteract specific infections and inflammation.
With respect to the use of LL-37 as a biomarker, the peptide was found to represent a T-cell autoantigen in the majority of patients with moderate-to-severe psoriasis and psoriatic arthritis (Lande et al., 2014; Fuentes-Duculan et al., 2017; Frasca et al., 2018). Abnormally high levels of LL-37 are also found in rosacea (Cribier, 2022) where it contributes to its pathophysiology by stimulating cytokine release, angiogenesis, chemotaxis and other pro-inflammatory events. Its presence is due to overexpression and release of the inactive hCAP18 precursor associated with increased levels of the processing enzyme kallikrein 5 from the stratum corneum of epidermidis, suggesting that inhibition of the enzyme might improve the clinical signs of rosacea. A proof of concept pilot study showed the first clinical evidence of the involvement of kallikrein in the pathogenesis of rosacea and its inhibition did correlated with clinical improvement of the disease (Yamasaki et al., 2007; Two et al., 2014; Thibaut de Ménonville et al., 2017).
A problem with the use of Type-A peptides like LL-37 as anti-infective agents is that being membranolytic they are quite cytotoxic and tend to have rather narrow therapeutic windows, even for use as topic agents. Redesigning them to improve this window can however change their mode of action and reduce their multifunctional effects. A great effort has been expended in this direction to develop LL-37 fragments and variants able to maintain or improve antimicrobial activity while reducing toxicity towards eukaryotic cells and susceptibility to degradation by proteases present in mammalian body fluids. Truncation or replacement of residues in the LL-37 can in fact improve its antibacterial capacity, as long as truncation does not significantly alter the overall charge and amphipathic features. For example, central fragments [e.g., LL-37 (7–27) or LL-37 (5–24)] maintain the antibacterial activity against both Gram-positive and Gram-negative bacteria while reducing haemolytic activity with respect to the native peptide (Braff et al., 2005; Thennarasu et al., 2010; Wang et al., 2012; Krishnamoorthy et al., 2023).
The most advanced clinical trial among the truncated variants of LL-37 is the peptide P60.4Ac that was developed from an overlapping synthetic library (Nell et al., 2006). This peptide, derived from the sequence 13-36 of LL-37 with some residue substitutions to improve amphipathicity, was positively tested for ototoxicity in guinea pigs and in vitro antimicrobial activity on reference bacterial and fungal strains. Subsequent studies in patients suffering of chronic suppurative otitis media (CSOM) found that the peptide was safe and well-tolerated when used as ototopical drops. A randomized, double blind, placebo-controlled, multicentre phase IIa study on more than 30 patients showed a successful treatment in 47% of cases vs. 6% in the placebo group (Peek et al., 2020), providing strong support to further develop the peptide for CSOM treatment.
An interesting LL-37 derivative is the peptide 17BIPHE2, based on the short (17–32) fragment, where the native Ile20 and 24 and Leu28 were changed to D-Leu, and Phe17 and 27 to biphenylalanine. Compared to LL-37 and its fragments, this peptide is highly resistant to various proteases present in mammalian body fluids, and displays an increased antimicrobial activity against multidrug-resistant bacteria and a high antibiofilm activity (Wang et al., 2014; Narayana et al., 2019). It was recently shown to be active in vitro against Neisseria gonorrhoeae and has spermicidal activity on human and mouse sperm via membrane permeabilization. In addition, multiple transcervical injections of the peptide in female mice did not affect the histological features of vagina, cervix and uterus, suggesting its use both as contraceptive and antimicrobial agent against sexually transmitted infections (Lee et al., 2022). Further examples are reported in Supplementary Table S2.
An interesting concept to reduce toxicity is to mimic their endogenous release from the CLD by incorporating a part of it in a pro-drug. In this respect, an artificial pro-form was rationally designed to protect the D-enantiomer of the shortened bovine cathelicidin BMAP18, to be released and activated only upon proteolytic cleavage by elastase. In this preclinical study, this prodrug showed a very low cytotoxicity and was correctly converted to D-BMAP18 in the presence of cystic fibrosis sputum as a model of a pathologic lung environment, then showing good antimicrobial activity, so it may be a good candidate for aerosol treatment of the CF lung (Degasperi et al., 2022).
Type-P cathelicidin HDPs (PrAMPs) are of significant interest for the development of new antimicrobial compounds due to their potent antimicrobial efficacy (although mainly effective against Gram-negative bacterial species) combined with a lower cytotoxicity than other structural types (Welch et al., 2020). As described in Section 2.4, they display a peculiar mode of action targeting protein synthesis through stalling bacterial ribosomes as their main antimicrobial mechanism (Mardirossian et al., 2014; 2018a; 2018b; Gagnon et al., 2016; Seefeldt et al., 2016), while destabilization of the bacterial membrane is mostly a secondary effect exerted at well above their active concentrations, unless their sequence is modified (Podda et al., 2006; Sola et al., 2020). During the past 3 decades a few Type-P cathelicidin HDPs have undergone extensive structure-activity relationship studies aimed at isolating the pharmacophore sequence by testing numerous fragments and variants, with the aim of reducing synthetic cost and promoting antimicrobial potency through residue substitutions (Panteleev et al., 2018; 2022; Sola et al., 2020; Bolosov et al., 2023; Benincasa et al., 2004; Guida et al., 2015; Mardirossian et al., 2019b). In particular, the application of SPOT-synthesis and of deep mutational scanning protocols have increased the potential of these studies exponentially, due to the possibility to synthesize large panels of peptides (Lai et al., 2019; Mardirossian et al., 2019a; 2020) or to express peptide variants directly in bacteria (Collins and Hackel, 2024). The antimicrobial potential and low toxicity of some Type-P cathelicidin HDPs, as evidenced by in vitro studies, has also been supported by in vivo studies in animal models of infection (Benincasa et al., 2010; Di Stasi et al., 2019). However, assessment remains in the pre-clinical phase and data on the pharmaco-kinetics and -dynamics of these compounds are still at the embryonic stage (Benincasa et al., 2015), which, with the lack of experimentation in non-murine animal models, has as yet prevented derivatives of this type of cathelicidin HDP from entering clinical studies.
The potential of Type-P cathelicidin HDPs to fight pathogens is not limited to exploitation of their antimicrobial activity per se, but can exploit their capacity to be actively internalized by susceptible bacteria, as well as some eukaryotic cells, as carriers for other types of molecules such as fluorophores or quite large cargo such as PEG, proteins and impermeant PNA, (Sadler et al., 2002; Benincasa et al., 2015; Hansen et al., 2016). They can also be used in this manner to enhance or restore the activity of antibiotics in resistant strains (Gambato et al., 2023).
5 Conclusion
Cathelicidins belong to an ancient, ubiquitous family of vertebrate HDP proforms that carry highly diverse multifunctional bioactive peptides that can be grouped into at least five different structural types. Numerous experimental observations and genetic information indicate that many species possess multiple cathelicidin genes and that the types of derived peptides are structurally diverse. These aspects open up potential access to a large repertoire of bioactive peptides with anti-infective and immunomodulatory activities. The presence of relatively well conserved proregions and a significant degree of syntenic conservation with common flanking genes, facilitates the search for novel cathelicidin sequences in the rapidly growing number of available vertebrate genomes. This process would enable targeted mining approaches and lend itself to machine learning. The features of numerous characterized cathelicidin HDPs can then be used for structure/activity prediction studies aimed at 1) dissecting the functional domains responsible for the pleiotropic activity of some of these peptides; 2) suggesting modifications that could improve their druggability (e.g., stabilized analogues and truncated derivatives that reduce the production costs); 3) exploiting the different mechanisms of action of each structural type – membrane or target specific - to modulate the spectrum of activity, 4) reducing the likelihood of resistance, and 5) improving cytotoxicity and pharmacodynamic profiles. In addition, the particular architecture of cathelicidins, with a conserved proregion that likely keeps the peptide inactive, and avoids sequestration and/or degradation until it reaches the site of infection, could point to a possible delivery method to reduce side effects and not only use them as topical agents. Although the success of anti-infective peptides derived from AMPs in clinical trials has generally been low, concerted efforts in recent years suggest that cathelicidins may be useful both as anti-infective drugs that also aid wound healing (as demonstrated by their application on DFUs) and/or as markers for different conditions.
Author contributions
AT: Conceptualization, Data curation, Writing–original draft, Writing–review and editing, Formal Analysis, Supervision, Validation. MG: Data curation, Writing–original draft, Writing–review and editing, Formal Analysis, Validation. AC: Writing–original draft, Writing–review and editing. SP: Writing–original draft, Writing–review and editing. MM: Writing–review and editing, Writing–original draft. MS: Writing–review and editing, Writing–original draft. DP: Data curation, Writing–review and editing, Formal Analysis, Validation. GM: Writing–review and editing, Data curation, Formal Analysis, Validation. RG: Writing–original draft, Writing–review and editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported MUR PRIN 2022 grants ERC LS6–2022EKWRH (AC), 2022XFFTH5 (MS) and 2022EKWRHB (MM).
Acknowledgments
This review is dedicated to prof. Domenico Romeo, a pioneer in cathelicidin studies and mentor to many of us.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fddsv.2024.1458057/full#supplementary-material
Abbreviations
AMP, antimicrobial peptide; CLD, cathelin-like domain; DFU, diabetic foot ulcers; hCAP18, human 18 kDa cathelicidin antimicrobial protein; HDP, host defence peptide; LPS, lipopolysaccharide; LTA, lipoteichoic acid; VDRE, vitamin D response element.
References
Agerberth, B., Gunne, H., Odeberg, J., Kogner, P., Boman, H. G., and Gudmundsson, G. H. (1995). FALL-39, a putative human peptide antibiotic, is cysteine-free and expressed in bone marrow and testis. Proc. Natl. Acad. Sci. U. S. A. 92, 195–199. doi:10.1073/pnas.92.1.195
Agerberth, B., Lee, J.-Y., Bergman, T., Carlquist, M., Boman, H. G., Mutt, V., et al. (1991). Amino acid sequence of PR-39. Isolation from pig intestine of a new member of the family of proline-arginine-rich antibacterial peptides. Eur. J. Biochem. 202, 849–854. doi:10.1111/j.1432-1033.1991.tb16442.x
Agier, J., Efenberger, M., and Brzezińska-Błaszczyk, E. (2015). Cathelicidin impact on inflammatory cells. Cent.-Eur. J. Immunol. 40, 225–235. doi:10.5114/ceji.2015.51359
Ahn, B., Jeon, H., Cho, H., Nagasundarapandian, S., and Park, C. (2022). Sequence polymorphisms of PR39 cathelicidins and extensive copy variations in commercial pig breeds. Gene 822, 146323. doi:10.1016/j.gene.2022.146323
Alford, M. A., Baquir, B., Santana, F. L., Haney, E. F., and Hancock, R. E. W. (2020). Cathelicidin host defense peptides and inflammatory signaling: striking a Balance. Front. Microbiol. 11, 1902. doi:10.3389/fmicb.2020.01902
Alibardi, L. (2014). Ultrastructural immunolocalization of chatelicidin-like peptides in granulocytes of normal and regenerating lizard tissues. Acta histochem. 116, 363–371. doi:10.1016/j.acthis.2013.08.014
Andersson, E., Sørensen, O. E., Frohm, B., Borregaard, N., Egesten, A., and Malm, J. (2002). Isolation of human cationic antimicrobial protein-18 from seminal plasma and its association with prostasomes. Hum. Reprod. 17, 2529–2534. doi:10.1093/humrep/17.10.2529
Ando, S., Mitsuyasu, K., Soeda, Y., Hidaka, M., Ito, Y., Matsubara, K., et al. (2010). Structure-activity relationship of indolicidin, a Trp-rich antibacterial peptide. J. Pept. Sci. 16, 171–177. doi:10.1002/psc.1217
Antal, D., Alimohammadi, S., Bai, P., Szöllősi, A. G., and Szántó, M. (2022). Antigen-presenting cells in psoriasis. Life 12, 234. doi:10.3390/life12020234
Batista Araujo, J., Sastre de Souza, G., and Lorenzon, E. N. (2022). Indolicidin revisited: biological activity, potential applications and perspectives of an antimicrobial peptide not yet fully explored. World J. Microbiol. Biotechnol. 38, 39. doi:10.1007/s11274-022-03227-2
Benincasa, M., Pelillo, C., Zorzet, S., Garrovo, C., Biffi, S., Gennaro, R., et al. (2010). The proline-rich peptide Bac7(1-35) reduces mortality from Salmonella typhimurium in a mouse model of infection. BMC Microbiol. 10, 178. doi:10.1186/1471-2180-10-178
Benincasa, M., Scocchi, M., Podda, E., Skerlavaj, B., Dolzani, L., and Gennaro, R. (2004). Antimicrobial activity of Bac7 fragments against drug-resistant clinical isolates. Peptides 25, 2055–2061. doi:10.1016/j.peptides.2004.08.004
Benincasa, M., Zahariev, S., Pelillo, C., Milan, A., Gennaro, R., and Scocchi, M. (2015). PEGylation of the peptide Bac7(1-35) reduces renal clearance while retaining antibacterial activity and bacterial cell penetration capacity. Eur. J. Med. Chem. 95, 210–219. doi:10.1016/j.ejmech.2015.03.028
Bhattacharjya, S., Zhang, Z., and Ramamoorthy, A. (2024). LL-37: structures, antimicrobial activity, and influence on amyloid-related diseases. Biomolecules 14, 320. doi:10.3390/biom14030320
Bolintineanu, D. S., Vivcharuk, V., and Kaznessis, Y. N. (2012). Multiscale models of the antimicrobial peptide protegrin-1 on gram-negative bacteria membranes. Int. J. Mol. Sci. 13, 11000–11011. doi:10.3390/ijms130911000
Bolosov, I. A., Panteleev, P. V., Balandin, S. V., Shamova, O. V., and Ovchinnikova, T. V. (2023). Structural and functional characteristics of the proline-rich antimicrobial peptide minibactenecin from leukocytes of domestic goat Capra hircus. Bull. Exp. Biol. Med. 174, 440–445. doi:10.1007/s10517-023-05725-1
Bowdish, D. M. E., Davidson, D. J., Scott, M. G., and Hancock, R. E. W. (2005). Immunomodulatory activities of small host defense peptides. Antimicrob. Agents Chemother. 49, 1727–1732. doi:10.1128/AAC.49.5.1727-1732.2005
Braff, M. H., Hawkins, M. A., Nardo, A. D., Lopez-Garcia, B., Howell, M. D., Wong, C., et al. (2005). Structure-function relationships among human cathelicidin peptides: dissociation of antimicrobial properties from host immunostimulatory activities. J. Immunol. 174, 4271–4278. doi:10.4049/jimmunol.174.7.4271
Broekman, D. C., Zenz, A., Gudmundsdottir, B. K., Lohner, K., Maier, V. H., and Gudmundsson, G. H. (2011). Functional characterization of codCath, the mature cathelicidin antimicrobial peptide from Atlantic cod (Gadus morhua). Peptides 32, 2044–2051. doi:10.1016/j.peptides.2011.09.012
Browne, K., Chakraborty, S., Chen, R., Willcox, M. D., Black, D. S., Walsh, W. R., et al. (2020). A new era of antibiotics: the clinical potential of antimicrobial peptides. Int. J. Mol. Sci. 21, 7047. doi:10.3390/ijms21197047
Carretero, M., Escámez, M. J., García, M., Duarte, B., Holguín, A., Retamosa, L., et al. (2008). In vitro and in vivo wound healing-promoting activities of human cathelicidin LL-37. J. Investig. Dermatol. 128, 223–236. doi:10.1038/sj.jid.5701043
Castellanos, F. X., Moreno-Santillán, D., Hughes, G. M., Paulat, N. S., Sipperly, N., Brown, A. M., et al. (2023). The evolution of antimicrobial peptides in Chiroptera. Front. Immunol. 14, 1250229. doi:10.3389/fimmu.2023.1250229
Chan, D. I., Prenner, E. J., and Vogel, H. J. (2006). Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim. Biophys. Acta BBA - Biomembr. 1758, 1184–1202. doi:10.1016/j.bbamem.2006.04.006
Chen, J., Falla, T. J., Liu, H., Hurst, M. A., Fujii, C. A., Mosca, D. A., et al. (2000). Development of protegrins for the treatment and prevention of oral mucositis: structure–activity relationships of synthetic protegrin analogues. Pept. Sci. 55, 88–98. doi:10.1002/1097-0282(2000)55:1<88::AID-BIP80>3.0.CO;2-K
Chen, Y., Cai, S., Qiao, X., Wu, M., Guo, Z., Wang, R., et al. (2017). As-CATH1-6, novel cathelicidins with potent antimicrobial and immunomodulatory properties from Alligator sinensis, play pivotal roles in host antimicrobial immune responses. Biochem. J. 474, 2861–2885. doi:10.1042/BCJ20170334
Cheng, Y., Prickett, M. D., Gutowska, W., Kuo, R., Belov, K., and Burt, D. W. (2015). Evolution of the avian β-defensin and cathelicidin genes. BMC Evol. Biol. 15, 188. doi:10.1186/s12862-015-0465-3
Choi, M., Cho, H.-S., Ahn, B., Prathap, S., Nagasundarapandian, S., and Park, C. (2022). Genomewide analysis and biological characterization of cathelicidins with potent antimicrobial activity and low cytotoxicity from three bat species. Antibiot. Basel Switz. 11, 989. doi:10.3390/antibiotics11080989
Choi, M.-K., Le, M. T., Cho, H., Soundrarajan, N., Jeon, H., Park, C. K., et al. (2014). Defining the genetic relationship of protegrin-related sequences and the in vivo expression of protegrins. FEBS J. 281, 5420–5431. doi:10.1111/febs.13072
Coelho, L. P., Santos-Júnior, C. D., and de la Fuente-Nunez, C. (2024). Challenges in computational discovery of bioactive peptides in ’omics data. Proteomics 24, e2300105. doi:10.1002/pmic.202300105
Collins, J., and Hackel, B. J. (2024). Sequence-activity mapping via depletion reveals striking mutational tolerance and elucidates functional motifs in Tur1a antimicrobial peptide. Protein Eng. Des. Sel. 37, gzae006. doi:10.1093/protein/gzae006
Cribier, B. (2022). Rosacea: treatment targets based on new physiopathology data. Ann. Dermatol. Vénéréologie 149, 99–107. doi:10.1016/j.annder.2021.11.001
Cudic, M., and Otvos Jr, L. (2002). Intracellular targets of antibacterial peptides. Curr. Drug Targets 3, 101–106. doi:10.2174/1389450024605445
Czechowicz, P., Jaśkiewicz, M., Neubauer, D., Gościniak, G., and Kamysz, W. (2021). Anticandidal activity of omiganan and its retro analog alone and in combination with fluconazole. Probiotics Antimicrob. Proteins 13, 1173–1182. doi:10.1007/s12602-021-09757-9
de Barros, E., Gonçalves, R. M., Cardoso, M. H., Santos, N. C., Franco, O. L., and Cândido, E. S. (2019). Snake venom cathelicidins as natural antimicrobial peptides. Front. Pharmacol. 10, 1415. doi:10.3389/fphar.2019.01415
Degasperi, M., Sgarra, R., Mardirossian, M., Pacor, S., Maschio, M., and Scocchi, M. (2022). Elastase-activated antimicrobial peptide for a safer pulmonary treatment of cystic fibrosis infections. Antibiotics 11, 319. doi:10.3390/antibiotics11030319
Deng, Q.-F., Chu, H., Wen, Z., and Cao, Y.-S. (2019). Vitamin D and urinary tract infection: a systematic review and meta-analysis. Ann. Clin. Lab. Sci. 49, 134–142.
D’Este, F., Benincasa, M., Cannone, G., Furlan, M., Scarsini, M., Volpatti, D., et al. (2016). Antimicrobial and host cell-directed activities of Gly/Ser-rich peptides from salmonid cathelicidins. Fish. Shellfish Immunol. 59, 456–468. doi:10.1016/j.fsi.2016.11.004
Dijksteel, G. S., Ulrich, M. M. W., Middelkoop, E., and Boekema, B. K. H. L. (2021). Review: lessons learned from clinical trials using antimicrobial peptides (AMPs). Front. Microbiol. 12, 616979. doi:10.3389/fmicb.2021.616979
Di Stasi, A., Bozzer, S., Pacor, S., de Pascale, L., Morici, M., Favero, L., et al. (2019). The proline-rich antimicrobial peptide B7-005: low tendency to elicit bacterial resistance, safe for human cells and effective in zebrafish embryo bacteraemia model. (submitted).
Djanani, A., Mosheimer, B., Kaneider, N. C., Ross, C. R., Ricevuti, G., Patsch, J. R., et al. (2006). Heparan sulfate proteoglycan-dependent neutrophil chemotaxis toward PR-39 cathelicidin. J. Inflamm. Lond. Engl. 3, 14. doi:10.1186/1476-9255-3-14
Dlozi, P. N., Gladchuk, A., Crutchley, R. D., Keuler, N., Coetzee, R., and Dube, A. (2022). Cathelicidins and defensins antimicrobial host defense peptides in the treatment of TB and HIV: pharmacogenomic and nanomedicine approaches towards improved therapeutic outcomes. Biomed. Pharmacother. 151, 113189. doi:10.1016/j.biopha.2022.113189
Fabisiak, A., Murawska, N., and Fichna, J. (2016). LL-37: cathelicidin-related antimicrobial peptide with pleiotropic activity. Pharmacol. Rep. P. R. 68, 802–808. doi:10.1016/j.pharep.2016.03.015
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi:10.1093/nar/gkr367
Frank, R. W., Gennaro, R., Schneider, K., Przybylski, M., and Romeo, D. (1990). Amino acid sequences of two proline-rich bactenecins. Antimicrobial peptides of bovine neutrophils. J. Biol. Chem. 265, 18871–18874. doi:10.1016/S0021-9258(17)30595-1
Franks, P. J., Barker, J., Collier, M., Gethin, G., Haesler, E., Jawien, A., et al. (2016). Management of patients with venous leg ulcers: challenges and current best practice. J. Wound Care 25, S1-S67–S67. doi:10.12968/jowc.2016.25.Sup6.S1
Frasca, L., Palazzo, R., Chimenti, M. S., Alivernini, S., Tolusso, B., Bui, L., et al. (2018). Anti-LL37 antibodies are present in psoriatic arthritis (PsA) patients: new biomarkers in PsA. Front. Immunol. 9, 1936. doi:10.3389/fimmu.2018.01936
Fritsche, T. R., Rhomberg, P. R., Sader, H. S., and Jones, R. N. (2008). Antimicrobial activity of omiganan pentahydrochloride against contemporary fungal pathogens responsible for catheter-associated infections. Antimicrob. Agents Chemother. 52, 1187–1189. doi:10.1128/AAC.01475-07
Fuentes-Duculan, J., Bonifacio, K. M., Hawkes, J. E., Kunjravia, N., Cueto, I., Li, X., et al. (2017). Autoantigens ADAMTSL5 and LL37 are significantly upregulated in active Psoriasis and localized with keratinocytes, dendritic cells and other leukocytes. Exp. Dermatol. 26, 1075–1082. doi:10.1111/exd.13378
Furlan, M., Rosani, U., Gambato, S., Irato, P., Manfrin, A., Mardirossian, M., et al. (2018). Induced expression of cathelicidins in trout (Oncorhynchus mykiss) challenged with four different bacterial pathogens. J. Pept. Sci. 24, e3089. doi:10.1002/psc.3089
Gagnon, M. G., Roy, R. N., Lomakin, I. B., Florin, T., Mankin, A. S., and Steitz, T. A. (2016). Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 44, 2439–2450. doi:10.1093/nar/gkw018
Gambato, S., Bellotto, O., Mardirossian, M., Di Stasi, A., Gennaro, R., Pacor, S., et al. (2023). Designing new hybrid antibiotics: proline-rich antimicrobial peptides conjugated to the aminoglycoside tobramycin. Bioconjug. Chem. 34, 1212–1220. doi:10.1021/acs.bioconjchem.2c00467
Gao, W., Xing, L., Qu, P., Tan, T., Yang, N., Li, D., et al. (2015). Identification of a novel cathelicidin antimicrobial peptide from ducks and determination of its functional activity and antibacterial mechanism. Sci. Rep. 5, 17260. doi:10.1038/srep17260
Gautier, R., Douguet, D., Antonny, B., and Drin, G. (2008). HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinforma. Oxf. Engl. 24, 2101–2102. doi:10.1093/bioinformatics/btn392
Ghosh, A., Kar, R. K., Jana, J., Saha, A., Jana, B., Krishnamoorthy, J., et al. (2014). Indolicidin targets duplex DNA: structural and mechanistic insight through a combination of spectroscopy and microscopy. ChemMedChem 9, 2052–2058. doi:10.1002/cmdc.201402215
Giles, F. J., Miller, C. B., Hurd, D. D., Wingard, J. R., Fleming, T. R., Sonis, S. T., et al. (2003). A phase III, randomized, double-blind, placebo-controlled, multinational trial of iseganan for the prevention of oral mucositis in patients receiving stomatotoxic chemotherapy (PROMPT-CT trial). Leuk. Lymphoma 44, 1165–1172. doi:10.1080/1042819031000079159
Gombart, A. F., Borregaard, N., and Koeffler, H. P. (2005). Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J. 19, 1067–1077. doi:10.1096/fj.04-3284com
Gombart, A. F., Saito, T., and Koeffler, H. P. (2009). Exaptation of an ancient Alu short interspersed element provides a highly conserved vitamin D-mediated innate immune response in humans and primates. BMC Genomics 10, 321. doi:10.1186/1471-2164-10-321
Graf, M., Mardirossian, M., Nguyen, F., Seefeldt, A. C., Guichard, G., Scocchi, M., et al. (2017). Proline-rich antimicrobial peptides targeting protein synthesis. Nat. Prod. Rep. 34, 702–711. doi:10.1039/c7np00020k
Grönberg, A., Mahlapuu, M., Ståhle, M., Whately-Smith, C., and Rollman, O. (2014). Treatment with LL-37 is safe and effective in enhancing healing of hard-to-heal venous leg ulcers: a randomized, placebo-controlled clinical trial. Wound Repair Regen. Off. Publ. Wound heal. Soc. Eur. Tissue Repair Soc. 22, 613–621. doi:10.1111/wrr.12211
Gubatan, J., Mehigan, G. A., Villegas, F., Mitsuhashi, S., Longhi, M. S., Malvar, G., et al. (2020). Cathelicidin mediates a protective role of vitamin D in ulcerative colitis and human colonic epithelial cells. Inflamm. Bowel Dis. 26, 885–897. doi:10.1093/ibd/izz330
Guida, F., Benincasa, M., Zahariev, S., Scocchi, M., Berti, F., Gennaro, R., et al. (2015). Effect of size and N-terminal residue characteristics on bacterial cell penetration and antibacterial activity of the proline-rich peptide Bac7. J. Med. Chem. 58, 1195–1204. doi:10.1021/jm501367p
Gupta, K., Kotian, A., Subramanian, H., Daniell, H., and Ali, H. (2015). Activation of human mast cells by retrocyclin and protegrin highlight their immunomodulatory and antimicrobial properties. Oncotarget 6, 28573–28587. doi:10.18632/oncotarget.5611
Hancock, R. E. W., Haney, E. F., and Gill, E. E. (2016). The immunology of host defence peptides: beyond antimicrobial activity. Nat. Rev. Immunol. 16, 321–334. doi:10.1038/nri.2016.29
Hansen, A. M., Bonke, G., Larsen, C. J., Yavari, N., Nielsen, P. E., and Franzyk, H. (2016). Antibacterial peptide nucleic acid–antimicrobial peptide (PNA–AMP) conjugates: antisense targeting of fatty acid biosynthesis. Bioconjug. Chem. 27, 863–867. doi:10.1021/acs.bioconjchem.6b00013
Hao, X., Yang, H., Wei, L., Yang, S., Zhu, W., Ma, D., et al. (2012). Amphibian cathelicidin fills the evolutionary gap of cathelicidin in vertebrate. Amino Acids 43, 677–685. doi:10.1007/s00726-011-1116-7
Hao, Z., Liu, G., Ren, L., Liu, J., Liu, C., Yang, T., et al. (2023). A self-healing multifunctional hydrogel system accelerates diabetic wound healing through orchestrating immunoinflammatory microenvironment. ACS Appl. Mat. Interfaces 15, 19847–19862. doi:10.1021/acsami.2c23323
Heilborn, J. D., Nilsson, M. F., Kratz, G., Weber, G., Sørensen, O., Borregaard, N., et al. (2003). The cathelicidin anti-microbial peptide LL-37 is involved in re-epithelialization of human skin wounds and is lacking in chronic ulcer epithelium. J. Investig. Dermatol. 120, 379–389. doi:10.1046/j.1523-1747.2003.12069.x
Helbing, C. C., Hammond, S. A., Jackman, S. H., Houston, S., Warren, R. L., Cameron, C. E., et al. (2019). Antimicrobial peptides from Rana [Lithobates] catesbeiana: gene structure and bioinformatic identification of novel forms from tadpoles. Sci. Rep. 9, 1529. doi:10.1038/s41598-018-38442-1
Hsu, C.-H., Chen, C., Jou, M.-L., Lee, A. Y.-L., Lin, Y.-C., Yu, Y.-P., et al. (2005). Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 33, 4053–4064. doi:10.1093/nar/gki725
Hu, B., Coulson, L., Moyer, B., and Price, P. A. (1995). Isolation and molecular cloning of a novel bone phosphoprotein related in sequence to the cystatin family of thiol protease inhibitors. J. Biol. Chem. 270, 431–436. doi:10.1074/jbc.270.1.431
Ishige, T., Hara, H., Hirano, T., Kono, T., and Hanzawa, K. (2017). Characterization of the cathelicidin cluster in the Japanese quail (Coturnix japonica). Anim. Sci. J. Nihon Chikusan Gakkaiho 88, 1249–1257. doi:10.1111/asj.12752
Jelodari, S., Daemi, H., Mohammadi, P., Verdi, J., J Al-Awady, M., Ai, J., et al. (2023). Assessment of the efficacy of an LL-37-encapsulated keratin hydrogel for the treatment of full-thickness wounds. ACS Appl. Bio Mat. 6, 2122–2136. doi:10.1021/acsabm.2c01068
Kahlenberg, J. M., and Kaplan, M. J. (2013). Little peptide, big effects: the role of LL-37 in inflammation and autoimmune disease. J. Immunol. 191, 4895–4901. doi:10.4049/jimmunol.1302005
Kannoth, S., Ali, N., Prasanth, G. K., Arvind, K., Mohany, M., Hembrom, P. S., et al. (2023). Transcriptome analysis of Corvus splendens reveals a repertoire of antimicrobial peptides. Sci. Rep. 13, 18728. doi:10.1038/s41598-023-45875-w
Kim, D., Soundrarajan, N., Lee, J., Cho, H., Choi, M., Cha, S.-Y., et al. (2017). Genomewide analysis of the antimicrobial peptides in Python bivittatus and characterization of cathelicidins with potent antimicrobial activity and low cytotoxicity. Antimicrob. Agents Chemother. 61. doi:10.1128/AAC.00530-17
Kin, N. W., Chen, Y., Stefanov, E. K., Gallo, R. L., and Kearney, J. F. (2011). Cathelin-related antimicrobial peptide differentially regulates T- and B-cell function. Eur. J. Immunol. 41, 3006–3016. doi:10.1002/eji.201141606
Kokryakov, V. N., Harwig, S. S., Panyutich, E. A., Shevchenko, A. A., Aleshina, G. M., Shamova, O. V., et al. (1993). Proteins: leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett. 327, 231–236. doi:10.1016/0014-5793(93)80175-t
Kolodziejczyk, R., Michalska, K., Hernandez-Santoyo, A., Wahlbom, M., Grubb, A., and Jaskolski, M. (2010). Crystal structure of human cystatin C stabilized against amyloid formation. FEBS J. 277, 1726–1737. doi:10.1111/j.1742-4658.2010.07596.x
Koo, H. B., and Seo, J. (2019). Antimicrobial peptides under clinical investigation. Pept. Sci. 111, e24122. doi:10.1002/pep2.24122
Kordiš, D., and Turk, V. (2009). Phylogenomic analysis of the cystatin superfamily in eukaryotes and prokaryotes. BMC Evol. Biol. 9, 266. doi:10.1186/1471-2148-9-266
Kościuczuk, E. M., Lisowski, P., Jarczak, J., Krzyżewski, J., Zwierzchowski, L., and Bagnicka, E. (2014). Expression patterns of β-defensin and cathelicidin genes in parenchyma of bovine mammary gland infected with coagulase-positive or coagulase-negative Staphylococci. BMC Vet. Res. 10, 246. doi:10.1186/s12917-014-0246-z
Krepel, S. A., and Wang, J. M. (2019). Chemotactic ligands that activate G-protein-coupled formylpeptide receptors. Int. J. Mol. Sci. 20, 3426. doi:10.3390/ijms20143426
Krishnamoorthy, R., Adhikari, P., and Anaikutti, P. (2023). Design, synthesis, and characterization of non-hemolytic antimicrobial peptides related to human cathelicidin LL-37. RSC Adv. 13, 15594–15605. doi:10.1039/d3ra02473c
Krizsan, A., Volke, D., Weinert, S., Sträter, N., Knappe, D., and Hoffmann, R. (2014). Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70 S ribosome. Angew. Chem. Int. Ed. 53, 12236–12239. doi:10.1002/anie.201407145
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi:10.1093/molbev/msx116
Lai, P., Tresnak, D. T., and Hackel, B. J. (2019). Identification and elucidation of proline-rich antimicrobial peptides with enhanced potency and delivery. Biotechnol. Bioeng. 116, 2439–2450. doi:10.1002/bit.27092
Lai, Y., and Gallo, R. L. (2009). AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 30, 131–141. doi:10.1016/j.it.2008.12.003
Lande, R., Botti, E., Jandus, C., Dojcinovic, D., Fanelli, G., Conrad, C., et al. (2014). The antimicrobial peptide LL37 is a T-cell autoantigen in psoriasis. Nat. Commun. 5, 5621. doi:10.1038/ncomms6621
Lastra, J. M. P. de la, Garrido-Orduña, C., A.Borges, A., Jiménez-Arias, D., García-Machado, F. J., Hernández, M., et al. (2018). “Bioinformatics discovery of vertebrate cathelicidins from the mining of available genomes,” in Drug discovery - concepts to market. (IntechOpen). doi:10.5772/intechopen.76631
Lazaridis, T., He, Y., and Prieto, L. (2013). Membrane interactions and pore formation by the antimicrobial peptide protegrin. Biophys. J. 104, 633–642. doi:10.1016/j.bpj.2012.12.038
Lee, J. Y., Yang, S.-T., Lee, S. K., Jung, H. H., Shin, S. Y., Hahm, K.-S., et al. (2008). Salt-resistant homodimeric bactenecin, a cathelicidin-derived antimicrobial peptide. FEBS J. 275, 3911–3920. doi:10.1111/j.1742-4658.2008.06536.x
Lee, S. G., Kiattiburut, W., Khongkha, T., Schinkel, S. C. B., Lunn, Y., Decker, A. P., et al. (2022). 17BIPHE2, an engineered cathelicidin antimicrobial peptide with low susceptibility to proteases, is an effective spermicide and microbicide against Neisseria gonorrhoeae. Hum. Reprod. Oxf. Engl. 37, 2503–2517. doi:10.1093/humrep/deac188
Levy, O. (1996). Antibiotic proteins of polymorphonuclear leukocytes. Eur. J. Haematol. 56, 263–277. doi:10.1111/j.1600-0609.1996.tb00714.x
Li, W., Tailhades, J., O’Brien-Simpson, N. M., Separovic, F., Otvos, L., Hossain, M. A., et al. (2014). Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. Amino Acids 46, 2287–2294. doi:10.1007/s00726-014-1820-1
Liang, L., and Sonis, S. T. (2024). Comparisons of successful and failed Phase III trials of drugs and biologicals tested for mitigation of oral mucositis in patients being treated with radiotherapy with or without concomitant chemotherapy for cancers of the head and neck. Drug Dev. Res. 85, e22188. doi:10.1002/ddr.22188
Linde, A., Lushington, G. H., Abello, J., and Melgarejo, T. (2013). Clinical relevance of cathelicidin in infectious disease. J. Clin. Cell. Immunol. doi:10.4172/2155-9899.S13-003
Loffredo, M. R., Savini, F., Bobone, S., Casciaro, B., Franzyk, H., Mangoni, M. L., et al. (2021). Inoculum effect of antimicrobial peptides. Proc. Natl. Acad. Sci. U. S. A. 118, e2014364118. doi:10.1073/pnas.2014364118
Mahlapuu, M., Håkansson, J., Ringstad, L., and Björn, C. (2016). Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 6, 194. doi:10.3389/fcimb.2016.00194
Mahlapuu, M., Sidorowicz, A., Mikosinski, J., Krzyżanowski, M., Orleanski, J., Twardowska-Saucha, K., et al. (2021). Evaluation of LL-37 in healing of hard-to-heal venous leg ulcers: a multicentric prospective randomized placebo-controlled clinical trial. Wound Repair Regen. Off. Publ. Wound heal. Soc. Eur. Tissue Repair Soc. 29, 938–950. doi:10.1111/wrr.12977
Maier, V. H., Dorn, K. V., Gudmundsdottir, B. K., and Gudmundsson, G. H. (2008). Characterisation of cathelicidin gene family members in divergent fish species. Mol. Immunol. 45, 3723–3730. doi:10.1016/j.molimm.2008.06.002
Mardirossian, M., Barrière, Q., Timchenko, T., Müller, C., Pacor, S., Mergaert, P., et al. (2018a). Fragments of the nonlytic proline-rich antimicrobial peptide Bac5 kill Escherichia coli cells by inhibiting protein synthesis. Antimicrob. Agents Chemother. 62, e00534-18. doi:10.1128/AAC.00534-18
Mardirossian, M., Grzela, R., Giglione, C., Meinnel, T., Gennaro, R., Mergaert, P., et al. (2014). The host antimicrobial peptide Bac71-35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem. Biol. 21, 1639–1647. doi:10.1016/j.chembiol.2014.10.009
Mardirossian, M., Pérébaskine, N., Benincasa, M., Gambato, S., Hofmann, S., Huter, P., et al. (2018b). The dolphin proline-rich antimicrobial peptide Tur1A inhibits protein synthesis by targeting the bacterial ribosome. Cell. Chem. Biol. 25, 530–539. doi:10.1016/j.chembiol.2018.02.004
Mardirossian, M., Sola, R., Beckert, B., Collis, D. W. P., Di Stasi, A., Armas, F., et al. (2019a). Proline-rich peptides with improved antimicrobial activity against E. coli, K. pneumoniae, and A. Baumannii. ChemMedChem 14, 2025–2033. doi:10.1002/cmdc.201900465
Mardirossian, M., Sola, R., Beckert, B., Valencic, E., Collis, D. W. P., Borišek, J., et al. (2020). Peptide inhibitors of bacterial protein synthesis with broad spectrum and SbmA-independent bactericidal activity against clinical pathogens. J. Med. Chem. 63, 9590–9602. doi:10.1021/acs.jmedchem.0c00665
Mardirossian, M., Sola, R., Degasperi, M., and Scocchi, M. (2019b). Search for shorter portions of the proline-rich antimicrobial peptide fragment bac5(1-25) that retain antimicrobial activity by blocking protein synthesis. ChemMedChem 14, 343–348. doi:10.1002/cmdc.201800734
Masso-Silva, J. A., and Diamond, G. (2014). Antimicrobial peptides from fish. Pharmaceuticals 7, 265–310. doi:10.3390/ph7030265
Miranda, E., Bramono, K., Yunir, E., Reksodiputro, M. H., Suwarsa, O., Rengganis, I., et al. (2023). Efficacy of LL-37 cream in enhancing healing of diabetic foot ulcer: a randomized double-blind controlled trial. Arch. Dermatol. Res. 315, 2623–2633. doi:10.1007/s00403-023-02657-8
Mookherjee, N., Brown, K. L., and Hancock, R. E. W. (2013). “Cathelicidins,” in Handbook of biologically active peptides (Academic Press), 77–84.
Moretta, A., Scieuzo, C., Petrone, A. M., Salvia, R., Manniello, M. D., Franco, A., et al. (2021). Antimicrobial peptides: a new hope in biomedical and pharmaceutical fields. Front. Cell. Infect. Microbiol. 11, 668632. doi:10.3389/fcimb.2021.668632
Morgera, F., Vaccari, L., Antcheva, N., Scaini, D., Pacor, S., and Tossi, A. (2009). Primate cathelicidin orthologs display different structures and membrane interactions. Biochem. J. 417, 727–735. doi:10.1042/BJ20081726
Murakami, M., Lopez-Garcia, B., Braff, M., Dorschner, R. A., and Gallo, R. L. (2004). Postsecretory processing generates multiple cathelicidins for enhanced topical antimicrobial defense. J. Immunol. 172, 3070–3077. doi:10.4049/jimmunol.172.5.3070
Nakagawa, Y., and Gallo, R. L. (2015). Endogenous intracellular cathelicidin enhances TLR9 activation in dendritic cells and macrophages. J. Immunol. 194, 1274–1284. doi:10.4049/jimmunol.1402388
Narayana, J. L., Mishra, B., Lushnikova, T., Golla, R. M., and Wang, G. (2019). Modulation of antimicrobial potency of human cathelicidin peptides against the ESKAPE pathogens and in vivo efficacy in a murine catheter-associated biofilm model. Biochim. Biophys. Acta Biomembr. 1861, 1592–1602. doi:10.1016/j.bbamem.2019.07.012
Nell, M. J., Tjabringa, G. S., Wafelman, A. R., Verrijk, R., Hiemstra, P. S., Drijfhout, J. W., et al. (2006). Development of novel LL-37 derived antimicrobial peptides with LPS and LTA neutralizing and antimicrobial activities for therapeutic application. Peptides 27, 649–660. doi:10.1016/j.peptides.2005.09.016
Niemeyer-van der Kolk, T., van der Wall, H., Hogendoorn, G. K., Rijneveld, R., Luijten, S., van Alewijk, D. C. J. G., et al. (2020). Pharmacodynamic effects of topical omiganan in patients with mild to moderate atopic dermatitis in a randomized, placebo-controlled, phase II trial. Clin. Transl. Sci. 13, 994–1003. doi:10.1111/cts.12792
Pag, U., Oedenkoven, M., Sass, V., Shai, Y., Shamova, O., Antcheva, N., et al. (2008). Analysis of in vitro activities and modes of action of synthetic antimicrobial peptides derived from an -helical “sequence template.”. J. Antimicrob. Chemother. 61, 341–352. doi:10.1093/jac/dkm479
Panteleev, P. V., Bolosov, I. A., Kalashnikov, A. À., Kokryakov, V. N., Shamova, O. V., Emelianova, A. A., et al. (2018). Combined antibacterial effects of goat cathelicidins with different mechanisms of action. Front. Microbiol. 9, 2983. doi:10.3389/fmicb.2018.02983
Panteleev, P. V., Safronova, V. N., Kruglikov, R. N., Bolosov, I. A., Bogdanov, I. V., and Ovchinnikova, T. V. (2022). A novel proline-rich cathelicidin from the alpaca Vicugna pacos with potency to combat antibiotic-resistant bacteria: mechanism of action and the functional role of the C-terminal region. Membranes 12, 515. doi:10.3390/membranes12050515
Pazgier, M., Ericksen, B., Ling, M., Toth, E., Shi, J., Li, X., et al. (2013). Structural and functional analysis of the pro-domain of human cathelicidin, LL-37. Biochemistry 52, 1547–1558. doi:10.1021/bi301008r
Peek, N. F. A. W., Nell, M. J., Brand, R., Jansen-Werkhoven, T., Hoogdalem, E. J. van, Verrijk, R., et al. (2020). Ototopical drops containing a novel antibacterial synthetic peptide: safety and efficacy in adults with chronic suppurative otitis media. PLOS ONE 15, e0231573. doi:10.1371/journal.pone.0231573
Peel, E., Cheng, Y., Djordjevic, J. T., O’Meally, D., Thomas, M., Kuhn, M., et al. (2021). Koala cathelicidin PhciCath5 has antimicrobial activity, including against Chlamydia pecorum. PLOS ONE 16, e0249658. doi:10.1371/journal.pone.0249658
Pérez de la Lastra, J. M., Asensio-Calavia, P., González-Acosta, S., Baca-González, V., and Morales-delaNuez, A. (2021). Bioinformatic analysis of genome-predicted bat cathelicidins. Molecules 26, 1811. doi:10.3390/molecules26061811
Podda, E., Benincasa, M., Pacor, S., Micali, F., Mattiuzzo, M., Gennaro, R., et al. (2006). Dual mode of action of Bac7, a proline-rich antibacterial peptide. Biochim. Biophys. Acta BBA - Gen. Subj. 1760, 1732–1740. doi:10.1016/j.bbagen.2006.09.006
Raj, P. A., Karunakaran, T., and Sukumaran, D. K. (2000). Synthesis, microbicidal activity, and solution structure of the dodecapeptide from bovine neutrophils. Biopolymers 53, 281–292. doi:10.1002/(SICI)1097-0282(20000405)53:4<281::AID-BIP1>3.0.CO;2-2
Rhie, A., McCarthy, S. A., Fedrigo, O., Damas, J., Formenti, G., Koren, S., et al. (2021). Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746. doi:10.1038/s41586-021-03451-0
Ritonja, A., Kopitar, M., Jerala, R., and Turk, V. (1989). Primary structure of a new cysteine proteinase inhibitor from pig leucocytes. FEBS Lett. 255, 211–214. doi:10.1016/0014-5793(89)81093-2
Romeo, D., Skerlavaj, B., Bolognesi, M., and Gennaro, R. (1988). Structure and bactericidal activity of an antibiotic dodecapeptide purified from bovine neutrophils. J. Biol. Chem. 263, 9573–9575. doi:10.1016/s0021-9258(19)81553-3
Rozek, A., Friedrich, C. L., and Hancock, R. E. (2000). Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry 39, 15765–15774. doi:10.1021/bi000714m
Rozek, A., Powers, J.-P. S., Friedrich, C. L., and Hancock, R. E. W. (2003). Structure-based design of an indolicidin peptide analogue with increased protease stability. Biochemistry 42, 14130–14138. doi:10.1021/bi035643g
Rubinchik, E., Dugourd, D., Algara, T., Pasetka, C., and Friedland, H. D. (2009). Antimicrobial and antifungal activities of a novel cationic antimicrobial peptide, omiganan, in experimental skin colonisation models. Int. J. Antimicrob. Agents 34, 457–461. doi:10.1016/j.ijantimicag.2009.05.003
Runti, G., Lopez Ruiz, M. d. C., Stoilova, T., Hussain, R., Jennions, M., Choudhury, H. G., et al. (2013). Functional characterization of SbmA, a bacterial inner membrane transporter required for importing the antimicrobial peptide bac7(1-35). J. Bacteriol. 195, 5343–5351. doi:10.1128/JB.00818-13
Ryge, T. S., Doisy, X., Ifrah, D., Olsen, J. E., and Hansen, P. R. (2004). New indolicidin analogues with potent antibacterial activity. J. Pept. Res. 64, 171–185. doi:10.1111/j.1399-3011.2004.00177.x
Sader, H. S., Fedler, K. A., Rennie, R. P., Stevens, S., and Jones, R. N. (2004). Omiganan pentahydrochloride (MBI 226), a topical 12-amino-acid cationic peptide: spectrum of antimicrobial activity and measurements of bactericidal activity. Antimicrob. Agents Chemother. 48, 3112–3118. doi:10.1128/AAC.48.8.3112-3118.2004
Sadler, K., Eom, K. D., Yang, J.-L., Dimitrova, Y., and Tam, J. P. (2002). Translocating proline-rich peptides from the antimicrobial peptide bactenecin 7. Biochemistry 41, 14150–14157. doi:10.1021/bi026661l
Sanchez, J.-F., Hoh, F., Strub, M.-P., Aumelas, A., and Dumas, C. (2002). Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein. Structure 10, 1363–1370. doi:10.1016/S0969-2126(02)00859-6
Schibli, D. J., Hwang, P. M., and Vogel, H. J. (1999). Structure of the antimicrobial peptide tritrpticin bound to micelles: a distinct membrane-bound peptide fold. Biochemistry 38, 16749–16755. doi:10.1021/bi990701c
Scocchi, M., Lüthy, C., Decarli, P., Mignogna, G., Christen, P., and Gennaro, R. (2009a). The proline-rich antibacterial peptide Bac7 binds to and inhibits in vitro the molecular chaperone DnaK. Int. J. Pept. Res. Ther. 15, 147–155. doi:10.1007/s10989-009-9182-3
Scocchi, M., Mardirossian, M., Runti, G., and Benincasa, M. (2016). Non-membrane permeabilizing modes of action of antimicrobial peptides on bacteria. Curr. Top. Med. Chem. 16, 76–88. doi:10.2174/1568026615666150703121009
Scocchi, M., Pallavicini, A., Salgaro, R., Bociek, K., and Gennaro, R. (2009b). The salmonid cathelicidins: a gene family with highly varied C-terminal antimicrobial domains. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 152, 376–381. doi:10.1016/j.cbpb.2009.01.003
Scocchi, M., Skerlavaj, B., Romeo, D., and Gennaro, R. (1992). Proteolytic cleavage by neutrophil elastase converts inactive storage proforms to antibacterial bactenecins. Eur. J. Biochem. 209, 589–595. doi:10.1111/j.1432-1033.1992.tb17324.x
Scocchi, M., Tossi, A., and Gennaro, R. (2011). Proline-rich antimicrobial peptides: converging to a non-lytic mechanism of action. Cell. Mol. Life Sci. CMLS 68, 2317–2330. doi:10.1007/s00018-011-0721-7
Seefeldt, A. C., Graf, M., Pérébaskine, N., Nguyen, F., Arenz, S., Mardirossian, M., et al. (2016). Structure of the mammalian antimicrobial peptide Bac7(1–16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 44, 2429–2438. doi:10.1093/nar/gkv1545
Selsted, M. E., Novotny, M. J., Morris, W. L., Tang, Y. Q., Smith, W., and Cullor, J. S. (1992). Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J. Biol. Chem. 267, 4292–4295. doi:10.1016/s0021-9258(18)42830-x
Shagaghi, N., Palombo, E. A., Clayton, A. H. A., and Bhave, M. (2016). Archetypal tryptophan-rich antimicrobial peptides: properties and applications. World J. Microbiol. Biotechnol. 32, 31. doi:10.1007/s11274-015-1986-z
Skerlavaj, B., Gennaro, R., Bagella, L., Merluzzi, L., Risso, A., and Zanetti, M. (1996). Biological characterization of two novel cathelicidin-derived peptides and identification of structural requirements for their antimicrobial and cell lytic activities. J. Biol. Chem. 271, 28375–28381. doi:10.1074/jbc.271.45.28375
Sola, R., Mardirossian, M., Beckert, B., Luna, L. S. D., Prickett, D., Tossi, A., et al. (2020). Characterization of cetacean proline-rich antimicrobial peptides displaying activity against ESKAPE pathogens. Int. J. Mol. Sci. 17, 7367. doi:10.3390/ijms21197367
Sørensen, O., Arnljots, K., Cowland, J. B., Bainton, D. F., and Borregaard, N. (1997). The human antibacterial cathelicidin, hCAP-18, is synthesized in myelocytes and metamyelocytes and localized to specific granules in neutrophils. Blood 90, 2796–2803. doi:10.1182/blood.v90.7.2796.2796_2796_2803
Sørensen, O., Bratt, T., Johnsen, A. H., Madsen, M. T., and Borregaard, N. (1999). The human antibacterial cathelicidin, hCAP-18, is bound to lipoproteins in plasma. J. Biol. Chem. 274, 22445–22451. doi:10.1074/jbc.274.32.22445
Staubitz, P., Peschel, A., Nieuwenhuizen, W. F., Otto, M., Götz, F., Jung, G., et al. (2001). Structure–function relationships in the tryptophan-rich, antimicrobial peptide indolicidin. J. Pept. Sci. 7, 552–564. doi:10.1002/psc.351
Steinstraesser, L., Lam, M. C., Jacobsen, F., Porporato, P. E., Chereddy, K. K., Becerikli, M., et al. (2014). Skin electroporation of a plasmid encoding hCAP-18/LL-37 host defense peptide promotes wound healing. Mol. Ther. J. Am. Soc. Gene Ther. 22, 734–742. doi:10.1038/mt.2013.258
Stie, J., Jesaitis, A. V., Lord, C. I., Gripentrog, J. M., Taylor, R. M., Burritt, J. B., et al. (2007). Localization of hCAP-18 on the surface of chemoattractant-stimulated human granulocytes: analysis using two novel hCAP-18-specific monoclonal antibodies. J. Leukoc. Biol. 82, 161–172. doi:10.1189/jlb.0906586
Storici, P., Tossi, A., Lenarcic, B., and Romeo, D. (1996). Purification and structural characterization of bovine cathelicidins, precursors of antimicrobial peptides. Eur. J. Biochem. 238, 769–776. doi:10.1111/j.1432-1033.1996.0769w.x
Sun, T., Zhan, B., and Gao, Y. (2015). A novel cathelicidin from Bufo bufo gargarizans Cantor showed specific activity to its habitat bacteria. Gene 571, 172–177. doi:10.1016/j.gene.2015.06.034
Tang, S., Peel, E., Belov, K., Hogg, C. J., and Farquharson, K. A. (2024). Multi-omics resources for the Australian southern stuttering frog (Mixophyes australis) reveal assorted antimicrobial peptides. Sci. Rep. 14, 3991. doi:10.1038/s41598-024-54522-x
Thennarasu, S., Tan, A., Penumatchu, R., Shelburne, C. E., Heyl, D. L., and Ramamoorthy, A. (2010). Antimicrobial and membrane disrupting activities of a peptide derived from the human cathelicidin antimicrobial peptide LL37. Biophys. J. 98, 248–257. doi:10.1016/j.bpj.2009.09.060
Thibaut de Ménonville, S., Rosignoli, C., Soares, E., Roquet, M., Bertino, B., Chappuis, J.-P., et al. (2017). Topical treatment of rosacea with ivermectin inhibits gene expression of cathelicidin innate immune mediators, LL-37 and KLK5, in reconstructed and ex vivo skin models. Dermatol. Ther. 7, 213–225. doi:10.1007/s13555-017-0176-3
Tomasinsig, L., Benincasa, M., Scocchi, M., Skerlavaj, B., Tossi, A., Zanetti, M., et al. (2010). Role of cathelicidin peptides in bovine host defense and healing. Probiotics Antimicrob. Proteins 2, 12–20. doi:10.1007/s12602-010-9035-6
Tomasinsig, L., Morgera, F., Antcheva, N., Pacor, S., Skerlavaj, B., Zanetti, M., et al. (2009). Structure dependence of biological activities for primate cathelicidins. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 15, 576–582. doi:10.1002/psc.1143
Tomasinsig, L., Skerlavaj, B., Papo, N., Giabbai, B., Shai, Y., and Zanetti, M. (2006). Mechanistic and functional studies of the interaction of a proline-rich antimicrobial peptide with mammalian cells. J. Biol. Chem. 281, 383–391. doi:10.1074/jbc.M510354200
Tomasinsig, L., and Zanetti, M. (2005). The cathelicidins--structure, function and evolution. Curr. Protein Pept. Sci. 6, 23–34. doi:10.2174/1389203053027520
Toney, J. H. (2002). Iseganan (IntraBiotics pharmaceuticals). Curr. Opin. Investig. Drugs 3, 225–228.
Tossi, A., Sandri, L., and Giangaspero, A. (2000). Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55, 4–30. doi:10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M
Tossi, A., Scocchi, M., Skerlavaj, B., and Gennaro, R. (1994). Identification and characterization of a primary antibacterial domain in CAP18, a lipopolysaccharide binding protein from rabbit leukocytes. FEBS Lett. 339, 108–112. doi:10.1016/0014-5793(94)80395-1
Tossi, A., Skerlavaj, B., D’Este, F., and Gennaro, R. (2017). “Structural and functional diversity of cathelicidins,” in Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. Editor G. Wang (Boston, MA: CABI), 20–48. doi:10.1079/9781786390394.0020
Trotti, A., Garden, A., Warde, P., Symonds, P., Langer, C., Redman, R., et al. (2004). A multinational, randomized phase iii trial of iseganan hcl oral solution for reducing the severity of oral mucositis in patients receiving radiotherapy for head-and-neck malignancy. Int. J. Radiat. Oncol. 58, 674–681. doi:10.1016/S0360-3016(03)01627-4
Turkoglu, O., Emingil, G., Eren, G., Atmaca, H., Kutukculer, N., and Atilla, G. (2017). Gingival crevicular fluid and serum hCAP18/LL-37 levels in generalized aggressive periodontitis. Clin. Oral Investig. 21, 763–769. doi:10.1007/s00784-016-1834-z
Two, A. M., Hata, T. R., Nakatsuji, T., Coda, A. B., Kotol, P. F., Wu, W., et al. (2014). Reduction in serine protease activity correlates with improved rosacea severity in a small, randomized pilot study of a topical serine protease inhibitor. J. Investig. Dermatol. 134, 1143–1145. doi:10.1038/jid.2013.472
Usachev, K. S., Kolosova, O. A., Klochkova, E. A., Yulmetov, A. R., Aganov, A. V., and Klochkov, V. V. (2016). Oligomerization of the antimicrobial peptide Protegrin-5 in a membrane-mimicking environment. Structural studies by high-resolution NMR spectroscopy. Eur. Biophys. J. 46, 293–300. doi:10.1007/s00249-016-1167-5
Uzzell, T., Stolzenberg, E. D., Shinnar, A. E., and Zasloff, M. (2003). Hagfish intestinal antimicrobial peptides are ancient cathelicidins. Peptides 24, 1655–1667. doi:10.1016/j.peptides.2003.08.024
Vaezi, Z., Roversi, D., Branca, D., and Stella, L. (2014). Testing the “sand in the gearbox” model: antimicrobial peptide effects on membrane dynamics. J. Pept. Sci. 20, S108. doi:10.1002/psc.2688
Valdez-Miramontes, C. E., De Haro-Acosta, J., Aréchiga-Flores, C. F., Verdiguel-Fernández, L., and Rivas-Santiago, B. (2021). Antimicrobial peptides in domestic animals and their applications in veterinary medicine. Peptides 142, 170576. doi:10.1016/j.peptides.2021.170576
Vandamme, D., Landuyt, B., Luyten, W., and Schoofs, L. (2012). A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell. Immunol. 280, 22–35. doi:10.1016/j.cellimm.2012.11.009
van der Does, A. M., Bergman, P., Agerberth, B., and Lindbom, L. (2012). Induction of the human cathelicidin LL-37 as a novel treatment against bacterial infections. J. Leukoc. Biol. 92, 735–742. doi:10.1189/jlb.0412178
van Dijk, A., Guabiraba, R., Bailleul, G., Schouler, C., Haagsman, H. P., and Lalmanach, A.-C. (2023). Evolutionary diversification of defensins and cathelicidins in birds and primates. Mol. Immunol. 157, 53–69. doi:10.1016/j.molimm.2023.03.011
van Harten, R., van Woudenbergh, E., van Dijk, A., and Haagsman, H. (2018). Cathelicidins: immunomodulatory antimicrobials. Vaccines 6, 63. doi:10.3390/vaccines6030063
van Hoek, M. L. (2014). Antimicrobial peptides in reptiles. Pharmaceuticals 7, 723–753. doi:10.3390/ph7060723
van Hoek, M. L., Prickett, M. D., Settlage, R. E., Kang, L., Michalak, P., Vliet, K. A., et al. (2019). The Komodo dragon (Varanus komodoensis) genome and identification of innate immunity genes and clusters. BMC Genomics 20, 684. doi:10.1186/s12864-019-6029-y
Veldhuizen, E. J. A., Schneider, V. A. F., Agustiandari, H., van Dijk, A., Tjeerdsma-van Bokhoven, J. L. M., Bikker, F. J., et al. (2014). Antimicrobial and immunomodulatory activities of PR-39 derived peptides. PLoS ONE 9, e95939. doi:10.1371/journal.pone.0095939
Wang, G., Epand, R. F., Mishra, B., Lushnikova, T., Thomas, V. C., Bayles, K. W., et al. (2012). Decoding the functional roles of cationic side chains of the major antimicrobial region of human cathelicidin LL-37. Antimicrob. Agents Chemother. 56, 845–856. doi:10.1128/AAC.05637-11
Wang, G., Hanke, M. L., Mishra, B., Lushnikova, T., Heim, C. E., Chittezham Thomas, V., et al. (2014). Transformation of human cathelicidin LL-37 into selective, stable, and potent antimicrobial compounds. ACS Chem. Biol. 9, 1997–2002. doi:10.1021/cb500475y
Wang, T.-T., Nestel, F. P., Bourdeau, V., Nagai, Y., Wang, Q., Liao, J., et al. (2004). Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. 173, 2909–2912. doi:10.4049/jimmunol.173.5.2909
Wang, Y., Agerberth, B., Löthgren, A., Almstedt, A., and Johansson, J. (1998). Apolipoprotein A-I binds and inhibits the human antibacterial/cytotoxic peptide LL-37. J. Biol. Chem. 273, 33115–33118. doi:10.1074/jbc.273.50.33115
Wang, Y., Wang, M., Shan, A., and Feng, X. (2020). Avian host defense cathelicidins: structure, expression, biological functions, and potential therapeutic applications. Poult. Sci. 99, 6434–6445. doi:10.1016/j.psj.2020.09.030
Wei, L., Yang, J., He, X., Mo, G., Hong, J., Yan, X., et al. (2013). Structure and function of a potent lipopolysaccharide-binding antimicrobial and anti-inflammatory peptide. J. Med. Chem. 56, 3546–3556. doi:10.1021/jm4004158
Welch, N. G., Li, W., Hossain, M. A., Separovic, F., O’Brien-Simpson, N. M., and Wade, J. D. (2020). (Re)Defining the proline-rich antimicrobial peptide family and the identification of putative new members. Front. Chem. 8, 607769. doi:10.3389/fchem.2020.607769
Whelehan, C. J., Barry-Reidy, A., Meade, K. G., Eckersall, P. D., Chapwanya, A., Narciandi, F., et al. (2014). Characterisation and expression profile of the bovine cathelicidin gene repertoire in mammary tissue. BMC Genomics 15, 128. doi:10.1186/1471-2164-15-128
White, J. H. (2010). Vitamin D as an inducer of cathelicidin antimicrobial peptide expression: past, present and future. J. Steroid Biochem. Mol. Biol. 121, 234–238. doi:10.1016/j.jsbmb.2010.03.034
White, J. H. (2018). Vitamin D deficiency and the pathogenesis of Crohn’s disease. J. Steroid Biochem. Mol. Biol. 175, 23–28. doi:10.1016/j.jsbmb.2016.12.015
Wu, J., Yang, J., Wang, X., Wei, L., Mi, K., Shen, Y., et al. (2018). A frog cathelicidin peptide effectively promotes cutaneous wound healing in mice. Biochem. J. 475, 2785–2799. doi:10.1042/BCJ20180286
Xhindoli, D., Pacor, S., Benincasa, M., Scocchi, M., Gennaro, R., and Tossi, A. (2016). The human cathelicidin LL-37 - a pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta 1858, 546–566. doi:10.1016/j.bbamem.2015.11.003
Xhindoli, D., Pacor, S., Guida, F., Antcheva, N., and Tossi, A. (2014). Native oligomerization determines the mode of action and biological activities of human cathelicidin LL-37. Biochem. J. 457, 263–275. doi:10.1042/BJ20131048
Xiao, Y., Cai, Y., Bommineni, Y. R., Fernando, S. C., Prakash, O., Gilliland, S. E., et al. (2006). Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 281, 2858–2867. doi:10.1074/jbc.M507180200
Xiao, Y., Lyu, W., Yang, H., Xu, X., Zhou, C., Lu, L., et al. (2020). Molecular characterization, mRNA gene expression, and antimicrobial activity of 2 new cathelicidin genes in goose. Poult. Sci. 99, 2983–2991. doi:10.1016/j.psj.2020.03.021
Yamasaki, K., Di Nardo, A., Bardan, A., Murakami, M., Ohtake, T., Coda, A., et al. (2007). Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat. Med. 13, 975–980. doi:10.1038/nm1616
Yamasaki, K., Schauber, J., Coda, A., Lin, H., Dorschner, R. A., Schechter, N. M., et al. (2006). Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J. 20, 2068–2080. doi:10.1096/fj.06-6075com
Yang, Y., Sanchez, J. F., Strub, M.-P., Brutscher, B., and Aumelas, A. (2003). NMR structure of the cathelin-like domain of the protegrin-3 precursor. Biochemistry 42, 4669–4680. doi:10.1021/bi027133c
Young-Speirs, M., Drouin, D., Cavalcante, P. A., Barkema, H. W., and Cobo, E. R. (2018). Host defense cathelicidins in cattle: types, production, bioactive functions and potential therapeutic and diagnostic applications. Int. J. Antimicrob. Agents 51, 813–821. doi:10.1016/j.ijantimicag.2018.02.006
Zaiou, M., Nizet, V., and Gallo, R. L. (2003). Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J. Investig. Dermatol. 120, 810–816. doi:10.1046/j.1523-1747.2003.12132.x
Zanetti, M. (2004). Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 75, 39–48. doi:10.1189/jlb.0403147
Zanetti, M. (2005). The role of cathelicidins in the innate host defenses of mammals. Curr. Issues Mol. Biol. 7, 179–196. doi:10.21775/cimb.007.179
Zanetti, M., Del Sal, G., Storici, P., Schneider, C., and Romeo, D. (1993). The cDNA of the neutrophil antibiotic Bac5 predicts a pro-sequence homologous to a cysteine proteinase inhibitor that is common to other neutrophil antibiotics. J. Biol. Chem. 268, 522–526. doi:10.1016/S0021-9258(18)54182-X
Zanetti, M., Gennaro, R., and Romeo, D. (1995). Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 374, 1–5. doi:10.1016/0014-5793(95)01050-o
Zanetti, M., Gennaro, R., Scocchi, M., and Skerlavaj, B. (2002). “Structure and biology of cathelicidins,” in The biology and pathology of innate immunity mechanisms. Editors Y. Keisari, and I. Ofek (Springer US), 203–218. Available at: http://link.springer.com/chapter/10.1007/0-306-46831-X_17 (Accessed July 15, 2015).
Zanetti, M., Litteri, L., Gennaro, R., Horstmann, H., and Romeo, D. (1990). Bactenecins, defense polypeptides of bovine neutrophils, are generated from precursor molecules stored in the large granules. J. Cell. Biol. 111, 1363–1371. doi:10.1083/jcb.111.4.1363
Zhang, L., Jie, H., Xiao, Y., Zhou, C., Lyu, W., and Bai, W. (2019). Genomic identification and expression analysis of the cathelicidin gene family of the forest musk deer. Anim. Open Access J. MDPI 9, 481. doi:10.3390/ani9080481
Zhong, L., Liu, J., Teng, S., and Xie, Z. (2020). Identification of a novel cathelicidin from the deinagkistrodon acutus genome with antibacterial activity by multiple mechanisms. Toxins 12, 771. doi:10.3390/toxins12120771
Zhou, L., Liu, X., Jin, P., and Li, Q. (2009). Cloning of the kininogen gene from Lampetra japonica provides insights into its phylogeny in vertebrates. J. Genet. Genomics Yi Chuan Xue Bao 36, 109–115. doi:10.1016/S1673-8527(08)60097-1
Zhu, S. (2008a). Did cathelicidins, a family of multifunctional host-defense peptides, arise from a cysteine protease inhibitor? Trends Microbiol. 16, 353–360. doi:10.1016/j.tim.2008.05.007
Zhu, S. (2008b). Positive selection targeting the cathelin-like domain of the antimicrobial cathelicidin family. Cell. Mol. Life Sci. 65, 1285–1294. doi:10.1007/s00018-008-8070-x
Zhu, S., and Gao, B. (2009). A fossil antibacterial peptide gives clues to structural diversity of cathelicidin-derived host defense peptides. FASEB J. 23, 13–20. doi:10.1096/fj.08-114579
Zhu, S., and Gao, B. (2017). Positive selection in cathelicidin host defense peptides: adaptation to exogenous pathogens or endogenous receptors? Heredity 118, 453–465. doi:10.1038/hdy.2016.117
Zhu, Y., Hao, W., Wang, X., Ouyang, J., Deng, X., Yu, H., et al. (2022). Antimicrobial peptides, conventional antibiotics, and their synergistic utility for the treatment of drug-resistant infections. Med. Res. Rev. 42, 1377–1422. doi:10.1002/med.21879
Zouboulis, C. C., Dessinioti, C., Tsatsou, F., and Gollnick, H. P. M. (2017). Anti-acne drugs in phase 1 and 2 clinical trials. Expert Opin. Investig. Drugs 26, 813–823. doi:10.1080/13543784.2017.1337745
Zughaier, S. M., Svoboda, P., and Pohl, J. (2014). Structure-Dependent immune modulatory activity of protegrin-1 analogs. Antibiotics 3, 694–713. doi:10.3390/antibiotics3040694
Keywords: cathelicidin, CRAMP, LL-37, PrAMP, host defence peptides, antimicrobial peptides, innate immunity
Citation: Tossi A, Gerdol M, Caporale A, Pacor S, Mardirossian M, Scocchi M, Prickett MD, Manzini G and Gennaro R (2024) Cathelicidins—a rich seam of antimicrobial peptides waiting for exploitation. Front. Drug Discov. 4:1458057. doi: 10.3389/fddsv.2024.1458057
Received: 01 July 2024; Accepted: 09 August 2024;
Published: 06 September 2024.
Edited by:
Tânia Martins Silva, Universidade do Porto, PortugalReviewed by:
Chankyu Park, Konkuk University, Republic of KoreaGill Diamond, University of Louisville, United States
Copyright © 2024 Tossi, Gerdol, Caporale, Pacor, Mardirossian, Scocchi, Prickett, Manzini and Gennaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alessandro Tossi, YXRvc3NpQHVuaXRzLml0
 Alessandro Tossi
Alessandro Tossi Marco Gerdol
Marco Gerdol Andrea Caporale2,3
Andrea Caporale2,3 Marco Scocchi
Marco Scocchi Renato Gennaro
Renato Gennaro