
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Drug Discov., 07 October 2024
Sec. Technologies and Strategies to Enable Drug Discovery
Volume 4 - 2024 | https://doi.org/10.3389/fddsv.2024.1416728
 Liam M. Ashander1
Liam M. Ashander1 Amanda L. Lumsden1
Amanda L. Lumsden1 Yuefang Ma1
Yuefang Ma1 Alwin Chun Rong Tan1
Alwin Chun Rong Tan1 Binoy Appukuttan1
Binoy Appukuttan1 Santhosh Daniel1,2
Santhosh Daniel1,2 Michael Z. Michael1,2
Michael Z. Michael1,2 Justine R. Smith1,2*
Justine R. Smith1,2*As a key host protein involved in cellular infection by the severe acute respiratory syndrome coronavirus (SARS-CoV-2), angiotensin converting enzyme (ACE)2 is an ideal target for antiviral drugs. Manipulation of transcription provides opportunity for graduated blockade that preserves physiological functions. We sought to develop a model system for evaluating manipulation of ACE2 gene transcription using human retinal pigment epithelium. Retinal pigment epithelial cell isolates were prepared from human posterior eyecups (n = 11 individual isolates). The cells expressed ACE2 transcript and protein, and expression was not induced by hypoxia mimetic dimethyloxaloylglycine, or inflammatory cytokine IL-1β. ACE2 gene transcription factors were predicted in silico and cross-referenced with the human retinal pigment epithelial cell transcriptome, and five candidate transcription factors were identified: ETS proto-oncogene 1 transcription factor (ETS1), nuclear factor I C (NFIC), nuclear receptor subfamily 2 group C member 1 (NR2C1), TEA domain transcription factor 1 (TEAD1), and zinc finger protein 384 (ZNF384). The candidates were individually targeted in cells by transfection with small interfering (si)RNA. Knockdowns reduced mean cellular expression of all the transcription factors in comparison to expression in cells transfected with control non-targeted siRNA. Mean cellular ACE2 transcript was reduced under the condition of NR2C1 knockdown, but not for ETS1, NFIC, TEAD1, and ZNF384 knockdowns. Our findings build on previous work demonstrating the potential for drugging gene transcription. Importantly, we show the value of human retinal pigment epithelium as a system for evaluating ACE2 transcriptional blockade, a possible approach for treating SARS-CoV-2 infection. Brief Research Report.
Measurements of excess mortality for over 70 countries and territories indicate that approximately 18.2 million people died in 2020 and 2021 as a result of the coronavirus disease 2019 (COVID-19) pandemic (COVID-19 Excess Mortality Collaborators, 2022). The pandemic has not yet ended, with new waves of infection continuing to arise (Giovanetti et al., 2023). A range of vaccines have been developed against infection with the causative virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Eroglu et al., 2021; Fiolet et al., 2022; Li et al., 2022). These vaccines are effective in inducing an immune response and lowering the risk of infection (Tregoning et al., 2021). For additional protection of individuals at risk of severe disease and those people who are not vaccinated, anti-viral drugs that target the replication of SARS-CoV-2 have been introduced into clinical care (Hui and Zumla, 2022; Lai et al., 2022; Nazir et al., 2024). In evaluating these major advances, a group of experts have called for diversification in anti-viral drug development, suggesting new therapeutics should include drugs that target host molecules involved in viral replication (Edwards et al., 2022).
Angiotensin converting enzyme (ACE)2 is a host molecule that is essential for the replicative lifecycle of SARS-CoV-2 (Scialo et al., 2020; Hu et al., 2021). This carboxypeptidase is a key component of the renin-angiotensin system that controls the body’s fluid and electrolyte balance, as well as the blood pressure (Clarke and Turner, 2012; Verano-Braga et al., 2020). Critically, ACE2 converts angiotensin (Ang) II to Ang (1-7), and Ang I to Ang (1-9) (Clarke and Turner, 2012; Verano-Braga et al., 2020). The molecule has additional substrates, and beyond the enzymatic activity, it has important roles in metabolism and inflammation (Aleksova et al., 2021). The spike (S) protein of SARS-CoV-2 recognizes membrane-bound ACE2 as the first step in viral infection of a human cell (Hu et al., 2021). Because of its specific interaction with the S protein, ACE2 is an appealing potential drug target for the treatment of COVID-19 (Mostafa-Hedeab, 2020; Singh et al., 2022; Suvarnapathaki et al., 2022; Vitiello and Ferrara, 2022). However, given the multiple important physiological roles of ACE2, there are valid concerns about inhibiting this molecule, including the potential to aggravate COVID-19 by compromising cardiovascular function (Cousin et al., 2021; Momtazi-Borojeni et al., 2021; Zhu et al., 2021).
Manipulating transcription provides the opportunity for graduated blockade of a target molecule (Ashander et al., 2016). As demonstration of this concept, we achieved a partial reduction in the level of intercellular adhesion molecule (ICAM)-1 on human endothelial cells by silencing the transcription factors, C2 calcium dependent domain containing 4B (C2CD4B) and interferon regulatory factor 1 (IRF1) (Haydinger et al., 2023; Ma et al., 2023). In the study described in this Brief Research Report, we developed a model system for testing the manipulation of ACE2 gene transcription. We predicted in silico transcription factors that might control ACE2 gene expression, and then evaluated the impact of small interfering (si)RNA-mediated knockdown of the candidates in human epithelial cells. We used retinal pigment epithelial cells obtained from human cadaveric donor eyes for the system for several reasons. Firstly, there are robust methods for the isolation and characterization of this cell population (Blenkinsop et al., 2013; Smith et al., 2020; Markert et al., 2022). Secondly, these cells are highly receptive to transfection (Feng et al., 2022; Hu X. et al., 2022b; Mattern et al., 2022). Thirdly, as we confirm in this work, human retinal pigment epithelial cells express ACE2 (Yuan et al., 2020; Eriksen et al., 2021; Albertos-Arranz et al., 2023).
Retinal pigment epithelial cell isolates were prepared from paired human cadaveric donor eyes obtained from the Eye Bank of South Australia (Adelaide, Australia). The isolation method has been detailed in previous publications (Ashander et al., 2019; Lie et al., 2019; Ashander et al., 2022), and yields cells that express cytokeratin-8, retinal pigment epithelium-specific protein 65, cellular retinaldehyde-binding protein, and zonula occludens 1, and not the mesenchymal differentiation marker, α-smooth muscle actin (Smith et al., 2020).
In brief, the retinal pigment epithelium-choroid was dissected from posterior eyecups and incubated with 0.5 mg/mL collagenase IA and 0.5 mg/mL collagenase IV (Merck-Sigma Aldrich, St Louis, MO) in Hanks’ Balanced Salt Solution (HBSS) for 30 min at 37°C and 5% CO2 in air, and subsequently rinsed with Dulbecco’s phosphate buffered saline (DPBS) with 2% fetal bovine serum (FBS, Bovogen Biologicals, Keilor East, Australia). Sheets of retinal pigment epithelium were gently scraped off Bruch’s membrane with a spatula at the dissecting microscope and suspended in fresh DPBS with 2% FBS. Suspended cell sheets were pelleted by centrifugation and resuspended in DPBS with 2% FBS, and subsequently layered onto 10% sucrose solution in DPBS with 2% FBS, and centrifuged to collect the cell sheets. Sheets were resuspended in 50% Minimum Essential Medium Eagle alpha modification (with sodium bicarbonate) (MEM), 25% Dulbecco’s Modified Eagles Medium (DMEM) and 25% F-12 with 1x N1 Medium Supplement, 1x Non-Essential Amino Acids Solution, 1x GlutaMAX Supplement, 0.25 mg/mL taurine, 0.02 μg/mL hydrocortisone, 0.013 ng/mL 3,3′,5-triiodo-L-thyronine sodium, 100 U/mL Penicillin-Streptomycin (all obtained from Merck-Sigma Aldrich or Thermo Fisher Scientific-Gibco, Grand Island, NY), and 2% FBS. Sheets were distributed in a dish to 50% covered, and cultured at 37°C and 5% CO2 in air until confluent. In some experiments, cells were treated with 1 mM dimethyloxaloylglycine (DMOG) (Merck-Sigma Aldrich) or 10 ng/mL human recombinant interleukin (IL)-1β (R&D Systems, Minneapolis, MN).
Primary human retinal pigment epithelial cells were plated in glass bottom chamber slides (Ibidi GmBH, Grafelfing, Germany) and grown to confluence at 37°C and 5% CO2 in air. Medium was removed, and monolayers were washed twice in DPBS prior to fixation for 10 min in 4% paraformaldehyde at room temperature. All subsequent steps were carried out in room temperature unless noted. After an additional three washes in non-Dulbecco’s phosphate buffered saline (PBS), monolayers were blocked for 1 h in PBS containing 5% normal rabbit serum (Vector Labs, Burlingame, MA), and then incubated for 1 h with 7.5 μg/mL polyclonal goat anti-human ACE2 antibody (R&D Systems, Waltham, MA: catalogue number AF933) or purified goat immunoglobulin (Ig)G (Vector Labs) in PBS containing 5% normal rabbit serum and 0.05% saponin. Primary antibody incubation was continued overnight at 4°C, after which monolayers were washed 3 times in PBS and incubated for 45 min with 2.5 μg/mL Alexa Fluor 488-conjugated rabbit anti-goat IgG (Thermo Fisher Scientific-Molecular Probes, Eugene, OR) in PBS containing 5% normal rabbit serum and 0.05% saponin. Monolayers were washed again in PBS and fixed for 10 min in 4% paraformaldehyde prior to counterstaining for 5 min with 300 nM 4′,6-diamidino-2-phenylindole (DAPI, Merck-Sigma Aldrich). After additional washes in PBS, stained monolayers were mounted in Fluoromount aqueous mounting medium (Merck-Sigma Aldrich) and imaged on the LSM 880 Fast Airyscan confocal microscope (Carl Zeiss AG, Oberkochen, Germany) under a 63x/1.4 objective with oil immersion. Images were processed using ZEN microscopy software v3.2 (Carl Zeiss AG).
Retinal pigment epithelial cell monolayers were treated with Buffer RLT (Qiagen, Hilden, Germany) containing 1:100 2-mercaptoethanol and subsequently frozen at −80°C. After thawing, RNA was extracted using the RNeasy Mini Kit (Qiagen) with optional on-column DNAse I digestion, according to the manufacturer’s protocol. Nucleic acid concentration was determined by spectrophotometry using the Nanodrop 2000 (Thermo Fisher Scientific, Wilmington, DE). The reverse transcription (RT) was carried out using iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad Laboratories, Hercules, CA) with 250 ng RNA input per reaction. Duplicate cDNA synthesis reactions were prepared for each sample and the resulting cDNA was pooled prior to the polymerase chain reaction (PCR).
Standard PCR was carried out on the T100 Touch Thermocycler (Bio-Rad Laboratories). In addition to nuclease-free water and 1X PCR buffer, each reaction contained 0.4 mM dNTP mix, 1.5 mM or 5 mM (ETS1) MgCl2, 0.25 μM or 0.8 μM (ETS1) each of forward and reverse primer, 1 unit of HotStarTaq Plus DNA polymerase (Qiagen), and 2 μL of neat cDNA template. Cycling conditions were as follows: 5-min pre-amplification hold at 95°C followed by 40 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 61°C (ETS1: 58°C), extension for 30 s at 72°C, and a post-amplification hold of 72°C for 5 min. The PCR product sizes were confirmed by 2% agarose gel electrophoresis. Ribosomal protein lateral stalk subunit P0 (RPLP0) was used as a reference gene.
To assess reduction of targeted transcripts by siRNA and the accompanying ACE2 gene expression changes, real-time quantitative PCR was performed on a CFX Connect Real-Time PCR System (Bio-Rad Laboratories). In addition to SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories) and nuclease-free water, each reaction contained 300 nM of the forward and the reverse primer, and 2 µL of undiluted cDNA. Cycling conditions included a pre-amplification hold of 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 61°C for 30 s, extension at 72°C for 30 s, and fluorescence reading after a 1 s hold at 75°C. Melting curves from 70°C to 95°C were performed for each run to confirm a single peak was produced for all primer sets, and amplicon sizes were confirmed by agarose gel electrophoresis. Relative expression of transcripts of interest, normalized to stable reference genes (i.e., coefficient of variation less than 0.25) peptidylprolyl isomerase A (PPIA) and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ), was determined by the Pfaffl method (Pfaffl, 2001). Primer efficiency ranged from 78.5% to 93.4% with the exception of YWHAZ, where efficiency was 106.1%, as determined by CFX Manager software v3.0 (Bio-Rad Laboratories) using standard curves generated by serial dilution of purified PCR products. All primer sequences and expected product sizes are shown in Supplementary Table S1. For primers designed in-house, the product was confirmed by sequencing.
Human ACE2 gene sequence was obtained from the National Center for Biotechnology Information (NCBI) Nucleotide database (NG_012575.1: Homo sapiens angiotensin I converting enzyme 2 (ACE2), RefSeqGene on chromosome X: https://www.ncbi.nlm.nih.gov/nuccore/NG_012575.1/). The regulatory sequence, defined as 3,000 bp of sequence immediately upstream of the ACE2 gene transcription start site, was interrogated for potential transcription factor binding sites using JASPAR 2020, release 8: https://jaspar2020.genereg.net. JASPAR is an open-access platform of ‘curated, non-redundant transcription factor-binding profiles stored as position frequency matrices for transcription factors across multiple species in six taxonomic groups’ (Fornes et al., 2020). The computation task was performed on the Vertebrata taxonomic group section of the JASPAR platform, setting H. sapiens as the search species. The list of predicted binding sites was cross-referenced with the human retinal pigment epithelial cell transcriptome, which we previously generated by RNA-sequencing, achieving 40.3 × 106 mean paired reads aligning to the human genome (Smith et al., 2020), to identify transcription factors that might impact ACE2 gene transcription in this cell population. The transcriptome is lodged in the NCBI Gene Expression Omnibus (Series GSE132091: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132091). The transcription factor candidates were prioritized for investigation in this work on the basis of: normalized read count of at least 100; minimum of three predicted binding sites; and activator function established by report in the peer-reviewed literature.
Primary human retinal pigment epithelial cells were grown to confluence in 12-well plates (growth area 3.8 cm2) at 37°C and 5% CO2 in air. Confluent monolayers were then treated with Silencer Select targeted siRNA (Supplementary Table S2) or the Negative Control No.1 siRNA (Thermo Fisher Scientific-Ambion, Austin, TX). The siRNA were diluted in Opti-MEM I reduced serum medium (Thermo Fisher Scientific-Gibco), and combined with Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific-Ambion). Each well received 200 μL of Opti-MEM I containing 12 pmol siRNA and 2 μL of lipofectamine, for a final medium volume of 1.2 mL per well. Monolayers were incubated for 24 h at 37°C and 5% CO2 in air, then siRNA-containing medium was replaced with 1 mL of fresh medium. Monolayers were incubated for a further 24–48 h, after which the medium was replaced with RLT lysis buffer in preparation for RNA extraction.
Data were analyzed using Prism versions 7.04 or 9.0.0 (GraphPad Software, La Jolla, CA). In comparisons of results for multiple human cell isolates, the two-tailed paired Student’s t-test was employed for two conditions and the repeated measures one-way analysis of variance (ANOVA) with Geisser-Greenhouse correction, and with Dunnett’s multiple comparisons test, was implemented for multiple conditions. Sample size determination and power analysis were not undertaken in this work which was planned as a feasibility study. Normality tests were not performed due to small sample numbers, following the recommendations stated in the Prism Statistics Guide. A statistically significant difference was defined by a p-value of less than 0.05 for all tests.
Use of human cadaveric donor eyes for this research was approved by the Southern Adelaide Clinical Human Research Ethics Committee (protocol number: 175.13).
Primary human retinal pigment epithelial cell isolates were prepared from paired eyecups of four men and seven women cadaveric donors, whose ages at death ranged from 46 to 78 years (median = 62 years). Time from death to processing of the human eyecups ranged from 14 to 41 h (median = 23 h).
Expression of ACE2 by human retinal pigment epithelial cells was investigated using isolates prepared from eyecups of multiple donors. Standard RT-PCR detected the ACE2 amplicon in RNA extracts obtained from each of six individual cell isolates (Figure 1A). Using indirect immunocytofluorescence and confocal microscopy, ACE2 protein was detected on monolayers of three different retinal pigment epithelial cell isolates; control monolayers immunolabelled with primary antibody directed against an irrelevant antigen showed no positive staining (Figure 1B). The real-time quantitative PCR showed over 3-fold variation in the level of transcript expression across cell isolates from six donors. ACE2 may be regulated by hypoxia and inflammatory cytokines in some settings (Clarke and Turner, 2012; Cousin et al., 2021). However, 24-h treatments with the hypoxia mimetic DMOG, or the inflammatory cytokine IL-1β, in standard concentrations, did not alter expression of ACE2 transcript in human retinal pigment epithelial cells (p > 0.05) (Figure 1C).
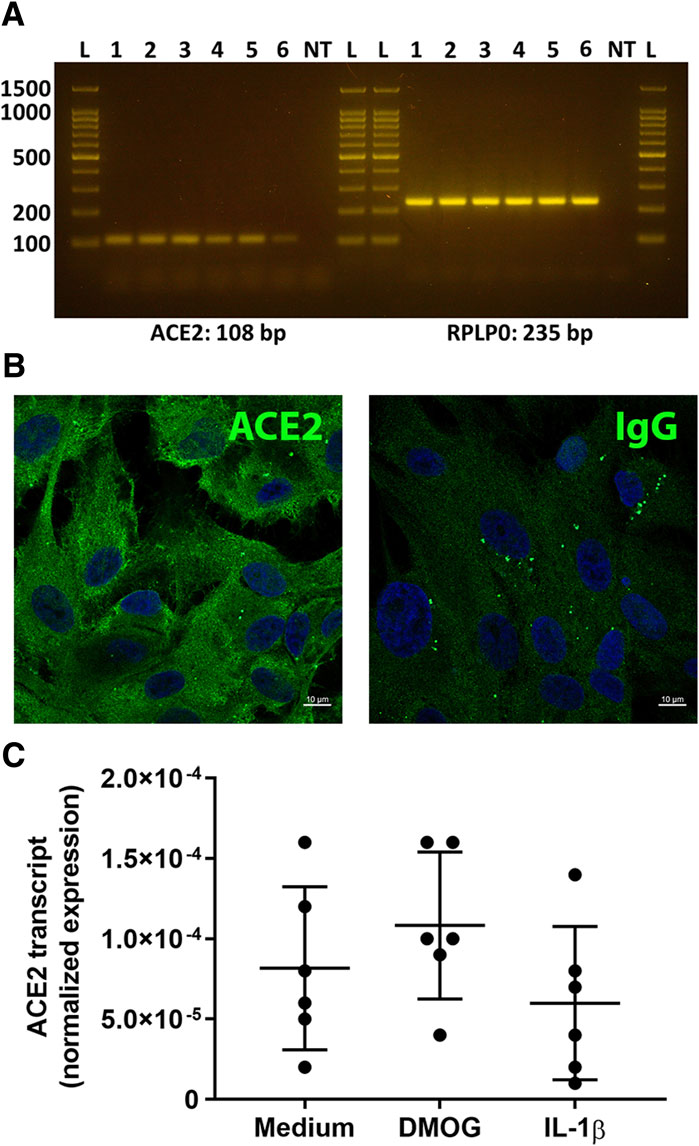
Figure 1. Expression of ACE2 by primary human retinal pigment epithelial cells. (A) Gel images show ACE2 and reference gene RPLP0 amplicons. Lanes: L = DNA ladder (indicating 100, 200, 500, 1,000, 1,500 base pair markers); 1-6 = primary human retinal pigment epithelial cell isolates from six individual donors; NT = no cDNA template control. Expected product size is stated in base pairs (bp) below gel image. (B) Fluorescence photomicrographs of human retinal pigment epithelial cells immunolabelled to detect the presence of ACE2, with negative control labelled with species-matched immunoglobulin (Ig)G. Alexa Fluor 488 (green) with DAPI nuclear counterstain (blue). Scale bar represents 10 microns. Representative of results obtained for three individual cell isolates. (C) Graph showing normalized expression of ACE2 transcripts in 6 cell isolates treated with dimethyloxaloylglycine (DMOG) or interleukin (IL)-1β, calculated relative to PPIA and YWHAZ. Circles represent individual cell isolates; crossbars indicate mean; and error bars indicate standard deviation. Data were analyzed by one-way ANOVA with Dunnett’s multiple comparison test applied to compare individual groups: there were no significant differences between control and treatment groups.
A search of 3,000 bp of sequence immediately upstream of the human ACE2 gene transcription start site on the JASPAR 2020 platform (Fornes et al., 2020) predicted a total of 255 potential cis-regulatory elements, which equated to binding sites for 93 different transcription factors (Supplementary Table S3). Cross-referencing this list with a human retinal pigment epithelial cell transcriptome, generated by RNA-sequencing (Smith et al., 2017), and prioritizing candidates on the basis of normalized read count, minimum predicted binding sites, and activator function, yielded five transcription factors: ETS proto-oncogene 1, transcription factor (ETS1, 12 cis-regulatory motifs), nuclear factor I C (NFIC, 4 cis-regulatory motifs), nuclear receptor subfamily 2 group C member 1 (NR2C1, 4 cis-regulatory motifs), TEA domain transcription factor 1 (TEAD1, 4 cis-regulatory motifs), and zinc finger protein 384 (ZNF384, 29 cis-regulatory motifs) (Table 1). Standard RT-PCR confirmed the expression of each of these transcription factors across human retinal pigment epithelial cell isolates from six donors (Figures 2A–E).
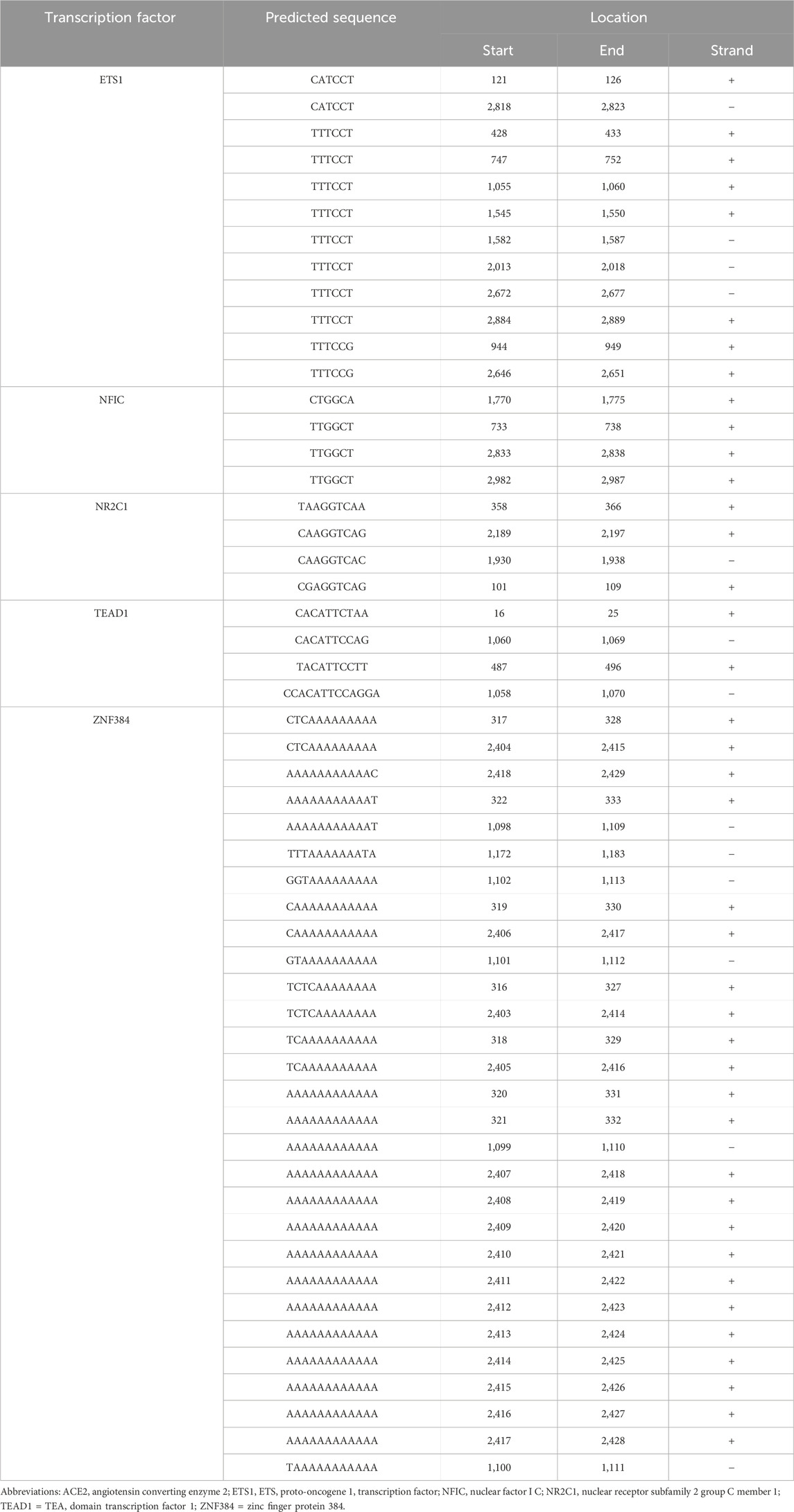
Table 1. Sequences and locations of putative transcription factor binding sites within the regulatory region of the human ACE2 gene. Gene sequences were obtained from the JASPAR 2020 database (Fornes et al., 2020).
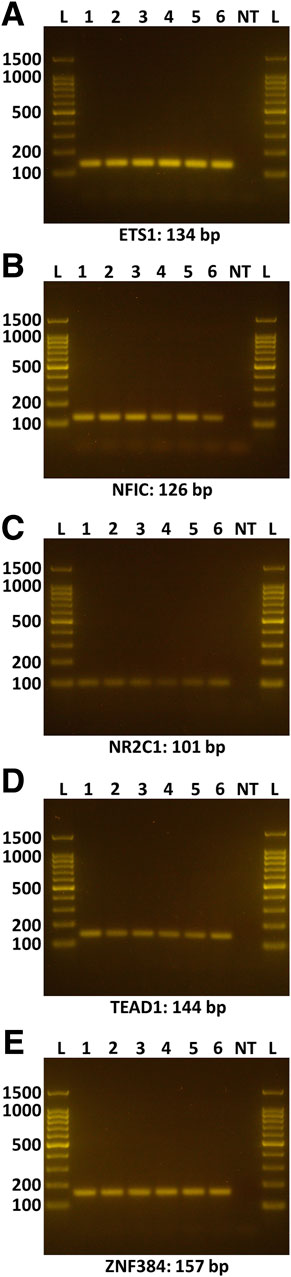
Figure 2. Expression of selected transcription factors in human retinal pigment epithelial cells. Gel images show amplicons representing the following transcription factors: (A) ETS1, (B) NFIC, (C) NR2C1, (D) TEAD1, (E) ZNF384. Lanes: L = DNA ladder (indicating 100, 200, 500, 1,000, 1,500 base pair markers); 1-6 = primary human retinal pigment epithelial cell isolates from six individual donors; NT = no cDNA template control. Expected product size is stated in base pairs (bp) below gel image.
Individual targeted knockdowns for the candidate transcription factors, plus positive control hepatocyte nuclear factor 1 alpha (HNF1α) (Pedersen et al., 2013), were performed in human retinal pigment epithelial cell isolates from five human donors using siRNA. This manipulation achieved highly significantly reduced mean expression across all the transcription factors (p ≤ 0.0014), averaging 74.6%–95.1% in comparison to expression in cells transfected with control non-targeted siRNA (Figures 3A–E). Levels of the ACE2 transcript were determined by real-time quantitative RT-PCR in these same transfected cell isolates. This demonstrated significantly reduced mean ACE2 expression under the condition of NR2C1 knockdown (p = 0.0059), as well as positive control HNF1α knock-down (p = 0.0064), and no significant changes in ACE2 expression for ETS1, NFIC, TEAD1 and ZNF384 knockdowns (p > 0.05) (Figure 3F). NR2C1 knockdown achieved an average 43.3% reduction in ACE2 transcript across the 5 cell isolates, similar to the 39.5% reduction for HNF1α knock-down (Figure 3G).
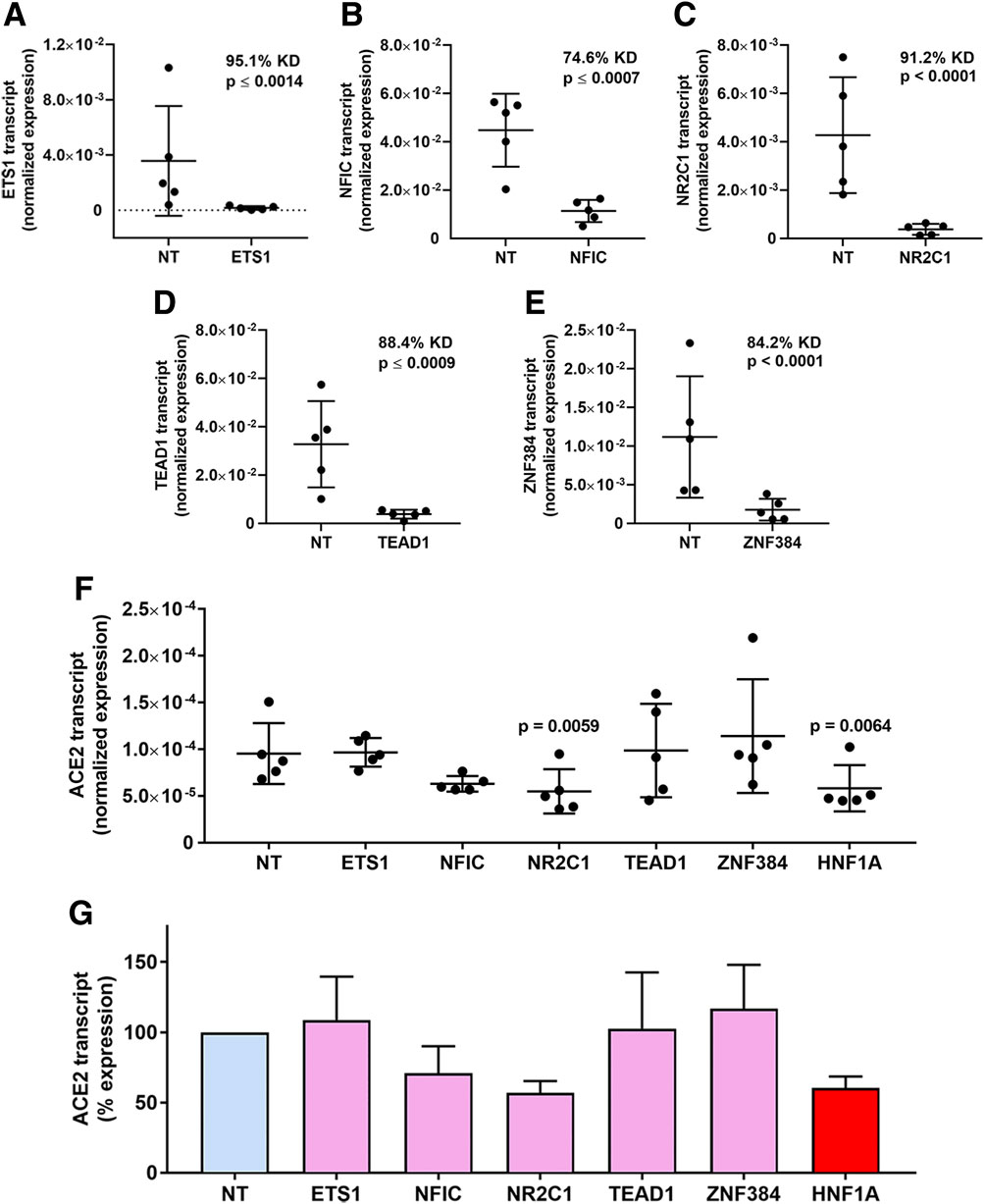
Figure 3. Transcription factor knockdown in human retinal pigment epithelial cells. (A–E) Graphs showing normalized expression of transcription factor transcripts, calculated relative to PPIA and YWHAZ, in human retinal pigment epithelial cell isolates treated with siRNA targeting (A) ETS1, (B) NFIC, (C) NR2C1, (D) TEAD1 and (E) ZNF384, or control non-targeted (NT) siRNA. Circles represent individual cell isolates (n = 4 monolayers per isolate); cross-bars indicate mean; and error bars indicate standard deviation. KD is knockdown effect expressed as percent. Data were analyzed by two-tailed paired Student’s t-test. (F) Graph showing normalized expression of ACE2 transcript, calculated relative to PPIA and YWHAZ, across the same samples studied in (A–E). Circles represent individual cell isolates; crossbars indicate mean; and error bars indicate standard deviation. Data were analyzed by one-way ANOVA with Dunnett’s multiple comparison test applied to compare individual groups. (G) Graph showing percentage changes in ACE2 transcript corresponding with normalized expression showed in (F). Bars indicate mean; and error bars indicate standard deviation.
As the receptor for the SARS-CoV-2 S protein, ACE2 is an appealing target for new anti-viral drugs to treat COVID-19 (Mostafa-Hedeab, 2020; Singh et al., 2022; Suvarnapathaki et al., 2022; Vitiello and Ferrara, 2022). In this Brief Research Report, we have shown expression of ACE2 transcript and protein in the retinal pigment epithelium across multiple primary human cell isolates. By cross-referencing a list of in silico-predicted transcription factors involved in ACE2 gene transcription with the human retinal pigment epithelial cell transcriptome, we identified and then confirmed five candidate transcription factors. We observed that specific knockdown of NR2C1 significantly reduced the level of ACE2 transcript in these cells. Our findings build on previous work demonstrating the potential for drugging gene transcription (Ashander et al., 2016; Ma et al., 2023), and also demonstrate the value of human retinal pigment epithelial cells as a testing system.
The transcription factor, NR2C1, a member of the nuclear receptor superfamily subgroup 2C, was originally cloned from human testis and named testicular nuclear receptor 2 (TR2) (Lin et al., 2017). Subsequently the transcription factor was found in epithelia, including the retinal pigment epithelium (Baker et al., 2016). NR2C1 has been implicated in early neuroretinal development, participating in phenotypic development of photoreceptors and amacrine cells (Mollema et al., 2011; Olivares et al., 2017). While not known to be involved in retinal disease, limited work suggests possible roles for NR2C1 in oncogenesis (Safe et al., 2014; Sun et al., 2021; Liu et al., 2023).
Other investigators have predicted transcription factor targets for influencing ACE2 gene expression in different model systems, typically drawing on in silico tools. One team used human lung and mouse tracheal RNA-sequencing datasets to identify binding sites for multiple transcription factors in the ACE2 gene promoter, although they did not interrogate any candidates in vitro (Barker and Parkkila, 2020). An independent team (Liang et al., 2022) predicted a series of transcription factors from chromatin immunoprecipitation (ChIP)-on-chip and ChIP-sequencing datasets, and showed that the candidate signal transducer and activator of transcription (STAT)3 influenced expression of the ACE2 gene in the immortalized human bronchial 16HBE cell line. Using reposited data generated from human and mouse non-specified epithelial cells, another group also predicted binding sites for STAT3 in the ACE2 gene regulatory region, as well as for STAT1, interferon regulatory factor (IRF)1, and IRF8 (Ziegler et al., 2020). Other researchers predicted specificity protein 1 and HNF4α as positive and negative regulators of ACE2 gene transcription, and confirmed these predictions in immortalized human alveolar epithelial cells and the A549 lung adenocarcinoma cell line using small molecule inhibitors and siRNA in promoter reporter assays and protein immunoassays (Han et al., 2024). Interestingly, an earlier study had identified HNF4α as an activator of ACE2 gene transcription in the HepG2 human hepatoma cell line (Niehof and Borlak, 2011).
Several groups have investigated ACE2 expression in human retinal cells (Yuan et al., 2020; Eriksen et al., 2021; Albertos-Arranz et al., 2023). Our observations made using multiple primary human retinal pigment epithelial cell isolates confirm the findings of studies undertaken with human adult eye cell isolates (Eriksen et al., 2021) and human retinal tissue sections (Albertos-Arranz et al., 2023), as well as a bioinformatics analysis of a range of public RNA-sequencing datasets (Yuan et al., 2020). Retinal pigment epithelial cells are readily isolated from non-diseased human cadaver eyes. Given the role of ACE2 genotype in the susceptibility to SARS-CoV-2 infection (Hattori et al., 2022; Hu P. et al., 2022a), the availability of multiple individual isolates for research studies is important. We also noted differences in the level of ACE2 gene expression between cell isolates. Although our work was not directed at retinal pathology in COVID-19, being primarily a respiratory disease, there are cases of retinopathy that suggest infection of the retinal pigment epithelium in vivo (Costa et al., 2021; Zhang and Stewart, 2021; Ng et al., 2024).
A novel short isoform of ACE2 that lacks most of the SARS-CoV-2 binding site, and is inducible by interferon and by rhinovirus, has been described by three independent groups (Ng et al., 2020; Onabajo et al., 2020; Blume et al., 2021). Human primary bronchial epithelial cells upregulate the transcript encoding the short ACE2 isoform following treatment with type I and type II interferons, and lung carcinoma cell lines Calu-3 and A549 respond similarly to type I interferon (Blume et al., 2021; Scagnolari et al., 2021). While this transcript is detectable in nasopharyngeal swabs from SARS-CoV-2-infected patients, and it shows a moderate positive correlation with the level of interferon-stimulated gene 15 transcript in these samples, the function of the short isoform of ACE2 remains unclear (Oliveto et al., 2022). Interestingly, the truncated transcript is present in hTERT-RPE1 cells, an immortalized human retinal pigment epithelial cell line, although these cells do not show stable protein expression (Blume et al., 2021). The primers used in the current study amplify a region common to known reference sequences for long and short ACE2 transcript variants and therefore would not differentiate between those encoding the short and long isoforms.
In conclusion, transcriptional blockade is receiving renewed attention as a therapeutic modality, particularly in those settings where it can be applied directly to cells involved in a disease (Henley and Koehler, 2021; Su and Henley, 2021). In COVID-19, which affects the respiratory tract, a locally applied, epithelial cell-directed transcription blocking agent is feasible. We have demonstrated the potential to use human retinal pigment epithelial cells as a straight-forward testing system for evaluating transcriptional blockade targeting ACE2. These primary epithelial cell isolates are readily generated from human cadaveric donor eyes, which are widely collected for use in corneal transplantation surgery (Gain et al., 2016), and thus more available than non-diseased human lung tissue. Moreover, interindividual differences in gene expression are captured by using multiple primary cell isolates as opposed to a single cell line.
The RNA-sequencing dataset used in this study is available in the NCBI Gene Expression Omnibus repository under accession number GSE132091: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132091.
The studies using primary human retinal cells were approved by the Southern Adelaide Clinical Human Research Ethics Committee (protocol 175.13). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation did not apply because the participants were deceased persons. Next-of-kin gave consent for the research use of eye tissue that would otherwise be discarded after removal of corneas for use in corneal transplantation surgeries.
LA: Conceptualization, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. AL: Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. YM: Investigation, Methodology, Writing–original draft, Writing–review and editing. AT: Formal Analysis, Investigation, Methodology, Writing–review and editing. BA: Conceptualization, Supervision, Writing–review and editing. SD: Conceptualization, Supervision, Writing–review and editing. MM: Conceptualization, Supervision, Writing–review and editing. JS: Conceptualization, Funding acquisition, Supervision, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Health and Medical Research Council (GNT1123684 and GNT1139857) and the Flinders Foundation.
The authors wish to thank Ms. Janet Matthews for administrative support in preparing this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fddsv.2024.1416728/full#supplementary-material
Albertos-Arranz, H., Martinez-Gil, N., Sanchez-Saez, X., Noailles, A., Monferrer Adsuara, C., Remoli Sargues, L., et al. (2023). Microglia activation and neuronal alterations in retinas from COVID-19 patients: correlation with clinical parameters. Eye Vis. (Lond) 10, 12. doi:10.1186/s40662-023-00329-2
Aleksova, A., Gagno, G., Sinagra, G., Beltrami, A. P., Janjusevic, M., Ippolito, G., et al. (2021). Effects of SARS-CoV-2 on cardiovascular system: the dual role of angiotensin-converting enzyme 2 (ACE2) as the virus receptor and homeostasis regulator-review. Int. J. Mol. Sci. 22, 4526. doi:10.3390/ijms22094526
Ashander, L. M., Appukuttan, B., Ma, Y., Gardner-Stephen, D., and Smith, J. R. (2016). Targeting endothelial adhesion molecule transcription for treatment of inflammatory disease: a proof-of-concept study. Mediat. Inflamm. 2016, 7945848. doi:10.1155/2016/7945848
Ashander, L. M., Lie, S., Ma, Y., Rochet, E., Washington, J. M., Furtado, J. M., et al. (2019). Neutrophil activities in human ocular toxoplasmosis: an in vitro study with human cells. Invest Ophthalmol. Vis. Sci. 60, 4652–4660. doi:10.1167/iovs.19-28306
Ashander, L. M., Lumsden, A. L., Dawson, A. C., Ma, Y., Ferreira, L. B., Oliver, G. F., et al. (2022). Infection of human retinal pigment epithelial cells with dengue virus strains isolated during outbreaks in Singapore. Microorganisms 10, 310. doi:10.3390/microorganisms10020310
Baker, J. L., Wood, B., Karpinski, B. A., LaMantia, A. S., and Maynard, T. M. (2016). Testicular receptor 2, Nr2c1, is associated with stem cells in the developing olfactory epithelium and other cranial sensory and skeletal structures. Gene Expr. Patterns 20, 71–79. doi:10.1016/j.gep.2015.12.002
Barker, H., and Parkkila, S. (2020). Bioinformatic characterization of angiotensin-converting enzyme 2, the entry receptor for SARS-CoV-2. PLoS One 15, e0240647. doi:10.1371/journal.pone.0240647
Blenkinsop, T. A., Salero, E., Stern, J. H., and Temple, S. (2013). The culture and maintenance of functional retinal pigment epithelial monolayers from adult human eye. Methods Mol. Biol. 945, 45–65. doi:10.1007/978-1-62703-125-7_4
Blume, C., Jackson, C. L., Spalluto, C. M., Legebeke, J., Nazlamova, L., Conforti, F., et al. (2021). A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat. Genet. 53, 205–214. doi:10.1038/s41588-020-00759-x
Clarke, N. E., and Turner, A. J. (2012). Angiotensin-converting enzyme 2: the first decade. Int. J. Hypertens. 2012, 307315. doi:10.1155/2012/307315
Costa, I. F., Bonifacio, L. P., Bellissimo-Rodrigues, F., Rocha, E. M., Jorge, R., Bollela, V. R., et al. (2021). Ocular findings among patients surviving COVID-19. Sci. Rep. 11, 11085. doi:10.1038/s41598-021-90482-2
Cousin, V. L., Giraud, R., and Bendjelid, K. (2021). Pathophysiology of COVID-19: everywhere you look you will see ACE2. Front. Med. (Lausanne) 8, 694029. doi:10.3389/fmed.2021.694029
COVID-19 Excess Mortality Collaborators (2022). Estimating excess mortality due to the COVID-19 pandemic: a systematic analysis of COVID-19-related mortality, 2020-21. Lancet 399, 1513–1536. doi:10.1016/S0140-6736(21)02796-3
Edwards, A. M., Baric, R. S., Saphire, E. O., and Ulmer, J. B. (2022). Stopping pandemics before they start: lessons learned from SARS-CoV-2. Science 375, 1133–1139. doi:10.1126/science.abn1900
Eriksen, A. Z., Moller, R., Makovoz, B., Uhl, S. A., tenOever, B. R., and Blenkinsop, T. A. (2021). SARS-CoV-2 infects human adult donor eyes and hESC-derived ocular epithelium. Cell Stem Cell 28, 1205–1220.e7. doi:10.1016/j.stem.2021.04.028
Eroglu, B., Nuwarda, R. F., Ramzan, I., and Kayser, V. (2021). A narrative review of COVID-19 vaccines. Vaccines (Basel) 10, 62. doi:10.3390/vaccines10010062
Feng, L., Li, H., Du, Y., Zhang, T., Zhu, Y., Li, Z., et al. (2022). Chaperonin-containing TCP1 subunit 5 protects against the effect of Mer receptor tyrosine kinase knockdown in retinal pigment epithelial cells by interacting with filamentous actin and activating the LIM-kinase 1/cofilin pathway. Front. Med. (Lausanne) 9, 861371. doi:10.3389/fmed.2022.861371
Fiolet, T., Kherabi, Y., MacDonald, C. J., Ghosn, J., and Peiffer-Smadja, N. (2022). Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review. Clin. Microbiol. Infect. 28, 202–221. doi:10.1016/j.cmi.2021.10.005
Fornes, O., Castro-Mondragon, J. A., Khan, A., van der Lee, R., Zhang, X., Richmond, P. A., et al. (2020). JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92. doi:10.1093/nar/gkz1001
Gain, P., Jullienne, R., He, Z., Aldossary, M., Acquart, S., Cognasse, F., et al. (2016). Global survey of corneal transplantation and eye banking. JAMA Ophthalmol. 134, 167–173. doi:10.1001/jamaophthalmol.2015.4776
Giovanetti, M., Branda, F., Cella, E., Scarpa, F., Bazzani, L., Ciccozzi, A., et al. (2023). Epidemic history and evolution of an emerging threat of international concern, the severe acute respiratory syndrome coronavirus 2. J. Med. Virol. 95, e29012. doi:10.1002/jmv.29012
Han, H., Luo, R. H., Long, X. Y., Wang, L. Q., Zhu, Q., Tang, X. Y., et al. (2024). Transcriptional regulation of SARS-CoV-2 receptor ACE2 by SP1. Elife 13, e85985. doi:10.7554/eLife.85985
Hattori, T., Saito, T., Okuya, K., Takahashi, Y., Miyamoto, H., Kajihara, M., et al. (2022). Human ACE2 genetic polymorphism affecting SARS-CoV and SARS-CoV-2 entry into cells. Microbiol. Spectr. 10, e0087022. doi:10.1128/spectrum.00870-22
Haydinger, C. D., Ashander, L. M., Tan, A. C. R., and Smith, J. R. (2023). Intercellular adhesion molecule 1: more than a leukocyte adhesion molecule. Biol. (Basel) 12, 743. doi:10.3390/biology12050743
Henley, M. J., and Koehler, A. N. (2021). Advances in targeting ‘undruggable’ transcription factors with small molecules. Nat. Rev. Drug Discov. 20, 669–688. doi:10.1038/s41573-021-00199-0
Hu, B., Guo, H., Zhou, P., and Shi, Z. L. (2021). Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 19, 141–154. doi:10.1038/s41579-020-00459-7
Hu, P., Bauer, V. L., Sawyer, S. L., and Diaz-Griffero, F. (2022a). Human ACE2 polymorphisms from different human populations modulate SARS-CoV-2 infection. Viruses 14, 1451. doi:10.3390/v14071451
Hu, X., Binter, M., Hufendiek, K., Tode, J., Framme, C., and Fuchs, H. (2022b). MiR-302d inhibits TGFB-induced EMT and promotes MET in primary human RPE cells. PLoS One 17, e0278158. doi:10.1371/journal.pone.0278158
Hui, D. S. C., and Zumla, A. (2022). Advances in the epidemiology, clinical features, diagnosis, clinical management and prevention of coronavirus disease 2019. Curr. Opin. Pulm. Med. 28, 166–173. doi:10.1097/MCP.0000000000000875
Lai, C. C., Wang, Y. H., Chen, K. H., Chen, C. H., and Wang, C. Y. (2022). The clinical efficacy and safety of anti-viral agents for non-hospitalized patients with COVID-19: a systematic review and network meta-analysis of randomized controlled trials. Viruses 14, 1706. doi:10.3390/v14081706
Li, M., Wang, H., Tian, L., Pang, Z., Yang, Q., Huang, T., et al. (2022). COVID-19 vaccine development: milestones, lessons and prospects. Signal Transduct. Target Ther. 7, 146. doi:10.1038/s41392-022-00996-y
Liang, L. J., Wang, D., Yu, H., Wang, J., Zhang, H., Sun, B. B., et al. (2022). Transcriptional regulation and small compound targeting of ACE2 in lung epithelial cells. Acta Pharmacol. Sin. 43, 2895–2904. doi:10.1038/s41401-022-00906-6
Lie, S., Rochet, E., Segerdell, E., Ma, Y., Ashander, L. M., Shadforth, A. M. A., et al. (2019). Immunological molecular responses of human retinal pigment epithelial cells to infection with Toxoplasma gondii. Front. Immunol. 10, 708. doi:10.3389/fimmu.2019.00708
Lin, S. J., Yang, D. R., Yang, G., Lin, C. Y., Chang, H. C., Li, G., et al. (2017). TR2 and TR4 orphan nuclear receptors: an overview. Curr. Top. Dev. Biol. 125, 357–373. doi:10.1016/bs.ctdb.2017.02.002
Liu, S., He, X., Di, Y., Li, Q., Li, F., Ma, Y., et al. (2023). NamiRNA-enhancer network of miR-492 activates the NR2C1-TGF-β/Smad3 pathway to promote epithelial-mesenchymal transition of pancreatic cancer. Carcinogenesis 44, 153–165. doi:10.1093/carcin/bgac102
Ma, Y., Ashander, L. M., Appukuttan, B., Ryan, F. J., Tan, A. C. R., Matthews, J. M., et al. (2023). Selective transcription factor blockade reduces human retinal endothelial cell expression of intercellular adhesion molecule-1 and leukocyte binding. Int. J. Mol. Sci. 24, 3304. doi:10.3390/ijms24043304
Markert, E. K., Klein, H., Viollet, C., Rust, W., Strobel, B., Kauschke, S. G., et al. (2022). Transcriptional comparison of adult human primary retinal pigment epithelium, human pluripotent stem cell-derived retinal pigment epithelium, and ARPE19 cells. Front. Cell Dev. Biol. 10, 910040. doi:10.3389/fcell.2022.910040
Mattern, L., Otten, K., Miskey, C., Fuest, M., Izsvak, Z., Ivics, Z., et al. (2022). Molecular and functional characterization of BDNF-overexpressing human retinal pigment epithelial cells established by sleeping beauty transposon-mediated gene transfer. Int. J. Mol. Sci. 23, 12982. doi:10.3390/ijms232112982
Mollema, N. J., Yuan, Y., Jelcick, A. S., Sachs, A. J., von Alpen, D., Schorderet, D., et al. (2011). Nuclear receptor Rev-erb alpha (Nr1d1) functions in concert with Nr2e3 to regulate transcriptional networks in the retina. PLoS One 6, e17494. doi:10.1371/journal.pone.0017494
Momtazi-Borojeni, A. A., Banach, M., Reiner, Z., Pirro, M., Bianconi, V., Al-Rasadi, K., et al. (2021). Interaction between coronavirus S-protein and human ACE2: hints for exploring efficient therapeutic targets to treat COVID-19. Angiology 72, 122–130. doi:10.1177/0003319720952284
Mostafa-Hedeab, G. (2020). ACE2 as drug target of COVID-19 virus treatment, simplified updated review. Rep. Biochem. Mol. Biol. 9, 97–105. doi:10.29252/rbmb.9.1.97
Nazir, F., Kombe, A. J. K., Khalid, Z., Bibi, S., Zhang, H., Wu, S., et al. (2024). SARS-CoV-2 replication and drug discovery. Mol. Cell Probes 77, 101973. doi:10.1016/j.mcp.2024.101973
Ng, H. W., Scott, D. A. R., Danesh-Meyer, H. V., Smith, J. R., McGhee, C. N., and Niederer, R. L. (2024). Ocular manifestations of COVID-19. Prog. Retin Eye Res. 102, 101285. doi:10.1016/j.preteyeres.2024.101285
Ng, K. W., Attig, J., Bolland, W., Young, G. R., Major, J., Wrobel, A. G., et al. (2020). Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option. Nat. Genet. 52, 1294–1302. doi:10.1038/s41588-020-00732-8
Niehof, M., and Borlak, J. (2011). HNF4alpha dysfunction as a molecular rational for cyclosporine induced hypertension. PLoS One 6, e16319. doi:10.1371/journal.pone.0016319
Olivares, A. M., Han, Y., Soto, D., Flattery, K., Marini, J., Mollema, N., et al. (2017). The nuclear hormone receptor gene Nr2c1 (Tr2) is a critical regulator of early retina cell patterning. Dev. Biol. 429, 343–355. doi:10.1016/j.ydbio.2017.05.021
Oliveto, G., Scagnolari, C., Frasca, F., Sorrentino, L., Matera, L., Nenna, R., et al. (2022). The non-functional ACE2 isoform, but not the SARS-CoV-2 receptor, is induced as an interferon-stimulated gene, in SARS-CoV-2 infected adults. Cytokine 158, 155997. doi:10.1016/j.cyto.2022.155997
Onabajo, O. O., Banday, A. R., Stanifer, M. L., Yan, W., Obajemu, A., Santer, D. M., et al. (2020). Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 52, 1283–1293. doi:10.1038/s41588-020-00731-9
Pedersen, K. B., Chhabra, K. H., Nguyen, V. K., Xia, H., and Lazartigues, E. (2013). The transcription factor HNF1α induces expression of angiotensin-converting enzyme 2 (ACE2) in pancreatic islets from evolutionarily conserved promoter motifs. Biochim. Biophys. Acta 1829, 1225–1235. doi:10.1016/j.bbagrm.2013.09.007
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. doi:10.1093/nar/29.9.e45
Safe, S., Jin, U. H., Hedrick, E., Reeder, A., and Lee, S. O. (2014). Minireview: role of orphan nuclear receptors in cancer and potential as drug targets. Mol. Endocrinol. 28, 157–172. doi:10.1210/me.2013-1291
Scagnolari, C., Bitossi, C., Viscido, A., Frasca, F., Oliveto, G., Scordio, M., et al. (2021). ACE2 expression is related to the interferon response in airway epithelial cells but is that functional for SARS-CoV-2 entry? Cytokine 140, 155430. doi:10.1016/j.cyto.2021.155430
Scialo, F., Daniele, A., Amato, F., Pastore, L., Matera, M. G., Cazzola, M., et al. (2020). ACE2: the major cell entry receptor for SARS-CoV-2. Lung 198, 867–877. doi:10.1007/s00408-020-00408-4
Singh, B., Singh, D., Verma, V., Yadav, R., and Kumar, R. (2022). Angiotensin-converting enzyme 2 as a potential therapeutic target for COVID-19: a review. J. Pharm. Anal. 12, 215–220. doi:10.1016/j.jpha.2021.12.003
Smith, J. R., Ashander, L. M., Ma, Y., Rochet, E., and Furtado, J. M. (2020). Model systems for studying mechanisms of ocular toxoplasmosis. Methods Mol. Biol. 2071, 297–321. doi:10.1007/978-1-4939-9857-9_17
Smith, J. R., Todd, S., Ashander, L. M., Charitou, T., Ma, Y., Yeh, S., et al. (2017). Retinal pigment epithelial cells are a potential reservoir for Ebola virus in the human eye. Transl. Vis. Sci. Technol. 6, 12. doi:10.1167/tvst.6.4.12
Su, B. G., and Henley, M. J. (2021). Drugging fuzzy complexes in transcription. Front. Mol. Biosci. 8, 795743. doi:10.3389/fmolb.2021.795743
Sun, G., Sun, K., and Shen, C. (2021). Human nuclear receptors (NRs) genes have prognostic significance in hepatocellular carcinoma patients. World J. Surg. Oncol. 19, 137. doi:10.1186/s12957-021-02246-x
Suvarnapathaki, S., Chauhan, D., Nguyen, A., Ramalingam, M., and Camci-Unal, G. (2022). Advances in targeting ACE2 for developing COVID-19 therapeutics. Ann. Biomed. Eng. 50, 1734–1749. doi:10.1007/s10439-022-03094-w
Tregoning, J. S., Flight, K. E., Higham, S. L., Wang, Z., and Pierce, B. F. (2021). Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 21, 626–636. doi:10.1038/s41577-021-00592-1
Verano-Braga, T., Martins, A. L. V., Motta-Santos, D., Campagnole-Santos, M. J., and Santos, R. A. S. (2020). ACE2 in the renin-angiotensin system. Clin. Sci. (Lond) 134, 3063–3078. doi:10.1042/CS20200478
Vitiello, A., and Ferrara, F. (2022). Pharmacotherapy based on ACE2 targeting and COVID-19 infection. Int. J. Mol. Sci. 23, 6644. doi:10.3390/ijms23126644
Yuan, J., Fan, D., Xue, Z., Qu, J., and Su, J. (2020). Co-expression of mitochondrial genes and ACE2 in cornea involved in COVID-19. Invest Ophthalmol. Vis. Sci. 61, 13. doi:10.1167/iovs.61.12.13
Zhang, Y., and Stewart, J. M. (2021). Retinal and choroidal manifestations of COVID-19. Curr. Opin. Ophthalmol. 32, 536–540. doi:10.1097/ICU.0000000000000801
Zhu, H., Zhang, L., Ma, Y., Zhai, M., Xia, L., Liu, J., et al. (2021). The role of SARS-CoV-2 target ACE2 in cardiovascular diseases. J. Cell Mol. Med. 25, 1342–1349. doi:10.1111/jcmm.16239
Ziegler, C. G. K., Allon, S. J., Nyquist, S. K., Mbano, I. M., Miao, V. N., Tzouanas, C. N., et al. (2020). SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016–1035.e19. e1-e11. doi:10.1016/j.cell.2020.04.035
Keywords: transcription, ACE2, NR2C1, retinal pigment epithelium, drugging
Citation: Ashander LM, Lumsden AL, Ma Y, Tan ACR, Appukuttan B, Daniel S, Michael MZ and Smith JR (2024) Brief research report: Transcriptional blockade of angiotensin converting enzyme 2 modelled in human retinal pigment epithelial cells. Front. Drug Discov. 4:1416728. doi: 10.3389/fddsv.2024.1416728
Received: 13 April 2024; Accepted: 29 August 2024;
Published: 07 October 2024.
Edited by:
Liang Xu, University of Kansas, United StatesReviewed by:
Marina Ramal Sánchez, University of Teramo, ItalyCopyright © 2024 Ashander, Lumsden, Ma, Tan, Appukuttan, Daniel, Michael and Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Justine R. Smith, anVzdGluZS5zbWl0aEBmbGluZGVycy5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.