
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Drug Discov., 17 July 2024
Sec. Anti-Infective Agents
Volume 4 - 2024 | https://doi.org/10.3389/fddsv.2024.1385460
This article is part of the Research TopicTackling Antimicrobial ResistanceView all 4 articles
Antimicrobial Resistance (AMR) is a critical global health challenge, and in this review article, we examine the limitations of traditional therapeutic methods and the emerging role of alternative therapies. By examining the reasons behind the failure of conventional treatments, including the inadequacy of one-drug-one-enzyme approaches, the complex evolution of AMR, and the impact of drug biotransformation, we better understand why conventional treatments failed. Moreover, the review discusses several alternative therapies, including RNA-based treatments, aptamers, peptide-based therapies, phage therapy, and probiotics, discussing their applications, advantages, and limitations. Additionally, we discuss the obstacles to develop these therapies, including funding shortages, regulatory barriers, and public perception. This comprehensive analysis aims to provide insight into the future of AMR, emphasizing the need for innovative strategies and practical approaches.
One of the 20th century’s most influential medical inventions is antibiotics, the most important class of pharmaceuticals. There is no doubt that antibiotics have proved to be a boon to human society in the fight against bacteria, saving millions of lives as a result (Ribeiro da Cunha et al., 2019). Microorganisms such as bacteria, fungi, parasites, and viruses develop antimicrobial resistance (AMR) when they become resistant to the antimicrobial drugs, such as antibiotics, that are used to treat them. With the rapid growth of AMR infection rates and the lack of new antimicrobial medications introduced to combat this problem, AMR has become one of the greatest concerns of the 21st century (Gajdacs and Albericio, 2019). The development of AMR presents significant challenges to traditional therapies. As pathogens evolve resistance mechanisms, once effective treatments become obsolete. This resistance arises from several factors. Such as over-prescription and misuse, incomplete treatment courses, global accessibility, agricultural use, lack of new antibiotics, hospital-acquired infections, environmental spread, genetic mutations, biofilm formation, etc. (Figure 1). Common instances of resistant pathogens include “Methicillin-resistant Staphylococcus aureus (MRSA) (Singh et al., 2023), “Vancomycin-resistant Enterococcus (VRE) (Singh et al., 2023), and “multidrug-resistant Mycobacterium tuberculosis (MDR-TB)” (Singha et al., 2024). The resistance mechanisms are varied, involving genetic changes that confer the ability to deactivate antimicrobials, modify drug targets, or prevent drug penetration.
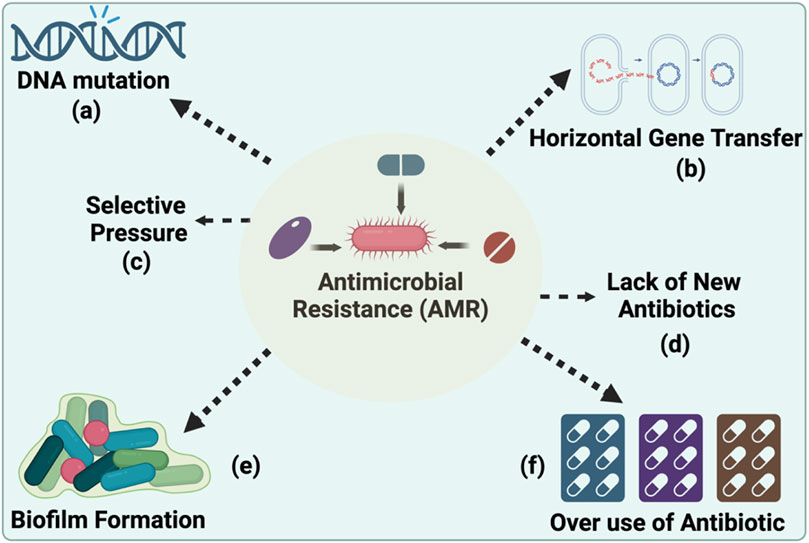
Figure 1. Key Drivers of Antimicrobial Resistance Development. (A) DNA mutations and (B) horizontal gene transfer as genetic mechanisms by which bacteria acquire and disseminate resistance. (C) Selective pressure, resulting from the (D) lack of novel antibiotics, is shown to favour the survival of resistant strains and (E) the role of biofilm formation in sheltering bacterial communities from antibiotic penetration and the impact of a lack of new antibiotics entering the market, which exacerbates the challenge of addressing resistant infections and (F) overuse and misuse of antibiotics, is shown to favour the survival of resistant strain. (Created using Biorender.com).
As the “Silent Pandemic,” AMR requires urgent attention and should be managed more effectively rather than being considered an issue for the future. By 2050, it is estimated that AMR may become the world’s leading cause of death without preventative measures. In 2019, more than 1.2 million people died directly as a result of AMR, and this figure is expected to rise to approximately 20 million deaths per year by 2050 if insufficient efforts are made to control AMR (Watkins and Bonomo, 2016). In the 21st century, we must focus on alternative therapies that do not rely on traditional antibiotics to bypass AMR. A number of studies have demonstrated the connection between COVID-19 and antimicrobial resistance (AMR), including the finding that severe cases often involved vanB clones of Enterococcus faecium in German intensive care units, and were accompanied by deaths due to infection with Enterobacter cloacae, which produces the New Delhi metallo-beta-lactamase. There was also an increase in the incidence of invasive fungal infections, exemplified by a deadly co-infection with resistant Candida glabrata and bacteria in Italy, as well as evidence that antimicrobial treatments for Aspergillus infections were ineffective (Singh and Sodhi, 2023). In order to monitor the spread of antibiotic resistance genes, whole-genome sequencing is one of the most popular tools (Sodhi et al., 2023).
Several alternative methods are available for avoiding the use of high doses of antibiotics to kill antibiotic-resistant bacteria: Phage Therapy, Probiotics, Antimicrobial Peptides (AMPs), Fecal Microbiota Transplantation (FMT), Host Defence Peptides, Metal Ions and Compounds, Antibody Therapies, Plant-Based Compounds, CRISPR-Cas Systems. An integrated approach to managing infections without contributing to the rise of AMR involves combining these alternative therapies with strategies to preserve the efficacy of existing anti-microbials. The purpose of this article is to summarize the fundamental mechanism behind AMR, limitations of routine therapeutic approaches, alternative therapies capable of combating AMR and its advantages and disadvantages, and future challenges.
The alarming issue of AMR is largely attributed to the failure of traditional one-drug one-enzyme therapeutics. This conventional approach to treating infections, where a single drug targets a specific bacterial enzyme or pathway, becomes ineffective as bacteria evolve resistance mechanisms (Prestinaci et al., 2015). This resistance can render standard antibiotics powerless, leading to treatment failures. Methicillin-resistant S. aureus (MRSA) illustrates how AMR undermines traditional, one-drug, one-enzyme treatments. Staphylococcus aureus infections were initially treated with penicillin, but as the bacteria developed resistance, methicillin was introduced as a penicillinase-resistant alternative. MRSA, characterized by the mecA gene that encodes a penicillin-binding protein with a low affinity for β-lactam antibiotics, including methicillin, rendered these treatments ineffective (Ali Alghamdi et al., 2023). As a result of the genetic evolution of MRSA, a broader issue in bacterial adaptation is highlighted, emphasizing the challenge of antibiotic resistance in previously treatable bacterial infections. The emergence of MRSA has had significant clinical implications. It requires alternative, often more expensive antibiotics, such as vancomycin or linezolid, which can have more side effects. Moreover, MRSA infections are associated with higher morbidity and mortality rates, especially in healthcare settings where patients are more vulnerable. This situation illustrates the broader challenge of AMR: as bacteria continue to develop resistance to existing drugs, the efficacy of standard antibiotic therapies is diminished, necessitating the development of new drugs and treatment strategies. Bacteria are complex organisms that receive and transfer DNA at an alarming frequency with seemingly little or no cost to themselves; contrary to previous dogma, the presence of an antibiotic has little impact on the transfer of mobile bacterial DNA but can select for newly acquired resistant mechanisms.
There are two main forms of antibiotic resistance: natural and acquired. When it comes to intrinsic resistance, it can either be innate (it can be expressed in the organisms) or mediated (the resistance genes are normally present in the bacteria, but they are activated to resistance levels after antibiotic treatment) (Reygaert, 2018) (Table 1). On the other hand, acquired resistance may result from bacteria acquiring genetic material through translation, conjugation, transposition (Lerminiaux and Cameron, 2019), or mutations in its own chromosomal DNA (Culyba et al., 2015) (Table 2).
AMR mechanisms based on the structure as mentioned in Table 3: 1) limitation of drug uptake; 2) drug target modification; 3) drug inactivation; and 4) drug efflux. Due to structural differences, Gram-negative bacteria can utilize above mentioned, all four mechanisms. In contrast, Gram-positive bacteria are less likely to utilize drug efflux mechanisms due to the absence of lipopolysaccharide in the outer membrane (Munita and Arias, 2016).
There is an inherent permeability shield created by the lipopolysaccharide (LPS) layer in most Gram-negative bacteria, making them more resistant to certain antibiotics than Gram-positive bacteria. For example, glycopeptide antibiotics, e.g., vancomycin, inhibits cell wall synthesis by binding to the D-Ala-D-Ala terminus of cell wall precursor units, is ineffective against Gram-negative bacteria because of the lack of penetration through the outer membrane, which is a prime illustration of the efficiency of this natural barrier (Munita and Arias, 2016) Modifying the outer membrane permeability strongly affects hydrophilic molecules, such as lactams, tetracyclines, and fluoroquinolones (Darby et al., 2023). In addition, alterations to envelope structure, such as porin loss or changes to phospholipid and fatty acid content of the cytoplasmic membrane, can affect the ability of a drug to penetrate the cell and contribute to the emergence of AMR (Delcour, 2009). Mycobacteria, produce a polysaccharide capsule coat that prevents hydrophilic molecules from entering the cell (Draper, 1998; Kumar et al., 2022). It has been shown that polar molecules have difficulties entering enterococci cell walls due to porin channel downregulation. A recent study suggests that reductions in porin expression significantly increase resistance to carbapenems among members of the Enterobacterales order, Acinetobacter spp., and Pseudomonas spp. (Iredell et al., 2016). Biofilms are another mechanism that aids bacteria in colonizing, resulting in the development of AMR. Due to the use of biofilm as a protection against antibiotics, P. aeruginosa has become a leading cause of morbidity and mortality in individuals with cystic fibrosis and immunocompromised individuals (Pang et al., 2019).
Many pathogenic bacteria exhibit antibiotic efflux as one of their most common resistance mechanisms (Webber and Piddock, 2003; Li et al., 2015). All bacterial efflux pumps are located on the inner membrane. Gram-negative bacteria have three components in their cell envelope, i.e., outer membrane, peptidoglycan layer, and inner membrane. Gram-positive bacteria have only two components in their cell envelope, i.e., peptidoglycan layer and inner membrane. Many antibiotics are actively transported out of the cell by bacterial efflux pumps, which contribute to Gram-negative bacteria’s intrinsic resistance. Six types of efflux pump families have been identified in bacteria, i.e., ATP-binding cassette (ABC) superfamily, major facilitator superfamily (MFS), small multidrug resistance (SMR) family, proteobacterial antimicrobial compound efflux (PACE) family, multidrug and toxin extrusion (MATE) family, and resistance-nodulation-cell division (RND) superfamily. AMR is associated with ATP-binding cassette efflux pumps (ABCs). Many substances, including antibiotics, are transported across cellular membranes using the energy from ATP hydrolysis. MacAB-TolC is an ABC family efflux pump contributing to AMR in Salmonella enterica serovar Typhimurium (Kudirkiene et al., 2018). Efflux pumps are known to confer resistance to macrolide antibiotics. By actively expelling these antibiotics from the bacterial cell, the MacAB-TolC system reduces their intracellular concentration, diminishing their effectiveness. These ABC efflux pumps can render certain antibiotics ineffective in pathogenic bacteria. Consequently, targeting these efflux pumps could be a potential strategy for overcoming AMR and improving antibiotic efficacy (Bogomolnaya et al., 2013). In host-pathogen communication, MFS transporters play an important role in adhesion, invasion, intracellular survival, and biofilm formation. Inhibiting MFS transporters is a promising strategy for combating drug resistance and reducing pathogenic virulence. Inactivating the MFS efflux pump, AbaF of A. baumannii reduces bacterial virulence in a Caenorhabditis elegans model (Sharma et al., 2017; Pasqua et al., 2021). The pathogen Neisseria gonorrhoeae (N. gonorrhoeae) causes gonorrhoea, a sexually transmitted disease. In N. gonorrhoeae, FarAB is a MFS pump, promotes resistance to long-chain fatty acids such as oleic, linoleic, and palmitic acids (Lee and Shafer, 1999). A foodborne pathogen, Listeria monocytogenes (L. monocytogenes), can penetrate the intestinal barrier and quickly spread to the liver and spleen. MdrM and MdrT are two MFS pumps that play a significant role in the pathophysiology of L. monocytogenes. MdrM and MdrT play a significant role in bile resistance and modulate the cytosolic surveillance pathway of innate immunity, contributing to the spread of bacteria and the invasion of tissues (Crimmins et al., 2008). The Small Multidrug Resistance (SMR) family is a group of efflux pumps found in bacteria that contribute to AMR by expelling various antimicrobial agents from the bacterial cell, thereby decreasing drug susceptibility. An example of an SMR family efflux pump involved in AMR is the EbrAB efflux system in Bacillus subtilis (Masaoka et al., 2000). This efflux pump has been shown to confer resistance to certain dyes, detergents, and disinfectants structurally similar to antimicrobial compounds. Another well-documented case is the QacA and QacB transporters in S. aureus, which confer resistance to quaternary ammonium compounds - a class of disinfectants commonly used in hospital settings. These examples illustrate the role of SMR family pumps in protecting bacteria from a range of chemically diverse antimicrobial substances, thus contributing to the broader problem of AMR (Oliveira-Tintino et al., 2023). The Proteobacterial Antimicrobial Compound Efflux (PACE) family of efflux pumps is a recent addition to the known efflux systems contributing to AMR. These pumps can expel a broad range of antimicrobial compounds, which contributes to the survival of bacteria in the presence of toxic substances, including antibiotics and disinfectants. An example of a PACE family efflux pump contributing to AMR is AceI, found in Acinetobacter baumannii. AceI provides resistance to various biocides, including chlorhexidine, a common disinfectant used in healthcare settings (Short et al., 2021). The Multidrug And Toxin Extrusion (MATE) family of efflux pumps is another group of transporters implicated in AMR. These pumps typically utilize the proton or sodium gradients across the bacterial cell membrane to extrude a wide variety of compounds, including antibiotics and toxins, from the cell. The MATE family efflux pump involved in AMR is NorM, which was first characterized in Vibrio parahaemolyticus (Stephen et al., 2022). NorM is capable of conferring resistance to a variety of compounds, including fluoroquinolones, a class of broad-spectrum antibiotics (Poole, 2000). This resistance occurs as NorM actively expels these antibiotics from the cell, lowering their intracellular concentration and thereby diminishing their efficacy. The ability of MATE pumps like NorM to reduce the effectiveness of antibiotics underlines the critical need to develop new therapeutic strategies to combat AMR. The Resistance-Nodulation-Cell Division (RND) superfamily of efflux pumps is a key player in bacterial AMR. These pumps are primarily found in Gram-negative bacteria and are known for their ability to expel a wide variety of substances, including antibiotics, detergents, dyes, and bile salts, which contribute significantly to the multidrug resistance phenotype. The RND efflux pump involved in AMR is the AcrAB-TolC system in Escherichia coli (Anes et al., 2015). This tripartite pump spans the entire cell envelope and can actively expel multiple classes of antibiotics, such as β-lactams, fluoroquinolones, and tetracyclines, from the bacterial cell. This efflux system significantly reduces the concentration of antibiotics within the bacterial cell, thereby protecting the bacteria from their bactericidal effects and contributing to the problem of AMR. The AcrAB-TolC system is a prime target for the development of new drugs designed to inhibit efflux pumps and restore antibiotic efficacy (Jang, 2023).
Bacteria can neutralize antibiotics through either degradation of the antibiotic molecule or chemical modification, rendering the drug ineffective. Chemical modification of the drug: Bacteria produce enzymes that attach different chemical groups to drugs. As a result, the antibiotic will not be able to bind to its target in the bacterial cell. The most effective method of drug inactivation through chemical group transfer is the transfer of phosphoryl, acetyl, and adenyl groups to the compound. Aminoglycosides, chloramphenicol, streptogramins, and fluoroquinolones are all examples of drugs that use acetylation as a mechanism of action. Adenylation and phosphorylation are thought to be the mechanisms by which aminoglycosides are targeted. An aminoglycoside modifying enzyme (AME) alters the hydroxyl or amino groups of the aminoglycoside molecule covalently, resulting in the inactivity of the aminoglycoside molecule, leading to the development of AMR (Munita and Arias, 2016).
Target modification is a prevalent strategy bacteria use to confer resistance to antibiotics (Reygaert, 2018). Alterations in penicillin-binding proteins (PBPs), both in their structure and quantity, contribute to β-lactam antibiotic resistance, affecting the drug’s ability to bind effectively (Bush and Bradford, 2016). For instance, the mecA gene in S. aureus leads to a PBP alteration that significantly diminishes or entirely inhibits the binding of β-lactam drugs (Foster, 2017). Similarly, resistance to macrolides, streptogramins, and lincosamides involves the erythromycin ribosome methylase (erm) gene family, which methylates 16S rRNA, thereby modifying the antibiotic binding site (Peterson and Kaur, 2018). Furthermore, mutations in DNA gyrase or topoisomerase IV, which are the targets of fluoroquinolones, can change the enzymes’ structures enough to prevent the drugs from attaching, resulting in resistance (Nainu et al., 2021).
AMR has no natural boundaries and has silently evolved into a global public health issue threatening populations from high, medium, and low-risk countries. The increased frequencies of AMR, especially among clinically significant pathogens such as Enterococcus species, S. aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, Enterobacter species, and E. coli, has put tremendous pressure on the healthcare, veterinary, and agriculture industries, making it one of the world’s most urgent public health concerns. Further, in a ‘One Health’ context, the consequences of the spread of AMR bacteria from food animals may profoundly impact both animal health and public health. Considering the global health impact of AMR bacteria and the need for new antibiotics, new strategies are being implemented to protect and treat (multidrug-resistant) MDR, (extensively drug-resistant) XDR, and (pandrug-resistant) PDR infections, as even so-called ‘antibiotics of last resort’ are becoming ineffective in clinical settings. To combat this problem, a few alternative approaches have been widely utilized in recent years (Table 4).
A strategy that also has the potential to generate novel antimicrobials is RNA silencing, naturally processed in bacteria, it was first described in 1985 and is now recognized as a regulator of a wide range of genes. A single m-RNA (antisense sequence) contains cis and trans sequences that are complementary and, upon binding, can reversibly block translation. Synthetic antisense sequences can potentially be developed to repress the translation of enzymes that enable bacteria to resist antibiotics (Good and Stach, 2011). RNA silencing is applied in the discovery of new antimicrobial compounds, determination of the stringency requirement for those targets, development of highly sensitized antimicrobial screens, and mode of action. Defeating AMR is a critical global issue that requires immediate attention. The development of alternative strategies to combat bacterial pathogens is of utmost importance. To address this issue, RNA-based therapies, such as synthetic sRNAs or (Clustered regularly interspaced short palindromic repeats) CRISPR guide RNAs, are attractive strategies (Parmeciano Di Noto et al., 2019). In particular, extended-spectrum beta-lactams, carbapenems, or colistin resistance genes can be targeted by both approaches. However, it is important to develop systems that are not only capable of delivering highly effective RNA elements but may also be rapidly modified upon the acquisition of new resistances by bacteria and thus limit their selection for MDR strains. Additionally, a combined system targeting several mRNAs in a coordinated manner would be more effective. RNA-based therapies represent a novel approach in combating AMR, utilizing the unique mechanisms of RNA molecules to target and inhibit bacterial growth. For instance, RNA interference (RNAi) can be employed to silence essential bacterial genes, with small interfering RNAs (siRNAs) designed to be complementary to bacterial mRNA, leading to its degradation and hindering bacterial survival. Similarly, antisense RNA (asRNA) therapies use synthetic RNA strands to block mRNA translation in bacteria, effectively preventing the production of key proteins. An example of this is the use of asRNA targeting essential genes in antibiotic-resistant strains of S. aureus, effectively reducing its virulence (Saberi et al., 2016). Furthermore, RNA silencing can also be applied to the development of antibacterial screening methods. As a result, genes of interest can be knocked down. Merck Research Laboratories (New Jersey, American) used a strain of S. aureus that expressed antisense RNA to screen 250,000 natural products for inhibitors of the FabF/FabH pathway, preventing the biosynthesis of fatty acids (Young et al., 2006). It is also possible to use RNA silencing to determine the mode of action of novel antibiotics; for instance, novel inhibitors of enoyl-acyl carrier protein reductase (Fabl) have been discovered using RNA silencing (Good and Stach, 2011). Additionally, the CRISPR-Cas system, adapted from bacterial immune defences, offers a way to specifically target and dismantle antibiotic resistance genes (Tao et al., 2022). The precision of these RNA-based methods allows for targeted attacks on resistant bacteria while minimizing the impact on beneficial microbiota, offering a promising alternative to traditional antibiotics. Despite their potential, challenges such as effective delivery and stability in the human body remain to be fully addressed for these therapies to become a mainstay in the fight against AMR. Virulence, resistance genes, and mobile genetic elements are appealing target candidates to combat antibiotic-resistant bacteria. It has been reported that some sRNAs are involved in antibiotic uptake (GcvB, RyhB, MicF, ErsA), drug efflux (DsrA RydC, SdsR, NrrF), biofilm formation (RprA OmrA/B, McaS, RybB, RydC), and modification of lipopolysaccharide and cell wall synthesis (MgrR, MicA, Sr006) (Felden and Cattoir, 2018; Tao et al., 2022). While most of these sRNAs have been extensively studied in E. coli and Salmonella, we need to understand the role of sRNA more extensively to use it as a new strategy in combating XDR and MDR bacteria (Dersch et al., 2017).
Over the past half-century, significant advances have been made in treating infectious diseases. However, many problems exist associated with developing effective therapies and dealing with antibiotic side effects and pathogen drug resistance. The development of aptamers could be an essential tool for expanding new therapeutic factors based on their ability to block pathogen microorganisms through binding of specifically to targets such as proteins and pathogens. In addition, they are simple, cheap, and have fewer side effects than conventional antibiotics. Aptamers, small single-stranded oligonucleotides (DNAs or RNAs), have emerged as powerful and flexible tools for therapeutic goals (Thiel and Giangrande, 2009). They offer several advantages over antibodies, including lower complexity and immunogenicity, higher affinity and specificity against various targets, flexibility, and stability (Zhou and Rossi, 2017). The antimicrobial activities of aptamers have been demonstrated in various studies, showing potential in addressing multidrug-resistant bacterial strains such as Salmonella. Aptamer G9 targets the polyphosphate kinase (PPK) gene, which is responsible for managing polyP metabolism and evading immune detection and is the key to the persistence of M. tuberculosis (Shum et al., 2011). Aptamers have been evaluated for their role in inhibiting biofilm formation, a major challenge in treating chronic infections, as they are resistant to current antibiotics and immune responses. A study has shown binding of specific aptamer to the flagella of S. choleraesuis, preventing biofilm formation (Ning et al., 2015). Furthermore, this study demonstrated that aptamer pre-treatment of S. choleraesuis biofilms could significantly reduce the amount of ampicillin needed for similar inhibition as well as prevent the development of antibiotic-resistant strains. As a result, the use of aptamers pre-treatment could eliminate the need for highly potent antibiotics in high doses. They have also been used to inhibit microbial toxins, which are important factors in bacterial pathogenesis. A key virulence factor produced by antibiotic resistance S. aureus is the alpha-toxin, which damages host epithelial tissue and targets cell membranes. To prevent alpha-toxin-mediated tissue damage, aptamers (AT-27, AT-33, AT-36, and AT-49) target the toxin (Okano, 1974). To explore aptamers as intracellular delivery vehicles, addressing the limitations of conventional antibiotic therapy, such as low intracellular drug concentrations and the development of multidrug resistance (Gopinath et al., 2016; Afrasiabi et al., 2020). Aptamer-nanoparticle conjugates are highlighted for their potential in antimicrobial activities and drug delivery. Additionally, aptamer-siRNA/miRNA conjugates are discussed for their promising modulatory effect on virulence, drug resistance, and pathogenesis (Afrasiabi et al., 2020). Aptamer-drug conjugates, like aptamer-ampicillin conjugate, have shown synergistic effects against biofilms, aiding drug penetration and reducing tolerance to drugs (Afrasiabi et al., 2020; Guan and Zhang, 2020).
Bacterial resistance to antibiotics and the rapid increase in infectious diseases due to AMR bacteria. Currently, antibiotic resistance and the associated infections are widespread and pose severe health and economic burdens globally, necessitating a massive demand for alternative treatment approaches. Additionally, many emerging alternative treatment options are at different stages of preclinical and clinical trials (Paul et al., 2023; Rossino et al., 2023). Antimicrobial peptides (AMPs), have an amino acid length ranging from 15 to 150, and hold promises as valuable therapeutic options against drug-resistant pathogens (Lazzaro et al., 2020). AMPs are low molecular weight oligopeptides that are found in plants, animals, and humans. They play a crucial role in the host’s innate immune response and have broad-spectrum antimicrobial activity. The AMPs hold promise as a viable therapeutic approach against drug-resistant pathogens. AMPs are oligopeptides with low molecular weight (Mba and Nweze, 2022). They have broad-spectrum antimicrobial activities against pathogenic microorganisms. AMPs are nonspecific and target components of microbes that facilitate immune response by acting as the first-line defence mechanisms against invading pathogenic microbes. The diversity and potency of AMPs make them good candidates for alternative use. They could be used alone or in combination with several other biomaterials for improved therapeutic activity. They can also be employed in vaccine production targeting drug-resistant pathogens (Mba and Nweze, 2022). This review covers the opportunities and advances in AMP discovery and development targeting AMR bacteria. Briefly, it presents an overview of the global burden of the AMR crisis, portraying the global magnitude, challenges, and consequences. After that, it critically and comprehensively evaluates the potential roles of AMPs in addressing the AMR crisis, highlighting the major potentials and prospects. Studies have shown that AMPs disrupt bacterial cell membranes, which can enhance the effectiveness of antibiotics on AMR strains. For example, peptides like P4-9 have been shown to increase cell membrane permeability in drug-resistant P. aeruginosa, enhancing the impact of antibiotics like novobiocin (Razquin-Olazaran et al., 2020). This effect, known as “Post-Antibiotic Effect-associated Permeabilization,” persists even after the peptide is removed. Another aspect is the use of D-enantiomeric peptides, such as DJK-6, which have been effective in preventing and eradicating biofilms in resistant strains of K. pneumoniae (Ribeiro et al., 2015) and P. aeruginosa (de la Fuente-Nunez et al., 2015). These also potentiate the activity of β-lactam antibiotics. Similarly, other AMPs like brevinin-1, derived from frog skin secretions, have broad-spectrum activity against pathogens including MRSA and Enterococcus faecalis (Mba and Nweze, 2022). Furthermore, the potential impact of modifying AMPs has been found useful with enhance therapeutic impacts. Such as the modified N6NH2 peptide exhibited improved stability and antimicrobial activity (Han et al., 2020). These peptides can act through various mechanisms, including cell permeabilization, interaction with DNA, and forming outer membrane vesicles. In conclusion, AMPs not only offer a promising avenue for combating antibiotic-resistant bacteria but also have potential applications in vaccine design due to their immunomodulatory properties. This makes them a valuable tool in addressing the global crisis of AMR. Moreover, various studies have shown the effect of AMP and their combination with other medication or antibiotics against clinical infections (Table 5).
Phage therapy, an innovative approach to combating AMR, involves bacteriophages (viruses that specifically infect bacteria) to treat bacterial infections, particularly those resistant to conventional antibiotics (Singha et al., 2023a). Phage therapy will become more popular in the twenty-first century since antibiotics kill bacteria in a broad spectrum, whereas bacteriophages are highly specific to their target bacteria. For example, in cases of drug-resistant S. aureus infections, phage therapy can be employed where specific phages are identified that selectively target and lyse these resistant bacterial cells, without harming beneficial microbiota (Yang et al., 2014; Pincus et al., 2015). In a recent study, a patient with multidrug-resistant Mycobacterium abscessus infection had been unable to respond to antibiotics, although cocktail of drugs can effectively eradicate drug-susceptible M. absecssus infections (Dedrick et al., 2023; Hatfull, 2023). Cystic fibrosis (CF) patients are particularly susceptible to chronic lung infections caused by AMR-Achromobacter. A recent study has shown that lytic phages could be an alternative method to combat AMR-Achromobacter infections (Winzig et al., 2022). This targeted action is particularly beneficial in combating biofilms, common in chronic wounds and notoriously resistant to standard antibiotic treatments. Phage therapy’s adaptability also allows it to evolve in response to bacterial resistance mechanisms, offering a dynamic solution to the growing challenge of AMR. In phage therapy, specific bacteriophages are selected for their ability to penetrate and disrupt these biofilms. For instance, phages like the Pseudomonas-specific phage ΦKZ have been studied for their efficacy against P. aeruginosa biofilms (Aghaee et al., 2021). These phages can invade the biofilm matrix, replicate within bacterial cells, and cause cell lysis, which not only kills the bacteria but can also disrupt the structure of the biofilm, making it more susceptible to antibiotics and the immune system. Clinical case reports and studies have shown that phage therapy can be effective in reducing P. aeruginosa density in biofilms and improving clinical outcomes, especially in cases where antibiotics alone were insufficient. This approach highlights the potential of phage therapy as a complementary treatment or alternative in cases where antibiotic resistance is a significant challenge. The available literature on the use of phages and phage-derived proteins for combating bacterial infections, specifically those of multidrug-resistant bacteria, increasingly shows promise for the prospect of phage therapy as either an alternative or a supplement to antibiotics. However, discrepancies in recent findings on the immunomodulatory effects, the host range, and the potential for horizontal gene transfer make it abundantly clear that we need a better understanding of the interaction between phage, microbiome, and human host before implementing phage therapy on a large scale.
Phage genome engineering involves extending their utility beyond their natural capabilities, such as by removing genes that promote lysogeny or introducing genes that enhance antibacterial activity (Sarkis et al., 1995; Pires et al., 2016; Hatoum-Aslan, 2018). To accomplish precise genome editing, advanced methods, such as recombination and CRISPR-Cas systems have been developed. As an example, the Bacteriophage Recombineering of Electroporated DNA (BRED) technique allows the production of recombinants without selection, which can be used to construct specific deletions, as demonstrated in phages that target Mycobacterium and E. coli (Marinelli et al., 2008; Feher et al., 2012; Shin et al., 2014; Pan et al., 2017; Singha et al., 2023b).
AMR is a critical issue, and natural products provide potential alternative therapies, offering a wealth of novel compounds with diverse antimicrobial properties. Various natural compounds, including those derived from plants, microbes, and marine organisms, have unique mechanisms that can combat resistant pathogens, often with a reduced likelihood of resistance developing. As an example, tea tree oil, obtained from Melaleuca alternifolia leaves, has been demonstrated to be effective against several bacteria and fungi, including some strains that are resistant to the oil (Carson et al., 2006). Curcumin, the active ingredient in turmeric, is another notable example. Besides fighting microbes directly, it has also shown promise in enhancing the efficacy of existing antibiotics and reducing bacterial resistance (Hussain et al., 2022). Furthermore, the antimicrobial potential of allicin, derived from garlic, and berberine, found in plants such as goldenseal, illustrates the wide variety of possibilities offered by natural products (Bhatwalkar et al., 2021; Hussain et al., 2022). Various compounds have been studied for their ability to combat infections, including those resistant to traditional antibiotics. Natural products can be used to address AMR, but there are challenges associated with their use. Variations in compound concentration, difficulties with standardization and mass production, and the need for extensive clinical trials to confirm efficacy and safety are significant challenges. However, these challenges also present opportunities for innovation in pharmaceutical development and healthcare practices. The integration of natural products into the mainstream medical field requires a multidisciplinary approach, involving botanists, chemists, pharmacologists, and clinicians. This collaborative effort is essential for the successful identification, extraction, and development of these natural compounds into safe and effective therapeutic agents. In addition to scientific research, regulatory frameworks must evolve to support the integration of natural products into treatment protocols, ensuring quality control and patient safety. Moreover, public awareness and education about the potential and limitations of natural products as alternative therapies are crucial for their acceptance and appropriate use (Rossino et al., 2023).
World Health Organization (WHO) released a list of innovative antibacterial approaches in April 2021, including the use of antibodies as a form of antibiotic therapy. Historically, serum antibodies were commonly used to treat infectious diseases before the widespread use of antibiotics (Bhatwalkar et al., 2021). Since antibiotic resistance is on the rise, there has been renewed interest in exploring vaccines for bacterial infections. Antibodies are therapeutic agents that harness the immune system to provide therapeutic benefits. As a result of their reduced clearance rate, lower toxicity, and increased longevity (the half-life of IgG antibodies is approximately 21 days), human monoclonal antibodies are especially advantageous. Furthermore, their high specificity enables them to target pathogens without disrupting the body’s normal bacterial flora. Numerous monoclonal antibodies are currently being developed, are undergoing clinical trials, and are showing promise against various pathogens (Motley et al., 2019). These include nosocomial infections such as S. aureus and P. aeruginosa, as well as challenging pathogens such as A. baumannii. A human monoclonal antibody developed by Trellis Biosciences inhibits biofilm formation among multiple bacteria by targeting the DNABII protein, an essential component (Zurawski and McLendon, 2020). Roche is also developing a conjugate of an antibody and an antibiotic that targets S. aureus (Peck et al., 2019). The conjugate enhances the effectiveness of the antibiotic by bringing it into proximity to the bacteria. MEDI3902, a bi-specific antibody targeting a surface polysaccharide of P. aeruginosa, inhibits its ability to form biofilms. This is another development by AstraZeneca PLC (Chung J. et al., 2023).
Moreover, recent clinical trials have evaluated suvratoxumab, a monoclonal antibody designed to treat ventilator-associated pneumonia caused by S aureus in patients using mechanical ventilation in intensive care settings. To reduce the harmful effects of this toxin, this antibody targets the toxin. Although many of these monoclonal antibodies are still in the early stages of clinical trials, a few shows potential for commercial development. To enhance the role of monoclonal antibodies in combating drug resistance, further research is essential to provide an alternative that is less likely to contribute to the growing problem of antibiotic resistance. Besides antibodies, studies have indicated cytokines, particularly interleukins and IFN-γ, are pivotal in enhancing the immune system response against intracellular pathogens, influencing the control of infections such as listeriosis and tuberculosis. Studies on mice show that IFN-γ boosts macrophage activity to suppress L. monocytogenes and Mycobacterium bovis, while anti-TNF antibodies can worsen infection outcomes (Li et al., 2020). This underscores the nuanced role of cytokines in AMR and their potential as therapeutic targets (Kaufmann and Flesch, 1990). Study has shown mucosa-associated invariant T (MAIT) cells play important roles against antimicrobial-resistant infections through their capacity to directly clear multidrug-resistant bacteria and overcome mechanisms of AMR (Leeansyah et al., 2021; Meermeier et al., 2022).
Probiotics are live microorganisms that produce antimicrobial metabolites and compete with microbes for nutrients. In contrast, antibiotics are active substances that directly fight pathogens. Among the most common strains used in human consumption are Lactobacillus and Bifidobacterium. Research has indicated that these organisms possess antimicrobial properties and are effective against several human pathogens. Recent research has demonstrated that probiotics have antimicrobial properties and enhanced immunological functions beyond gut health (Mazziotta et al., 2023; Petrariu et al., 2023). Several mechanisms are involved in their action, including the production of inhibitory substances such as bacteriocins and hydrogen peroxide, competition for adhesion sites, and competition for nutrients. Research has demonstrated that probiotics inhibit pathogenic bacteria and modulate the immune system (Neal-McKinney et al., 2012; Sikorska and Smoragiewicz, 2013; Markowiak and Slizewska, 2017). It includes increasing macrophage activity and modifying cytokine profiles to increase phagocytic activity (Jain et al., 2008; Dong et al., 2012; Zhao et al., 2012; Plaza-Diaz et al., 2017). Probiotics have a wide range of benefits, including anti-mutagenic (Yu and Li, 2016; Casas-Solis et al., 2020), anti-carcinogenic (Wollowski et al., 2001; Devaraj et al., 2019; Casas-Solis et al., 2020), and antidiarrheal properties (Liu et al., 2017; Devaraj et al., 2019), in addition to stimulating the immune system (Casas-Solis et al., 2020) and managing skin conditions such as atopic dermatitis and acne (Rather et al., 2016; Huang et al., 2017; Lise et al., 2018). Probiotics may be an adjunct strategy for controlling and preventing infectious diseases. For example, certain strains of Bifidobacterium can inhibit the growth of S. aureus, one of the most important pathogens associated with systemic and peri-implant infections (Lahtinen et al., 2007). A notable characteristic of multi-resistant bacteria such as MRSA is the ability of probiotics to disrupt and prevent the formation of bacterial biofilms. Probiotics can kill S. aureus through quorum-sensing inhibition, while Bacillus sp. produces antifungal lipopeptides, such as fengycin (Chung and Raffatellu, 2019). Pediococcus acidilactici has been shown to mitigate the virulence of P. aeruginosa by inhibiting motility and biofilm formation (Lee et al., 2020), and in the context of S. aureus biofilms on titanium surfaces, strains of Lactobacillus brevis and Bifidobacterium bifidum demonstrated significant inhibitory effects.
Additionally, probiotic lactobacilli have been implicated in reducing MRSA biofilm formation through various mechanisms, including competition and production of bacteriocin-like substances (Sikorska and Smoragiewicz, 2013). Probiotics against Helicobacter pylori infection are gaining much attention to enhance antibiotic regimens for eradication and mitigating conditions such as diarrhoea. Further, some Lactobacillus strains have been shown to prevent the formation of biofilms and disrupt those that have already formed. Furthermore, probiotics have been proposed to manage COVID-19-associated intestinal dysbiosis, with the potential to regulate the gut microbiota and minimize the risk of secondary infection (Xu et al., 2020). In infectious diarrhoea, the synergistic effects of probiotics and prebiotics have enhanced anti-rotavirus responses, reducing the intensity and duration of rotavirus-related diarrhoea.
Metals play a dual role with bacteria, essential for survival yet toxic at high levels, driving interest in metal-based compounds as potential antimicrobials amid rising antimicrobial resistance (AMR). Metalloantibiotics, including metal ions, nanoparticles, and complexes, though not yet in clinical use, show promise for novel antibiotic developments. Research has highlighted the significance of essential metals for bacterial processes and the immune system’s strategies to alter metal homeostasis to combat infections. Strategies leveraging specific bacterial metal uptake pathways, such as metallophores and siderophores, alongside the antimicrobial properties of metals like silver, copper, and zinc, offer innovative approaches to targeting bacterial infections. Metalloantibiotics, particularly those with investigated actions like rhenium and ruthenium complexes, demonstrate diverse structures and mechanisms, presenting a potential breakthrough in antibiotic therapy. Additionally, applications in antimicrobial photodynamic therapy and diagnostic imaging highlight the versatile benefits of metal complexes in addressing AMR. Metal-mediated antibacterial therapy is most commonly used with copper, whose history dates back to ancient Greece where it was used to treat lung disease and to purify water (Frei et al., 2023). It remains widely used in products to prevent bacterial growth, owing to its stability and cost-effectiveness compared to silver, and its ability to readily bind to bacterial surface groups. It also shows potential synergy with other drugs, enhancing the efficacy against resistant bacteria. The action of copper is hypothesized to involve the generation of hydroxyl radicals from its ions in an aqueous environment, leading to bacterial cell damage (Imlay et al., 1988; Sodhi et al., 2023). Copper has shown high inhibitory activity for Gram-positive bacteria. Copper ions also inactivate enzymes and impair membrane function, notably in E. coli (Macomber and Imlay, 2009). The study indicating copper complexed with antibiotics such as ciprofloxacin would be more efficient in against P. aeruginosa (Tewes et al., 2019). Cu(II) is especially studied for its effectiveness against diseases like tuberculosis and cancer (Macomber and Imlay, 2009). Quinolone-based copper complexes that target DNA gyrase, an enzyme crucial for DNA replication, have been examined, offering new avenues for antibacterial compounds against resistant strains. Additionally, a complex of Cu(II) with nalidixic acid showcased broader antibacterial activity compared to the antibiotic alone. The reduced polarity of such complexes due to chelation allows for better membrane penetration and thus enhanced bacterial cell growth inhibition.
Plant-derived pharmaceuticals are an important source of addressing multidrug resistance (MDR). There is potential for these plants to produce novel antibiotics and chemosensitizers to combat multidrug resistance, by focusing on the secondary metabolites they produce, which may offer new, biocompatible therapeutic options. Using traditional knowledge from practices such as Ayurveda and Traditional Chinese Medicine, ethnobotany and ethnopharmacology are vital for discovering beneficial medicinal plants and developing new drugs. Secondary metabolites of plants, which are essential for defence, have shown promise for therapeutic use, possibly offering protection against the development of resistance to some synthetic drugs. As herbal medicines become more widely recognized for their efficacy, lower side effects, and cost-effectiveness, they are becoming increasingly popular. The use of medicinal plants to develop drugs against resistant microbes (Table 6), however, requires continued research and international collaboration.
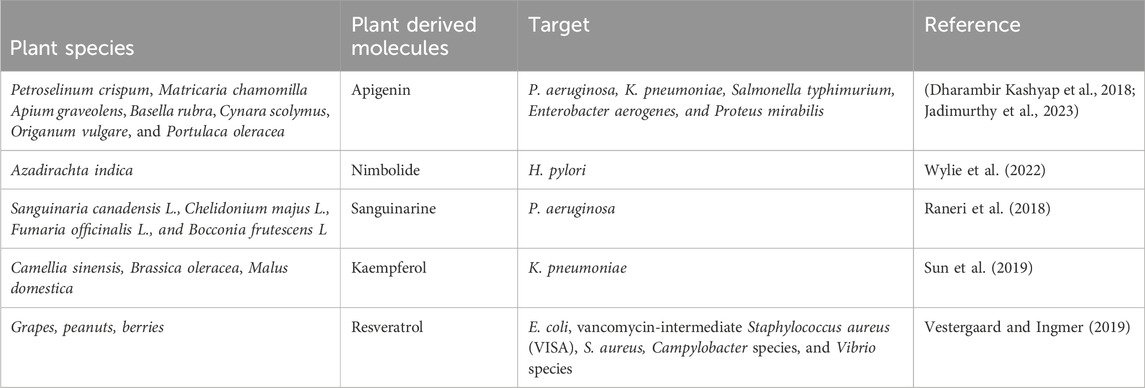
Table 6. List of phytochemicals that have demonstrated antibacterial activity against clinically important microorganisms.
In a recent study, Fecal Microbiota Transplantation (FMT) was found to be effective in decolonizing various common resistant pathogens, such as vancomycin-resistant E. faecium and K. pneumoniae carbapenemase (Caballero et al., 2015; Mrazek et al., 2019). In another study shown, mice treated with fecal microbiota transplantation (FMT) from murine and human donors showed significant reductions in multidrug-resistant P. aeruginosa within 7 days of the transplant. This study also suggests that donors from the same species may offer greater colonization resistance. According to Stalenhoef et al., a patient after one FMT session failed to eradicate an ESBL-producing multi-antibiotic-resistant E. coli strain, but was able to eradicate MDR P. aeruginosa within 3 months (Stalenhoef et al., 2017). Furthermore, Biernat et al. reported that their first patient was able to eliminate all infections after three weekly FMT sessions, while the second patient was still unable to remove MDR E coli and Citrobacter freundii after four weekly FMT sessions (Biernat et al., 2020).
A biofilm disruption agent is a substance that interferes with the structure and function of a biofilm, the protective layer that bacteria form to protect themselves against external threats, including antibiotics. These agents can effectively treat antibiotic-resistant infections (AMR) as they break down the biofilm matrix, exposing the bacteria to antibiotics and the immune system, thus enhancing treatment effectiveness. Dispersin B is an enzyme that specifically targets and breaks down polysaccharide intercellular adhesin (PIA), an essential component of biofilms formed by Staphylococcus epidermidis and S. aureus (Lister and Horswill, 2014; Nguyen et al., 2020). Dispersin B disrupts the biofilm by degrading PIA, making bacteria more susceptible to antibiotics and immune response (Abdelkader et al., 2023). Methicillin-resistant S. aureus (MRSA), a type of AMR bacteria, is particularly susceptible to this treatment. C-2-decenoic acid (C2DA), a medium-chain fatty acid produced by P. aeruginosa, and D-amino acids have emerged as potential disruptors of biofilms in various bacteria, including antibiotic-resistant strains like MRSA (Ali Alghamdi et al., 2023). It has been demonstrated that C2DA inhibits and disperses biofilms in both Gram-positive and Gram-negative bacteria, suggesting its dual effectiveness in inhibiting biofilm initiation and promoting its dispersion (Jennings et al., 2012; Chung and Toh, 2014). C2DA has been demonstrated to enhance antibiotic effectiveness against biofilms, including those by Jennings et al. (2012) and Davies and Marques (2009), although it does not eradicate them completely, indicating the need for combination therapy to achieve the best results (Jennings et al., 2012). In addition, Kolodkin-Gal et al. have observed that D-amino acids have the ability to dismantle biofilms by integrating into bacterial cell walls and disrupting the structural integrity of the biofilm matrix (Kolodkin-Gal et al., 2010).
Alternative therapeutics hold promise for offering new solutions in the fight against infections, especially in the face of growing antibiotic resistance. However, they are not a magic bullet and face several challenges before widespread adoption. The future will likely see a combination of optimized antibiotics and well-researched alternatives used strategically to combat different types of infections and minimize resistance development. It is important to remember that research in this field is ongoing, and the landscape of potential alternatives is constantly evolving. Here, we summarize a general overview, and specific advantages and disadvantages may vary depending on the individual therapeutic approach being considered.
1. Potential for targeting diverse resistance mechanisms: Unlike traditional antibiotics that often target one specific bacterial process, alternative therapies can aim at different pathways involved in resistance, potentially overcoming broader spectrum resistance (Park et al., 2011).
2. Reduced selective pressure for further resistance development: Since they often work with different mechanisms, alternative therapies may exert less selective pressure on bacteria, potentially slowing down the emergence of new resistance (Takahashi et al., 2004).
3. Enhanced immune system involvement: Some alternative approaches, like phage therapy or immunomodulatory strategies, can stimulate the body’s own defence mechanisms, offering a more holistic approach to fighting infections (Howard et al., 1977).
4. Reduced side effects and potentially fewer drug interactions: Alternative therapies can potentially offer benefits over antibiotics in terms of side effects, especially for patients with pre-existing conditions or concerns about antibiotic-associated adverse events (Lei et al., 2019).
5. Potential for addressing non-bacterial infections: Some alternative approaches, like phage therapy or CRISPR-based interventions, might have applications beyond bacterial infections, offering broader utility in the fight against microbial resistance (Laverty et al., 2011).
1. Limited clinical data and evidence: Many alternative therapies are still in early stages of research and development, lacking robust clinical data and established efficacy compared to conventional antibiotics.
2. Difficulties in targeting specific bacteria: Some alternative therapies might struggle to selectively target specific bacterial pathogens, potentially harming beneficial bacteria in the microbiome and leading to other complications.
3. Cost and accessibility challenges: Developing, manufacturing, and delivering some alternative therapies might be more expensive than traditional antibiotics, raising concerns about accessibility, especially in resource-limited settings.
4. Regulatory uncertainties and approval processes: Regulatory frameworks for approving alternative therapies might not be clear or established, leading to delays and uncertainties in bringing them to market.
5. Lack of standardization and quality control: Ensuring consistent quality and effectiveness across different formulations or producers of alternative therapies can be a challenge, requiring robust quality control measures.
Overall, while alternative therapies offer promising avenues for tackling antibiotic drug resistance, significant challenges remain before they can fully replace or complement traditional antibiotics.
AMR is a ticking time bomb in the medical world, and despite several advantages and motivations, developing alternative therapies presents a multitude of challenges. Here are some key hurdles to overcome.
1. Targeting different mechanisms: Unlike conventional antibiotics that directly kill bacteria, alternatives often target different, more complex mechanisms, like interfering with resistance pathways or modulating the microbiome (Alaoui Mdarhri et al., 2022). This can be difficult to achieve effectively and selectively.
2. Limited understanding of resistance: The sheer diversity and rapid evolution of bacterial resistance mechanisms make it hard to predict how effectively new therapies will work and for how long these will survive without generating resistance (McCubbin et al., 2021).
3. Lack of robust preclinical models: Testing alternative therapies often requires sophisticated models that accurately mimic human infections and resistance profiles. Developing and validating such models is time-consuming and expensive.
4. Safety and efficacy concerns: New therapies need to be thoroughly tested for safety and efficacy in humans, which can be a lengthy and costly process (Saarela, 2019). Additionally, ensuring effectiveness against diverse resistant strains adds another layer of complexity.
1. Low commercial viability: Compared to drugs for chronic diseases, antibiotics are used for shorter durations, limiting their profit potential for pharmaceutical companies. This disincentivizes investment in research and development.
2. Regulatory challenges: Regulatory pathways for approving alternative therapies are not well-established, leading to delays and uncertainties. Governments and agencies need to adapt their processes to accommodate these innovative approaches.
3. Lack of awareness and infrastructure: Knowledge and adoption of alternative therapies among healthcare professionals and communities need to be improved. Building infrastructure and addressing logistical challenges for delivery, especially in resource-limited settings, is crucial.
1. Public perception and overuse: Misuse and unnecessary antibiotic use fuel the emergence and spread of resistance. Addressing public perception and promoting responsible antibiotic use remains a critical aspect.
2. Collaboration and knowledge sharing: Effective development and implementation of alternative therapies require collaboration between researchers, clinicians, policymakers, and industry players. Sharing knowledge and resources openly is essential.
Despite these challenges, the urgent need for effective solutions against AMR drives continuous research and development. Exploring diverse approaches like phage therapy, antimicrobial peptides, CRISPR-based interventions, and immunomodulatory strategies holds promise for the future. Continued research, development, and collaboration between researchers, clinicians, and policymakers are crucial to overcome these hurdles and harness the potential of these innovative approaches in effectively combating AMR (Robinson et al., 2016).
The fight against drug-resistant bacteria enters a new chapter with growing research into promising non-antibiotic therapies. These approaches hold immense potential to combat the alarming rise of antibiotic resistance, offering a potent alternative to conventional weapons. However, we must tread cautiously. While these therapies present highly sophisticated modes of action, extensive clinical trials are still needed to solidify their efficacy and safety. Therefore, implementing them as routine treatments demands careful consideration. In addition to treating AMR bacteria with alternative therapies, we should also consider strategies for sustainable remediation of antibiotics utilizing waste products, biochar, and microbial fuel cells (Kushneet Kaur Sodhi, 2022). Ultimately, a deep understanding of resistance patterns and mechanisms of action is crucial. This knowledge will empower clinicians to tailor antimicrobial treatment, paving the way for personalized and precise medicine.
BS: Conceptualization, Writing–original draft, Writing–review and editing. VSi: Formal Analysis, Funding acquisition, Resources, Validation, Writing–original draft, Writing–review and editing. VSo: Conceptualization, Data curation, Formal Analysis, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. VSi acknowledges South African Medical Research Council (SAMRC) and the South African National Research Foundation for their support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdelkader, J., Alelyani, M., Alashban, Y., Alghamdi, S. A., and Bakkour, Y. (2023). Modification of dispersin B with cyclodextrin-ciprofloxacin derivatives for treating staphylococcal. Molecules 28, 5311. doi:10.3390/molecules28145311
Afrasiabi, S., Pourhajibagher, M., Raoofian, R., Tabarzad, M., and Bahador, A. (2020). Therapeutic applications of nucleic acid aptamers in microbial infections. J. Biomed. Sci. 27 (6), 6. doi:10.1186/s12929-019-0611-0
Aghaee, B. L., Khan Mirzaei, M., Alikhani, M. Y., Mojtahedi, A., and Maurice, C. F. (2021). Improving the inhibitory effect of phages against Pseudomonas aeruginosa isolated from a burn patient using a combination of phages and antibiotics. Viruses 13, 334. doi:10.3390/v13020334
Alaoui Mdarhri, H., Benmessaoud, R., Yacoubi, H., Seffar, L., Guennouni Assimi, H., Hamam, M., et al. (2022). Alternatives therapeutic approaches to conventional antibiotics: advantages, limitations and potential application in medicine. Antibiot. (Basel) 11, 1826. doi:10.3390/antibiotics11121826
Ali Alghamdi, B., Al-Johani, I., Al-Shamrani, J. M., Musamed Alshamrani, H., Al-Otaibi, B. G., Almazmomi, K., et al. (2023). Antimicrobial resistance in methicillin-resistant staphylococcus aureus. Saudi J. Biol. Sci. 30, 103604. doi:10.1016/j.sjbs.2023.103604
Anes, J., McCusker, M. P., Fanning, S., and Martins, M. (2015). The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol. 6, 587. doi:10.3389/fmicb.2015.00587
Arreola, R., Quintero-Fabián, S., López-Roa, R. I., Flores-Gutiérrez, E. O., Reyes-Grajeda, J. P., Carrera-Quintanar, L., et al. (2015). Immunomodulation and anti-inflammatory effects of garlic compounds. J. Immunol. Res. 2015, 401630. doi:10.1155/2015/401630
Bhatwalkar, S. B., Mondal, R., Krishna, S. B. N., Adam, J. K., Govender, P., and Anupam, R. (2021). Antibacterial properties of organosulfur compounds of garlic (allium sativum). Front. Microbiol. 12, 613077. doi:10.3389/fmicb.2021.613077
Biazzo, M., and Deidda, G. (2022). Fecal microbiota transplantation as new therapeutic avenue for human diseases. J. Clin. Med. 11, 4119. doi:10.3390/jcm11144119
Biernat, M. M., Urbaniak-Kujda, D., Dybko, J., Kapelko-Słowik, K., Prajs, I., and Wróbel, T. (2020). Fecal microbiota transplantation in the treatment of intestinal steroid-resistant graft-versus-host disease: two case reports and a review of the literature. J. Int. Med. Res. 48, 300060520925693. doi:10.1177/0300060520925693
Bogomolnaya, L. M., Andrews, K. D., Talamantes, M., Maple, A., Ragoza, Y., Vazquez-Torres, A., et al. (2013). The ABC-type efflux pump MacAB protects Salmonella enterica serovar typhimurium from oxidative stress. mBio 4, e00630–e00613. doi:10.1128/mBio.00630-13
Bush, K., and Bradford, P. A. (2016). β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb. Perspect. Med. 6, a025247. doi:10.1101/cshperspect.a025247
Bush, K., and Bradford, P. A. (2020). Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 33, e00047. doi:10.1128/CMR.00047-19
Caballero, S., Carter, R., Ke, X., Sušac, B., Leiner, I. M., Kim, G. J., et al. (2015). Distinct but spatially overlapping intestinal niches for vancomycin-resistant Enterococcus faecium and carbapenem-resistant Klebsiella pneumoniae. PLoS Pathog. 11, e1005132. doi:10.1371/journal.ppat.1005132
Carson, C. F., Hammer, K. A., and Riley, T. V. (2006). Melaleuca alternifolia (Tea Tree) oil: a review of antimicrobial and other medicinal properties. Clin. Microbiol. Rev. 19, 50–62. doi:10.1128/CMR.19.1.50-62.2006
Casas-Solis, J., Huizar-Lopez, M. D. R., Irecta-Najera, C. A., Pita-Lopez, M. L., and Santerre, A. (2020). Immunomodulatory effect of Lactobacillus casei in a murine model of colon carcinogenesis. Probiotics Antimicrob. Proteins 12, 1012–1024. doi:10.1007/s12602-019-09611-z
Chung, C., Kudchodkar, S. B., Park, Y. K., Xu, Z., Pardi, N., et al. (2023a). Expanding the reach of monoclonal antibodies: a review of synthetic nucleic acid delivery in immunotherapy. Antibodies (Basel) 12, 46. doi:10.3390/antib12030046
Chung, J., Eisha, S., Park, S., Morris, A. J., and Martin, I. (2023b). How three self-secreted biofilm exopolysaccharides of Pseudomonas aeruginosa, psl, pel, and alginate, can each Be exploited for antibiotic adjuvant effects in cystic fibrosis lung infection. Int. J. Mol. Sci. 24, 8709. doi:10.3390/ijms24108709
Chung, L. K., and Raffatellu, M. (2019). Probiotic fengycins dis(Agr)ee with Staphylococcus aureus colonization. Cell. Res. 29, 93–94. doi:10.1038/s41422-018-0126-3
Chung, P. Y., and Toh, Y. S. (2014). Anti-biofilm agents: recent breakthrough against multi-drug resistant Staphylococcus aureus. Pathog. Dis. 70, 231–239. doi:10.1111/2049-632X.12141
Crimmins, G. T., Herskovits, A. A., Rehder, K., Sivick, K. E., Lauer, P., Dubensky, T. W., et al. (2008). Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc. Natl. Acad. Sci. U. S. A. 105, 10191–10196. doi:10.1073/pnas.0804170105
Culyba, M. J., Mo, C. Y., and Kohli, R. M. (2015). Targets for combating the evolution of acquired antibiotic resistance. Biochemistry 54, 3573–3582. doi:10.1021/acs.biochem.5b00109
Darby, E. M., Trampari, E., Siasat, P., Gaya, M. S., Alav, I., Webber, M. A., et al. (2023). Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 21, 280–295. doi:10.1038/s41579-022-00820-y
Dedrick, R. M., Smith, B. E., Cristinziano, M., Freeman, K. G., Jacobs-Sera, D., Belessis, Y., et al. (2023). Phage therapy of Mycobacterium infections: compassionate use of phages in 20 patients with drug-resistant mycobacterial disease. Clin. Infect. Dis. 76, 103–112. doi:10.1093/cid/ciac453
de la Fuente-Nunez, C., Reffuveille, F., Mansour, S. C., Reckseidler-Zenteno, S. L., Hernández, D., Brackman, G., et al. (2015). D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 22, 196–205. doi:10.1016/j.chembiol.2015.01.002
Delcour, A. H. (2009). Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 1794, 808–816. doi:10.1016/j.bbapap.2008.11.005
Dempsey, E., and Corr, S. C. (2022). Lactobacillus spp. for gastrointestinal health: current and future perspectives. Front. Immunol. 13, 840245. doi:10.3389/fimmu.2022.840245
Dersch, P., Khan, M. A., Muhlen, S., and Gorke, B. (2017). Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 8, 803. doi:10.3389/fmicb.2017.00803
Devaraj, N. K., Suppiah, S., Veettil, S. K., Ching, S. M., Lee, K. W., Menon, R. K., et al. (2019). The effects of probiotic supplementation on the incidence of diarrhea in cancer patients receiving radiation therapy: a systematic review with meta-analysis and trial sequential analysis of randomized controlled trials. Nutrients 11, 2886. doi:10.3390/nu11122886
Dharambir Kashyap, A. S., Singh Tuli, H., Sak, K., Garg, V. K., Singh Buttar, H., Setzer, W. N., et al. (2018). Apigenin: a natural bioactive flavone-type molecule with promising therapeutic function. J. Funct. Foods 48, 457–471. doi:10.1016/j.jff.2018.07.037
Dong, H., Rowland, I., and Yaqoob, P. (2012). Comparative effects of six probiotic strains on immune function in vitro. Br. J. Nutr. 108, 459–470. doi:10.1017/S0007114511005824
Draper, P. (1998). The outer parts of the mycobacterial envelope as permeability barriers. Front. Biosci. 3, D1253–D1261. doi:10.2741/a360
Feher, T., Karcagi, I., Blattner, F. R., and Posfai, G. (2012). Bacteriophage recombineering in the lytic state using the lambda red recombinases. Microb. Biotechnol. 5, 466–476. doi:10.1111/j.1751-7915.2011.00292.x
Felden, B., and Cattoir, V. (2018). Bacterial adaptation to antibiotics through regulatory RNAs. Antimicrob. Agents Chemother. 62, e02503. doi:10.1128/AAC.02503-17
Fishovitz, J., Hermoso, J. A., Chang, M., and Mobashery, S. (2014). Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 66, 572–577. doi:10.1002/iub.1289
Foster, T. J. (2017). Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 41, 430–449. doi:10.1093/femsre/fux007
Frei, A., Verderosa, A. D., Elliott, A. G., Zuegg, J., and Blaskovich, M. A. T. (2023). Metals to combat antimicrobial resistance. Nat. Rev. Chem. 7, 202–224. doi:10.1038/s41570-023-00463-4
Fu, Y., Zhang, F., Zhang, W., Chen, X., Zhao, Y., Ma, J., et al. (2007). Differential expression of bla(SHV) related to susceptibility to ampicillin in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 29, 344–347. doi:10.1016/j.ijantimicag.2006.10.015
Gajdacs, M., and Albericio, F. (2019). Antibiotic resistance: from the bench to patients. Antibiot. (Basel) 8, 129. doi:10.3390/antibiotics8030129
Good, L., and Stach, J. E. (2011). Synthetic RNA silencing in bacteria - antimicrobial discovery and resistance breaking. Front. Microbiol. 2, 185. doi:10.3389/fmicb.2011.00185
Gopinath, S. C., Lakshmipriya, T., Chen, Y., Arshad, M. K. M., Kerishnan, J. P., Ruslinda, A. R., et al. (2016). Cell-targeting aptamers act as intracellular delivery vehicles. Appl. Microbiol. Biotechnol. 100, 6955–6969. doi:10.1007/s00253-016-7686-2
Guan, B., and Zhang, X. (2020). Aptamers as versatile ligands for biomedical and pharmaceutical applications. Int. J. Nanomedicine 15, 1059–1071. doi:10.2147/IJN.S237544
Han, H., Wang, Z., Teng, D., Mao, R., Hao, Y., et al. (2020). Improved stability and activity of a marine peptide-N6NH2 against edwardsiella tarda and its preliminary application in fish. Mar. Drugs 18, 650. doi:10.3390/md18120650
Hartman, B., and Tomasz, A. (1981). Altered penicillin-binding proteins in methicillin-resistant strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 19, 726–735. doi:10.1128/AAC.19.5.726
Hatfull, G. F. (2023). Phage therapy for nontuberculous mycobacteria: challenges and opportunities. Pulm. Ther. 9, 91–107. doi:10.1007/s41030-022-00210-y
Hatoum-Aslan, A. (2018). Phage genetic engineering using CRISPR(-)Cas systems. Viruses 10, 335. doi:10.3390/v10060335
Hibstu, Z., Belew, H., Akelew, Y., and Mengist, H. M. (2022). Phage therapy: a different approach to fight bacterial infections. Biologics 16, 173–186. doi:10.2147/BTT.S381237
Hollenbeck, B. L., and Rice, L. B. (2012). Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 3, 421–433. doi:10.4161/viru.21282
Hosnedlova, B., Kabanov, D., Kepinska, M., B Narayanan, V. H., Parikesit, A. A., Fernandez, C., et al. (2022). Effect of biosynthesized silver nanoparticles on bacterial biofilm changes in S. aureus and E. coli. Nanomater. (Basel) 12, 2183. doi:10.3390/nano12132183
Howard, F. M., Flynn, D. M., Bradley, J. M., Noone, P., and Szawatkowski, M. (1977). Outbreak of necrotising enterocolitis caused by Clostridium butyricum. Lancet 2, 1099–1102. doi:10.1016/s0140-6736(77)90546-3
Huang, R., Ning, H., Shen, M., Li, J., Zhang, J., and Chen, X. (2017). Probiotics for the treatment of atopic dermatitis in children: a systematic review and meta-analysis of randomized controlled trials. Front. Cell. Infect. Microbiol. 7, 392. doi:10.3389/fcimb.2017.00392
Hussain, Y., Alam, W., Ullah, H., Dacrema, M., Daglia, M., Khan, H., et al. (2022). Antimicrobial potential of curcumin: therapeutic potential and challenges to clinical applications. Antibiot. (Basel) 11, 322. doi:10.3390/antibiotics11030322
Imlay, J. A., Chin, S. M., and Linn, S. (1988). Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240, 640–642. doi:10.1126/science.2834821
Iredell, J., Brown, J., and Tagg, K. (2016). Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ 352, h6420. doi:10.1136/bmj.h6420
Jadimurthy, R., Jagadish, S., Nayak, S. C., Kumar, S., Mohan, C. D., and Rangappa, K. S. (2023). Phytochemicals as invaluable sources of potent antimicrobial agents to combat antibiotic resistance. Life 13, 948. doi:10.3390/life13040948
Jahangiri, A., Neshani, A., Mirhosseini, S. A., Ghazvini, K., Zare, H., and Sedighian, H. (2021). Synergistic effect of two antimicrobial peptides, Nisin and P10 with conventional antibiotics against extensively drug-resistant Acinetobacter baumannii and colistin-resistant Pseudomonas aeruginosa isolates. Microb. Pathog. 150, 104700. doi:10.1016/j.micpath.2020.104700
Jain, S., Yadav, H., and Sinha, P. R. (2008). Stimulation of innate immunity by oral administration of dahi containing probiotic Lactobacillus casei in mice. J. Med. Food 11, 652–656. doi:10.1089/jmf.2006.0132
Jang, S. (2023). AcrAB-TolC, a major efflux pump in Gram negative bacteria: toward understanding its operation mechanism. BMB Rep. 56, 326–334. doi:10.5483/BMBRep.2023-0070
Jangra, M., Raka, V., and Nandanwar, H. (2020). In vitro evaluation of antimicrobial peptide tridecaptin M in combination with other antibiotics against multidrug resistant acinetobacter baumannii. Molecules 25, 3255. doi:10.3390/molecules25143255
Jennings, J. A., Courtney, H. S., and Haggard, W. O. (2012). Cis-2-decenoic acid inhibits S. aureus growth and biofilm in vitro: a pilot study. Clin. Orthop. Relat. Res. 470, 2663–2670. doi:10.1007/s11999-012-2388-2
Kapoor, G., Saigal, S., and Elongavan, A. (2017). Action and resistance mechanisms of antibiotics: a guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 33, 300–305. doi:10.4103/joacp.JOACP_349_15
Kaufmann, S. H., and Flesch, I. E. (1990). Cytokines in antibacterial resistance: possible applications for immunomodulation. Lung 168 (Suppl. l), 1025–1032. doi:10.1007/BF02718240
Kolodkin-Gal, I., Romero, D., Cao, S., Clardy, J., Kolter, R., and Losick, R. (2010). D-amino acids trigger biofilm disassembly. Science 328, 627–629. doi:10.1126/science.1188628
Kosciuczuk, E. M., Lisowski, P., Jarczak, J., Strzałkowska, N., Jóźwik, A., Horbańczuk, J., et al. (2012). Cathelicidins: family of antimicrobial peptides. A review. Mol. Biol. Rep. 39, 10957–10970. doi:10.1007/s11033-012-1997-x
Krawczyk-Balska, A., and Markiewicz, Z. (2016). The intrinsic cephalosporin resistome of Listeria monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics. J. Appl. Microbiol. 120, 251–265. doi:10.1111/jam.12989
Kudirkiene, E., Andoh, L. A., Ahmed, S., Herrero-Fresno, A., Dalsgaard, A., Obiri-Danso, K., et al. (2018). The use of a combined bioinformatics approach to locate antibiotic resistance genes on plasmids from whole genome sequences of Salmonella enterica serovars from humans in Ghana. Front. Microbiol. 9, 1010. doi:10.3389/fmicb.2018.01010
Kumar, S., Khan, M. Z., Khandelwal, N., Chongtham, C., Singha, B., Dabla, A., et al. (2022). Mycobacterium tuberculosis transcription factor EmbR regulates the expression of key virulence factors that aid in ex vivo and in vivo survival. mBio 13, e0383621. doi:10.1128/mbio.03836-21
Kushneet Kaur Sodhi, C. K. S. (2022). Recent development in the sustainable remediation of antibiotics: a review. Total Environ. Res. Themes, 3–4. doi:10.1016/j.totert.2022.100008
Kyriakidis, I., Vasileiou, E., Pana, Z. D., and Tragiannidis, A. (2021). Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens 10, 373. doi:10.3390/pathogens10030373
Lahtinen, S. J., Jalonen, L., Ouwehand, A. C., and Salminen, S. J. (2007). Specific Bifidobacterium strains isolated from elderly subjects inhibit growth of Staphylococcus aureus. Int. J. Food Microbiol. 117, 125–128. doi:10.1016/j.ijfoodmicro.2007.02.023
Laverty, G., Gorman, S. P., and Gilmore, B. F. (2011). The potential of antimicrobial peptides as biocides. Int. J. Mol. Sci. 12, 6566–6596. doi:10.3390/ijms12106566
Lazzaro, B. P., Zasloff, M., and Rolff, J. (2020). Antimicrobial peptides: application informed by evolution. Science 368, eaau5480. doi:10.1126/science.aau5480
Lee, D. H., Kim, B. S., and Kang, S. S. (2020). Bacteriocin of pediococcus acidilactici HW01 inhibits biofilm formation and virulence factor production by Pseudomonas aeruginosa. Probiotics Antimicrob. Proteins 12, 73–81. doi:10.1007/s12602-019-09623-9
Lee, E. H., and Shafer, W. M. (1999). The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33, 839–845. doi:10.1046/j.1365-2958.1999.01530.x
Leeansyah, E., Boulouis, C., Kwa, A. L. H., and Sandberg, J. K. (2021). Emerging role for MAIT cells in control of antimicrobial resistance. Trends Microbiol. 29, 504–516. doi:10.1016/j.tim.2020.11.008
Lei, J., Sun, L., Huang, S., Zhu, C., He, J., et al. (2019). The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 11, 3919–3931.
Lerminiaux, N. A., and Cameron, A. D. S. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65, 34–44. doi:10.1139/cjm-2018-0275
Li, M. C., Lu, J., Lu, Y., Xiao, T. Y., Liu, H. C., Lin, S. Q., et al. (2021). rpoB mutations and effects on rifampin resistance in Mycobacterium tuberculosis. Infect. Drug Resist 14, 4119–4128. doi:10.2147/IDR.S333433
Li, X., Korner, H., and Liu, X. (2020). Susceptibility to intracellular infections: contributions of TNF to immune defense. Front. Microbiol. 11, 1643. doi:10.3389/fmicb.2020.01643
Li, X. Z., Plesiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi:10.1128/CMR.00117-14
Lise, M., Mayer, I., and Silveira, M. (2018). Use of probiotics in atopic dermatitis. Rev. Assoc. Med. Bras. 64, 997–1001. doi:10.1590/1806-9282.64.11.997
Lister, J. L., and Horswill, A. R. (2014). Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 4, 178. doi:10.3389/fcimb.2014.00178
Liu, M. M., Li, S. T., Shu, Y., and Zhan, H. Q. (2017). Probiotics for prevention of radiation-induced diarrhea: a meta-analysis of randomized controlled trials. PLoS One 12, e0178870. doi:10.1371/journal.pone.0178870
Macomber, L., and Imlay, J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106, 8344–8349. doi:10.1073/pnas.0812808106
Marinelli, L. J., Piuri, M., Swigonová, Z., Balachandran, A., Oldfield, L. M., van Kessel, J. C., et al. (2008). BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS One 3, e3957. doi:10.1371/journal.pone.0003957
Markowiak, P., and Slizewska, K. (2017). Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 9, 1021. doi:10.3390/nu9091021
Masaoka, Y., Ueno, Y., Morita, Y., Kuroda, T., Mizushima, T., and Tsuchiya, T. (2000). A two-component multidrug efflux pump, EbrAB, in Bacillus subtilis. J. Bacteriol. 182, 2307–2310. doi:10.1128/JB.182.8.2307-2310.2000
Mazziotta, C., Tognon, M., Martini, F., Torreggiani, E., and Rotondo, J. C. (2023). Probiotics mechanism of action on immune cells and beneficial effects on human health. Cells 12, 184. doi:10.3390/cells12010184
Mba, I. E., and Nweze, E. I. (2022). Antimicrobial peptides therapy: an emerging alternative for treating drug-resistant bacteria. Yale J. Biol. Med. 95, 445–463.
McCubbin, K. D., Anholt, R. M., de Jong, E., Ida, J. A., Nóbrega, D. B., Kastelic, J. P., et al. (2021). Knowledge gaps in the understanding of antimicrobial resistance in Canada. Front. Public Health 9, 726484. doi:10.3389/fpubh.2021.726484
Meermeier, E. W., Zheng, C. L., Tran, J. G., Soma, S., Worley, A. H., Weiss, D. I., et al. (2022). Human lung-resident mucosal-associated invariant T cells are abundant, express antimicrobial proteins, and are cytokine responsive. Commun. Biol. 5, 942. doi:10.1038/s42003-022-03823-w
Midgley, J. E., and Smith, R. J. (1973). The effect of trimethoprim on macromolecular synthesis in Escherichia coli. Ribosome maturation in RCstr and RCrel strains. Biochem. J. 136, 235–247. doi:10.1042/bj1360235
Morroni, G., Bressan, R., Fioriti, S., D'Achille, G., Mingoia, M., Cirioni, O., et al. (2021). Antimicrobial activity of aztreonam in combination with old and new β-lactamase inhibitors against MBL and ESBL Co-producing gram-negative clinical isolates: possible options for the treatment of complicated infections. Antibiot. (Basel) 10, 1341. doi:10.3390/antibiotics10111341
Motley, M. P., Banerjee, K., and Fries, B. C. (2019). Monoclonal antibody-based therapies for bacterial infections. Curr. Opin. Infect. Dis. 32, 210–216. doi:10.1097/QCO.0000000000000539
Mouneimne, H., Robert, J., Jarlier, V., and Cambau, E. (1999). Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43, 62–66. doi:10.1128/AAC.43.1.62
Mrazek, K., Bereswill, S., and Heimesaat, M. M. (2019). Fecal microbiota transplantation decreases intestinal loads of multi-drug resistant Pseudomonas aeruginosa in murine carriers. Eur. J. Microbiol. Immunol. (Bp) 9, 14–22. doi:10.1556/1886.2019.00002
Munita, J. M., and Arias, C. A. (2016). Mechanisms of antibiotic resistance. Microbiol. Spectr. 4. doi:10.1128/microbiolspec.VMBF-0016-2015
Nainu, F., Masyita, A., Bahar, M. A., Raihan, M., Prova, S. R., Mitra, S., et al. (2021). Pharmaceutical prospects of bee products: special focus on anticancer, antibacterial, antiviral, and antiparasitic properties. Antibiot. (Basel) 10, 822. doi:10.3390/antibiotics10070822
Neal-McKinney, J. M., Lu, X., Duong, T., Larson, C. L., Call, D. R., Shah, D. H., et al. (2012). Production of organic acids by probiotic lactobacilli can be used to reduce pathogen load in poultry. PLoS One 7, e43928. doi:10.1371/journal.pone.0043928
Nguyen, H. T. T., Nguyen, T. H., and Otto, M. (2020). The staphylococcal exopolysaccharide PIA - biosynthesis and role in biofilm formation, colonization, and infection. Comput. Struct. Biotechnol. J. 18, 3324–3334. doi:10.1016/j.csbj.2020.10.027
Ning, Y., Cheng, L., Ling, M., Feng, X., Chen, L., Wu, M., et al. (2015). Efficient suppression of biofilm formation by a nucleic acid aptamer. Pathog. Dis. 73, ftv034. doi:10.1093/femspd/ftv034
Okano, K. (1974). An autopsy case of epidemic hemorrhagic fever in Osaka and its epidemiological surveys (author's transl). Rinsho Byori 22, 379–383.
Oliveira-Tintino, C. D. M., Tintino, S. R., Justino de Araújo, A. C., Dos Santos Barbosa, C. R., Ramos Freitas, P., Araújo Neto, J. B. d., et al. (2023). Efflux pump (QacA, QacB, and QacC) and beta-lactamase inhibitors? An evaluation of 1,8-naphthyridines against Staphylococcus aureus strains. Molecules 28, 1819. doi:10.3390/molecules28041819
Pallee Shree, C. K. S., Kaur Sodhi, K., Surya, J. N., and Biofilms, D. K. S. (2022). Understanding the structure and contribution towards bacterial resistance in antibiotics. Med. Microecology 16. doi:10.1016/j.medmic.2023.100084
Pan, Y. J., Lin, T. L., Chen, C. C., Tsai, Y. T., Cheng, Y. H., Chen, Y. Y., et al. (2017). Klebsiella phage φk64-1 encodes multiple depolymerases for multiple host capsular types. J. Virol. 91, e02457. doi:10.1128/JVI.02457-16
Pang, Z., Raudonis, R., Glick, B. R., Lin, T. J., and Cheng, Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192. doi:10.1016/j.biotechadv.2018.11.013
Park, S. C., Park, Y., and Hahm, K. S. (2011). The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int. J. Mol. Sci. 12, 5971–5992. doi:10.3390/ijms12095971
Parmeciano Di Noto, G., Molina, M. C., and Quiroga, C. (2019). Insights into non-coding RNAs as novel antimicrobial drugs. Front. Genet. 10, 57. doi:10.3389/fgene.2019.00057
Pasqua, M., Bonaccorsi di Patti, M. C., Fanelli, G., Utsumi, R., Eguchi, Y., Trirocco, R., et al. (2021). Host - bacterial pathogen communication: the wily role of the multidrug efflux pumps of the MFS family. Front. Mol. Biosci. 8, 723274. doi:10.3389/fmolb.2021.723274
Paul, M., Dishon-Benattar, Y., Dickstein, Y., and Yahav, D. (2023). Optimizing patient recruitment into clinical trials of antimicrobial-resistant pathogens. JAC Antimicrob. Resist 5, dlad005. doi:10.1093/jacamr/dlad005
Peck, M., Rothenberg, M. E., Deng, R., Lewin-Koh, N., She, G., Kamath, A. V., et al. (2019). A phase 1, randomized, single-ascending-dose study to investigate the safety, tolerability, and pharmacokinetics of DSTA4637S, an anti-Staphylococcus aureus thiomab antibody-antibiotic conjugate, in healthy volunteers. Antimicrob. Agents Chemother. 63, e02588. doi:10.1128/AAC.02588-18
Perewari, D. O., Otokunefor, K., and Agbagwa, O. E. (2022). Tetracycline-resistant genes in Escherichia coli from clinical and nonclinical sources in rivers state, Nigeria. Int. J. Microbiol. 2022, 9192424. doi:10.1155/2022/9192424
Peterson, E., and Kaur, P. (2018). Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 9, 2928. doi:10.3389/fmicb.2018.02928
Petrariu, O. A., Barbu, I. C., Niculescu, A. G., Constantin, M., Grigore, G. A., Cristian, R. E., et al. (2023). Role of probiotics in managing various human diseases, from oral pathology to cancer and gastrointestinal diseases. Front. Microbiol. 14, 1296447. doi:10.3389/fmicb.2023.1296447
Pincus, N. B., Reckhow, J. D., Saleem, D., Jammeh, M. L., Datta, S. K., and Myles, I. A. (2015). Strain specific phage treatment for Staphylococcus aureus infection is influenced by host immunity and site of infection. PLoS One 10, e0124280. doi:10.1371/journal.pone.0124280
Pinto, R. M., Soares, F. A., Reis, S., Nunes, C., and Van Dijck, P. (2020). Innovative strategies toward the disassembly of the EPS matrix in bacterial biofilms. Front. Microbiol. 11, 952. doi:10.3389/fmicb.2020.00952
Pires, D. P., Cleto, S., Sillankorva, S., Azeredo, J., and Lu, T. K. (2016). Genetically engineered phages: a review of advances over the last decade. Microbiol. Mol. Biol. Rev. 80, 523–543. doi:10.1128/MMBR.00069-15
Plaza-Diaz, J., Ruiz-Ojeda, F. J., Vilchez-Padial, L. M., and Gil, A. (2017). Evidence of the anti-inflammatory effects of probiotics and synbiotics in intestinal chronic diseases. Nutrients 9, 555. doi:10.3390/nu9060555
Poole, K. (2000). Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44, 2233–2241. doi:10.1128/AAC.44.9.2233-2241.2000
Portelinha, J., and Angeles-Boza, A. M. (2021). The antimicrobial peptide gad-1 clears Pseudomonas aeruginosa biofilms under cystic fibrosis conditions. Chembiochem 22, 1646–1655. doi:10.1002/cbic.202000816
Prestinaci, F., Pezzotti, P., and Pantosti, A. (2015). Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob. Health 109, 309–318. doi:10.1179/2047773215Y.0000000030
Raneri, M., Pinatel, E., Peano, C., Rampioni, G., Leoni, L., Bianconi, I., et al. (2018). Pseudomonas aeruginosa mutants defective in glucose uptake have pleiotropic phenotype and altered virulence in non-mammal infection models. Sci. Rep. 8, 16912. doi:10.1038/s41598-018-35087-y
Rather, I. A., Bajpai, V. K., Kumar, S., Lim, J., Paek, W. K., and Park, Y. H. (2016). Probiotics and atopic dermatitis: an overview. Front. Microbiol. 7, 507. doi:10.3389/fmicb.2016.00507
Razquin-Olazaran, I., Shahrour, H., and Martinez-de-Tejada, G. (2020). A synthetic peptide sensitizes multi-drug resistant Pseudomonas aeruginosa to antibiotics for more than two hours and permeabilizes its envelope for twenty hours. J. Biomed. Sci. 27, 85. doi:10.1186/s12929-020-00678-3
Reygaert, W. C. (2018). An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 4, 482–501. doi:10.3934/microbiol.2018.3.482
Ribeiro, S. M., de la Fuente-Núñez, C., Baquir, B., Faria-Junior, C., Franco, O. L., and Hancock, R. E. W. (2015). Antibiofilm peptides increase the susceptibility of carbapenemase-producing Klebsiella pneumoniae clinical isolates to β-lactam antibiotics. Antimicrob. Agents Chemother. 59, 3906–3912. doi:10.1128/AAC.00092-15
Ribeiro da Cunha, B., Fonseca, L. P., and Calado, C. R. C. (2019). Antibiotic discovery: where have we come from, where do we go? Antibiot. (Basel) 8, 45. doi:10.3390/antibiotics8020045
Robinson, T. P., Bu, D. P., Carrique-Mas, J., Fèvre, E. M., Gilbert, M., Grace, D., et al. (2016). Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 110, 377–380. doi:10.1093/trstmh/trw048
Rossino, G., Marchese, E., Galli, G., Verde, F., Finizio, M., Serra, M., et al. (2023). Peptides as therapeutic agents: challenges and opportunities in the green transition era. Molecules 28, 7165. doi:10.3390/molecules28207165
Ruiz-Ripa, L., Feßler, A. T., Hanke, D., Eichhorn, I., Azcona-Gutiérrez, J. M., Pérez-Moreno, M. O., et al. (2020). Mechanisms of linezolid resistance among enterococci of clinical origin in Spain-detection of optrA- and cfr(D)-Carrying E. faecalis. Microorganisms 8, 1155. doi:10.3390/microorganisms8081155
Saarela, M. H. (2019). Safety aspects of next generation probiotics. Curr. Opin. Food Sci. 30, 8–13. doi:10.1016/j.cofs.2018.09.001
Saberi, F., Kamali, M., Najafi, A., Yazdanparast, A., and Moghaddam, M. M. (2016). Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications. Cell. Mol. Biol. Lett. 21 (6), 6. doi:10.1186/s11658-016-0007-z
Sabnis, A., Hagart, K. L., Klöckner, A., Becce, M., Evans, L. E., Furniss, R. C. D., et al. (2021). Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife 10, e65836. doi:10.7554/eLife.65836
Sarkis, G. J., Jacobs, W. R., and Hatfull, G. F. (1995). L5 luciferase reporter mycobacteriophages: a sensitive tool for the detection and assay of live mycobacteria. Mol. Microbiol. 15, 1055–1067. doi:10.1111/j.1365-2958.1995.tb02281.x
Sharma, A., Sharma, R., Bhattacharyya, T., Bhando, T., and Pathania, R. (2017). Fosfomycin resistance in Acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter-AbaF. J. Antimicrob. Chemother. 72, 68–74. doi:10.1093/jac/dkw382
Shin, H., Lee, J. H., Yoon, H., Kang, D. H., and Ryu, S. (2014). Genomic investigation of lysogen formation and host lysis systems of the Salmonella temperate bacteriophage SPN9CC. Appl. Environ. Microbiol. 80, 374–384. doi:10.1128/AEM.02279-13
Short, F. L., Liu, Q., Shah, B., Clift, H. E., Naidu, V., Li, L., et al. (2021). The Acinetobacter baumannii disinfectant resistance protein, AmvA, is a spermidine and spermine efflux pump. Commun. Biol. 4, 1114. doi:10.1038/s42003-021-02629-6
Shum, K. T., Lui, E. L. H., Wong, S. C. K., Yeung, P., Sam, L., Wang, Y., et al. (2011). Aptamer-mediated inhibition of Mycobacterium tuberculosis polyphosphate kinase 2. Biochemistry 50, 3261–3271. doi:10.1021/bi2001455
Sikorska, H., and Smoragiewicz, W. (2013). Role of probiotics in the prevention and treatment of meticillin-resistant Staphylococcus aureus infections. Int. J. Antimicrob. Agents 42, 475–481. doi:10.1016/j.ijantimicag.2013.08.003
Singh, C. K., and Sodhi, K. K. (2023). Antimicrobial resistance in the time of COVID-19. Appl. Microbiol. 3, 1388–1391. doi:10.3390/applmicrobiol3040093
Singh, C. K., Sodhi, K. K., and Mubarak, M. S. (2023). Editorial: new drugs, approaches, and strategies to combat antimicrobial resistance. Front. Pharmacol. 14, 1295623. doi:10.3389/fphar.2023.1295623
Singha, B., Singh, R. B., Venkataraman, R., Nair, T., Rosenn, E. H., and Soni, V. (2023a). Gut microbiome associated dysbiosis: limited regimens and expanding horizons of phage therapy. Aspects Mol. Med. 2, 100029.
Singha, B., Behera, D., Khan, M. Z., Singh, N. K., Sowpati, D. T., Gopal, B., et al. (2023b). The unique N-terminal region of Mycobacterium tuberculosis sigma factor A plays a dominant role in the essential function of this protein. J. Biol. Chem. 299, 102933. doi:10.1016/j.jbc.2023.102933
Singha, B., Murmu, S., Nair, T., Rawat, R. S., Sharma, A. K., and Soni, V. (2024). Metabolic rewiring of Mycobacterium tuberculosis upon drug treatment and antibiotics resistance. Metabolites 14, 63. doi:10.3390/metabo14010063
Sodhi, K. K., Singh, C. K., Kumar, M., and Singh, D. K. (2023). Whole-genome sequencing of Alcaligenes sp. strain MMA: insight into the antibiotic and heavy metal resistant genes. Front. Pharmacol. 14, 1144561. doi:10.3389/fphar.2023.1144561
Stalenhoef, J. E., Terveer, E. M., Knetsch, C. W., Van't Hof, P. J., Vlasveld, I. N., Keller, J. J., et al. (2017). Fecal microbiota transfer for multidrug-resistant gram-negatives: a clinical success combined with microbiological failure. Open Forum Infect. Dis. 4, ofx047. doi:10.1093/ofid/ofx047
Stephen, J., Lekshmi, M., Ammini, P., Kumar, S. H., and Varela, M. F. (2022). Membrane efflux pumps of pathogenic Vibrio species: role in antimicrobial resistance and virulence. Microorganisms 10, 382. doi:10.3390/microorganisms10020382
Sun, Z., Li, Q., Hou, R., Sun, H., Tang, Q., Wang, H., et al. (2019). Kaempferol-3-O-glucorhamnoside inhibits inflammatory responses via MAPK and NF-κB pathways in vitro and in vivo. Toxicol. Appl. Pharmacol. 364, 22–28. doi:10.1016/j.taap.2018.12.008
Takahashi, M., Taguchi, H., Yamaguchi, H., Osaki, T., Komatsu, A., and Kamiya, S. (2004). The effect of probiotic treatment with Clostridium butyricum on enterohemorrhagic Escherichia coli O157:H7 infection in mice. FEMS Immunol. Med. Microbiol. 41, 219–226. doi:10.1016/j.femsim.2004.03.010
Tao, S., Chen, H., Li, N., and Liang, W. (2022). The application of the CRISPR-cas system in antibiotic resistance. Infect. Drug Resist 15, 4155–4168. doi:10.2147/IDR.S370869
Tewes, F., Bahamondez-Canas, T. F., and Smyth, H. D. C. (2019). Efficacy of ciprofloxacin and its copper complex against Pseudomonas aeruginosa biofilms. AAPS PharmSciTech 20, 205. doi:10.1208/s12249-019-1417-9
Thiel, K. W., and Giangrande, P. H. (2009). Therapeutic applications of DNA and RNA aptamers. Oligonucleotides 19, 209–222. doi:10.1089/oli.2009.0199
Vestergaard, M., and Ingmer, H. (2019). Antibacterial and antifungal properties of resveratrol. Int. J. Antimicrob. Agents 53, 716–723. doi:10.1016/j.ijantimicag.2019.02.015
Wagner, T. M., Pontinen, A. K., Al Rubaye, M., Sundsfjord, A., and Hegstad, K. (2024). Adaptive cell wall thickening in Enterococcus faecalis is associated with decreased vancomycin susceptibility. Clin. Microbiol. Infect. 30, 396.e1–396.e5. doi:10.1016/j.cmi.2023.12.002
Wang, H., Chen, Y., Wang, L., Liu, Q., Yang, S., and Wang, C. (2023). Advancing herbal medicine: enhancing product quality and safety through robust quality control practices. Front. Pharmacol. 14, 1265178. doi:10.3389/fphar.2023.1265178
Watkins, R. R., and Bonomo, R. A. (2016). Overview: global and local impact of antibiotic resistance. Infect. Dis. Clin. North Am. 30, 313–322. doi:10.1016/j.idc.2016.02.001
Webber, M. A., and Piddock, L. J. (2003). The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 51, 9–11. doi:10.1093/jac/dkg050
Weisblum, B. (1995). Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39, 577–585. doi:10.1128/AAC.39.3.577
Winzig, F., Gandhi, S., Lee, A., Würstle, S., Stanley, G. L., Capuano, I., et al. (2022). Inhaled bacteriophage therapy for multi-drug resistant achromobacter. Yale J. Biol. Med. 95, 413–427.
Wollowski, I., Rechkemmer, G., and Pool-Zobel, B. L. (2001). Protective role of probiotics and prebiotics in colon cancer. Am. J. Clin. Nutr. 73, 451S-455S–455S. doi:10.1093/ajcn/73.2.451s
Wylie, M. R., Windham, I. H., Blum, F. C., Wu, H., and Merrell, D. S. (2022). In vitro antibacterial activity of nimbolide against Helicobacter pylori. J. Ethnopharmacol. 285, 114828. doi:10.1016/j.jep.2021.114828
Xu, K., Cai, H., Shen, Y., Ni, Q., Chen, Y., Hu, S., et al. (2020). Management of COVID-19: the zhejiang experience. Zhejiang Da Xue Xue Bao Yi Xue Ban. 49, 147–157. doi:10.3785/j.issn.1008-9292.2020.02.02
Yang, H., Zhang, Y., Yu, J., Huang, Y., Zhang, X. E., and Wei, H. (2014). Novel chimeric lysin with high-level antimicrobial activity against methicillin-resistant Staphylococcus aureus in vitro and in vivo. Antimicrob. Agents Chemother. 58, 536–542. doi:10.1128/AAC.01793-13
Yin, X., Heeney, D., Srisengfa, Y., Golomb, B., Griffey, S., and Marco, M. (2018). Bacteriocin biosynthesis contributes to the anti-inflammatory capacities of probiotic Lactobacillus plantarum. Benef. Microbes 9, 333–344. doi:10.3920/BM2017.0096
Young, K., Jayasuriya, H., Ondeyka, J. G., Herath, K., Zhang, C., Kodali, S., et al. (2006). Discovery of FabH/FabF inhibitors from natural products. Antimicrob. Agents Chemother. 50, 519–526. doi:10.1128/AAC.50.2.519-526.2006
Yu, A. Q., and Li, L. (2016). The potential role of probiotics in cancer prevention and treatment. Nutr. Cancer 68, 535–544. doi:10.1080/01635581.2016.1158300
Zairi, A., Ferrières, L., Latour-Lambert, P., Beloin, C., Tangy, F., Ghigo, J. M., et al. (2014). In vitro activities of dermaseptins K4S4 and K4K20S4 against Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa planktonic growth and biofilm formation. Antimicrob. Agents Chemother. 58, 2221–2228. doi:10.1128/AAC.02142-13
Zarzecka, U., Zakrzewski, A. J., Chajecka-Wierzchowska, W., and Zadernowska, A. (2022). Linezolid-resistant Enterococcus spp. isolates from foods of animal origin-the genetic basis of acquired resistance. Foods 11, 975. doi:10.3390/foods11070975
Zhao, Q., Mutukumira, A., Lee, S. J., Maddox, I., and Shu, Q. (2012). Functional properties of free and encapsulated Lactobacillus reuteri DPC16 during and after passage through a simulated gastrointestinal tract. World J. Microbiol. Biotechnol. 28, 61–70. doi:10.1007/s11274-011-0792-5
Zhou, J., and Rossi, J. (2017). Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 16, 181–202. doi:10.1038/nrd.2016.199
Zhou, M., Wang, L., Wang, Z., Kudinha, T., Wang, Y., Xu, Y., et al. (2022). Molecular characterization of penicillin-binding Protein2x, 2b and 1a of Streptococcus pneumoniae causing invasive pneumococcal diseases in China: a multicenter study. Front. Microbiol. 13, 838790. doi:10.3389/fmicb.2022.838790
Keywords: antimicrobial resistance (AMR), alternative therapies, biofilm disruption, RNA-based treatments aptamers, peptide-based therapies, phage therapy, probiotics, drug biotransformation
Citation: Singha B, Singh V and Soni V (2024) Alternative therapeutics to control antimicrobial resistance: a general perspective. Front. Drug Discov. 4:1385460. doi: 10.3389/fddsv.2024.1385460
Received: 12 February 2024; Accepted: 26 June 2024;
Published: 17 July 2024.
Edited by:
Tânia Martins Silva, Universidade do Porto, PortugalReviewed by:
Ramona Iseppi, University of Modena and Reggio Emilia, ItalyCopyright © 2024 Singha, Singh and Soni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vijay Soni, dmlzMjAzMkBtZWQuY29ybmVsbC5lZHU=; Vinayak Singh, dmluYXlhay5zaW5naEB1Y3QuYWMuemE=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.