- 1Special Centre for Molecular Medicine, Jawaharlal Nehru University, New Delhi, India
- 2Department of Pathology and Laboratory Medicine, University of Kansas Medical Center, Kansas City, KS, United States
More than sesquicentennial years of malarial research, however the unique malarial parasite, Plasmodium still bewilders us with its atypical characteristic features. Elimination strategies, deeper knowledge of the parasite biology and pathways can help combat this global health concern that affects ∼250 million people annually. In this review, we unveil an unusual phenomenon observed in the parasite proteome, N-terminal extensions in proteins and highlight that the proteases that may be involved in their processing events, are potential candidates to target this pathogen. Plasmodium encodes larger proteins as compared to its eukaryotic counterparts with homology regions present in the C-terminus of the protein. In contrast, the function of unusual extensions in the N-terminus remains mostly elusive. This novelty observed in Plasmodium proteins is collated here with a focus on replication proteins. The plausible functions and prevalence of these extensions, despite the reduction in genome size, through the parasite evolution are also mentioned. We hypothesize that these extensions, propagated via the energy consuming cellular processes in the otherwise host-dependent obligate parasite, are beneficial to the parasite in ways that are yet to be explored. Consequently, targeting the proteolytic processing of these proteins and the involved proteases would serve as a new drug development regimen to tackle the emerging resistance in parasites to existing antimalarials.
Introduction
The deadly parasitic Plasmodium infection malaria, affects approximately 250 million individuals annually worldwide. The WHO reported 619,000 deaths due to this global menace in 2021 (Organization, 2022). Umpteen strategies and stringent measures have been devised globally to eradicate this mosquito vector transmitted disease. Malaria-related symptoms include fever and chills; the severe form results in coma and system failure. Among the five protozoa species, P. falciparum and P. vivax are the culprits of maximum malarial deaths (Basu and Sahi, 2017). This fatal parasite has been prevalent since ancient history and is known to complete its complex life cycle in both mosquito (definitive host) and humans (secondary host) (Garcia, 2010; Lee et al., 2019). A deeper understanding of the parasite biology, rapid diagnostic tests, treatment approaches and prevention strategies have proved quite remarkable in tackling this disease. However, global malaria elimination and the ways to overcome the alarmingly rise in drug resistance have become exigent. Moreover, the inability of any malarial vaccine to induce a consistent immune response has raised concerns (Saha et al., 2016). Genomic data, genetic polymorphisms and profound knowledge of the parasitic pathways can reduce the global burden on public health. Potential drug target screening, along with the development of novel anti-malarials through meticulous research, can assist in achieving the global malaria eradication plan (Joshi, 2003; Garrido-Cardenas et al., 2019; Kumpornsin et al., 2019).
Apicomplexan parasites have diverse hosts and need to replicate their genomes within different niches, sometimes under complicated stress conditions. This uphill battle is accomplished with the help of dedicated replication machinery and gene regulators. The appropriate timing and regulation of replication, multiple nuclear division and cell division are the vital factors for these parasites to thrive in different environments. Technological and genomic advances have aided researchers to understand the complex genomic, transcriptional, proteomic and metabolic details of parasite. This information is helpful for the development of drug-targeted intervention approaches. Genomic analyses have revealed that these parasites have reduced genome size and lesser number of genes as compared to their ancestors. The crafty nature of the parasite is to hijack the host proteins and scavenge the host nutrients, thus making up for the lineage-specific losses. Plasmodium, a member of the apicomplexan parasite family is also dependent on human host for its survival and has evolved through endosymbiosis (Francia and Striepen, 2014; Deshmukh et al., 2016; Swapna and Parkinson, 2017).
Any functional protein can be divided into two segments: N and C terminus. The N-terminus carries critical information that determines the fate of the protein within the cell and also acts as intracellular postal codes. Here we review a novel insight depicting an unusual phenomenon in Plasmodium proteins taking replication proteins as our model. Plasmodium proteome exhibits unique N-terminal extensions in proteins as compared to their eukaryotic counterparts (Figure 1A) (Coppi et al., 2005; Gupta et al., 2008; Deshmukh et al., 2012; Miao et al., 2013; Espinosa et al., 2015; Osman et al., 2015; Seliverstov et al., 2015; Yusuf et al., 2015; Sharma et al., 2018; Robert-Paganin et al., 2019; Bhowmick et al., 2020). Phylogeny tree analysis revealed divergence among the Plasmodium proteins taken here as model (ORC1, ORC5 and GCN5), from S. cerevisiae and H. sapiens as indicated by branch lengths (Figure 1B). We anticipate that this novelty, despite reduced genome size, is a classical means adopted by the parasite as a regulation strategy to synthesize multi-functional proteins and carry specific information for protein localization (Figure 1C). Thus, the proteases responsible for processing of these proteins become important for their function that may lead to exploring possibilities to use them as potential drug targets. These proteases may have many substrates in the malaria parasite and, in turn, affect many essential pathways of the parasite biology. Among several substrates some could be proteins with unusual N-terminal extensions, serving as substrates to specific proteases and ultimately undergo proteolytic processing to deliver multifunctional proteins. The proteases involved in the processing can belong to either class of proteases: Serine, Threonine, Aspartic, Cysteine and Metalloproteases. Therefore, identifying and targeting the specific class of protease for such processing presents an attractive way of targeting this deadly parasite.
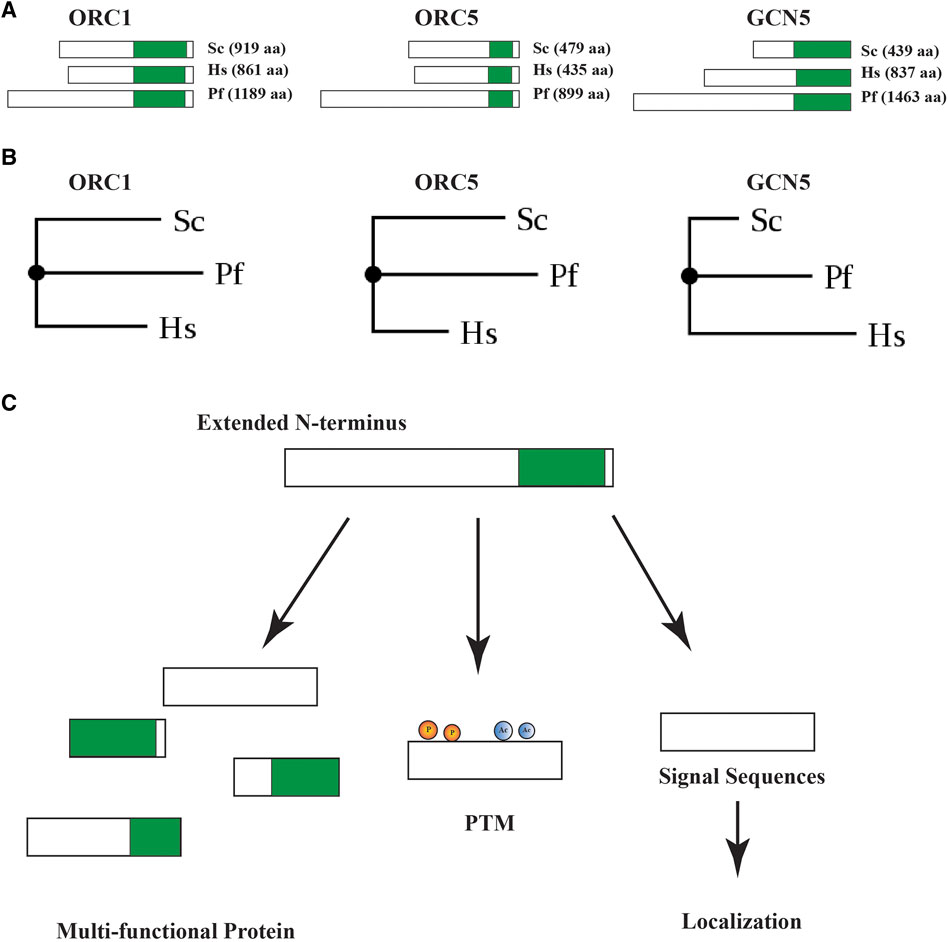
FIGURE 1. (A) Pictorial representation of Saccharomyces, Human and Plasmodium ORC1, ORC5 and GCN5 proteins depicting unusually long N-terminal extensions in Plasmodium proteins. The conserved regions in proteins with respect to Plasmodium proteins are shown in green color. (B) Phylogeny tree depicting the divergence in Saccharomyces, Human and Plasmodium ORC1, ORC5 and GCN5. PhyML 3.0 (Guindon et al., 2010) was used to generate the tree by using the alignment file in PHYLIP format (Dereeper et al., 2008). (C) Diagrammatic representation of multiple effects of the unusual N-terminal extensions in Plasmodium proteins.
N-terminal extension idiosyncrasy through the evolutionary process
Proteins with long N-terminus extensions (NTEs) that are associated with DNA replication are not ubiquitous, but few are present in different species. In eukaryotes, subunits of Minichromosome Maintenance protein helicase complex; MCM2, MCM4 and MCM6 possess large disordered NTEs. Whereas, MCMs in archaea lack these NTEs (Riera et al., 2017). Additionally, the mammalian Cdt1 comprises a long extension at N-terminus, that is, advantageous to the protein structure. The N-terminus of Cdt1 binds to MCM2, whereas MCM4 and MCM6 bind at the C-terminus. A long loop at the N-terminus attaches it to the C-terminus of Cdt1, making the complex of MCM-Cdt1 intact (Wei et al., 2010; Liu et al., 2012). Moreover, the N-terminal extension distinguishes human mitochondrial single-stranded DNA binding protein from its bacterial counterparts (Yang et al., 1997; Oliveira and Kaguni, 2010). Recent findings highlight the presence of long NTEs in coronaviruses. SARS-CoV Nsp12 RNA-dependent RNA polymerase contains a unique N-terminus extension. The NTE has a nidovirus RdRp-associated nucleotidyl transferase (NiRAN) and a kinase-like domain that assists in its binding to co-factors nsp7 and nsp8 (Kirchdoerfer and Ward, 2019). Long NTEs are observed in SARS-CoV-2, the causative agent of the pandemic. Long extensions of nsp8a and nsp8b provide binding sites for an array of replication factors (Chen et al., 2020).
In prokaryotes, the majority of the repeat containing proteins (RCPs) are uncharacterized. Those characterized function as enzymes, transport proteins, structural proteins, and in transcription/translation. Contrastingly, in eukaryotes (Human, Drosophila and C. elegans) irrespective of the type of repeat, RCPs are mostly associated with DNA, RNA or chromatin (Faux et al., 2005). RCPs are located at the C-terminus as well as the middle region of the protein but, they are preferentially positioned at the N-terminus (Alba and Guigo, 2004). Plasmodium falciparum genome is unique as many of the proteins encoded are oddly rich in repetitive and low complexity sequences. Approximately one in every 467 nucleotides, repetitive sequences occur in P. falciparum (Tan et al., 2010). During the asexual blood-stage, the insertion and deletion of these repeats are highly dynamic (Hamilton et al., 2017). Gene sequences having these repeats are variable among different Plasmodium strains (Moser et al., 2020).
Many of the Plasmodium proteins with repeats are intrinsically disordered (Huntley and Golding, 2000; Feng et al., 2006). Intrinsically disordered proteins are involved in cellular processes and due to their flexible nature, they function as “molecular springs” (Dunker et al., 2001). As explained above, since these repeats are disordered in most cases, they have no functional importance. But with an evolutionary point of view, they play a very different role. These repeats contract and expand via strand slippage and unequal crossing over events during DNA replication and meiosis, adds to their ability to create more variation among strains (Gemayel et al., 2010). This expansion or contraction leads to significant changes in the activity of a protein or in the formation of a protein with novel function (Davies et al., 2017). Plasmodium genome has a high frequency of low complexity regions (LCRs) that evolve rapidly (Pizzi and Frontali, 2001; Zilversmit et al., 2010). Further, single amino acid repeats are abundantly found in the genome. 25% of the proteome contains asparagine repeats (Muralidharan et al., 2011). Like poly-Gln repeat, poly-Asn repeats also tend to form aggregates at higher temperatures (Halfmann et al., 2011). Considering the fact that protein aggregation is toxic to the cell, the presence of such a large number of protein aggregates gives an evolutionary disadvantage to Plasmodium. However, the Plasmodium chaperones like Hsp110c protects the organism from such life-threatening situation (Muralidharan et al., 2012).
N-terminal extensions in Plasmodium and functional enigma
Proteins in P. falciparum parasite are longer in size, on average, as compared to other species. Plasmodium proteome analysis reveals that the homologous sequences within the proteins (when compared with similar proteins from other systems) reside exclusively in the C-terminus of the protein and the N-terminus has non-homologous, unique extensions which may/may not have a specific function to be performed. The critical role played by these unusual extensions remains elusive and needs to be further characterized to understand the parasite pathways and biology in a profound manner. Furthermore, the long N-terminus extensions mostly consist of repetitive sequences such as poly-asparagine tracts, found in approximately one-fourth of the P. falciparum proteins (Davies et al., 2017). These low-complexity regions help the parasite to escape the host immune complex and enable protein-protein interactions. It was assumed that these repeat regions do not confer any advantage/disadvantage to the parasites. However, recent evidences have shown that they are critical for parasite protein function and can cause changes in the protein folding, biochemistry or generate novel functional domains (Davies et al., 2017). Moreover, these repeats are highly dynamic and are known to have expanded evolutionary.
The manifestation of the clinical symptoms of malaria occur due to the continuous multiplication of the parasite in blood (Chang et al., 2008). Due to presence of the parasite, the host cell undergoes constant remodeling. This coup is achieved by Plasmodium through defenestration of numerous proteins from the parasite into the erythrocyte. Most of the proteins, including the integral membrane proteins, are fused with signal sequences to facilitate their entry into the ER membrane. Apart from the signal sequence, there is an additional downstream sequence, the vacuolar targeting signal (PEXEL). This signal aids in the trafficking of the protein from the parasitophorous vacuole to the host cell. It is located 20–30 amino acids downstream of the signal sequence. Exceptions to this process are also present within the parasite as there are proteins that lack the PEXEL sequence but are exported to the host cell. The PEXEL sequences are recognized by an integral membrane associated aspartic protease plasmepsin V. Chaperone HSP101 and PTEX150 aid in the translocation of the cleaved proteins across the parasitophorous vacuole membrane (PVM) (Beck et al., 2014; Gabriela et al., 2022). Phosphatidylinositol-3-phosphate (PI(3)P) binding in ER by the Host-targeting signal, prior to and independent of plasmepsin V action, facilitates protein export. The cleavage by protease plasmepsin V may be facilitated by PI(3)P binding (Bhattacharjee et al., 2012).
Many of the Plasmodium proteins display a unique property of possessing an unusually long N-terminus. FIKK8, a member of the protein kinases FIKK subfamily, is reported to contain N-terminal extension and conserved kinase domains within the C-terminus. The N-terminal extension is an integral component of the FIKK architecture. It consists of auto-phosphorylation components that may be crucial for regulation purposes, but the functional significance is unknown (Osman et al., 2015). Another example of a protein with essential N-terminal extensions is CDP-DAG synthase. The full length 78 kDa protein is cleaved into a 28 kDa unusually long asparagine-rich N-terminal extension and a 50 kDa fragment consisting of conserved C-terminal cytidylyltransferase domain. The N-terminus fragment is trafficked to the parasitophorous vacuole and is a peripheral membrane protein, however, the function remains unknown. Intriguingly, Myosin A tail-interacting protein (MTIP) and MyoA have unstructured “N-terminal extension” that consists of phosphorylation sites that regulate the output of the motor complex. The disordered N-terminal extension of MTIP is crucial for its interaction with GAP45, a component of the glideosome complex (Douse et al., 2012; Yusuf et al., 2015; Robert-Paganin et al., 2019). The conserved key acetyltransferase PfMYST, expressed in two isoforms in the parasite, consists of chromodomain, MYST acetyltransferase domain and zinc finger domain (Shekhar et al., 2022). Interestingly, N-terminal extensions are present in PfMYST, upstream of the chromodomain. The function of this extension is unknown, but it is reported to be heavily acetylated with multiple acetylation sites within this extended region (Miao et al., 2013).
The essential histone acetyltransferase, PfGCN5 regulates gene expression in the parasite. This chromatin-remodeling enzyme is 170 kDa long and the conserved bromodomain (BrD) and acetyltransferase domain (GNAT) reside within the C-terminus region. The N-terminus of this protein is unusually long and the functional significance of this extension is not yet known. The full-length protein is subjected to a novel proteolytic processing event via cysteine protease-like enzyme that results in the generation of multiple fragments of the protein or the mature peptides including ∼45 kDa C-terminal fragment containing the HAT domain. This proteolytic cleavage is crucial for the in vivo activity of PfGCN5 (Bhowmick et al., 2020).
Thus, N-terminal extensions have been reported in multiple proteins belonging to different pathways in the Plasmodium parasite. One can understand its necessity for the proteins exported out of the parasite and targeted to a particular organelle within the parasite that it accommodates the vacuolar targeting sequence and the signal sequence. However, unusually long N-terminus is found in numerous proteins that are not exported out of the parasite like the proteins involved in DNA replication. The presence of these unusual long N-terminal extensions in the machinery of this highly conserved process of replication is quite astonishing that may reflect the unique properties of this deadly parasite.
Prevalence of extensions in replication proteins
The fundamental cellular life process DNA replication is executed with the help of a closely monitored machinery. The parasite circulates between two hosts and replication occurs five times in the Plasmodium life cycle (Arnot and Gull, 1998; Mitra et al., 2012). The unusual features of schizogony, asynchronous replication, multiple rounds of replication, differences in the machinery components and dynamics distinguish the parasite from other eukaryotic species. In Plasmodium, the time taken for genome duplication and the resultant number of daughter cells varies significantly among the different rounds of replication. It could be less than 4 min during gametogenesis to approximately 17 h for sporogony (Stanojcic et al., 2017; Matthews et al., 2018). Interestingly, some of the reported inhibitors of DNA replication and the involved proteins are quite efficacious in killing drug-resistant parasites as well. Acriflavine targeting PfGyrase B is a potent therapeutic against both CQ-resistant and sensitive parasites (Dana et al., 2014). Further, CQ-resistant parasites exhibited more sensitivity towards the replication inhibitors, pyrrolobenzimidazole RWJ68198 and pyridinylimidazole RWJ67657 (Brumlik et al., 2011).
The replicative machinery components include ORC proteins, MCMs, PCNAs and DNA polymerases similar to eukaryotes. ORC1, the first subunit of the complex to be identified in P. falciparum in both sexual and asexual stages, binds to ARS like sequences (PfARS) (Agarwal et al., 2017). PfORC1 is homologous with its eukaryotic counterparts (yeast and human) and consists of conserved sequences of the Cdc6/Cdc18/ORC1 superfamily (Li and Cox, 2003; Mehra et al., 2005). PfORC1, the largest subunit of origin recognition complex (ORC), contains an extensively long N-terminus region with potential phosphorylation sites. This unusually long extension shows no sequence homology with any other ORC1 protein in eukaryotes, whereas the C-terminus of the protein is conserved. The N-terminus of yeast ORC1 protein is involved in the interaction with different sirtuins and plays a role in transcriptional silencing (Triolo and Sternglanz, 1996). BAH domain (bromo adjacent homology domain) is present in the N-terminus of human ORC1 and assists in chromatin binding (Noguchi et al., 2006). Interestingly, the extended region in the PfORC1 N-terminus has a significant number of charged amino acids, indicating its involvement in interaction with other proteins. PfORC1 is a nuclear protein as it contains two NLS sequences and a leucine zipper motif in its N-terminus. The N-terminus of PfORC1 localises at the nuclear periphery and has an affinity towards the telomeric regions (TARES) and var gene promoters (Deshmukh et al., 2012). In association with Sir2 (histone deacetylase), PfORC1 plays a role in the silencing of var genes by creating a heterochromatin like silencing zone. The study established a crucial role of the N-terminus region, comprising unusual extensions, in var gene regulation, that is, important for the parasite virulence and pathogenicity. Thus, the single protein behaves as a multi-functional protein in which the C-terminus plays essential roles in DNA replication and the N-terminus containing unusual extension is associated with var gene regulation.
ORC5 has also been characterized and it exhibits colocalisation with ORC1 and PCNA, establishing its function as a replication protein (Gupta et al., 2008). PfORC5 also has homology with its eukaryotic counterparts at C-terminus only and is a critical component of the fundamental process of replication. Like other ORC members, PfORC5 also has an unusual N-terminal extension consisting of asparagine/aspartic acid/lysine-rich repetitive sequences (Gupta et al., 2008). The length of the conserved C-terminus is ∼440 amino acids which is comparable with the eukaryotic ORC5 proteins (∼450 residues). The full length PfORC5 protein is ∼899 residues long and therefore, the extra residues apart from the conserved C-terminus comprise the long N-terminal extension. The function of these extra residues constituting the unique N-terminal extension is not known yet and requires further detailed studies.
Another ORC subunit homologue ORC2, has been identified in P. falciparum. A novel unconventional protein trafficking pathway involving the ER association of this protein followed by translocation to the nucleus has been proposed (Sharma et al., 2018). PfORC2 has a NLS sequence in its long N-terminus, and it shows conserved residues in the C-terminus. The full length ORC2 protein undergoes processing and its N-terminal is cleaved by a SPP like peptidase in the ER. Post processing, a smaller C-terminal fragment travels to the nucleus via Golgi where it performs essential functions in DNA replication (Sharma et al., 2018). The remaining subunits of ORC are yet to be explored in P. falciparum.
Followed by the initiator proteins, recruitment of helicase ring completes the pre-RC formation. As in eukaryotes, P. falciparum also has 6 MCM subunits, MCM2-7 that comprise the signature MCM motif, WALKER-A domain and zinc-finger motif. MCM2, 6 and 7 have been characterized and the remaining subunits are not yet studied (Patterson et al., 2006). The transition of pre-RC to replication fork formation is critical to the replication process to continue. The required enzyme for the primer synthesis during the elongation step, primase, is annotated in the P. falciparum genome and has 15 introns and codes for a 53 kDa protein that has conserved motifs similar to eukaryotic primases. Moreover, PfPrimase has regions homologous to DNA and RNA polymerases and reverse transcriptases (Prasartkaew et al., 1996).
Eukaryotes have five polymerases, namely DNA polymerase alpha (α), DNA polymerase gamma (γ), DNA polymerase delta (δ), DNA polymerase beta (β) and DNA polymerase epsilon (ε). In P. falciparum, Pol α and Pol β from the crude extract and Pol γ from mitochondria have been characterized. Pol δ has also been studied in detail (Abu-Elheiga et al., 1990; Fox and Bzik, 1991; Ridley et al., 1991; Chavalitshewinkoon et al., 1993; Horrocks et al., 1996; Chavalitshewinkoon-Petmitr et al., 2000; Hubscher et al., 2000; Nunthawarasilp et al., 2007; Vasuvat et al., 2016). Remarkably, P. falciparum encodes two PCNAs, PCNA1 and PCNA2 that have been reported to play essential roles in increasing replication processivity and DNA damage repair response (Kilbey et al., 1993; Horrocks et al., 1996; Li et al., 2002; Patterson et al., 2002; Mitra et al., 2015; Banu et al., 2018; Pradhan et al., 2019). Further, the sliding clamp loader, RFC and the eukaryotic single-stranded DNA-binding protein, replication protein A (RPA), have been well characterized and studied in detail (Voss et al., 2002; Gangwar et al., 2009; Jirage et al., 2010). The homolog of topoisomerase II has also been identified in P. falciparum (Cheesman et al., 1994).
Replication proteins are the “sine quo non” of the Plasmodium life cycle. The role of long N-terminus in replication initiation proteins (ORC proteins) still remains elusive. It raises the question about its necessity as they do not require to be exported out of the cell. Further, we analyzed sequences of the different proteins involved in DNA replication of P. falciparum. The sequence analysis of the essential chromatin modifying enzyme, GCN5 known to regulate parasite gene expression, was also performed. The presence of long N-terminus and low complexity repeats were screened using bioinformatics tools such as ScanProsite and Delta Blast (Table 1). It was observed that apart from ORC proteins described above (PfORC1, PfORC2 and PfORC5), PfORC4 (putative), PfCdt1, RFC subunit1, PfRPA1, DNA polymerase α, DNA ligase I and GCN5 possess long N-terminus extensions. As mentioned earlier, PfORC1, PfORC2, PfORC5 and GCN5 have already been reported to comprise unusual N-terminus extensions, hence validate our findings using the bioinformatics tools. Among these proteins, excluding the asparagine repeats, ORC1 has serine residue repeats, ORC1 and ORC2 have lysine repeats. Nuclear localization signal is present in ORC1, ORC2, ORC5 and GCN5 (Table 1).
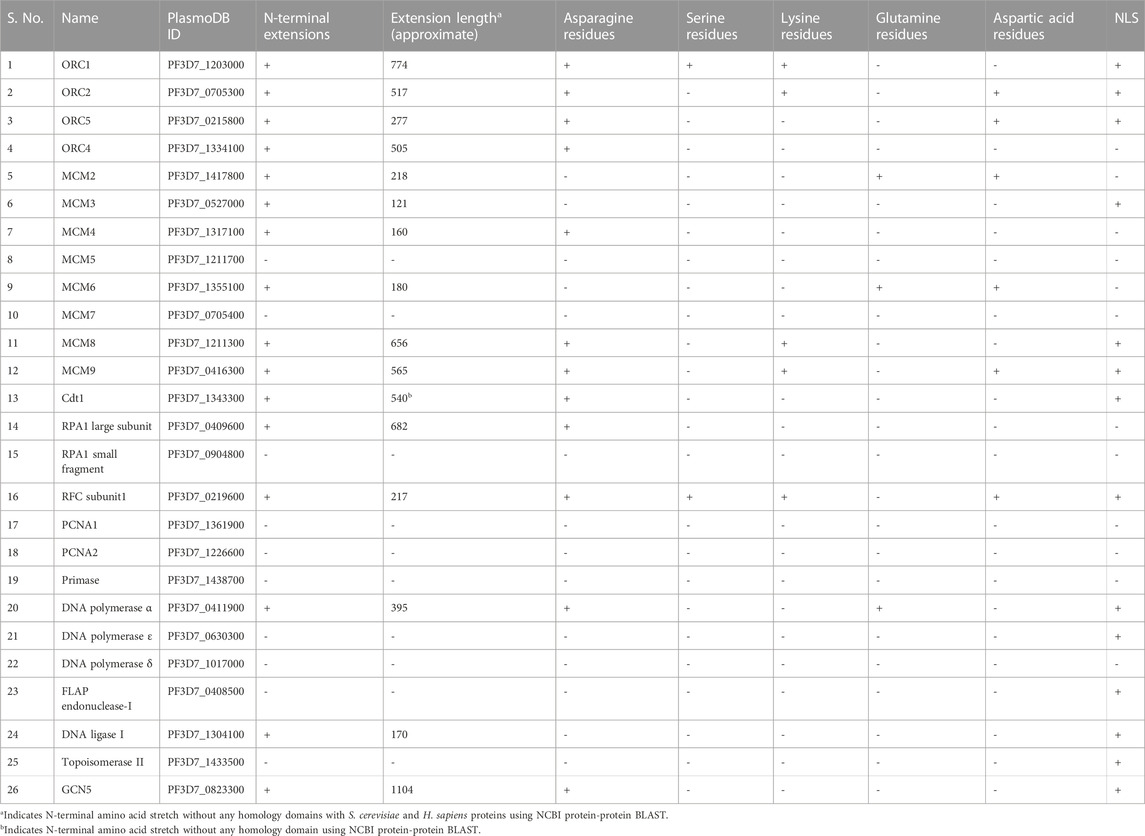
TABLE 1. P. falciparum replication proteins and the essential histone acetyltransferase, GCN5 are listed. The presence of N-terminal extensions and the prevalence of the respective amino acids within these extensions are mentioned. NLS was predicted using NLStradamus (Nguyen Ba et al., 2009).
It is interesting to note that the replication proteins analyzed and listed in Table 1, in the proteins with unusual N-terminal extensions, asparagine repeats are highly prevalent within these extensions. These unique extensions are an excellent candidate for further detailed studies to reveal the need of these long terminus and repeats in the parasite proteins within the cell. Similarly, longer N-terminus extensions are also found in the apicoplast-targeted proteins in both T. gondii and P. falciparum. Some of these larger extensions are processed to expose the N-terminal apicoplast translocation signals. Additionally, these extensions are thought to be involved in regulating the gene expression via protein-protein interactions and facilitating transport across membranes (Seliverstov et al., 2015).
Furthermore, the presence of N-terminal extensions within replication proteins in Plasmodium, but not in other eukaryotic species, hints at parasite specific roles. It is possible that these longer replication proteins undergo proteolytic processing, as has been reported for PfOrc2, to produce smaller fragments in order to carry out the conserved process of replication efficiently. The translocation and formation of a larger complex comprising these longer proteins might impede origin firing and prevent the completion of replication, known to occur five times at remarkably rapid rates within the parasite. Thus, the smaller fragments generated by means of proteolytic processing would adhere to the exceptionally swift replication rates and cell cycle dynamics in the parasite. Moreover, the long N terminal extensions and further processing to create separate proteins would keep the genome size minimal as two or more proteins can be made without the need of separate 5′and 3′sequences for each protein thus, curtailing cellular energy expenses.
Proteolytic processing events and proteases as drug targets
As described above, proteins with unusual N-terminal extensions are processed to yield multifunctional proteins. Proteolytic processing of the proteins is catalyzed by proteases that hydrolyze the peptide bond. Based on the catalytic mechanism employed, these proteases are differentiated into different classes that include serine, cysteine, threonine, aspartic and metallo proteases (Figure 2). The pioneer work highlighting the importance of these proteolytic events was the protein activation by limited proteolysis, in blood coagulation mechanism. Following which, several other processes of cellular regulation through proteolytic processing were discovered including DNA replication, cell-cycle progression, hemostasis, cell proliferation and death, immune response, tissue remodeling and wound healing (Turk, 2006). Several inhibitors targeting a proteolytic event by specific class of protease have been developed for treatment of acute lung injury, hepatitis C, cancer, AIDS, diabetes and myocardial infarction. Angiotensin-converting enzyme (ACE) inhibitors that target the zinc metalloproteinase and inhibitors targeting aspartic protease Renin are used to treat heart attack, heart failure and hypertension.
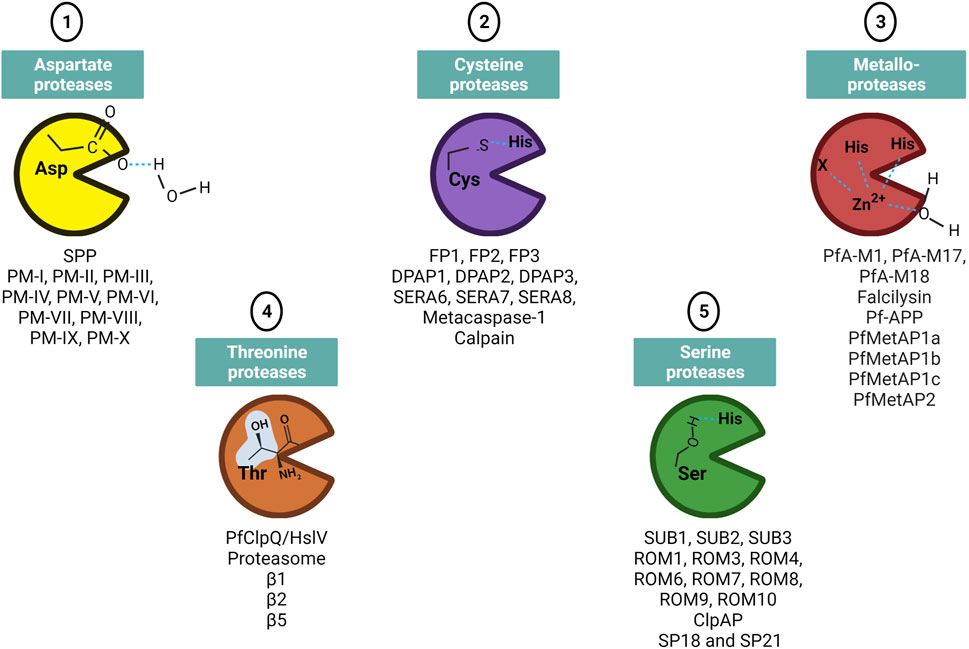
FIGURE 2. Different classes of protease families present in Plasmodium. The residue involved in catalytic site are mentioned on the protease structure shown as Pac-man. Few of the identified proteases in each protease class are mentioned below the Pac-man. Figure created with BioRender.com.
In Plasmodium, few proteases have been characterized and shown to be potential drug targets (Deu, 2017). However, the total number of putative proteases identified via genomic approaches and data mining studies range from 115–137 (Cai et al., 2011). Apart from the widely studied and druggable targets, aspartic and cysteine proteases plasmepsins and falcipains respectively, several other proteases play important roles in parasite processes and may serve as potent drug targets (Roy, 2017). A putative PPPDE protease with papain-like fold, annotated as PF3D7_0919200 can be an attractive target for antimalarial development as papain-family proteases have roles in cell cycle regulation and deubiquitination (Iyer et al., 2004). The malarial calpain is critical for progression of parasite cell cycle and growth but the exact biological role in the parasite remains unexplored. Its high divergence from human calpains underscores it as a potential drug target (Russo et al., 2009). Further, metacaspases, M1 alanyl aminopeptidase, M17 leucine aminopeptidase, apicoplast-targeted proteases including ClpP endopeptidase family can be studied extensively for drug designing purposes (Wu et al., 2003; Le Chat et al., 2007; Meslin et al., 2007; Kuang et al., 2009; El Bakkouri et al., 2010; Rathore et al., 2010; Skinner-Adams et al., 2010). The proteolytic cleavage of N-terminal domain of CSP is a critical process for invasion in hepatocyte by sporozoites. This process is mediated by cysteine class of protease and inhibition by specific inhibitors prevented infection (Coppi et al., 2005; Espinosa et al., 2015). Subtilisin-like protease 1 (SUB1) has multiple substrates involved in egress and invasion. Cysteine protease dipeptidyl aminopeptidase 3 (DPAP3) is involved in RBC invasion and DPAP2 shows role in parasite egress. The action of Aspartyl protease plasmepsin IX is linked with RBC invasion. Hemoglobin degradation is carried by multiple proteases including plasmepsins I-IV, falcipains 2 and 3 and metalloprotease falcilysin. Several proteases are also involved in the trafficking of majority of proteins either to host or to specific organelle in the parasite and include PM-V, aspartyl SPP.
The proteolytic processing of PfGCN5 is essential for its nuclear function, and a cysteine protease mediates this crucial processing. The protease is a Falcipain and the processing event takes place in the vicinity of food vacuole. These falcipains have role in hemoglobin degradation in food vacuole, erythrocyte invasion and rupture. Interestingly, they have role in activation of pro-plasmepsins (Abugri et al., 2022). Thus, targeting the proteolytic event of PfGCN5 by developing specific inhibitors against falcipain would also hamper the above-mentioned processes and in turn will kill the parasites. Indeed, the inhibitors of falcipain exhibit potent antimalarial activity. NP1024, a nonpeptidic inhibitor and a natural fluorescent probe of falcipain-2 demonstrates antimalarial activity and can also be employed in diagnosis (Zhu et al., 2020). The structure of Falcipain-2 in complex with an inhibitor belonging to (E)-chalcone family of molecules opens up new strategy for anti-malarial drug design by using this biologically appropriate molecule as a scaffold (Machin et al., 2019). Several other potent molecules targeting falcipain include both peptide and non-peptide inhibitors and exhibit antimalarial activity or provide a structural basis to utilize them for antimalarial designing (Kerr et al., 2009; Ettari et al., 2010; Bertoldo et al., 2015; Chen et al., 2017; Salas-Sarduy et al., 2017; Hernandez-Gonzalez et al., 2018; Melo et al., 2018; Nizi et al., 2018; Royo et al., 2018; Ettari et al., 2021). However, the structurally homologous human cysteine-proteases of the cathepsin family, are off-target to these inhibitors leading to their poor selectivity. Additionally, the peptide inhibitors cannot be administered orally and degrade rapidly in vivo thus limiting their potential as drug candidates. Therefore, “structural signatures” of a particular falcipain and development of biologically potent small molecule inhibitors are required to develop remarkable antimalarials.
Similarly, the processing of Orc2 can be targeted by developing small molecule inhibitors against the SPP-like protease (of the aspartyl class) which is involved in the processing event. Targeting this aspartyl class of protease will inhibit the processing of this replication protein and thus affect the replication process that occurs five times in these highly proliferating parasites. The known aspartic protease inhibitor, Pepstatin inhibits this class of proteases and blocks P. falciparum development (Bailly et al., 1992). Interestingly, E-64 (cysteine protease inhibitor) and pepstatin exhibited synergistic effects when administered together.
Further, the proteases involved in the possible processing of proteins with extensions as mentioned in Table 1 will first require proper identification of the type of protease class involved followed by screening and validation of potent small molecule inhibitors with antimalarial activity. The processing of these proteins with unusual N-terminal extensions occurs mostly in Food vacuole or ER where the proteases generally reside in the parasites. Thus, targeting these two organelles either alone or in combination will affect the proteolytic processing of many of these proteins in turn affecting the critical processes of this pathogen.
Therefore, identifying and targeting the specific class of protease presents an attractive way of targeting this deadly parasite. These proteases are responsible for protein quality control and processing to yield multifunctional domains essential for proper parasite development. Interestingly, Plasmodium proteases have unique “structural signatures” that can be employed for developing specific and effective anti-malarials (Mishra et al., 2019). The ubiquitous nature of these proteases further accentuates proteases as potential drug targets.
Pros and cons of proteases as drug targets
The ubiquitous and highly diverse mechanistic nature of proteases makes them attractive drug targets. The different class of proteases operate via distinct class specific mechanism structurally and functionally. Moreover, these proteases have specific substrates that can be exploited for therapeutic purposes by developing substrate mimicking inhibitors. As discussed above, the proteases are involved in proteolytic processing of multiple proteins and thus contribute to the parasite development, progression, egress, nutrient uptake, protein trafficking and homeostasis, organelle biogenesis and host invasion. The involvement of proteases in these critical and a number of processes underscores proteases as prime targets for antimalarial therapy. Not just proteins with N-terminal extensions but a particular protease can cleave other protein substrates that play important roles in the parasite. Thus, affecting multiple processes and pathways further adds to the potential of targeting proteases for antimalarial therapeutics. Apart from the above-mentioned advantages of using proteases as drug targets, a combination of inhibitors targeting one or more proteases will be a potent approach. For example, E-64 (the cysteine protease inhibitor) and pepstatin (the aspartic protease inhibitor) exhibited synergistic effect together and blocked parasite development (Bailly et al., 1992). Novel combinations of already existing antimalarial and protease inhibitors can also be explored further which would also minimize the development of emerging antimalarial resistance in the parasites. Interestingly, it has been reported that inhibition of the essential Plasmodium PfUSP protease increased the efficacy of artemisinin-based drugs (Arora et al., 2023). Additionally, treatment with antiretroviral protease inhibitors enhanced sensitivity to chloroquine in CQ-resistant malaria parasites both in vitro and in vivo (He et al., 2009).
A thorough biological, biochemical and chemical validation of the inhibitors is crucial for its development as antimalarial agent. These validation approaches include investigating the inhibition of the specific target, uptake by cells, bioavailability and negligible secondary effects. Homology with host proteases can lead to off-targeting of the general non-specific inhibitors designed to target proteases in the parasite. This can impact the host and ultimately lead to detrimental effects on the host. Therefore, a stringent bioinformatics and structural analysis of the respective Plasmodium protease to develop specific inhibitor against the protease of interest is extremely critical to avoid off targets. Additionally, redundancy among host proteases and higher concentration of proteases within host cells can be exploited to design and select inhibitors in a way such that the host cells show less sensitivity as compared to parasites at a particular concentration (Selzer et al., 1999). Further, the high-resolution crystal structures of proteases and “structural signatures” identified in Plasmodium proteases will pave way for the development of parasite-specific inhibitors.
Discussion
Comparative genomic analyses and large-scale sequencing datasets have paved the way for a better understanding of the parasite biology, that is, critical for the pathogen eradication. The abundance of unusual extensions in the N-terminal region of proteins in the Plasmodium proteome is thought-provoking. Interestingly, most of these extensions comprise of asparagine repeats. These repeats, accounting for 25% of the Plasmodium proteome, is known to have expanded evolutionary and undergone positive selection pressure. Several hypotheses have come up with the putative functions of these highly prevalent repeats, but none seem likely to answer the ubiquitous nature of these repeats (Muralidharan and Goldberg, 2013). Similarly, the function of these unique N-terminal extensions, not observed in the eukaryotic counterparts, remains elusive and is not yet characterized fully. We reckon that these extensions, as observed in the replication proteins, serve as substrates to different proteases in order to generate shorter fragments to facilitate nuclear translocation, multiple origin firing and carry out replication of the Plasmodium mega-genome swiftly. Additionally, the generation of multiple proteins upon processing minimizes the genome size and energy expenditure by averting the need of separate 5′and 3′sequences for each protein. Intriguingly, the N-terminus of the replication protein PfOrc1 comprising N-terminal extensions is involved in var gene regulation whereas the C-terminus functions in DNA replication highlighting multiple functions of a protein.
Therefore, despite the reduced gene content due to the deletion of introns and unwanted genes in Plasmodium, the expansion and prevalence of these N-terminal extensions throughout the parasite genome is quite intriguing. The putative functions of these extensions include specific information for protein localization, processing events, serve as adapters for recruiting interacting partners, or behave as a separate functional protein altogether upon possible proteolytic processing (Figure 1C). How these extra set of residues at the N-terminus of parasite proteins benefit the parasite and influence its function and phenotype remains to be investigated thoroughly to combat this deadly infection. The role of proteases in processing of these proteins with N-terminal extension is crucial and thus a strategy to develop specific inhibitors against these parasitic proteases is exigent. Targeting the involved proteases that mostly reside either in the digestive food vacuole and ER will impede proteolytic processing of these N-terminal extension containing proteins leading to obstruction of critical processes of this pathogen (Figure 3). Additionally, the use of protease inhibitors in the treatment of HIV and COVID-19 infections further encourages the development of protease inhibitors as antimalarial therapeutics. Detailed structural analysis of the parasite protease will also reveal parasite-specific sites that can be targeted for generation of parasite-specific inhibitors. Moreover, targeting one or more proteases in combinatorial mode would serve as a potential antimalarial therapy to overcome the developing cases of resistance to currently available antimalarial drugs.
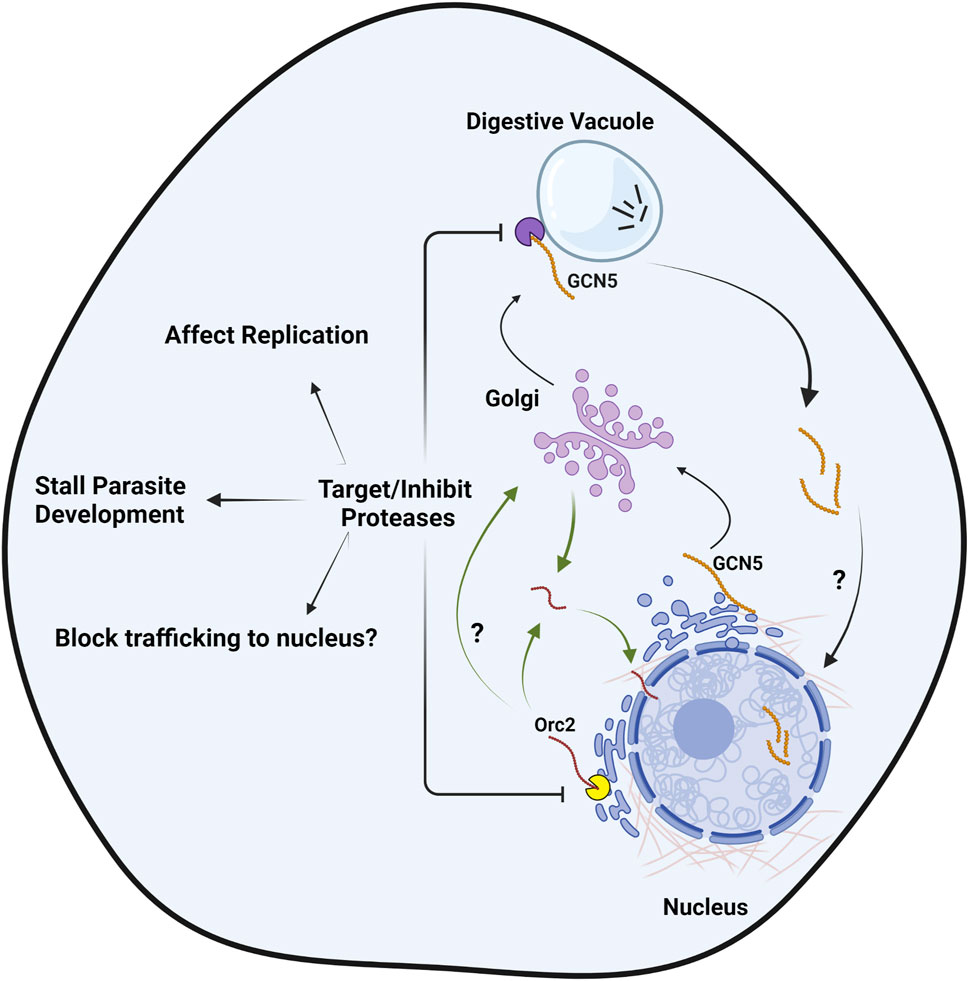
FIGURE 3. Targeting proteases to impede the proteolytic processing of proteins with N-terminal extensions, a unique phenomenon in Plasmodium. The processing of PfOrc2 (in ER) and PfGCN5 (in vicinity of digestive vacuole) is taken as a model here to show the novel targeting approach against aspartate (yellow Pac-man) and cysteine class proteases (purple Pac-man) involved in the processing events respectively, for developing antimalarial therapeutics. Following processing, PfGCN5 enters nucleus and PfOrc2 trafficks to nucleus either in Golgi-dependent or independent pathway. Vesicles are not shown in the figure for clarity. Figure created with BioRender.com.
Author contributions
Conceptualization: AT, AS, and SD; writing, review and editing: AT, AS, and SD; funding acquisition: SD. All authors contributed to the article and approved the submitted version.
Acknowledgments
SD acknowledges SERB, DST, Govt. of India for providing J. C. Bose fellowship (JCB/2021/000029) to carry forward this work. Jawaharlal Nehru University, New Delhi is acknowledged for providing infrastructural support. AT acknowledges CSIR and AS acknowledges UGC, India for fellowships.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
Pf, plasmodium falciparum; NTE, N-terminal extensions; WHO, world health organization; ER, endoplasmic reticulum; ORC, origin recognition complex; ORC, origin recognition complex; GCN5, general control nonderepressible 5; PTM, post-translational modifications; HAT, histone acetyltransferase; MCM, minichromosome maintenance protein.
References
Abu-Elheiga, L., Spira, D. T., and Bachrach, U. (1990). Plasmodium falciparum: Properties of an alpha-like DNA polymerase, the key enzyme in DNA synthesis. Exp. Parasitol. 71, 21–26. doi:10.1016/0014-4894(90)90004-v
Abugri, J., Ayariga, J., Sunwiale, S. S., Wezena, C. A., Gyamfi, J. A., Adu-Frimpong, M., et al. (2022). Targeting the Plasmodium falciparum proteome and organelles for potential antimalarial drug candidates. Heliyon 8, e10390. doi:10.1016/j.heliyon.2022.e10390
Agarwal, M., Bhowmick, K., Shah, K., Krishnamachari, A., and Dhar, S. K. (2017). Identification and characterization of ARS-like sequences as putative origin(s) of replication in human malaria parasite Plasmodium falciparum. FEBS J. 284, 2674–2695. doi:10.1111/febs.14150
Alba, M. M., and Guigo, R. (2004). Comparative analysis of amino acid repeats in rodents and humans. Genome Res. 14, 549–554. doi:10.1101/gr.1925704
Arnot, D. E., and Gull, K. (1998). The Plasmodium cell-cycle: Facts and questions. Ann. Trop. Med. Parasitol. 92, 361–365. doi:10.1080/00034989859357
Arora, P., Narwal, M., Thakur, V., Mukhtar, O., Malhotra, P., and Mohmmed, A. (2023). A Plasmodium falciparum ubiquitin-specific protease (PfUSP) is essential for parasite survival and its disruption enhances artemisinin efficacy. Biochem. J. 480, 25–39. doi:10.1042/BCJ20220429
Bailly, E., Jambou, R., Savel, J., and Jaureguiberry, G. (1992). Plasmodium falciparum: Differential sensitivity in vitro to E-64 (cysteine protease inhibitor) and pepstatin A (aspartyl protease inhibitor). J. protozoology 39, 593–599. doi:10.1111/j.1550-7408.1992.tb04856.x
Banu, K., Mitra, P., Subbarao, N., and Dhar, S. K. (2018). Role of tyrosine residue (Y213) in nuclear retention of PCNA1 in human malaria parasite Plasmodium falciparum. FEMS Microbiol. Lett. 365. doi:10.1093/femsle/fny182
Basu, S., and Sahi, P. K. (2017). Malaria: An update. Indian J. Pediatr. 84, 521–528. doi:10.1007/s12098-017-2332-2
Beck, J. R., Muralidharan, V., Oksman, A., and Goldberg, D. E. (2014). PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature 511, 592–595. doi:10.1038/nature13574
Bertoldo, J. B., Chiaradia-Delatorre, L. D., Mascarello, A., Leal, P. C., Cordeiro, M. N., Nunes, R. J., et al. (2015). Synthetic compounds from an in house library as inhibitors of falcipain-2 from Plasmodium falciparum. J. enzyme inhibition Med. Chem. 30, 299–307. doi:10.3109/14756366.2014.920839
Bhattacharjee, S., Stahelin, R. V., Speicher, K. D., Speicher, D. W., and Haldar, K. (2012). Endoplasmic reticulum PI(3)P lipid binding targets malaria proteins to the host cell. Cell 148, 201–212. doi:10.1016/j.cell.2011.10.051
Bhowmick, K., Tehlan, A., SunitaSudhakar, R., Kaur, I., Sijwali, P. S., Krishnamachari, A., et al. (2020). Plasmodium falciparum GCN5 acetyltransferase follows a novel proteolytic processing pathway that is essential for its function. J. Cell Sci. 133, jcs236489. doi:10.1242/jcs.236489
Brumlik, M. J., Nkhoma, S., Kious, M. J., Thompson, G. R., Patterson, T. F., Siekierka, J. J., et al. (2011). Human p38 mitogen-activated protein kinase inhibitor drugs inhibit Plasmodium falciparum replication. Exp. Parasitol. 128, 170–175. doi:10.1016/j.exppara.2011.02.016
Cai, H., Kuang, R., Gu, J., and Wang, Y. (2011). Proteases in malaria parasites - a phylogenomic perspective. Curr. genomics 12, 417–427. doi:10.2174/138920211797248565
Chang, H. H., Falick, A. M., Carlton, P. M., Sedat, J. W., DeRisi, J. L., and Marletta, M. A. (2008). N-terminal processing of proteins exported by malaria parasites. Mol. Biochem. Parasitol. 160, 107–115. doi:10.1016/j.molbiopara.2008.04.011
Chavalitshewinkoon, P., de Vries, E., Stam, J. G., Franssen, F. F., van der Vliet, P. C., and Overdulve, J. P. (1993). Purification and characterization of DNA polymerases from Plasmodium falciparum. Mol. Biochem. Parasitol. 61, 243–253. doi:10.1016/0166-6851(93)90070-e
Chavalitshewinkoon-Petmitr, P., Chawprom, S., Naesens, L., Balzarini, J., and Wilairat, P. (2000). Partial purification and characterization of mitochondrial DNA polymerase from Plasmodium falciparum. Parasitol. Int. 49, 279–288. doi:10.1016/s1383-5769(00)00057-x
Cheesman, S., McAleese, S., Goman, M., Johnson, D., Horrocks, P., Ridley, R. G., et al. (1994). The gene encoding topoisomerase II from Plasmodium falciparum. Nucleic acids Res. 22, 2547–2551. doi:10.1093/nar/22.13.2547
Chen, J., Malone, B., Llewellyn, E., Grasso, M., Shelton, P. M. M., Olinares, P. D. B., et al. (2020). “Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex,”. bioRxiv : the preprint server for biology.
Chen, W., Huang, Z., Wang, W., Mao, F., Guan, L., Tang, Y., et al. (2017). Discovery of new antimalarial agents: Second-generation dual inhibitors against FP-2 and PfDHFR via fragments assembely. Bioorg. Med. Chem. 25, 6467–6478. doi:10.1016/j.bmc.2017.10.017
Coppi, A., Pinzon-Ortiz, C., Hutter, C., and Sinnis, P. (2005). The Plasmodium circumsporozoite protein is proteolytically processed during cell invasion. J. Exp. Med. 201, 27–33. doi:10.1084/jem.20040989
Dana, S., Prusty, D., Dhayal, D., Gupta, M. K., Dar, A., Sen, S., et al. (2014). Potent antimalarial activity of acriflavine in vitro and in vivo. ACS Chem. Biol. 9, 2366–2373. doi:10.1021/cb500476q
Davies, H. M., Nofal, S. D., McLaughlin, E. J., and Osborne, A. R. (2017). Repetitive sequences in malaria parasite proteins. FEMS Microbiol. Rev. 41, 923–940. doi:10.1093/femsre/fux046
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic acids Res. 36, W465–W469. doi:10.1093/nar/gkn180
Deshmukh, A. S., Agarwal, M., and Dhar, S. K. (2016). Regulation of DNA replication proteins in parasitic protozoans: Possible role of CDK-like kinases. Curr. Genet. 62, 481–486. doi:10.1007/s00294-015-0562-2
Deshmukh, A. S., Srivastava, S., Herrmann, S., Gupta, A., Mitra, P., Gilberger, T. W., et al. (2012). The role of N-terminus of Plasmodium falciparum ORC1 in telomeric localization and var gene silencing. Nucleic acids Res. 40, 5313–5331. doi:10.1093/nar/gks202
Deu, E. (2017). Proteases as antimalarial targets: Strategies for genetic, chemical, and therapeutic validation. FEBS J. 284, 2604–2628. doi:10.1111/febs.14130
Douse, C. H., Green, J. L., Salgado, P. S., Simpson, P. J., Thomas, J. C., Langsley, G., et al. (2012). Regulation of the Plasmodium motor complex: Phosphorylation of myosin A tail-interacting protein (MTIP) loosens its grip on MyoA. J. Biol. Chem. 287, 36968–36977. doi:10.1074/jbc.M112.379842
Dunker, A. K., Lawson, J. D., Brown, C. J., Williams, R. M., Romero, P., Oh, J. S., et al. (2001). Intrinsically disordered protein. J. Mol. Graph. Model. 19, 26–59. doi:10.1016/s1093-3263(00)00138-8
El Bakkouri, M., Pow, A., Mulichak, A., Cheung, K. L., Artz, J. D., Amani, M., et al. (2010). The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum. J. Mol. Biol. 404, 456–477. doi:10.1016/j.jmb.2010.09.051
Espinosa, D. A., Gutierrez, G. M., Rojas-Lopez, M., Noe, A. R., Shi, L., Tse, S. W., et al. (2015). Proteolytic cleavage of the Plasmodium falciparum circumsporozoite protein is a target of protective antibodies. J. Infect. Dis. 212, 1111–1119. doi:10.1093/infdis/jiv154
Ettari, R., Bova, F., Zappala, M., Grasso, S., and Micale, N. (2010). Falcipain-2 inhibitors. Med. Res. Rev. 30, 136–167. doi:10.1002/med.20163
Ettari, R., Previti, S., Di Chio, C., and Zappala, M. (2021). Falcipain-2 and falcipain-3 inhibitors as promising antimalarial agents. Curr. Med. Chem. 28, 3010–3031. doi:10.2174/0929867327666200730215316
Faux, N. G., Bottomley, S. P., Lesk, A. M., Irving, J. A., Morrison, J. R., de la Banda, M. G., et al. (2005). Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 15, 537–551. doi:10.1101/gr.3096505
Feng, Z. P., Zhang, X., Han, P., Arora, N., Anders, R. F., and Norton, R. S. (2006). Abundance of intrinsically unstructured proteins in P. falciparum and other apicomplexan parasite proteomes. Mol. Biochem. Parasitol. 150, 256–267. doi:10.1016/j.molbiopara.2006.08.011
Fox, B. A., and Bzik, D. J. (1991). The primary structure of Plasmodium falciparum DNA polymerase delta is similar to drug sensitive delta-like viral DNA polymerases. Mol. Biochem. Parasitol. 49, 289–296. doi:10.1016/0166-6851(91)90072-e
Francia, M. E., and Striepen, B. (2014). Cell division in apicomplexan parasites. Nat. Rev. Microbiol. 12, 125–136. doi:10.1038/nrmicro3184
Gabriela, M., Matthews, K. M., Boshoven, C., Kouskousis, B., Jonsdottir, T. K., Bullen, H. E., et al. (2022). A revised mechanism for how Plasmodium falciparum recruits and exports proteins into its erythrocytic host cell. PLoS Pathog. 18, e1009977. doi:10.1371/journal.ppat.1009977
Gangwar, D., Kalita, M. K., Gupta, D., Chauhan, V. S., and Mohmmed, A. (2009). A systematic classification of Plasmodium falciparum P-loop NTPases: Structural and functional correlation. Malar. J. 8, 69. doi:10.1186/1475-2875-8-69
Garrido-Cardenas, J. A., Gonzalez-Ceron, L., Manzano-Agugliaro, F., and Mesa-Valle, C. (2019). Plasmodium genomics: An approach for learning about and ending human malaria. Parasitol. Res. 118, 1–27. doi:10.1007/s00436-018-6127-9
Gemayel, R., Vinces, M. D., Legendre, M., and Verstrepen, K. J. (2010). Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44, 445–477. doi:10.1146/annurev-genet-072610-155046
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi:10.1093/sysbio/syq010
Gupta, A., Mehra, P., and Dhar, S. K. (2008). Plasmodium falciparum origin recognition complex subunit 5: Functional characterization and role in DNA replication foci formation. Mol. Microbiol. 69, 646–665. doi:10.1111/j.1365-2958.2008.06316.x
Halfmann, R., Alberti, S., Krishnan, R., Lyle, N., O'Donnell, C. W., King, O. D., et al. (2011). Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol. Cell 43, 72–84. doi:10.1016/j.molcel.2011.05.013
Hamilton, W. L., Claessens, A., Otto, T. D., Kekre, M., Fairhurst, R. M., Rayner, J. C., et al. (2017). Extreme mutation bias and high AT content in Plasmodium falciparum. Nucleic acids Res. 45, 1889–1901. doi:10.1093/nar/gkw1259
He, Z., Chen, L., You, J., Qin, L., and Chen, X. (2009). Antiretroviral protease inhibitors potentiate chloroquine antimalarial activity in malaria parasites by regulating intracellular glutathione metabolism. Exp. Parasitol. 123, 122–127. doi:10.1016/j.exppara.2009.06.008
Hernandez-Gonzalez, J. E., Salas-Sarduy, E., Hernandez Ramirez, L. F., Pascual, M. J., Alvarez, D. E., Pabon, A., et al. (2018). Identification of (4-(9H-fluoren-9-yl) piperazin-1-yl) methanone derivatives as falcipain 2 inhibitors active against Plasmodium falciparum cultures. Biochimica biophysica acta General Subj. 1862, 2911–2923. doi:10.1016/j.bbagen.2018.09.015
Horrocks, P., Jackson, M., Cheesman, S., White, J. H., and Kilbey, B. J. (1996). Stage specific expression of proliferating cell nuclear antigen and DNA polymerase delta from Plasmodium falciparum. Mol. Biochem. Parasitol. 79, 177–182. doi:10.1016/0166-6851(96)02657-6
Hubscher, U., Nasheuer, H. P., and Syvaoja, J. E. (2000). Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci. 25, 143–147. doi:10.1016/s0968-0004(99)01523-6
Huntley, M., and Golding, G. B. (2000). Evolution of simple sequence in proteins. J. Mol. Evol. 51, 131–140. doi:10.1007/s002390010073
Iyer, L. M., Koonin, E. V., and Aravind, L. (2004). Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle 3, 1440–1450. doi:10.4161/cc.3.11.1206
Jirage, D., Chen, Y., Caridha, D., O'Neil, M. T., Eyase, F., Witola, W. H., et al. (2010). The malarial CDK Pfmrk and its effector PfMAT1 phosphorylate DNA replication proteins and co-localize in the nucleus. Mol. Biochem. Parasitol. 172, 9–18. doi:10.1016/j.molbiopara.2010.03.009
Joshi, H. (2003). Markers for population genetic analysis of human plasmodia species, P. falciparum and P. vivax. J. vector borne Dis. 40, 78–83.
Kerr, I. D., Lee, J. H., Pandey, K. C., Harrison, A., Sajid, M., Rosenthal, P. J., et al. (2009). Structures of falcipain-2 and falcipain-3 bound to small molecule inhibitors: Implications for substrate specificity. J. Med. Chem. 52, 852–857. doi:10.1021/jm8013663
Kilbey, B. J., Fraser, I., McAleese, S., Goman, M., and Ridley, R. G. (1993). Molecular characterisation and stage-specific expression of proliferating cell nuclear antigen (PCNA) from the malarial parasite, Plasmodium falciparum. Nucleic acids Res. 21, 239–243. doi:10.1093/nar/21.2.239
Kirchdoerfer, R. N., and Ward, A. B. (2019). Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 10, 2342. doi:10.1038/s41467-019-10280-3
Kuang, R., Gu, J., Cai, H., and Wang, Y. (2009). Improved prediction of malaria degradomes by supervised learning with SVM and profile kernel. Genetica 136, 189–209. doi:10.1007/s10709-008-9336-9
Kumpornsin, K., Kochakarn, T., and Chookajorn, T. (2019). The resistome and genomic reconnaissance in the age of malaria elimination. Dis. models Mech. 12, dmm040717. doi:10.1242/dmm.040717
Le Chat, L., Sinden, R. E., and Dessens, J. T. (2007). The role of metacaspase 1 in Plasmodium berghei development and apoptosis. Mol. Biochem. Parasitol. 153, 41–47. doi:10.1016/j.molbiopara.2007.01.016
Lee, W. C., Russell, B., and Renia, L. (2019). Sticking for a cause: The falciparum malaria parasites cytoadherence paradigm. Front. Immunol. 10, 1444. doi:10.3389/fimmu.2019.01444
Li, J. L., and Cox, L. S. (2003). Characterisation of a sexual stage-specific gene encoding ORC1 homologue in the human malaria parasite Plasmodium falciparum. Parasitol. Int. 52, 41–52. doi:10.1016/s1383-5769(02)00079-x
Li, J. L., Warren, A. V., and Cox, L. S. (2002). Identification of a second proliferating cell nuclear antigen in the human malarial pathogen Plasmodium falciparum. Int. J. Parasitol. 32, 1683–1692. doi:10.1016/s0020-7519(02)00162-5
Liu, C., Wu, R., Zhou, B., Wang, J., Wei, Z., Tye, B. K., et al. (2012). Structural insights into the Cdt1-mediated MCM2-7 chromatin loading. Nucleic acids Res. 40, 3208–3217. doi:10.1093/nar/gkr1118
Machin, J. M., Kantsadi, A. L., and Vakonakis, I. (2019). The complex of Plasmodium falciparum falcipain-2 protease with an (E)-chalcone-based inhibitor highlights a novel, small, molecule-binding site. Malar. J. 18, 388. doi:10.1186/s12936-019-3043-0
Matthews, H., Duffy, C. W., and Merrick, C. J. (2018). Checks and balances? DNA replication and the cell cycle in Plasmodium. Parasites vectors 11, 216. doi:10.1186/s13071-018-2800-1
Mehra, P., Biswas, A. K., Gupta, A., Gourinath, S., Chitnis, C. E., and Dhar, S. K. (2005). Expression and characterization of human malaria parasite Plasmodium falciparum origin recognition complex subunit 1. Biochem. biophysical Res. Commun. 337, 955–966. doi:10.1016/j.bbrc.2005.09.131
Melo, P. M. S., El Chamy Maluf, S., Azevedo, M. F., Paschoalin, T., Budu, A., Bagnaresi, P., et al. (2018). Inhibition of Plasmodium falciparum cysteine proteases by the sugarcane cystatin CaneCPI-4. Parasitol. Int. 67, 233–236. doi:10.1016/j.parint.2017.12.005
Meslin, B., Barnadas, C., Boni, V., Latour, C., De Monbrison, F., Kaiser, K., et al. (2007). Features of apoptosis in Plasmodium falciparum erythrocytic stage through a putative role of PfMCA1 metacaspase-like protein. J. Infect. Dis. 195, 1852–1859. doi:10.1086/518253
Miao, J., Lawrence, M., Jeffers, V., Zhao, F., Parker, D., Ge, Y., et al. (2013). Extensive lysine acetylation occurs in evolutionarily conserved metabolic pathways and parasite-specific functions during Plasmodium falciparum intraerythrocytic development. Mol. Microbiol. 89, 660–675. doi:10.1111/mmi.12303
Mishra, M., Singh, V., and Singh, S. (2019). Structural insights into key Plasmodium proteases as therapeutic drug targets. Front. Microbiol. 10, 394. doi:10.3389/fmicb.2019.00394
Mitra, P., Banu, K., Deshmukh, A. S., Subbarao, N., and Dhar, S. K. (2015). Functional dissection of proliferating-cell nuclear antigens (1 and 2) in human malarial parasite Plasmodium falciparum: Possible involvement in DNA replication and DNA damage response. Biochem. J. 470, 115–129. doi:10.1042/BJ20150452
Mitra, P., Deshmukh, A. S., and Dhar, S. K. (2012). DNA replication during intra-erythrocytic stages of human malarial parasite Plasmodium falciparum. Curr. Sci. 102, 725–740.
Moser, K. A., Drabek, E. F., Dwivedi, A., Stucke, E. M., Crabtree, J., Dara, A., et al. (2020). Strains used in whole organism Plasmodium falciparum vaccine trials differ in genome structure, sequence, and immunogenic potential. Genome Med. 12, 6. doi:10.1186/s13073-019-0708-9
Muralidharan, V., and Goldberg, D. E. (2013). Asparagine repeats in Plasmodium falciparum proteins: Good for nothing? PLoS Pathog. 9, e1003488. doi:10.1371/journal.ppat.1003488
Muralidharan, V., Oksman, A., Iwamoto, M., Wandless, T. J., and Goldberg, D. E. (2011). Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag. Proc. Natl. Acad. Sci. U. S. A. 108, 4411–4416. doi:10.1073/pnas.1018449108
Muralidharan, V., Oksman, A., Pal, P., Lindquist, S., and Goldberg, D. E. (2012). Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat. Commun. 3, 1310. doi:10.1038/ncomms2306
Nguyen Ba, A. N., Pogoutse, A., Provart, N., and Moses, A. M. (2009). NLStradamus: A simple hidden markov model for nuclear localization signal prediction. BMC Bioinforma. 10, 202. doi:10.1186/1471-2105-10-202
Nizi, E., Sferrazza, A., Fabbrini, D., Nardi, V., Andreini, M., Graziani, R., et al. (2018). Peptidomimetic nitrile inhibitors of malarial protease falcipain-2 with high selectivity against human cathepsins. Bioorg. Med. Chem. Lett. 28, 1540–1544. doi:10.1016/j.bmcl.2018.03.069
Noguchi, K., Vassilev, A., Ghosh, S., Yates, J. L., and DePamphilis, M. L. (2006). The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 25, 5372–5382. doi:10.1038/sj.emboj.7601396
Nunthawarasilp, P., Petmitr, S., and Chavalitshewinkoon-Petmitr, P. (2007). Partial purification and characterization of DNA polymerase beta-like enzyme from Plasmodium falciparum. Mol. Biochem. Parasitol. 154, 141–147. doi:10.1016/j.molbiopara.2007.04.011
Oliveira, M. T., and Kaguni, L. S. (2010). Functional roles of the N- and C-terminal regions of the human mitochondrial single-stranded DNA-binding protein. PloS one 5, e15379. doi:10.1371/journal.pone.0015379
Osman, K. T., Lou, H. J., Qiu, W., Brand, V., Edwards, A. M., Turk, B. E., et al. (2015). Biochemical characterization of FIKK8-A unique protein kinase from the malaria parasite Plasmodium falciparum and other apicomplexans. Mol. Biochem. Parasitol. 201, 85–89. doi:10.1016/j.molbiopara.2015.06.002
Patterson, S., Robert, C., Whittle, C., Chakrabarti, R., Doerig, C., and Chakrabarti, D. (2006). Pre-replication complex organization in the atypical DNA replication cycle of Plasmodium falciparum: Characterization of the mini-chromosome maintenance (MCM) complex formation. Mol. Biochem. Parasitol. 145, 50–59. doi:10.1016/j.molbiopara.2005.09.006
Patterson, S., Whittle, C., Robert, C., and Chakrabarti, D. (2002). Molecular characterization and expression of an alternate proliferating cell nuclear antigen homologue, PfPCNA2, in Plasmodium falciparum. Biochem. biophysical Res. Commun. 298, 371–376. doi:10.1016/s0006-291x(02)02436-1
Pizzi, E., and Frontali, C. (2001). Low-complexity regions in Plasmodium falciparum proteins. Genome Res. 11, 218–229. doi:10.1101/gr.gr-1522r
Pradhan, S., Kalia, I., Roy, S. S., Singh, O. P., Adak, T., Singh, A. P., et al. (2019). Molecular characterization and expression profile of an alternate proliferating cell nuclear antigen homolog PbPCNA2 in Plasmodium berghei. IUBMB life 71, 1293–1301. doi:10.1002/iub.2036
Prasartkaew, S., Zijlstra, N. M., Wilairat, P., Overdulve, J. P., and de Vries, E. (1996). Molecular cloning of a Plasmodium falciparum gene interrupted by 15 introns encoding a functional primase 53 kDa subunit as demonstrated by expression in a baculovirus system. Nucleic acids Res. 24, 3934–3941. doi:10.1093/nar/24.20.3934
Rathore, S., Sinha, D., Asad, M., Bottcher, T., Afrin, F., Chauhan, V. S., et al. (2010). A cyanobacterial serine protease of Plasmodium falciparum is targeted to the apicoplast and plays an important role in its growth and development. Mol. Microbiol. 77, 873–890. doi:10.1111/j.1365-2958.2010.07251.x
Ridley, R. G., White, J. H., McAleese, S. M., Goman, M., Alano, P., de Vries, E., et al. (1991). DNA polymerase delta: Gene sequences from Plasmodium falciparum indicate that this enzyme is more highly conserved than DNA polymerase alpha. Nucleic acids Res. 19, 6731–6736. doi:10.1093/nar/19.24.6731
Riera, A., Barbon, M., Noguchi, Y., Reuter, L. M., Schneider, S., and Speck, C. (2017). From structure to mechanism-understanding initiation of DNA replication. Genes and Dev. 31, 1073–1088. doi:10.1101/gad.298232.117
Robert-Paganin, J., Robblee, J. P., Auguin, D., Blake, T. C. A., Bookwalter, C. S., Krementsova, E. B., et al. (2019). Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism. Nat. Commun. 10, 3286. doi:10.1038/s41467-019-11120-0
Roy, K. K. (2017). Targeting the active sites of malarial proteases for antimalarial drug discovery: Approaches, progress and challenges. Int. J. Antimicrob. agents 50, 287–302. doi:10.1016/j.ijantimicag.2017.04.006
Royo, S., Schirmeister, T., Kaiser, M., Jung, S., Rodriguez, S., Bautista, J. M., et al. (2018). Antiprotozoal and cysteine proteases inhibitory activity of dipeptidyl enoates. Bioorg. Med. Chem. 26, 4624–4634. doi:10.1016/j.bmc.2018.07.015
Russo, I., Oksman, A., Vaupel, B., and Goldberg, D. E. (2009). A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc. Natl. Acad. Sci. U. S. A. 106, 1554–1559. doi:10.1073/pnas.0806926106
Saha, A., Chauhan, S., and Bagchi, T. (2016). Effect of recombinant malarial antigen on monocyte functionality. Trans. R. Soc. Trop. Med. Hyg. 110, 480–486. doi:10.1093/trstmh/trw049
Salas-Sarduy, E., Guerra, Y., Covaleda Cortes, G., Aviles, F. X., and Chavez Planes, M. A. (2017). Identification of tight-binding plasmepsin II and falcipain 2 inhibitors in aqueous extracts of marine invertebrates by the combination of enzymatic and interaction-based assays. Mar. drugs 15, 123. doi:10.3390/md15040123
Seliverstov, A. V., Zverkov, O. A., Istomina, S. N., Pirogov, S. A., and Kitsis, P. S. (2015). Comparative analysis of apicoplast-targeted protein extension lengths in apicomplexan parasites. BioMed Res. Int. 2015, 452958. doi:10.1155/2015/452958
Selzer, P. M., Pingel, S., Hsieh, I., Ugele, B., Chan, V. J., Engel, J. C., et al. (1999). Cysteine protease inhibitors as chemotherapy: Lessons from a parasite target. Proc. Natl. Acad. Sci. U. S. A. 96, 11015–11022. doi:10.1073/pnas.96.20.11015
Sharma, R., Sharma, B., Gupta, A., and Dhar, S. K. (2018). Identification of a novel trafficking pathway exporting a replication protein, Orc2 to nucleus via classical secretory pathway in Plasmodium falciparum. Biochimica biophysica acta Mol. Cell Res. 1865, 817–829. doi:10.1016/j.bbamcr.2018.03.003
Shekhar, S., Bhowmick, K., and Dhar, S. K. (2022). Role of PfMYST in DNA replication in Plasmodium falciparum. Exp. Parasitol. 242, 108396. doi:10.1016/j.exppara.2022.108396
Skinner-Adams, T. S., Stack, C. M., Trenholme, K. R., Brown, C. L., Grembecka, J., Lowther, J., et al. (2010). Plasmodium falciparum neutral aminopeptidases: New targets for anti-malarials. Trends Biochem. Sci. 35, 53–61. doi:10.1016/j.tibs.2009.08.004
Stanojcic, S., Kuk, N., Ullah, I., Sterkers, Y., and Merrick, C. J. (2017). Single-molecule analysis reveals that DNA replication dynamics vary across the course of schizogony in the malaria parasite Plasmodium falciparum. Sci. Rep. 7, 4003. doi:10.1038/s41598-017-04407-z
Swapna, L. S., and Parkinson, J. (2017). Genomics of apicomplexan parasites. Crit. Rev. Biochem. Mol. Biol. 52, 254–273. doi:10.1080/10409238.2017.1290043
Tan, J. C., Tan, A., Checkley, L., Honsa, C. M., and Ferdig, M. T. (2010). Variable numbers of tandem repeats in Plasmodium falciparum genes. J. Mol. Evol. 71, 268–278. doi:10.1007/s00239-010-9381-8
Triolo, T., and Sternglanz, R. (1996). Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381, 251–253. doi:10.1038/381251a0
Turk, B. (2006). Targeting proteases: Successes, failures and future prospects. Nat. Rev. Drug Discov. 5, 785–799. doi:10.1038/nrd2092
Vasuvat, J., Montree, A., Moonsom, S., Leartsakulpanich, U., Petmitr, S., Focher, F., et al. (2016). Biochemical and functional characterization of Plasmodium falciparum DNA polymerase δ. Malar. J. 15, 116. doi:10.1186/s12936-016-1166-0
Voss, T. S., Mini, T., Jenoe, P., and Beck, H. P. (2002). Plasmodium falciparum possesses a cell cycle-regulated short type replication protein A large subunit encoded by an unusual transcript. J. Biol. Chem. 277, 17493–17501. doi:10.1074/jbc.M200100200
Wei, Z., Liu, C., Wu, X., Xu, N., Zhou, B., Liang, C., et al. (2010). Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J. Biol. Chem. 285, 12469–12473. doi:10.1074/jbc.C109.094599
Wu, Y., Wang, X., Liu, X., and Wang, Y. (2003). Data-mining approaches reveal hidden families of proteases in the genome of malaria parasite. Genome Res. 13, 601–616. doi:10.1101/gr.913403
Yang, C., Curth, U., Urbanke, C., and Kang, C. (1997). Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat. Struct. Biol. 4, 153–157. doi:10.1038/nsb0297-153
Yusuf, N. A., Green, J. L., Wall, R. J., Knuepfer, E., Moon, R. W., Schulte-Huxel, C., et al. (2015). The Plasmodium class XIV myosin, MyoB, has a distinct subcellular location in invasive and motile stages of the malaria parasite and an unusual light chain. J. Biol. Chem. 290, 12147–12164. doi:10.1074/jbc.M115.637694
Zhu, L., Shan, L., Zhu, J., Li, L., Li, S., Wang, L., et al. (2020). Discovery of a natural fluorescent probe targeting the Plasmodium falciparum cysteine protease falcipain-2. Sci. China Life Sci. 63, 1016–1025. doi:10.1007/s11427-019-1581-2
Keywords: Plasmodium falciparum, N-terminal extensions (NTE), proteases, replication, antimalarials, proteolytic processing
Citation: Tehlan A, Saha A and Dhar SK (2023) Targeting proteases and proteolytic processing of unusual N-terminal extensions of Plasmodium proteins: parasite peculiarity. Front. Drug Discov. 3:1223140. doi: 10.3389/fddsv.2023.1223140
Received: 15 May 2023; Accepted: 28 June 2023;
Published: 13 July 2023.
Edited by:
Brijesh Rathi, University of Delhi, IndiaReviewed by:
Prakasha Kempaiah, Mayo Clinic Florida, United StatesKailash C. Pandey, National Institute of Malaria Research (ICMR), India
Vinoth Rajendran, Pondicherry University, India
Copyright © 2023 Tehlan, Saha and Dhar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suman Kumar Dhar, c2tkaGFyMjAwMkB5YWhvby5jby5pbg==, c2tkaGFyQG1haWwuam51LmFjLmlu
†These authors have contributed equally to this work
 Ankita Tehlan
Ankita Tehlan Abhik Saha
Abhik Saha Suman Kumar Dhar
Suman Kumar Dhar