
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Drug Discov., 26 August 2022
Sec. Anti-Infective Agents
Volume 2 - 2022 | https://doi.org/10.3389/fddsv.2022.925825
This article is part of the Research TopicDevelopment of COVID-19 Therapies: Lessons Learnt and Ongoing EffortsView all 13 articles
 Emmanuel O Ogbadoyi1,2,3*
Emmanuel O Ogbadoyi1,2,3* Ndagi Umar1,2,4
Ndagi Umar1,2,4The disease which is today known as COVID-19 is caused by severe acute respiratory. Syndrome coronavirus 2 (SARS-COV-2), was first reported in Wuhan, China in December 2019. The disease has claimed well over six million lives from over 500 million cases. Vaccine hesitancy militates against successful mass vaccination. There is the rapid emergence of new SARS-COV-2 variants, constituting a challenge to the effectiveness of vaccines. Moreover, none of the available vaccines offers 100% protection and even the protection offered is of short duration necessitating booster doses to be taken. Moving forward, the development of plant-based edible vaccines will be a remarkable strategic approach to overcome vaccine hesitancy and improve vaccine uptake. So far only about nine drugs for COVID-19 treatment have approvals by either or both the European Medicines Agency and the FDA. While drug repurposing to address the emerging need in the early period of the COVID-19 pandemic has been contextually very useful, investment in it remains relatively low for commercial reasons arising from patenting issues. Embarking on new drug discovery and development strategies targeting both the virus and host factors is a very appealing option. Targeting druggable targets that are present across viruses, particularly the coronaviruses, for drug discovery and development represents an important strategy for pandemic preparedness. Natural products are an important reservoir of chemical scaffolds with huge potential for the discovery of novel chemical entities for development of novel therapeutics. Phytopharming is an available technology that can be used for mass and accelerated production of therapeutic molecules that will be required within short periods of time as is the case in pandemic outbreaks. Nanotechnology provides excellent platforms for formulating multivalent vaccines and pan-viral medicines for the treatment of COVID-19. Taken together, this review discusses the potential for the development of therapeutics by using the tools of biocomputing, nanotechnology, and phytopharming for accelerated therapeutic development to achieve effective COVID-19 treatment and associated complications, including new and emerging variants of SARS-COV-2 and other viral pandemics that may emerge or re-emerge.
The ongoing Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) which started in the city of Wuhan in Hubei province, China with the earliest onset of symptoms on 1 December 2019 (Liu et al., 2020) has today spread to about 226 countries and territories worldwide. The disease which was initially diagnosed as viral pneumonia (Huang et al., 2020; Zhu et al., 2020) was referred to as Wuhan pneumonia by the press (Liu et al., 2020).
The virus was initially termed 2019 novel coronavirus (2019-nCoV) on 12 January 2020 and a month later, the World Health Organization officially named the disease coronavirus disease 2019 (COVID-19) and went on to declare it a pandemic on 11 March 2020. According to the World Health Organization, globally there are 497,960,492 confirmed cases with 6,181,850 deaths as of 12 April 2022. Horton (2020) described COVID- 19 as a triple crisis–medical, economic, and psychological.
Generally, diseases are controlled through the use of both pharmaceutical and non-pharmaceutical approaches. COVID-19 is not an exception to this generalization. COVID-19 prevention is through a combination of vaccination, prophylactic or preventive treatment, and non-pharmaceutical measures of social distancing, quarantine, isolating infected persons and patients, lockdowns, avoiding crowding, wearing protective face masks, regular washing of hands with soap, use of hand sanitizers, and not touching the face with hands, particularly the mouth, nose and the eyes which are ready and easy viral entry points into the body. Implementation of these were very challenging, particularly in low- and middle-income countries and among the unscientifically minded populations. This was most common with the use of face masks where: materials that will readily allow entry of the virus are used for the face masks; many people touched the masks that may have been infected with the virus with their bare hands thereby possibly carrying the viruses in their hands and then touching their faces with unwashed and unsanitized hands; inappropriate disposal of used face masks which become potential sources of spread of the virus within the population as children and even some adults pick up discarded used masks from roadsides and waste bins (this is common in poor resource settings and among individuals with a low level of education). The pharmaceutical approaches involve the use of therapeutics, which in a broad sense can be described as medications or remedies taken to prevent, treat, or remediate a health problem. Therapeutics include but are not limited to, vaccines, monoclonal antibodies, drugs, functional foods, and nutraceuticals.
The field of vaccinology came into being in 1796 with the discovery of the smallpox vaccine by Edward Jenner in which he used the whole live organism that causes smallpox, cowpox virus, to develop the vaccine. This was followed by the formulation of the polio vaccine using the killed or inactivated polio virus to develop the polio vaccine. Then there came the live attenuated vaccines in which the infectious agent is neither killed nor inactivated but rendered non-infective. Some examples include measles, mumps, and rubella vaccine (MMR). Following this are the toxoid vaccines which are based on inactivated toxins of pathogenic microorganisms such as tetanus and diphtheria. In more recent times, subunit vaccines (an example is the HPV vaccine), in which components of the pathogen antigen to which immune response is stimulated are used in developing the vaccines. The components of the pathogen antigen can also be produced as recombinants and used for vaccine production. Subunit vaccines have excellent safety profiles but are generally less immunogenic compared with inactivated or attenuated vaccines and require much stronger adjuvants for enhanced immunogenicity. Other forms of vaccine platforms include conjugate vaccines in which poorly immunogenic surface molecules, such as polysaccharides of many bacteria, are synthesized and conjugated to strongly immunogenic proteins and are then used for vaccine development. Some examples include HiB, meningitis C, and pneumococcal vaccines; the next generation vaccines: vectored vaccines which involve putting vaccine antigens inside replication-deficient microorganisms such as Adenoviruses and modified Vaccinia Ankara capable of triggering an immune response and the antigen is then vectored into the host cells; The genome-based approach (nucleic acid approach) which is based on the use of DNA or RNA to make desired antigens in the host thereby prompting an immune response; Reverse vaccinology in which vaccine candidates are selected on the basis of predicted immunogenicity emanating from sequence information obtained from the use of modern sequencing technology. This was successfully used in developing a vaccine against meningococcus B, which, till then, was difficult to be achieved using conventional vaccine technologies (Rappuolia et al., 2021); Structural vaccinology (structure-based antigen design) which can also be considered as a variant of reverse vaccinology uses information on the structure of antigenic epitopes and protein conformation in the design of vaccines. Some examples include the design of a single meningococcal antigen containing the epitopes of three antigenic variants of the same molecule, and a host of others (reviewed by Rappuolia et al., 2021). The stages involved in traditional vaccine discovery and development are shown in Figure 1.
Structural vaccinology is novel and innovative and particularly potentially very useful in antiviral vaccine development to combat very challenging viruses that traditional approaches have failed or may fail to produce effective vaccines. Essentially, the process involves four stages: determining the atomic structure of the antigen or antigen-antibody complex; remodeling the antigen or the epitope by reverse molecular engineering; incorporating the re-engineered antigen or epitope into one of the vaccine platforms; and testing the safety and efficacy of the candidate vaccine in vivo (Anasir and Poh 2019).
In summarizing, three broad platforms can be used for the design of vaccines: the whole organism (virus or bacterium), the parts of the organism (subunits and recombinants), and the genetic material (DNA or RNA). These are illustrated in Figure 2.
Broadly, drug discovery involves the following approaches: Screening extracts of natural origin for bioactivity and then isolating bioactive compounds from potent extracts for drug development. Opium and taxol are a good testimony to this; Random synthesis and screening in which several organic compounds are synthesized and pharmacologically screened for therapeutic potentials. The efficiency of this approach has been greatly enhanced by automated high throughput screening technology and combinatorial chemistry which has greatly accelerated synthetic methods, enabling the synthesis of a huge library of compounds that can then be screened for bioactivity. However, this approach is not a successful path to drug discovery; Rational drug design is dependent on several determinants such as the pathophysiology of the disease against which the drug is being developed. This uses the tools of genomics, structural biology, synthetic biology, medicinal chemistry, computational biology/bioinformatics, and cheminformatics. Rational drug design is a very efficient and successful approach as the innovative tools mentioned can significantly reduce the time for the discovery process. The steps involved in the drug discovery and development process are summarized in Figure 3.
Drug repurposing, though not drug discovery per se, is a useful approach to obtaining drugs for meeting unmet medical treatment needs.
The world was unprepared to immediately and adequately mitigate the catastrophic consequences of the outbreak of COVID-19. Since there was no specific treatment for the disease at the onset, the most appropriate control method was to develop vaccines for mass immunization to prevent transmission and bring the disease under control. The response was massive as researchers in academia, research institutes, and industries worked together with manufacturing companies to produce various vaccines using eleven different platforms (Table 1). These platforms can be classified into conventional vaccines (live attenuated virus, inactivated virus, virus-like particle, and protein subunit) and next-generation vaccines (Gene-based vaccine platforms based on viral vectors, DNA, and RNA). The international coordinated efforts led to the unprecedented scientific achievement of getting a vaccine (Comirnaty vaccine of Pfizer/BioNTech) for use in just about a year of the pandemic outbreak and many others followed (Table 2). As of 20 May 2022, there were 157 vaccines in clinical development (Table 1) and 198 in preclinical development (World Health Organization, 2021). The percentage of vaccines in each platform is summarized in Figure 4. Two vaccine platforms, mRNA and viral vectors particularly stand out in the fight against COVID-19. Despite this remarkable progress, there are still serious challenges of limited efficacy (Table 2), waning protection over short periods of time, and rapid emergence of new variants that may further reduce the efficacy of existing vaccines. These inadequacies can be overcome by developing platforms for polyvalent vaccines using CRISPR-engineered viral vectors and/or nanomedicine. Furthermore, the availability of vaccines alone cannot prevent the disease unless people get vaccinated. However, vaccine hesitancy continues to remain a serious challenge militating against vaccine acceptance and uptake by the general population. The SAGE working group on vaccine hesitancy describes vaccine hesitancy as the “delay in acceptance or refusal of vaccination despite the availability of vaccination services” (MacDonald, 2015). Vaccine hesitancy, driven largely by misinformation and disinformation, has been described as one of the top ten global health threats (Scheres and Kuszewski, 2019). Although globally a total of 11,250,782,214 vaccine doses had been administered as of 5 April 2022, the dream of herd immunity is far from being realized. This is compounded by “vaccine nationalism and regionalism” and a lack of production capacity in many countries, particularly the low- and middle-income countries. For instance, there are only 24 countries out of the total of 235 countries and territories in the world that produce World Health Organization prequalified vaccines and none of the 24 countries is in Africa with twelve countries leading vaccine production as shown in Table 3. However, in February 2022, the technology for the production of mRNA vaccines was given to six countries in Africa (Egypt, Kenya, Nigeria, Senegal, South Africa, and Tunisia) to enable the production of mRNA vaccines in these countries.
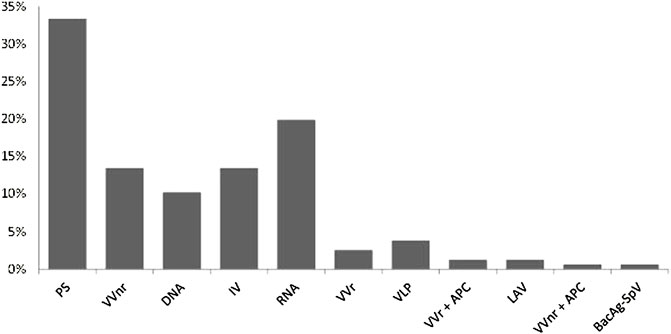
FIGURE 4. COVID-19 vaccines in clinical devolopment,the devolopmental platforms (x axis), and the percentage of vaccines in each devoloment platform (y axis). Key: PS=Protein subunit; VVnr= Viral Vector, non-replicating; DNA; IV= Inactivated virus; RNA; VVr = Viral Vector, replicating; VLP = Virus Like Particle; VVr + APC = VVr + Antigen Presenting Cell; LAV = Live Attenuated Virus; VVnr+ APC + VVnr + Antigen Presenting Cell; BacAg-SpV = Bacterial antigen-spore expression Vector. Based on data obtained from WHO (2022) at https://www.who.int/publications/m/item/draft-landscape-of covid-19-candidate-vaccines.
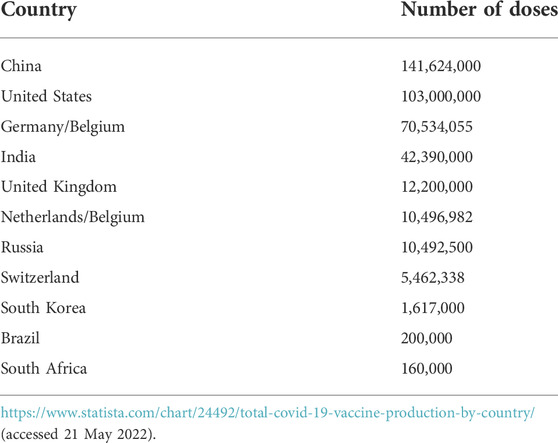
TABLE 3. A total number of COVID-19 vaccine doses produced by the top 12 producing countries as of 03 March 2021.
Although the rollout of vaccines and non-pharmaceutical interventions are significantly helping to contain the COVID-19 pandemic, treatment remains a major control measure. However, it is an understatement to say that COVID-19 chemotherapeutic armory is grossly inadequate at the moment. There is huge potential to successfully address this inadequacy by providing solutions for COVID-19 treatment using drug repurposing and traditional drug discovery approaches.
The outbreak of the COVID-19 pandemic can be said to have taken the world unaware and unprepared to contain it and there was more or less pandemonium at the onset as there was neither vaccine nor any specific treatment. While scientists and researchers immediately began efforts in collaborative vaccine development, intensified and concerted efforts were also made by scientists and researchers to evaluate pre-existing approved drugs for efficacy against COVID-19 so that they could be used for the treatment of the disease in what is known as drug repurposing (also referred to as repositioning, re-profiling, re-tasking, indication expansion, indication shift, or rescue of drugs). Drug repurposing has also been described as establishing new medical uses for already known drugs, including approved, discontinued, shelved, and experimental drugs (Talevi and Bellera, 2020). Therefore, in response to the immense pressure the disease placed on world health systems, the World Health Organization rationally established an international “Solidarity” clinical trial to accelerate the finding of an effective treatment for the disease (World Health Organization, 2020). What followed were clinical trials of pre-existing multiple antiviral medications that had been used for SARS-CoV, MERS-CoV, and antimalarials (Li and De Clercq, 2020). The initial repurposing efforts involved remdesivir, the malaria medications hydroxychloroquine and chloroquine, the combination of HIV drugs called Kaletra, consisting of lopinavir and ritonavir, and other combinations, including interferon beta-1a (Savi et al., 2020). Candidates for clinical trials came from the deployment of available screening techniques of current pharmacopoeia to reveal novel drug indications of already established drugs (Jarada et al., 2020).
The methodologies used for drug repurposing can be broadly categorized into three depending on available information in the context of pharmacological, toxicological, and biological activity. They include those based on the: drug, drug target, and disease/therapy. Three fundamental steps are involved before a potential drug for repurposing can be developed and marketed. These are: identifying the candidate drug; evaluating mechanistically, the drug effects in preclinical models; and evaluating the efficacy of candidate drugs in phase II clinical trials (Khataniar et al., 2022). There are two alternative but complementary approaches that can and have been used for drug repurposing: an experimental approach which is also referred to as the activity-based approach, and the computational methods (in silico) approach (Lipinski, 2011; Senanayake, 2020; Dhaneshwar and Bhasin, 2021; Naasani, 2021; Ng et al., 2021; Khataniar et al., 2022). Whichever approach is chosen, the process begins with the selection of the drug for repurposing followed by the identification of the drug target(s). From what is known about the biology of SARS-CoV-2, the drug targets frequently mentioned are: Spike (S) protein receptor binding domain (RBD), RNA-dependent RNA polymerases (RdRps), helicase; SARS-CoV Chymotrypsin-like cysteine protease, 3CLpro (also known as MPRO); papain-like cysteine protease (PLpro), which are all important for viral replication and they are therefore viral-based targets. This has been reviewed in the literature (Jang et al., 2021; Nawaz, 2021; Yang and Rao, 2021; Khataniar et al., 2022) and will not be discussed in detail in the current review. There are also host factors that can be targeted for therapeutic intervention and they include, but are not limited to: transmembrane protease serine 2 (TMPRSS2) for viral entry; NF-κB, IL-1β, and IL-6, interferon-gamma (IFN-γ), tumor necrosis factor-alpha (TNF-α), and Janus kinase (JAK) for immunomodulation and cytokine storm prevention and control; superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GSPHx), reduced glutathione (GSH), glutathione reductase (GR) and lipid peroxidation (LPO) for mitigating against oxidative stress.
In the experimental approaches, either phenotypic or target-based screening can be done to identify potent drug candidates for validation, optimization, selection, and repositioning. Phenotypic screening is followed by target deconvolution (retrospective identification of molecular targets) using appropriate deconvolution strategies involving binding assays for identification of target interactions, in which techniques such as affinity chromatography and mass spectrometry, and other techniques such as three-hybrid systems, phage, and mRNA display, protein and “reverse transfected” cell microarrays, and biochemical suppression are frequently used. The targets can then be validated by functional studies that use methods such as RNA interference and protein overexpression.
Although drug repurposing has the advantages of saving time and cost (Pushpakom et al., 2018) and therefore a very compelling approach to drug discovery in emergencies as was the case in the early phase of COVID-19 drug discovery, it is not without shortcomings. The shortcomings include intellectual property and economic considerations (legal and commercial barriers); data and compound availability; and fear of exhausting the repurposing space (Talevi and Bellera, 2020). In spite of these challenges, remarkable success stories abound, including repurposed drugs for COVID-19 treatment (Remdesivir has got FDA approval) and some others have been granted emergency use authorization.
Computational methods are today used in both traditional drug discovery processes and drug repurposing and it is extensively discussed in another section of this review. Suffice it to mention here that the most common techniques used in computational methods include target-based, network or pathway-based, knowledge-based, signature-based, and artificial-intelligence-based techniques which rely largely on docking and molecular dynamics simulations.
Apart from drug repurposing, there have also been efforts in developing antibody therapies using blood samples obtained from recovered patients. These involve the isolation and identification of antibodies including neutralizing monoclonal antibodies and nanobodies for the treatment of COVID-19 (Huo et al., 2020; Renn et al., 2020; Adamson et al., 2021).
The trajectories for drug repurposing and traditional drug discovery are essentially the same, differing mainly in speed and cost. While drug repurposing was going on, scientists across the globe were intensifying strategic basic research efforts to accumulate knowledge about SARS-CoV-2 and the pathophysiology of COVID-19 to allow for the commencement of the drug discovery process specifically for COVID-19. Technologies generally involved in drug discovery include various screening techniques such as phenotypic, CRISPR, and target-based screening; structure-based drug discovery; and Biocomputing/bioinformatics. Typically, the drug discovery and development process involve the following key steps: target (a biological molecule, usually a protein but can also be DNA or RNA or gene whose function is to be modulated to bring about a desired or beneficial therapeutic effect to the patient) discovery, identification, and validation; assay (investigative procedures for the qualitative evaluation of the effects of a compound on identified molecular, cellular, or biochemical targets) development; screening to obtain molecules or extracts with a promising activity which are referred to as hits; counter-screening to identify compounds that may interfere with the primary assay used in the primary screen and to eliminate cytotoxic compounds; hit-to-lead and lead optimization to obtain a compound with suitable properties that can be modified to produce a drug candidate. The drug candidate then goes into development and progresses into clinical trials.
Whatever the drug discovery approach used, challenges abound. A major factor militating against anti-SARS-CoV-2 drug discovery is the requirement for Category three containment level which is expensive and difficult to maintain and therefore not within the reach of many scientists, particularly in poor resource settings. Screening of compounds against live SARS-CoV-2 is therefore problematic. The alternatives are to use ex vivo systems such as pseudoviruses and/or perform inhibition studies against SARS-CoV-2 enzymes such as RNA-dependent RNA polymerases (RdRps) and proteases or block critical protein-protein interactions. SARS-CoV-2 is not an exception to drug resistance which is a major problem in antiviral therapy generally. This is particularly so with respect to RNA viruses as a consequence of the error-prone nature of RNA-dependent RNA polymerases (RdRps), the rapid rate of viral replication, and the high frequency of recombination events. The deployment of nanotechnology to formulate multi-component and multi-target nanoparticle cocktails will produce medicines to which resistance will be much less likely.
Natural products are compounds or substances produced by living organisms which include plants, animals, and microorganisms. The use of natural products for the treatment of diseases dates back to prehistoric times. Scientists are digging into nature to harness the “goldmine” of healing molecules to meet various unmet medical treatment needs. Natural products continue to be major sources of prototypes of antimicrobial and antiviral drugs (Adalja and Inglesby, 2019) and represent viable sources of therapeutic alternatives for many diseases (González-Maldonado et al., 2022). It is estimated that over 70% of all existing pharmaceutical products are of natural product origin (Wangchuk, 2018; Abd et al., 2019). In the United States, approximately 118 of the top 150 prescription drugs are based on natural sources (Chen et al., 2016). Several classes of natural compounds such as flavonoids, alkaloids, peptides, and others have been tested against COVID-19 (Antonio et al., 2020; Verma et al., 2020; Antonio et al., 2021; González-Maldonado et al., 2022) with promising outcomes.
Medicinal plants are globally recognized as valuable sources of new medicines with up to 80% of people in developing countries relying on herbal medicines for their primary health care needs. Additionally, more than 25% of medicines prescribed in developed countries are derived from wild plant species (reviewed by Chen et al., 2016). At the dawn of the 21st century, 11% of the 252 drugs considered basic and essential by the WHO were exclusive of flowering plant origin (Veeresham, 2012). Some plants known to have antiviral and immunomodulatory properties include Glycyrrhiza glabra, Azadirachta indica, Andrographis paniculata, Calotropis gigantea, Ocimum sanctum, Curcuma longa, Withania somnifera, Zingiber officinale, Allium sativum, Tinospora cordifolia, Moringa oleifera (Ganjhu et al., 2015; Tiwari et al., 2018). There are a number of plant-specific compounds such as lignans, saponins, alkaloids, flavonoids (kaempferol, luteolin, apigenin, baicalin, quercetin, catechins), and polysulphates (sulphated polysaccharides) that are known to destroy the nucleocapsid and genetic material, inhibit viral entry and inhibit the replication of viruses such as dengue, herpes simplex virus (HSV), hepatitis C virus (HCV), influenza, chikungunya, SARS, etc. (Dhama et al., 2018). Exploiting medicinal plants for the development of COVID-19 therapeutics is therefore a very attractive option.
In the early days of the heat of the COVID-19 pandemic when no specific treatment was available, many countries explored the potential of phytochemicals obtained from medicinal plants and herbs for treating COVID-19 patients (Aanouz et al., 2020; Divya et al., 2020; Jahan and Onay, 2020; Qamar et al., 2020; Xu and Zhang, 2020). Significant progress has already been made in COVID-19 drug discovery from medicinal plants. Polyphenols have been shown to inhibit coronaviruses (Mani et al., 2020) while virtual screening has demonstrated the strong binding potential of absinthin, quercetin 3-glucuronide-7-glucoside, and quercetin 3-vicianoside to SARS-CoV-2 main protease (Mpro) and angiotensin-converting enzyme 2 (Joshi et al., 2020). It is documented in the literature that 83 phytocompounds with anti-COVID-19 activity, the most potent being the alkaloid, lycorine, lignan, savinin, and a total of 39 herbal medicines (listed by WHO and European Medicines agency, EMA) which are indicated for “respiratory diseases” that were evaluated for efficacy as an adjuvant symptomatic therapy for COVID-19 the results of which are shown in Table 4 (Silveira et al., 2020). Thymoquinone obtained from natural Nigella sativa has also been documented to be potentially useful for the treatment of COVID-19 (Abdelrahim et al., 2022). Additionally, several plant-derived bioactive compounds from traditional herbal medicine include andrographolide, panduratin A, baicalein, digoxin, and digitoxin, have demonstrated potent SARS-CoV-2 antiviral activity comparable with some repurposed FDA-approved drugs (Liana and Phanumartwiwath, 2021). Methanolic extract of Stachytarpheta cayennensis remarkably inhibited SARS-CoV-2 entry (González-Maldonado et al., 2022). The Lamiaceae family members, Zingiber officinale, and Glycyrrhiza spp. are known to be particularly good as sources of medicines for the treatment of COVID-19.

TABLE 4. Herbal medicines and efficacy levels as adjuvant symptomatic therapies in early COVID-19 (Silveira et al., 2020).
While the descriptions of the huge potentials of natural products-derived anti-COVID-19 medicines are heartwarming, only Lianhuaqingwen, a Chinese herbal mixture of 11 medicinal species containing 61 compounds (Wang et al., 2016) has recommendation (by the Chinese National Health Commission) for clinical application to treat or manage COVID-19 (Yang et al., 2020). The herbal mixture inhibited SARS-CoV-2 replication in a dose-dependent manner with an IC50 of 411.2 μg/ml. Furthermore, the mixture was able to suppress the release of pro-inflammatory cytokines (TNF-a, IL-6, CCL-2/MCP-1, and CXCL-10/IP-10) in a dose-dependent manner (Runfeng et al., 2020).
Terpenoids, lectins, glycoproteins, lentinan, galactomannan, and polysaccharides from mushrooms (Agaricus subrufescens Peck, Agaricus blazei Murill, Cordyceps sinensis (Berk.) Sacc., Ganoderma lucidum (Curtis.) P. Karst., Grifola frondosa (Dicks.) Gray, Hericium erinaceus (Bull.) Pers., Inonotus obliquus (Arch. Ex Pers.) Pilát., Lentinula edodes (Berk.) Pegler, Pleurotus ostreatus (Jacq.) P. Kumm., Poria cocos F.A. Wolf, and Trametes versicolor (L.) Lloyd are promising prophylactic or therapeutic agents against COVID-19 (Arunachalam et al., 2022). The terpene, erylosides B from Red Sea invertebrate inhibits SARS-CoV-2 main protease (MPro) and identified as a promising anti-COVID-19 drug lead (Ibrahim et al., 2021).
Novavax vaccine consisting of proteins has been created in cultures of Fall armyworm (Spodoptera frugiperda) cells where the cells produce protein spikes that coat SARS-CoV-2 when infected with an engineered virus. The vaccine is achieving high success rates against all main variants of the virus and is entering authorization processes around the world. Peptides from animal venoms (snake, scorpion, frog, insect venoms) are rich sources of antiviral drugs.
Flavonoids, phlorotannins, alkaloids, terpenoids, peptides, lectins, polysaccharides, lipids and others substances of marine origin have been shown to exert desirable effects on coronaviruses penetration and entry into the cell, replication of the viral nucleic acid and release of the virion from the cell also can act on the host's cellular targets (Zaporozhets and Besednova, 2020). Similarly, several classes of compounds from various marine organisms (diverse sponges and algae and bacteria) have been shown to affect various virulence factors of SARS-CoV-2 and also induce the innate immune response and downregulate human ACE-2 (Geahchan et al., 2021).
As good and attractive as drug repurposing and other drug discovery processes are, drug discovery from natural sources is even more attractive and very compelling. This is because of the rich chemical diversity and the huge potential for the discovery of novel chemical scaffolds for discovery of novel drugs. The application of computational methods has made the discovery process much more robust and it is described in the section that follows.
The availability of many untapped floral and faunal bioactive compounds (phytochemicals) in Africa and gradual understanding of the use of bioinformatics tools in drug discovery, drug design, and computer applications in a genetic study is opening a new chapter of scientific understanding (gradual shift) from conventional laboratory investigation to in silico approach to drug discovery and molecular biology. In most African drug research centers, computer-aided drug design (CADD) is now being used to facilitate the process of drug discovery. Though the method is predictable the precision and its efficiency have been ascertained by various researchers. In the last decade, bioinformatics-based models of specific biomolecules have offered rapid and inexpensive methods for the discovery of effective viral therapies. In the presence of a target biomolecule, these models are capable of predicting inhibitor candidates in a structure-based manner. The inhibitors can also be used to predict the specific target (ligand-based) if enough data are presented to a model and it can aid the search for a drug or vaccine candidate by identifying patterns within the data. The availability of enormous data from African bioactive compounds (phytomedicines) provides a baseline for which bioinformatics tools can be used to explore inhibitors of various micro-organisms including viruses like SARS-CoV-2. Database, like phytochemical database, management system has been successfully developed and used in in silico drug design.
The phytomedicines derived from medicinal plants have proven to be a rich source of diverse chemical agents that have been used as drugs and supplements in the millennium (Mahmud et al., 2022). The dependence of Africans on phytomedicines as the first line in the treatment of disease conditions and perhaps the consumption of various vegetables with medicinal properties must have contributed to the low incidence of SARS-CoV-2 in most African countries relative to European countries despite a devastating prediction from World Health Organisation (WHO). It is therefore important to explore the efficacy of different African phytochemicals as inhibitors of SARS-CoV-2 using an already established phytochemical database and bioinformatics tools. In recent times, statistics of newly approved drugs by the United States Food and Drug Administration (FDA) shows that Phytomedicines account for a large number of the approved drugs used as general tonics, antioxidants, cell protectives, and immune stimulants (Kandeel et al., 2020) in the management of SARS-CoV-2, even in this era of combinatorial chemical drugs (Mahmud et al., 2022). Information relating to biological activity, molecular weight, and molecular structure of phytomedicines are deposited in the various databases. Few phytomedicine databases include:
Phytochemdb (Mahmud et al., 2022) is a database that is manually managed and compiles 525 lists of plants and their corresponding 8093 phytochemicals (Mahmud et al., 2022). It is a comprehensive database that gathers most of the information about medicinal plants in one platform, which is considered to be very beneficial to the work of researchers on medicinal plants. “Phytochemdb” is available for free at https://phytochemdb.com/.
This is an evidence-based database for research on natural medicine. It is a cataloged database that provides information on traditional medicinal plants with natural compounds that includes potential bio-target information (Hu et al., 2016). Comprehensive information on a plant-compound-protein relationship can be accessed from the TarNet platform. TarNet is freely available at http://www.herbbol.org:8001/tarnet.
ETM-DB is the largest web-based integrated resource of Ethiopian traditional medicine (Bultum et al., 2019), freely accessible, and provides traditional herbal medicine entities and their relationships in well-structured forms including references (Bultum et al., 2019). The ETM-DB website interface is user-friendly and allows the users to search the entities using various options provided by the search menu. ETM-DB is expected to expedite the process of drug discovery and development and also promote in silico research from Ethiopian natural products leveraging information on the chemical composition and related human target gene/proteins. Phytochemicals from this database can be virtually screened against different targets of SARS-CoV-2. Therefore, this database is key in the discovery of antiviral drugs including RNA viruses such as SARS-Cov-2. The current version of ETM-DB is openly accessible at http://biosoft.kaist.ac.kr/etm.
This is a database of selected highly potent and diverse natural product libraries from African medicinal plants (Ntie-Kang et al., 2013). AfroDb is said to represent the largest ‘‘drug-like’’ and diverse collection of 3D structures of natural products (NPs) covering the geographical region of the entire African continent (Ntie-Kang et al., 2013). The database is readily accessible and can be used in the integrated virtual screening program. The huge information on phytochemicals in this database can be leveraged in the discovery of antiviral drugs. This drug bank could serve as a reservoir of potent molecules active against most viral infections including SARS-CoV-2 and its variants. Since it is possible to predict the variants of SARS-CoV-2 by inducing mutation on SARS-CoV-2 main targets using in silico method, the targets from these variants can be used as reference targets for the virtual screening (target-based virtual screening) or the molecules from the database can be screened against targets (ligand-based virtual screening). AfroDb is therefore a drug bank in which artificial intelligence (bioinformatics tools) can be used to explore huge information on African phytochemicals to aid in drug discovery and drug design, particularly in the pre and post pandemic period.
Computer-aided drug design (CADD) has helped to facilitate the process of drug discovery and development by minimizing the cost and time (Gurung et al., 2021). The availability of a drug data bank (phytochemical database) also enhances the process of drug discovery. In the search for antiviral drugs, two important methods of computer-aided drug design (CADD) is key to the discovery of potent anti Covid-19 drugs (Gurung et al., 2021): the ligand-based and structure-based virtual screening. Molecular docking and molecular dynamic simulation are important techniques in structure-based drug design whereas ligand-based drug design includes pharmacophore modeling, quantitative structure-activity relationships (QSARs), and artificial intelligence (AI) (Gurung et al., 2021). The CADD plays a significant role in the design and discovery of promising drug candidates against various drug targets implicated in the pathogenesis of SARS-CoV-2 (Gurung et al., 2021).
The availability of the three-dimensional crystal structure of the therapeutic target proteins and exploration of the binding site or active site residues forms the basis of structure-based drug design (SBDD) (Batool et al., 2019). This approach is highly selective and effectively fast in the identification of lead molecules and their optimization which has led to a better understanding of diseases at a molecular level (Lionta et al., 2014). Some of the common methods used in SBDD include structure-based virtual screening (SBVS), molecular docking, and molecular dynamics (MD) simulations (Gurung et al., 2021). A lot of information can be extracted from these methods, some of which include assessment of binding energetics, principal component analysis (PCA), dynamics cross-correlation, protein-ligand interactions, and conformational changes in the receptor upon ligand binding (Batool et al., 2019). SBDD is a computational technique has greatly helped in the discovery of several drugs available on the market today. For example, the discovery of Amprenavir as a potential inhibitor of the human immunodeficiency virus (HIV) protease using the crystallized protein model and molecular dynamics (MD) simulations (Adamson et al., 2009; Liao and Nicklaus, 2010). Others include thymidylate synthase inhibitor, an anticancer agent, and Raltitrexed implicated in the treatment of HIV-infected cancer patients using the SBDD approach (Anderson, 2003; Medina-Moreno et al., 2019). Norfloxacin a topoisomerases II and IV inhibitor was also discovered through SBVS (Batool et al., 2019). In recent time, SBVS conducted using different protein targets of SARS-CoV-2 and phytochemical database revealed high binding energy and very good interaction with the residues of the active site target protein such as 3-Chymotrypsin-like proteases (3CLpro) of SARS-CoV-2 (Gyebi et al., 2021). Some of the phytochemical agents that show good binding energy and ligand-residue interaction are shown in Table 5.

TABLE 5. Binding energies of some phytochemicals docked in the active sites of 3-chymotrypsin-like proteases of coronaviruses (Gyebi et al., 2021).
These plants, native to most of the African countries, demonstrate the reservoir of phytochemicals in Africa with potent inhibitory activity against target proteins in SARS-CoV-2. The basic steps involved in Covid-19 drug discovery using bioinformatics tools (SBDD) consist of the preparation of target structure, identification of the ligand-binding site, compound library preparation, molecular docking and scoring functions, molecular dynamic simulation, and binding free energy calculation (Gurung et al., 2021).
Ligand-based drug design is a computer-aided drug design technique that is widely used in drug discovery and design. It is employed when the three-dimensional structure of the target receptor is not available. The information obtained from a set of active compounds against a specific target receptor is useful in the identification of physicochemical and structural properties that are responsible for the specific biological activity which is based on the structure-activity relationship (Prathipati et al., 2007; Gurung et al., 2021). The commonest techniques used in the ligand-based virtual screening approach include pharmacophore modeling, quantitative structure-activity relationships (QSARs), and artificial intelligence (AI).
Artificial intelligence (AI) is a machine learning intelligence that depends on the ability of computers to learn from existing data (Gurung et al., 2021). AI has been used in various computational modeling methods to predict the biological activities and toxicity profiles of drug molecules (Patel et al., 2014). In addition, AI has also been widely used in the prediction of protein folding, protein-protein interaction, virtual screening, Quantitative Structural Activity Relationship (QSAR), evaluation of absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of the drug molecule, and de novo drug design (Gurung et al., 2021).
There are two major methods of AI that are widely used in rational drug design: machine learning (ML) and deep learning (DL) (Cortes and Vapnik, 1995). A support vector machine (SVM) is an ML algorithm that has been extensively used in drug discovery (Cortes and Vapnik, 1995). Others include Random Forest (RF) (Breiman, 2001) and Naive Bayesian (NB) (Sammut and Webb, 2017). Deep learning methods include convolutional neural network (CNN), deep neural network (DNN), recurrent neural network (RNN), autoencoder, and restricted Boltzmann machine (RBN) (Gurung et al., 2021). In summary, artificial intelligence has been widely used in drug discovery and vaccine development (Arshadi et al., 2020). As has been documented (Keshavarzi et al., 2022), this advancement in therapeutics research is critical for the current situation of pandemic and urgent need for the SARS-COV-2 therapy discovery for the following reasons (1) deep learning has an automatic feature extraction ability that can support models with better accuracy and deliver good reliable results (2) deep learning models demonstrate the generative ability that can be utilized to create more druggable molecules and better epitope prediction, (3) reduced chances of failure in the drug pipeline, and (4) because of the novelty of the virus the data related to its therapies remain scarce, which is a suitable scenario for knowledge transfer while leveraging on the learned knowledge from previous tasks (e.g., TranscreenTM) (Salem et al., 2020). Transfer learning is very effective in the transferring of learned knowledge and parameters from a secondary task with big data available to the task at hand (Weiss et al., 2016). Therefore, the use of deep learning in the discovery of therapies for SARS-COV-2 is necessary for a timely and accurate response to the viral pandemic (Arshadi et al., 2020).
With the increase in the number of confirmed positive and death cases from SARS-CoV-2 infection with related evidence of viral mutation, computer-aided drug design (CADD) is the most viable and reliable technique in drug and medicinal research because of its attributed time saving and cost reduction in the design of therapeutic agents (Gurung et al., 2021; Ojha et al., 2021). In addition, the high impact of the pandemic resulting from COVID-19 infection and the relative lack of approved drugs create room for drugs to be repurposed within a short period of time. The CADD enhances this method by facilitating the discovery of new drugs or repurposing FDA-approved drugs whose safety and adverse effects are already known (Basak et al., 2021; Gurung et al., 2021). Since SARS-CoV-2, an RNA virus, poses a high mutation rate, the genome may hinder disease prevention and treatment (Gurung et al., 2021). CADD can be used efficiently to induce mutation on the existing targets, allowing for in silico prediction of the possible SARS-CoV-2 variant and subsequently developing potent molecules against these variants that are likely to cause a future pandemic (Gurung et al., 2021; Sharma et al., 2021). This is one of the major advantages (strength) of CADD in the discovery of COVID-19 drugs. Therefore, CADD is an asset in the drug discovery and development process. However, the CADD method is limited by the inability to validate its lead compound through clinical trials before market approval (Ojha et al., 2021). The molecular understanding of the disease pathogenesis of COVID-19 are still been strengthened, and the existence of the limited data on variants of COVID-19 can have a major impact on the precision and accuracy of CADD methods such as artificial intelligence (Ojha et al., 2021).
In spite of the odds, tremendous opportunities exist to overcome the challenges by integrating advances in science and technology such as Genomics (including CRISPR-Cas system) Phytopharming/Molecular farming, Biocomputing, and nanomedicine to develop novel and innovative antiviral therapies with broad-spectrum activities against human pathogenic viruses of the present and those that may emerge or re-emerge in the future.
Phytopharming or plant molecular farming is the production of biopharmaceuticals in plants using the tools of plant metabolic engineering and plant biotechnology. It is intended to overcome the limitations of high operating cost, prolonged production time, low yield, chances of contamination with pathogenic microorganisms, and limited posttranslational modifications of the current platforms that use bacterial systems, microbial eukaryotes, insect cells, and mammalian cells (Shanmugaraj et al., 2020). The use of plants as a platform for the production of diagnostic reagents and pharmaceutical proteins has been around for well over 30 years (Schillberg et al., 2019; Fischer and Buyel, 2020). The Israeli biotech company Protalix and Pfizer got FDA approval for the first drug, Elelyso (taliglucerase alfa), developed in plant cells (cells from carrot) in 2012. In February 2022, the Canadian biotech company, Medicago, got approval for its two-dose COVID-19 vaccine, the first world’s plant-based COVID-19 vaccine (The Pharmaeutical Journal, 2022). The rapidity with which this feat was achieved is outstanding, taking just over 2 weeks. This is what the executive vice president of Medicago, Marc-André D’Aoust said. “From the moment we had the sequence on our computer, to the moment we [had the] first purified product, it took 19 days”. Comparing this speed with five to 6 months for a conventional egg-grown vaccine, the plant-based approach will be of great advantage in potential future pandemics. This is particularly useful in poor and developing countries where the cost of the infrastructure of conventional platforms for the production of biopharmaceuticals is out of reach. Living plants have therefore effectively become bioreactors for the production of biopharmaceuticals. It may well be that plants may offer the only platform that can be used to manufacture COVID-19 diagnostic reagents and therapeutics at scale in a timeframe of weeks, compared with months or even years for cell-based systems (Capell et al., 2020). Lico and other colleagues in Italy made a case for molecular farming to complement conventional methods of production for the rapid and scalable supply of protein antigens as reagents and vaccine candidates, antibodies for virus detection and passive immunotherapy, other therapeutic proteins, and virus-like particles as novel vaccine platforms to meet the urgent therapeutic needs imposed by COVID-19 (Lico et al., 2020).
The concept is not different from molecular farming except that in this case the vaccine is produced in edible crops such as tomatoes, banana, potatoes, rice, carrot, corn, cucumber, lettuce, and spinach. Tomato has the advantages of having excellent biomass, being easy to transform and the whole plant can be generated within a short period. Tomato has therefore been described as a green vaccine factory (Sohrab, 2020). Sohrab and other colleagues listed the advantages of plant-based vaccines to include: oral use as fruits and vegetables; obtainable as capsules from dried leaf tissue powder; no requirement for adjuvants to enhance immune responses; mucosal immunity elicited by orally-introduced antigens; bulk production on site is easy and can be transported and stored at less cost and without cold chain requirement; not administered by injection and therefore no need for a trained medical person; easy to express, separate and purify; they can be stored as seeds and oils and dried tissue without any refrigeration; They do not have any risk of microbial contamination and disease spread; There is the possibility of enhanced compliance, especially in children (Sohrab et al., 2017). Significant progress has been made as scientists from the Centre for Genomics and Bioinformatics of the Academy of Sciences at Uzbekistan have developed a tomato-based vaccine against COVID-19 as of 2021. Seedlings in the laboratory are grown in the form of a vaccine from seedlings in the Centre after 2 months, and people who eat these tomatoes are expected to produce antibodies against the virus (Abdulkerimov, 2021).
Nanomedicine is a branch of medicine that applies nanotechnology, which is essentially the manipulation and manufacture of materials and devices that are in the size range of 1–100 nm, to the diagnosis, prevention, treatment, and monitoring (follow-ups) of diseases and also for medical imaging and the repair and regeneration of biological systems (regenerative medicine). It uses the properties developed by a material at its nanometric scale which is different from those of the same material at a bigger scale in terms of physics, chemis,try or biology to achieve the many intended aims of nanomedicine applications. At the nano-scale, the surface-to-volume ratio is such that the surface properties become an intrinsic parameter of the potential actions of a particle or material. Therefore, coating the nanoparticles and functionalizing their surfaces enhance the biocompatibility of the particle and its circulation time in the blood, as well as ensuring a highly selective binding to the target of choice. The design and development of nanoparticles containing drug(s) (nanoparticulate nanomedicines) for drug delivery is an aspect of nanomedicine that has attracted tremendous attention. Nanoparticulate nanomedicines are designed and developed to deliver drugs through various mechanisms such as solubilization, passive targeting, active targeting, and triggered release which will ultimately increase therapeutic efficacy, decrease the therapeutically effective dose, and/or reduce the risk of systemic side effects (Hua et al., 2018). The nanomaterials of choice will include, but not limited to, liposomes (Figure 5), solid lipid nanoparticles (SLNs), nanostructured lipid carriers (NLCs), and polymeric nanoparticles. As of 2012, the global nanomedicine market stood at about 78 billion dollars (Viseu, 2022). In 2022, this stands at 159.43 billion dollars and this is expected to rise to $291.15 billion by 2027 (Market Data Forcast, 2022). This means that the market outlook for investors is bright and attractive and funding may not be a problem. Moreover, any investment in health is an economic investment because of the intricate relationship between health and the national economy.
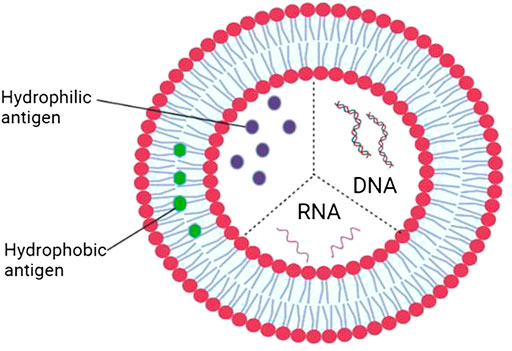
FIGURE 5. The application of nanotechnology in multivalent vaccine devolopment using liposome as the encapsulating biomaterial.
Development of nanomedicine-based therapeutics against a variety of viral infections, including hepatitis B virus, human immunodeficiency virus, respiratory syncytial virus, influenza virus, and coronaviruses are already known. The attraction for the use of nanoparticles in nanomedicine is in their unique physicochemical properties: the small size which is also the scale of many biological mechanisms in the human body allows them to easily cross intracellular barriers/membranes to access new sites of delivery and interact with a variety of biological components of similar size, interacting with DNA or small proteins at different levels, in blood or within organs, tissues or cells; the surface polarity which can be modified by various functional groups to increase their binding efficacy and stability and reduce aggregation and precipitation (Dutta, 2022, ETPN, 2022).
Metallic nanoparticles such as iron-oxide, copper-oxide, and silver nanoparticles with antiviral properties can be used to entrap and inactivate SARS-CoV-2. These particles disrupt the cell membrane, damage proteins, and DNA, form free radicals, inhibit biofilm formation, and/or exert heavy metal toxicity in viruses thereby destroying the viruses. Iron-oxide nanoparticles have been shown to interact with the spike protein of SARS-CoV-2, altering its structural conformation. So have lipid nanoparticles loaded with SARS-CoV-2-specific small-interfering RNAs (siRNAs) for targeted delivery to the lungs to suppress viral replication and prevent the establishment of infection and disease. Poly lactic-co-glycolic acid polymer-based nanosponge coated with human lung epithelial cell and macrophage membrane to mimic the cellular physiology required for SARS-CoV-2 host cell entry is a good target for SARS-CoV-2. The binding of SARS-CoV-2 to this synthetic cellular nanosponge leads to the neutralization of the virus and infection of cells is blocked.
Apart from targeting SARS-CoV-2 directly, nanoparticulate nanomedicines can also be used to modulate the immune system in such a way that hyperinflammation and the consequent cytokine storm in COVID-19 patients can be prevented. Graphene-oxide nanoparticles are known to increase the levels of macrophages and T cells thereby enhancing adaptive immune response and viral clearance. Similarly nano-diamonds induce anti-inflammatory macrophages to reduce hyperinflammation. In the same way, carbon and graphene sheets can be modified to eliminate cytokines and interleukins (pro-inflammatory mediators) from the blood (reviewed by Dutta, 2022).
Natural products represent a large reservoir of chemicals from which new chemical scaffolds can be obtained for the discovery of novel drugs, not only for COVID-19 but for other infectious diseases, including emerging and re-emerging diseases. Monotherapy in which one drug to one target approach is used is no longer a viable option. A strategic therapeutic approach in which a multicomponent and multi-target pan-viral therapy is developed for the treatment of COVID-19 and associated complications and future viral pandemics is a very compelling option and the drug discovery and development community should embrace it. Nanomedicine presents an excellent platform to achieve this as different drugs each with different targets can be loaded into a drugs-nanoparticle complex formulation as shown in Figure 5. This is also useful for the development of multivalent vaccines for activity against existing human pathogenic viruses and those that may emerge or re-emerge in the future. Plant molecular farming is undoubtedly a very useful technology for relatively cheap, fast, and large-scale production of biopharmaceuticals and relevant stakeholders should ensure that the required infrastructure for this is put in place in countries across the world for pandemic preparedness. After the “hype” about edible vaccines in the 90s, it is now becoming a reality and should be vigorously pursued for COVID-19 vaccine production particularly in poor resource settings, and quick response in the future should there be an outbreak of a pandemic. Edible vaccines will be an important strategy to significantly reduce vaccine hesitancy. However, worries about GMO foods are a potential problem but strong advocacy may be able to remove or significantly reduce these worries. Finally, it is important to sustain and improve on the coordinated global effort by all stakeholders in the development of COVID-19 therapeutics so that the several decades of accumulated knowledge from previous epidemics and pandemics can be effectively combined with the tremendous advances in science and technology to develop therapeutics of the present and the future.
NU wrote the Biocomputing and Bioinformatics section while EO wrote the other portions of the manuscript.
Work in EO’s laboratory is supported by the West African Network for Infectious Diseases ACEs (WANIDA) and the Africa Centre of Excellence for Mycotoxin and Food Safety (ACEMFS).
The authors are grateful to Israel Ogbadoyi for his assistance with the graphics.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aanouz, I., Belhassan, A., El-Khatabi, K., Lakhlifi, T., El-Ldrissi, M., and Bouachrine, M. (2020). Moroccan Medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. J. Biomol. Struct. Dyn. 39, 2971–2979. doi:10.1080/07391102.2020.1758790
Abd, E. N. H., Abd El-Aziz, F. A., Abouelmagd, S. A., Abd El-Hamid, B. N., and Hetta, H. F. (2019). Spidroin in carbopol-based gel promotes wound healing in earthworm's skin model. Drug Dev. Res. 80, 1051–1061. doi:10.1002/ddr.21583
Abdelrahim, M., Esmail, A., Al Saadi, N., Zsigmond, E., Al Najjar, E., Bugazia, D., et al. (2022). Thymoquinone’s antiviral effects: It is time to be proven in the covid-19 pandemic era and its omicron variant surge. Front. Pharmacol. 13, 848676. doi:10.3389/fphar.2022.848676
Abdulkerimov, B. (2021). Uzbek Scientists working on edible vaccine against COVID-19. Available at: https://www.aa.com.tr/en/asia-pacific/uzbek-scientists-working-on-edible-vaccine-against-covid-19/2348236 (accessed 20 April 2022).
Adalja, A., and Inglesby, T. (2019). Broad-spectrum antiviral agents: a crucial pandemic tool. Expert Rev. anti. Infect. Ther. 17, 467–470. doi:10.1080/14787210.2019.1635009
Adamson, C. S., Chibale, K., Goss, R. J. M., Jaspars, M., Newman, D. J., and Dorrington, R. A. (2021). Antiviral drug discovery: preparing for the next pandemic. Chem. Soc. Rev. 50, 3647–3655. doi:10.1039/d0cs01118e
Adamson, C. S., Salzwedel, K., and Freed, E. O. (2009). Virus maturation as a new HIV-1 therapeutic target. Expert Opin. Ther. Targets 13, 895–908. doi:10.1517/14728220903039714
Anasir, M. I., and Poh, C. L. (2019). Structural vaccinology for viral vaccine design. Front. Microbiol. 10, 738. doi:10.3389/fmicb.2019.00738
Anderson, A. C. (2003). The process of structure-based drug design. Chem. Biol. 10, 787–797. doi:10.1016/j.chembiol.2003.09.002
Antonio, A. S., Wiedemann, L., and Veiga-Junior, V. (2020). Natural products' role against COVID-19. RSC Adv. 10, 23379–23393. doi:10.1039/d0ra03774e
Antonio, A. S., Wiedemann, L. S. M., Galante, E. B. F., Guimar∼aes, A. C., Matharu, A. S., Veiga-Junior, V. F., et al. (2021). Efficacy and sustainability of natural products in COVID-19 treatment development: opportunities and challenges in using agro-industrial waste from citrus and apple. Heliyon 7, e07816. doi:10.1016/j.heliyon.2021.e07816
Arshadi, A. K., Webb, J., Salem, M., Cruz, E., Calad-Thomson, S., Ghadirian, N., et al. (2020). Artificial intelligence for COVID-19 drug discovery and vaccine development. Front. Artif. Intell. 3, 65. doi:10.3389/frai.2020.00065
Arunachalam, K., Sasidharan, S. P., and Yang, X. (2022). A concise review of mushrooms antiviral and immunomodulatory properties that may combat against COVID-19. Food Chem. Adv. 1, 100023.doi:10.1016/j.focha.2022.100023
Batool, M., Ahmad, B., and Choi, S. (2019). A structure-based drug discovery paradigm. Int. J. Mol. Sci. 20, 2783. doi:10.3390/ijms20112783
Bultum, L. E., Woyessa, A. M., and Lee, D. (2019). ETM-DB: Integrated Ethiopian traditional herbal medicine and phytochemicals database. BMC Complement. Altern. Med. 19, 212. doi:10.1186/s12906-019-2634-1
Capell, T., Twyman, R. M., Armario-Najera, V., Ma, J. K.-C., Schillberg, S., Christou, P., et al. (2020). Potential applications of plant biotechnology against SARS-CoV-2. Trends Plant Sci. 25 (7), 635–643. doi:10.1016/j.tplants.2020.04.009
Chen, S-L., Yu, H., Luo, H-M., Wu, Q., Li, C-F., Steinmetz, A., et al. (2016). Conservation and sustainable use of medicinal plants: problems, progress, and prospects. Chin. Med. 11, 37. doi:10.1186/s13020-016-0108-7
Cortes, C., and Vapnik, V. (1995). Support-vector networks. Mach. Learn. 20, 273–297. doi:10.1007/BF00994018
Dhama, K., Karthik, K., Khandia, R., Munjal, A., Tiwari, R., Rana, R., et al. (2018). Medicinal and therapeutic potential of herbs and plant metabolites/extracts countering viral pathogens—current knowledge and future prospects. Curr. Drug Metab. 19, 236–263. doi:10.2174/1389200219666180129145252
Dhaneshwar, S., and Bhasin, B. (2021). Drug repurposing: An emerging tool for drug reuse, recycling and discovery. Curr. Drug Res. Rev. 13, 101–119. doi:10.2174/2589977513666210211163711
Divya, M., Vijayakumar, S., Chen, J., Vaseeharan, B., and Durán-Lara, E. F. (2020). South Indian medicinal plant has the ability to combat against deadly viruses along with COVID-19? Microb. Pathog. 148, 104277.doi:10.1016/j.micpath.2020.104277
Dutta, S. S. (2022). Nanomedicine and COVID-19 news-medical. Available at: https://www.news-medical.net/health/Nanomedicine-and-COVID-19.aspx. (Accessed April 20, 2022)
Fears, J. R. (2004). The plague under Marcus Aurelius and the decline and fall of the Roman empire. Infect. Dis. Clin. North Am. 18 (1), 65–77. doi:10.1016/S0891-5520(03)00089-8
Fischer, R., and Buyel, J. F. (2020). Molecular farming – the slope of enlightenment. Biotechnol. Adv. 40, 107519. doi:10.1016/j.biotechadv.2020.107519
Ganjhu, R. K., Mudgal, P. P., Maity, H., Dowarha, D., Devadiga, S., Nag, S., et al. (2015). Herbal plants and plant preparations as remedial approach for viral diseases. Virusdisease 26, 225–236. doi:10.1007/s13337-015-0276-6
Geahchan, S., Ehrlich, H., and Rahman, M. A. (2021). The anti-viral applications of marine resources for COVID-19 treatment: An overview. Mar. Drugs 19, 409. doi:10.3390/md19080409
González-Maldonado, P., Alvarenga, N., Burgos-Edwards, A., Flores-Giubi, M. E., Barúa, J. E., Romero-Rodríguez, M. C., et al. (2022). Screening of natural products inhibitors of SARS-CoV-2 entry. Molecules 27, 1743. doi:10.3390/molecules27051743
Gurung, A. B., Ali, M. A., Lee, J., Farah, M. A., and Al-Anazi, K. M. (2021). An updated review of computer-aided drug design and its application to COVID-19. Biomed. Res. Int. 2021, 8853056. doi:10.1155/2021/8853056
Gyebi, G. A., Elfiky, A. A., Ogunyemi, O. M., Ibrahim, I. M., Adegunloye, A. P., Adebayo, J. O., et al. (2021). Structure-based virtual screening suggests inhibitors of 3-Chymotrypsin-Like Protease of SARS-CoV-2 from Vernonia amygdalina and Occinum gratissimum. Comput. Biol. Med. 136, 104671. doi:10.1016/j.compbiomed.2021.104671
Horton, Richard (2020). After COVID-19—Is an “alternate society” possible? Lancet 395 (10238), 1682. doi:10.1016/S0140-6736(20)31241-1
Hu, R., Ren, G., Sun, G., and Sun, X. (2016). TarNet: An evidence-based database for natural medicine research. PLoS One 11, e0157222. doi:10.1371/journal.pone.0157222
Hua, S., de Matos, M. B. C., Metselaar, J. M., and Storm, G. (2018). Current trends and challenges in the clinical translation of nanoparticulate nanomedicines: Pathways for translational development and commercialization. Front. Pharmacol. 9, 790. doi:10.3389/fphar.2018.00790
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. doi:10.1016/S0140-6736(20)30183-5
Huo, J., Bas, A. L., Ruza, R. R., Duyvesteyn, H. M. E., Mikolajek, H., Malinauskas, T., et al. (2020). Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 27, 846–854. doi:10.1038/s41594-020-0469-6
Ibrahim, M. A. A., Abdelrahman, A. H. M., Mohamed, T. A., Atia, M. A. M., Al-Hammady, M. A. M., Abdeljawaad, K. A. A., et al. (2021). Silico Mining of Terpenes from Red-Sea Invertebrates for SARS-CoV-2, 26, 2082. doi:10.3390/molecules26072082Main protease (mpro) inhibitorsMolecules
Jahan, I., and Onay, A. (2020). Potentials of plant-based substance to inhabit and probable cure for the COVID-19. Turk. J. Biol. 44, 228–241. doi:10.3906/biy-2005-114
Jang, W. D., Jeon, S., Kim, S., and Lee, S. Y. (2021). Drugs repurposed for COVID-19 by virtual screening of 6218 drugs and cell-based assay. Proc. Natl. Acad. Sci. U. S. A. 118, e2024302118. doi:10.1073/pnas.2024302118
Jarada, T. N., Rokne, J. G., and Alhajj, R. (2020). A review of computational drug repositioning: Strategies, approaches, opportunities, challenges, and directions. J. Cheminform. 12, 46. doi:10.1186/s13321-020-00450-7
Joshi, T., Joshi, T., Sharma, P., Mathpal, S., Pundir, H., Bhatt, V., et al. (2020). In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur. Rev. Med. Pharmacol. Sci. 24, 4529–4536. doi:10.26355/eurrev_202004_21036
Kandeel, M., Kitade, Y., and Almubarak, A. (2020). Repurposing FDA-approved phytomedicines, natural products, antivirals and cell protectives against SARS-CoV-2 (COVID-19) RNA-dependent RNA polymerase. PeerJ 8, e10480. doi:10.7717/peerj.10480
Khataniar, A., Pathak, U., Rajkhowa, S., and Jha, A. N. (2022). A comprehensive review of drug repurposing strategies against known drug targets of COVID-19. COVID 2, 148–167. doi:10.3390/covid2020011
Li, G., and De Clercq, E. (2020). Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 19, 149. doi:10.1038/d41573-020-00016-0
Liana, D., and Phanumartwiwath, A. (2021). Leveraging knowledge of Asian herbal medicine and its active compounds as COVID-19 treatment and prevention. J. Nat. Med. 76, 20–37. doi:10.1007/s11418-021-01575-1
Liao, C., and Nicklaus, M. C. (2010). Computer tools in the discovery of HIV-1 integrase inhibitors. Future Med. Chem. 2, 1123–1140. doi:10.4155/fmc.10.193
Lico, C., Santi, L., Baschieri, S., Noris, E., Marusic, C., Donini, M., et al. (2020). Plant molecular farming as a strategy against COVID-19 – the Italian perspective. Front. Plant Sci. 11, 609910. doi:10.3389/fpls.2020.609910
Lionta, E., Spyrou, G., Vassilatis, D., and Cournia, Z. (2014). Structure-based virtual screening for drug discovery: Principles, applications and recent advances. Curr. Top. Med. Chem. 14, 1923–1938. doi:10.2174/1568026614666140929124445
Lipinski, C. (2011). “Drug repurposing,” in Drug discovery today: Therapeutic strategies (Amsterdam, Netherlands: Elsevier).
Liu, Y-C., Kuo, R-L., and Shih, S-R. (2020). COVID-19: The first documented coronavirus pandemic in history. Biomed. J. 43, 328–333. doi:10.1016/j.bj.2020.04.007
MacDonald, N. E. (2015). Vaccine hesitancy: Definition, scope and determinants. Vaccine 33 (34), 4161–4164. doi:10.1016/j.vaccine.2015.04.036
Mahmud, S., Paul, G. K., Biswas, S., Kazi, T., Mahbub, S., Mita, M. A., et al. (2022). Phytochemdb: a platform for virtual screening and computer-aided drug designing. Database. 2022, baac002. doi:10.1093/database/baac002
Mani, J. S., Johnson, J. B., Steel, J. C., Broszczak, D. A., Neilsen, P. M., Walsh, K. B., et al. (2020). Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 284, 197989. doi:10.1016/j.virusres.2020.197989
Market Data Forecast (2022). Nanomedicine market. Available at: https://www.marketdataforecast.com/market-reports/nanomedicine-market (accessed 18 April 2022).
Medina-Moreno, S., Zapata, J. C., Cottrell, M. L., Le, N. M., Tao, S., Bryant, J., et al. (2019). Disparate effects of cytotoxic chemotherapy on the antiviral activity of antiretroviral therapy: Implications for treatments of HIV-infected cancer patients. Antivir. Ther. 24, 177–186. doi:10.3851/IMP3285
Naasani, I. (2021). COMPARE analysis, a bioinformatic approach to accelerate drug repurposing against covid-19 and other emerging epidemics. SLAS Discov. 26, 345–351. doi:10.1177/2472555220975672
Nawaz, S. (2021). COVID-19, SARS-CoV-2, origin, transmission and treatment aspects, a brief review. Infect. Disord. Drug Targets 21, e270421186673. doi:10.2174/1871526520666201006163641
Ng, Y. L., Salim, C. K., and Chu, J. J. H. (2021). Drug repurposing for COVID-19: Approaches, challenges and promising candidates. Pharmacol. Ther. 228, 107930. doi:10.1016/j.pharmthera.2021.107930
Ntie-Kang, F., Zofou, D., Babiaka, S. B., Meudom, R., Scharfe, M., Lifongo, L. L., et al. (2013). AfroDb: a select highly potent and diverse natural product library from african medicinal plants. PLoS One 8, e78085. doi:10.1371/journal.pone.0078085
Patel, H. M., Noolvi, M. N., Sharma, P., Jaiswal, V., Bansal, S., Lohan, S., et al. (2014). Quantitative structure-activity relationship (QSAR) studies as strategic approach in drug discovery. Med. Chem. Res. 23, 4991–5007. doi:10.1007/s00044-014-1072-3
Prathipati, P., Dixit, A., and Saxena, A. (2007). Computer-aided drug design: Integration of structure-based and ligand-based approaches in drug design. Curr. Comput. Aided-Drug Des. 3, 133–148. doi:10.2174/157340907780809516
Pushpakom, S., Iorio, F., Eyers, P. A., Escott, K. J., Hopper, S., Wells, A., et al. (2018). Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 18, 41–58. doi:10.1038/nrd.2018.168
Rappuolia, R., De Gregorioa, E., Del Giudicea, G., Phogata, S., Pecettaa, S., Pizzaa, M., et al. (2021). Vaccinology in the post-COVID-19 era. Proc. Natl. Acad. Sci. U. S. A. 118 (3), e2020368118. doi:10.1073/pnas.2020368118
Renn, A., Fu, Y., Hu, X., Hall, M. D., and Simeonov, A. (2020). Fruitful neutralizing antibody pipeline brings hope to defeat SARS-cov-2. Trends Pharmacol. Sci. 41, 815–829. doi:10.1016/j.tips.2020.07.004
Runfeng, L., Yunlong, H., Jicheng, H., Weiqi, P., Qinhai, M., Yongxia, S., et al. (2020). Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol. Res. 156, 104761. doi:10.1016/j.phrs.2020.104761
Salem, M., Khormali, A., Arshadi, A. K., Webb, J., and Yuan, J. S. (2020). TranScreen: Transfer learning on graph-based anti-cancer virtual screening model. Big Data Cogn. Comput. 4, 16. doi:10.3390/bdcc4030016
Sammut, C., and Webb, G. I. (2017). Encyclopedia of machine learning and data mining. New York, NY: Springer.
Savi, C. D., Hughes, D. L., and Kvaerno, L. (2020). Quest for a COVID-19 cure by repurposing small-molecule drugs: Mechanism of action, clinical development, synthesis at scale, and outlook for supply. Org. Process Res. Dev. 24, 940–976. doi:10.1021/acs.oprd.0c00233
Scheres, J., and Kuszewski, K. (2019). The Ten Threats to Global Health in 2018 and 2019. A welcome and informative communication of WHO to everybody. Zesz. Nauk. Ochr. Zdrowia. Zdr. Publiczne i Zarzadzanie 17 (1), 2–8. doi:10.4467/20842627oz.19.001.11297
Schillberg, S., Raven, N., Spiegel, H., Rasche, S., and Buntru, M. (2019). Critical analysis of the commercial potential of plants for the production of recombinant Proteins.of plants for the production of recombinant proteins. Front. Plant Sci. 10, 720. doi:10.3389/fpls.2019.00720
Senanayake, S. L. (2020). Drug repurposing strategies for COVID-19. Future Drug Discov. 2, 2. doi:10.4155/fdd-2020-0010
Shanmugaraj, B., Bulaon, C. J. I., and Phoolcharoen, W. (2020). Plant molecular farming: A viable platform for recombinant biopharmaceutical production. Plants 9, 842. doi:10.3390/plants9070842
Sharma, T., Abohashrh, M., Baig, M. H., Dong, J. J., Alam, M. M., Ahmad, I., et al. (2021). Screening of drug databank against WT and mutant main protease of SARS-CoV-2: Towards finding potential compound for repurposing against COVID-19. Saudi J. Biol. Sci. 28, 3152–3159. doi:10.1016/j.sjbs.2021.02.059
Silveira, D., Prieto-Garcia, J. M., Boylan, F., Estrada, O., Fonseca-Bazzo, Y. M., Jamal, C. M., et al. (2020). COVID-19: Is there evidence for the use of herbal medicines as adjuvant symptomatic therapy? Front. Pharmacol. 11, 581840. doi:10.3389/fphar.2020.581840
Sohrab, S. S. (2020). An edible vaccine development for coronavirus disease 2019: the concept. Clin. Exp. Vaccine Res. 9, 164–168. doi:10.7774/cevr.2020.9.2.164
Sohrab, S. S., Suhail, M., Kamal, M. A., Husen, A., and Azhar, E. I. (2017). Recent development and future prospects of plant-based vaccines. Curr. Drug Metab. 18, 831–841. doi:10.2174/1389200218666170711121810
Talevi, A., and Bellera, C. L. (2020). Challenges and opportunities with drug repurposing: Finding strategies to find alternative uses of therapeutics. Expert Opin. Drug Discov. 15, 397–401. doi:10.1080/17460441.2020.1704729
The Pharmaceutical Journal (2022). Going plant-based: how molecular farming could revolutionise drug development. Pharm. J. 308 (7959), 1–8. doi:10.1211/PJ.2022.1.132683
Tiwari, R., Latheef, S. K., Ahmed, I., Iqbal, H., Bule, M. H., Dhama, K., et al. (2018). Herbal immunomodulators—a remedial panacea for designing and developing effective drugs and medicines: Current scenario and future prospects. Curr. Drug Metab. 19, 264–301. doi:10.2174/1389200219666180129125436
Veeresham, C. (2012). Natural products derived from plants as a source of drugs. J. Adv. Pharm. Technol. Res. 3, 200–201. doi:10.4103/2231-4040.104709
Verma, S., Twilley, D., Esmear, T., Oosthuizen, C. B., Reid, A-M., Nel, M., et al. (2020). Anti-SARS-CoV natural products with the potential to inhibit SARS-CoV-2 (COVID-19). Front. Pharmacol. 11, 561334. doi:10.3389/fphar.2020.561334
Viseu, A. (2022). Nanomedicine. Available at: https://www.britannica.com/science/nanomedicine (Accessed 16 April 2022).
Wang, C. H., Zhong, Y., Zhang, Y., Liu, J. P., Wang, Y. F., Jia, W. N., et al. (2016). A network analysis of the Chinese medicine Lianhua-Qingwen formula to identify its main effective components. network analysis of the Chinese medicine Lianhua-Qingwen formula to identify its main effective components. Mol. Biosyst. 12 (2), 606–613. doi:10.1039/C5MB00448A
Wangchuk, P. (2018). Therapeutic applications of natural products in herbal medicines, biodiscovery programs, and biomedicine. J. Biol. Act. Prod. Nat. 8 (1), 1–20. doi:10.1080/22311866.2018.1426495
Weiss, K., Khoshgoftaar, T. M., and Wang, D. D. (2016). A survey of transfer learning. J. Big Data 3, 9. doi:10.1186/s40537-016-0043-6
World Health Organization (2021). HIV/AIDS, key facts. Available at: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (accessed April 12, 2022).
World Health Organization (2020). “Solidarity” clinical trial for COVID-19 treatments. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments (accessed 13 April 2022).
Xu, J., and Zhang, Y. (2020). Traditional Chinese medicine treatment of COVID-19. Complement. Ther. Clin. Pract. 39, 101165. doi:10.1016/j.ctcp.2020.101165
Yang, H. T., and Rao, Z. H. (2021). Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 19, 685–700. doi:10.1038/s41579-021-00630-8
Yang, Y., Islam, M. S., Wang, J., Li, Y., and Chen, X. (2020). Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): A review and perspective. Int. J. Biol. Sci. 16 (10), 1708–1717. doi:10.7150/ijbs.45538
Zaporozhets, T. S., and Besednova, Nataliya N. (2020). Biologically active compounds from marine organisms in the strategies for combating coronaviruses. AIMS Microbiol. 6 (4), 470–494. doi:10.3934/microbiol.2020028
Keywords: COVID-19, SARS-CoV-2, Drug discovery, Natural products, Computational methods
Citation: Ogbadoyi EO and Umar N (2022) The challenges and opportunities for the development of COVID-19 therapeutics and preparing for the next pandemic. Front. Drug. Discov. 2:925825. doi: 10.3389/fddsv.2022.925825
Received: 21 April 2022; Accepted: 15 July 2022;
Published: 26 August 2022.
Edited by:
Bruno Villoutreix, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Walaa A. Negm, Tanta University, EgyptCopyright © 2022 Ogbadoyi and Umar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emmanuel O. Ogbadoyi, ZW9nYmFkb3lpQGZ1dG1pbm5hLmVkdS5uZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.