- Collaborations Pharmaceuticals, Inc., Raleigh, NC, United States
While we currently have multiple highly effective vaccines approved for use against SARS-CoV-2 in the USA and other countries, there are far fewer small molecule antivirals approved to date. The emergence of the latest SARS-CoV-2 variant, Omicron which is heavily mutated in the spike protein, is also raising concerns about the effectiveness of these current vaccines and increasing the call for more therapeutic options. At the time of writing only remdesivir is approved by the FDA while molnupiravir (already approved in the United Kingdom) and Paxlovid (PF-07321332) have emergency use authorizations from the FDA. Repurposed molecules, such as dexamethasone and baricitinib, have been authorized for emergency use in some countries and are used in combination with remdesivir. After 2 years we are only now starting to see the progression of further molecules through animal models to assess their efficacy before clinical trials. As datasets accumulate from both in vitro and in vivo animal efficacy models, this may allow us to understand the physicochemical properties necessary for antiviral activity and enable the search for additional antivirals. We now summarize 25 small molecule drugs that are either approved, in the process of approval or in the pipeline for COVID which have both in vitro and in vivo data. We demonstrate that these drugs are structurally diverse and cover a wide chemistry space. This information may aid our understanding of what it takes to be a promising treatment for COVID-19 and propose how to discover antivirals faster and more efficiently for the next pandemic.
Introduction
At the time of writing, we are still in the midst of a major a global health crisis caused by the virus Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) that was originally reported in Wuhan, China in late 2019 (Coronaviridae, 2020; Wu et al., 2020). This virus causes the disease COVID-19 3 and shares aspects of pathology and pathogenesis with the earlier Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) (Liu et al., 2020a). SARS-CoV-2, SARS-CoV and MERS-CoV belong to the same family (Coronaviridae) and genus (Betacoronavirus). SARS-CoV-2 results in cough, loss of smell and taste, respiratory distress and pneumonia as well as a host of other symptoms (Pan et al., 2020) including extrapulmonary events characterized by a sepsis-like disease collectively called 2019 coronavirus disease (COVID-19) (WHO, 2020). SARS-CoV-2 directly interacts with angiotensin converting enzyme 2 (ACE2) receptor in many cell types (Brann et al., 2020; Bunyavanich et al., 2020; Sungnak et al., 2020; Whitcroft and Hummel, 2020). SARS-CoV-2 rapidly spread worldwide prompting the World Health Organization to declare the outbreak a pandemic, with more than 1.5 million cases confirmed in less than 100 days.4 The high infection rate has caused considerable stress on global healthcare systems leading to over 6 M deaths from COVID-19 at the time of writing (January 2022, World Health Organization COVID-19 dashboard).
In the USA, there are 3 vaccines available to protect against SARS-CoV-2 (Huang et al., 2021; Kyriakidis et al., 2021; Rehman et al., 2021), and globally there are over 20 vaccines currently approved (Craven, 2021). COVID-19 continues to represent an ongoing public health crisis for which vaccines represent our first line of defense. The recent identification in South Africa (and subsequently in other countries) of a new strain B.1.1.529 named Omicron as a variant of concern due to its heavily mutated nature with over 30 changes to the spike protein alone suggests it may reduce vaccine efficacy (Callaway, 2021) although those who received boosters may be better protected. This rapidly developing scenario would suggest the urgent need for other therapeutic approaches to address this and future variants.
There are limited options when it comes to small molecule antivirals, with only remdesivir being FDA approved currently in the US. Several other already approved drugs were quickly touted by the popular press, such as hydroxychloroquine and ivermectin, based on either limited in vitro or clinical data and subsequent clinical trials have demonstrated their resounding lack of efficacy (Galan et al., 2021; Vallejos et al., 2021). There have been extensive repurposing efforts since the pandemic began and numerous computational approaches have proposed drugs to be tested. Much of this early work has been reviewed by us and others previously (Ekins et al., 2020; Muratov et al., 2021). As the general public have observed that vaccines for COVID-19 were developed rapidly in months, there is the unrealistic expectation that antiviral small molecule drugs can also be developed as rapidly. Unfortunately, those in the industry accept that it normally takes a decade or more for a small molecule to progress through the various stages from drug discovery to the clinic at a cost in excess of $1 billion (Paul et al., 2010). Repurposing already FDA approved drugs may be expected to progress much more rapidly.
We were keen to evaluate small molecules which have both in vitro activity and have been tested in vivo against one of the various SARS-CoV-2 animal models (mouse, hamster or non-human primate etc.) (Muñoz-Fontela et al., 2020). While this evaluation is likely not comprehensive and because of the fast-moving nature of COVID-19 research it will almost certainly be rapidly outdated. Our goal is therefore to understand the classes of molecules that have shown promise to date. Obviously, there are major pharmaceutical companies, with seemingly unlimited resources and capabilities, involved in identifying molecules (e.g., Pfizer) or licensing them from others (e.g., Merck). Those outside of these larger pharmaceutical companies in either smaller companies or increasingly in academia need to find a way to collaborate with those who have the capabilities to test molecules under BSL3 conditions in vitro and in vivo. This will require different skill sets such as coordinating complex, multidimensional projects and may include multiple international partners which may add other issues related to funding and intellectual property.
While thousands of papers (nearly 200,000 articles in PubMed at the time of writing) have been written relating to COVID-19, it would be impossible to compress this knowledge into a single review. For example, there are also likely thousands of clinical trials globally, which is outside the scope of this effort. Instead, we will describe in vitro screening efforts and the molecules derived from these screens that progressed to in vivo models as this is more manageable and valuable for future drug discovery efforts.
In Vitro Screening
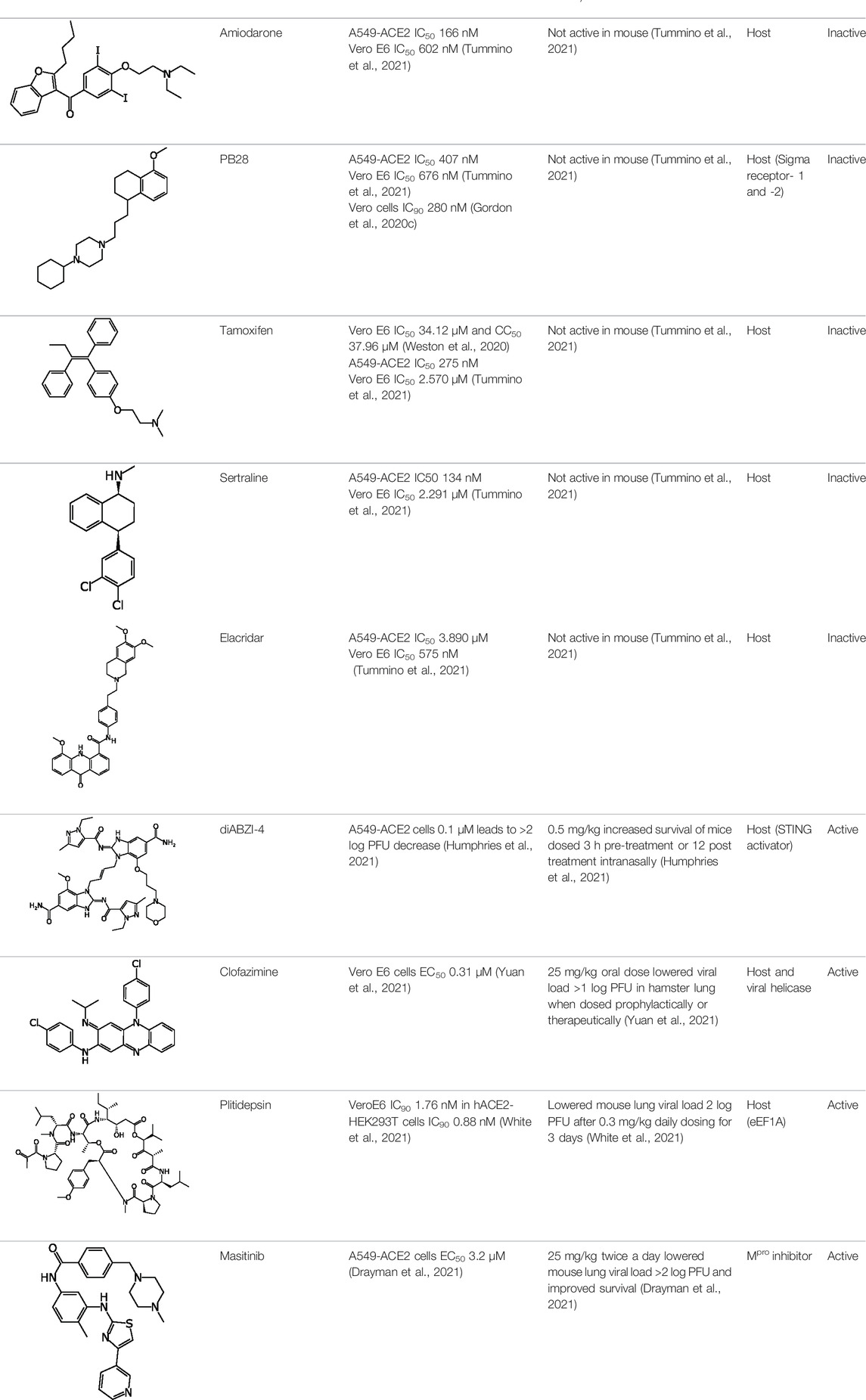
Early in the SARS-CoV-2 pandemic many of the repurposing efforts used FDA approved drugs that had previously been shown to have antiviral activity against other related viruses. Several of these drugs had low μM activity and a selectivity index (SI) greater than 10 in Vero cells, including nitazoxanide (EC50 2.12 μM), remdesivir (EC50 0.77 μM, and chloroquine (EC50 1.13 μM). This work alone represented one of the earliest articles describing the use of remdesivir and chloroquine (Wang et al., 2020a). While chloroquine was identified early on, the derivative hydroxychloroquine was in multiple clinical trials in China by February 2020. It is also worth noting that the in vitro activity of hydroxychloroquine (EC50 4.51 μM) was not as potent as chloroquine when assessed at four different multiplicities of infection (Liu et al., 2020b). However, it is likely this work generated significant interest in this compound that led to the subsequent numerous clinical trials. Other groups confirmed this activity in Vero cells and also demonstrated activity in Caco-2 but not Calu-3 cells (Clementi et al., 2020). Several groups showed similar activity for remdesivir in Vero cells (EC50 1.65 μM), with increased activity in human epithelial cultures (EC50 0.01 μM) and Calu-3 (EC50 0.28 μM) (Pruijssers et al., 2020). Additional molecules were identified including eight artemisinins and lumefantrine (EC50 23.50 μM) which were tested in Vero cells and time of addition studies suggested this was working post entry (Cao et al., 2020). While not a focus of the current analysis, natural products were also tested in vitro, such as lycorine (EC50 0.31 μM) and oxysophoridine (EC50 0.18 μM), and many of these had activity in Vero cells with increased potency over drugs like gemcitabine (EC50 1.24 μM) and chloroquine (EC50 1.38 μM) (Zhang et al., 2020a). Larger collections of molecules were also tested in Vero cells which resulted in identification of niclosamide (IC50 0.28 μM) and ciclesonide (IC50 4.33 μM) as hits (Jeon et al., 2020). Additionally, the antiviral tilorone (IC50 4 μM) was identified as an early hit (Jeon et al., 2020) and has previously been shown to have similar activity against MERS (Ekins and Madrid, 2020) and Ebola (Ekins et al., 2015a) [as has remdesivir (de Wit et al., 2020)]. An earlier preprint (Jeon et al., 2020) also included the antimalarial pyronaridine (IC50 31 μM). The FDA approved antiparasitic, ivermectin (IC50 2.8 μM) was also shown to have in vitro activity in Vero cells, which likely also sparked early interest in this molecule (Caly et al., 2020). 12,000 clinical stage or FDA approved compounds in the ReFRAME library were screened against Vero cells. 21 hits were identified with promising dose response relationships in Vero cells including apilimod (EC50 0.023 μM) and clofazimine (EC50 0.310 μM) (Riva et al., 2020). Further, the PIKfyve kinase inhibitor apilimod was tested in 293T cells (EC50 0.012 μM) and Huh-7 cells (0.088 μM) (Riva et al., 2020). A second group demonstrated how SARS-CoV-2 modified phosphorylation in infected cells and proposed kinase inhibitors as important (Bouhaddou et al., 2020) including apilimod which showed activity in Vero (IC50 < 0.08 μM) and in A549 cells (IC50 0.007 μM) (Bouhaddou et al., 2020). A protein interaction map identified FDA and clinical stage compounds binding to sigma-1 and 2 receptors which act as host factors, with the most potent compound identified in Vero cells being PB28 (IC90 280 nM) (Gordon et al., 2020a). Much of this early in vitro screening work was in Vero cells, and when promising compounds are tested in human cells, they may have very different activity likely due to the lack of the host protein TMPRSS2 (Shulla et al., 2011; Hoffmann et al., 2020; Shang et al., 2020).
Other cell types have also been used for larger screens, such as a quantitative HTS in Huh7 cells, which tested 1425 compounds and identified 11 novel compounds with activity IC50 < 1 μM including lactoferrin which showed potent activity (IC50 308 nM) (Mirabelli et al., 2020). An enzyme-linked immunosorbent assay (ELISA) and cell viability assay screen of 1528 compounds led to 19 hits, out of which 4 were the most active in Vero cells and included cetilistat (EC50 1.13 μM), diiodhydroyquinoline (EC50 1.38 μM), abiraterone acetate (EC50 1.94 μM) and bexarotene (EC50 2.01 μM) (Yuan et al., 2020). A screen of the Prestwick library in hPSC lung organoids identified imatinib (EC50 4.86 μM), mycophenolic acid (EC50 0.15 μM) and quinacrine (EC50 2.83 μM) (Han et al., 2020). This was followed by testing in mice treated with these 3 drugs infected with SARS-CoV-2 pseudovirus and showed significant decreases in infected cells (Han et al., 2020). This is of interest for several reasons, one being that others had not observed in vitro SARS-CoV-2 activity for quinacrine in Vero cells (Jeon et al., 2020). Quinacrine and tilorone have also previously been demonstrated to possess activity against Ebola infected HeLa cells (Ekins et al., 2015a) and not Vero cells (Lane et al., 2019a) and we more recently have tested these compounds in several cell types infected with SARS-CoV-2 (Puhl et al., 2021a). Target-based screens have also been performed, with a FRET-based screen of Mpro which assessed 10,000 compounds, finding 7 primary hits and one of these being ebselen (IC50 0.67 μM) which also had activity in Vero cells (EC50 4.67 μM) (Jin et al., 2020a). It should be noted that from all this predominantly in vitro testing, very few compounds have progressed to in vivo animal models of SARS-CoV-2 infection.
Antivirals of Most Interest
Remdesivir
There are currently few small-molecule drugs approved for COVID-19 (Hall et al., 2021), including remdesivir (Eastman et al., 2020), which as described above originally demonstrated activity in Vero cells (Wang et al., 2020a; Pruijssers et al., 2020), human epithelial cells and in Calu-3 cells (Pruijssers et al., 2020) infected with SARS-CoV-2 prior to clinical testing. Remdesivir represents a “repurposed prodrug” which was originally developed for Hepatitis C virus, then repurposed for treating Ebola virus (EBOV) and has since reached clinical trials for EBOV (Mulangu et al., 2019). Remdesivir’s target specificity and potency towards RNA polymerase was noted early on (Gordon et al., 2020b), as it causes irreversible chain termination. A primary human lung epithelium infection model and a lung organoid model were used to show remdesivir could suppress viral infection (Mulay et al., 2020). Remdesivir was therefore repurposed very quickly (Wang et al., 2020a; Pruijssers et al., 2020) obtaining an emergency use authorization and then FDA approval in less than a year (Eastman et al., 2020). Subsequently, there have been many clinical studies for remdesivir, but the effectiveness of this drug is far from comprehensive (Wang et al., 2020b; Goldman et al., 2020; Spinner et al., 2020; Barratt-Due et al., 2021) and yet still it is the only small molecule approved by the FDA (while molnupiravir and paxlovid have emergency use authorizations) for use alone against COVID-19. This antiviral is severely limited by its requirement to be administered I.V. and its use is therefore restricted to a hospital setting. We are aware of remdesivir oral formulations being tested so these may overcome the limitations in future.
Molnupiravir
Molnupiravir (EIDD-2801, MK4482) is a prodrug that was identified as an inhibitor of influenza A and respiratory syncytial virus acting as an RNA mutagen and like remdesivir was initially developed as a hepatitis C inhibitor in the early 2000s. This was shown early on in the pandemic to be active in vitro against SARS-CoV-2 and progressed to in vivo testing in mice and hamster (Sheahan et al., 2020; Rosenke et al., 2021; Wahl et al., 2021). We are not aware of any clinical trial (NCT04392219) data that has been peer reviewed yet for this molecule although Merck have obtained emergency use authorization from the FDA. Molnupiravir was initially reported to have reduced the risk of hospitalization or death by approximately 50% compared to placebo for patients with mild or moderate COVID-19 (NCT04575597) (Anon, 2021), although this was recently adjusted to 30% (Anon, 2021b) and may impact its ultimate approval. Molnupiravir is approved in Britain for use in people with mild to moderate COVID-19 and at least one risk factor for developing severe illness, such as obesity, older age diabetes, and heart disease.
Paxlovid
The rapid development of the potent Mpro inhibitor PF-07321332 and clinical testing demonstrates the capabilities of a major pharma. However, it is worth pointing out that its development also began nearly 20 years earlier from a potent Mpro inhibitor for SARS-CoV-1. This compound was also a known P-glycoprotein substrate requiring it to be tested in vitro in Vero E6 cells with a P-gp inhibitor as these cells express high levels of this efflux transporter (Owen et al., 2021). The molecule is also a substrate for CYP3A4. Clinically this compound is used in combination with the protease inhibitor ritonavir to inhibit its metabolism and has been branded as Paxlovid (Owen et al., 2021). Like for molnupiravir, we are not aware of any clinical trial data that has been peer reviewed and published yet for this molecule at the time of writing, although Pfizer have also obtained an FDA emergency use authorization. This drug has been reported to reduce the risk of hospitalization or death by 89% compared to placebo in non-hospitalized high-risk adults with COVID-19 in interim analysis of phase 2/3 EPIC-HR study in which no deaths were reported in patients who received Paxlovid compared to 10 deaths in patients who received placebo. One of the major limitations of this drug is its complex synthesis and limited supply of the clinical material. Therefore, efforts to develop inhibitors that are more readily synthesized may be ideal and there is considerable activity around developing additional Mpro inhibitors such as GC376 (Dampalla et al., 2021).
PF-00835231
PF-00835231 is potent inhibitor of Mpro, which binds covalently to the protease and is administered i. v. PF-00835231 is an analog of rupintrivir, a human rhinovirus (HRV) Mpro inhibitor. PF-07304814 is a phosphate prodrug that is rapidly converted in vivo to the active moiety, PF-00835231, which exhibits high selectivity over human proteases, acts as a broad-spectrum protease inhibitor and demonstrates potent antiviral activity in vivo (Boras et al., 2021). PF-07304814 exhibits an encouraging preclinical profile that has the ADME, safety, and once converted to PF-00835231, SARS-CoV-2 antiviral activity to support progression to the clinic as a COVID-19 single-agent antiviral treatment. The favorable profile of PF-07304814 enabled the rapid progression to clinical trials (NCT04627532 and NCT04535167) (Boras et al., 2021).
Favipiravir
Favipiravir is an approved antiviral in Japan for pandemic influenza and has also demonstrated some activity against Ebola and other viruses in vivo animal models, suggesting a broad-spectrum activity. Like molnupiravir, favipiravir leads to mutations in the viral RNA (Driouich et al., 2021). Favipiravir is not potent and often requires high doses leading to some toxicity in animal models. To date most of these SARS-CoV-2 in vivo studies have been in hamster (Kaptein et al., 2020; Driouich et al., 2021; Touret et al., 2021).
Dexamethasone
As inflammation is one of the hallmarks in COVID-19 and particularly severe in hospitalized patients, the role of anti-inflammatory agents such as steroids was studied early on. By July 2020 a clinical trial had showed the effectiveness of dexamethasone by decreasing mortality in patients requiring supplemental oxygen or mechanical ventilation (Group et al., 2021). More recent trials in patients with moderate to severe infection against SARS-CoV-19 showed survival benefits of using dexamethasone when given in addition to the standard of care (Tomazini et al., 2020). We are not aware of any in vitro or in vivo studies for this compound that enabled its progression to clinical trials.
Fluvoxamine
Like dexamethasone, other molecules have apparently progressed to clinical trials without apparent in vitro or in vivo testing against SARS-CoV-2. One such molecule is the selective serotonin reuptake inhibitor fluvoxamine, which is used to treat obsessive compulsive disorder and depression, with promising recent clinical trial results against SARS-CoV-2. This molecule has been shown to bind the sigma receptor, reduces inflammation and protects against septic shock in mice (Rosen et al., 2019). The first small trial was a double blind randomized fully remote contactless clinical trial with 80 patients treated with fluvoxamine 100 mg and 72 patients with a placebo, dosed 3 times a day. Patients on fluvoxamine had lower odds of clinical deterioration (Lenze et al., 2020). The most recent study described a clinical trial performed in Brazil with 741 patients given fluvoxamine 100 mg twice daily for 10 days. Amongst high-risk patients with early diagnosed COVID-19 hospitalization was reduced (Reis et al., 2021). There are several likely mechanisms for fluvoxamine against SARS-CoV-2 (Sukhatme et al., 2021). One of them is that sigma 1 receptor agonists like fluvoxamine and fluoxetine are lysosomotropic (Hallifax and Houston, 2007; Kazmi et al., 2013). Given the lysosomal egress of β-coronaviruses from infected cells, lysosomotropic drugs like fluvoxamine could have antiviral effects in the virus laden lysosomes (Homolak and Kodvanj, 2020).
Pyronaridine
A machine learning model was used to identify pyronaridine tetraphosphate (Ekins et al., 2015b) for testing against EBOV and subsequently this molecule inhibited EBOV and Marburg in vitro as well as demonstrating significant efficacy in the mouse-adapted EBOV (ma-EBOV) model (Ekins et al., 2018; Lane et al., 2019a; Lane et al., 2019b). Pyronaridine was identified as a possible virus entry inhibitor (Lane and Ekins, 2020). Pyronaridine tetraphosphate is used as an antimalarial in several countries as part of a combination therapy with artesunate (Pyramax). Pyronaridine alone also demonstrated significant activity in the guinea pig-adapted model of EBOV infection (Lane et al., 2020a). It has been recently shown that this compound possesses in vitro activity against SARS-CoV-2 (Bae et al., 2020; Jeon et al., 2020; Puhl et al., 2021a) and pyronaridine is in a clinical trial administered in combination with artesunate. Using A549-ACE2 cells, which support SARS-CoV2 growth to about 107 PFU/ml, pyronaridine showed SARS-CoV-2 inhibition demonstrating IC50 0.23 μM and a good selectivity index and binding to SARS-CoV-2 spike RBD (Kd 0.62 μM) (Puhl et al., 2021a) while more recently it has been shown to inhibit Plpro (IC50 1.8 μM) (Puhl et al., 2021b). Pyronaridine is of particular interest as the Cmax data for pyronaridine in our previous mouse pharmacokinetics studies (i.p. dosing) suggested that plasma levels that are above the average IC50 observed for SARS-CoV-2 inhibition in vitro (Lane et al., 2019b) can be reached with dosing well below the maximum tolerated dose. Pyronaridine also has excellent in vitro ADME properties with a long half-life that makes a single dose treatment possible (Lane et al., 2019b; Puhl et al., 2021a). We have recently assessed the in vivo efficacy of pyronaridine in a K18-hACE2 mouse model of COVID-19 (Puhl et al., 2021b) where it resulted in a decreased viral load and improved lung histopathology. Cytokine and chemokine analysis showed increased INF-1β levels and decreased IL-6, CXCL1, CCL4 (Puhl et al., 2021b) (Table 1).
Kinase Inhibitors
Many FDA-approved kinase inhibitors have previously been proposed as broad-spectrum antiviral therapies (Baranov et al., 2020) as they have multiple host protein targets required for the viral life cycle, replication, and infection of multiple virus types. Kinase inhibitors also possess anti-inflammatory and cytokine inhibitory activity properties which may address the lung damage from respiratory virus infections (Baranov et al., 2020). One of the earliest molecules to be computationally repurposed using a knowledge graph approach was the AAK1 and JAK1/2 kinase inhibitor baricitinib (Richardson et al., 2020). The mechanism was eventually validated in vitro and in human patients (Stebbing et al., 2020; Lenz et al., 2021) as well as in combination with hydroxychloroquine (Titanji et al., 2021). This molecule was granted an FDA emergency use authorization in combination with remdesivir. Subsequently, protein kinase inhibitors have been proposed for treating SARS-CoV-2 and have demonstrated in vitro (Riva et al., 2020; Weisberg et al., 2020; Baranov et al., 2020; Hoang et al., 2021; Zhang et al., 2020b; Zhao et al., 2020) and in vivo activity while several are also in clinical trials (Raghuvanshi and Bharate, 2021; Drayman et al., 2021). We also recently screened a panel of 45 kinase inhibitors in a model of SARS-CoV-2 in the β-coronavirus murine hepatitis virus (MHV), a model of SARS-CoV-2 infection, and identified 10 compounds with activity (Puhl et al., 2021c). 2 compounds demonstrated activity against SARS-CoV-2, HCov-229E and MHV (entrectinib and vandetanib) for which the main mechanism remains to be elucidated (Puhl et al., 2021c). Imatinib, masitinib and vandetanib are all kinase inhibitors that have similar low µM in vitro activity against SARS-CoV-2. Out of these, only masitinib (Drayman et al., 2021) showed a decrease in viral load in mice, while vandetanib reduced lung inflammation in mice and altered cytokine levels indicative that this might be useful to address the cytokine storm in COVID-19 (Puhl et al., 2021c). Imatinib did not decrease viral load in hamsters infected with SARS-CoV-2 (Touret et al., 2021) (Table 1) and clinically it did not decrease the time to discontinuation of ventilation and supplemental oxygen from more than 48 consecutive hours in patients with COVID-19 that required supplemental oxygen (Aman et al., 2021).
Quaternary Ammonium Compounds
A text mining approach undertaken by our team and collaborators recently described hundreds of molecules that have been identified with antiviral effects against coronaviruses in the literature (Baker et al., 2020). One of these was cetylpyridinium chloride, the quaternary ammonium compound used in mouthwashes and nasal sprays which destroys the viral capsid upon direct contact (Baker et al., 2020). Several pilot clinical trials have also suggested the utility of this and other mouthwashes to destroy the virus (Eduardo et al., 2021; Seneviratne et al., 2021). Others have shown a more than 98% reduction of SARS-CoV-2 S pseudovirion entry in 293/hACE2 cells when the cells were treated with lysosomotropic agents increasing endosomal pH, such as ammonium chloride and bafilomycin A (Ou et al., 2021).
Phospholidosis
A recent article (Tummino et al., 2021) focused on compounds originally identified from a repurposing screen that showed no relationship between sigma 1 receptor potency and SARS-CoV-2 antiviral activity. They demonstrated that for cationic amphiphilic drugs (CADs), phospholipidosis was observed in cells and correlated strongly with their in vitro antiviral activity. We recently discussed (Lane and Ekins, 2021) this study and pointed out that compounds with a basic pKa (>6.5) and cLogP of >2 tend to be lysosomotropic and accumulate in the lysosomes (Nadanaciva et al., 2011), which is a key for many phospholipidosis-inducing compounds. 4 phospholipidosis-inducing drugs (amiodarone, sertraline, PB28 and tamoxifen–Table 1) that showed in vitro activity were tested in a 3-days mouse efficacy model for SARS-CoV-2 infection and these did not show efficacy as measured by viral load (and neither did elacridar a compound that is not a CAD) (Tummino et al., 2021). We pointed out that phospholipidosis may not even be relevant in the mouse model for SARS-CoV-2 as the authors showed amiodarone offers neither antiviral protection nor hallmarks of phospholipidosis. Others have described how CADs accumulate in different subcellular compartments (e.g., mitochondria and lysosomes) (Vater et al., 2017a; Vater et al., 2017b) and basic amines may also lead to accumulation in these compartments and change the pH, inhibiting entry for some viruses. This approach has been proposed as a strategy to decrease SARS-CoV-2 viral infection (Gorshkov et al., 2021). In summary, many CADs have antiviral activity and induce phospholipidosis during chronic treatment and yet this toxicity is reversible and therefore manageable (Salata et al., 2017).
Drug Transporters
CADs are also of interest because of potential for interfering with human drug transporters. Cationic compounds with SARS-CoV-2 antiviral activity (chloroquine, hydroxychloroquine and quinacrine) inhibited OCT and MATE transporters in vitro (Martinez-Guerrero et al., 2021). An independent study has also evaluated 25 drugs used in COVID-19 clinical trials to assess the potential for drug-drug interactions (Yee et al., 2021). Transporters can also be targeted to reach viral sanctuary sites such as the brain and testes. As an example, the human equilibrative nucleoside transporters 1 and 2 (ENT) are of interest because their substrates may gain entry to the testes and other sites. We are at the early stages for understanding these structure-inhibitor and structure-substrate relationships for these transporters (Miller et al., 2021a; Miller et al., 2021b; Miller et al., 2021c) and most recently evaluated remdesivir and molnupiravir (Miller et al., 2021a). This illustrated how these transporters may have a role in the efficacy of these compounds and how it may differ for each based on the affinity for these transporters.
Targeting the Cytokine Storm
It has been demonstrated that SARS-CoV-2 causes an imbalance in the human immune system which may lead to a cytokine and chemokine storm (Costela-Ruiz et al., 2020) impacting PDGF, VEGF (Huang et al., 2020), IL-6 (Herold et al., 2020; Wang et al., 2021), IL‐8, IL‐10 (Wang et al., 2021), TNF‐α (Wang et al., 2021) and IFN-α and -β (Molony et al., 2017; Hu et al., 2018; Zhang et al., 2020c; Xia et al., 2020; Yuen et al., 2020). Furthermore, an impaired type I interferon response has already been observed in COVID-19 (Hadjadj et al., 2020), which is followed by increased circulating levels of IL-6 and TNF-α. This may also result in acute respiratory distress syndrome (ARDS), coagulation disorders, and eventually multiple organ failure (Costela-Ruiz et al., 2020; Chen et al., 2021). Disease severity is linked to a highly dysregulated innate immune response, which is broadly characterized by a delayed interferon I (IFN-I) and IFN-III response relative to symptom onset and possibly peak virus replication, and the production of an exuberant inflammatory response (Lowery et al., 2021), exacerbated proinflammatory cytokine production and in extensive cellular infiltrates in the respiratory tract, resulting in lung pathology (Lowery et al., 2021). Gene expression in human lung only mice demonstrated interferon-stimulated genes and inflammatory cytokines including IL6, CXCL8, CXCL10, TNF and CCL5 were induced from infected lung tissue (Wahl et al., 2021). Hence, in this mouse model, SARS-CoV-2 causes an upregulation of the innate immune response.
Most recently, a randomized, placebo-controlled trial of Janus-kinase inhibition using tofacitinib, has been reported to improve COVID-19 survival, in the presence of background glucocorticoid treatment (received by 89% of patients) (Guimaraes et al., 2021). Antivirals that can dampen the cytokine storm in a selective manner would provide a useful therapeutic approach for treating patients, for example vandetanib (Puhl et al., 2021c) (Table 1).
Physiochemical Properties of Antivirals
What should the perfect COVID drug look like? In an ideal world, a molecule that directly addresses one or more viral target as well as having host effects to modulate the cytokine storm would be considered promising. We have focused on a small set of compounds which are predominantly already approved drugs or drug candidates, that we have separated into those that demonstrated some degree of in vivo efficacy in various animal models for SARS-CoV-2 and those that do not. Interestingly, 17 out of 25 molecules displayed activity against host targets or mechanisms (Table 1). Our criteria are broad so that we can capture molecules that may have a direct antiviral effect or a host effect. Therefore, viral load reduction alone was not a solo-criteria and in some cases, molecules showed improvements in histopathology alone, and these were considered active. Clearly there has been some concern around CADs and whether they represent a waste of resources based on a small in vitro: in vivo analysis (Tummino et al., 2021). Our analysis of physiochemical properties for these 25 drugs that have been tested in vitro and in vivo (Table 2) clearly shows a wide range of logP and pKa values, such that there are molecules that comply with requirements for phospholipidosis [basic pKa (>6.5) and cLogP of >2) and CADs (cLogP (≥3) and pKa (≥7.4)] in both the in vivo active and inactive groups (Table 2). This perhaps provides a larger and more convincing dataset than this earlier study (Tummino et al., 2021) to demonstrate why we should pursue CADs as antivirals alongside other classes of molecules.
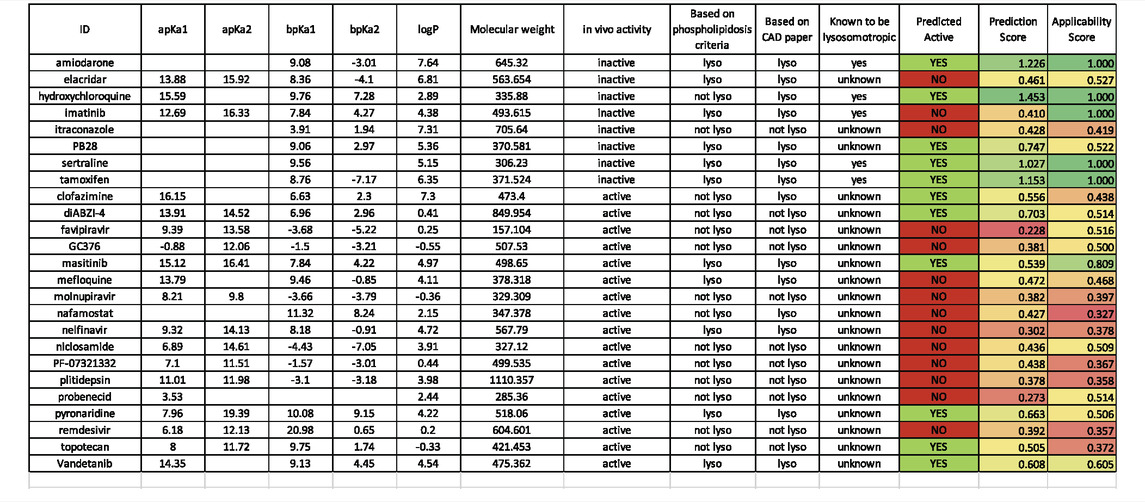
TABLE 2. A. Calculated and predicted physicochemical properties for SAR-CoV-2 inhibitors. pKa and LogP calculated with ChemAxon software (Budapest, Hungary). Predictions using a machine learning model (Lane et al., 2020b) for lysosomotropic activity are highlighted in the last 3 columns, CAD paper = (Tummino et al., 2021) known lysosomotropic = (Nadanaciva et al., 2011; Kazmi et al., 2013).
Computational Approaches to Guide Drug Discovery: Machine Learning Models
To date we have collated hundreds of drugs that have in vitro data against this virus primarily in Vero cells (Wang et al., 2020a; Liu et al., 2020b; Jeon et al., 2020; Jin et al., 2020b). This has enabled machine learning models and even with these relatively modest datasets we have shown that such models can be used to select additional compounds for testing (Gawriljuk et al., 2021). In addition, we can use these molecules that have been tested to date to visualize the in vivo in active and inactive molecules (Figure 1A) and these appear to show the coverage of this property space is relatively even and not clustered in any particular area. Similarly, the in vivo active and inactive molecules are well dispersed in the thousands of molecules in the SuperDrug database (Figure 1B). Selecting compounds close to the active molecules in these property spaces may be one way to help select additional compounds for future testing. Other sources of in vitro data are available as groups have screened libraries such as the NIH NCATS OpenData portal (Anon, 2022) and this resource could be used for modeling. In addition, as datasets are built up specifically for antiviral targets like Mpro, PLpro or others, these could be used for target specific machine leaning models that could be combined with the whole cell models.
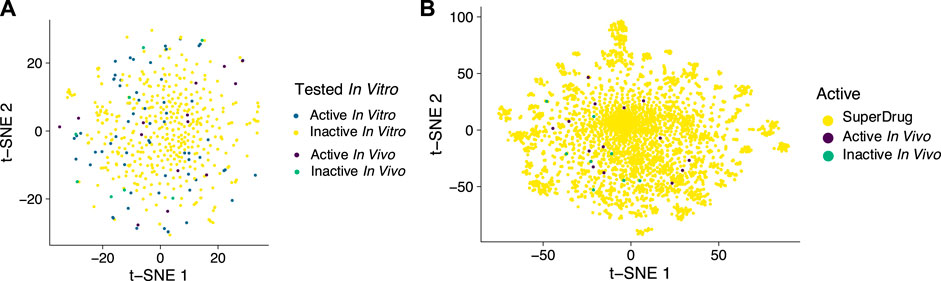
FIGURE 1. t-SNE plots using 1024-bit ECFP6 fingerprints as input features to visualize compounds tested in vivo versus (A). compounds tested in vitro and used for a machine learning model, (B). versus approved drugs in SuperDrug to illustrate the coverage of molecular property space.
Impact Beyond COVID-19
Several in vivo studies of small molecule drugs have described the direct measurement of cytokine and chemokine levels or gene expression patterns as an attempt to understand these drugs and their effects on inflammation. The utility of this is that we may be able to identify molecules with a specific pattern of increased or decreased cytokine levels that may be the mirror image for biomarkers for other lung or other diseases. This is analogous to the connectivity map (CMap), which brings together data on genes and thousands of drugs and disease states used in several repurposing projects to identify new uses for old drugs (Lamb et al., 2006; Lamb, 2007; Zimmer et al., 2010; Subramanian et al., 2017). There is therefore the potential to find new drugs that could be potentially useful for these other (lung) diseases which may not currently have viable treatments or with limited treatment options (e.g. lung fibrosis).
Addressing COVID-19 Symptoms
Several of the symptoms of COVID-19 include impacts on the peripheral nervous system. Olfactory dysfunction was described early on as well as the diagnosis and management of these symptoms (Whitcroft and Hummel, 2020). Loss of the sense of smell (hyposmia/anosmia) and/or taste (hypogeusia/ageusia) have been widely reported (Lechien et al., 2020; Mao et al., 2020), can predict SARS-CoV-2 infection and have been added to the list of major symptoms (Menni et al., 2020) as well as the Center for disease Control and Prevention’s website of symptoms (CDC, 2020). 64–67% of those testing positive (>7,000) in a study in the US and United Kingdom described a loss of smell and taste (Menni et al., 2020). Surveys have shown that taste and smell dysfunction may be an early symptom of COVID-19 in over 50% of those questioned (Mercante et al., 2020) and another study reported resolution of these symptoms in 48.7% of patients within 4 weeks of onset (Boscolo-Rizzo et al., 2020). While at first glance these may not seem as severe as other symptoms of the virus such as fever and cough, they can be long-lived based on what we know of other viral infections (Suzuki et al., 2007). There are considerable ongoing efforts in drug and vaccine discovery for COVID-19 (with hundreds of drugs in various stages of research and clinical trials ongoing), yet there is relatively little discussion of how SARS-CoV-2 might be causing these specific symptoms (Bilinska and Butowt, 2020) or even whether these could be targets for pharmaceutical intervention. To date, there have not been proposals to mitigate the taste and smell symptoms as a treatment strategy. Besides SARS-CoV-2 directly interacting with angiotensin converting enzyme 2 (ACE2) receptor in the nasal epithelium (Whitcroft and Hummel, 2020), nasal goblet cells (Sungnak et al., 2020) and olfactory mucosa (Brann et al., 2020) there have been few alternative suggestions of how the virus might be causing these symptoms or how to treat them. For example, decreased IL-6 improved smell and taste in COVID-19 patients (Cazzolla et al., 2020). SARS-CoV-2 infection of non-neuronal cell types has been proposed to lead to olfactory dysfunction in COVID-19 patients (Brann et al., 2020). One group has recently proposed that ACE2-independent pathways may be involved such that alternative viral receptors may yet be identified (Bilinska and Butowt, 2020). Significant differences in the level of gene expression of ACE2 in different age groups (Bunyavanich et al., 2020) may explain differences in susceptibility. There is considerable previous discussion for the side effects of drugs including the impact of ageing (Schiffman, 1997; Schiffman et al., 2002) and anosmia and agusia (Schiffman and Doty, 2015; Schiffman, 2018) diseases, which include drug treatment for other diseases (including other viruses such as influenza) (Schiffman, 1983a; Schiffman, 1983b). The exact mechanisms by which SARS-CoV-2 and other neurotropic or neuro-invasive viruses impair these chemical senses are not yet fully understood (Bilinska and Butowt, 2020; Xydakis et al., 2020). Interestingly, while many papers have discussed the role of GPCR’s in the role of taste and smell (Meunier et al., 2020), there has been no discussion on whether these receptors themselves could be directly or indirectly affected by SARS-CoV-2. Further research into the likely many mechanisms responsible for chemosensory losses may provide insights into the virus and provide new knowledge for the development of treatments. This would also point to the need for more investment in this research area. Clearly, prior to COVID-19 few patients were seen with sensorineural viral anosmia which limited clinical research. We now have an abundance of research subjects (Xydakis et al., 2020) and it would be important to take advantage of this situation as it could inform how we address future coronaviruses. Certainly, there are many other important symptoms associated with COVID-19 gathering some public attention such as hair loss (trichodynia and telogen effluvium) that need to be addressed (Rossi et al., 2021; Sharquie and Jabbar, 2021; Starace et al., 2021). Small molecules addressing these many COVID-19 symptoms may also provide future generations of antivirals that can further differentiate themselves from remdesivir, molnupiravir and paxlovid.
The Need for Global Collaborations
One of the challenges we identified from the very outset of the pandemic was identifying laboratories that could perform the in vitro and in vivo testing of efficacy for small molecules. Many of the collaborators we had previously worked with for different viruses were setting up testing against SAR-CoV-2, with much of the work done in Vero cells initially. We ran into the challenge of using Vero cells with 3 compounds identified previously for Ebola virus, which had more cytotoxicity in Vero versus human cell lines such as HeLa (Bae et al., 2020). There was a long lag time before human cell line models became accessible for testing with SARS-CoV-2. Subsequently, we have been keen to assess a molecules’ activity against as many cell lines as possible (Puhl et al., 2021a). The potential for P-glycoprotein to have a role in effluxing compounds out of Vero cells as noted by Pfizer (Owen et al., 2021) is one issue, another might be the lack of an interferon response in Vero cells. As an example, pyronaridine does not appear to be a P-gp substrate based on testing in Caco-2 cells (Lane et al., 2019b) so this may be having a lesser impact. From our experience we found the lack of a clear coordinated US (and for that matter global) response has led to silo-ing of capabilities such that there are initiatives that could probably assist companies in developing small molecule antivirals, but these are relatively difficult to gain entry to. There were many US government agencies as well as philanthropic organizations and websites that promoted calls for molecules and research for COVID (e.g. NIAID, BARDA etc). After numerous submissions in response to government agencies, foundations, and requests for funding there was little in the way of responses. This could be because of the strict filter implemented in order to wade through the massive numbers of applications or it could be the criteria set for antivirals has been set too high e.g., direct acting molecules were likely preferred over host targeting molecules, or molecules with known mechanisms of action were preferred over those with no known mechanism. We have been fortunate to be able to coordinate testing and collaborations with many other global academic laboratories, driven by the shared desire to find molecules that could be rapidly brought to patients, rather than by a financial return. We are also starting to see others share their experiences of COVID drug discovery such as the COVID Moonshot and we can likely learn from these for future efforts (von Delft et al., 2021).
Future Prospects: Accelerating the Drug Discovery Pipeline
After spending a significant amount of our time over the past 2 years on COVID-19 drug discovery what have we learnt? First, few laboratories still have a comprehensive drug-discovery pipeline for the disease. While many academic laboratories are experts at the biology or the animal model development, they lack compounds and are reliant on big pharmaceutical companies to supply them. This requires academics to access pharmaceutical companies who may already have molecules with antiviral activity. This relationship is symbiotic: the academics likely get funded for the experiments and the industry obtains the data they need from key opinion leaders. Other approaches are available in the USA such as the NIAID antiviral testing capabilities which contracts out the testing to academic laboratories. When COVID-19 was identified, in vitro testing in Vero cells was available after several months followed by Caco-2 and Calu-3 cells. We have been able to additionally obtain testing against A549 cells at several academic laboratories in the USA and Brazil. Being able to submit molecules for testing against a panel of human and animal cells from the outset would be ideal in future and potentially speed up identification of molecules and perhaps filter out compounds that may be less useful. We initially, could not move pyronaridine forward until we could also demonstrate the molecule was active against SARS-CoV-2 in human cells, just as it showed activity differences between Vero and Hela cells for EBOV. There also needs to be a clear pathway for anyone to route their molecules through an in vitro-in vivo profiling service provided globally. Even now this does not really exist with each country likely focused on their own researchers. In the USA, there is a patchwork of government agencies such as BARDA, NIH etc. each advertising resources wanting to identify small molecules and using websites to solicit information. This is a one-way street and submitting information does not guarantee a response. What is needed is a truly global initiative with a transparent pathway from the very outset to counter the next pandemic virus.
One wonders how much has been financially invested by researchers and small companies without any guarantee of funding or reward, in the hope that they can hit the lottery and find a molecule that they can then convince a large pharma company to license. For every small company that licenses a drug to a major pharma there are likely many hundreds that are currently striving to get to that point. Increasingly, the bar will continue to be set higher. Remdesivir is a relatively low bar to overcome (e.g., find a drug that can demonstrate efficacy that is not administered i. v.). Molnupiravir will be harder (find a drug that likely will not have the mutation concerns or require less frequent dosing), and paxlovid harder still (find a drug as potent, orally delivered that does not interfere with CYP3A4). After these there will likely be combinations of these and other drugs as we have seen for HIV and hepatitis C. Already there have been efforts to predict synergistic combinations (Jin et al., 2021)We will then probably move into finding drugs that specifically address some of the symptoms like loss of smell and taste, decrease inflammation as well as impact long COVID. This will unfortunately require a long wait for those patients suffering from these symptoms now.
We will likely see these newer drugs brought to market more rapidly than any other drugs to date because of the unmet need. This in turn will set expectations for faster review for drugs for other diseases like cancer which have generally been expedited, but COVID-19 has clearly identified the ‘need for speed’ in drug discovery. There is also a need for efficiency and there was certainly redundancy of testing as many groups tried the same or similar compounds or experiments. This may point to a need for some degree of coordination of efforts being needed. As the Omicron variant suggests, we will need to move very quickly to keep up with this moving target of COVID-19. The next pandemic could be worse so we will need to be ready with broad spectrum antivirals. We cannot afford to rest on our laurels as often happens after outbreaks make the news (e.g. Ebola, Zika, flu). Our global medicine cabinet needs more antivirals targeting different mechanisms and we therefore should not be too selective about avoiding compounds that may appear undesirable such as CADs or impact cytokines. These characteristics of molecules may turn out to be desirable features. Recent publications while this was in review, include additional compounds like cannabidiol which has an EC50 ∼1 μM and can significantly inhibit viral replication in mouse lungs when treated for a week before infection (Nguyen et al., 2022). The mechanism appears to be via induction of the interferon pathway. We would expect to see many other molecules identified that may possess promising direct antiviral or host effects, but these may have to overcome considerable hurdles to compete with the drugs that are either approved or emergency authorized currently. The situation with COVID-19 is dynamic and we have also seen the recent removal of authorizations for two monoclonal antibodies by the FDA as they are infective against Omicron. This also points to the need for continual drug discovery and development of antivirals for COVID-19 and why it may be important to understand the physicochemical properties and potential mechanisms of molecules that have been identified to date, so we can learn from them.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
We kindly acknowledge NIH funding R44GM122196 from NIH NIGMS as well as our many collaborators on the studies described. Susan S. Schiffman and Stephen H Wright are thanked for stimulating discussions. Mindy Davis and others are gratefully acknowledged for assistance with the NIAID virus screening capabilities. Collaborations Pharmaceuticals, Inc. has utilized the non-clinical and pre-clinical services program offered by the National Institute of Allergy and Infectious Diseases.
Conflict of Interest
SE is CEO, AP, TRL, FU are employees of Collaborations Pharmaceuticals, Inc. Collaborations Pharmaceuticals, Inc. has obtained FDA orphan drug designations for pyronaridine, tilorone and quinacrine for use against Ebola. CPI have also filed a provisional patent for use of these molecules against Marburg and other viruses.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Puhl, A. C., Gomes, G. F., Damasceno, S., Fritch, E. J., Levi, J. A., Johnson, N. J., et al. Vandetanib Reduces Inflammatory Cytokines and Ameliorates COVID-19 in Infected Mice. BIORXIV/2021/472155 (2021)
Aman, J., Duijvelaar, E., Botros, L., Kianzad, A., Schippers, J. R., Smeele, P. J., et al. (2021). Imatinib in Patients with Severe COVID-19: a Randomised, Double-Blind, Placebo-Controlled, Clinical Trial. Lancet Respir. Med. 9 (9), 957–968. doi:10.1016/S2213-2600(21)00237-X
Anon, (2021b). “Merck and Ridgeback Biotherapeutics Provide Update on Results from MOVe-OUT Study of Molnupiravir, an Investigational Oral Antiviral Medicine,” in At Risk Adults with Mild-To-Moderate COVID-19. Available at https://www.merck.com/news/merck-and-ridgeback-biotherapeutics-provide-update-on-results-from-move-out-study-of-molnupiravir-an-investigational-oral-antiviral-medicine-in-at-risk-adults-with-mild-to-moderate-covid-19/.
Anon, (2021a). Merck and Ridgeback’s Investigational Oral Antiviral Molnupiravir Reduced the Risk of Hospitalization or Death by Approximately 50 Percent Compared to Placebo for Patients with Mild or Moderate COVID-19 in Positive Interim Analysis of Phase 3 Study. Available at https://www.merck.com/news/merck-and-ridgebacks-investigational-oral-antiviral-molnupiravir-reduced-the-risk-of-hospitalization-or-death-by-approximately-50-percent-compared-to-placebo-for-patients-with-mild-or-moderat/.
Anon, (2022). NCATS OpenData portal in. Available at https://opendata.ncats.nih.gov/covid19/index.html.
Bae, J.-Y., Lee, G. E., Park, H., Cho, J., Kim, Y.-E., Lee, J.-Y., et al. (2020). Pyronaridine and Artesunate Are Potential Antiviral Drugs against COVID-19 and Influenza. bioRxiv 0728, 225102. doi:10.1101/2020.07.28.225102
Baker, N., Williams, A. J., Tropsha, A., and Ekins, S. (2020). Repurposing Quaternary Ammonium Compounds as Potential Treatments for COVID-19. Pharm. Res. 37, 104. doi:10.1007/s11095-020-02842-8
Baranov, M. V., Bianchi, F., and van den Bogaart, G. (2020). The PIKfyve Inhibitor Apilimod: A Double-Edged Sword against COVID-19. Cells 10 (1), 30. doi:10.3390/cells10010030
Barratt-Due, A., Olsen, I. C., Nezvalova-Henriksen, K., Kåsine, T., Lund-Johansen, F., Hoel, H., et al. (2021). Evaluation of the Effects of Remdesivir and Hydroxychloroquine on Viral Clearance in COVID-19. Ann. Intern. Med. 174 (9), 1261–1269. doi:10.7326/M21-0653
Bilinska, K., and Butowt, R. (2020). Anosmia in COVID-19: A Bumpy Road to Establishing a Cellular Mechanism. ACS Chem. Neurosci. doi:10.1021/acschemneuro.0c00406
Boras, B., Jones, R. M., Anson, B. J., Arenson, D., Aschenbrenner, L., Bakowski, M. A., et al. (2021). Preclinical Characterization of an Intravenous Coronavirus 3CL Protease Inhibitor for the Potential Treatment of COVID19. Nat. Commun. 12 (1), 6055. doi:10.1038/s41467-021-26239-2
Boscolo-Rizzo, P., Borsetto, D., Fabbris, C., Spinato, G., Frezza, D., Menegaldo, A., et al. (2020). Evolution of Altered Sense of Smell or Taste in Patients with Mildly Symptomatic COVID-19. JAMA Otolaryngol. Head Neck Surg. doi:10.1001/jamaoto.2020.1379
Bouhaddou, M., Memon, D., Meyer, B., White, K. M., Rezelj, V. V., Correa Marrero, M., et al. (2020). The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 182 (3), 685–712. doi:10.1016/j.cell.2020.06.034
Brann, D. H., Tsukahara, T., Weinreb, C., Lipovsek, M., Van den Berge, K., Gong, B., et al. (2020). Non-neuronal Expression of SARS-CoV-2 Entry Genes in the Olfactory System Suggests Mechanisms Underlying COVID-19-Associated Anosmia. Sci. Adv. 6, eabc5801. doi:10.1126/sciadv.abc5801
Brunaugh, A. D., Seo, H., Warnken, Z., Ding, L., Seo, S. H., and Smyth, H. D. C. (2021). Development and Evaluation of Inhalable Composite Niclosamide-Lysozyme Particles: A Broad-Spectrum, Patient-Adaptable Treatment for Coronavirus Infections and Sequalae. PLoS One 16 (2), e0246803. doi:10.1371/journal.pone.0246803
Bunyavanich, S., Do, A., and Vicencio, A. (2020). Nasal Gene Expression of Angiotensin-Converting Enzyme 2 in Children and Adults. JAMA 323, 2427. doi:10.1001/jama.2020.8707
Caceres, C. J., Cardenas-Garcia, S., Carnaccini, S., Seibert, B., Rajao, D. S., Wang, J., et al. (2021). Efficacy of GC-376 against SARS-CoV-2 Virus Infection in the K18 hACE2 Transgenic Mouse Model. Sci. Rep. 11 (1), 9609. doi:10.1038/s41598-021-89013-w
Callaway, E. (2021). Heavily Mutated Omicron Variant Puts Scientists on Alert. Nature 600, 21. doi:10.1038/d41586-021-03552-w
Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., and Wagstaff, K. M. (2020). The FDA-Approved Drug Ivermectin Inhibits the Replication of SARS-CoV-2 In Vitro. Antiviral Res. 178, 104787. doi:10.1016/j.antiviral.2020.104787
Cao, R., Hu, H., Li, Y., Wang, X., Xu, M., Liu, J., et al. (2020). Anti-SARS-CoV-2 Potential of Artemisinins In Vitro. ACS Infect. Dis. 6 (9), 2524–2531. doi:10.1021/acsinfecdis.0c00522
Cazzolla, A. P., Lovero, R., Lo Muzio, L., Testa, N. F., Schirinzi, A., Palmieri, G., et al. (2020). Taste and Smell Disorders in COVID-19 Patients: Role of Interleukin-6. ACS Chem. Neurosci. 11 (17), 2774–2781. doi:10.1021/acschemneuro.0c00447
Chen, R., Lan, Z., Ye, J., Pang, L., Liu, Y., Wu, W., et al. (2021). Cytokine Storm: The Primary Determinant for the Pathophysiological Evolution of COVID-19 Deterioration. Front. Immunol. 12, 589095. doi:10.3389/fimmu.2021.589095
Clementi, N., Criscuolo, E., Diotti, R. A., Ferrarese, R., Castelli, M., Dagna, L., et al. (2020). Combined Prophylactic and Therapeutic Use Maximizes Hydroxychloroquine Anti-SARS-CoV-2 Effects In Vitro. Front. Microbiol. 11, 1704. doi:10.3389/fmicb.2020.01704
Coronaviridae, V. (2020). The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-nCoV and Naming it SARS-CoV-2. Nat. Microbiol. 5 (4), 536–544. doi:10.1038/s41564-020-0695-z
Costela-Ruiz, V. J., Illescas-Montes, R., Puerta-Puerta, J. M., Ruiz, C., and Melguizo-Rodríguez, L. (2020). SARS-CoV-2 Infection: The Role of Cytokines in COVID-19 Disease. Cytokine Growth Factor. Rev. 54, 62–75. doi:10.1016/j.cytogfr.2020.06.001
Dampalla, C. S., Zheng, J., Perera, K. D., Wong, L.-Y. R., Meyerholz, D. K., Nguyen, H. N., et al. (2021). Postinfection Treatment with a Protease Inhibitor Increases Survival of Mice with a Fatal SARS-CoV-2 Infection. Proc. Natl. Acad. Sci. USA 118 (29), e2101555118. doi:10.1073/pnas.2101555118
de Wit, E., Feldmann, F., Cronin, J., Jordan, R., Okumura, A., Thomas, T., et al. (2020). Prophylactic and Therapeutic Remdesivir (GS-5734) Treatment in the Rhesus Macaque Model of MERS-CoV Infection. Proc. Natl. Acad. Sci. USA 117, 6771–6776. doi:10.1073/pnas.1922083117
Drayman, N., DeMarco, J. K., Jones, K. A., Azizi, S.-A., Froggatt, H. M., Tan, K., et al. (2021). Masitinib Is a Broad Coronavirus 3CL Inhibitor that Blocks Replication of SARS-CoV-2. Science 373, 931–936. doi:10.1126/science.abg5827
Driouich, J.-S., Cochin, M., Lingas, G., Moureau, G., Touret, F., Petit, P.-R., et al. (2021). Favipiravir Antiviral Efficacy against SARS-CoV-2 in a Hamster Model. Nat. Commun. 12 (1), 1735. doi:10.1038/s41467-021-21992-w
Eastman, R. T., Roth, J. S., Brimacombe, K. R., Simeonov, A., Shen, M., Patnaik, S., et al. (2020). Remdesivir: A Review of its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 6 (5), 672–683. doi:10.1021/acscentsci.0c00489
Eduardo, F. d. P., Corrêa, L., Heller, D., Daep, C. A., Benitez, C., Malheiros, Z., et al. (2021). Salivary SARS-CoV-2 Load Reduction with Mouthwash Use: A Randomized Pilot Clinical Trial. Heliyon 7 (6), e07346. doi:10.1016/j.heliyon.2021.e07346
Ekins, S., Freundlich, J. S., Clark, A. M., Anantpadma, M., Davey, R. A., and Madrid, P. (2015). Machine Learning Models Identify Molecules Active against the Ebola Virus In Vitro. F1000Res 4, F1000Res1091. doi:10.12688/f1000research.7217.2
Ekins, S., Freundlich, J. S., Clark, A. M., Anantpadma, M., Davey, R. A., and Madrid, P. (2015). Machine Learning Models Identify Molecules Active against the Ebola Virus In Vitro. F1000Res 4, F1000Res1091. doi:10.12688/f1000research.7217.3
Ekins, S., Lingerfelt, M. A., Comer, J. E., Freiberg, A. N., Mirsalis, J. C., O'Loughlin, K., et al. (2018). Efficacy of Tilorone Dihydrochloride against Ebola Virus Infection. Antimicrob. Agents Chemother. 62 (2). doi:10.1128/AAC.01711-17
Ekins, S., and Madrid, P. B. (2020). Tilorone, a Broad-Spectrum Antiviral for Emerging Viruses. Antimicrob. Agents Chemother. 64 (5), e00440–20. doi:10.1128/AAC.00440-20
Ekins, S., Mottin, M., Ramos, P. R. P. S., Sousa, B. K. P., Neves, B. J., Foil, D. H., et al. (2020). Déjà Vu: Stimulating Open Drug Discovery for SARS-CoV-2. Drug Discov. Today 25 (5), 928–941. doi:10.1016/j.drudis.2020.03.019
Galan, L. E. B., Santos, N. M. D., Asato, M. S., Araújo, J. V., de Lima Moreira, A., Araújo, A. M. M., et al. (2021). Phase 2 Randomized Study on Chloroquine, Hydroxychloroquine or Ivermectin in Hospitalized Patients with Severe Manifestations of SARS-CoV-2 Infection. Pathog. Glob. Health 115 (4), 235–242. doi:10.1080/20477724.2021.1890887
Gawriljuk, V. O., Zin, P. P. K., Puhl, A. C., Zorn, K. M., Foil, D. H., Lane, T. R., et al. (2021). Machine Learning Models Identify Inhibitors of SARS-CoV-2. J. Chem. Inf. Model. 61 (9), 4224–4235. doi:10.1021/acs.jcim.1c00683
Goldman, J. D., Lye, D. C. B., Hui, D. S., Marks, K. M., Bruno, R., Montejano, R., et al. (2020). Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. N. Engl. J. Med. 383 (19), 1827–1837. doi:10.1056/NEJMoa2015301
Gordon, C. J., Tchesnokov, E. P., Woolner, E., Perry, J. K., Feng, J. Y., Porter, D. P., et al. (2020). Remdesivir Is a Direct-Acting Antiviral that Inhibits RNA-dependent RNA Polymerase from Severe Acute Respiratory Syndrome Coronavirus 2 with High Potency. J. Biol. Chem. 295 (20), 6785–6797. doi:10.1074/jbc.RA120.013679
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., O’Meara, M. J., et al. (2020). A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. bioRxiv. doi:10.1101/2020.03.22.002386
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020). A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature 583 (7816), 459–468. doi:10.1038/s41586-020-2286-9
Gorshkov, K., Chen, C. Z., Bostwick, R., Rasmussen, L., Tran, B. N., Cheng, Y.-S., et al. (2021). The SARS-CoV-2 Cytopathic Effect Is Blocked by Lysosome Alkalizing Small Molecules. ACS Infect. Dis. 7 (6), 1389–1408. doi:10.1021/acsinfecdis.0c00349
Group, R. C., Horby, P., Lim, W. S., Emberson, J. R., Mafham, M., Bell, J. L., et al. (2021). Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 384 (8), 693–704. doi:10.1056/NEJMoa2021436
Guimaraes, P. O., Quirk, D., Furtado, R. H., Maia, L. N., Saraiva, J. F., Antunes, M. O., et al. (2021). Tofacitinib in Patients Hospitalized with Covid-19 Pneumonia. N. Engl. J. Med. 385 (5), 406–415. doi:10.1056/NEJMoa2101643
Hadjadj, J., Yatim, N., Barnabei, L., Corneau, A., Boussier, J., Smith, N., et al. (2020). Impaired Type I Interferon Activity and Inflammatory Responses in Severe COVID-19 Patients. Science 369 (6504), 718–724. doi:10.1126/science.abc6027
Hall, M. D., Anderson, J. M., Anderson, A., Baker, D., Bradner, J., Brimacombe, K. R., et al. (2021). Report of the National Institutes of Health SARS-CoV-2 Antiviral Therapeutics Summit. J. Infect. Dis. 224 (Suppl. ment_1), S1–S21. doi:10.1093/infdis/jiab305
Hallifax, D., and Houston, J. B. (2007). Saturable Uptake of Lipophilic Amine Drugs into Isolated Hepatocytes: Mechanisms and Consequences for Quantitative Clearance Prediction. Drug Metab. Dispos 35 (8), 1325–1332. doi:10.1124/dmd.107.015131
Han, Y., Yang, L., Duan, X., Duan, F., Nilsson-Payant, B. E., Yaron, T. M., et al. (2020). Identification of Candidate COVID-19 Therapeutics Using hPSC-Derived Lung Organoidsids. bioRxiv. doi:10.1101/2020.05.05.079095
Herold, T., Jurinovic, V., Arnreich, C., Lipworth, B. J., Hellmuth, J. C., von Bergwelt-Baildon, M., et al. (2020). Elevated Levels of IL-6 and CRP Predict the Need for Mechanical Ventilation in COVID-19. J. Allergy Clin. Immunol. 146 (1), 128e4–136. doi:10.1016/j.jaci.2020.05.008
Ho, J. S. Y., Mok, B. W., Campisi, L., Jordan, T., Yildiz, S., Parameswaran, S., et al. (2021). TOP1 Inhibition Therapy Protects against SARS-CoV-2-Induced Lethal Inflammation. Cell 184 (10), 2618–2632. doi:10.1016/j.cell.2021.03.051
Hoang, T. N., Pino, M., Boddapati, A. K., Viox, E. G., Starke, C. E., Upadhyay, A. A., et al. (2021). Baricitinib Treatment Resolves Lower-Airway Macrophage Inflammation and Neutrophil Recruitment in SARS-CoV-2-Infected Rhesus Macaques. Cell 184 (2), 460–475. doi:10.1016/j.cell.2020.11.007
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181 (2), 271e8–280. doi:10.1016/j.cell.2020.02.052
Homolak, J., and Kodvanj, I. (2020). Widely Available Lysosome Targeting Agents Should Be Considered as Potential Therapy for COVID-19. Int. J. Antimicrob. Agents 56 (2), 106044. doi:10.1016/j.ijantimicag.2020.106044
Hu, R., Xia, C. Q., Butfiloski, E., and Clare-Salzler, M. (2018). Effect of High Glucose on Cytokine Production by Human Peripheral Blood Immune Cells and Type I Interferon Signaling in Monocytes: Implications for the Role of Hyperglycemia in the Diabetes Inflammatory Process and Host Defense against Infection. Clin. Immunol. 195, 139–148. doi:10.1016/j.clim.2018.06.003
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. The Lancet 395 (10223), 497–506. doi:10.1016/S0140-6736(20)30183-5
Huang, H.-Y., Wang, S.-H., Tang, Y., Sheng, W., Zuo, C.-J., Wu, D.-W., et al. (2021). Landscape and Progress of Global COVID-19 Vaccine Development. Hum. Vaccin. Immunother. 17, 3276–3280. doi:10.1080/21645515.2021.1945901
Humphries, F., Shmuel-Galia, L., Jiang, Z., Wilson, R., Landis, P., Ng, S.-L., et al. (2021). A Diamidobenzimidazole STING Agonist Protects against SARS-CoV-2 Infection. Sci. Immunol. 6 (59), eabi9002. doi:10.1126/sciimmunol.abi9002
Hung, H. C., Ke, Y. Y., Huang, S. Y., Huang, P. N., Kung, Y. A., Chang, T. Y., et al. (2020). Discovery of M Protease Inhibitors Encoded by SARS-CoV-2. Antimicrob. Agents Chemother. 64 (9). doi:10.1128/AAC.00872-20
Jan, J. T., Cheng, T. R., Juang, Y. P., Ma, H. H., Wu, Y. T., Yang, W. B., et al. (2021). Identification of Existing Pharmaceuticals and Herbal Medicines as Inhibitors of SARS-CoV-2 Infection. Proc. Natl. Acad. Sci. U S A. 118 (5). doi:10.1073/pnas.2021579118
Jeon, S., Ko, M., Lee, J., Choi, I., Byun, S. Y., Park, S., et al. (2020). Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob. Agents Chemother. 64, e00819–20. doi:10.1128/AAC.00819-20
Jin, W., Stokes, J. M., Eastman, R. T., Itkin, Z., Zakharov, A. V., Collins, J. J., et al. (2021). Deep Learning Identifies Synergistic Drug Combinations for Treating COVID-19. Proc. Natl. Acad. Sci. U S A. 118 (39). doi:10.1073/pnas.2105070118
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. (2020). Structure of Mpro from SARS-CoV-2 and Discovery of its Inhibitors. Nature 582 (7811), 289–293. doi:10.1038/s41586-020-2223-y
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. (2020). Structure of M(pro) from SARS-CoV-2 and Discovery of its Inhibitors. Nature 582, 289–293. doi:10.1038/s41586-020-2223-y
Kaptein, S. J. F., Jacobs, S., Langendries, L., Seldeslachts, L., Ter Horst, S., Liesenborghs, L., et al. (2020). Favipiravir at High Doses Has Potent Antiviral Activity in SARS-CoV-2−infected Hamsters, whereas Hydroxychloroquine Lacks Activity. Proc. Natl. Acad. Sci. USA 117 (43), 26955–26965. doi:10.1073/pnas.2014441117
Kazmi, F., Hensley, T., Pope, C., Funk, R. S., Loewen, G. J., Buckley, D. B., et al. (2013). Lysosomal Sequestration (Trapping) of Lipophilic Amine (Cationic Amphiphilic) Drugs in Immortalized Human Hepatocytes (Fa2N-4 Cells). Drug Metab. Dispos 41 (4), 897–905. doi:10.1124/dmd.112.050054
Kyriakidis, N. C., López-Cortés, A., González, E. V., Grimaldos, A. B., and Prado, E. O. (2021). SARS-CoV-2 Vaccines Strategies: a Comprehensive Review of Phase 3 Candidates. NPJ Vaccin. 6 (1), 28. doi:10.1038/s41541-021-00292-w
Lamb, J., Crawford, E. D., Peck, D., Modell, J. W., Blat, I. C., Wrobel, M. J., et al. (2006). The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 313 (5795), 1929–1935. doi:10.1126/science.1132939
Lamb, J. (2007). The Connectivity Map: a New Tool for Biomedical Research. Nat. Rev. Cancer 7 (1), 54–60. doi:10.1038/nrc2044
Lane, T. R., Comer, J. E., Freiberg, A. N., Madrid, P. B., and Ekins, S. (2019). Repurposing Quinacrine against Ebola Virus InfectionIn Vivo. Antimicrob. Agents Chemother. 63, e01142–19. doi:10.1128/AAC.01142-19
Lane, T. R., Dyall, J., Mercer, L., Goodin, C., Foil, D. H., Zhou, H., et al. (2020). Repurposing Pyramax® for the Treatment of Ebola Virus Disease: Additivity of the Lysosomotropic Pyronaridine and Non-lysosomotropic Artesunate. Antiviral Res. 182, 104908. doi:10.1016/j.antiviral.2020.104908
Lane, T. R., and Ekins, S. (2021). Defending Antiviral Cationic Amphiphilic Drugs that May Cause Drug-Induced Phospholipidosis. J. Chem. Inf. Model. 61, 4125–4130. doi:10.1021/acs.jcim.1c00903
Lane, T. R., and Ekins, S. (2020). Toward the Target: Tilorone, Quinacrine, and Pyronaridine Bind to Ebola Virus Glycoprotein. ACS Med. Chem. Lett. 11, 1653–1658. doi:10.1101/2020.05.26.11818210.1021/acsmedchemlett.0c00298
Lane, T. R., Massey, C., Comer, J. E., Anantpadma, M., Freundlich, J. S., Davey, R. A., et al. (2019). Repurposing the Antimalarial Pyronaridine Tetraphosphate to Protect against Ebola Virus Infection. Plos Negl. Trop. Dis. 13 (11), e0007890. doi:10.1371/journal.pntd.0007890
Lane, T. R., Massey, C., Comer, J. E., Freiberg, A. N., Zhou, H., Dyall, J., et al. (2020). Pyronaridine Tetraphosphate Efficacy against Ebola Virus Infection in guinea Pig. Antiviral Res. 181, 104863. doi:10.1016/j.antiviral.2020.104863
Lechien, J. R., Chiesa-Estomba, C. M., De Siati, D. R., Horoi, M., Le Bon, S. D., Rodriguez, A., et al. (2020). Olfactory and Gustatory Dysfunctions as a Clinical Presentation of Mild-To-Moderate Forms of the Coronavirus Disease (COVID-19): a Multicenter European Study. Eur. Arch. Otorhinolaryngol. doi:10.1007/s00405-020-05965-1
Lenz, H.-J., Richardson, P., and Stebbing, J. (2021). The Emergence of Baricitinib: A Story of Tortoises versus Hares. Clin. Infect. Dis. 72 (7), 1251–1252. doi:10.1093/cid/ciaa940
Lenze, E. J., Mattar, C., Zorumski, C. F., Stevens, A., Schweiger, J., Nicol, G. E., et al. (2020). Fluvoxamine vs Placebo and Clinical Deterioration in Outpatients with Symptomatic COVID-19. JAMA 324 (22), 2292–2300. doi:10.1001/jama.2020.22760
Li, K., Meyerholz, D. K., Bartlett, J. A., and McCray, P. B. (2021). The TMPRSS2 Inhibitor Nafamostat Reduces SARS-CoV-2 Pulmonary Infection in Mouse Models of COVID-19. mBio 12 (4), e0097021. doi:10.1128/mBio.00970-21
Liesenborghs, L., Spriet, I., Jochmans, D., Belmans, A., Gyselinck, I., Teuwen, L. A., et al. (2021). Corrigendum to "itraconazole for COVID-19: Preclinical Studies and a Proof-Of-Concept Randomized Clinical Trial Laurens. EBioMedicine 69, 103454. doi:10.1016/j.ebiom.2021.103454
Liesenborghs, L., Spriet, I., Jochmans, D., Belmans, A., Gyselinck, I., Teuwen, L. A., et al. (2021). Itraconazole for COVID-19: Preclinical Studies and a Proof-Of-Concept Randomized Clinical Trial. EBioMedicine 66, 103288. doi:10.1016/j.ebiom.2021.103288
Liu, J., Cao, R., Xu, M., Wang, X., Zhang, H., Hu, H., et al. (2020). Hydroxychloroquine, a Less Toxic Derivative of Chloroquine, Is Effective in Inhibiting SARS-CoV-2 Infection In Vitro. Cell Discov 6, 16. doi:10.1038/s41421-020-0156-0
Liu, J., Zheng, X., Tong, Q., Li, W., Wang, B., Sutter, K., et al. (2020). Overlapping and Discrete Aspects of the Pathology and Pathogenesis of the Emerging Human Pathogenic Coronaviruses SARS‐CoV, MERS‐CoV, and 2019‐nCoV. J. Med. Virol. 92, 491–494. doi:10.1002/jmv.25709
Lowery, S. A., Sariol, A., and Perlman, S. (2021). Innate Immune and Inflammatory Responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe 29 (7), 1052–1062. doi:10.1016/j.chom.2021.05.004
Mao, L., Jin, H., Wang, M., Hu, Y., Chen, S., He, Q., et al. (2020). Neurologic Manifestations of Hospitalized Patients with Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. doi:10.1001/jamaneurol.2020.1127
Martinez, D. R., Schafer, A., Leist, S. R., Li, D., Gully, K., Yount, B., et al. (2021). Prevention and Therapy of SARS-CoV-2 and the B.1.351 Variant in Mice. Cell Rep 36 (4), 109450. doi:10.1016/j.celrep.2021.109450
Martinez-Guerrero, L., Zhang, X., Zorn, K. M., Ekins, S., and Wright, S. H. (2021). Cationic Compounds with SARS-CoV-2 Antiviral Activity and Their Interaction with Organic Cation Transporter/Multidrug and Toxin Extruder Secretory Transporters. J. Pharmacol. Exp. Ther. 379, 96–107. In press. doi:10.1124/jpet.121.000619
Menni, C., Valdes, A. M., Freidin, M. B., Sudre, C. H., Nguyen, L. H., Drew, D. A., et al. (2020). Real-time Tracking of Self-Reported Symptoms to Predict Potential COVID-19. Nat. Med. doi:10.1038/s41591-020-0916-2
Mercante, G., Ferreli, F., De Virgilio, A., Gaino, F., Di Bari, M., Colombo, G., et al. (2020). Prevalence of Taste and Smell Dysfunction in Coronavirus Disease 2019. JAMA Otolaryngol. Head Neck Surg. doi:10.1001/jamaoto.2020.1155
Meunier, N., Briand, L., Jacquin-Piques, A., Brondel, L., and Penicaud, L. (2020). COVID 19-Induced Smell and Taste Impairments: Putative Impact on Physiology. Front. Physiol. 11, 625110. doi:10.3389/fphys.2020.625110
Miller, S. R., Lane, T. R., Zorn, K. M., Ekins, S., Wright, S. H., and Cherrington, N. J. (2021). Multiple Computational Approaches for Predicting Drug Interactions with Human Equilibrative Nucleoside Transporter 1. Drug Metab. Dispos 49 (7), 479–489. doi:10.1124/dmd.121.000423
Miller, S. R., McGrath, M. E., Zorn, K. M., Ekins, S., Wright, S. H., and Cherrington, N. J. (2021). Remdesivir and EIDD-1931 Interact with Human Equilibrative Nucleoside Transporters 1 and 2: Implications for Reaching SARS-CoV-2 Viral Sanctuary Sites. Mol. Pharmacol. 100, 548–557. In press. doi:10.1124/molpharm.121.000333
Miller, S. R., Zhang, X., Hau, R. K., Jilek, J. L., Jennings, E. Q., Galligan, J. J., et al. (2021). Predicting Drug Interactions with Human Equilibrative Nucleoside Transporters 1 and 2 Using Functional Knockout Cell Lines and Bayesian Modeling. Mol. Pharmacol. 99 (2), 147–162. doi:10.1124/molpharm.120.000169
Mirabelli, C., Wotring, J. W., Zhang, C. J., McCarty, S. M., Fursmidt, R., Frum, T., et al. (2020). Morphological Cell Profiling of SARS-CoV-2 Infection Identifies Drug Repurposing Candidates for COVID-19. bioRxiv. doi:10.1101/2020.05.27.117184
Molony, R. D., Nguyen, J. T., Kong, Y., Montgomery, R. R., Shaw, A. C., and Iwasaki, A. (2017). Aging Impairs Both Primary and Secondary RIG-I Signaling for Interferon Induction in Human Monocytes. Sci. Signal. 10 (509). doi:10.1126/scisignal.aan2392
Mulangu, S., Dodd, L. E., Davey, R. T., Tshiani Mbaya, O., Proschan, M., Mukadi, D., et al. (2019). A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 381 (24), 2293–2303. doi:10.1056/NEJMoa1910993
Mulay, A., Konda, B., Garcia, G., Yao, C., Beil, S., Sen, C., et al. (2020). SARS-CoV-2 Infection of Primary Human Lung Epithelium for COVID-19 Modeling and Drug Discovery. bioRxiv. doi:10.1101/2020.06.29.174623
Muñoz-Fontela, C., Dowling, W. E., Funnell, S. G. P., Gsell, P.-S., Riveros-Balta, A. X., Albrecht, R. A., et al. (2020). Animal Models for COVID-19. Nature 586 (7830), 509–515. doi:10.1038/s41586-020-2787-6
Muratov, E. N., Amaro, R., Andrade, C. H., Brown, N., Ekins, S., Fourches, D., et al. (2021). A Critical Overview of Computational Approaches Employed for COVID-19 Drug Discovery. Chem. Soc. Rev. 50, 9121–9151. doi:10.1039/d0cs01065k
Murray, J., Hogan, R. J., Martin, D. E., Blahunka, K., Sancilio, F. D., Balyan, R., et al. (2021). Probenecid Inhibits SARS-CoV-2 Replication In Vivo and In Vitro. Sci. Rep. 11 (1), 18085. doi:10.1038/s41598-021-97658-w
Nadanaciva, S., Lu, S., Gebhard, D. F., Jessen, B. A., Pennie, W. D., and Will, Y. (2011). A High Content Screening Assay for Identifying Lysosomotropic Compounds. Toxicol. Vitro 25 (3), 715–723. doi:10.1016/j.tiv.2010.12.010
Nguyen, L. C., Yang, D., Nicolaescu, V., Best, T. J., Gula, H., Saxena, D., et al. (2022). Cannabidiol Inhibits SARS-CoV-2 Replication through Induction of the Host ER Stress and Innate Immune Responses. Sci. Adv. eabi6110.
Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., et al. (2021). Author Correction: Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and its Immune Cross-Reactivity with SARS-CoV. Nat. Commun. 12 (1), 2144. doi:10.1038/s41467-021-22614-1
Owen, D. R., Allerton, C. M. N., Anderson, A. S., Aschenbrenner, L., Avery, M., Berritt, S., et al. (2021). An Oral SARS-CoV-2 M Pro Inhibitor Clinical Candidate for the Treatment of COVID-19. Science 374, 1586–1593. doi:10.1126/science.abl4784
Pan, Y., Guan, H., Zhou, S., Wang, Y., Li, Q., Zhu, T., et al. (2020). Initial CT Findings and Temporal Changes in Patients with the Novel Coronavirus Pneumonia (2019-nCoV): a Study of 63 Patients in Wuhan, China. Eur. Radiol. 30, 3306–3309. doi:10.1007/s00330-020-06731-x
Paul, S. M., Mytelka, D. S., Dunwiddie, C. T., Persinger, C. C., Munos, B. H., Lindborg, S. R., et al. (2010). How to Improve R&D Productivity: the Pharmaceutical Industry's Grand challenge. Nat. Rev. Drug Discov. 9 (3), 203–214. doi:10.1038/nrd3078
Pruijssers, A. J., George, A. S., Schäfer, A., Leist, S. R., Gralinksi, L. E., Dinnon, K. H., et al. (2020). Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cel Rep. 32 (3), 107940. doi:10.1016/j.celrep.2020.107940
Puhl, A. C., Fritch, E. J., Lane, T. R., Tse, L. V., Yount, B. L., Sacramento, C. Q., et al. (2021). Repurposing the Ebola and Marburg Virus Inhibitors Tilorone, Quinacrine, and Pyronaridine: In Vitro Activity against SARS-CoV-2 and Potential Mechanisms. ACS Omega 6 (11), 7454–7468. doi:10.1021/acsomega.0c05996
Puhl, A. C., Gomes, G. F., Damasceno, S., Godoy, A. S., Noske, G. D., Nakamura, A. M., et al. (2021). Pyronaridine Protects against SARS-CoV-2 in Mouse. bioRxiv 0930, 462449. doi:10.1101/2021.09.30.462449
Raghuvanshi, R., and Bharate, S. B. (2021). Recent Developments in the Use of Kinase Inhibitors for Management of Viral Infections. J. Med. Chem. 65, 893–921. doi:10.1021/acs.jmedchem.0c01467
Rehman, M. F. U., Fariha, C., Anwar, A., Shahzad, N., Ahmad, M., Mukhtar, S., et al. (2021). Novel Coronavirus Disease (COVID-19) Pandemic: A Recent Mini Review. Comput. Struct. Biotechnol. J. 19, 612–623. doi:10.1016/j.csbj.2020.12.033
Reis, G., Dos Santos Moreira-Silva, E. A., Silva, D. C. M., Thabane, L., Milagres, A. C., Ferreira, T. S., et al. (2021). Effect of Early Treatment with Fluvoxamine on Risk of Emergency Care and Hospitalisation Among Patients with COVID-19: the TOGETHER Randomised, Platform Clinical Trial. Lancet Glob. Health. doi:10.1016/S2214-109X(21)00448-4
Richardson, P., Griffin, I., Tucker, C., Smith, D., Oechsle, O., Phelan, A., et al. (2020). Baricitinib as Potential Treatment for 2019-nCoV Acute Respiratory Disease. The Lancet 395 (10223), e30–e31. doi:10.1016/S0140-6736(20)30304-4
Riva, L., Yuan, S., Yin, X., Martin-Sancho, L., Matsunaga, N., Pache, L., et al. (2020). Discovery of SARS-CoV-2 Antiviral Drugs through Large-Scale Compound Repurposing. Nature 586, 113–119. doi:10.1038/s41586-020-2577-1
Rosen, D. A., Seki, S. M., Fernández-Castañeda, A., Beiter, R. M., Eccles, J. D., Woodfolk, J. A., et al. (2019). Modulation of the Sigma-1 Receptor-IRE1 Pathway Is Beneficial in Preclinical Models of Inflammation and Sepsis. Sci. Transl. Med. 11 (478). doi:10.1126/scitranslmed.aau5266
Rosenke, K., Hansen, F., Schwarz, B., Feldmann, F., Haddock, E., Rosenke, R., et al. (2021). Orally Delivered MK-4482 Inhibits SARS-CoV-2 Replication in the Syrian Hamster Model. Nat. Commun. 12 (1), 2295. doi:10.1038/s41467-021-22580-8
Rosenke, K., Jarvis, M. A., Feldmann, F., Schwarz, B., Okumura, A., Lovaglio, J., et al. (2020). Hydroxychloroquine Prophylaxis and Treatment Is Ineffective in Macaque and Hamster SARS-CoV-2 Disease Models. JCI Insight 5 (23). doi:10.1172/jci.insight.143174
Rossi, A., Magri, F., Sernicola, A., Michelini, S., Caro, G., Muscianese, M., et al. (2021). Telogen Effluvium after SARS-CoV-2 Infection: A Series of Cases and Possible Pathogenetic Mechanisms. Skin Appendage Disord. 21, 1–5. doi:10.1159/000517223
Salata, C., Calistri, A., Parolin, C., Baritussio, A., and Palù, G. (2017). Antiviral Activity of Cationic Amphiphilic Drugs. Expert Rev. Anti-infective Ther. 15 (5), 483–492. doi:10.1080/14787210.2017.1305888
Schiffman, S. S. (2015). “Influence of Drugs on Taste Function,” in Handbook of Olfaction and Gustation. Editor R. L. Doty (John Wiley & Sons). doi:10.1002/9781118971758.ch40
Schiffman, S. S. (2018). Influence of Medications on Taste and Smell. World J. Otorhinolaryngol. Head Neck Surg. 4 (1), 84–91. doi:10.1016/j.wjorl.2018.02.005
Schiffman, S. S. (1983). Taste and Smell in Disease (First of Two Parts). N. Engl. J. Med. 308 (21), 1275–1279. doi:10.1056/NEJM198305263082107
Schiffman, S. S. (1983). Taste and Smell in Disease (Second of Two Parts). N. Engl. J. Med. 308 (22), 1337–1343. doi:10.1056/NEJM198306023082207
Schiffman, S. S. (1997). Taste and Smell Losses in normal Aging and Disease. JAMA 278 (16), 1357–1362. doi:10.1001/jama.1997.03550160077042
Schiffman, S. S., Zervakis, J., Graham, B. G., and Westhall, H. L. (2002). “Age-related Chemosensory Losses: Effect of Medications,” in Chemistry of Taste. Editor P. G. D. Paredes (Washington DC: American Chemical Society). doi:10.1021/bk-2002-0825.ch008
Seneviratne, C. J., Balan, P., Ko, K. K. K., Udawatte, N. S., Lai, D., Ng, D. H. L., et al. (2021). Efficacy of Commercial Mouth-Rinses on SARS-CoV-2 Viral Load in Saliva: Randomized Control Trial in Singapore. Infection 49 (2), 305–311. doi:10.1007/s15010-020-01563-9
Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., et al. (2020). Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 117 (21), 11727–11734. doi:10.1073/pnas.2003138117
Sharquie, K. E., and Jabbar, R. I. (2021). COVID-19 Infection Is a Major Cause of Acute Telogen Effluvium. Ir J. Med. Sci. doi:10.1007/s11845-021-02754-5
Sheahan, T. P., Sims, A. C., Zhou, S., Graham, R. L., Pruijssers, A. J., Agostini, M. L., et al. (2020). An Orally Bioavailable Broad-Spectrum Antiviral Inhibits SARS-CoV-2 in Human Airway Epithelial Cell Cultures and Multiple Coronaviruses in Mice. Sci. Transl. Med. 12 (541), eabb5883. doi:10.1126/scitranslmed.abb5883
Shulla, A., Heald-Sargent, T., Subramanya, G., Zhao, J., Perlman, S., and Gallagher, T. (2011). A Transmembrane Serine Protease Is Linked to the Severe Acute Respiratory Syndrome Coronavirus Receptor and Activates Virus Entry. J. Virol. 85 (2), 873–882. doi:10.1128/JVI.02062-10
Spinner, C. D., Gottlieb, R. L., Criner, G. J., Arribas López, J. R., Cattelan, A. M., Soriano Viladomiu, A., et al. (2020). Effect of Remdesivir vs Standard Care on Clinical Status at 11 Days in Patients with Moderate COVID-19. JAMA 324 (11), 1048–1057. doi:10.1001/jama.2020.16349
Starace, M., Iorizzo, M., Sechi, A., Alessandrini, A. M., Carpanese, M., Bruni, F., et al. (2021). Trichodynia and Telogen Effluvium in COVID-19 Patients: Results of an International Expert Opinion Survey on Diagnosis and Management. JAAD Int. 5, 11–18. doi:10.1016/j.jdin.2021.07.006
Stebbing, J., Krishnan, V., Bono, S., Ottaviani, S., Casalini, G., Richardson, P. J., et al. (2020). Mechanism of Baricitinib Supports Artificial Intelligence‐predicted Testing in COVID ‐19 Patients. EMBO Mol. Med. 12 (8), e12697. doi:10.15252/emmm.202012697
Subramanian, A., Narayan, R., Corsello, S. M., Peck, D. D., Natoli, T. E., Lu, X., et al. (2017). A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171 (6), 1437–1452. doi:10.1016/j.cell.2017.10.049
Sukhatme, V. P., Reiersen, A. M., Vayttaden, S. J., and Sukhatme, V. V. (2021). Fluvoxamine: A Review of its Mechanism of Action and its Role in COVID-19. Front. Pharmacol. 12 (763). doi:10.3389/fphar.2021.652688
Sungnak, W., Huang, N., Huang, N., Bécavin, C., Berg, M., Queen, R., et al. (2020). SARS-CoV-2 Entry Factors Are Highly Expressed in Nasal Epithelial Cells Together with Innate Immune Genes. Nat. Med. 26 (5), 681–687. doi:10.1038/s41591-020-0868-6
Suzuki, M., Saito, K., Min, W. P., Vladau, C., Toida, K., Itoh, H., et al. (2007). Identification of Viruses in Patients with Postviral Olfactory Dysfunction. Laryngoscope 117 (2), 272–277. doi:10.1097/01.mlg.0000249922.37381.1e
Titanji, B. K., Farley, M. M., Mehta, A., Connor-Schuler, R., Moanna, A., Cribbs, S. K., et al. (2021). Use of Baricitinib in Patients with Moderate to Severe Coronavirus Disease 2019. Clin. Infect. Dis. 72 (7), 1247–1250. doi:10.1093/cid/ciaa879
Tomazini, B. M., Maia, I. S., Cavalcanti, A. B., Berwanger, O., Rosa, R. G., Veiga, V. C., et al. (2020). Effect of Dexamethasone on Days Alive and Ventilator-free in Patients with Moderate or Severe Acute Respiratory Distress Syndrome and COVID-19. JAMA 324 (13), 1307–1316. doi:10.1001/jama.2020.17021
Touret, F., Driouich, J.-S., Cochin, M., Petit, P. R., Gilles, M., Barthélémy, K., et al. (2021). Preclinical Evaluation of Imatinib Does Not Support its Use as an Antiviral Drug against SARS-CoV-2. Antiviral Res. 193, 105137. doi:10.1016/j.antiviral.2021.105137
Tummino, T. A., Rezelj, V. V., Fischer, B., Fischer, A., O’Meara, M. J., Monel, B., et al. (2021). Drug-induced Phospholipidosis Confounds Drug Repurposing for SARS-CoV-2. Science 373 (6554), 541–547. doi:10.1126/science.abi4708
Vallejos, J., Zoni, R., Bangher, M., Villamandos, S., Bobadilla, A., Plano, F., et al. (2021). Ivermectin to Prevent Hospitalizations in Patients with COVID-19 (IVERCOR-COVID19) a Randomized, Double-Blind, Placebo-Controlled Trial. BMC Infect. Dis. 21 (1), 635. doi:10.1186/s12879-021-06348-5
Vater, M., Möckl, L., Gormanns, V., Fademrecht, C. S., Mallmann, A. M., Ziegart-Sadowska, K., et al. (2017). Erratum: Corrigendum: New Insights into the Intracellular Distribution Pattern of Cationic Amphiphilic Drugs. Sci. Rep. 7, 46011. doi:10.1038/srep46011
Vater, M., Möckl, L., Gormanns, V., Schultz Fademrecht, C., Mallmann, A. M., Ziegart-Sadowska, K., et al. (2017). New Insights into the Intracellular Distribution Pattern of Cationic Amphiphilic Drugs. Sci. Rep. 7, 44277. doi:10.1038/srep44277
von Delft, F., Calmiano, M., Chodera, J., Griffen, E., Lee, A., London, N., et al. (2021). A white-knuckle Ride of Open COVID Drug Discovery. Nature 594 (7863), 330–332. doi:10.1038/d41586-021-01571-1
Wahl, A., Gralinski, L. E., Johnson, C. E., Yao, W., Kovarova, M., Dinnon, K. H., et al. (2021). SARS-CoV-2 Infection Is Effectively Treated and Prevented by EIDD-2801. Nature 591 (7850), 451–457. doi:10.1038/s41586-021-03312-w
Wang, J., Yang, X., Li, Y., Huang, J.-a., Jiang, J., and Su, N. (2021). Specific Cytokines in the Inflammatory Cytokine Storm of Patients with COVID-19-Associated Acute Respiratory Distress Syndrome and Extrapulmonary Multiple-Organ Dysfunction. Virol. J. 18 (1), 117. doi:10.1186/s12985-021-01588-y
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. (2020). Remdesivir and Chloroquine Effectively Inhibit the Recently Emerged Novel Coronavirus (2019-nCoV) In Vitro. Cell Res 30 (3), 269–271. doi:10.1038/s41422-020-0282-0
Wang, Y., Zhang, D., Du, G., Du, R., Zhao, J., Jin, Y., et al. (2020). Remdesivir in Adults with Severe COVID-19: a Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial. The Lancet 395 (10236), 1569–1578. doi:10.1016/S0140-6736(20)31022-9
Weisberg, E., Parent, A., Yang, P. L., Sattler, M., Liu, Q., Liu, Q., et al. (2020). Repurposing of Kinase Inhibitors for Treatment of COVID-19. Pharm. Res. 37 (9), 167. doi:10.1007/s11095-020-02851-7
Weston, S., Coleman, C. M., Haupt, R., Logue, J., Matthews, K., Li, Y., et al. (2020). Broad Anti-coronavirus Activity of Food and Drug Administration-Approved Drugs against SARS-CoV-2 In Vitro and SARS-CoV In Vivo. J. Virol. 94 (21). doi:10.1128/JVI.01218-20
Whitcroft, K. L., and Hummel, T. (2020). Olfactory Dysfunction in COVID-19: Diagnosis and Management. JAMA. doi:10.1001/jama.2020.83910.1001/jama.2020.8391
White, K. M., Rosales, R., Yildiz, S., Kehrer, T., Miorin, L., Moreno, E., et al. (2021). Plitidepsin Has Potent Preclinical Efficacy against SARS-CoV-2 by Targeting the Host Protein eEF1A. Science 371 (6532), 926–931. doi:10.1126/science.abf4058
Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., et al. (2020). Author Correction: A New Coronavirus Associated with Human Respiratory Disease in China. Nature 580 (7803), E7. doi:10.1038/s41586-020-2202-3
Xia, H., Cao, Z., Xie, X., Zhang, X., Chen, J. Y., Wang, H., et al. (2020). Evasion of Type I Interferon by SARS-CoV-2. Cel Rep 33 (1), 108234. doi:10.1016/j.celrep.2020.108234
Xydakis, M. S., Dehgani-Mobaraki, P., Holbrook, E. H., Geisthoff, U. W., Bauer, C., Hautefort, C., et al. (2020). Smell and Taste Dysfunction in Patients with COVID-19. Lancet Infect. Dis. doi:10.1016/S1473-3099(20)30293-0
Yee, S. W., Vora, B., Oskotsky, T., Zou, L., Jakobsen, S., Enogieru, O. J., et al. (2021). Drugs in COVID‐19 Clinical Trials: Predicting Transporter‐Mediated Drug‐Drug Interactions Using In Vitro Assays and Real‐World Data. Clin. Pharmacol. Ther. 110 (1), 108–122. doi:10.1002/cpt.2236
Yuan, S., Chan, J. F. W., Chik, K. K. H., Chan, C. C. Y., Tsang, J. O. L., Liang, R., et al. (2020). Discovery of the FDA-Approved Drugs Bexarotene, Cetilistat, Diiodohydroxyquinoline, and Abiraterone as Potential COVID-19 Treatments with a Robust Two-Tier Screening System. Pharmacol. Res. 159, 104960. doi:10.1016/j.phrs.2020.104960
Yuan, S., Yin, X., Meng, X., Chan, J. F., Ye, Z. W., Riva, L., et al. (2021). Clofazimine Broadly Inhibits Coronaviruses Including SARS-CoV-2. Nature 593 (7859), 418–423. doi:10.1038/s41586-021-03431-4
Yuen, C. K., Lam, J. Y., Wong, W. M., Mak, L. F., Wang, X., Chu, H., et al. (2020). SARS-CoV-2 Nsp13, Nsp14, Nsp15 and Orf6 Function as Potent Interferon Antagonists. Emerg. Microbes Infect. 9 (1), 1418–1428. doi:10.1080/22221751.2020.1780953
Zhang, Q., Bastard, P., Liu, Z., Le Pen, J., Moncada-Velez, M., Chen, J., et al. U. T. C. I. Group (2020). Inborn Errors of Type I IFN Immunity in Patients with Life-Threatening COVID-19. Science 370 (6515). doi:10.1126/science.abd4570
Zhang, Q., Chen, C. Z., Swaroop, M., Xu, M., Wang, L., Lee, J., et al. (2020). Heparan Sulfate Assists SARS-CoV-2 in Cell Entry and Can Be Targeted by Approved Drugs In Vitro. Cel Discov 6 (1), 80. doi:10.1038/s41421-020-00222-5
Zhang, Y.-N., Zhang, Q.-Y., Li, X.-D., Xiong, J., Xiao, S.-Q., Wang, Z., et al. (2020). Gemcitabine, Lycorine and Oxysophoridine Inhibit Novel Coronavirus (SARS-CoV-2) in Cell Culture. Emerging Microbes & Infections 9 (1), 1170–1173. doi:10.1080/22221751.2020.1772676
Zhao, H., Mendenhall, M., and Deininger, M. W. (2020). Imatinib Is Not a Potent Anti-SARS-CoV-2 Drug. Leukemia 34 (11), 3085–3087. doi:10.1038/s41375-020-01045-9
Zimmer, M., Lamb, J., Ebert, B. L., Lynch, M., Neil, C., Schmidt, E., et al. (2010). The Connectivity Map Links Iron Regulatory Protein-1-Mediated Inhibition of Hypoxia-Inducible Factor-2a Translation to the Anti-inflammatory 15-deoxy-delta12,14-prostaglandin J2. Cancer Res. 70 (8), 3071–3079. doi:10.1158/0008-5472.CAN-09-2877
Keywords: SARS-CoV-2, antiviral, cytokine storm, COVID-19, drug discovery pipeline, repurposing
Citation: Puhl AC, Lane TR, Urbina F and Ekins S (2022) The Need for Speed and Efficiency: A Brief Review of Small Molecule Antivirals for COVID-19. Front. Drug. Discov. 2:837587. doi: 10.3389/fddsv.2022.837587
Received: 16 December 2021; Accepted: 27 January 2022;
Published: 30 March 2022.
Edited by:
Bruno Villoutreix, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
José L Medina-Franco, National Autonomous University of Mexico, MexicoOlivier Terrier, UMR5308 Centre International de Recherche en Infectiologie (CIRI), France
Giuseppe Felice Mangiatordi, Italian National Research Council, Italy
Copyright © 2022 Puhl, Lane, Urbina and Ekins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sean Ekins, c2VhbkBjb2xsYWJvcmF0aW9uc3BoYXJtYS5jb20=
 Ana C. Puhl
Ana C. Puhl Thomas R. Lane
Thomas R. Lane Fabio Urbina
Fabio Urbina Sean Ekins
Sean Ekins