
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Drug Deliv. , 05 July 2024
Sec. CNS Drug Delivery
Volume 4 - 2024 | https://doi.org/10.3389/fddev.2024.1433453
 Sarah F. Hathcock1
Sarah F. Hathcock1 Hallie E. Knight1
Hallie E. Knight1 Emma G. Tong1
Emma G. Tong1 Alexandra E. Meyer1
Alexandra E. Meyer1 Henry D. Mauser1
Henry D. Mauser1 Nadine Vollmuth1
Nadine Vollmuth1 Brandon J. Kim1,2,3,4*
Brandon J. Kim1,2,3,4*The blood-brain barrier (BBB) is comprised of specialized brain endothelial cells (BECs) that contribute to maintaining central nervous system (CNS) homeostasis. BECs possess properties such as an array of multi-drug efflux transporters that eject various drugs and toxins, preventing their entry into the CNS. Together, it is estimated that these efflux transporters can eject up to 98% of known xenobiotic compounds. P-glycoprotein (P-gp) is a promiscuous efflux transporter at the BBB and can efflux up to 90 various substrates, representing a major hurdle in CNS drug delivery for therapeutic interventions. This necessitates the study of P-gp to discover drugs that are non-substrates of P-gp as well as to identify novel P-gp inhibitors. Here we report the generation of P-gp overexpressing BECs under the endogenous promoter control that could be used in the screening of P-gp substrates. These cells could provide utility in the design of drugs or identification of novel inhibitors.
The blood-brain barrier (BBB) is comprised of highly specialized brain endothelial cells (BECs) that maintain central nervous system (CNS) homeostasis by tightly regulating the passage of molecules from the circulation (Engelhardt and Sorokin, 2009; Abbott et al., 2010; Redzic, 2011). BECs are unique compared to peripheral endothelial cells as they possess complex tight junctions, exhibit low rates of endocytosis and transcytosis, lack fenestrations in the capillaries, and express multi-drug efflux transporters (Engelhardt and Sorokin, 2009; Abbott et al., 2010; Redzic, 2011). P-glycoprotein (P-gp/MDR1) is a major efflux transporter found at the BBB and other tissues such as the gut and the blood-placental barrier (Mahar et al., 2002; Bauer et al., 2005; Perriere et al., 2005; van Assema et al., 2012; Davis et al., 2014). Generally, P-gp excludes small lipophilic molecules and some large molecules and proteins such as the amyloid-
hCMEC/D3 cells were maintained in EndoGRO MV (Millipore) media as previously described (Weksler et al., 2005; Espinal et al., 2022). Briefly, cells were passaged onto new flasks coated with 1% rat tail collagen (RTC) at 80% confluency. Generation of P-gp overexpressing hCMEC/D3 cells (hCMEC/D3-MDR1) was initiated by passing naïve hCMEC/D3s into media containing 0.5 μg/mL puromycin (AdipoGen cat# AG-CN2-0078-M025). Surviving cells were passed for five passages in 0.5 μg/mL puromycin before switching the media to 1 μg/mL puromycin. hCMEC/D3-MDR1s were stocked, frozen, and maintained in 1 μg/mL puromycin. All cells were incubated at 37°C + 5% CO2 in EndoGRO MV media with or without puromycin as described above, and media was changed on cells to EndoGRO MV without puromycin 1 day before any downstream applications.
Transendothelial electrical resistance (TEER) was measured using an EVOM II (World Precision Instruments) and the chopstick electrodes associated. hCMEC/D3 or hCMEC/D3-MDR1s were seeded onto Corning Transwells (3460) coated in 1% RTC and grown for 2 days at 37°C + 5% CO2 in EndoGRO MV media with or without puromycin as described above. Measurements were taken each day following seeding for 5 days.
hCMEC/D3s or hCMEC/D3-MDR1s were seeded onto 1% RTC coated 24-well plates in triplicate and grown for 4–5 days at 37°C + 5% CO2 in EndoGRO MV media with or without puromycin as described above. RNA was collected and purified using the Machery-Nagel NucleoSpin RNA kit (Fisher cat# 740955.250) and concentrations were measured using a Nanodrop 2000. Matching amounts of purified RNA were converted to cDNA using the qScript kit (Quantabio cat# 95047-500) and diluted 1:10 in nuclease-free water to be used for qPCR. SYBR green (Thermo Fisher cat# A25743) qPCR was performed on cDNA and measured using a QuantStudio3 (Thermo Fisher). Delta-delta CT calculations were made using 18S as a housekeeping gene. Data are expressed as fold change compared to naïve cells.
hCMEC/D3s or hCMEC/D3-MDR1s were seeded onto 1% RTC coated 25 cm2 flasks and grown for 4–5 days at 37°C + 5% CO2 in EndoGRO MV media with or without puromycin as described above. Cells were washed 3 times with PBS and removed from the flask by applying 1 mL Accutase™ (Stemcell Technologies cat# 07920) and incubating at 37°C + 5% CO2 for 10 min. Cells were collected with 4 mL of fresh EndoGRO MV media and pelleted at 500 × g for 10 min. Cells were fixed with 1% formaldehyde in PBS for 15 min at RT. Cells were then pelleted and washed twice with wash buffer (5% bovine serum albumin (BSA) + 0.1% Triton-X in PBS) to block and permeabilize for staining of total P-gp. Cells were resuspended in 1 mL wash buffer and cell density was estimated using a Countess 3 Automated Cell Counter. Samples were stained in wash buffer with anti-P-gp (Thermo Fisher cat# MA5-13854 [F4]) or an IgG1 isotype (Bio X Cell cat# BE0083) at .05 μg/million cells, and gently agitated overnight at 4°C. The following day, cells were washed twice in wash buffer and stained with a fluorescent secondary antibody (anti-mouse Alexa fluor 488, Thermo cat# A11001) at 1:5000 in wash buffer for 1 h at room temperature. Cells were then washed in wash buffer and resuspended in PBS. Data was collected on an Attune NxT Flow Cytometer, and histograms and dot plots were generated using the Attune NxT Software.
hCMEC/D3s and hCMEC/D3-MDR1s were seeded onto 1% RTC coated 24-well plates and grown for 4–5 days at 37°C + 5% CO2. Samples were lysed in RIPA buffer containing protease inhibitor cocktail and protein abundance was measured by bicinchoninic acid (BCA) assay (Thermo Fisher cat# 23227). Equal amounts of protein were then loaded on to 4%–12% Tris-glycine SDS-PAGE gels followed by transfer to a nitrocellulose membrane. Membranes were then stained by Ponceau S stain and imaged prior to blocking in 5% milk in Tris-buffered saline +0.1% Tween (TBST) for 1 h at room temperature. Primary antibodies diluted in 5% milk in TBST were then applied and allowed to incubate with the membrane overnight at 4°C. The following day, membranes were washed three times in TBST for 5 minutes each, followed by incubation with appropriate secondary antibodies (anti-mouse HRP Jackson ImmunoResearch cat# 115-035-003) (1:1000) for at least 1 h at room temperature. Following incubation, membranes were washed three times in TBST for 5 minutes each and imaged using enhanced chemiluminescence substrate (Thermo Fisher cat# 34577) and an iBright imager. Primary antibodies used: anti-VE-Cadherin (Santa Cruz cat# sc-52752 [BV9]) (1:1000), anti-GLUT1 (Thermo Fisher cat# MA5-11315 [SPM-498]) (1:1000), anti-P-glycoprotein (Thermo Fisher cat# PA5-28801]) (1:1000), anti-PECAM (CD31) (Thermo Fisher cat#BMS137 [Gi18]) (1:1000).
Substrate accumulation assays were performed as described previously (Stebbins et al., 2016; Kim et al., 2019). For P-gp substrate accumulation, Rhodamine 123 (R123) (Sigma-Aldrich 83702) was utilized to measure relative P-gp function. Briefly, cells were washed in Hank’s Balanced Salt Solution (with Calcium and Magnesium) (Thermo Fisher 14065056) and incubated with or without the inhibitors cyclosporine A (CsA) (Sigma Aldrich C1832) or Valspodar (PSC833) (R&D systems 40-421) at 10 μM for 1 h at 37°C + 5% CO2. Following pre-incubation, cells were then treated with 10 μM R123 with or without inhibitors and incubated for 2 h at 37°C + 5% CO2. Following incubation, cells were washed twice in ice-cold PBS and lysed in RIPA buffer. Samples were then measured using a SpectraMax iD3 plate reader (Molecular Devices), and BCA assays (Thermo Fisher cat# 23227) were then conducted on samples from each well. Fluorescence accumulation measurements were normalized to protein amounts.
hCMEC/D3s and hCMEC/D3-MDR1s were seeded onto 1% RTC coated 24-well plates and grown for 4–5 days at 37°C + 5% CO2. Cells were then imaged on a Nikon Ti2 microscope using a 20X objective and images were processed using ImageJ software (scale bar represents 50 µm).
GraphPad Prism version 10.2.1 was used for all statistics calculations. For pair-wise comparisons, Student’s t tests were performed. Statistical significance was accepted if p < 0.05.
hCMEC/D3 cells are typically maintained in EndoGRO MV media. To generate hCMEC/D3-MDR1, we chose to select for highly expressing P-gp cells by subjecting cells to the cytotoxic P-gp substrate puromycin. This strategy has been utilized previously to selectively purify BECs from other cell types in the brain as BECs expressing P-gp will efflux the toxin and survive (Demeuse et al., 2004; Perriere et al., 2005; Calabria et al., 2006). To select for highly expressing P-gp cells, hCMEC/D3 cells were passaged into flasks containing 0.5 μg/mL puromycin and allowed to expand. After at least five passages in 0.5 μg/mL, these were then passaged into 1 μg/mL puromycin (Figure 1A). hCMEC/D3 and hCMEC/D3-MDR1 possess similar morphology at low and high densities (Figure 1B). Expression of endogenous P-gp was measured by qPCR, which demonstrated that hCMEC/D3-MDR1 significantly upregulate ABCB1 (P-gp) compared to naïve hCMEC/D3s (Figure 1C). Examination of P-gp protein by flow cytometry (Figure 1D, Supplementary Figure S1) and immunoblot show increased levels of P-gp protein abundance in hCMEC/D3-MDR1 when compared to naïve hCMEC/D3s (Figures 1E, F). Levels of TEER remained similar between naïve hCMEC/D3 and hCMEC/D3-MDR1s once confluence was reached (Supplementary Figure S2A). Other markers of endothelial identity were slightly decreased in the hCMEC/D3-MDR1s versus the naïve hCMEC/D3s, indicating a partial loss of endothelial identity (Supplementary Figures S2B, C). Taken together these data demonstrate that the hCMEC/D3-MDR1s can be maintained in puromycin and overexpress P-gp without any exogenous genetic manipulation.
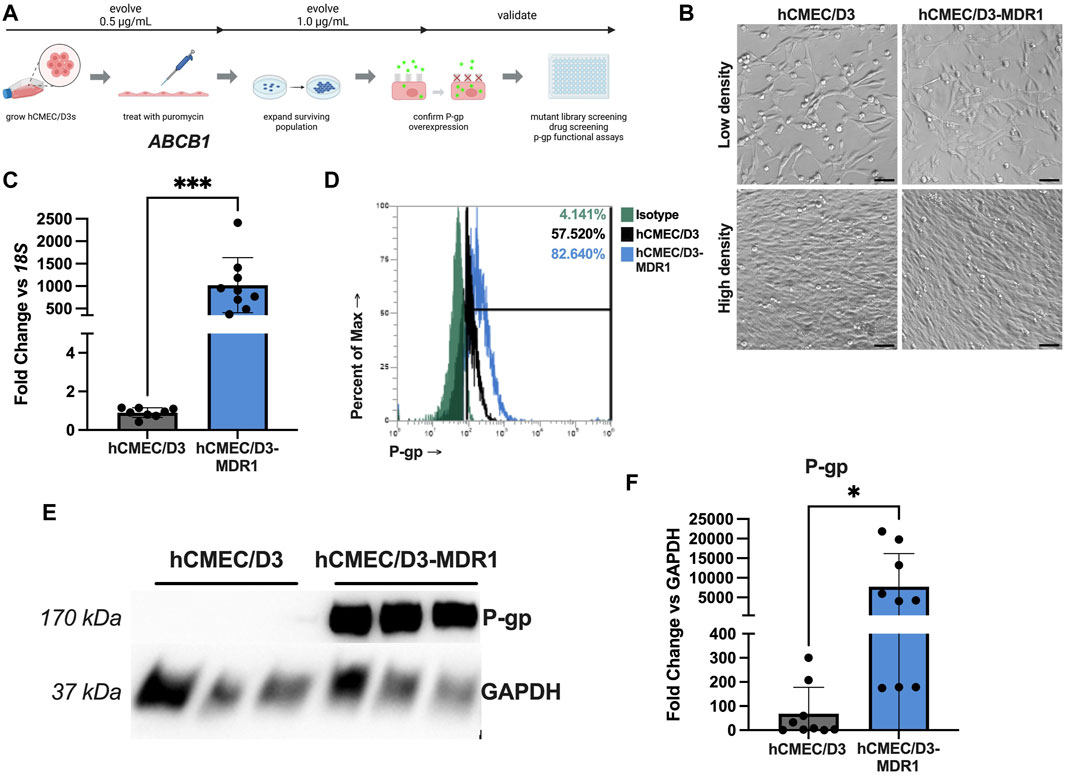
Figure 1. hCMEC/D3-MDR1 can be maintained in puromycin media and overexpress P-gp (A) Schematic of selection of hCMEC/D3-MDR1 and downstream applications (Created with Biorender.com), (B) Differential interference contrast microscopy of hCMEC/D3s versus hCMEC/D3-MDR1s at low and high densities reveal similar morphology between cell types, (C) qPCR of ABCB1 in hCMEC/D3s (gray) versus hCMEC/D3-MDR1s (blue) reveal upregulation of P-gp transcripts in hCMEC/D3-MDR1, (D) Flow cytometry reveal increased total P-gp in hCMEC/D3-MDR1 (blue = P-gp; green = isotype) versus hCMEC/D3 (black = P-gp) populations, (E) Western blot of P-gp in hCMEC/D3s versus hCMEC/D3-MDR1s reveal increased P-gp abundance in hCMEC/D3-MDR1 quantified in (F). Scale bars represent 50 µm. Student’s t-test was performed; *p < 0.05, ***p < 0.001. Experiments were performed on three distinct passages and in technical triplicate (n = 8 or 9) (C–F); representative images are shown.
We have demonstrated that hCMEC/D3-MDR1 overexpress P-gp (Figure 1). Next, to assess if this overexpression leads to a functional exclusion of substrates, we applied a well characterized substrate accumulation assay. Typically, P-gp excludes Rhodamine 123 (R123) from cells; however, if P-gp is inhibited with pharmacological inhibitors such as cyclosporine A (CsA) or the second-generation P-gp inhibitor Valspodar (PSC833), R123 is able to accumulate inside cells. To determine if another transporter, Breast Cancer Resistance Protein (BCRP), was functionally changed during the generation of hCMEC/D3-MDR1, we employed a similar substrate accumulation assay using the BCRP substrate Hoechst and inhibitor Ko143. Overall accumulation of R123 in hCMEC/D3 versus hCMEC/D3-MDR1 reveal that hCMEC/D3-MDR1 exclude R123 significantly more efficiently than hCMEC/D3 (Figure 2A). We also found no change in Hoechst exclusion between cell types (Figure 2B). We found that hCMEC/D3s minimally exclude R123 even in the absence of CsA or PSC833 (Figures 2C, D). However, hCMEC/D3-MDR1s significantly exclude R123 in the absence of an inhibitor and accumulate R123 in the presence of both inhibitors, suggesting that functional P-gp is highly expressed in hCMEC/D3-MDR1 (Figures 2C, D). Finally, we found that hCMEC/D3s and hCMEC/D3-MDR1s exclude Hoechst at similar rates, even in the presence of Ko143 (Figure 2E). These data suggest that hCMEC/D3-MDR1 overexpress functional P-gp and that the generation of hCMEC/D3-MDR1 did not impact BCRP expression or function. Therefore, we conclude that the hCMEC/D3-MDR1 will have particular utility in the screening for P-gp specific inhibitors or substrates.
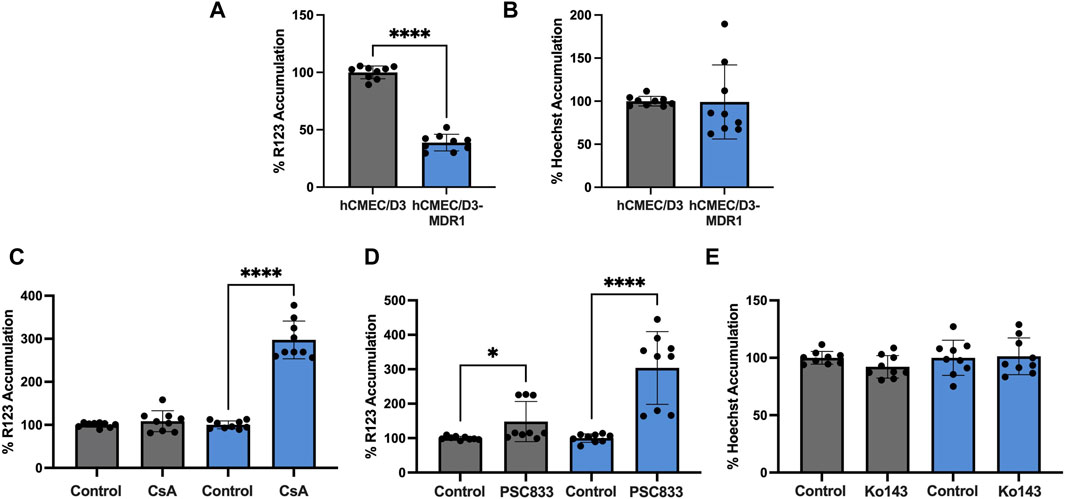
Figure 2. hCMEC/D3-MDR1 display increased P-gp specific function (A) Substrate accumulation of 10 µM R123 is significantly diminished in hCMEC/D3-MDR1 (blue) versus hCMEC/D3 (gray) indicating increased total P-gp mediated efflux, (B) Substrate accumulation of Hoechst does not differ between hCMEC/D3-MDR1s and hCMEC/D3s, indicating no change in BCRP mediated exclusion, substrate accumulation of R123 in the presence of P-gp inhibitors 10 µM cyclosporine A (C) or 10 µM PSC833 (D) indicate an increased dynamic range in P-gp inhibition in hCMEC/D3-MDR1s versus hCMEC/D3s, (E) Substrate accumulation of 10 µM Hoechst in the presence of BCRP inhibitor 1 µM Ko143 indicate no difference in the dynamic range of BCRP inhibition of hCMEC/D3-MDR1s versus hCMEC/D3s. All data are expressed as percent accumulation of control. Student’s t-test was performed; *p < 0.05, ****p < 0.0001. Experiments were performed on three distinct passages and in technical triplicate (n = 9).
Drug delivery to the CNS remains a major hurdle to overcome for the treatment of neurological disorders (Azad et al., 2015; Bhunia et al., 2023; Chatterjee et al., 2023; Cox et al., 2023; Ronaldson and Davis, 2024). The BBB restricts the majority of small molecules from entering the CNS due in part to the expression of functional efflux transporters (Azad et al., 2015; Bhunia et al., 2023; Chatterjee et al., 2023; Cox et al., 2023; Ronaldson and Davis, 2024). P-gp is a major efflux transporter present in BECs that contributes to the efflux of many drugs and toxins, preventing their entry to the CNS (Azad et al., 2015; Chatterjee et al., 2023; Cox et al., 2023). Here we report on a cell line that could be used to screen drugs and potential P-gp inhibitors (Figure 1A) that is of BEC origin and controls P-gp under the endogenous promoter.
Since the cytotoxic puromycin is a substrate for P-gp and has been utilized to purify BECs from primary tissue, we selectively expanded hCMEC/D3s that highly express P-gp and protect themselves from puromycin’s toxic impacts (Demeuse et al., 2004; Perriere et al., 2005; Calabria et al., 2006). These hCMEC/D3-MDR1s highly express functional P-gp and retain most characteristics of the parent hCMEC/D3 cell line (Figures 1, 2). Other P-gp overexpressing cells such as the MDCK-MDR1 cell line use exogenous promoters and genes to achieve this goal (Braun et al., 2000; Liu and Zeng, 2008; Lippmann et al., 2012; Cecchelli et al., 2014). These models are highly useful as they generate polarized monolayers, and when grown on transwells, can indicate whether a particular drug is a P-gp substrate as measured by drug concentrations between the apical and basolateral sides. Additionally, as these cells provide a polarized monolayer, they can mimic the properties of many cellular barriers, primarily epithelial. hCMEC/D3 cells are of endothelial origin and since they do not possess extremely tight barrier properties they may not be the most useful for drug delivery studies (Weksler et al., 2005; Helms et al., 2016). While the hCMEC/D3-MDR1s also do not possess tight barrier properties, they could still prove useful in the screening of potential P-gp inhibitors on endothelial cells. Furthermore, although endothelial properties are slightly altered in hCMEC/D3-MDR1 (Supplementary Figure S2), functional efflux transport still remains (Figure 2), preserving the utility of the model. hCMEC/D3-MDR1s control P-gp expression under the endogenous promoter and are of endothelial origin. Other endothelial models that demonstrate P-gp function include induced pluripotent stem-cell (iPSC) derived brain-like endothelial cell models (iBECs) (Pastan et al., 1988; Liu and Zeng, 2008). However, there are some iPSC lines that, even after differentiation, do not seem to possess robust transporter function (Ozgur et al., 2023; Nielsen et al., 2024). iBECs that do possess efflux transporter function demonstrate a roughly 50%–100% increase in R123 substrate accumulation when subjected to CsA or PSC833 (Lippmann et al., 2012; Lippmann et al., 2014; Stebbins et al., 2016; Hollmann et al., 2017; Kim et al., 2019; Neal et al., 2019). In contrast, the hCMEC/D3-MDR1s exhibit a dynamic range in the same assay of nearly 300% (Figure 2), potentially increasing the signal-to-noise ratio required to screen for novel P-gp inhibitors on endothelial cells. Overall, we report that the hCMEC/D3-MDR1 cell line robustly expresses functional P-gp and could be used for the screening of drug libraries or microbial mutant libraries for novel P-gp inhibitors in a native endothelial cell.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
SH: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. HK: Investigation, Methodology, Writing–review and editing. ET: Investigation, Methodology, Writing–review and editing. AM: Investigation, Methodology, Writing–review and editing. HM: Data curation, Writing–review and editing, Formal Analysis, Methodology. NV: Methodology, Writing–review and editing. BK: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by NIH/NINDS R15NS131921 to BK.
All diagrams created with Biorender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fddev.2024.1433453/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | (A) Representative flow cytometry with gating for unstained population reveals a larger shift into the gate for P-gp positive hCMEC/D3-MDR1s versus hCMEC/D3s with total mean gated percentages represented in (B) Student's t test was performed; * p < 0.05. Experiments were performed on three distinct passages and in technical triplicate (n = 9).
SUPPLEMENTARY FIGURE S2 | hCMEC/D3-MDR1 display diminished endothelial identity (A) TEER remains at similar levels between hCMEC/D3s (gray) and hCMEC/D3-MDR1 (blue) at confluence, (B) Western blots of VE-Cadherin, CD31, and GLUT1 reveal trending diminished abundance of CD31 and VE-Cadherin and similar levels of GLUT1 in hCMEC/D3-MDR1s versus hCMEC/D3s quantified in (C) Student's t test was performed; **** p < 0.0001. Experiments were performed on three distinct passages and in technical triplicate (n = 9); representative images are shown.
Abbott, N. J., Patabendige, A. A., Dolman, D. E., Yusof, S. R., and Begley, D. J. (2010). Structure and function of the blood-brain barrier. Neurobiol. Dis. 37 (1), 13–25. doi:10.1016/j.nbd.2009.07.030
Amin, M. L. (2013). P-Glycoprotein inhibition for optimal drug delivery. Drug Target Insights 7, 27–34. doi:10.4137/DTI.S12519
Azad, T. D., Pan, J., Connolly, I. D., Remington, A., Wilson, C. M., and Grant, G. A. (2015). Therapeutic strategies to improve drug delivery across the blood-brain barrier. Neurosurg. Focus 38 (3), E9. doi:10.3171/2014.12.FOCUS14758
Bauer, B., Hartz, A. M., Fricker, G., and Miller, D. S. (2005). Modulation of p-glycoprotein transport function at the blood-brain barrier. Exp. Biol. Med. (Maywood) 230 (2), 118–127. doi:10.1177/153537020523000206
Bhunia, S., Kolishetti, N., Vashist, A., Yndart Arias, A., Brooks, D., and Nair, M. (2023). Drug delivery to the brain: recent advances and unmet challenges. Pharmaceutics 15 (12), 2658. doi:10.3390/pharmaceutics15122658
Braun, A., Hammerle, S., Suda, K., Rothen-Rutishauser, B., Gunthert, M., Kramer, S. D., et al. (2000). Cell cultures as tools in biopharmacy. Eur. J. Pharm. Sci. 11 (Suppl. 2), S51–S60. doi:10.1016/s0928-0987(00)00164-0
Calabria, A. R., Weidenfeller, C., Jones, A. R., de Vries, H. E., and Shusta, E. V. (2006). Puromycin-purified rat brain microvascular endothelial cell cultures exhibit improved barrier properties in response to glucocorticoid induction. J. Neurochem. 97 (4), 922–933. doi:10.1111/j.1471-4159.2006.03793.x
Cecchelli, R., Aday, S., Sevin, E., Almeida, C., Culot, M., Dehouck, L., et al. (2014). A stable and reproducible human blood-brain barrier model derived from hematopoietic stem cells. PLoS One 9 (6), e99733. doi:10.1371/journal.pone.0099733
Chatterjee, S., Deshpande, A. A., and Shen, H. (2023). Recent advances in the in vitro and in vivo methods to assess impact of P-glycoprotein and breast cancer resistance protein transporters in central nervous system drug disposition. Biopharm. Drug Dispos. 44 (1), 7–25. doi:10.1002/bdd.2345
Cordon-Cardo, C., O'Brien, J. P., Casals, D., Rittman-Grauer, L., Biedler, J. L., Melamed, M. R., et al. (1989). Multidrug-resistance gene (P-glycoprotein) is expressed by endothelial cells at blood-brain barrier sites. Proc. Natl. Acad. Sci. U. S. A. 86 (2), 695–698. doi:10.1073/pnas.86.2.695
Cox, B., Nicolai, J., and Williamson, B. (2023). The role of the efflux transporter, P-glycoprotein, at the blood-brain barrier in drug discovery. Biopharm. Drug Dispos. 44 (1), 113–126. doi:10.1002/bdd.2331
Davis, T. P., Sanchez-Covarubias, L., and Tome, M. E. (2014). P-glycoprotein trafficking as a therapeutic target to optimize CNS drug delivery. Adv. Pharmacol. 71, 25–44. doi:10.1016/bs.apha.2014.06.009
Demeuse, P., Fragner, P., Leroy-Noury, C., Mercier, C., Payen, L., Fardel, O., et al. (2004). Puromycin selectively increases mdr1a expression in immortalized rat brain endothelial cell lines. J. Neurochem. 88 (1), 23–31. doi:10.1046/j.1471-4159.2003.02071.x
Engelhardt, B., and Sorokin, L. (2009). The blood-brain and the blood-cerebrospinal fluid barriers: function and dysfunction. Semin. Immunopathol. 31 (4), 497–511. doi:10.1007/s00281-009-0177-0
Espinal, E. R., Matthews, T., Holder, B. M., Bee, O. B., Humber, G. M., Brook, C. E., et al. (2022). Group B streptococcus-induced macropinocytosis contributes to bacterial invasion of brain endothelial cells. Pathogens 11 (4), 474. doi:10.3390/pathogens11040474
Helms, H. C., Abbott, N. J., Burek, M., Cecchelli, R., Couraud, P. O., Deli, M. A., et al. (2016). In vitro models of the blood-brain barrier: an overview of commonly used brain endothelial cell culture models and guidelines for their use. J. Cereb. Blood Flow. Metab. 36 (5), 862–890. doi:10.1177/0271678X16630991
Hollmann, E. K., Bailey, A. K., Potharazu, A. V., Neely, M. D., Bowman, A. B., and Lippmann, E. S. (2017). Accelerated differentiation of human induced pluripotent stem cells to blood-brain barrier endothelial cells. Fluids Barriers CNS 14 (1), 9. doi:10.1186/s12987-017-0059-0
Kim, B. J., McDonagh, M. A., Deng, L., Gastfriend, B. D., Schubert-Unkmeir, A., Doran, K. S., et al. (2019). Streptococcus agalactiae disrupts P-glycoprotein function in brain endothelial cells. Fluids Barriers CNS 16 (1), 26. doi:10.1186/s12987-019-0146-5
Lippmann, E. S., Al-Ahmad, A., Azarin, S. M., Palecek, S. P., and Shusta, E. V. (2014). A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources. Sci. Rep. 4, 4160. doi:10.1038/srep04160
Lippmann, E. S., Azarin, S. M., Kay, J. E., Nessler, R. A., Wilson, H. K., Al-Ahmad, A., et al. (2012). Derivation of blood-brain barrier endothelial cells from human pluripotent stem cells. Nat. Biotechnol. 30 (8), 783–791. doi:10.1038/nbt.2247
Liu, Y., and Zeng, S. (2008). Advances in the MDCK-MDR1 cell model and its applications to screen drug permeability. Yao Xue Xue Bao 43 (6), 559–564.
Mahar, D. K. M., Humphreys, J. E., Webster, L. O., Wring, S. A., Shampine, L. J., Serabjit-Singh, C. J., et al. (2002). Passive permeability and P-glycoprotein-mediated efflux differentiate central nervous system (CNS) and non-CNS marketed drugs. J. Pharmacol. Exp. Ther. 303 (3), 1029–1037. doi:10.1124/jpet.102.039255
Neal, E. H., Marinelli, N. A., Shi, Y., McClatchey, P. M., Balotin, K. M., Gullett, D. R., et al. (2019). A simplified, fully defined differentiation scheme for producing blood-brain barrier endothelial cells from human iPSCs. Stem Cell Rep. 12 (6), 1380–1388. doi:10.1016/j.stemcr.2019.05.008
Nielsen, A. K. R., Lilieholm-Rongren, L., Schmid, B., Holst, B., Brodin, B., and Saaby, L. (2024). Generation of an iPSC-line (BIONi010C-48) with restored P-glycoprotein functionality following transfection with the human MDR1 gene in the AAVS1 locus. Stem Cell Res. 76, 103348. doi:10.1016/j.scr.2024.103348
Ozgur, B., Puris, E., Brachner, A., Appelt-Menzel, A., Oerter, S., Balzer, V., et al. (2023). Characterization of an iPSC-based barrier model for blood-brain barrier investigations using the SBAD0201 stem cell line. Fluids Barriers CNS 20 (1), 96. doi:10.1186/s12987-023-00501-9
Pastan, I., Gottesman, M. M., Ueda, K., Lovelace, E., Rutherford, A. V., and Willingham, M. C. (1988). A retrovirus carrying an MDR1 cDNA confers multidrug resistance and polarized expression of P-glycoprotein in MDCK cells. Proc. Natl. Acad. Sci. U. S. A. 85 (12), 4486–4490. doi:10.1073/pnas.85.12.4486
Perriere, N., Demeuse, P., Garcia, E., Regina, A., Debray, M., Andreux, J. P., et al. (2005). Puromycin-based purification of rat brain capillary endothelial cell cultures. Effect on the expression of blood-brain barrier-specific properties. J. Neurochem. 93 (2), 279–289. doi:10.1111/j.1471-4159.2004.03020.x
Redzic, Z. (2011). Molecular biology of the blood-brain and the blood-cerebrospinal fluid barriers: similarities and differences. Fluids Barriers CNS 8 (1), 3. doi:10.1186/2045-8118-8-3
Ronaldson, P. T., and Davis, T. P. (2024). Blood-brain barrier transporters: a translational consideration for CNS delivery of neurotherapeutics. Expert Opin. Drug Deliv. 21 (1), 71–89. doi:10.1080/17425247.2024.2306138
Schinkel, A. H. (1999). P-Glycoprotein, a gatekeeper in the blood-brain barrier. Adv. Drug Deliv. Rev. 36 (2-3), 179–194. doi:10.1016/s0169-409x(98)00085-4
Stebbins, M. J., Wilson, H. K., Canfield, S. G., Qian, T., Palecek, S. P., and Shusta, E. V. (2016). Differentiation and characterization of human pluripotent stem cell-derived brain microvascular endothelial cells. Methods 101, 93–102. doi:10.1016/j.ymeth.2015.10.016
Thiebaut, F., Tsuruo, T., Hamada, H., Gottesman, M. M., Pastan, I., and Willingham, M. C. (1989). Immunohistochemical localization in normal tissues of different epitopes in the multidrug transport protein P170: evidence for localization in brain capillaries and crossreactivity of one antibody with a muscle protein. J. Histochem Cytochem 37 (2), 159–164. doi:10.1177/37.2.2463300
van Assema, D. M., Lubberink, M., Bauer, M., van der Flier, W. M., Schuit, R. C., Windhorst, A. D., et al. (2012). Blood-brain barrier P-glycoprotein function in Alzheimer's disease. Brain 135 (Pt 1), 181–189. doi:10.1093/brain/awr298
Keywords: blood-brain barrier, P-glycoprotein, efflux transport, drug screening, brain endothelial cells, puromycin, drug delivery
Citation: Hathcock SF, Knight HE, Tong EG, Meyer AE, Mauser HD, Vollmuth N and Kim BJ (2024) Induction of P-glycoprotein overexpression in brain endothelial cells as a model to study blood-brain barrier efflux transport. Front. Drug Deliv. 4:1433453. doi: 10.3389/fddev.2024.1433453
Received: 15 May 2024; Accepted: 10 June 2024;
Published: 05 July 2024.
Edited by:
Anika M. S. Hartz, University of Kentucky, United StatesReviewed by:
Anuska V. Andjelkovic, University of Michigan, United StatesCopyright © 2024 Hathcock, Knight, Tong, Meyer, Mauser, Vollmuth and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brandon J. Kim, QmpraW00QHVhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.