
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Digit. Health, 14 June 2023
Sec. Health Informatics
Volume 5 - 2023 | https://doi.org/10.3389/fdgth.2023.1186516
This article is part of the Research TopicHealthcare Text Analytics: Unlocking the Evidence from Free Text, Volume IIIView all 8 articles
 Murray Cutforth1
Murray Cutforth1 Hannah Watson1
Hannah Watson1 Cameron Brown2
Cameron Brown2 Chaoyang Wang1
Chaoyang Wang1 Stuart Thomson1
Stuart Thomson1 Dickon Fell1
Dickon Fell1 Vismantas Dilys1
Vismantas Dilys1 Morag Scrimgeour1
Morag Scrimgeour1 Patrick Schrempf1
Patrick Schrempf1 James Lesh1
James Lesh1 Keith Muir2
Keith Muir2 Alexander Weir1
Alexander Weir1 Alison Q O’Neil1,3*
Alison Q O’Neil1,3*
Introduction: Thrombolysis treatment for acute ischaemic stroke can lead to better outcomes if administered early enough. However, contraindications exist which put the patient at greater risk of a bleed (e.g. recent major surgery, anticoagulant medication). Therefore, clinicians must check a patient's past medical history before proceeding with treatment. In this work we present a machine learning approach for accurate automatic detection of this information in unstructured text documents such as discharge letters or referral letters, to support the clinician in making a decision about whether to administer thrombolysis.
Methods: We consulted local and national guidelines for thrombolysis eligibility, identifying 86 entities which are relevant to the thrombolysis decision. A total of 8,067 documents from 2,912 patients were manually annotated with these entities by medical students and clinicians. Using this data, we trained and validated several transformer-based named entity recognition (NER) models, focusing on transformer models which have been pre-trained on a biomedical corpus as these have shown most promise in the biomedical NER literature.
Results: Our best model was a PubMedBERT-based approach, which obtained a lenient micro/macro F1 score of 0.829/0.723. Ensembling 5 variants of this model gave a significant boost to precision, obtaining micro/macro F1 of 0.846/0.734 which approaches the human annotator performance of 0.847/0.839. We further propose numeric definitions for the concepts of name regularity (similarity of all spans which refer to an entity) and context regularity (similarity of all context surrounding mentions of an entity), using these to analyse the types of errors made by the system and finding that the name regularity of an entity is a stronger predictor of model performance than raw training set frequency.
Discussion: Overall, this work shows the potential of machine learning to provide clinical decision support (CDS) for the time-critical decision of thrombolysis administration in ischaemic stroke by quickly surfacing relevant information, leading to prompt treatment and hence to better patient outcomes.
An acute stroke is a clinical emergency that requires prompt assessment and management. Around 85% of strokes are ischaemic (1), as opposed to haemorrhagic, requiring timely treatment by thrombolysis (intravenous clot busting medication) and/or thrombectomy (surgical mechanical clot retrieval). Not all ischaemic stroke patients are eligible for thrombolysis or thrombectomy; the decision is based upon historical and current patient factors, alongside imaging features. The remaining 15% of haemorrhagic strokes may require neurosurgical intervention but must not be treated with thrombolysis. We focus in this paper on thrombolysis; the faster the “door-to-needle time” with thrombolysis treatment, the better the chance of a good functional outcome for the patient (2). Current guidelines from the National Institute for Health and Care Excellence (NICE) state that treatment with a thrombolytic agent should be administered within 4.5 h post symptom-onset (3).
There are well-defined indications and, importantly, contraindications to thrombolysis that must be checked for all patients. Indications relate to the potential benefit of treatment, for instance, the pre- and post-stroke levels of independence. Contraindications to thrombolysis relate to the risk of bleeding, for instance, recent major surgery, anticoagulant medication, or a history of intracranial haemorrhage. Therefore, inappropriate treatment with thrombolysis can lead to catastrophic patient outcomes. The task of obtaining and reviewing all the relevant clinical information is complex and time-critical, leading to risks for both eligible and ineligible thrombolysis candidates, namely delayed treatment and missed contraindications respectively.
Information about indications and contraindications for thrombolysis comes from a variety of sources. Much of the information comes from the patient evaluation at the point of care, e.g. the patient history for the acute stroke event and the physical examination findings, as well as any immediate imaging results. However, a significant proportion of the required clinical information relates to the past medical history (e.g. recent major surgery, anticoagulant medication). The patient’s (electronic) health record can therefore be an important source of information, containing rich descriptions of past medical history in unstructured text documents such as discharge letters or referral letters. Automated surfacing of relevant information from the patient record could support the clinician in reviewing this information more quickly.
Named entity recognition (NER) is a well-studied information extraction task from the field of natural language processing (NLP). The task is to extract spans, i.e. subsections of a text, which refer to particular named entities. For example, in the general domain the entity set could be {person, location, organisation}. In our case, the list of relevant entities was compiled with reference to two sets of thrombolysis eligibility criteria: national guidelines from NICE (3) and a local checklist from Queen Elizabeth University Hospital (Table 1). This yielded a total of 86 entities, ranging from subarachnoid-haemorrhage to visual-disturbance. We classify the entities according to the following five categories: Diagnosis, Symptom, Social History, Medication, Treatment.
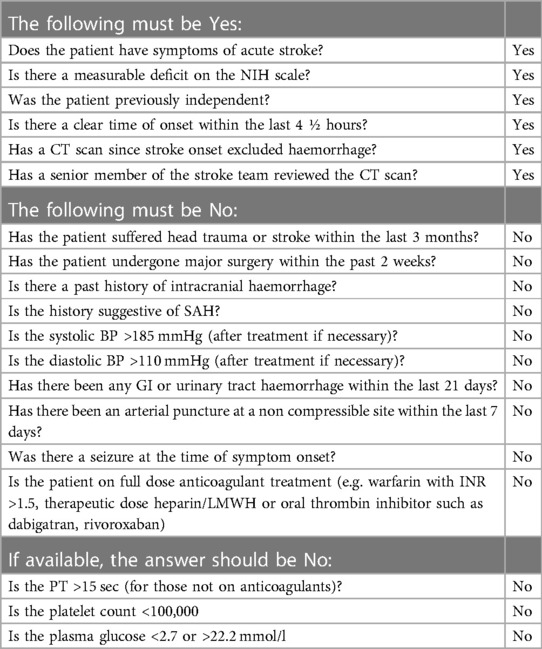
Table 1. The eligibility checklist for thrombolysis administration in use at Queen Elizabeth University Hospital, Glasgow, UK. This illustrates the scale of the criteria that must be satisfied and the information retrieval task required prior to administering thrombolysis.
In this work we measure and analyse the efficacy of transformer-based methods for NER in clinical text, training and validating on a large-scale dataset of unstructured clinical documents for the real-world problem of timely thrombolysis treatment in acute stroke patients. Data from almost 3,000 stroke patients was collected and annotated, as shown in the overview in Figure 1. We aim to evaluate if deep learning can provide accurate and robust performance for a clinical decision support (CDS) task. To illustrate the output of our work, Figure 2 shows the operation of our NER model on a synthesised discharge letter, and Figure 3 shows how the information from the NER model may be presented to the clinician to aid rapid understanding of contraindications.
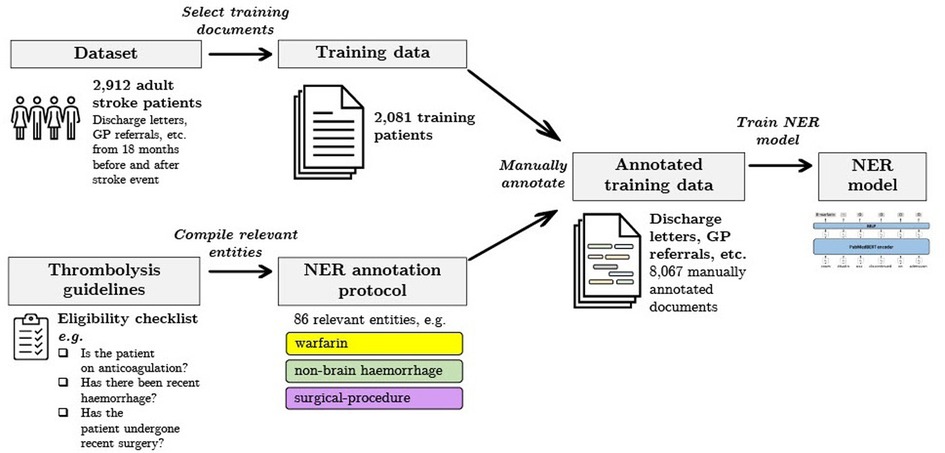
Figure 1. Summary of our data selection and annotation process. Our dataset contains discharge letters, clinic letters, GP referrals and endoscopy reports from 2,912 patients. The training set comprises 2,081 patients and the remainder are reserved for the held-out test set. Using both local and national guidelines, clinical knowledge was used to compile the relevant entities for thrombolysis decision. The NER model is trained to recognise 86 entities relevant to the thrombolysis eligibility checklist. Data was manually annotated by a team of clinicians and medical students.
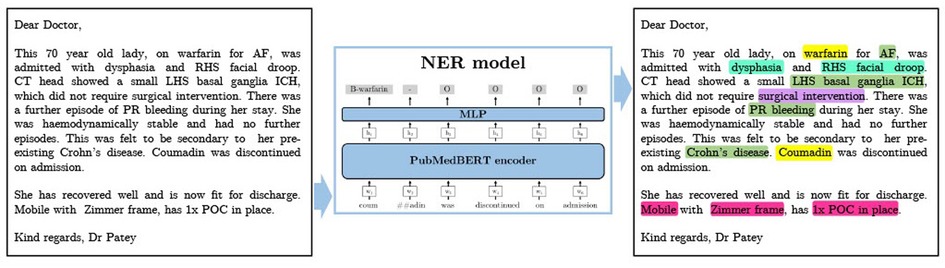
Figure 2. A synthesised discharge letter on which we demonstrate the application of our method. The output shows the entities that should be detected. Colour coding reflects category of entity (Green=Diagnosis, Turquoise=Symptom, Pink=Social History, Yellow=Medication, Purple=Treatment). AF, atrial fibrillation; ICH, intracranial haemorrhage; POC, package of care; PR, per rectum; RHS, right hand side; SAH, subarachnoid haemorrhage.
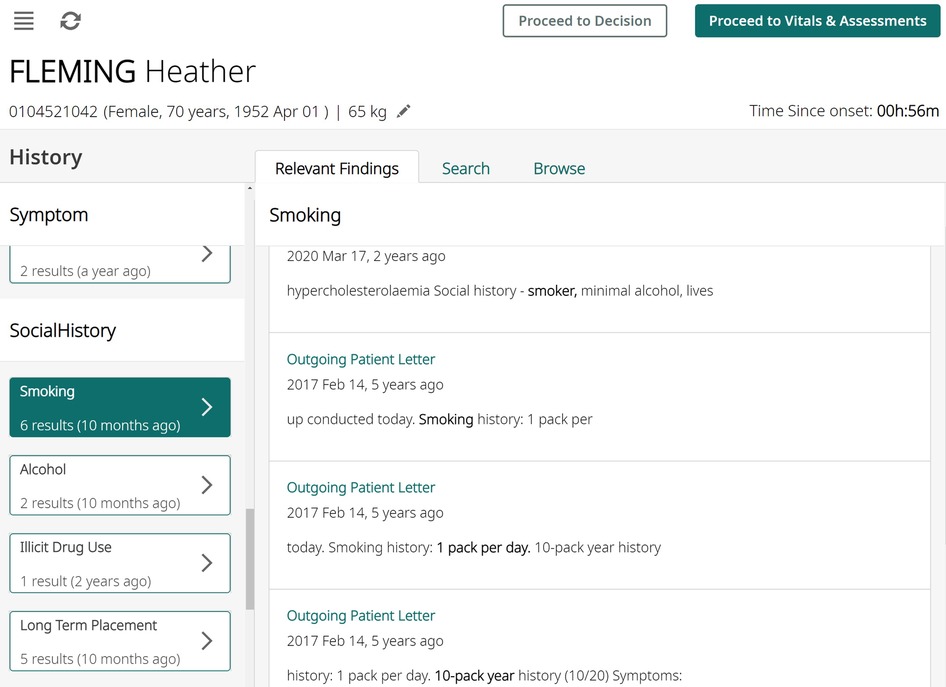
Figure 3. An illustration of how the algorithm results might be presented to clinicians using synthesised data. All entity occurrences are presented on the left pane, and then shown with context on the right pane, aiding rapid understanding of any contraindications.
We first review existing systems which aim to aid in thrombolysis CDS, and then subsequently review biomedical named entity recognition methods. In common with other NLP sub-fields, biomedical NER has advanced significantly in recent years due to the application of large pre-trained transformer architectures (4).
Of the research groups working on CDS in the stroke domain, some identify and display contraindications to thrombolysis at the point of care (5, 6), whilst others focus on predicting outcomes in the case that thrombolysis is administered versus not administered (7–9). A recent review of machine learning methods for selecting patients who might benefit most from thrombolysis treatment is provided in (10). These methods are complimentary to our contraindication-finding approach. Most closely related to our approach is a small-scale feasibility study that designed a user interface to specifically highlight contraindications to thrombolysis by matching Unified Medical Language System (UMLS) (11) concepts between a thrombolysis eligibility checklist, and a stroke patient EHR (5). In contrast, our study is focused on the extraction of relevant concepts, and unlike (5) we use clinical domain expertise to curate a set of relevant entities and perform a detailed examination of the performance of various NER methods. To the best of our knowledge, none of the solutions described above are currently used in clinical practice.
Biomedical NER is considered to be a slightly harder task than general domain NER due to the prevalence of abbreviations, synonymy, and morphological complexity as a result of the use of unusual characters such as Greek letters, digits, and punctuation (12). Clinical text poses an additional challenge, since clinicians frequently write in shorthand and may not always employ correct grammar. A number of public datasets exist which have allowed the development of different NER techniques, including JNLPBA (13), BC5CDR (14), NCBI (15), i2b2 (16), and MedMentions (17). Early methods included the application of dictionaries/gazetteers (18), or handcrafted features with probabilistic graphical models such as a conditional random field (19). The first generation of end-to-end deep neural architectures were based on character-level and/or word-level recurrent neural networks, often combined with a probabilistic graphical model to predict the final tag sequence, typified by the early work of (20).
Most recently, researchers in the biomedical NER field have focused on the application of large pre-trained transformer encoder models, based on Bidirectional Encoder Representations from transformers (BERT) (21). In this approach, a transformer model comprising encoder blocks only is pre-trained on a large corpus with two unsupervised NLP tasks: masked language modelling and next sentence prediction. A fully connected layer is then used to map the contextual word embeddings from the output of the BERT encoder to NER class logits, and the full model is fine-tuned using a supervised dataset. BERT-based NER approaches have been shown to outperform previous approaches (12).
The BERT-based NER approach has been extended in various ways. Some approaches aim to increase domain relevance. For instance, numerous studies have shown that biomedical NER tasks benefit from pre-training and vocabulary selection on a biomedical text corpus such as PubMed,1 yielding domain-specific models such as SciBERT (22), BioBERT (23), BioMed-RoBERTa (24), and PubMedBERT (25). Other studies have investigated integrating the Unified Medical Language System (UMLS) (11) biomedical knowledge graph with BERT architectures, such as (26–28), generally yielding a modest performance increase.
Other approaches target architectural improvements. For instance, BERT models have been combined with BiLSTMs in a two-stage proposal/refinement method in (29). Scaling up the model size and careful tuning of hyperparameters also gave modest improvements in (30). Finally, an encoder-decoder transformer architecture (text-to-text model) has shown strong performance on biomedical NLP tasks in (31).
Pre-training on biomedical data and using a biomedical vocabulary gives a consistent gain in performance. For example, on the NCBI-disease dataset, PubMedBERT (25) obtains an F1 of 87.8% versus the top scoring model with 90.4%, while general-domain BERT obtains 85.6%. On the BC5-chem dataset (14) PubMedBERT scores 93.3% versus the current top result of 95.0%, while general-domain BERT obtains 89.2%.
We use data obtained through a collaboration with the Industrial Centre for Artificial Intelligence Research in Digital Diagnostics (iCAIRD),2 for which we obtained ethical approval.3 The data was sourced from hospitals in the Greater Glasgow & Clyde (GG&C) area in Scotland and comprises all adult patients who were diagnosed with a stroke in the period 1 Jan 2013 to 31 Dec 2018. The data is pseudonymised and we accessed it onsite at the West of Scotland Safe Haven within NHS Greater Glasgow and Clyde via the Safe Haven Artificial Intelligence Platform (SHAIP) (32). Note that since we requested data for 18 months on either side of the stroke event, many of the text documents arose from other non-stroke clinical events.
This dataset contains approximately 50 K documents from 10 K patients. Documents were a mixture of General Practice referrals, Intermediate Discharge letters (IDLs), Final Discharge letters (FDLs), Outpatient clinic letters (OPCLs), Emergency Department letters, and Endoscopy reports. All documents comprise unstructured (free text) data, and the content was generally written by doctors, either general practitioners (GPs) or hospital doctors from various specialties encountered by patients during the 18-month period preceding and following the stroke event.
For the purpose of the study, we annotated a subset of 8,067 documents from 2,912 patients, which we split at the outset into 2,081 training patients and a held-out test set of 831 patients. The training split was then randomly subdivided into five folds to be used for cross validation and ensembling.
We designed an annotation protocol and then conducted an extensive annotation effort involving 8 medical students and 4 clinicians with 2–10 years of clinical experience each. Annotation was performed using the brat rapid annotation tool (33). All annotators attended a refresher teaching session on stroke and a training session on the protocol prior to beginning annotation. Additionally, throughout the process, annotators were able to ask questions regarding annotation work, with questions and answers made visible to all annotators.
Through the duration of the annotation period, we performed regular quality checks on each annotator by interleaving a common subset of documents through each annotator’s allocated folder of documents, and computing agreement with consensus gold standard annotations created by the two lead clinicians who developed the protocol and were responsible for ongoing updates (H.W. and C.B., with 5 years and 10 years of clinical experience respectively). Approximately 5% of the annotated documents were repeatedly annotated in this way. In Section 3.1, an analysis of the human annotator error relative to the gold standard is presented.
The annotation protocol was collaboratively designed between Canon Medical Research Europe, the University of Glasgow and Deep Cognito (34) We identified which entities to annotate based on the local Queen Elizabeth University Hospital clinical guidelines and national clinical guidelines from NICE (3). Table 2 describes the full set of 86 entities which emerged from this process. Entities are grouped under 5 categories: Diagnosis, Symptom, Social History, Medication, Treatment.
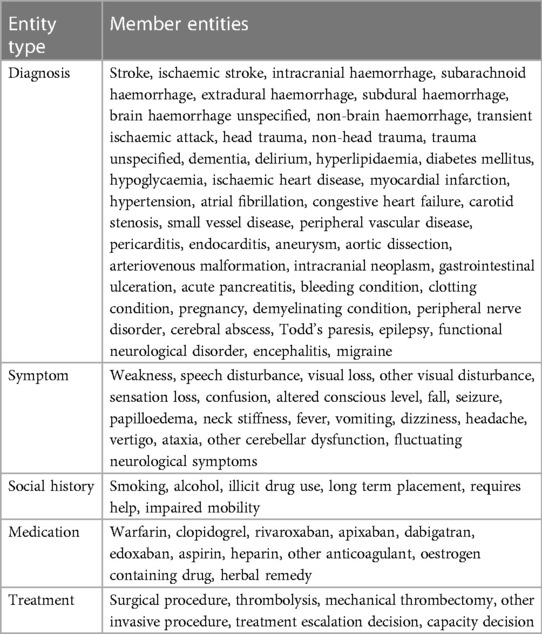
Table 2. Exhaustive list of stroke-related entities that require annotation alongside their synonyms or equivalents as per the annotation protocol.
The protocol aims to contain sufficient detail to ensure consistency between annotators. A list of synonyms is provided for each entity and these lists were updated over the course of the annotation process, e.g. for the entity congestive heart failure possible synonyms are “Left ventricular systolic dysfunction,” “LVSD,” “cardiac decompensation” and “LV dysfunction.” Instructions are also provided on how to determine the extent of the annotation text span, e.g. for the entity ischaemic stroke the span might be “Apparent R MCA infarct” i.e. omit the word “apparent” but contain the anatomical qualifiers. In addition, examples of correct and incorrect spans were provided to disambiguate difficult cases, e.g. for the entity Confusion a correct span would be “altered mental status” and an incorrect span would be “memory loss.”
In total, 8,067 documents were annotated. A breakdown of the patient demographics and number of annotations by document type, data split, and entity category are presented in Tables 3–5. There is a 55%–45% train-test split at the level of the annotated documents. All document types were annotated approximately in proportion to their frequency in our dataset.
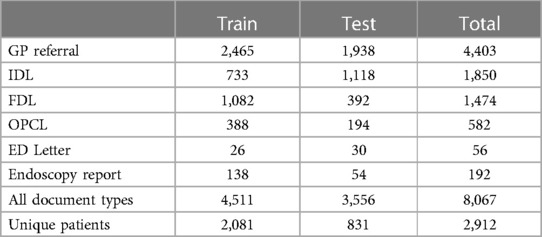
Table 4. Number of annotated documents (and corresponding unique patients), showing the prevalence of different document types in our dataset.
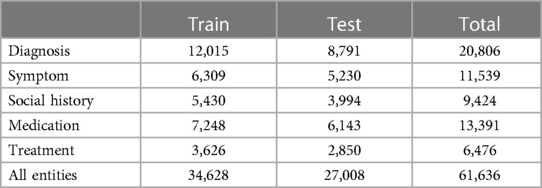
Table 5. Number of annotated entities. We labelled 86 different entities drawn from five categories, and instances are grouped in this table according to the entity categories.
We now describe the NER algorithms which are evaluated in this work. Our main approach is based on a token-level transformer classifier.
As a baseline, we have implemented a naive exact string search method. In this approach, the training data is used to compile a list of possible strings for every label (ambiguous cases are ignored). Predictions are then generated by searching for occurrences of each string, and then applying the corresponding label to that span.
A second, more sophisticated baseline is the transition-based model implemented in the spaCy library (35). This approach uses fixed word embeddings and a convolutional neural network, and is designed to be an effective and efficient general purpose NER method.
The main approach applied to the NER problem in this work is a token-level classifier based on the BERT architecture (21). We used a number of different pre-trained weights and vocabularies from the literature: PubMedBERT (25) and BioMed-RoBERTa (24) are trained on biomedical papers from PubMed, while SciBERT (22) is trained on general scientific papers from Semantic Scholar). A separate label is assigned to each token in the model input by passing the contextual embedding of each input token to a multilayer perceptron (MLP) of 3 layers with 512 nodes. A probability distribution over the output classes is obtained by taking the softmax over the logits. In order to classify arbitrary length spans, an Inside-Outside-Beginning (IOB) tag scheme (36) is used. Under this scheme, the first token of an entity span should be assigned the -entity tag and any subsequent tokens assigned -entity tags, and any tokens not relating to entities should be assigned tags. Therefore, given entity classes, the model chooses from possible tags (in our case, 86 entities leads to 173 IOB classes). All trainable parameters in the model are optimised using the Adam optimiser (37) with a categorical cross-entropy loss function.
A number of model hyperparameters were explored, using a validation fold consisting of 20% of the training set. In Table 6 we present the optimal values which were found, as well as the search bounds. Where applicable, optimal parameters were found using the hyperopt Python package (39).
Model ensembling is a well-known technique to obtain a modest performance improvement by taking the average prediction over a group of classifiers (40); ensembling causes random errors arising from individual classifiers to be smoothed out. We constructed an ensemble classifier using five variants of the best transformer model variant, trained on different subsets of the training set, according to a 5-fold cross validation split. The ensemble predictions were obtained by averaging the logits and taking the argmax over classes, allowing the confidence of individual models to be taken into account.
In Section 3, we evaluate the performance of models using precision, recall, and F1. In the NER setting, these metrics can be computed in either a strict or lenient fashion. With strict matching, a predicted entity span must exactly match the ground truth span and label to count as a true positive, whereas with lenient matching the labels must match but a partial overlap between the spans is sufficient. For example, for the phrase “Patient on warfarin” (tokenised to [“patient,” “on,” “warfarin”]), the prediction [O, B-warfarin, I-warfarin] with ground truth [O, O, B-warfarin] would count as a lenient match but not a strict match.
In order to better understand the performance of the NER methods on different labels, we examine three label properties, taking inspiration from the work in (41) which introduces the concepts of “name regularity” and “context regularity”. Here, we propose corresponding numeric definitions to measure each property for a given label in a given annotated dataset. The relationship between label properties and performance is presented in Section 3.2. The three properties examined for each label are:
1. Training set frequency. This is simply the number of instances of a particular entity label in the training set.
2. Name regularity. This is a measure of the similarity of all training set spans which are labelled as a particular entity, defined as:
where is the total number of spans for the given label, and is the number of unique spans. The name regularity has a range of , taking a value of 1 when every span is identical, and a value of 0 when every span is different.
3. Semantic context regularity. This is a measure of the similarity of the context surrounding each entity mention. For each entity example, we take the 5 tokens on either side of the entity span, and compute the average of their word embeddings.4 For sample , we call this mean embedding . The semantic context similarity is then defined as:
In other words, this is a measurement of the similarity of the context around each entity mention, using the cosine similarity between the average of the word embeddings of the context. It has a range of , where a higher value indicates a more similar context.
In Table 7, quantitative results on our held-out test set are presented. Uncertainty estimates were obtained by training 5 models using different random seeds and different training sets (using a five-fold cross validation split of the training set), and hence reflect the variance due to the model training procedure and due to the training set sample. Overall, these tables show that BERT-based models outperform our exact string search baseline, and that out of PubMedBERT (25), SciBERT (22), and BioMedRoBERTa (24), the PubMedBERT model performs best. A significant improvement in performance is observed using an ensemble of PubMedBERT models, particularly in the model precision.
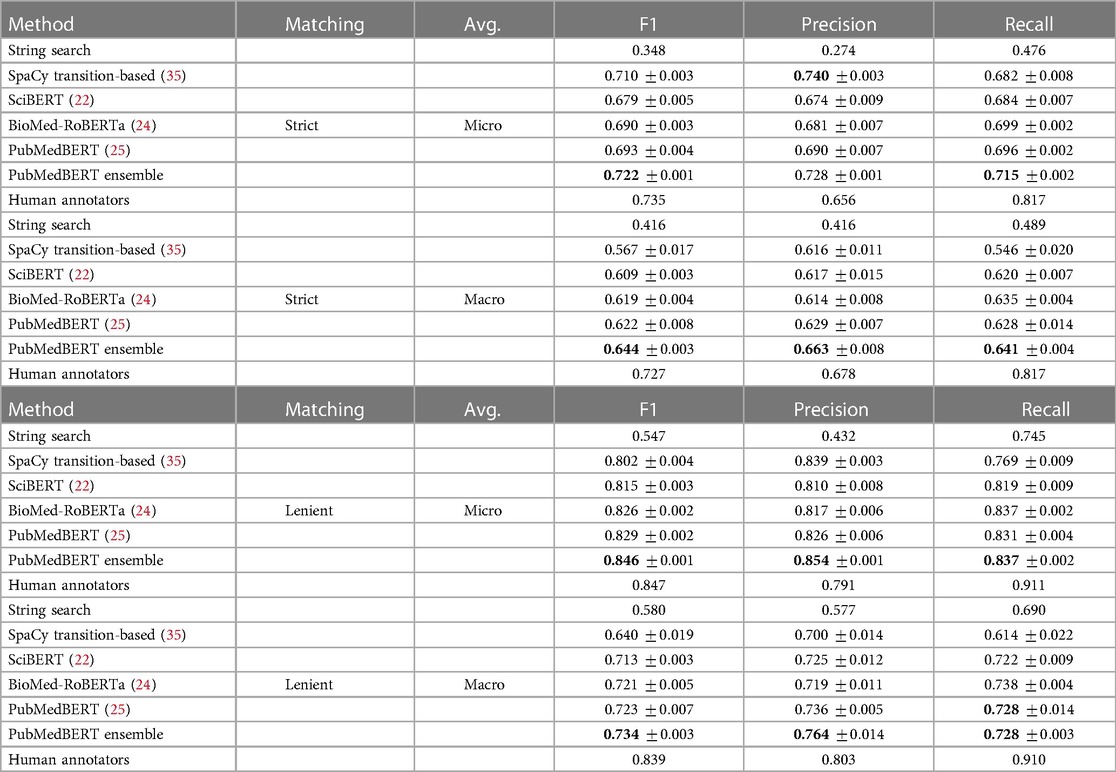
Table 7. NER results on held-out test set using strict (upper table) and lenient (lower table) span matching, where strict matching requires an exact match of span boundaries and label, while lenient matching requires an exact match of label, and overlap in span boundaries. Best results indicated in bold. Estimated human annotation performance computed on a subset of 5% of documents shown for reference.
For reference, we compare model prediction accuracy to the performance of our human annotators by comparing to the gold standard annotations on a common subset of documents which were annotated by all annotators. In total, there were 292 gold standard documents containing 2,870 spans. We can evaluate annotator performance on this set of documents using the quantitative metrics described in Section 2.3. Although the numbers are not directly comparable due to having been computed on different sets of documents, the ensemble model is approaching our estimate of human annotator micro F1. The ensemble model compares much less favourably in macro F1 to the human annotators, suggesting that the algorithm has difficulty learning how to label rare classes such as todds-paresis or peripheral-nerve-disorder.
The fact that the PubMedBERT-based models are approaching our estimate of the annotation error implies that quantitative evaluation using these annotations may not be reliable. In order to understand this issue, a further manual evaluation was undertaken by a clinician; results are presented in Section 3.3.
We further investigate the types of errors made by the PubMedBERT-based models (which performed best in Table 8), comparing to errors made by the baseline string search approach. Then, we examine the relationship between properties of individual labels and their performance. The large number of labels present in our NER problem means that the distribution of per-label scores carries information which we can analyse to understand what makes this NER problem difficult for the models under consideration.
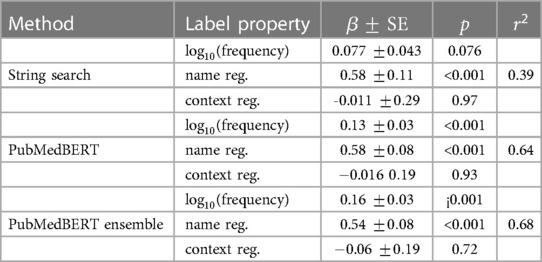
Table 8. Multiple linear regression analysis for each model in which the independent variables are the three label properties, and the dependent variable is the lenient per–label F1. A scatter plot of each variable independent of the others is shown in Figure 5.
The distribution of lenient F1 scores across each label is presented in Figure 4, showing that there is a relatively wide spread of performance between different labels. The best three labels for the ensemble method are aspirin (0.968), warfarin (0.965), and edoxaban (0.949), while the worst5 three labels are functional-neurological-disorder (0.095), cerebral-abscess (0.118), and neck-stiffness (0.2). Medications generally have a small set of possible surface forms, so these results suggest, as might be expected, that labels with fewer possible variants (i.e. higher name regularity) are easier to detect. Indeed, the best three labels have an average name regularity of 0.95, versus 0.17 for the bottom three.
In Figure 5 and Table 8 the effect of three label properties (described in Section 2.4) on performance for each model is investigated. Multiple linear regression was performed, treating the property as an independent variable and the per-label lenient F1 as the dependent variable. For all models, name regularity had the largest effect size. Suprisingly, the number of training set examples only had only a weak positive effect, and context regularity independent of performance. However, the relatively low values show that these three label properties are still insufficient to explain label performance.
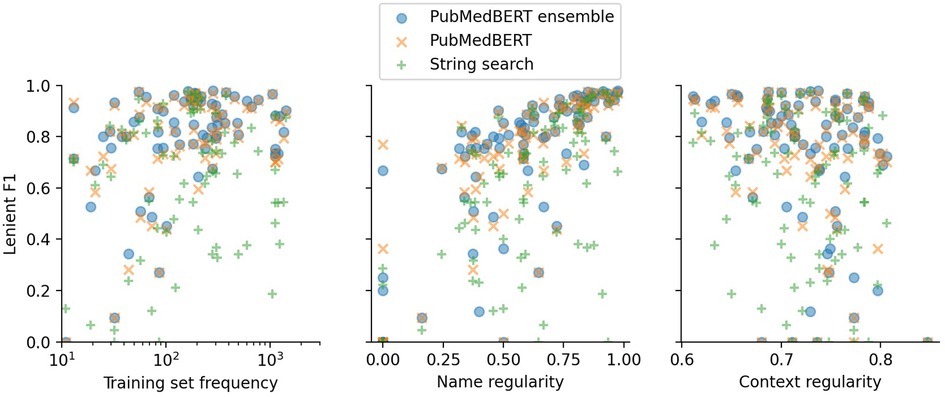
Figure 5. Lenient micro F1 versus different label properties. The -value of each point shows the F1 score for a particular label using a particular model. Multiple linear regression analysis on this data is presented in Table 8.
Given that on certain metrics there is negligible difference between the estimate of human annotator error and the model error, the metrics computed against the ground truth may not reliably demonstrate clinically important differences. Therefore, in order to further investigate the differences in performance between the models, we conducted a manual clinical error analysis of the results.
A random subset of 310 documents from the held-out test set (this corresponds to approximately 10% of the test set) was selected for the manual error analysis. The corresponding human ground truth was then combined with the predictions from each model in turn, and the two sets of annotations were combined into a single view and displayed within our in-house text annotation software. A junior doctor (C.W.) with 3 years of clinical experience then reviewed all predictions, and either marked as correct, or if not then corrected as necessary. The evaluator was blinded to the source of the annotations (i.e. model prediction or human annotation) to reduce bias. This evaluation permits direct comparison between the precision of the model predictions and the human annotations, albeit on a much-reduced subset of the test set.
The manually-measured precisions are shown in Table 9. The precision of the ensemble model is notably much improved compared to the single PubMedBERT model with the number of pure false positive errors (type 1) decreased by two-thirds, and marginally better than the human annotators. The estimated precision of the human annotators is significantly higher than previously estimated using the gold standard documents in Section 3.1 (0.791 versus 0.944). This likely reflects the style of evaluation, where the evaluator is not making decisions independently of the annotator but rather rating visible annotations, and therefore in edge cases may tend to be generous. We also note that the evaluation document subsets are different so the metrics are not directly comparable.
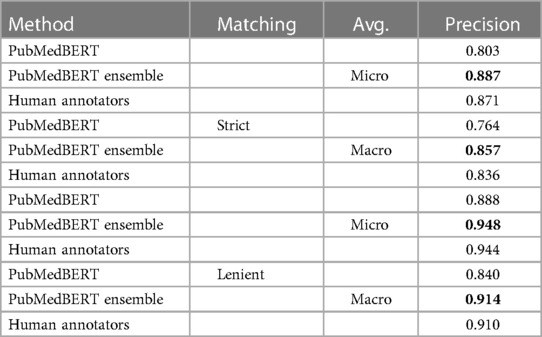
Table 9. Manual evaluation of precision of selected models and human annotations on a subset of 310 documents from the test set.
Our results show that contemporary NER approaches are able to perform well on the task of locating relevant entities for the thrombolysis decision. Evaluation on the test set suggests that the ensemble of five PubMedBERT models achieves an almost identical F1 score to the human annotators (0.846 vs 0.847), which is exciting. However, further inspection shows the model has a better micro precision (0.854 vs. 0.791) but a worse micro recall (0.837 vs. 0.911) than the human annotators, which from a clinical point of view is not desirable, since false negatives are more serious than false positives when searching for contraindications. It is also notable that the micro and macro averages are similar for human annotators, whereas the machine-learning systems experience a drop of approximately 0.1 on macro averaged metrics, suggesting that rare labels are handled less effectively. However, the multiple linear regression analysis showed that in fact it was name regularity rather than training set frequency which played the most important role in determining performance i.e. rare spans are more problematic than rare labels. This point was reinforced by our observation that all transformer-based models experienced a significant drop in recall between the in-dict (i.e. spans which were also present in the training set) and out-dict portions of the test set (results not shown here). This suggests that name memorisation still plays a significant role for these models, and recognition of context or knowledge of synonymy learned during biomedical pre-training is not yet fully utilised. Name memorisation could be tackled through data augmentation techniques in which synonyms are gathered from knowledge sources such as UMLS (11). This type of data augmentation has already been implemented for NER with BERT models in (43), finding up to a 7% improvement in micro-F1, with the benefit falling off as the dataset size increased.
Compared to standard biomedical NER problems in the literature, this application features a large number (86) of fine-grained labels which allowed us to do inter-label analysis. It was shown that there is a large variation in performance between labels (inter-quartile range on per-label performance for ensemble model is 0.23). In the limiting case of many classes, this problem becomes very similar to entity linking—the task of linking entity mentions to knowledge graph nodes. A cross encoder refinement model from the two-stage models (44) which have been applied in entity linking may be useful to improve performance on some labels. The worst-performing labels may require special treatment, such as synthesized training data, in a future clinical application.
As previously stated, on some metrics the ensemble model obtained comparable performance to our estimate of the human annotator error. Manual analysis showed that the ensemble model predictions often surpass the original human annotations as measured by overall F1, but that the error types are different; human errors are more likely to be false negatives or protocol noncompliance, while model errors are more likely to be false positives. This suggests that review and improvement of the ground truth is required in order to make further improvements to the model, both to improve the standard of the training data and to improve our ability to identify and measure model improvements.
In this paper, we have considered only clinical free text data. However, structured data is also available both within the documents that we were working with and from other data types in the EHR. Information that might be expressed in structured format includes the patient’s current medications, diagnoses, recent procedures, or recent lab test results. Structured data is generally an easier and more standardised data format to parse (likely not requiring machine learning), and any clinical decision support system should consider this alongside the free-text data, integrating entity detections from both sources.
A limitation of this work is that the models were trained and evaluated on documents from both before and after the index stroke event. As the intended use for stroke CDS is on pre-stroke documents only, the performance in this context may change relative to the results in this work, however we believe that such a change is likely to be minor due to the similarity of the language between these cases, and the fact that 36% of the patients in our dataset experienced multiple strokes.
Finally, we remark that at present, our system does not directly answer the individual thrombolysis eligibility checklist items as this requires more information than just the existence of an entity. For instance, answering the question “Has the patient undergone major surgery in the last two weeks?” would involve first locating any occurrences of the surgery entity (using the NER model presented in this work), but also extracting any important modifiers which apply to each entity (e.g. negation and timeframe). The NER-only system presented here is a first step towards such a fully automated system, that leaves decision-making in the hands of the clinician who must judge for themselves the meaning of the surfaced information about relevant entities.
This work represents the first text-focused clinical decision support system for acute stroke treatment. Clinical guidelines were translated into a set of 86 entities relevant to the thrombolysis decision, and a large dataset of unstructured clinical letters of acute stroke patients was annotated for spans relating to these entities. Multiple transformer-based NER approaches were trained and evaluated. An ensemble of five PubMedBERT models obtained the best results (lenient micro F1 0.846/macro F1 0.734). This model was comparable to our estimate of human annotator performance (lenient micro F1 0.847/macro F1 0.839). One of the unusual aspects of our NER application was the large number of fine-grained labels, and a detailed error analysis showed that the name regularity was the strongest predictor of model performance on a given label. Finally, a further manual evaluation showed that the ensemble model outperformed a single PubMedBERT model by an even larger margin than suggested by the test set ground truth, due to annotation errors.
To the best of our knowledge, this work is the first text-focused decision support system for acute stroke treatment. Further, our system is the first step towards a clinical decision support system providing a recommendation for patients’ eligibility for thrombolysis in acute stroke care.
An important avenue for future work is to adopt a data-centric approach and develop review and correction techniques to improve the accuracy of human annotation ground truth. This will improve the standard of the training data and improve our ability to identify and measure model improvements, in order to enable further accuracy gains for this clinically critical task. Furthermore, in future work the entities extracted from unstructured data using the methods presented here should be integrated with entities extracted from any structured data which is already present, such as medication lists.
The datasets presented in this article are not readily available because the data used in this study was obtained through the Industrial Centre for AI Research in Digital Diagnostics collaboration and comes from NHS Greater Glasgow and Clyde. It was accessed through the Canon Safe Haven Artificial Intelligence Platform tool. Due to patient confidentiality, the data used in this study is only accessible on application to the West of Scotland Safe Haven. Requests to access the datasets should be directed to https://icaird.com/.
HW, CB, KM, AW and AON contributed to the conception and design of the study. HW, CB, MC, VD, CW, MS and PS organised the manual annotation. MC, MS, JL trained and evaluated NER algorithms. MC, CW, ST and DF contributed the further manual evaluation of the ensemble. MC, HW and AON wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
This work is part of the Industrial Centre for AI Research in Digital Diagnostics (iCAIRD) which is funded by Innovate UK on behalf of UK Research and Innovation (UKRI) project number 104690.
We thank the West of Scotland Safe Haven at NHS Greater Glasgow and Clyde for their assistance in creating this dataset. We would also like to acknowledge assistance of Canon Medical Research Europe Limited in providing the Canon Safe Haven Artificial Intelligence Platform (SHAIP) tool, assisting with the deidentification of data and the provision of a secure machine learning workspace. We are indebted to Azad Deghan of DeepCognito for his assistance in creating the annotation protocol, and to the medical students from the University of Glasgow (Marcus Boyd, Panchami Chandukudlu, Emma Christie, Vivienne Evans, Ramandeep Gill, Barbora Krivankova, Keziah Lewis and Khai Syuen Chew) and junior doctors undertaking a year of industrial research at Canon Medical Research Europe (Dr William Clackett and Dr Giovana Klefti) for annotating the data used in this study.
Authors MC, HW, CW, ST, DF, VD, MS, PS, JL, AW, AON were employed by Canon Medical Research Ltd. while working on this project.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1https://pubmed.ncbi.nlm.nih.gov/download/
3West of Scotland Safe Haven ethical approval number GSH19NE004.
4using the en_core_web_lg GloVe (42) word embeddings from the spaCy (35) package.
5Excluding 5 labels which the model failed to learn. These labels had F1 scores of 0, and on average appeared only 4.5 times each in the training set.
1. Zerna C, Thomalla G, Campbell BC, Rha JH, Hill MD. Current practice, future directions in the diagnosis, acute treatment of ischaemic stroke. Lancet. (2018) 392:1247–56. doi: 10.1016/S0140-6736(18)31874-9
2. Goyal M, Almekhlafi M, Dippel DW, Campbell BCV, Muir K, Demchuk AM, et al. Rapid alteplase administration improves functional outcomes in patients with stroke due to large vessel occlusions. Stroke. (2019) 50:645–51. doi: 10.1161/STROKEAHA.118.021840
3. NICE. Stroke and transient ischaemic attack in over 16s: diagnosis and initial management (2019). Available from: https://www.nice.org.uk/guidance/conditions-and-diseases/cardiovascular-conditions/stroke-and-transient-ischaemic-attack.
4. Perera N, Dehmer M, Emmert-Streib F. Named entity recognition and relation detection for biomedical information extraction. Front Cell Dev Biol. (2020) 8. doi: 10.3389/fcell.2020.00673
5. Sung SF, Chen K, Wu DP, Hung LC, Su YH, Hu YH. Applying natural language processing techniques to develop a task-specific emr interface for timely stroke thrombolysis: a feasibility study. Int J Med Inform. (2018) 112:149–57. doi: 10.1016/j.ijmedinf.2018.02.005
6. Sun MC, Chan JA. A clinical decision support tool to screen health records for contraindications to stroke thrombolysis–a pilot study. BMC Med Inform Decis Mak. (2015) 15:1–7. doi: 10.1186/s12911-015-0229-4
7. Xu H, Pang J, Yang X, Li M, Zhao D. Using predictive process monitoring to assist thrombolytic therapy decision-making for ischemic stroke patients. BMC Med Inform Decis Mak. (2020) 20:1–10. 10.1186%2Fs12911-020-1111-6 31906929
8. Chung CC, Hong CT, Huang YH, Su ECY, Chan L, Hu CJ, et al. Predicting major neurologic improvement, long-term outcome after thrombolysis using artificial neural networks. J Neurol Sci. (2020) 410:116667. doi: 10.1016/j.jns.2020.116667
9. Flynn D, Nesbitt DJ, Ford GA, McMeekin P, Rodgers H, Price C, et al. Development of a computerised decision aid for thrombolysis in acute stroke care. BMC Med Inform Decis Mak. (2015) 15:1–15. doi: 10.1186/s12911-014-0127-1
10. Shao H, Chen X, Ma Q, Shao Z, Du H, Chan LWC. The feasibility and accuracy of machine learning in improving safety and efficiency of thrombolysis for patients with stroke: Literature review and proposed improvements. Front Neurol. (2022) 13:934929. doi: 10.3389/fneur.2022.934929
11. Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. (2004) 32:D267–70. doi: 10.1093/nar/gkh061
12. Cariello MC, Lenci A, Mitkov R. A comparison between named entity recognition models in the biomedical domain. Proceedings of the Translation and Interpreting Technology Online Conference. INCOMA Ltd. (2021). p. 76–84.
13. Kim JD, Ohta T, Tsuruoka Y, Tateisi Y, Collier N. Introduction to the bio-entity recognition task at JNLPBA. Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications. Citeseer (2004). p. 70–5.
14. Li J, Sun Y, Johnson RJ, Sciaky D, Wei CH, Leaman R, et al. Biocreative V CDR task corpus: a resource for chemical disease relation extraction. Database (2016) 2016:baw068, doi: 10.1093/database/baw068
15. Doğan RI, Leaman R, Lu Z. NCBI disease corpus: a resource for disease name recognition and concept normalization. J Biomed Inform. (2014) 47:1–10. doi: 10.1016/j.jbi.2013.12.006
16. Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. (2011) 18:552–6. doi: 10.1136/amiajnl-2011-000203
17. Mohan S, Li DMM. A large biomedical corpus annotated with UMLS concepts. Automated Knowledge Base Construction (AKBC) (2019).
18. Yang Z, Lin H, Li Y. Exploiting the performance of dictionary-based bio-entity name recognition in biomedical literature. Comput Biol Chem. (2008) 32(4):287–91. doi: 10.1016/j.compbiolchem.2008.03.008
19. Li L, Zhou R, Huang D. Two-phase biomedical named entity recognition using CRFs. Comput Biol Chem. (2009) 33:334–8. doi: 10.1016/j.compbiolchem.2009.07.004
20. Collobert R, Weston J, Bottou L, Karlen M, Kavukcuoglu K, Kuksa P. Natural language processing (almost) from scratch. J Mach Learn Res. (2011) 12:2493–537. doi: 10.48550/arXiv.1103.0398
21. Devlin J, Chang MW, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. NAACL HLT 2019 - 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies - Proceedings of the Conference. Vol. 1 (2019). p. 4171–86.
22. Beltagy I, Lo K, Cohan A. SciBERT: a pretrained language model for scientific text. Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP) (2019). p. 3615–20.
23. Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. Biobert: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. (2020) 36:1234–40. doi: 10.1093/bioinformatics/btz682
24. Gururangan S, Marasović A, Swayamdipta S, Lo K, Beltagy I, Downey D, et al. Don’t stop pretraining: adapt language models to domains and tasks [Preprint] (2020). Available at: https://arxiv.org/2arXiv:2004.10964.
25. Gu Y, Tinn R, Cheng H, Lucas M, Usuyama N, Liu X, et al. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans Comput Healthc (HEALTH). (2021) 3:1–23. doi: 10.1145/3458754
26. Yuan Z, Liu Y, Tan C, Huang S, Huang F. Improving biomedical pretrained language models with knowledge (KeBioLM) (2021). Available from: https://doi.org/10.18653/v1/2021.bionlp-1.20.
27. He Y, Zhu Z, Zhang Y, Chen Q, Caverlee J. Infusing disease knowledge into bert for health question answering, medical inference and disease name recognition (2020). p. 4604–14. Available from: https://doi.org/doi:10.18653/v1/2020.emnlp-main.372.
28. Michalopoulos G, Wang Y, Kaka H, Chen H, Wong A. UmlsBERT: clinical domain knowledge augmentation of contextual embeddings using the unified medical language system metathesaurus. Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (2021). p. 1744–53.
29. Jeong M, Kang J. Enhancing label consistency on document-level named entity recognition [Preprint] (2022). Available at: https://arxiv.org/arXiv:2210.12949.
30. Shin HC, Zhang Y, Bakhturina E, Puri R, Patwary M, Shoeybi M, et al. BioMegatron: larger biomedical domain language model (2020). p. 4700–6. Available from: https://doi.org/10.18653/v1/2020.emnlp-main.379.
31. Phan LN, Anibal JT, Tran H, Chanana S, Bahadroglu E, Peltekian A, et al. Scifive: a text-to-text transformer model for biomedical literature [Preprint] (2021). Available at: https://arxiv.org/2106.03598.
32. Wilde K, Anderson L, Boyle M, Pinder A, Weir A. Introducing a new trusted research environment – the safe haven artificial platform (SHAIP). Int J Popul Data Sci. (2022) 7. doi: 10.23889/ijpds.v7i3.2056.
33. brat rapid annotation tool (2020). Available from: http://brat.nlplab.org/.
34. DeepCognito (2021). Available from: https://deepcognito.com/.
35. Honnibal M, Montani I. spaCy 2: natural language understanding with Bloom embeddings, convolutional neural networks and incremental parsing (2017). To appear.
36. Ramshaw LA, Marcus MP. Text chunking using transformation-based learning. Natural language processing using very large corpora. Springer (1999). p. 157–176.
38. Li X, Sun X, Meng Y, Liang J, Wu F, Li J. Dice loss for data-imbalanced NLP tasks. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics (2020). p. 465–76.
39. Bergstra J, Yamins D, Cox D. Making a science of model search: hyperparameter optimization in hundreds of dimensions for vision architectures. International Conference on Machine Learning. PMLR (2013). p. 115–23.
41. Lin H, Lu Y, Tang J, Han X, Sun L, Wei Z, et al. A rigorous study on named entity recognition: can fine-tuning pretrained model lead to the promised land? EMNLP (1) (2020). p. 7291–300.
42. Pennington J, Socher R, Manning CD. Glove: global vectors for word representation. Empirical Methods in Natural Language Processing (EMNLP) (2014). p. 1532–43.
43. Dai X, Adel H. An analysis of simple data augmentation for named entity recognition [Preprint] (2020). https://arxiv.org/2010.11683.
Keywords: clinical decision support (CDS), acute stroke, thrombolysis, machine learning (ML), named entity recognition (NER)
Citation: Cutforth M, Watson H, Brown C, Wang C, Thomson S, Fell D, Dilys V, Scrimgeour M, Schrempf P, Lesh J, Muir K, Weir A and O’Neil AQ (2023) Acute stroke CDS: automatic retrieval of thrombolysis contraindications from unstructured clinical letters. Front. Digit. Health 5:1186516. doi: 10.3389/fdgth.2023.1186516
Received: 14 March 2023; Accepted: 15 May 2023;
Published: 14 June 2023.
Edited by:
Angus Roberts, King’s College London, United KingdomReviewed by:
Karthik Adapa, University of North Carolina at Chapel Hill, United States© 2023 Cutforth, Watson, Brown, Wang, Thomson, Fell, Dilys, Scrimgeour, Schrempf, Lesh, Muir, Weir and O'Neil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alison Q. O’Neil YWxpc29uLm9uZWlsQG1yZS5tZWRpY2FsLmNhbm9u
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.