Abstract
Introduction:
Drug-drug interaction (DDI) may lead to adverse reactions in patients, thus it is important to extract such knowledge from biomedical texts. However, previously proposed approaches typically focus on capturing sentence-aspect information while ignoring valuable knowledge concerning the whole corpus. In this paper, we propose a Multi-aspect Graph-based DDI extraction model, named DDI-MuG.
Methods:
We first employ a bio-specific pre-trained language model to obtain the token contextualized representations. Then we use two graphs to get syntactic information from input instance and word co-occurrence information within the entire corpus, respectively. Finally, we combine the representations of drug entities and verb tokens for the final classification
Results:
To validate the effectiveness of the proposed model, we perform extensive experiments on two widely used DDI extraction dataset, DDIExtraction-2013 and TAC 2018. It is encouraging to see that our model outperforms all twelve state-of-the-art models.
Discussion:
In contrast to the majority of earlier models that rely on the black-box approach, our model enables visualization of crucial words and their interrelationships by utilizing edge information from two graphs. To the best of our knowledge, this is the first model that explores multi-aspect graphs to the DDI extraction task, and we hope it can establish a foundation for more robust multi-aspect works in the future.
1. Introduction
According to statistics from the U.S. Centers of Disease Control and Prevention, from 2015 to 2018, 48.6% of Americans used at least one prescription drug in 30 days.1 More seriously, 20% of the elderly took more than 10 drugs simultaneously (1). However, drug-drug interaction (DDI) may occur when patients take multiple drugs, resulting in reduced drug effectiveness or even, possibly, adverse drug reactions (ADRs) (2). Therefore, the study of DDI extraction can be considerably important to patients’ healthcare, as well as clinical research. Currently, a number of drug databases, such as DailyMed (3), TWOSIDES (4) and DrugBank (5) can be used for retrieving DDI knowledge directly. However, with the exponential growth in biomedical literature, huge amounts of the most current and valuable knowledge remain hidden in biomedical literature (1). Thus, the development of an automatic tool to extract DDI is an urgent need.
During the past few years, various deep learning-based approaches, such as (6–14) have been proposed to extract DDI knowledge. Recently, (15) proposed an Long Short-Term Memory(LSTM)-based RNN model with two distinct additional layers, i.e., bottom RNN and top RNN, for the DDI extraction. It is worth noting that compared with LSTM, Graph Neural Networks (GNNs) can better deal with complex structural knowledge. Based on this, Li and Ji (8) combined a Bio-specific BERT (16) and Graph Convolutional Network (GCN) (17) to capture contextualized representation together with syntactic knowledge. Shi et al. (13) adopted the Graph Attention Network (GAT) (18) on an enhanced dependency graph to obtain higher-level drug representations for DDI extraction. However, as examples in Table 1, all the previous models only pay attention to the sentence-aspect features and do not even exploit the corpus knowledge, which could cause essential clues to be overlooked.
Table 1
| Model | Sentence (semantic) | Sentence (syntactic) | Corpus |
|---|---|---|---|
| AB-LSTM (19) | GloVe (20) | No | No |
| DCNN (6) | Order embedding (21) | No | No |
| ASDP-LSTM (7) | Word2Vec (22) | Dependency parse | No |
| RHCNN (23) | Bio-word emb. (24) | Dependency parse | No |
| GCNN-DDI (25) | Bio-word emb. (24) | Dependency parse | No |
| BERTChem-DDI (10) | BioBERT (26) | No | No |
| BERTDesc-DDI (11) | SciBERT (27) | No | No |
| DDI-MuG (Ours) | PubMedBERT(28) | Dependency parse | PMI |
Summary of previous neural network-based models and our proposed model.
To alleviate the issues mentioned above, in this work, we propose a multi-aspect graphs-based DDI extraction model, DDI-MuG, which can make use of the information in both sentence and corpus aspects. First, we use PubMedBERT to obtain sentence semantic representation. We then apply a GCN with an average pooling layer to capture syntactic features from the input instance, and another GCN with average pooling is employed to model the word co-occurrence in the corpus level simultaneously. After that, attentive pooling is used to integrate and obtain the optimal feature from the output of PubMedBERT and both sentence-aspect and corpus-aspect graphs. Finally, we employ a fully connected neural network in the output layer for the classification. Our proposed model is evaluated on two benchmark datasets: DDIExtraction-2013 (29) and TAC 2018 corpora (30). Experimental results show that our proposed model improves the performance of DDI extraction effectively.
To recap, the main contributions of our work can be summarized as follows:
We propose a novel neural model, named DDI-MuG, to exploit information from sentence-aspect and corpus-aspect graphs. As far as we know, this is the first model that utilizes multi-aspect graphs for the DDI extraction task.
We explore the effectiveness of different components in DDI-MuG. Experimental results indicate that knowledge from multi-aspect graphs is complementary, and their effective combination can largely improve performance.
We evaluate the proposed model on two benchmark datasets and achieve new state-of-the-art performance on both of them.
The rest of the paper is organized as follows. First, we introduce the background in Section 1. Then, several related works are introduced in Section 2. Next, in Section 3, we explain the framework in the proposed model in detail. We then describe the two benchmark datasets, evaluation metrics, and parameter setting in Section 4. Section 5 presents the experimental results and discussion, and finally, we conclude this work in Section 6.
2. Related works
Knowledge in many applications is exceedingly complex for a single-aspect network to learn robust representations. Multi-aspect networks have thus emerged naturally in different fields. Khan and Blumenstock (31) developed a multi-aspect GCNs model to consider different aspects of phone networks for poverty research. They employed subspace analysis and a manifold ranking procedure in order to merge multiple views and prune the graph, respectively. Liu et al. (32) first constructed semantic-based, syntactic-based, and sequential-based text graphs, and then utilized an inter-graph propagation to coordinate heterogeneous information among graphs. In order to exploit richer sources of graph edge information, Gong and Cheng (33) resorted to multi-dimensional edge weights to encode edge directions. Similarly, Huang et al. (34) used multi-dimensional edge weights to exploit multiple attributes, adapting the edge weights before entering into the next layer. In order to improve the prediction accuracy of social trust evaluation, Jiang et al. (35) assigned different attention coefficients to multi-aspect graphs in online social networks. Recently, Zhang et al. (36) constructed MA-GNNs, which utilize multiple aspect-aware graphs to improve recommendation performance. This model disentangles user preferences into different aspects and constructs multiple aspect-aware graphs to learn aspect-based user preferences.
3. Methods
The architecture of the proposed model is illustrated in Figure 1. First, we obtain the contextual semantic representation of the input instances by PubMedBERT. Then, a sentence-aspect graph is constructed to encode the syntactic feature from the dependency path, while a corpus-aspect graph is used to explore word co-occurrence within the entire corpus. Based on the vocabulary and instances analysis, we find that the part-of-speech (POS) tag of words, especially words corresponding to verbs, might be helpful for the final representation. Therefore, we subsequently feed the representations of verbs and drug entities from PubMedBERT, together with the two graphs, into an attentive pooling layer to distinguish important features from all representations. Finally, a fully connected layer with softmax is employed to perform the classification. The process is described in the following subsections in detail.
Figure 1

The proposed model architecture. This example is selected from DDIExtraction-2013 dataset. Two drugs are labelled in bold. As the space is limited, only part of the edges is shown in the word co-occurrence-based graph.
3.1. Encoding sentences with PubMedBERT
PubMedBERT was pre-trained on 14 million biomedical abstracts with 3.2 billion words from scratch. Given an input sentence with drug entities and , we convert each word into word pieces and then feed them into PubMedBERT. After the PubMedBERT calculation, we employ average pooling to aggregate vectorial representations of word pieces as the word representations. We denote the two drugs and verbs representations by , , and respectively.
3.2. Graph construction
Considering a graph with n nodes, the node i at the th layer is updated based on the representation of all neighbourhood nodes in the th layer as follows:
Here, represents the normalized adjacency matrix, and is the adjacency matrix with added self-connections. is the diagonal node degree matrix with . is the node embedding matrix at the lth layer, n is the number of nodes, indicates the dimension of the node features. Finally, denotes a layer-specific trainable weight matrix, and is a nonlinear function.
For each input instance, we encode a dependency graph from the current instance and a word co-occurrence over the entire corpus.
3.2.1. Sentence-aspect dependency graph
Dependency parser is widely used in relation classification tasks with the aim of exploring the syntactic information of sentences. We apply the Stanford dependency parser (37) to extract dependency syntactic information. Figure 2 shows the dependency relation of the input text in Figure 1. The connection from coadministered to colestipol means that coadministered is the head word of colestipol, and “nsubjpass” denotes the “passive nominal subject” dependency relation between the two words. We use the word embedding from PubMedBERT as the initial node representations, and set edge weights as 0 or 1 to indicate if two nodes are connected in the dependency path.
Figure 2
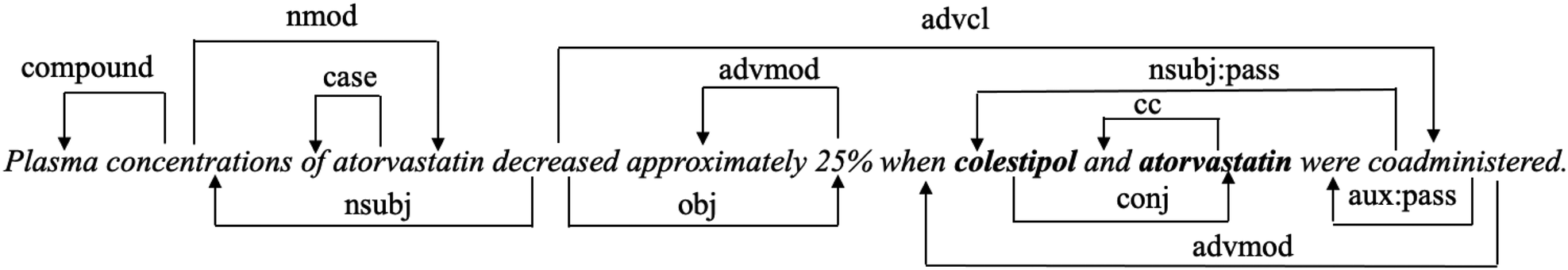
An example of dependency relation. Two drugs are labelled in bold.
Let the node representations in lth layer of the dependency graph be . We apply two graph convolutional layers to update each node, thus the updated is expressed as follows:
Then, an average pooling layer is applied to get the syntactic-based sentence embedding. Let be the updated node representations obtained from graph convolutional layers, the output of dependency graph, , is shown as:
We denote the outputs of drug and verbs representations as , , and , respectively.
3.2.2. Corpus-aspect word co-occurrence graph
Information on the co-occurrence of words indicates the connection between them, such as whether they form as a common phrase or provide clues for classification tasks. Firstly, we first lemmatize each word with Natural Language Toolkit (NLTK).2 Then we connect all word pairs in the graph, and employ point-wise mutual information (PMI) (38), a word associations measure, to store the word correlation information as an edge weight as follows:
The PMI between any two words is calculated as:where are words, is the number of examples in a fixed sliding window that contains both words, is the number of instances in the sliding window that contain word i, and is the total number of sliding windows. It is worth noting that the entire input sentence is set as the sliding window. Suppose there are 31,738 instances in the corpus, and the word of “decrease” and “coadminister” appear 1,821 and 953 times, respectively, and that they occur 27 times together in the whole corpus. Based on Formula 5 to 6, the PMI between these two words is -4.8. A positive PMI value corresponds to a high correlation between two words, while a negative value means that the two words have a small probability or no probability of occurrence. When two words have a negative PMI value, we view them as non-co-occurring and set their edge weight as 0.
Suppose the node representations in lth layer is . Similar to the dependency graph, the updated is shown as:
After an average pooling layer was utilized to get the word co-occurrence-based embedding, the graph is expressed as:where is the updated lth node representation from graph convolutional layers.
Drug and verbs representations, denotes by , , and , are extracted from and used as input for the next layer.
3.3. Attentive pooling layer
So far, given two drug entities and verbs, we have obtained rich feature representations from PubMedBERT and two graphs. As each instance has a different number of verbs, we apply an attentive pooling to get a fixed-length representation for verbs. In detail, this pooling mechanism computes the weights of feature vectors by using an attention mechanism, allowing it to learn the most significant feature effectively. Let and be the combined representation of drug entities from PubMedBERT and the two graphs, and be the corresponding verbs representation:where [;] denotes concatenation. These three representations are fed into the attentive pooling layer separately as follows:where is the learning parameter, is the attention weights. , and are the representation of the two drugs and verbs as the output of the attentive pooling layer.
3.4. Fully connected and softmax layer
In this layer, the updated representation of two drugs and verbs are concatenated as , and a nonlinear activation function tanh is then applied over into a fully connected layer. Finally, we deploy a softmax with a dropout layer to get the probability score for each class. The process is expressed as follows:where is the output of the fully connected layer, and are the softmax matrix and the bias parameter, respectively.
4. Experiments
In our experiments, two public DDI extraction corpora, i.e., DDIExtraction-2013 and TAC 2018, were used to evaluate the proposed model. This section introduces the two corpora in detail and then presents the evaluation metrics and parameters setting.
4.1. DDIExtraction-2013 dataset
We obtained the corpus from the challenge SemEval-2013 Task 9 (
39). This corpus is the major dataset that can be used to evaluate and compare the performance of DDI extraction models. It contains manually annotated sentences from 175 abstracts in MedLine,,
3and 730 abstracts in DrugBank.
4There are four kinds of positive interaction types:
Advice, Effect, Mechanism, Int. If the two drugs are unrelated, their relations are labelled as
Negative. The definitions of the five types are as follows:
Advice: a recommendation or advice regarding the simultaneous use of two drugs is described between two drugs.
Effect: an effect or a pharmacodynamic mechanism is described between two drugs.
Mechanism: a pharmacokinetic mechanism is described between two drugs.
Int: a DDI occurs between two drugs, but no additional information is provided.
Negative: there is no interaction between two drugs.
The original corpus suffers from a serious data imbalance problem. For example, the ratio of
Intto
Negativeinstances in the training set is 1:123.7, which heightens the difficulty of classifying drug pairs that hold
Intrelations, and continually affects the overall performance. To alleviate this data imbalance issue, many negative examples are filtered out in earlier studies, e.g., (
2,
6,
19,
40–
42). To ensure that the experimental results can be compared fairly with other baseline models, we adopted three rules in (
6) to remove negative instances:
If both drugs have the same name, remove the corresponding instances. The assumption is that drug will not interact with itself.
If one drug is a particular case or an abbreviation of the other, filter out the corresponding instances. Several patterns, such as “DRUG-A (DRUG-B)” and “DRUG-A such as DRUG-B”, are used to identify such cases.
If both drugs appear in the same coordinate structure, filter out the corresponding instances. Also, we use some pre-defined patterns, like “DRUG-A, , DRUG-B”, to filter out such instances.
summarizes the statistics and divisions of this corpora.
Table 2
| Training | Test | ||||
|---|---|---|---|---|---|
| Original | Filtered | Original | Filtered | ||
| Positive | Advice | 826 | 824 | 221 | 221 |
| Effect | 1,687 | 1,676 | 360 | 358 | |
| Mechanism | 1,319 | 1,309 | 302 | 301 | |
| Int | 188 | 187 | 96 | 96 | |
| Negative | 23,772 | 19,342 | 4,737 | 3,896 | |
| Overall | 27,792 | 23,338 | 5,716 | 4,872 | |
The statistics of DDIExtraction-2013 corpus.
4.2. TAC 2018 corpus
One of the tasks in “Drug-Drug Interaction Extraction from Drug Labels” track of the Text Analysis Conference (TAC) 2018
5was to detect and extract DDIs from structured product labellings (SPLs). The organizers provided a set of 22 SPLs for training (Training-22). Two other datasets containing 57 and 66 SPLs were provided as test sets. The organizers also provided an additional 180 SPLs (NLM-180) to supplement the training set. Interactions in this corpus are classified into one of the following three types:
Pharmacokinetic: This type includes phrases that demonstrate changes in physiological functions (30), such as decrease exposure, increased bioavailability.
Pharmacodynamic: This type includes phrases that describe the effects of the drugs, e.g., blood pressure lowering.
Unspecified: This type corresponds to caution phrases, e.g., avoid use.
As the original corpus is in XML format, we use the dataset in the KLncLSTMsentClf model (43) to train and evaluate our proposed model. In total, we obtain 6,436 training sentences by merging the training-22 and NLM-180 corpora. The two test sets contain 8,205 and 4,256 sentences, respectively.
4.3. Evaluation metrics
precision(P), recall(R) and F-score(F) are the major evaluation metrics in the DDI extraction task. In this paper, we adopt the standard micro-average precision, recall and F-score to evaluate the performance, and the formulas are listed as follows:
TP (true positive) represents the number of correctly classified positive instances, FP (false positive) denotes the number of negative instances that are misclassified as positive instances, and FN (false negative) is the number of positive instances that are misclassified as negative ones.
4.4. Parameters setting
In our experiment, PyTorch library (
44) is used as the computational framework. As there is no development or validation set in the original corpus, we randomly select 20% of the training dataset as the validation set to adjust the model parameters and the remaining 80% as the training set. The parameters used are shown as follows:
Maximal length .
Embedding size of PubMedBERT .
Hidden layer dimension of dependency and co-occurrence graph & .
Mini-batch size 32.
Dropout rate .
Learning rage .
Number of epoch 10.
5. Results and discussion
5.1. Results on DDIExtraction-2013
5.1.1. Comparison with baseline methods
We compare the performance of our DDI-MuG with 11 baseline methods. The comparison results of different models are shown in Table 3. The highest value is labelled in bold, and the second highest value is marked underline. In general, deep neural network-based approaches achieve better performance than statistical ML-based methods. It demonstrates the capability and potential of utilizing neural network in DDI extraction tasks. A notable exception is that the F1-score of SVM-DDI (40) is slightly higher than the AB-LSTM model (19). This might be due to SVM-DDI (40) benefiting from rich and complex lexical and syntactic handcraft features. It can be seen that our DDI-MuG obtains the best overall performances in view of precision and F1 score. In terms of the performances for all four types, DDI-MuG performs best on Advice, Mechanism and Int, and obtain the second best performance on Effect. It is worth noting that all methods achieve relatively low performance on Int. This discrepancy might be caused by the insufficient training samples of Int, which leads to these models to be underfitting.
Table 3
| Methods | Breakdown F1 | Overall performance | |||||
|---|---|---|---|---|---|---|---|
| Advice | Effect | Mechanism | Int | Precision | Recall | F1 | |
| Statistical ML-based methods | |||||||
| UTurKu (45) | 0.630 | 0.600 | 0.582 | 0.507 | 0.732 | 0.499 | 0.594 |
| WBI (46) | 0.632 | 0.610 | 0.618 | 0.510 | 0.642 | 0.579 | 0.609 |
| FBK-irst (47) | 0.692 | 0.628 | 0.679 | 0.547 | 0.646 | 0.656 | 0.651 |
| SVM-DDI (40) | 0.725 | 0.662 | 0.693 | 0.483 | – | – | 0.670 |
| Deep neural network-based methods | |||||||
| AB-LSTM (19) | 0.697 | 0.683 | 0.681 | 0.542 | 0.678 | 0.659 | 0.669 |
| DCNN (6) | 0.777 | 0.693 | 0.702 | 0.464 | 0.757 | 0.647 | 0.698 |
| Joint AB-LSTM (19) | 0.794 | 0.676 | 0.763 | 0.431 | 0.734 | 0.697 | 0.715 |
| ASDP-LSTM (7) | 0.803 | 0.718 | 0.740 | 0.543 | 0.741 | 0.718 | 0.729 |
| RHCNN (23) | 0.805 | 0.734 | 0.782 | 0.589 | 0.773 | 0.737 | 0.754 |
| GCNN-DDI (25) | 0.835 | 0.758 | 0.794 | 0.514 | 0.801 | 0.740 | 0.770 |
| DREAM (13) | 0.848 | 0.761 | 0.816 | 0.551 | 0.823 | 0.747 | 0.783 |
| Our methods | |||||||
| DDI-MuG(with word. graph) | 0.893 | 0.812 | 0.871 | 0.599 | 0.868 | 0.805 | 0.835 |
| DDI-MuG(with dep. graph) | 0.900 | 0.826 | 0.865 | 0.583 | 0.842 | 0.835 | 0.839 |
| DDI-MuG | 0.907 | 0.823 | 0.893 | 0.606 | 0.870 | 0.824 | 0.847 |
Performance comparisons on DDIExtraction-2013 Corpus. The highest value is labelled in bold, and the second highest value is marked underline.
Then, we find the contributions of multi-aspect graphs to the proposed model. By removing in turn the sentence-aspect dependency graph and corpus-aspect word co-occurrence graph, our method reduces to DDI-MuG(with word. graph) and DDI-MuG(with dep. graph), respectively. From Table 3, we can see that the F1-score of DDI-MuG(with dep. graph) is higher than the F1-score of DDI-MuG(with word. graph), which proves that the syntactic features are indeed valuable for identifying the interaction relation between two drugs. Overall, it can be seen that the F1-score of DDI-MuG surpass the DDI-MuG(with word. graph) and DDI-MuG(with dep. graph) by 0.012 and 0.008, separately. This indicates that multi-aspect graphs are complementary to each other and together can serve as an appropriate supplement to contextual information.
5.1.2. Impact of pre-trained embedding
To evaluate the efficiency of the pre-trained language model, we conduct experiments of replacing PubMedBERT with other similar models. As shown in Table 4, the four bio-specific models, i.e., BioBERT, SciBERT, ouBioBERT (48), and PubMedBERT, led to improvement over standard BERT. DDI-MuG by PubMedBERT achieves the best result for the reason that it was pre-trained on biomedical texts from scratch.
Table 4
| Pre-trained embedding | P | R | f F1 |
|---|---|---|---|
| DDI-MuG(by BERT) | 0.801 | 0.790 | 0.795 |
| DDI-MuG(by BioBERT) | 0.843 | 0.816 | 0.829 |
| DDI-MuG(by SciBERT) | 0.839 | 0.825 | 0.832 |
| DDI-MuG(by ouBioBERT) | 0.850 | 0.826 | 0.838 |
| DDI-MuG(by PubMedBERT) | 0.870 | 0.824 | 0.847 |
The effect of pre-trained embedding. The highest value is labelled in bold.
5.1.3. Error analysis
In addition, to present the above achievements, it is necessary to discuss the limitations of our approach. One common type of error is that the four kinds of positive instances are often misclassified as negative instances. This is due to the imbalanced data that small instance categories are misclassified as large instance categories. There is another notable error that 34.4% of
Inttype instances are misclassified as
Effecttype. This is because some
Intinstances have similar semantics to
Effectinstances. For example, in the following two instances:
“arbiturates may decrease the effectiveness of oral contraceptives, certain antibiotics, quinidine, theophylline, corticosteroids, anticoagulants, and beta blockers.”
“sulfoxone may increase the effects of barbiturates, tolbutamide, and uricosurics.”
The words decrease and increase are the clues for identifying interactions in the two semantically close sentences. However, the first instance belongs to the Int type, while the second belongs to Effect. The number of Int instances is far smaller than the number of Effect instances, which also leads to the occurrence of this kind of mistake.
5.1.4. Are verb representations really helpful?
In our previous vocabulary and instances analysis, we found that in the DDIExtraction-2013 corpus, when instances contain the words inhibit, increased, decreased, there is a great possibility that the drug pair has the Mechanism relation. On the other hand, when instances contain avoided, recommended or administered, the drug pair is likely to have the Advice relation.
Thus, to further investigate how the verbs are important for the final classification, we studied the effect of extracting DDI only from the drug information without using the verbs knowledge. Table 5 shows the comparison of the performance with and without the verb information. This result indicates verb representation can serve as a supplement to improve the model performance.
Table 5
| Precision | Recall | F-score | |
|---|---|---|---|
| DDI-MuG(drug-only) | 0.863 | 0.823 | 0.843 |
| DDI-MuG(all) | 0.870 | 0.824 | 0.847 |
The comparison of with or without verbs information. The highest value is labelled in bold.
5.2. Results on TAC 2018
5.2.1. Comparison with baseline model
Since we use the same dataset as KLncLSTMsentClf (43), we view it as the baseline model. From Table 6, we can see that our proposed model achieves better results in both two test sets, which indicates the transferability of our proposed model.
Table 6
| Dataset | Model | P | R | F1 |
|---|---|---|---|---|
| Test1 | KLncLSTMsentClf | 0.470 | 0.620 | 0.530 |
| Test1 | DDI-MuG(with word. graph) | 0.717 | 0.712 | 0.715 |
| Test1 | DDI-MuG(with dep. graph) | 0.688 | 0.718 | 0.703 |
| Test1 | DDI-MuG(all) | 0.721 | 0.728 | 0.723 |
| Test2 | KLncLSTMsentClf | 0.490 | 0.670 | 0.567 |
| Test2 | DDI-MuG(with word. graph) | 0.710 | 0.726 | 0.718 |
| Test2 | DDI-MuG(with dep. graph) | 0.713 | 0.730 | 0.721 |
| Test2 | DDI-MuG(all) | 0.717 | 0.743 | 0.729 |
Comparison with baseline models on the TAC 2018 corpus. The highest value is labelled in bold.
6. Conclusions
In this paper, we propose DDI-MuG, a novel multi-aspect graphs framework for DDI extraction tasks. Concretely, a bio-specific pre-trained language model, PubMedBERT, is first employed to encode the context information of each word from the aspect of sentence semantic information. Then, two graphs are utilized to explore sentence syntactic and corpus word co-occurrence information, respectively. After that, an attentive pooling mechanism is employed to update the representations of drug entities and verbs. Finally, by feeding the concatenated representation of the two drugs and verbs into a fully connected and softmax classifier, the interaction between the two drugs is obtained. Extensive comparison experiments with baseline models on two public datasets verify the effectiveness of multi-aspect graphs in the DDI extraction task.
In addition, Most previous models are based on the black-box concept that makes the prediction without showing how the model did so. However, with our proposed model, we can visualise the important words and its word-word relationship of the final classification by using the edge information in both dependency and co-occurrence graphs.
For future work, there are at least two directions that could be considered. Firstly, the performance on categories with small training samples, like Int in the DDIExtraction-2013 corpora, is unsatisfactory. The solution of contrastive learning can be explored. Secondly, drug knowledge from external databases could be integrated with the architecture for richer drug representations.
Statements
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://github.com/zhangyijia1979/hierarchical-RNNs-model-for-DDI-extraction/tree/master/DDIextraction2013.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
JY: Conceptualization, Data Curation, Validation, Writing - Original Draft; YD: Methodology, Implementation, Writing - Review; SL: Methodology, Implementation, Writing - Review; JP: Resources, Supervision; SCH (Corresponding Author): Conceptualization, Methodology, Resources, Supervision, Writing - Review & Editing. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1.^ https://www.cdc.gov/nchs/data/hus/2019/039-508.pdf
References
1.
Zhang T Leng J Liu Y . Deep learning for drug–drug interaction extraction from the literature: a review. Brief Bioinformatics. (2020) 21:1609–27. 10.1093/bib/bbz087
2.
Zhu Y Li L Lu H Zhou A Qin X . Extracting drug-drug interactions from texts with biobert, multiple entity-aware attentions. J Biomed Inform. (2020) 106:103451. 10.1016/j.jbi.2020.103451
3.
Barrière C Gagnon M . Drugs, disorders: from specialized resources to web data. In: Workshop on Web Scale Knowledge Extraction, 10th International Semantic Web Conference. Bonn, Germany: Springer (2011).
4.
Tatonetti NP Ye PP Daneshjou R Altman RB . Data-driven prediction of drug effects, interactions. Sci Transl Med. (2012) 4:125ra31. 10.1126/scitranslmed.3003377
5.
Wishart DS Feunang YD Guo AC Lo EJ Marcu A Grant JR , et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. (2017) 46:D1074–82. 10.1093/nar/gkx1037
6.
Liu S Tang B Chen Q Wang X . Drug-drug interaction extraction via convolutional neural networks. Comput Math Methods Med (2016) 2016:1–8. 10.1155/2016/6918381
7.
Zhang Y Zheng W Lin H Wang J Yang Z Dumontier M . Drug–drug interaction extraction via hierarchical RNNs on sequence, shortest dependency paths. Bioinformatics. (2018) 34:828–35. 10.1093/bioinformatics/btx659
8.
Li D Ji H . Syntax-aware multi-task graph convolutional networks for biomedical relation extraction. In: Proceedings of the Tenth International Workshop on Health Text Mining, Information Analysis (LOUHI 2019). Hong Kong: Association for Computational Linguistics (2019). p. 28–33.
9.
Ren Y Fei H Ji D . Drug-drug interaction extraction using a span-based neural network model. In: 2019 IEEE International Conference on Bioinformatics, Biomedicine (BIBM). San Diego, CA, US: IEEE (2019). p. 1237–9.
10.
Mondal I . BERTChem-DDI: improved drug-drug interaction prediction from text using chemical structure information. In: Proceedings of Knowledgeable NLP: the First Workshop on Integrating Structured Knowledge, Neural Networks for NLP. Suzhou, China: Association for Computational Linguistics (2020). p. 27–32.
11.
Asada M Miwa M Sasaki Y . Using drug descriptions and molecular structures for drug–drug interaction extraction from literature. Bioinformatics. (2020) 37:1739–46. 10.1093/bioinformatics/btaa907
12.
Fatehifar M Karshenas H . Drug-drug interaction extraction using a position and similarity fusion-based attention mechanism. J Biomed Inform. (2021) 115:103707. 10.1016/j.jbi.2021.103707
13.
Shi Y Quan P Zhang T Niu L . Dream: drug-drug interaction extraction with enhanced dependency graph and attention mechanism. Methods. (2022) 203:152–9. 10.1016/j.ymeth.2022.02.002
14.
Huang L Lin J Li X Song L Zheng Z Wong K-C . EGFI: drug–drug interaction extraction and generation with fusion of enriched entity and sentence information. Brief Bioinformatics. (2021) 23:bbab451. 10.1093/bib/bbab451
15.
Salman M Munawar HS Latif K Akram MW Khan SI Ullah F . Big data management in drug–drug interaction: a modern deep learning approach for smart healthcare. Big Data Cogn Comput. (2022) 6:30. 10.3390/bdcc6010030
16.
Devlin J Chang M-W Lee K Toutanova K . BERT: Pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers). Minneapolis, Minnesota: Association for Computational Linguistics (2019). p. 4171–86.
17.
Kipf TN Welling M . Semi-supervised classification with graph convolutional networks. In: 5th International Conference on Learning Representations. Toulon, France: OpenReview.net (2017).
18.
Veličković P Cucurull G Casanova A Romero A Liò P Bengio Y . Graph attention networks. In: International Conference on Learning Representations. Vancouver, BC, Canada: ICLR (2018).
19.
Sahu SK Anand A . Drug-drug interaction extraction from biomedical texts using long short-term memory network. J Biomed Inform. (2018) 86:15–24. 10.1016/j.jbi.2018.08.005
20.
Pennington J Socher R Manning C . GloVe: Global vectors for word representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). Doha, Qatar: Association for Computational Linguistics (2014). p. 1532–43.
21.
Lai S Liu K He S Zhao J . How to generate a good word embedding. IEEE Intell Syst. (2016) 31:5–14. 10.1109/MIS.2016.45
22.
Mikolov T Yih W-t. Zweig G . Linguistic regularities in continuous space word representations. In: Proceedings of the 2013 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Atlanta, Georgia: Association for Computational Linguistics (2013). p. 746–51.
23.
Sun X Dong K Ma L Sutcliffe R He F Chen S , et al. Drug-drug interaction extraction via recurrent hybrid convolutional neural networks with an improved focal loss. Entropy. (2019) 21:37. 10.3390/e21010037
24.
Pyysalo S Ginter F Moen H Salakoski T Ananiadou S . Distributional semantics resources for biomedical text processing. Proceedings of Languages in Biology and Medicine. Tokyo, Japan: Languages in Biology and Medicine (2013). p. 39–44.
25.
Xiong W Li F Yu H Ji D . Extracting drug-drug interactions with a dependency-based graph convolution neural network. In: 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). Los Alamitos, CA, USA: IEEE Computer Society (2019). p. 755–9.
26.
Jinhyuk L Wonjin Y Kim . BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. (2019) 36:1234–40. 10.1093/bioinformatics/btz682
27.
Beltagy I Lo K Cohan A . Scibert: pretrained language model for scientific text. In: Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing, the 9th International Joint Conference on Natural Language Processing. Hong Kong, China: Association for Computational Linguistics (2019). p. 3615–20.
28.
Gu Y Tinn R Cheng H Lucas M Usuyama N Liu X , et al. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans. Comput. Healthcare. (2021) 3:1–23. 10.1145/3458754
29.
Herrero-Zazo M Segura-Bedmar I Martínez P Declerck T . The DDI corpus: an annotated corpus with pharmacological substances, drug–drug interactions. J Biomed Inform. (2013) 46:914–20. 10.1016/j.jbi.2013.07.011
30.
Demner-Fushman D Fung KW Do P Boyce RD Goodwin TR . Overview of the TAC 2018 drug-drug interaction extraction from drug labels track. Theory and applications of categories. Gaithersburg, Maryland, US: Mount Allison University (2018).
31.
Khan MR Blumenstock JE . Multi-GCN: Graph convolutional networks for multi-view networks, with applications to global poverty. Proc AAAI Conf Artif Intell. (2019) 33:606–13. 10.1609/aaai.v33i01.3301606
32.
Liu X You X Zhang X Wu J Lv P . Tensor graph convolutional networks for text classification. Proc AAAI Conf Artif Intell. (2020) 34:8409–16. 10.1609/AAAI.V34I05.6359
33.
Gong L Cheng Q . Exploiting edge features for graph neural networks. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). IEEE; City: Long Beach, CA, US: IEEE (2019). p. 9203–11.
34.
Huang Z Li X Ye Y Ng MK , MR-GCN: Multi-relational graph convolutional networks based on generalized tensor product. In: Bessiere C, editor. Proceedings of the Twenty-Ninth International Joint Conference on Artificial Intelligence, IJCAI-20. International Joint Conferences on Artificial Intelligence Organization (2020). p. 1258–64.
35.
Jiang N Jie W Li J Liu X Jin D . GATrust: a multi-aspect graph attention network model for trust assessment in OSNs. IEEE Trans Knowl Data Eng. (2022) 18:1. 10.1109/TKDE.2022.3174044
36.
Zhang C Xue S Li J Wu J Du B Liu D , et al. Multi-aspect enhanced graph neural networks for recommendation. Neural Netw. (2023) 157:90–102. 10.1016/j.neunet.2022.10.001
37.
Chen D Manning C . A fast and accurate dependency parser using neural networks. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). Doha, Qatar: Association for Computational Linguistics (2014). p. 740–50.
38.
Turney P . Mining the web for synonyms: PMI-IR versus LSA on TOEFL. In: Proceedings of the Twelfth European Conference on Machine Learning. Freiburg, Germany: Springer (2001). p. 491–02.
39.
Segura-Bedmar I Martínez P Herrero-Zazo M . SemEval-2013 task 9: extraction of drug-drug interactions from biomedical texts (DDIExtraction 2013). In: Second Joint Conference on Lexical and Computational Semantics (*SEM), Volume 2: Proceedings of the Seventh International Workshop on Semantic Evaluation (SemEval 2013). Atlanta, Georgia, USA: Association for Computational Linguistics (2013). p. 341–50.
40.
Kim S Liu H Yeganova L Wilbur WJ . Extracting drug–drug interactions from literature using a rich feature-based linear kernel approach. J Biomed Inform. (2015) 55:23–30. 10.1016/j.jbi.2015.03.002
41.
Zhao Z Yang Z Luo L Lin H Wang J . Drug drug interaction extraction from biomedical literature using syntax convolutional neural network. Bioinformatics. (2016) 32:3444–53. 10.1093/bioinformatics/btw486
42.
Wang W Yang X Yang C Guo X Zhang X Wu C . Dependency-based long short term memory network for drug-drug interaction extraction. BMC Bioinf. (2017) 18:578. 10.1186/s12859-017-1962-8
43.
Baruah G Kolla M . Klicklabs at the TAC 2018 drug-drug interaction extraction from drug labels track. In: Proceedings of the 2018 Text Analysis Conference, TAC 2018; 2018 Nov 13–14; Gaithersburg, MD, USA. NIST (2018).
44.
Paszke A Gross S Massa F Lerer A Bradbury J Chanan G , et al. PyTorch: an imperative style, high-performance deep learning library. Red Hook, NY, USA: Curran Associates Inc (2019).
45.
Björne J Kaewphan S Salakoski T . UTurku: drug named entity recognition and drug-drug interaction extraction using SVM classification and domain knowledge. In Second Joint Conference on Lexical and Computational Semantics (*SEM), Volume 2: Proceedings of the Seventh International Workshop on Semantic Evaluation (SemEval 2013). Atlanta, Georgia, USA: Association for Computational Linguistics (2013). p. 651–9.
46.
Thomas P Neves M Rocktäschel T Leser U . WBI-DDI: drug-drug interaction extraction using majority voting. In: Second Joint Conference on Lexical and Computational Semantics (*SEM), Volume 2: Proceedings of the Seventh International Workshop on Semantic Evaluation (SemEval 2013). Atlanta, Georgia, USA: Association for Computational Linguistics (2013). p. 628–35.
47.
Chowdhury MFM Lavelli A . FBK-irst: a multi-phase kernel based approach for drug-drug interaction detection and classification that exploits linguistic information. In: Second Joint Conference on Lexical and Computational Semantics (*SEM), Volume 2: Proceedings of the Seventh International Workshop on Semantic Evaluation (SemEval 2013). Atlanta, Georgia, USA: Association for Computational Linguistics (2013). p. 351–5.
48.
[Dataset] Wada S Takeda T Manabe S Konishi S Kamohara J Matsumura Y . A pre-training technique to localize medical bert and to enhance biomedical bert (2020). arXiv preprint arXiv:2005.07202.
Summary
Keywords
drug-drug interactions, relation extraction, deep learning, multi-aspect graphs, graph neural network
Citation
Yang J, Ding Y, Long S, Poon J and Han SC (2023) DDI-MuG: Multi-aspect graphs for drug-drug interaction extraction. Front. Digit. Health 5:1154133. doi: 10.3389/fdgth.2023.1154133
Received
30 January 2023
Accepted
03 April 2023
Published
24 April 2023
Volume
5 - 2023
Edited by
Angus Roberts, King’s College London, United Kingdom
Reviewed by
Chengkun Wu, National University of Defense Technology, China, Haridimos Kondylakis, Foundation for Research and Technology (FORTH), Greece
Updates
Copyright
© 2023 Yang, Ding, Long, Poon and Han.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
* Correspondence: Soyeon Caren Han caren.han@sydney.edu.au; caren.han@uwa.edu.au
Specialty Section: This article was submitted to Health Informatics, a section of the journal Frontiers in Digital Health
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.