
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Dent. Med. , 19 February 2025
Sec. Periodontics
Volume 6 - 2025 | https://doi.org/10.3389/fdmed.2025.1543535
 Tomoyuki Nukaga1
Tomoyuki Nukaga1 Eitoyo Kokubu2
Eitoyo Kokubu2 Kazuko Okamoto-Shibayama2
Kazuko Okamoto-Shibayama2 Yuichiro Kikuchi2
Yuichiro Kikuchi2 Masahiro Furusawa1
Masahiro Furusawa1 Takashi Muramatsu3
Takashi Muramatsu3 Kazuyuki Ishihara2*
Kazuyuki Ishihara2*
Background: Periodontitis is caused by the dysbiosis of subgingival plaque, and Treponema denticola is the pathogen associated with this disease. Bacteriocins are involved in interbacterial competition during dysbiosis. In our previous study, three potential bacteriocin ABC transporter genes (tepA1-B1, tepA2-B2, and tepA3-B3) of T. denticola were investigated. Upstream of tepA1-B1, three genes annotated as bacteriocin-type signal domain proteins are located. However, the role of these proteins in T. denticola remains unclear. In the present study, these bacteriocin-type signal domain proteins were characterized to elucidate their putative roles in T. denticola.
Methods: Gene clusters surrounding bacteriocin-type signal domain protein genes were compared in silico. The expression of proteins and transporters was evaluated using real-time quantitative reverse transcription PCR (qRT-PCR). Bacteriocin-type signal domain proteins were detected using immunoblot analysis. The expression of bacteriocin-like proteins was investigated by co-culturing with Treponema vincentii.
Results: The DNA sequences of the bacteriocin-type signal domain protein genes and upstream lipoprotein genes were highly conserved. Expression of the bacteriocin-type signal domain protein and tepA1 was slightly higher in the mid-log phase than in the stationary phase and was reduced upon co-culture with T. vincentii. Bacteriocin ABC transporter gene tepA1 was expressed independently of tepA2 and tepA3. Immunoblot analysis detected bacteriocin-like proteins in culture supernatants. However, bactericidal activity was not detected in the culture supernatant of T. denticola.
Conclusion: Three tandem lipoprotein-bacteriocin-type signal domain protein genes may have originated from duplication. Bacteriocin-type signal domain proteins are expressed under unstimulated conditions and are secreted by T. denticola cells.
Periodontal disease is an inflammation of the periodontal tissue caused by subgingival plaque bacteria (1). A major etiologic agent is a microbiome alteration called dysbiosis, which is the shift in the composition of the microbiome to one with a large number of virulent microorganisms. Treponema denticola is an anaerobic, spiral-shaped microorganism (2) isolated from chronic periodontitis lesions in high abundance, and is a major disease pathogen (3, 4). A recent study identified T. denticola as an index strain for dysbiosis (5).
A large number of microbial taxa have been detected in dental plaque (6). Competition and symbiosis between microorganisms play key roles in their survival in dental plaques (7). In multispecies biofilms, bacteria produce toxic components, such as H2O2 and bacteriocins, to inhibit other microorganisms (8). T. denticola must evade the attack from other bacteria and communicate to other bacteria during dysbiosis. Bacteriocins are important weapons to overcome competitors (9) and many gram-positive bacteria, including Streptococcus mutans and Streptococcus salivarius, produce these molecules (10, 11). Bacteriocin-like effects have been reported for several periodontopathic bacteria (12–14). In a recent metagenomic analysis of the periodontitis microbiome, bacteriocins were reported as the second most abundant biosynthetic gene cluster (15). However, the specific genes involved in the interactions between periodontopathic bacteria remain unclear. Among the genome of periodontopathic bacteria, T. denticola has the largest number of annotations of “bacteriocin.” In our previous study, tepA2, whose deduced amino acid sequence is similar to that of the bacterial immunity protein (ImmA) of S. mutans was detected in the genome sequence of T. denticola. tepA2 is a putative bacteriocin ABC transporter, an ATP-binding/permease protein, and TepB2, which is located downstream of tepA2, is a putative bacteriocin ABC transporter, a bacteriocin-binding protein (16). These proteins might contain an ABC transporter in the cytoplasmic membrane of T. denticola. Additionally, two sets of other genes annotated as bacteriocin ABC transporters (tepA1-tepB1, and tepA3-tepB3) were detected. Among the three bacteriocin ABC transporters, the tepA1-tebB1 region has three upstream genes encoding bacteriocin-type signal domain proteins (bacteriocin-like proteins TDE_0416, TDE_0422, and TDE_0424) (Figure 1). These genes contain a bacteriocin-type signaling domain, although their functions remain unknown. Exported proteins may play a role in bacterial interactions.
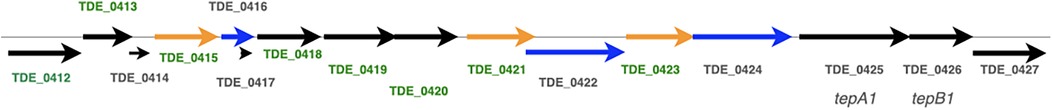
Figure 1. Bacteriocin-type signal domain proteins and flanking region. TDE_0416, TDE_0422, and TDE_0424 contained bacteriocin signal sequences. TDE_0425 and TDE_0426 are potential bacteriocin ABC transporters.
In the current study, we investigated the function of these cluster genes, focusing on the genes encoding bacteriocin-like proteins upstream of tepA1 to investigate the fate of proteins and their role in T. denticola.
The similarity of the genes flanking the bacteriocin-like protein genes against the genes and proteins in the database of the National Center for Biotechnology Information was determined using blast search (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignments were performed using ClustalW (17) and phylogenetic trees were constructed using the neighbor-joining method in GENETYX_MAC v.21 (NIHON SERVER, Tokyo, Japan). For a comparison of protein structures, the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk) was used.
The strains used in this study are listed in Table 1. These strains were obtained from the American Type Culture Collection or culture collection in our laboratory. T denticola, Treponema socranskii, and Treponema vincentii were maintained in TYGVS medium (20) containing tryptone (Becton Dickinson, Sparks, MD, USA), yeast extract (Becton Dickinson), gelatin (Becton Dickinson), volatile fatty acids, and rabbit serum (Nihon biotest, Tokyo, Japan) under anaerobic conditions (80% N2, 10% H2, and 10% CO2) in an anaerobic chamber (ANX-3, Hirasawa, Tokyo, Japan). To detect the bacteriocin-type signal domain protein of T. denticola in the culture supernatant, T. denticola was inoculated into TYGVHS medium, which is a TYGVS medium containing 2% horse serum (Nihon Biotest, Tokyo, Japan) instead of 10% rabbit serum. For T. denticola KT-3, 40 µg/mL erythromycin was added to the TYGVS medium. Aggregatibacter actinomycetemcomitans, Actinomyces, and Streptococcus were maintained on Tryptic soy agar (Becton Dickinson Sparks, MD) containing hemin (5 µg/mL), menadione (0.5 µg/mL), and 10% defibrinated horse blood (Nihon Biotest).
To detect the gene encoding the bacteriocin-like protein in T. denticola strains, Southern blotting was performed as described previously (16). Genomic DNA of T. denticola was isolated using a Gentra Pure Gene Kit (Qiagen, Tokyo, Japan). Genomic DNA was digested with Hind III, separated on a 1% agarose gel, and transferred onto a Nytran N blotting membrane (Cytiba, Tokyo, Japan). A digoxygenin-labeled probe (504 bp) was prepared using a Thermal Cycler C1000 (Bio-Rad Laboratories, Hercules, CA, USA) with primer pairs 416_probeF and 416_probeR listed in Table 2.
To determine the expression of bacteriocin-like proteins, the mRNA levels of TDE_0416, TDE_0422, and TDE_0424 were evaluated using real-time quantitative reverse transcription PCR (qRT-PCR). T. denticola ATCC 35405 was harvested at the mid-log or stationary phase, and total RNA was extracted using TRIzol reagent (Sigma-Aldrich, St. Louis, MO). cDNA was synthesized using the SuperScript First-Strand Synthesis cDNA kit (Invitrogen, Carlsbad, CA, USA). The expression levels of TDE_0416, TDE_0422, and TDE_0424 were determined by qRT-PCR using the primers and TaqMan probes listed in Table 2. Five microliters of cDNA were added in a 45 µL reaction mixture containing 25 μL of 2× TaqMan Universal PCR Master Mix, 5 μL of forward primer, reverse primer, and the TaqMan probe listed in Table 2. The cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min each using the 7500 Real-Time PCR system (Applied Biosystems, Foster, CA). 16S rRNA was used as an internal control. The expression levels of each gene were normalized using the relative standard curve method.
To investigate the relationship between the expression of the three bacteriocin ABC transporters, T. denticola ATCC 35405 and a tepA2-deficient mutant were harvested at the mid-log or stationary phase, and the expression of tepA1, B1, A3, and B3 was evaluated using qRT-PCR, as described above. The primers and TaqMan probes used for qRT-PCR analysis are listed in Table 2.
Antibodies were prepared as described previously (21). To prepare polyclonal antibodies that recognize bacteriocin-like proteins, the peptide GKPDGYEGKEGQRG, corresponding to the 89th–102nd residues of the deduced amino acid sequence of TDE_0424, which is also present in TDE_0416 and TDE_0422, was used. A Cys residue was conjugated to the N-terminus of the sequence and synthesized using F-moc chemistry. The peptide was conjugated to keyhole limpet hemocyanin using maleimidobenzoic acid N-hydroxysuccinimide ester. The conjugate (0.15 mg) was used to immunize the rabbits (six biweekly injections), and blood was collected 12 weeks after the initial immunization. Immunization and bleeding were performed by Eurofins Genomics (Tokyo, Japan). Antibody reactivity was confirmed by ELISA, using the peptide GKPDGYEGKEGQRG as the antigen.
To determine whether bacteriocin-type signal domain proteins were released from the cells, T. denticola was cultured in TYGVHS medium under anaerobic conditions for 4 days. The culture supernatant of T. denticola was centrifuged at 12,000×g for 15 min. The outer sheath fraction of T. denticola was isolated as described previously (21). The culture supernatants and outer sheath fractions were separated with 10%–20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), as described previously (21). The separated proteins were transferred using the Trans-Blot Turbo system (Bio-Rad, Hercules, CA, USA), and bacteriocin-type signal domain proteins were detected using the primary antibody described above (1/1,000) and a peroxidase-conjugated secondary antibody against anti-rabbit IgG (1/1,000, Bio-Rad) with a snap i.d. 2.0 (EMD Millipore Corporation, Billerica, MA). Signals were developed using a TMB Membrane Peroxidase Substrate (KPL, Gaithersburg, MD, USA).
T. denticola ATCC 35405 and T. vincentii ATCC 35580 were cultured in TYGVS medium separately at 37°C under anaerobic conditions for 2 days. When the absorbance at 660 nm reached approximately 0.3–0.6, 20 ml of each culture was added. The mixed and original cultures were incubated for 2 h. After incubation, the expression levels of TDE_0416, TDE_0422, and TDE_0424, and tepA1 in both mixed and single cultures were assessed as described above.
The bacteriocin activity was screened according to a previous report with minor modifications (22). T. denticola ATCC 35405 was precultured in TYGVS medium for 3 days and 10 µl of culture was spotted on TYGVS agar, which contains 0.8% Noble agar (Becton Dickinson, Sparks, MD, USA) and incubated anaerobically for 5 days. Actinomyces viscosus ATCC 15987, Aggregatibacter actinomycetemcomitans Y4, S. mutans MT8148R, Streptococcus sanguinis ATCC 10556, and Streptococcus oralis ATCC 10557 were precultured in Tryptic soy broth overnight at 37°C under anaerobic conditions. T. socranskii ATCC 35536 and T. vincentii ATCC 35580 were precultured in TYGVS broth at 37°C under anaerobic conditions for 3 days. After incubation, cell density was adjusted to OD660 = 0.2, mixed with molten TYGVS agar at 45°C, and overlayed on TYGVS agar, on which T. denticola was spotted. The plate was incubated at 37°C anaerobically for an additional 48 h and the presence of an inhibition zone by T. denticola was evaluated.
Gene expression was compared using the Student's t-test. For the effect of co-culture and expression of bacteriocin ABC transporter-like proteins, normality and homogeneity of variance were tested before applying the Student's t-test. Statistical significance was set at p < 0.05.
Three open reading frames encoding the bacteriocin-type signal domain proteins TDE_0416, TDE_0422, and TDE_0424 were located upstream of tepA1 (TDE_0425, Figure 1). At the DNA level, TDE_416 shared 98% and 96% identity with 501 and 367 bases in TDE_0422 and TDE_0424, respectively. TDE_0422 shared 94% identity with TDE_0424 in 1,714 bases. The deduced amino acid sequences of all open reading frames contained bacteriocin-like signal peptides with double glycine at the 17–18th residues (Figure 2). TDE_0422 and TDE_0424 contain 614 and 607 residues, respectively, whereas TDE_0416 contains 167. TDE_0416 has 98% identity in 167 amino acid residues and 84% identity in 167 amino acid residues with TDE_0422 and TDE_0424, respectively. TDE_0422 shares 89% identity in 616 amino acid residues with TDE_0424. The 459th–522nd amino acid residues of TDE_0422 and the 452nd–515th residues of TDE_0424 were similar to the fibronectin-binding autotransporter adhesin ShdA (PRK15319).
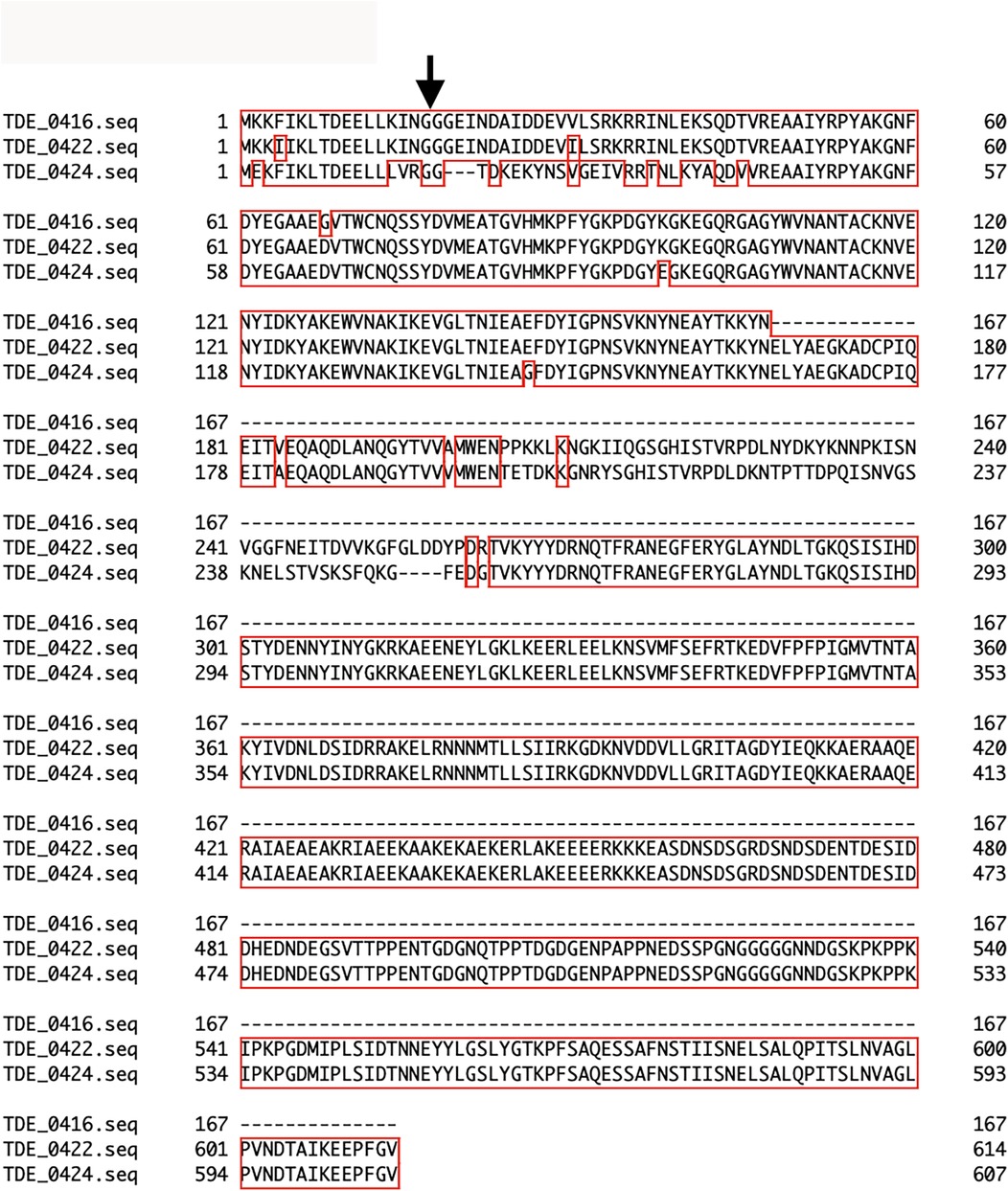
Figure 2. Multiple amino acid sequence alignments of bacteriocin-type signal domain proteins. The arrow indicates a double glycine, which is typical of bacteriocin signal peptides. Alignment of TDE_0416, TDE_0422, and TDE_0424 was performed using Genetyx-Mac 21.2.0.
Upstream of tepA1, there were eight open reading frames: TDE_0412, TDE_0413, TDE_0415, TDE_0418, TDE_0419, TDE_0420, TDE_0421, and TDE_0423, annotated as Treponema-clustered lipoproteins (Figure 1). Known signal sequences of lipoproteins were identified from these sequences, except TDE_0412, using SignalP program (https://services.healthtech.dtu.dk/service.php?SignalP-6.0). TDE_0413 and TDE_0418 were similar to the signal peptide regions of lipoproteins. However, no extensive homology was observed. Other genes were predicted to encode lipoproteins. The amino acid sequence of TDE_0415 showed 78% identity in 404 residues and 83% identity in 422 residues with TDE_0421 and TDE_0423, respectively (Figure 3). The similarity between TDE_0415 and other clustered proteins was less than 30%. The evolutionary tree of the DNA sequences indicated that TDE_0415, TDE_0421, and TDE_0423, located immediately upstream of the three bacteriocin-type signal domain proteins, were closely related (Figure 4). At the DNA level, TDE_0415 shared 85% identity in 1,278 bases and 89% identity in 1,270 bases with TDE_0421 and TDE_0423, respectively. These results suggest that the tandemly arranged genes encoding lipoprotein and bacteriocin-like proteins, TDE_0415-TDE_0416, TDE_0421-TDE_0422, and TDE_0423-TDE_0424, were derived from a common ancestor gene.
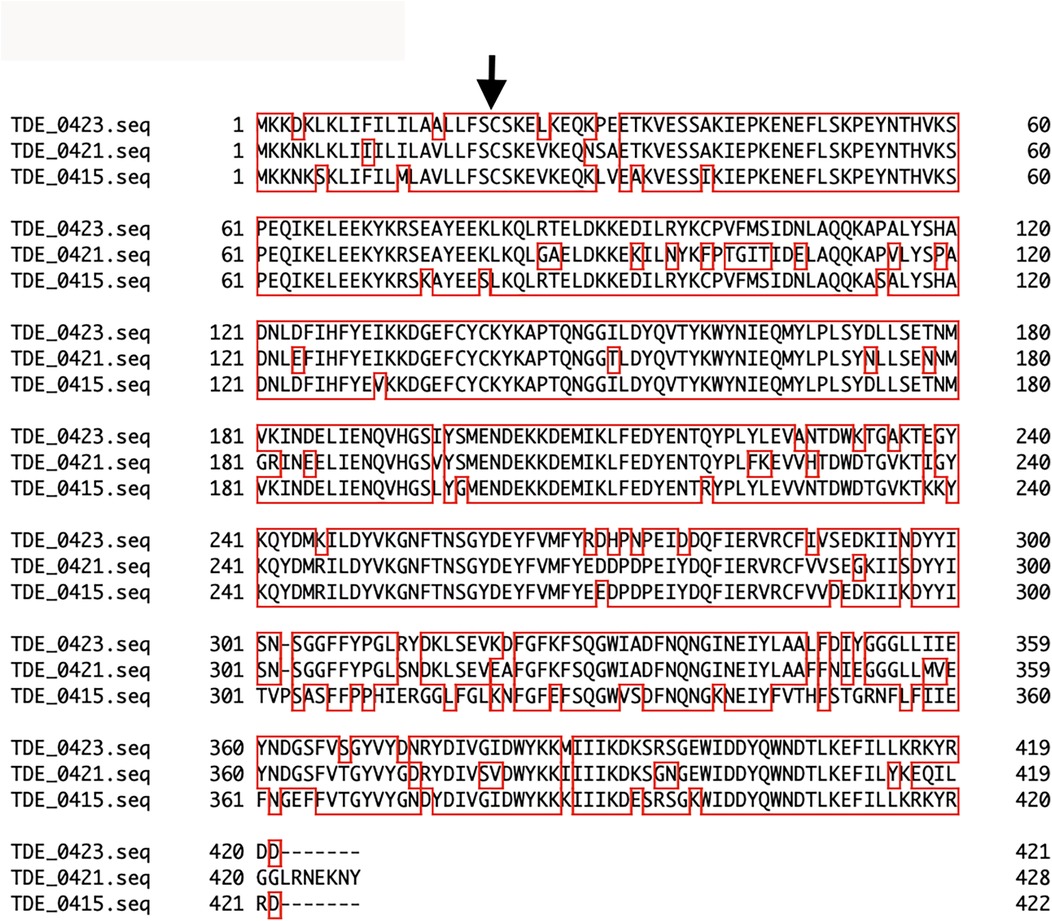
Figure 3. Amino acid sequence alignment analysis of clustered lipoproteins. The arrow indicates the cleavage site of the signal peptidase for lipoproteins as determined using the SignalP program. Alignment of TDE_0415, TDE_0421, and TDE_0423 was performed using Genetyx-Mac 21.2.0.
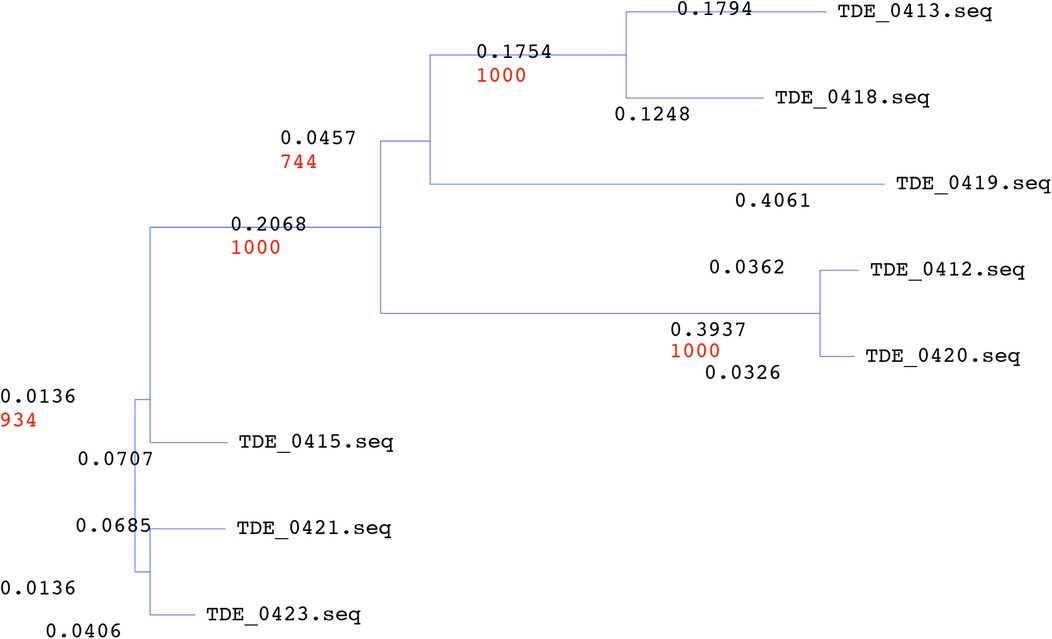
Figure 4. Evolutionary tree of Treponema denticola clustered with lipoproteins. The evolutionary tree was organized using the NJ method with 1,000 boost trap trials.
To investigate the prevalence of bacteriocin-type signal domain proteins among T. denticola strains, Southern blotting was performed using TDE_0416 as a probe. The results showed that bacteriocin-like proteins are common among T. denticola strains (Figure 5). In the AlphaFold Protein Structure Database, the outer membrane efflux protein of Magnetococcus marinus and two fimbrial proteins of Bacteroidetes bacteria were detected as structure similarity cluster by Foldseek search using TDE_0424.
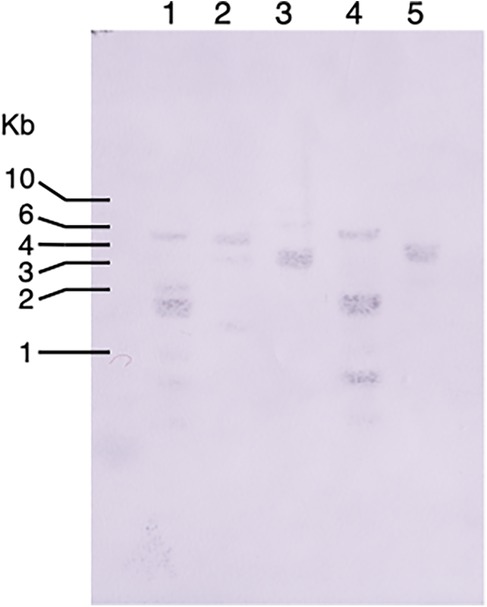
Figure 5. Southern blotting of bacteriocin-type signal domain proteins in T. denticola strains. The genomic DNA of T. denticola was digested with Hind III and the TDE_0416 fragment (504 bp) was used as a probe. Lanes: 1: T. denticola ATCC 33520; 2: T. denticola ATCC 33521; 3: T. denticola ATCC 35405; 4: T. denticola ATCC35405; 5: T. denticola GM1.
T. denticola was harvested in the mid-log and stationary phases to determine the expression of bacteriocin-type signal domain protein genes. The relative expression of bacteriocin-like proteins to 16S rRNA [mean ± standard deviation (SD)] of TDE_0416, TDE_0422, TDE_0424, and tepA1 were 1.27 ± 0.014, 0.67 ± 0.003, 0.71 ± 0.013, and 0.11 ± 0.001, respectively. The expression of all genes was slightly decreased in the stationary phase compared to the mid-log phase, but the difference was too small to have a biological effect (Figure 6).
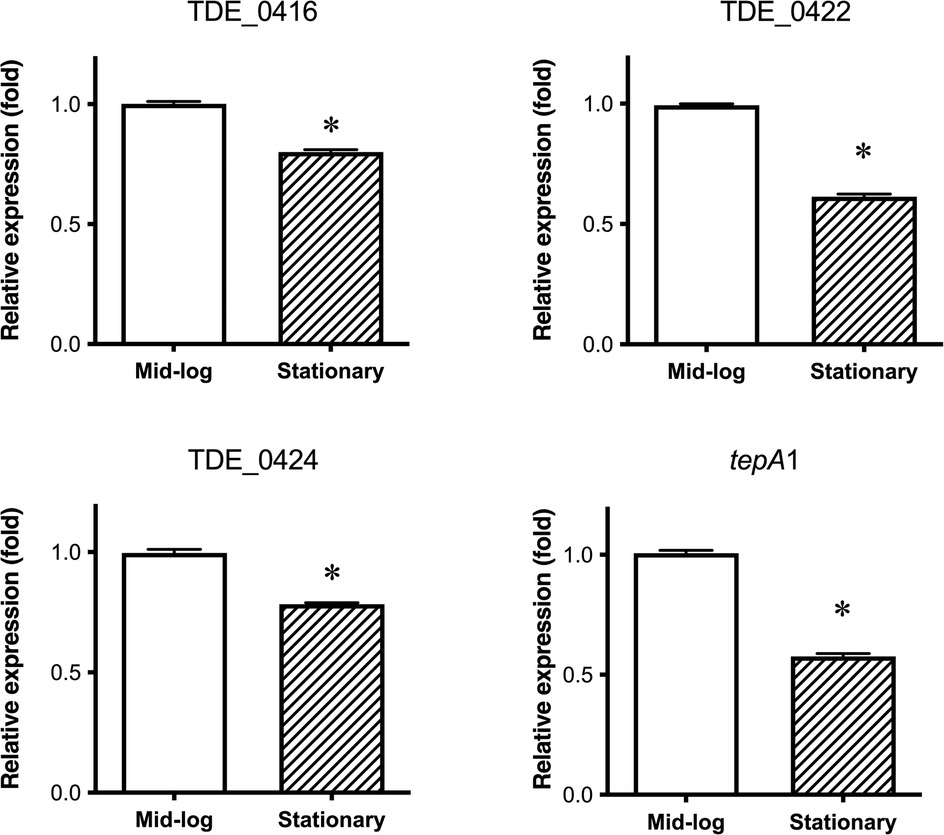
Figure 6. Expression of bacteriocin-type signal domain proteins and transporters in different growth phases. Mid-log: expression in mid-log phase; Stationary: expression in stationary phase. Each dataset is shown as the average fold of mid-log phase ± SD. Representative data (n = 3) from three experiments are provided (*P < 0.05).
Co-culturing T. denticola ATCC 35405 with T. vincentii ATCC 35580 highlighted the effect of competitive strains on the expression of bacteriocin-like proteins. The expression of TDE_0416, TDE_0422, TDE_0424, and TDE_0425 in T. denticola decreased after co-culture with T. vincentii (Figure 7). Expression of these genes was not detected in a single culture of T. vincentii.
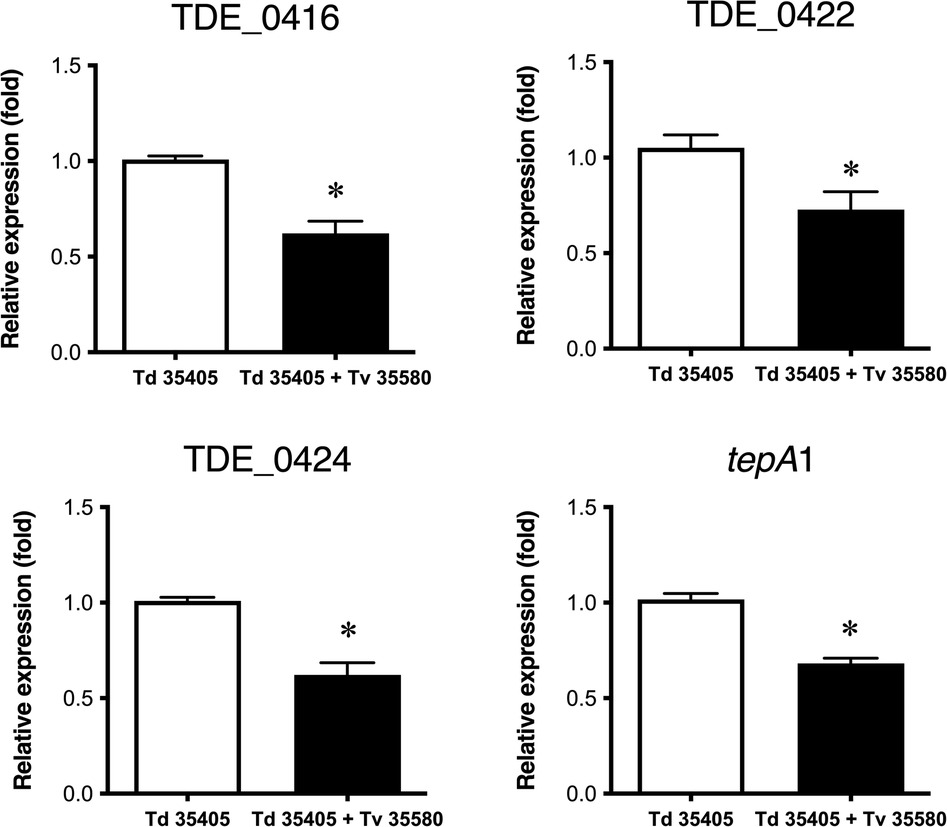
Figure 7. Effect of co-culture on the expression of bacteriocin-type signal domain proteins. Td 35405: single culture of T. denticola ATCC 35405; Td 35405 + Tv 35580: co-culture of T. denticola ATCC 35405 and T. vincentii ATCC 35580. Each dataset is shown as the fold value of the T. denticola mono culture ± SD (n = 8, *P < 0.05).
To confirm the secretion of bacteriocin-type signal domain proteins from T. denticola, an immunoblot analysis was performed using the culture supernatant and outer sheath of T. denticola ATCC 35405 and an antibody against the bacteriocin-type signal domain proteins (Figure 8). Bands of approximately 61, 43, and 14 kDa were detected in the culture supernatants. The 61 kDa band had a molecular mass close to that calculated from the amino acid sequences of TDE_0422 and TDE_0424 (66,549.77 and 65,688.48, respectively), and the 14 kDa was close to that of TDE_0416 (16,932.39). The 43 kDa protein may be a degraded 61 kDa protein. No bands were detected in the TYGVHS medium. No band was detected in the outer sheath fraction (data not shown), indicating that the bacteriocin-type signal domain proteins were not released into the vesicles. These results indicated that bacteriocin-type signal domain proteins were secreted by T. denticola cells.
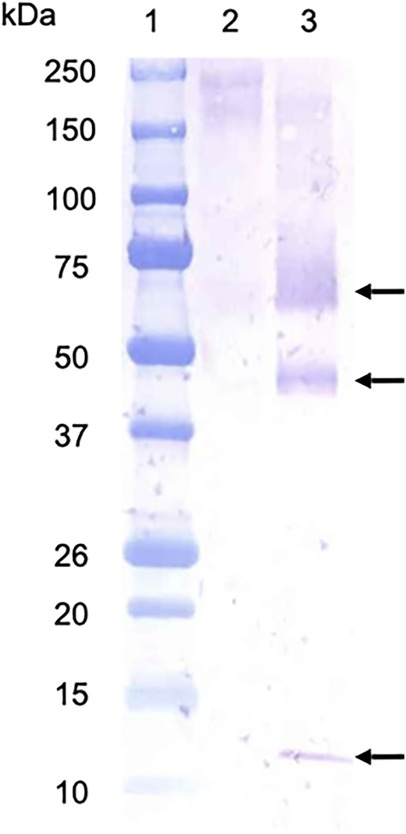
Figure 8. Immunoblot analysis of the culture supernatant of T. denticola. Culture supernatant; lane 1, molecular marker; lane 2, TYGVHS medium; lane 3: culture supernatants of T. denticola in the same medium. Arrows indicate bands detected using antiserum against bacteriocin-like protein.
In the genome sequence of T. denticola ATCC 35405 (https://www.ncbi.nlm.nih.gov/nuccore/AE017226.1/), three bacteriocin-type signal domain proteins (TDE_0416, TDE_0422, and TDE_0424) are located in the same region, whereas only tepA1 and tepB1 are located downstream of the bacteriocin-like protein genes. The other two bacteriocin ABC transporter genes (tepA1-B1 and tepA3-B3) are located in a different area without a bacteriocin-like protein gene (16). To investigate the involvement of bacteriocin ABC transporter genes in the export of bacteriocin-like proteins, we evaluated the expression of tepA1, tepB1, tepA3, and tepB3 in a T. denticola tepA2 gene-inactivated mutant (16) using qRT-PCR. As shown in Figure 9, the expression of tepA3 and B3 increased in the tepA2 mutant, whereas the expression of tepA1 and B1 was unaffected.
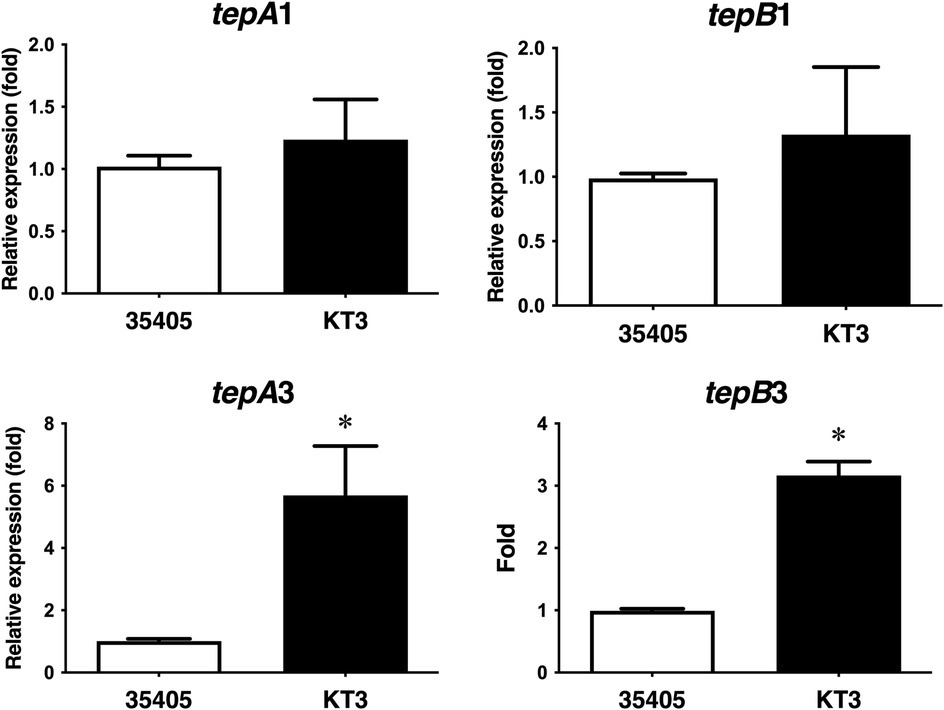
Figure 9. Expression of bacteriocin ABC transporter-like proteins in T. denticola ATCC 35405 and T. denticola KT-3 (tepA2 mutant). Expression of tepA1, B1, A3, and B3 is shown as the average fold of wild-type ± SD (n = 9, P < 0.05).
To clarify whether bacteriocin-type signal domain proteins possess bactericidal activity, the effects of the culture supernatant on the growth of oral bacteria were evaluated. No growth inhibition was observed for A. viscosus ATCC 15987, A. actinomycetemcomitans Y4, S. mutans MT8148R, S. sanguinis ATCC 10556, S. oralis ATCC 10557, T. socranskii ATCC 35536, or T. vincentii ATCC 35580 (data not shown). To investigate the function of the bacteriocin-like protein, we tried to replaced TDE_0423-TDE_0424 with ermB via homologous recombination; however, a TDE_0423-TDE_0424 mutant was not obtained.
T. denticola has three genes that encode bacteriocin-like proteins (16). These genes and their upstream lipoproteins exhibit high DNA sequence identity. The proteins were constitutively expressed, with a slight decrease in expression during the stationary phase (Figure 6). Although these proteins were secreted into the culture medium, no bactericidal effects were observed.
These three sets of lipoprotein-bacteriocin-like proteins exhibited high DNA sequence identity. Given the high similarity of the three sequences and their locations, it is likely that these gene sets resulted from the duplication of ancestral genes, a phenomenon commonly observed in oral bacteria. In Porphyromonas gingivalis, the catalytic domains of arg-gingipain and Hgp44, the C-terminal hemagglutinin/adhesion, are duplicated (23, 24) and Mfa5, a component of the Mfa1 fimbriae, is also duplicated (25). Tandemly arranged genes such as gtfB and gtfC, are thought to originate via tandem duplication (26, 27). Gene duplications can influence the oligomeric state of the protein (28). Gene duplications are a significant contributor to functional diversity (29). Additionally, bacteriocin-like proteins were detected in all the tested T. denticola strains. These findings suggest that these proteins play a physiological role in T. denticola and that the three lipoproteins upstream of the protein may be involved in their maturation or function.
The bacteriocin-like signal sequences of TDE_0416, TDE_0422, and TDE_0424, and downstream bacteriocin ABC transporter genes strongly suggested their secretion. Immunoblotting results showed that the sizes of the largest and smallest bands detected in the culture supernatant of T. denticola were consistent with those of the bacteriocin-like protein (Figure 8). The vesicles released from many bacteria, including T. denticola, export bacterial proteins (30, 31). The vesicles in T. denticola contain an outer sheath component of T. denticola such as the major outer sheath protein (31). The antibody against bacteriocin-like protein did not react with any proteins in the outer sheath fraction. These results suggest that bacteriocin-like proteins are secreted by bacteriocin ABC transporters. Three bacteriocin ABC transporter-like genes were detected in T. denticola ATCC 35405. Conversely, no bacteriocin-type signal domain protein was detected around tepA2-B2 and tepA3-B3. To investigate the relationship among these three ABC transporters, the expression of these genes was evaluated in the tepA2 mutant. Expression of tepA3 increased in the tepA2 mutant, whereas tepA1 expression remained unaffected (Figure 9). These results suggest that transporters organized by tepA2 and tepB2 cooperate with those organized by tepA3 and tepB3 and that these four proteins are not involved in the secretion of the bacteriocin-like protein.
Bacteriocin-like proteins showed similarity to the C-terminal domain of ShedA, which forms a beta-barrel structure in the outer membrane (32). The proteins were suggested to be bacteriocins, as their signal peptide resembles that of bacteriocins, and the region showing similarity to the beta-barrel region of ShedA may affect the structure of the outer or cytoplasmic membranes. To investigate the activity of these proteins, we attempted to inactivate TDE_0423 and TDE_0424; however, no mutants were obtained. The DNA sequence of 5′ of TDE_0423 showed high similarity with that of 3′ of TDE_0424. This may have interfered with homologous recombination. Because bacteriocin activity is typically restricted to the gram status of the producer, T. vincentii and A. actinomycetemcomitans were included as indicator strains for the evaluation of bacteriocin activity. In addition, gram-positive bacteria (A. viscosus, S. mutans, S. sanguinis, and S. oralis) were added as indicator strains. The culture supernatant of T. denticola ATCC 35405 showed no bactericidal activity against any of the oral bacteria tested. One explanation for this is the culture conditions used. The degree of bactericidal activity against sensitive bacteria is sometimes influenced by specific pH values (33) or by the presence of chemical agents that weaken cell wall integrity (34). The culture conditions may not have been optimal for producing bacteriocin-like proteins. Additionally, the spectrum of bactericidal activity may not have included the tested strains.
Another possibility is that these proteins have functions beyond bactericidal effects. Their amino acid sequences were not similar to any bacteriocin except for a double glycine, which is typical of the signal peptide of bacteriocins (35). A previous study identified double glycine-containing signal peptides as precursors of competence-stimulating peptides in Streptococcus pneumoniae (36). In S. mutans, first gene of ciaRH operonen coding a double-glycine (GG)-containing peptide acts as a calcium sensing signalling peptide (37). The mRNAs of bacteriocin-like proteins were expressed in T. denticola, and their levels slightly decreased in the mid-log phase compared to those in the log phase. Protein expression was reduced by co-culturing with T. vincentii (Figure 7), indicating that protein expression was regulated by interactions with other microorganisms. These proteins were secreted from T. denticola cell, and Foldseek analysis showed that structure similarity cluster to the bacteriocin-like protein was membrane protein and a fimbrial protein. It is possible that these proteins are membrane-associated or bind to receptor proteins. Dirix et al. reported that many possible GG motif-containing peptides obtained from a search of gram-negative bacterial genomes showed structural similarities to bacteriocins and peptide hormones (35). These proteins may be peptide hormones. Currently, no information concerning the similarity between the bacteriocin-like proteins and other functional proteins is available. Further analyses of gene expression under various physiological conditions and after gene inactivation are required to clarify the roles of these proteins.
In conclusion, our results show that bacteriocin-like proteins were expressed under unstimulated conditions and secreted from T. denticola cells. The expression of these proteins was reduced by co-culturing with T. vincentii, suggesting that physiological conditions, such as interactions with other bacteria, regulate their expression. The results of the present study shed light on the mechanism of dysbiosis by revealing novel interactions between T. denticola and other periodontopathic bacteria and contribute to the understanding of the etiology of periodontitis.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
TN: Data curation, Investigation, Writing – original draft. EK: Data curation, Writing – original draft, Formal Analysis. KO-S: Data curation, Writing – original draft, Resources. YK: Data curation, Writing – original draft, Formal Analysis. MF: Methodology, Writing – review & editing. TM: Writing – review & editing, Conceptualization. KI: Conceptualization, Writing – review & editing, Data curation, Funding acquisition.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Tokyo Dental College and JSPS KAKENHI (grant number 15K11023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kinane DF, Stathopoulou PG, Papapanou PN. Periodontal diseases. Nat Rev Dis Primers. (2017) 3:17038. doi: 10.1038/nrdp.2017.38
2. Ishihara K. Virulence factors of Treponema denticola. Periodontol 2000. (2010) 54(1):117–35. doi: 10.1111/j.1600-0757.2009.00345.x
3. Ellen RP, Galimanas VB. Spirochetes at the forefront of periodontal infections. Periodontol 2000. (2005) 38:13–32. doi: 10.1111/j.1600-0757.2005.00108.x
4. Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. (2012) 6(6):1176–85. doi: 10.1038/ismej.2011.191
5. Chen T, Marsh PD, Al-Hebshi NN. SMDI: an index for measuring subgingival microbial dysbiosis. J Dent Res. (2022) 101(3):331–8. doi: 10.1177/00220345211035775
6. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. (2010) 192(19):5002–17. doi: 10.1128/JB.00542-10
7. Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. (2018) 26(3):229–42. doi: 10.1016/j.tim.2017.09.008
8. Merritt J, Qi F. The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol. (2012) 27(2):57–69. doi: 10.1111/j.2041-1014.2011.00634.x
9. Cotter PD, Ross RP, Hill C. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol. (2013) 11(2):95–105. doi: 10.1038/nrmicro2937
10. Heng NC, Tagg JR, Tompkins GR. Competence-dependent bacteriocin production by Streptococcus gordonii DL1 (challis). J Bacteriol. (2007) 189(4):1468–72. doi: 10.1128/JB.01174-06
11. Ye D, Liu Y, Li J, Zhou J, Cao J, Wu Y, et al. Competitive dynamics and balance between Streptococcus mutans and commensal streptococci in oral microecology. Crit Rev Microbiol. (2024):1–12. doi: 10.1080/1040841X.2024.2389386
12. Apolonio AC, Carvalho MA, Bemquerer MP, Santoro MM, Pinto SQ, Oliveira JS, et al. Purification and partial characterization of a bacteriocin produced by Eikenella corrodens. J Appl Microbiol. (2008) 104(2):508–14. doi: 10.1111/j.1365-2672.2007.03565.x
13. Kaewsrichan J, Douglas CW, Nissen-Meyer J, Fimland G, Teanpaisan R. Characterization of a bacteriocin produced by Prevotella nigrescens ATCC 25261. Lett Appl Microbiol. (2004) 39(5):451–8. doi: 10.1111/j.1472-765X.2004.01608.x
14. Lucia LF, Farias FF, Eustaquio CJ, Auxiliadora M, Carvalho R, Alviano CS, et al. Bacteriocin production by Actinobacillus actinomycetemcomitans isolated from the oral cavity of humans with periodontal disease, periodontally healthy subjects and marmosets. Res Microbiol. (2002) 153(1):45–52. doi: 10.1016/s0923-2508(01)01285-2
15. Koohi-Moghadam M, Watt RM, Leung WK. Multi-site analysis of biosynthetic gene clusters from the periodontitis oral microbiome. J Med Microbiol. (2024) 73(10). doi: 10.1099/jmm.0.001898
16. Tanaka-Kumazawa K, Kikuchi Y, Sano-Kokubun Y, Shintani S, Yakushiji M, Kuramitsu HK, et al. Characterization of a potential ABC-type bacteriocin exporter protein from Treponema denticola. BMC Oral Health. (2016) 17(1):18. doi: 10.1186/s12903-016-0243-7
17. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. (2007) 23(21):2947–8. doi: 10.1093/bioinformatics/btm404
18. Nakao R, Hirayama S, Yamaguchi T, Senpuku H, Hasegawa H, Suzuki T, et al. A bivalent outer membrane vesicle-based intranasal vaccine to prevent infection of periodontopathic bacteria. Vaccine. (2023) 41(30):4369–83. doi: 10.1016/j.vaccine.2023.05.058
19. Kokubu E, Kinoshita E, Ishihara K. Inhibitory effects of lingonberry extract on oral streptococcal biofilm formation and bioactivity. Bull Tokyo Dent Coll. (2019) 60(1):1–9. doi: 10.2209/tdcpublication.2018-0007
20. Numata Y, Kikuchi Y, Sato T, Okamoto-Shibayama K, Ando Y, Miyai-Murai Y, et al. Novel transcriptional regulator OxtR1 regulates potential ferrodoxin in response to oxygen stress in Treponema denticola. Anaerobe. (2024) 87:102852. doi: 10.1016/j.anaerobe.2024.102852
21. Miyai-Murai Y, Okamoto-Shibayama K, Sato T, Kikuchi Y, Kokubu E, Potempa J, et al. Localization and pathogenic role of the cysteine protease dentipain in Treponema denticola. Mol Oral Microbiol. (2023) 38(3):212–23. doi: 10.1111/omi.12406
22. Yonezawa H, Kuramitsu HK. Genetic analysis of a unique bacteriocin, smb, produced by Streptococcus mutans GS5. Antimicrob Agents Chemother. (2005) 49(2):541–8. doi: 10.1128/AAC.49.2.541-548.2005
23. Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. Construction and characterization of arginin-specific cysteine proteinase (arg-gingipain)-deficient mutants of Porphyromonas gingivalis. J Biol Chem. (1995) 270:23619–26. doi: 10.1074/jbc.270.40.23619
24. Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. (2003) 4(6):397–407. doi: 10.2174/1389203033487036
25. Hasegawa Y, Nagano K. Porphyromonas gingivalis FimA and Mfa1 fimbriae: current insights on localization, function, biogenesis, and genotype. Jpn Dent Sci Rev. (2021) 57:190–200. doi: 10.1016/j.jdsr.2021.09.003
26. Fujiwara T, Terao Y, Hoshino T, Kawabata S, Ooshima T, Sobue S, et al. Molecular analyses of glucosyltransferase genes among strains of Streptococcus mutans. FEMS Microbiol Lett. (1998) 161(2):331–6. doi: 10.1111/j.1574-6968.1998.tb12965.x
27. Simpson CL, Cheetham NWH, Jacques NA. Four glucosyltransferases, GtfJ, GtfK, GtfL and GtfM, from Streptococcus salivarius ATCC 25975. Microbiology (Reading). (1995) 141(Pt 6):1451–60. doi: 10.1099/13500872-141-6-1451
28. Mallik S, Tawfik DS, Levy ED. How gene duplication diversifies the landscape of protein oligomeric state and function. Curr Opin Genet Dev. (2022) 76:101966. doi: 10.1016/j.gde.2022.101966
29. Serres MH, Kerr AR, McCormack TJ, Riley M. Evolution by leaps: gene duplication in bacteria. Biol Direct. (2009) 4:46. doi: 10.1186/1745-6150-4-46
30. Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. (2010) 64:163–84. doi: 10.1146/annurev.micro.091208.073413
31. Veith PD, Glew MD, Gorasia DG, Chen D, O'Brien-Simpson NM, Reynolds EC. Localization of outer membrane proteins in Treponema denticola by quantitative proteome analyses of outer membrane vesicles and cellular fractions. J Proteome Res. (2019) 18(4):1567–81. doi: 10.1021/acs.jproteome.8b00860
32. Kingsley RA, Keestra AM, de Zoete MR, Baumler AJ. The ShdA adhesin binds to the cationic cradle of the fibronectin 13FnIII repeat module: evidence for molecular mimicry of heparin binding. Mol Microbiol. (2004) 52(2):345–55. doi: 10.1111/j.1365-2958.2004.03995.x
33. Bahrami S, Andishmand H, Pilevar Z, Hashempour-Baltork F, Torbati M, Dadgarnejad M, et al. Innovative perspectives on bacteriocins: advances in classification, synthesis, mode of action, and food industry applications. J Appl Microbiol. (2024) 135(11). doi: 10.1093/jambio/lxae274
34. Stevens KA, Sheldon BW, Klapes NA, Klaenhammer TR. Nisin treatment for inactivation of Salmonella species and other gram-negative bacteria. Appl Environ Microbiol. (1991) 57(12):3613–5. doi: 10.1128/aem.57.12.3613-3615.1991
35. Dirix G, Monsieurs P, Dombrecht B, Daniels R, Marchal K, Vanderleyden J, et al. Peptide signal molecules and bacteriocins in gram-negative bacteria: a genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides. (2004) 25(9):1425–40. doi: 10.1016/j.peptides.2003.10.028
36. Pestova EV, Havarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. (1996) 21(4):853–62. doi: 10.1046/j.1365-2958.1996.501417.x
Keywords: Treponema denticola, periodontitis, bacteriocin, clustered lipoprotein, bacteriocin ABC transporter
Citation: Nukaga T, Kokubu E, Okamoto-Shibayama K, Kikuchi Y, Furusawa M, Muramatsu T and Ishihara K (2025) Characterization of clustered bacteriocin-type signal domain protein genes in Treponema denticola. Front. Dent. Med 6:1543535. doi: 10.3389/fdmed.2025.1543535
Received: 11 December 2024; Accepted: 28 January 2025;
Published: 19 February 2025.
Edited by:
Carlos M. Ardila, University of Antioquia, ColombiaReviewed by:
Qiao Fang, University of Illinois at Chicago, United StatesCopyright: © 2025 Nukaga, Kokubu, Okamoto-Shibayama, Kikuchi, Furusawa, Muramatsu and Ishihara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuyuki Ishihara, aXNoaWhhcmFAdGRjLmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.