
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Dent. Med. , 25 May 2022
Sec. Pediatric Dentistry
Volume 3 - 2022 | https://doi.org/10.3389/fdmed.2022.871185
This article is part of the Research Topic The Development of the Oral Microbiome in Children View all 4 articles
Early childhood caries (ECC) is a common chronic infectious disease of childhood with a complex etiology and many contributing risk factors. Its prevalence is greater in certain racial and ethnic minority groups and populations with low socioeconomic status. Among the species of bacteria that contribute to the progression of dental caries, Streptococcus mutans (SM) has long been considered a primary etiological agent of the disease. We report here on the genotypic diversity, commonality, and fidelity of mother-child transmission of S. mutans in mother-child dyads in two high-risk populations.
Methods: Thirty-eight mother-child dyads from a Southeast Iowa population and 40 dyads from a Northern Plains American Indian Tribe were selected for a comparison of S. mutans genotype profiles within and between populations. S. mutans was isolated from whole mouth plaque samples collected from each subject. DNA was extracted and AP-PCR using OPA2 primer was performed. Amplified DNA was electrophoresed and images of the resulting patterns were compared via GelCompar®IIv6.5 software.
Results: Thirty unique S. mutans genotypes were identified from the 1,638 S. mutans isolates analyzed. Fifteen genotypes (50%) were seen in both cohorts, while 11 were unique to the American Indian (AI) cohort and 4 were unique to the Southeast Iowa (SEI) population. Within the AI cohort, 61.5% (16/26) of the S. mutans genotypes identified were seen in ≥2 individuals and 14 of the 26 (53.8%) were seen in ≥2 dyads. In the SEI cohort, 78.9% (15/19) of the S. mutans genotypes identified were seen in ≥2 individuals and 13 of the 19 (68.4%) were present in ≥2 dyads. Fifty-seven percent of AI children and 23% of SEI children displayed fidelity of mother-child transmission of ≥1 S. mutans genotype.
Conclusion: In comparing the populations, we observed large variation in genotypic diversity and fidelity of mother-child transmission, while the amount of commonality seen in both cohorts was similarly high in both groups. This study furthers our understanding of the genotypic diversity of S. mutans in both of these populations and provides a basis for further comparison to other populations at greater risk for developing ECC.
Dental caries is the most common chronic disease of childhood (1). Tooth decay present in children under the age of 6 years old is designated early childhood caries (ECC), or in its more acute form, severe early childhood caries (S-ECC). Children impacted by ECC/S-ECC are at greater risk for cavitation of permanent dentition and other health issues, such as nutritional deficiencies and below average growth (2–6). While ECC is present in all populations, it is a substantial public health challenge in certain racial and ethnic minority groups. Hispanic children have almost twice the amount of untreated decay as Caucasian children (20 and 11% respectively) (7). The highest rate of untreated decay, by far, is found in American Indian and Alaska Native (AI/AN) children at 43%, almost four times the amount reported in Caucasian children (7). There has been a national trend indicating a decrease in untreated decay in children over the past decade. While this trend has also been observed in minority populations, the disproportionate burden of disease in these populations remains (8). Although a full understanding of the underlying causes of the increased rates of ECC/S-ECC in minority and indigenous populations has not been reached, it has been posited that many risk factors contribute to the progression and severity of the disease. These risks include behavioral choices (e.g., sugar intake and oral health hygiene routine), oral microflora, sociodemographic characteristics (e.g., household income, parental education and crowded living conditions), limited access to care, environmental (e.g., fluoridated/non-fluoridated water source) and host factors (5, 7–10).
Mutans streptococci (MS) have been the focus of much caries research. Numerous studies support the belief that this group of bacteria play a pivotal role in the development of dental caries and that MS levels may be a potential indicator of ECC risk (11–17). Species within the mutans streptococci group, specifically Streptococcus mutans, have long been considered primary etiological agents of dental caries with time of colonization in children associated with severity of disease (18–21). Many studies have investigated genotypic diversity of S. mutans within various high-risk populations in an effort to understand the oral health disparities present in low-SES (socio-economic status) and racial/ethnic minority groups (22–25). While much work has been done looking at S. mutans genotypes identified in individual populations, there is a lack of comparisons between populations displaying high rates of ECC/S-ECC.
In our laboratory, we have investigated S. mutans genotypic diversity via arbitrarily primed PCR (AP-PCR) in mother-child dyads within two distinct high-risk populations – one located in Southeast Iowa and the second a Northern Plains American Indian Tribe. Studies have been conducted and reported on concerning initial colonization of S. mutans in American Indian children, including some analyses of genotypic diversity and transmission seen during initial acquisition (birth through 16 months of age) (26). Previous investigations have also examined the genotypic diversity seen in 2- to 5-year old children in this same Southeast Iowa population (mean age = 3.6 years) and compared it to what is seen in 3-year-old children in the American Indian cohort (27). Our focus to date has been on children. Therefore, we have a paucity of data looking at genotypic diversity of MS within and across families. A more comprehensive analysis of the overall diversity, commonality, and fidelity of transmission of S. mutans genotypes found in both cohorts is needed. Herein, we compare all S. mutans genotypes identified in a subset of mother-child dyads from both populations.
Descriptions of populations and details of recruitment, consent, sample collection, sample processing, S. mutans isolation and identification, DNA extraction, AP-PCR and dendrogram analysis of S. mutans isolates have all been described in previous laboratory publications (26–29). Brief descriptions of study procedures are presented here. More detailed descriptions can be found in the previously published manuscripts.
Details regarding this study population recruitment, sample collection, sample processing and genotyping have been published previously (26, 28, 29).
Multiple methods were utilized to recruit Northern Plains American Indian women who were pregnant or had recently given birth (n = 239) for a longitudinal birth cohort study. The proposed study was described to pregnant women during prenatal visits with Indian Health Service (IHS). It was advertised in local papers and on local radio stations. Recruitment was also done by local dental clinic staff. The onsite research team was directed by a senior dental hygienist in the tribe and all team members were American Indian.
Of the 239 dyads recruited for the AI study, a subset of 80 were randomly selected for microbiological analyses. Due to the fact that the AI study was longitudinal, S. mutans isolates (n = 1,401) from multiple time points were analyzed for all mother-child dyads. Selection of mother-child dyads for this study was partially a convenience sampling that included the subjects from the previously completed comparison of SEI and AI children. We also took into consideration what dyads had all available samples processed for genotyping. This resulted in the selection of 40 dyads (50% of the subset; 17% of all dyads recruited) for comparison in this study. For the AI cohort, isolates from multiple time points throughout the study were analyzed. For AI children with S. mutans detected within the study period (n = 35), the mother's S. mutans isolates were genotyped at the baseline visit (1 month of age ±30 days), the visit when mutans streptococci presence was established in the child (child age varied), and the final visit (36 months of age ±30 days). S. mutans was not detected in 3 of the mothers.
Samples were collected from each mother and child at 8 time points from birth to 36 months of age – baseline, 4, 8, 12, 16, 22, 29, and 36 months (±30 days). Whole mouth plaque samples were collected by swabbing all tooth surfaces with a sterile cotton swab and placing swabs into tubes containing TSB-YE (Tryptic Soy Broth supplemented with 0.5% Yeast Extract) (Difco, Sparks, MD, USA) with 10% glycerol. Prior to tooth eruption in children, the oral mucosa and tongue were swabbed. Samples were refrigerated until shipment via FedEX overnight in temperature controlled Saf-T-Temp™ packaging (Saf-T-Pak, Hanover, MD, USA) to the microbiology labs in the Iowa Institute for Oral health Research at the College of Dentistry, University of Iowa.
Swab samples were vortexed (3 min), followed by sonication (1 min) before sample dilution and spiral plating. Samples were plated on Mitis-Salivarius-Kanamycin-Bacitracin agar (MSKB) (Difco, Sparks, MD, USA) using an Autoplate® Spiral Plating System (Advanced Instruments, Inc., Norwood, MA, USA) for determination of S. mutans counts. Following incubation (37°C, 5%CO2, 72–96 h), ten colonies displaying S. mutans colony morphology were selected from the MSKB plate. If fewer than 10 colonies were present, all available colonies were isolated. Identification was confirmed utilizing a previously described protocol that combines both morphological identification and PCR targeting the gtfB gene (28). Isolates were frozen in TSB-YE (10% glycerol) and stored at -80°C.
At the onset of the American Indian study, DNA was extracted using the Epicenter® MasterPure™ Gram Positive DNA Purification Kit (Epicenter, Madison, WI, USA) modified as described in a previous publication (26). However, the volume of samples being processed for isolation and identification of mutans streptococci (MS) necessitated the development of a more rapid, but equally effective, protocol for DNA extraction and S. mutans identification, as detailed in a previous manuscript (28). Quality of extracted DNA was verified using a Nanodrop® spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Details regarding this study population recruitment, sample collection, sample processing and genotyping have been published previously (27).
The cohort for the SEI population was recruited through two locations: the University of Iowa Muscatine Pediatric Dental Clinic and Muscatine Head Start. Children between 2 and 5 years of age (mean = 3.6 years) were recruited (n = 190). Nearly 45% of children recruited were Hispanic. The majority of the remaining children were Caucasian. Recruitment was completed in person by the study coordinator.
As with the AI population, selection of mother-child dyads for this study was a convenience sampling based on previous comparisons of the children. Thirty-eight dyads (20% of all dyads recruited) were selected for comparison here. The SEI study was a cross-sectional study, therefore S. mutans isolates (n = 237) were analyzed from one sample per subject. In addition to the child isolates previously analyzed, all S. mutans isolates from the mothers in the SEI cohort were analyzed. Eleven mothers in the SEI dyads did not have detectable levels of S. mutans at the time of sampling.
Whole mouth plaque samples were collected on swabs that were placed into tubes containing reduced transport media (composition per liter: 0.045% KH2PO4, 0.045% K2HPO4, 0.09% NaCl, 0.09% (NH4)2SO4, 0.018% MgSO4, 0.038% EDTA, 0.04% Na3CO3, 0.02% dithiothreitol). Samples were placed on ice and transported to the microbiology labs.
Swab samples were processed and S. mutans isolated using the same method as in the American Indian study. Identification of isolates as S. mutans was confirmed by fermentation profile (mannitol, raffinose, salicin, and sorbitol) and arginine decarboxylase activity.
For the SEI dyads, the modified protocol for the Epicenter® MasterPure™ Gram Positive DNA Purification Kit (Epicenter, Madison, WI, USA) (26) was used for DNA extraction. DNA quality was verified by Nanodrop® spectrophotometry.
Genotypic diversity was examined in the same way for both populations. Arbitrarily Primed PCR (AP-PCR) using the OPA-2 primer was done on all S. mutans isolates. The positive control for both studies was S. mutans ATCC 25175. AP-PCR was set up and amplified products electrophoresed and imaged as previously described (26). Curve based cluster analysis using the Pearson correlation and Unweighted Pair Group Method using Arithmetic Averages (UPGMA) was used to generate dendrograms to assess genotypic diversity and strain relatedness (GelCompar®IIv6.5, Applied Maths, Austin, TX, USA).
Diversity of S. mutans genotypes was evaluated by determining the number of unique S. mutans genotypes (GT) identified in individual subjects and within mother-child dyads (Figure 1). A total of 1,638 SM (AI = 1,401; SEI = 237) isolates were analyzed in this study. The number of S. mutans genotypes seen in individuals in the SEI population ranged from 0 to 2. When looking at mothers and children separately, the range was 0–2 in mothers and 1–2 in children. In the AI population, the range of S. mutans genotypes per individual was 0–6 (mothers: 0–5, children: 0–6). When looked at as mother-child dyads, the range of S. mutans genotypes per mother-child pair was 1–3 in the SEI population and 1–8 in the AI population.

Figure 1. Diversity of S. mutans (SM) genotypes in subjects with S. mutans detected. Aggregate data, divided by population, showing the number of individual subjects and dyads displaying a specific number of unique S. mutans genotypes. Data are (A) children only, (B) mothers only, and (C) mother-child dyads (total number of genotypes found within the pair).
Fidelity of transmission of S. mutans genotypes from mother to child was also evaluated. In the 38 mother-child pairs analyzed from the SEI cohort, 23% (8/35) of children shared a genotype with his/her mother and 77% (27/35) did not. Transmission status could not be determined (no S. mutans genotype profile available for mother) for the remaining 3 children in this cohort. The percent of children that shared at least 1 genotyped with his/her mother in the AI cohort was 57% (23/40). The remaining 43% (17/40) did not display fidelity of transmission of any S. mutans genotypes.
Thirteen of the S. mutans genotypes identified showed fidelity of transmission in at least 1 mother-child dyad (Table 1). Additional genotypes were found in both mothers and children, but not within both subjects of a mother-child dyad. Genotypes 1 and 8 were the genotypes most commonly acquired by children from their mothers.
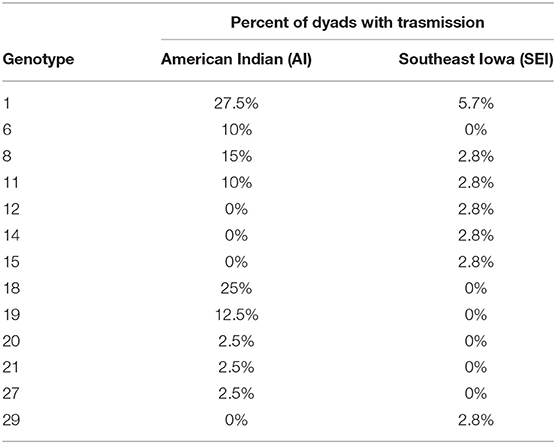
Table 1. Genotypes displaying fidelity of transmission from mother to child in ≥1 mother-child dyad.
A total of 30 unique S. mutans genotypes were identified within the study population (Figure 2). Of these 30 genotypes, 26 were seen in the AI population and 19 in the SEI cohort. Analysis of commonality, the sharing of genotypes by at least 2 people (30), within and between the two populations, showed that half (15/30) of the S. mutans genotypes identified were present in both populations. Eleven S. mutans genotypes were unique to the AI cohort and 4 were unique to the SEI cohort. Within the AI cohort, 61.5% (16/26) of the S. mutans genotypes identified were seen in ≥2 individuals and 14 of the 26 (53.8%) were seen in ≥2 dyads. In the SEI cohort, 78.9% (15/19) of the S. mutans genotypes identified were seen in ≥2 individuals and 13 of the 19 (68.4%) were present in ≥2 dyads. The three most common S. mutans genotypes in this study (GT1, GT8, and GT11) were seen in both populations and were identified in 35.82, 23.13, and 16.42% of all individuals, respectively (Table 2).
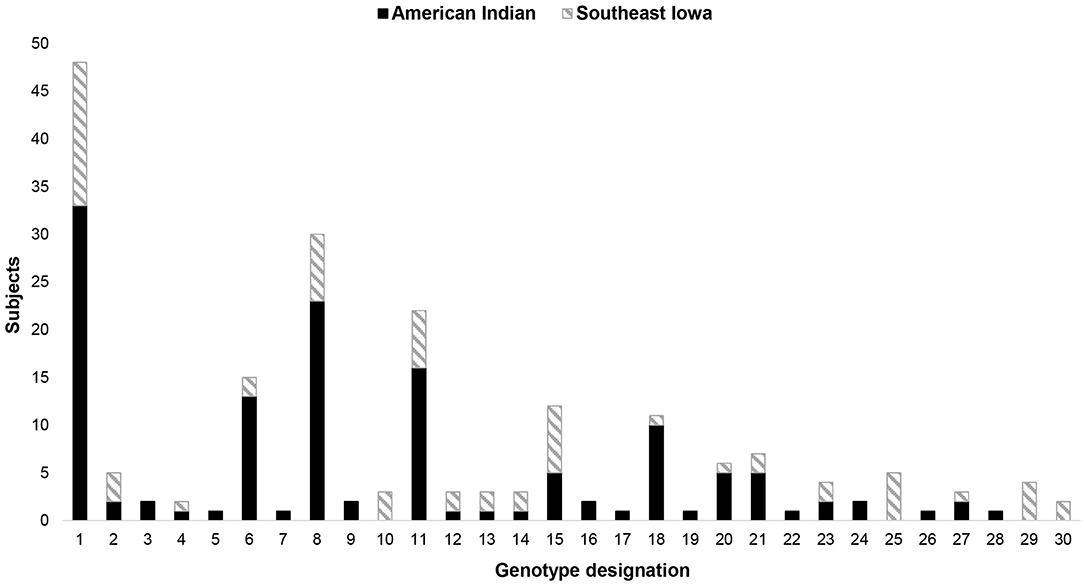
Figure 2. Dendrogam analysis of the S. mutans isolated from both populations. Thirty unique genotypes identified – 15 found in both populations, 11 found only in the AI cohort, and 4 found only in the SEI cohort.
While each cohort within this study can be classified as a Midwest population, they are both geographically and culturally distinct from each other. The populations were selected for study because, demographically, they were at risk for higher incidence of early childhood caries and, especially in the AI cohort, severe early childhood caries. The children from both populations displayed a higher incidence of caries than that of children in the same age group within the general U.S. population (9).
Genotypic diversity was the most challenging comparison to make in this study. The AI mothers and children were part of a longitudinal birth cohort study, so there were multiple samples to isolate S. mutans from for most subjects. The SEI study was cross-sectional, so S. mutans was only isolated from samples collected at one time point. This resulted in significantly more S. mutans isolates to analyze from the AI population. There were a total of 1,638 S. mutans isolates analyzed in this study, of which, 1,401 were from AI subjects and 237 were from SEI subjects.
Many studies have reported on genotypic diversity in both longitudinal and cross-sectional studies of various populations. Previous investigations have shown in cross-sectional studies, a range of 1–5 S. mutans genotypes per subject, with some reporting as few as 1–2 genotypes per subject (23, 24, 31–33). In a 7-year longitudinal birth cohort study, Lindquist et al. identified 2–7 S. mutans genotypes in children and 1–5 in mothers over the course of the study (34). While some longitudinal studies report slightly higher S. mutans genotypic diversity than cross-sectional studies, most subjects within the longitudinal studies displayed ≤ 3 unique genotypes per subject at a single time point (30, 34, 35). Similarly, the mothers and children in the AI population reported on here displayed a greater amount of genotypic diversity than subjects in the SEI population, but none displayed more than 3 unique genotypes at any single time point. The majority of studies discussed here utilized AP-PCR for the genotyping method, but rep-PCR (repetitive element sequence-based PCR) and RFLP (restriction fragment length polymorphism) analysis were also used. Variation in the method of genotyping could have an impact on results. Ages of participants also varied in these studies. Of the cross-sectional studies, two worked with pre-school aged children (3–5 years), two with kindergarten aged children (5–6 years), and one worked with adults (18–28 years). For the longitudinal studies, two worked with birth cohorts and one with children who were kindergarten aged (5–6 years) at the start of the study. While we recognize that it is challenging to compare the results from these two studies, overall, our data presented here were consistent to previously-published cross-sectional and longitudinal investigations and represent a first step toward comparing S. mutans diversity in multiple populations.
It is possible that we are underestimating genotypic diversity in the SEI population and additional genotypes may have been identified with sampling at additional time points. However, the high rate of commonality observed in this population and the fact that the results were consistent with other studies that have found lower rates of genotypic diversity within some populations (24, 32, 33), suggests that our data were a good assessment of overall genotypic diversity of S. mutans in this population.
Reported rates of vertical S. mutans transmission vary greatly between studied populations. The reasons for the differences in vertical transmission are not known. Rates as low as 21% and as high as 81% have been reported (35–38). We found that the transmission rates in both of our populations varied considerably. Fifty-seven percent (23/40) of AI mother-child dyads displayed fidelity of transmission of at least one S. mutans genotype. In the SEI dyads, only 24% (10/42) of children shared ≥1 genotype with their mother. The percentage of dyads in the AI and SEI cohorts that did not display fidelity of transmission (42% and 69% respectively) included: 1) dyads where both subjects had S. mutans, but did not share a genotype and 2) dyads where either mother or child exhibited no detectable S. mutans. In the AI cohort, there were five dyads where the child had no detectable S. mutans and three dyads where the mother had no detectable S. mutans. In the SEI cohort, there were eleven dyads where there was no detectable S. mutans in the mother's sample. There are many variables that may affect rates of vertical and horizontal transmission, including, but not limited to, dietary habits (e.g., prolonged bottle feeding, sugar intake, sharing food), behavioral and oral health practices (e.g., poor oral hygiene routines, lack of regular dental care), level of S. mutans colonization in the oral cavities of caregivers and crowded living conditions (39, 40).
Many studies in fact support the idea that, while maternal transmission is an important contributing factor to the establishment of S. mutans within a child's oral microflora, it may not be the primary source of S. mutans colonization in children at risk for ECC and S-ECC. In one Alabama population, 41% of mother-child pairs showed some level of maternal transmission; however, acquisition of S. mutans from non-maternal sources was observed in 74% of the pairs (41). Zhan et al. reported that 90% of children in their study harbored genotypes of non-maternal origin and the overall rate of maternal transmission was only 21% (36). We report here similar findings. Low percentages of children in either population exhibited SM genotypes that all matched those in their mothers (AI: 20%, SEI: 24%). A majority of children had ≥1 SM genotype that was different from those isolated from their mothers (AI: 77.1%, SEI: 69%), suggesting acquisition from other sources. No children in the SEI cohort had both maternal and non-maternal sources of S. mutans colonization, while 69% of AI children displayed both vertical and apparent horizontal transmission of S. mutans. Determining the source of SM colonization in young children is complex as there appear to be multiple variables that influence directly or indirectly transmission and acquisition events. Assessing the impact of a complex set of variables that may interact in ways that are unknown in transmission and acquisition of MS will require additional investigations.
The high rate of commonality (30) of S. mutans genotypes observed between families in both of these cohorts (53.8% AI, 68.4% SEI) provided additional support to the idea that both vertical and horizontal transmission events played a key role in establishing the genotypic profile of S. mutans in individuals within both of these populations. As with transmission, there has been a high degree of variance seen across populations in the amount of sharing of SM genotypes observed between individuals. Many studies that include data on sharing of genotypes between unrelated study subjects report little to no horizontal transmission observed (33–35, 42). In contrast, Cheon et al. found that 92.5% of the children in their study shared at least 1 genotype with another child and that the most prevalent genotype was identified in 36% of the children (30). In our AI cohort, 100% of the individuals that had S. mutans colonization (n=72) shared ≥1 genotype with at least one individual from another mother-child dyad within that cohort, while 93.1% (67/72) shared ≥1 genotype with ≥1 individual in the SEI population. There were 28 AI subjects (38.9%) that harbored ≥1 genotype that was unique to that population. In our SEI cohort, 90.9% (60/66) of subjects with S. mutans colonization (n=66) shared ≥1 genotype with at least one individual from another mother-child dyad within that cohort, while 84.8% (56/66) shared ≥1 genotype with ≥1 individual in the AI population. There were 14 individuals from the SEI cohort (21.2%) that harbored a genotype unique to that population.
We acknowledge that there are limitations and challenges to comparing these two populations due to differences in study design. However, as stated in the discussion, we do believe that the data obtained is a fair representation of the genotypic diversity of each population. Since the focus of this manuscript is a comparison of findings within populations, not side-by-side comparisons of individual mother child dyads, we feel it reasonable to make the following observations when looking at the data from both populations. Large variation between these high caries-risk populations was observed in genotypic diversity and fidelity of transmission from mother to child, whereas the amount of commonality and source of S. mutans colonization (maternal/non-maternal) numbers were more similar. The importance of these varied SM genotype profiles and acquisition patterns – vertical vs. horizontal – on colonization in these caries high-risk children is unknown. It is conceivable that specific SM genotypes are pivotal in caries development, or specific profiles of SM genotypes create conditions in the oral cavities of these children that, combined with other variables such as diet, behavior, crowded living conditions, etc. could set the stage for the development of early childhood and severe early childhood caries. This and future studies comparing multiple populations at increased risk of early childhood caries will contribute to gaining a better understanding of how these variables interact and will provide valuable information for development of new preventive and therapeutic treatment approaches to improve this area of health disparity.
The datasets presented in this article are not readily available and would need approval from the American Indian Tribe. Requests to access the datasets should be directed to ZGF2aWQtZHJha2VAdWlvd2EuZWR1.
Consent was obtained for all subjects who participated in both studies. For both populations, approval was obtained from the University of Iowa Institutional Review Board (IRB). Additionally, for the American Indian cohort, approval was required and obtained from both the Tribal Research Review Board and Aberdeen Area IRB.
AV worked on processing samples, AP-PCR and dendrogram analyses of S. mutans isolates, data collection, and manuscript writing. DL worked on processing samples, data collection, and AP-PCR. JW was the Primary Investigator for the Southeast Iowa study and was involved in all aspects of that study. DD was the Primary Investigator for the American Indian study and was involved in overseeing all aspects of the study, including manuscript revision. All authors contributed to the article and approved the submitted version.
This study was supported by NIH grant R01-DE017736 and Roy J. Carver Charitable Trust grant 06-2336.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Theron Layer for his work on this study.
1. General S. Oral health: the silent epidemic. Public Health Rep. (2010) 125:158–9. doi: 10.1177/003335491012500202
2. Peretz B, Ram D, Azo E, Efrat Y. Preschool caries as an indicator of future caries: a longitudinal study. Pediatr Dent. (2003) 25:114–8.
3. Acs G, Lodolini G, Kaminsky S, Cisneros GJ. Effect of nursing caries on body weight in a pediatric population. Pediatr Dent. (1992) 14:302–5.
4. Ayhan H, Suskan E, Yildirim S. The effect of nursing or rampant caries on height, body weight and head circumference. J Clin Pediatr Dent. (1996) 20:209–12.
5. AAPD AAoPD. Policy on Early Childhood Caries (ECC): classifications, consequences, and preventive strategies. Pediatr Dent. (2017) 39:59–61.
6. Zaror C, Matamala-Santander A, Ferrer M, Rivera-Mendoza F, Espinoza-Espinoza G, Martinez-Zapata MJ. Impact of early childhood caries on oral health-related quality of life: a systematic review and meta-analysis. Int J Dent Hyg. (2022) 20:120–35. doi: 10.1111/idh.12494
7. Phipps K, Ricks, TL,. The Oral Health of American Indian Alaska Native Children Aged 1-5 Years: Results of the 2014 IHS Oral Health Survey (2015). Available online at: https://www.ihs.gov/doh/documents/ihs_data_brief_1-5_year-old.pdf
8. Phipps, K, Ricks, TL, Mork, N, Lozon, T. The Oral Health of American Indian and Alaska Native Children Aged 1-5 Years: Results of the 2018-19 IHS Oral Health Survey. Available online at: https://www.ihs.gov/doh/documents/surveillance/2018-19%20Data%20Brief%20of%201-5%20Year-Old%20AI-AN%20Preschool%20Children.pdf: U.S. Department of Health and Human Services Indian Health Service Division of Oral Health
9. Edelstein BL, Chinn CH. Update on disparities in oral health and access to dental care for America's children. Acad Pediatr. (2009) 9:415–9. doi: 10.1016/j.acap.2009.09.010
10. Warren JJ, Blanchette D, Dawson DV, Marshall TA, Phipps KR, Starr D, et al. Factors associated with dental caries in a group of American Indian children at age 36 months. Commun Dent Oral Epidemiol. (2016) 44:154–61. doi: 10.1111/cdoe.12200
11. Tanzer JM, Livingston J, Thompson AM. The microbiology of primary dental caries in humans. J Dent Educ. (2001) 65:1028–37. doi: 10.1002/j.0022-0337.2001.65.10.tb03446.x
12. Karpinski TMS, Szkaradkiewicz A. Microbiology of dental caries. J Biol Earth Sci. (2013) 3:M21–M4.
13. Parisotto TM, Steiner-Oliveira C, Silva CM, Rodrigues LK, Nobre-dos-Santos M. Early childhood caries and mutans streptococci: a systematic review. Oral Health Prev Dent. (2010) 8:59–70.
14. Edelstein BL, Ureles SD, Smaldone A. Very high salivary Streptococcus mutans predicts caries progression in young children. Pediatr Dent. (2016) 38:325–30.
15. Villhauer A, Lynch D, Postler T, Dawson D, Drake D. Mutans streptococci and lactobacilli: colonization patterns and genotypic characterization of cariogenic bacterial species in American Indian Children. Front Dent Med. (2021) 2. doi: 10.3389/fdmed.2021.740900
16. Babaeekhou L, Ghane M, Ezatzade F, Eftekhari Toroghi S. Streptococcus mutans and Streptococcus sobrinus distribution in the saliva and plaque of Iranian population: Higher prevalence of S. mutans serotypes f and k. Int J Dent Hyg. (2021) 19:193–200. doi: 10.1111/idh.12485
17. Kemthong Mitrakul BA. Piamporn Thammachat. Quantitative analysis of Streptococcus mutans, streptococcus sobrinus and streptococcus sanguinis and their association with early childhood caries. J Clin Diagn Res. (2020) 14:ZC23–ZC8. doi: 10.7860/JCDR/2020/43086.13513
18. Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. (1986) 50:353–80. doi: 10.1128/mr.50.4.353-380.1986
19. Banas JA. Delaying Streptococcus mutans colonization in children leads to reduced caries experience. J Evid Based Dent Pract. (2013) 13:67–9. doi: 10.1016/j.jebdp.2013.04.007
20. Laitala M, Alanen P, Isokangas P, Soderling E, Pienihakkinen K. A cohort study on the association of early mutans streptococci colonisation and dental decay. Caries Res. (2012) 46:228–33. doi: 10.1159/000337303
21. Johansson I, Witkowska E, Kaveh B, Lif Holgerson P, Tanner AC. The microbiome in populations with a low and high prevalence of caries. J Dent Res. (2016) 95:80–6. doi: 10.1177/0022034515609554
22. Momeni SS, Whiddon J, Cheon K, Ghazal T, Moser SA, Childers NK. Genetic diversity and evidence for transmission of Streptococcus mutans by DiversiLab rep-PCR. J Microbiol Methods. (2016) 128:108–17. doi: 10.1016/j.mimet.2016.07.010
23. Pieralisi FJ, Rodrigues MR, Segura VG, Maciel SM, Ferreira FB, Garcia JE, et al. Genotypic diversity of Streptococcus mutans in caries-free and caries-active preschool children. Int J Dent. (2010) 2010:824976. doi: 10.1155/2010/824976
24. Cheon K, Moser SA, Whiddon J, Osgood RC, Momeni S, Ruby JD, et al. Genetic diversity of plaque mutans streptococci with rep-PCR. J Dent Res. (2011) 90:331–5. doi: 10.1177/0022034510386375
25. Crall JJ, Vujicic M. Children's oral health: progress, policy development, and priorities for continued improvement. Health Aff (Millwood). (2020) 39:1762–9. doi: 10.1377/hlthaff.2020.00799
26. Lynch DJ, Villhauer AL, Warren JJ, Marshall TA, Dawson DV, Blanchette DR, et al. Genotypic characterization of initial acquisition of Streptococcus mutans in American Indian children. J Oral Microbiol. (2015) 7:27182. doi: 10.3402/jom.v7.27182
27. Villhauer AL, Lynch DJ, Warren JJ, Dawson DV, Blanchette DR, Drake DR. Genotypic characterization and comparison of Streptococcus mutans in American Indian and Southeast Iowa children. Clin Exp Dent Res. (2017) 3:235–43. doi: 10.1002/cre2.94
28. Villhauer AL, Lynch DJ, Drake DR. Improved method for rapid and accurate isolation and identification of Streptococcus mutans and Streptococcus sobrinus from human plaque samples. J Microbiol Methods. (2017) 139:205–9. doi: 10.1016/j.mimet.2017.06.009
29. Drake D, Dawson D, Kramer K, Schumacher A, Warren J, Marshall T, et al. Experiences with the Streptococcus mutans in Lakota Sioux (SMILeS) Study: Risk factors for Caries in American Indian Children 0-3 Years. J Health Dispar Res Pract. (2015) 8:123–32.
30. Cheon K, Moser SA, Wiener HW, Whiddon J, Momeni SS, Ruby JD, et al. Characteristics of Streptococcus mutans genotypes and dental caries in children. Eur J Oral Sci. (2013) 121:148–55. doi: 10.1111/eos.12044
31. Zhou Q, Qin X, Qin M, Ge L. Genotypic diversity of Streptococcus mutans and Streptococcus sobrinus in 3-4-year-old children with severe caries or without caries. Int J Paediatr Dent. (2011) 21:422–31. doi: 10.1111/j.1365-263X.2011.01145.x
32. Tabchoury CP, Sousa MC, Arthur RA, Mattos-Graner RO, Del Bel Cury AA, Cury JA. Evaluation of genotypic diversity of Streptococcus mutans using distinct arbitrary primers. J Appl Oral Sci. (2008) 16:403–7. doi: 10.1590/S1678-77572008000600009
33. Domejean S, Zhan L, DenBesten PK, Stamper J, Boyce WT, Featherstone JD. Horizontal transmission of mutans streptococci in children. J Dent Res. (2010) 89:51–5. doi: 10.1177/0022034509353400
34. Lindquist B, Emilson CG. Colonization of Streptococcus mutans and Streptococcus sobrinus genotypes and caries development in children to mothers harboring both species. Caries Res. (2004) 38:95–103. doi: 10.1159/000075932
35. Klein MI, Florio FM, Pereira AC, Hofling JF, Goncalves RB. Longitudinal study of transmission, diversity, and stability of Streptococcus mutans and Streptococcus sobrinus genotypes in Brazilian nursery children. J Clin Microbiol. (2004) 42:4620–6. doi: 10.1128/JCM.42.10.4620-4626.2004
36. Zhan L, Tan S, Den Besten P, Featherstone JD, Hoover CI. Factors related to maternal transmission of mutans streptococci in high-risk children-pilot study. Pediatr Dent. (2012) 34:e86–91.
37. Teanpaisan R, Chaethong W, Piwat S, Thitasomakul S. Vertical transmission of mutans streptococci and lactobacillus in Thai families. Pediatr Dent. (2012) 34:e24–9.
38. Mitchell SC, Ruby JD, Moser S, Momeni S, Smith A, Osgood R, et al. Maternal transmission of mutans Streptococci in severe-early childhood caries. Pediatr Dent. (2009) 31:193–201.
39. Assiry AA. Transmission and colonization of Streptococcus mutans in children. Rev Med Microbiol. (2018) 29:116–9. doi: 10.1097/MRM.0000000000000140
40. Kaan AMM, Kahharova D, Zaura E. Acquisition and establishment of the oral microbiota. Periodontol 2000. (2021) 86:123–41. doi: 10.1111/prd.12366
41. Moser SA, Mitchell SC, Ruby JD, Momeni S, Osgood RC, Whiddon J, et al. Repetitive extragenic palindromic PCR for study of Streptococcus mutans diversity and transmission in human populations. J Clin Microbiol. (2010) 48:599–602. doi: 10.1128/JCM.01828-09
Keywords: Streptococcus mutans, early childhood caries, oral health disparities, genotyping, AP-PCR
Citation: Villhauer A, Lynch D, Warren J and Drake D (2022) Assessment of Diversity and Fidelity of Transmission of Streptococcus mutans Genotypes in American Indian and Southeast Iowa Mother-Child Dyads. Front. Dent. Med. 3:871185. doi: 10.3389/fdmed.2022.871185
Received: 07 February 2022; Accepted: 07 April 2022;
Published: 25 May 2022.
Edited by:
Sreekanth Kumar Mallineni, Majmaah University, Saudi ArabiaReviewed by:
Ahmad Faisal Ismail, International Islamic University Malaysia, MalaysiaCopyright © 2022 Villhauer, Lynch, Warren and Drake. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Drake, ZGF2aWQtZHJha2VAdWlvd2EuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.