- 1Department of Oral Microbiology, Faculty of Dentistry, Mahidol University, Bangkok, Thailand
- 2Integrative Computational Bioscience (ICBS) Center, Mahidol University, Nakorn Pathom, Thailand
- 3Department of Biochemistry, Faculty of Science, Mahidol University, Bangkok, Thailand
- 4Department of Microbiology, Faculty of Science, Mahidol University, Bangkok, Thailand
- 5Systems Biology of Diseases (SyBiD) Research Unit, Faculty of Science Mahidol University, Bangkok, Thailand
- 6Department of Microbiology and Research Unit on Oral Microbiology and Immunology, Faculty of Dentistry, Chulalongkorn University, Bangkok, Thailand
- 7Research Laboratory of Biotechnology, Chulabhorn Research Institute, Bangkok, Thailand
A comprehensive understanding of dental pulp cellular compositions and their molecular responses to infection are crucial for the advancement of regenerative dentistry. Here, we presented a pilot study of single-cell transcriptomic profiles of 6,810 pulpal cells isolated from a sound human maxillary third molar and three carious teeth with enamel and deep dental caries. We observed altered immune cell compositions of the dental pulp in deep, but not enamel ones. Differential expression analysis revealed up-regulation of several pro-inflammatory, anti-inflammatory, and mineralization-related genes in the immune and stromal cells of the deep dental caries. Making use of an algorithm for predicting cell-to-cell interactions from single-cell transcriptomic profiles, we showed an increase in cell-cell interactions between B cells, plasma cells and macrophages, and other cell types in deep dental caries, including those between TIMP1 (odontoblasts)—CD63 (myeloid cells), and CCL2 (macrophages)—ACKR1 (endothelial cells). Collectively, our work highlighted the single-cell level gene regulations and intercellular interactions in the dental pulps in health and disease.
Introduction
Dental caries is caused by the invasion of acid-producing oral bacteria into the enamel and dentin (1). When the carious lesions progress toward the dentin-pulp interface, the microbial composition that diffuses through the dentinal tubules could trigger the protective host responses by stromal and immune cells residing inside the pulpal tissue, resulting in the generation of tertiary dentin by odontoblasts to wall off the infection (2). With the removal of infectious tooth structure and proper tooth restoration, tissue homeostasis could be regained (1). However, stronger stimuli as in persistent deep carious lesions could lead to the death of odontoblasts, leading to the recruitment of cells from the pulp stem cell pool to the affected sites. These cells then differentiate into odontoblast-like cells that secrete the amorphous reparative dentin (2). At this stage where the inflammation outbalanced the healing, the disease could manifest as irreversible pulpitis and, in some cases, to pulp necrosis (1).
Endodontic treatment has been shown to achieve a satisfactory alternative to dental extraction as it can preserve the endodontically treated teeth in the oral cavity (3). The major drawback of this therapeutic modality is, however, the loss of tooth vitality. More recently, vital pulp therapy has gained much attention, especially on the treatment of immature permanent teeth (4). With the advancement of regenerative dentistry, biomaterials as well as stem cell therapy, which have been utilized to dampen the pulpal inflammation while preserving pulpal vitality (5). Nonetheless, there are currently limited indications for vital pulp therapy and regenerative endodontics (6).
An intricate balance of inflammation and regeneration is important for the defense against acid-producing bacteria that cause dental caries and the resolution of the inflammation afterwards (7). Therefore, a complete understanding of how the stromal and immune cells in the dental pulp function during physiologic and pathological conditions is needed. Multicolor flow cytometry has been used extensively to characterize the heterogeneous population of mesenchymal stem cells (MSCs) residing in the human dental pulp (8, 9), while the studies on the immune cell populations are much more limited. A previous study utilized flow cytometry analysis to show that immune cells, including neutrophils, T cells, macrophages and natural killer (NK) cells, account for only 1% of the whole pulpal population (10, 11). Meanwhile, B cells were reported to infiltrate in inflamed pulp tissues but not in healthy pulp (12, 13). Nonetheless, flow cytometry analysis is limited by the number of predetermined cell surface markers, making the true heterogeneity that includes rare cell types and functional subgroups remain undiscovered.
Transcriptomic studies have been used effectively to investigate the global changes of gene expression between healthy and inflamed pulpal tissues (14). The study has revealed that genes in the immune and inflammatory response pathways were upregulated in dental caries, while those in biosynthesis pathways were downregulated (14). Yet, the major drawback of “bulk” transcriptomes is the average gene expression levels from different cell types in the tissues, which are dominated by highly abundant cell types masking the information from minor but potentially significant cell types in dental pulp, such as immune cells. Single-cell RNA-sequencing (scRNA-seq) has been proven a powerful tool for the study of complex cell systems by providing comprehensive information on each cell response. A recent study has highlighted the utilization of scRNA-seq to delineate the heterogeneity of residential cells in dental pulp during the development of human teeth (15). Other work has provided an in-depth characterization and comparison of MSCs in the dental pulp and periodontal tissues (16). Single-cell transcriptomics has also allowed the discovery of pericyte subsets that showed the potential for the development to mature odontoblasts (17). However, as these studies focused on the tooth development and stem cell characterization, and only healthy dental pulp tissues were analyzed.
In our study, we employed scRNA-seq to explore the cell populations as well as their transcriptomic profiles during homeostasis and in presence of dental caries. Differential gene expression analysis was performed on each characterized cell type to provide insights into the global transcriptomic changes in healthy and diseased dental pulp, while preserving the cell identities. Lastly, potential interactions between cell types were explored using a computational prediction algorithm based on the transcriptomic profiles at the single-cell resolution.
Materials and Methods
Tooth Collection and Tissue Dissociation
Permanent upper third molars were collected from patients between 20 and 36 years of age receiving dental extraction at the Oral and Maxillofacial Surgery Department, Faculty of Dentistry, Mahidol University, Thailand, under the ethical review exemption from the Mahidol University Faculty of Dentistry and Pharmacy Institutional Review Board (MU-DT/PY-IRB 2019/068.0811). An upper maxillary molar without clinical or radiographic evidence of dental caries was classified according to the International Caries Detection and Assessment System (ICDAS). The tooth with ICAS 0 was classified as “sound.” An upper third maxillary molar with arrested dental caries not extending beyond enamel was classified as “enamel caries” (ICDAS 2). Two upper third maxillary molars from two different donors with dental caries with clinically distinct cavity, involving inner 1/3 of the dentine were classified as “deep dental caries” (ICDAS 6) (see Figure 1A; Supplementary Table 1). Freshly extracted molars were kept in 1X Phosphate buffer saline solution (PBS)containing 1% penicillin/streptomycin (GibcoTM) on ice and immediately transported to the laboratory for tissue dissociation within two hours from the extraction of the first tooth. Dental pulp tissues were dissociated from the teeth mechanically, followed by enzymatic digestion as previously described in (18). Briefly, the dental pulp tissue was washed with PBS before treated with 3 mg/ml collagenase (Stemcell technologies, USA) and 1U dispase (Stemcell technologies, USA) for 1 h at 37°C with a vertexing interval of 15 min. The dissociated cells were washed twice with PBS before single-cell library preparation.
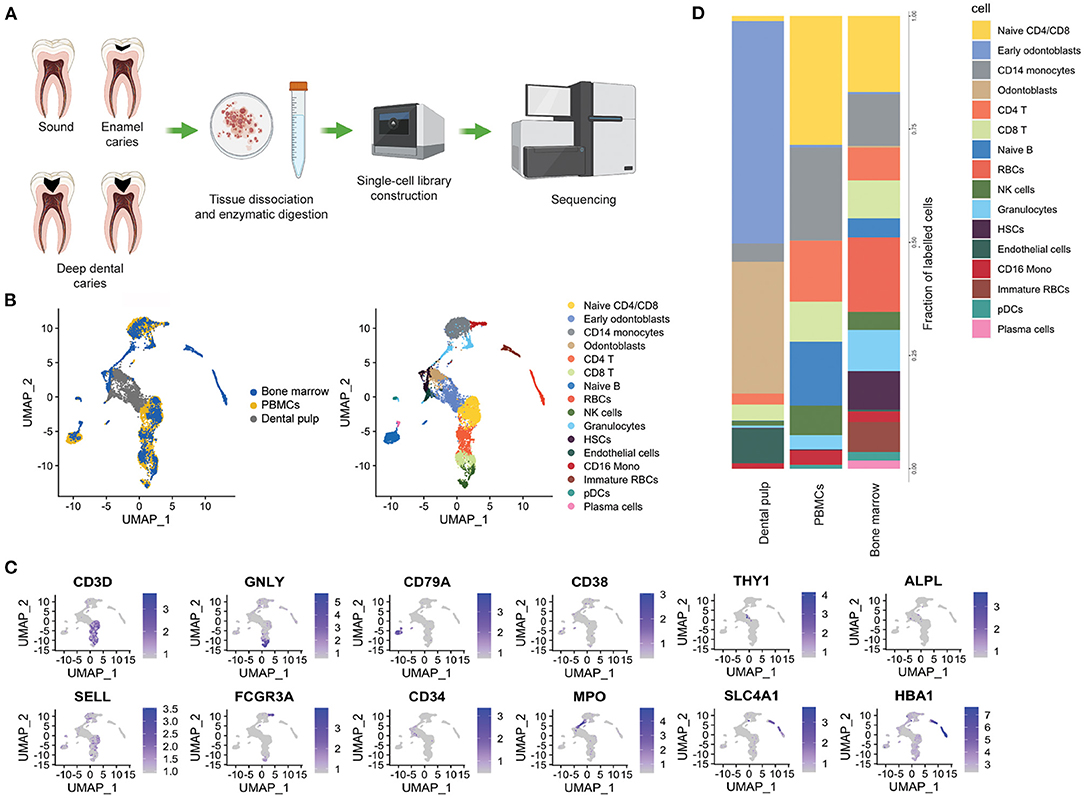
Figure 1. Tissue distribution of stromal and immune cells in the dental pulp. (A) Schematic representation of experimental design. (B) Integrated UMAP from the healthy bone marrow and PBMCs (from publicly available data) and dental pulp samples (sound and enamel caries, from this study). (C) Normalized expression levels of the key maker genes used for cell type identification. (D) Stacked bar plots showing the proportion of cell type within dental pulps, PBMCs and bone marrow.
Single-Cell Library Generation and Sequencing
Single-cell libraries were generated according to the Chromium Single-cell 3' Reagent Kits V2 User guide (CG00052 RevF). Cell suspension was reconstituted in 0.04% BSA PBS to achieve the cell concentration of 500–1,000 cells/ul, before passing through the 40 μM strainer twice. Gel Bead-In Emulsion (GEM) was created using the Chromium Single-cell 3' Reagent Kits V2, following the manufacturer's protocol (10x genomics, USA). GEMs were recovered by Recovery Agent (10x Genomics), followed by cDNA synthesis and library preparation. The single-cell libraries were submitted for sequencing on the Hiseq system (Illumina, USA) at Macrogen Inc., Korea, with the sequencing depth of at least 50,000 reads/cells as recommended by the manufacturer. Each library was subsequently demultiplexed using the i7 indices. Raw reads were mapped against the human reference genome GRCh38 version 2020-A using CellRanger version 5.0.0 (10x Genomics).
Data Quality Control
The gene-cell matrices generated by Cell Ranger were imported into R-studio (19) for further analysis. The cells that have the number of expressed genes lower than 200 genes were discarded and the genes that were detected in fewer than 3 cells were removed. The ambient RNA was estimated and regressed by SoupX version 1.5.0 (20). The cell doublets were then removed by DoubletFinder version 2.0.3 (21), with 3.9% doublet formation rate for 5,000 targeted cells, according to the 10x Genomics user manual. The matrices were further processed by the Seurat package version 3.2.3 (22). Low quality cells defined by the expression of mitochondrial genes exceeding 25% were removed.
Data Integration and Visualization
To further assess the cell population and function of each dental pulp cell library, each gene-cell matrix was normalized using the “LogNormalize” function in the Seurat V3 package (22). Two thousand genes were used as the anchored genes for data integration across samples using the “Integrate” function. The integrated Seurat object underwent PCA analysis with 30 principal components (PCs) selected and visualized using Uniform Manifold Approximation and Projection (UMAP) (23).
Differential Gene Expression
We used the Wilcoxon Rank Sum test under the functions FindMarkers and FindAllMarkers of Seurat V3 (22), to compare between clusters of interest, as well as between different carious stages. The genes that expressed in at least 10% of the cells were included in a cluster. DEseq2 was utilized for average differential gene expression analysis. P-value computed by DEseq2 version 1.26.0 (24) below 0.05 was considered statistically significant.
Cell-Cell Communication
The potential cell-cell communications were predicted by the level of expressions of ligands and receptors in each cell type as identified by NATMI: Ligand-Receptor Interactions (connectomeDB2020) (25). The total edge-count and mean expression levels of ligand and receptor pairs were used for the downstream analysis.
Results
Tissue Distribution of Structural and Immune Cells in the Dental Pulp
To investigate the similarities between the cells found in the dental pulp and the immune and structural cells, we first compared the cell composition of the dental pulp of our study, to that of publicly available healthy peripheral blood mononuclear cells (PBMCs) available from the 10x genomics website: https://www.10xgenomics.com/resources/datasets) and of the bone marrow (26) datasets (Figure 1B). We identified a population of fibroblasts by the expression of THY1, MCAM and that of odontoblasts by the expression of ALPL, OMD, and CCL2, which were exclusively found in the dental pulp (Figures 1B,C). On the other hand, the immune cells such as T cells (CD3D), NK cells (GNLY and FCGR3A) cells and monocytes (CD14) were found in all the three tissues (Figures 1B,D). Very low frequencies of B cells and plasma cells were found in the dental pulps of the sound tooth and the tooth with enamel caries, as compared with in the PBMCs and bone marrow (Figures 1B,D). Interestingly, we were also able to identify hematopoietic stem cells (HSCs) based on the expression of CD34, ENG and VCAM1 in both the dental pulp and bone marrow samples, but not in the PBMCs (Figures 1B–D).
The Cell Populations in Dental Pulp Were Altered in Response to Deep but Not Enamel Caries
We re-integrated the single-cell transcriptomic data from sound tooth, enamel caries and deep dental caries for more in-depth analysis of subpopulations within the dental pulp samples. Cell type annotation was performed by the expression markers as represented in Figure 2B; Supplementary Figure 1. Fourteen cell populations were identified in the sound tooth and enamel caries, while 16 were identified in the deep dental caries (Figure 2A). Of note, in our study we identified fibroblasts by the expression of THY1, and MCAM, which were also considered markers for mesenchymal stem cells (MSCs) in earlier studies (9, 27). We found heterogeneous populations of odontoblasts with varying degrees of gene expression associated with odontogenesis, such as OMD, ALPL and DMP1 (Figure 2A; Supplementary Figure 1) (28). Endothelial cells were identified by the expression of SELE and PECAM1 and hematopoietic stem cells (HSCs) in the adjacent cluster were annotated by the expression of CD34, ENG and VCAM1 (Figures 2A,B; Supplementary Figure 1). Immune cells such as T cells, NK cells, macrophages and the CD16+ monocytes and small number of plasma cells were found in all tooth conditions (Figures 2A,C,D). However, plasma cells were enriched in the deep dental caries, while B cells and the CD103+ dendritic cells (DCs) were only present in this population (Figures 2C,D), suggesting their potential roles in responding to bacterial infection.
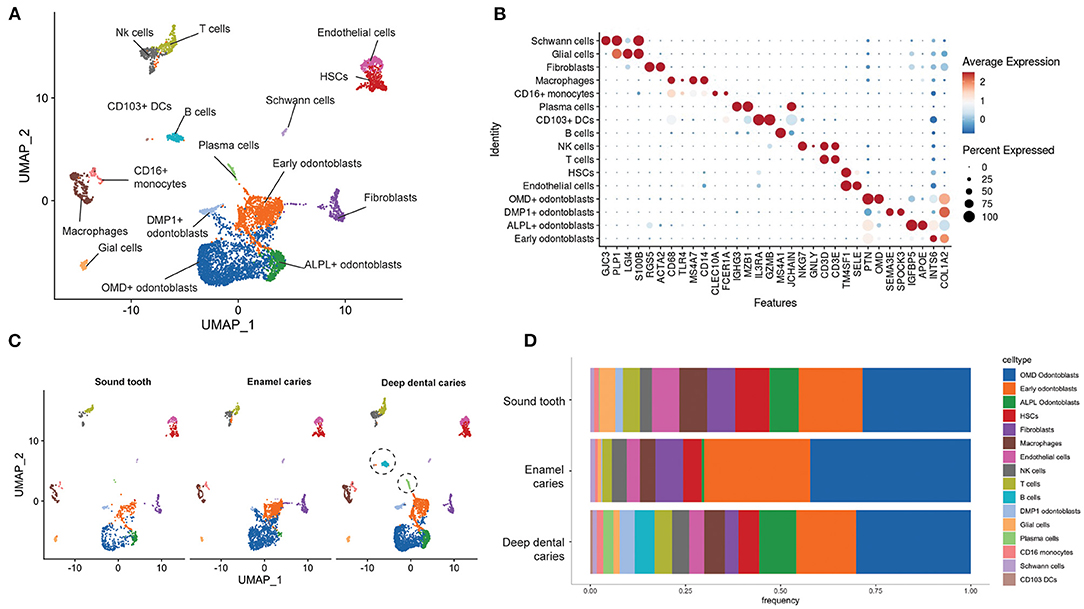
Figure 2. The cell population in dental pulp in sound, enamel, and deep carious teeth. (A) Integrated UMAP of the dental pulp samples: sound, enamel caries and deep dental caries. (B) The average expression levels of the marker genes used in the cell type annotation. (C) UMAP plots showing different distributions of cell populations in the sound, enamel, and deep dental caries. Dashed circles indicated the enrichment of B cells, plasma cells and CD103+ DCs in the deep dental caries (D) Stacked bar plots comparing the percentages of cell populations from the sound, arrested and deep dental caries.
Stromal and Immune Cells in the Dental Pulp Expressed Immune Pro-inflammatory, Anti-inflammatory, and Regenerative Genes in Deep Dental Caries
We next sought to identify the differentially expressed genes (DEGs) in the presence of deep dental caries, as compared to the sound tooth and enamel caries. Global differential gene expression analysis showed the upregulation of B cell and plasma cell-related genes such as IGHG1, IGHG2, IGLC1 and IGKC in the deep dental caries, which fell in line with the enriched B-cell and plasma cell numbers (Figure 3A). To gain better insights into how individual cell-types responded to deep dental caries, we performed differential gene expression (DEG) analysis on every cell type identified. We found changes in the expression levels of both cell-type specific and cell-type shared transcripts that could be characterized into three major groups: pro-inflammatory, anti-inflammatory, and regenerative genes.
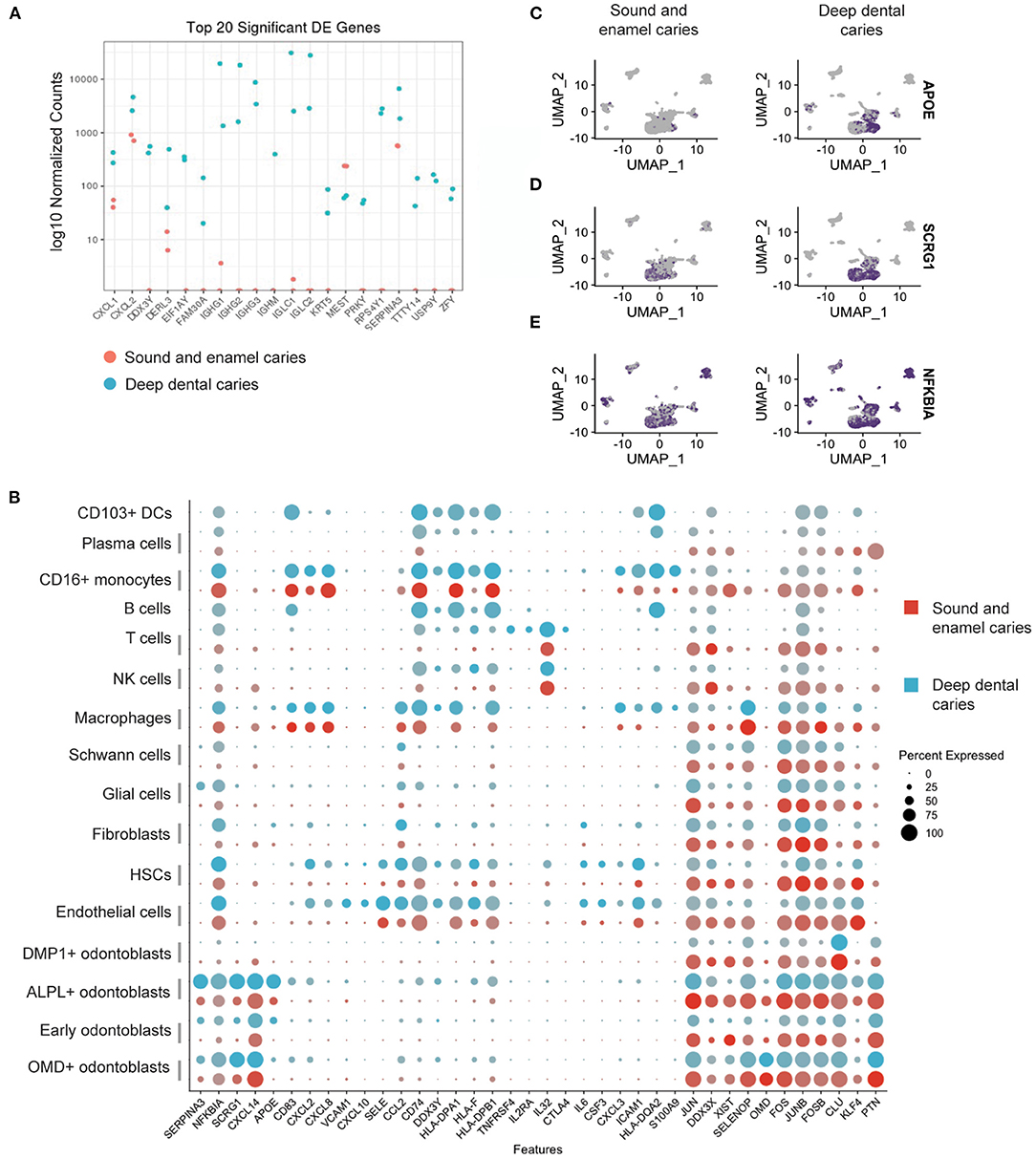
Figure 3. Differential gene expression analysis in dental pulp from sound tooth and enamel caries vs. deep dental caries. (A) Pseudo-bulk DEG analysis showing average log normalized counts of the top 20 DEGs. (B) Dot plots showing expression levels of representative genes. (C–E) Feature plots showing the expression levels of a representative regulatory gene, APOE (C), mineralization gene, SCRG1 (D) and pro-inflammatory gene, NFKBIA (E).
The upregulation of genes involved in inflammation and the activation of immune responses, which includes NFKBIA and CXCL2, was shared among fibroblasts, odontoblasts, and HSCs (Figures 3B,E). HSCs also showed the upregulation of proinflammatory mediators, including IL6 and CXCL8 specifically in the deep dental caries, while the endothelial cell activation signatures showed the upregulation of genes associated with chemoattraction and leukocyte adhesion, such as VCAM-1, SELE, CCL2 and CXCL10 (Figure 3B). Macrophage activation was also evident by the upregulation of genes related to antigen presentation such as HLA-DPA1, HLA-DPB1, HLA-DQA2 and CD74 (Figure 3B).
Anti-inflammatory and regenerative roles of the resident cells have been investigated in the resolution of the inflammation in pulps (29, 30). In the current study, we were able to identify the upregulation of SERPINA3, a protein known to inhibit neutrophil function (31) and APOE, known to suppress macrophage's proinflammatory responses to lipopolysaccharide (LPS) (Figures 3B,C) (32). Moreover, SCRG1, a gene associated with the stimulation of bone formation by mesenchymal stem cells (33), was also upregulated in the OMD+ and ALPL+ odontoblasts (Figures 3B,D). Several genes associated with T cell activation such as CD74 and IL32, showed the increase in expression levels in the deep dental caries (Figure 3B). Interestingly, we also found the upregulation of immune regulatory genes such as IL2RA, an important molecule for regulatory T cell (Treg) function in specifically the deep dental caries (Figure 3B). The full list of DEG in the presence of deep dental caries, as compared with the healthy tooth and the tooth with enamel caries, is provided in Supplementary Table 2.
Predicted Cell-Cell Communications in the Dental Pulp From Single-Cell RNA-Seq Data
The cell-to-cell interactions between stromal and immune cells have been suggested to mediate the protection and maintain homeostasis in the dental pulp, although the exact mechanisms have yet to be elucidated (7). MSCs and odontoblasts have been known to express chemokine receptors that have immuno-modulatory potential (7, 34). However, how they interact with the immune cells in healthy and inflamed pulps remains largely unknown. Here, we utilized NATMI (25) to predict cell-cell interactions between each cell type based on the expression of ligands and receptors in a sound tooth and teeth with enamel caries, as compared to deep dental caries.
We compared the overall enrichment of ligand-receptor interactions in the deep dental caries with sound tooth and enamel caries, as reflected by the difference in the number of edge-counts (Figures 4A,B). As expected, the interactions between B cells, CD103+ DCs and all cell types were enriched, as these cells were only found in the deep dental caries (Figures 4A,B). Moreover, the number of interactions between macrophages and other cell types were enriched in the deep dental caries, compared to the sound tooth and enamel caries (Figure 4B).
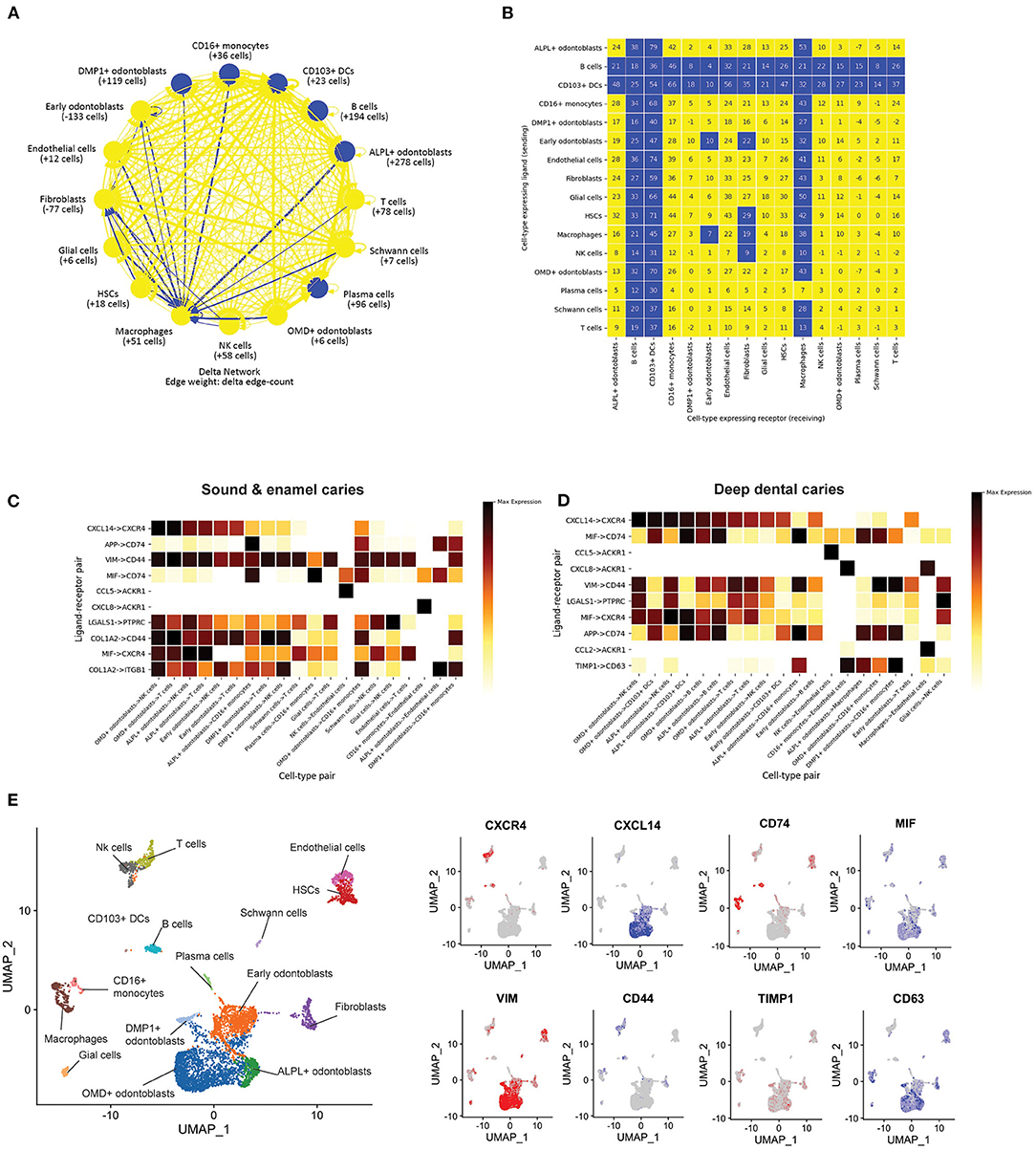
Figure 4. Predicted cell-cell communication in the dental pulp. (A) Interaction networks between cells in different tissues/cell types in our dental pulp samples. Arrows were drawn from ligand expressing cells to receptor expressing cells. Blue lines indicate higher numbers of interactions in the deep dental caries as compared to the sound tooth/enamel caries. (B) The difference in total number of edges between ligands and receptors between each cell type. Blue boxes indicate significantly higher numbers of interactions in dental caries. (C) Ligand-receptor pairs with the highest level of expression in sound tooth combined enamel caries and (D) deep dental caries. Darker shades represent higher levels of expression (E) Expression levels of representative ligands (red) and receptors (blue), projected on UMAP.
Next, we compared the levels of expression of ligand-receptors expressed by each cell type in sound tooth/enamel caries, and deep dental caries. Interestingly, several common ligand-receptor pairs were identified in both the sound/enamel caries and deep dental caries (Figures 4C,D). The predicted ligand-receptor interactions with the highest expression levels include those between the odontoblasts and immune cells, such as CXCL14 in odontoblasts and CXCR4 in immune cells (Figures 4C–E), which were suggested to be involved in the migration of immune cells (35). Another example is the MIF-CD74 interaction. MIF was expressed in the early odontoblasts, ALPL+ odontoblasts, and OMD+ odontoblasts, while CD74 was expressed in antigen presenting cells such as macrophages, CD16+ monocytes and CD103+ DCs (Figures 4C–E). This interaction is known for its role in the inhibition of inflammatory responses (36), but whether it is essential for the regulation of pulpal inflammation is unknown. Our result suggested that odontoblasts and immune cells potentially interact for tissue homeostasis and in response to dental caries.
The ligand receptor pairs exclusively found in the deep dental caries include TIMP1-CD63 in odontoblasts and myeloid cells (Figure 4D). While TIMP1 was known for its extracellular matrix remodeling (37), CD63 was reported to inhibit odontoblastic death in dental caries (38). However, how the interaction between TIMP1 and CD63 affects the pathogenesis of pulpal inflammation is not currently known. Another enriched ligand receptor pair exclusive to deep dental caries is CCL2 on macrophages and ACKR1 on endothelial cells (Figure 4D). ACKR1 was known to be expressed in vascular endothelial cells and functions in promoting or dampening inflammation upon interacting with several pro-inflammatory cytokines of the CC and CXC families (39).
Discussion
A better understanding of composition and intercellular communication in dental pulp would provide a foundation to regenerative dentistry in terms of clinical decision and therapeutic intervention. The characterization of dental pulp residential cells in our study has revealed that the dental pulp harbors HSCs as characterized by the expression of CD34. HSCs are multipotent progenitors for various blood cells residing mainly in the bone marrow and umbilical cord (40). Much interest has been given to MSCs; however, little is known about the role of HSCs in the dental pulp. Here, we have shown that in the deep dental caries that HSCs served as a source of IL-6 and CXCL8 expression, pro-inflammatory cytokines reported in pulpal inflammation (40). Whether HSCs play an important role in the overall inflammatory status of dental pulp, remains to be further elucidated.
The balance between inflammation, resolution, and regeneration, are essential steps in the control of infection and promotion of homeostasis in dental caries (1). Here, we have reported that the three processes may occur simultaneously in response to deep dental caries. However, one of the major limitations of this study was the limited number of samples. Hence, this experimental pilot study should be considered exploratory rather than a comprehensive investigation. A follow up study would be needed to validate the candidate genes of interest described in this study, to confirm veracity in situ. Moreover, the correlation between each biological marker to the clinical manifestation of pulpal disease would provide better insights into the molecular profiles that signal the stage of inflammation that could lead to a better tool for disease prognosis. Further study should include the clinical diagnosis, such as reversible or irreversible pulpitis to correlate with the molecular profiles for the discovery of candidate markers that aid the clinical decision making.
The ligand-receptor prediction tools allowed us to explore potentially new insights into the cell-cell interactions among different cell types in the dental pulp. We have shown that macrophages are among the cell types with a significant increase in the number of interactions with other cell types. Several studies have also shown that the interactions between odontoblasts and macrophages are essential to produce tertiary dentine through the Wnt signaling pathway (41). Interestingly, among the most highly expressed ligand-receptor interactions enriched in our deep dental caries samples, we found the TIMP1-CD63 interaction between odontoblasts and myeloid cells, including macrophage. TIMP1 has been shown to negatively regulate mineralization during the osteogenesis process through the Wnt/β-catenin signaling pathway (42). It therefore remains to be seen how this TIMP1-CD63 interaction affects tertiary dentinogenesis.
Another limitation in our study is the inability to effectively capture granulocytes. Granulocytes, especially neutrophils, have been known to be a challenge in single-cell transcriptomics, as they are sensitive to degradation, and they process relatively lower levels of transcripts compared to other white blood cells (43). As neutrophils are one of the most important players in inflammatory responses in dental pulp (44, 45), future studies should attempt to capture the neutrophils by different single-cell library construction platforms, such as BD Rhapsody (https://www.bdbiosciences.com/en-us/products/instruments/single-cell-multiomics-systems/rhapsody-express) or Seq-well (46).
Taken together, we have provided a pilot survey of dental pulp transcriptomic profiles in the healthy teeth and dental caries, which can serve as a platform for further in-depth and larger-scale investigation. To our knowledge, we are the first to compare the composition of dental pulp, perform the differential gene expression analysis, and predict the cell-cell interactions in health and disease. We have proposed potential candidates of genes that could be further validated for their potential as molecular markers for better clinical decision making and disease prognosis.
Data Availability Statement
The data presented in the study is deposited in the Gene Expression Omnibus, accession number GSE185222. The computational codes used in this study can be found at https://github.com/vclabsysbio/scRNAseq_Dentalpulp.
Ethics Statement
The studies involving human participants were reviewed and approved by Mahidol University Faculty of Dentistry and Pharmacy Institutional Review Board (MU-DT/PY-IRB 2019/068.0811). Written informed consent for participation was not required for this study in accordance with national legislation and institutional requirements.
Author Contributions
AO and VC: conceptualization. AO, SN, and VC: methodology. AO, SN, and BS: data analysis. VC and RS: reagents and materials. AO, VC, PM, and OM: writing. VC, PM, OM, and RS: supervision. VC: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by the Newton Advanced Fellowship to VC through the Royal Society (NA160153) and the Thailand Research Fund (DBG60800003). VC laboratory is supported by the Program Management Unit for National Competitiveness Enhancement (PMU-C) (C10F640057) and the mid-career researcher grant from National Research Council of Thailand (NRCT) and Mahidol University (NRCT5-RSA63015-24). AO is supported by Mahidol University Faculty of Dentistry Grant no. DTRS-PG-2022-02 (2022). BS is supported by the Development and Promotion of Science and Technology Talents Project (DPST).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Ms. Jantarika Kumar Arora for her support in bioinformatic analyses and to Dr. Jittranan Kaewprag for her insights in endodontics.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdmed.2021.806294/full#supplementary-material
Supplementary Table 1. Sample characterization and cell numbers.
Supplementary Table 2. Extended list of DEGs between sound/enamel caries and deep dental caries.
Supplementary Figure 1. Feature plots showing the expression levels of additional markers for the cell-type annotation as in Figure 2A.
References
1. Farges J-C, Alliot-Licht B, Renard E, Ducret M, Gaudin A, Smith AJ, et al. Dental pulp defence and repair mechanisms in dental caries. Mediators Inflamm. (2015) 2015:230251. doi: 10.1155/2015/230251
2. Galler KM, Weber M, Korkmaz Y, Widbiller M, Feuerer M. Inflammatory response mechanisms of the dentine–pulp complex and the periapical tissues. Int J Mol Sci. (2021) 22:1480. doi: 10.3390/ijms22031480
3. Kojima K, Inamoto K, Nagamatsu K, Hara A, Nakata K, Morita I, et al. Success rate of endodontic treatment of teeth with vital and nonvital pulps. A meta-analysis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. (2004) 97:95–9. doi: 10.1016/j.tripleo.2003.07.006
4. Hanna SN, Perez Alfayate R, Prichard J. Vital pulp therapy an insight over the available literature and future expectations. Eur Endod J. (2020) 5:46–53. doi: 10.14744/eej.2019.44154
5. Lin LM, Rosenberg PA. Repair and regeneration in endodontics. Int Endod J. (2011) 44:889–906. doi: 10.1111/j.1365-2591.2011.01915.x
6. Dentistry AAoP. Pulp Therapy for Primary and Immature Permanent Teeth. The Reference Manual of Pediatric Dentistry. Chicago, IL: American Academy of Pediatric Dentistry (2020). p. 384–92.
7. Leprince JG, Zeitlin BD, Tolar M, Peters OA. Interactions between immune system and mesenchymal stem cells in dental pulp and periapical tissues. Int Endod J. (2012) 45:689–701. doi: 10.1111/j.1365-2591.2012.02028.x
8. Ducret M, Fabre H, Degoul O, Atzeni G, McGuckin C, Forraz N, et al. Immunophenotyping reveals the diversity of human dental pulp mesenchymal stromal cells in vivo and their evolution upon in vitro amplification. Front Physiol. (2016) 7:512. doi: 10.3389/fphys.2016.00512
9. Ducret M, Farges JC, Pasdeloup M, Perrier-Groult E, Mueller A, Mallein-Gerin F, et al. Phenotypic identification of dental pulp mesenchymal stem/stromal cells subpopulations with multiparametric flow cytometry. Methods Mol Biol. (2019) 1922:77–90. doi: 10.1007/978-1-4939-9012-2_8
10. Mangkornkarn C, Steiner JC, Bohman R, Lindemann RA. Flow cytometric analysis of human dental pulp tissue. J Endod. (1991) 17:49–53. doi: 10.1016/S0099-2399(06)81607-9
11. Gaudin A, Renard E, Hill M, Bouchet-Delbos L, Bienvenu-Louvet G, Farges JC, et al. Phenotypic analysis of immunocompetent cells in healthy human dental pulp. J Endod. (2015) 41:621–7. doi: 10.1016/j.joen.2015.01.005
12. Hahn CL, Falkler WA Jr, Siegel MA. A study of T and B cells in pulpal pathosis. J Endod. (1989) 15:20–6. doi: 10.1016/S0099-2399(89)80093-7
13. Bruno KF, Silva JA, Silva TA, Batista AC, Alencar AH, Estrela C. Characterization of inflammatory cell infiltrate in human dental pulpitis. Int Endod J. (2010) 43:1013–21. doi: 10.1111/j.1365-2591.2010.01757.x
14. McLachlan JL, Smith AJ, Bujalska IJ, Cooper PR. Gene expression profiling of pulpal tissue reveals the molecular complexity of dental caries. Biochim Biophys Acta. (2005) 1741:271–81. doi: 10.1016/j.bbadis.2005.03.007
15. Krivanek J, Soldatov RA, Kastriti ME, Chontorotzea T, Herdina AN, Petersen J, et al. Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nat Commun. (2020) 11:4816. doi: 10.1038/s41467-020-18512-7
16. Pagella P, de Vargas Roditi L, Stadlinger B, Moor AE, Mitsiadis TA. A single-cell atlas of human teeth. iScience. (2021) 24:102405. doi: 10.1016/j.isci.2021.102405
17. Yianni V, Sharpe PT. Transcriptomic profiling of dental pulp pericytes: an RNAseq approach. Front Dent Med. (2020) 1:6. doi: 10.3389/fdmed.2020.00006
18. Tirino V, Paino F, De Rosa A, Papaccio G. Identification, isolation, characterization, and banking of human dental pulp stem cells. In: Singh SR, editor. Somatic Stem Cells: Methods and Protocols. Totowa, NJ: Humana Press (2012). p. 443–63. doi: 10.1007/978-1-61779-815-3_26
19. RStudio Team,. RStudio: Integrated Development for R. RStudio. (2020). Available online at: http://www.rstudio.com/ (accessed October 31, 2021).
20. Young MD, Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience. (2020) 9:giaa151. doi: 10.1093/gigascience/giaa151
21. McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. (2019) 8:329–37.e4. doi: 10.1016/j.cels.2019.03.003
22. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell. (2019) 177:1888–902.e21. doi: 10.1016/j.cell.2019.05.031
23. Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. (2018). doi: 10.1038/nbt.4314
24. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
25. Hou R, Denisenko E, Ong HT, Ramilowski JA, Forrest ARR. Predicting cell-to-cell communication networks using NATMI. Nat Commun. (2020) 11:5011. doi: 10.1038/s41467-020-18873-z
26. Oetjen KA, Lindblad KE, Goswami M, Gui G, Dagur PK, Lai C, et al. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight. (2018) 3:e124928. doi: 10.1172/jci.insight.124928
27. Ledesma-Martínez E, Mendoza-Núñez VM, Santiago-Osorio E. Mesenchymal stem cells derived from dental pulp: a review. Stem Cells Int. (2016) 2016:4709572. doi: 10.1155/2016/4709572
28. Lin W, Gao L, Jiang W, Niu C, Yuan K, Hu X, et al. The role of osteomodulin on osteo/odontogenic differentiation in human dental pulp stem cells. BMC Oral Health. (2019) 19:22. doi: 10.1186/s12903-018-0680-6
29. Farges J-C, Alliot-Licht B, Baudouin C, Msika P, Bleicher F, Carrouel F. Odontoblast control of dental pulp inflammation triggered by cariogenic bacteria. Front Physiol. (2013) 4:326. doi: 10.3389/fphys.2013.00326
30. Farges J-C, Carrouel F, Keller J-F, Baudouin C, Msika P, Bleicher F, et al. Cytokine production by human odontoblast-like cells upon Toll-like receptor-2 engagement. Immunobiology. (2011) 216:513–7. doi: 10.1016/j.imbio.2010.08.006
31. Kalsheker NA. Alpha 1-antichymotrypsin. Int J Biochem Cell Biol. (1996) 28:961–4. doi: 10.1016/1357-2725(96)00032-5
32. Ali K, Middleton M, Puré E, Rader DJ. Apolipoprotein E suppresses the type I inflammatory response in vivo. Circ Res. (2005) 97:922–7. doi: 10.1161/01.RES.0000187467.67684.43
33. Aomatsu E, Takahashi N, Sawada S, Okubo N, Hasegawa T, Taira M, et al. Novel SCRG1/BST1 axis regulates self-renewal, migration and osteogenic differentiation potential in mesenchymal stem cells. Sci Rep. (2014) 4:3652. doi: 10.1038/srep03652
34. Demircan PC, Sariboyaci AE, Unal ZS, Gacar G, Subasi C, Karaoz E. Immunoregulatory effects of human dental pulp-derived stem cells on T cells: comparison of transwell co-culture and mixed lymphocyte reaction systems. Cytotherapy. (2011) 13:1205–20. doi: 10.3109/14653249.2011.605351
35. Hara T, Tanegashima K. CXCL14 antagonizes the CXCL12-CXCR4 signaling axis. Biomol Conc. (2014) 5:167–73. doi: 10.1515/bmc-2014-0007
36. Farr L, Ghosh S, Moonah S. Role of MIF Cytokine/CD74 receptor pathway in protecting against injury and promoting repair. Front Immunol. (2020) 11:1273. doi: 10.3389/fimmu.2020.01273
37. Ries C. Cytokine functions of TIMP-1. Cell Mol Life Sci. (2014) 71:659–72. doi: 10.1007/s00018-013-1457-3
38. Wang HS, Yang FH, Wang YJ, Pei F, Chen Z, Zhang L. Odontoblastic exosomes attenuate apoptosis in neighboring cells. J Dent Res. (2019) 98:1271–8. doi: 10.1177/0022034519869580
39. Bonecchi R, Graham GJ. Atypical chemokine receptors and their roles in the resolution of the inflammatory response. Front Immunol. (2016) 7:224. doi: 10.3389/fimmu.2016.00224
40. Lee JY, Hong S-H. Hematopoietic stem cells and their roles in tissue regeneration. Int J Stem Cells. (2020) 13:1–12. doi: 10.15283/ijsc19127
41. Neves VCM, Yianni V, Sharpe PT. Macrophage modulation of dental pulp stem cell activity during tertiary dentinogenesis. Sci Rep. (2020) 10:20216.
42. Liang T, Gao W, Zhu L, Ren J, Yao H, Wang K, et al. TIMP-1 inhibits proliferation and osteogenic differentiation of hBMSCs through Wnt/β-catenin signaling. Biosci Rep. (2019) 39:BSR20181290. doi: 10.1042/BSR20181290
43. Qi J, D'Souza D, Dawson T, Geanon D, Stefanos H, Marvin R, et al. Multimodal single-cell characterization of the human granulocyte lineage. bioRxiv. doi: 10.1101/2021.06.12.448210
44. Cavalcanti BN, Rode Sde M, França CM, Marques MM. Pulp capping materials exert an effect on the secretion of IL-1β and IL-8 by migrating human neutrophils. Braz Oral Res. (2011) 25:13–8. doi: 10.1590/S1806-83242011000100003
45. Holder MJ, Wright HJ, Couve E, Milward MR, Cooper PR. Neutrophil extracellular traps exert potential cytotoxic and proinflammatory effects in the dental pulp. J Endod. (2019) 45:513–20.e3. doi: 10.1016/j.joen.2019.02.014
Keywords: dental pulp, caries, immune responses, single-cell transcriptomics, cell-cell interactions, regenerative dentistry
Citation: Opasawatchai A, Nguantad S, Sriwilai B, Matangkasombut P, Matangkasombut O, Srisatjaluk R and Charoensawan V (2022) Single-Cell Transcriptomic Profiling of Human Dental Pulp in Sound and Carious Teeth: A Pilot Study. Front. Dent. Med. 2:806294. doi: 10.3389/fdmed.2021.806294
Received: 31 October 2021; Accepted: 30 November 2021;
Published: 05 January 2022.
Edited by:
Val Yianni, King's College London, United KingdomReviewed by:
Vitor Neves, King's College London, United KingdomPaul Roy Cooper, University of Otago, New Zealand
Copyright © 2022 Opasawatchai, Nguantad, Sriwilai, Matangkasombut, Matangkasombut, Srisatjaluk and Charoensawan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anunya Opasawatchai, YW51bnlhLm9wYUBtYWhpZG9sLmVkdQ==; Varodom Charoensawan, dmFyb2RvbS5jaGFAbWFoaWRvbC5hYy50aA==
 Anunya Opasawatchai
Anunya Opasawatchai Sarintip Nguantad
Sarintip Nguantad Benjamaporn Sriwilai
Benjamaporn Sriwilai Ponpan Matangkasombut
Ponpan Matangkasombut Oranart Matangkasombut
Oranart Matangkasombut Ratchapin Srisatjaluk1
Ratchapin Srisatjaluk1 Varodom Charoensawan
Varodom Charoensawan