- School of Dental Medicine, University of Pittsburgh, Pittsburgh, PA, United States
The goal of this study was to test if oral health outcomes are associated with the same genetic markers in Black and White individuals who did not have cancer. From a total of 6,100 subjects from the Dental Registry and DNA Repository project, 1,042 individuals who self-identified as White and 266 as Black without a history of cancer were included in this analysis. Genotyping data from IRE1—rs196929, RHEB—rs2374261 and rs1109089, AXIN2—rs2240308 and rs11867417, and RPTOR—rs4396582, present in cell regulatory pathways, were analyzed. We ran separate analyses in self-reported Black and White groups to reduce possible confounding effects of population stratification. Internal diagnostic codes from our dental registry were converted into Phecodes in order to run the analysis using the PheWAS package, installed in R Studio software. Periodontitis was associated with RHEB in both Black and White patients, with the minor allele increasing the likelihood of developing periodontitis in the White group and yielding a protective effect in the Black individuals. The presence of ulcers and gingivitis were associated with RPTOR and AXIN2, respectively, in the White group, but an association was not detected for the Black group. On the other hand, phenotypes such as dental fracture, diseases of the tongue, attrition, erosion, abrasion, fordyce granules, and torus and exostosis were uniquely associated with the Black group. Periodontitis associated with RHEB in both Black and White patients, and associations found in Black individuals may be the result of social disparities that lead to higher levels of stress, and these observed differences require further study.
Introduction
Disparities in oral health are profound in the US, despite the improvements that in general happened in the last 50 years. Socioeconomic status, encountering racism, geographic origin (race/ethnicity), sex, age, geographic location, lifestyle behaviors (tobacco use, alcohol use, dietary choices), and years of education are variables that correlate with oral health disparities. Disparities in dental caries, periodontitis, and oral cancer by geographic origin are well-documented (1–3).
In Pittsburgh, which is the largest city of the Appalachian region, an area with some of the worse health indicators in the country, we created a registry of individuals that have comprehensive oral health clinical descriptions linked to biological (saliva) samples (4). With this resource, we tested the hypothesis that the same genes would show associations to different phenotypes depending on the disparities affecting individual ethnic groups. Immunosuppression and treatment for different types of cancer usually impact oral health outcomes. We excluded cancer diagnosed individuals because they constitute a group of patients that have a whole set of specific oral treatment needs that are different from individuals who do not have a history of cancer, avoiding confounder effects.
We studied genes that previously showed a correlation with orofacial phenotypes, including dental phenotypes affecting enamel and dentin, and soft tissue phenotypes such as diseases of the tongue, lips, and gums. Our preliminary data showed that genes present in regulatory pathways such as the mammalian target of rapamycin (mTOR) and endoplasmic reticulum (ER) stress pathways associated with one or more dental phenotypes (5). IRE1, RHEB, and RPTOR are present in pathways involved in cell proliferation, differentiation, protein synthesis, and inflammation, and AXIN2 has been associated with craniofacial phenotypes such as cleft lip and palate and tooth agenesis, reported in previous studies performed by our group (6) and others (7–11). Single nucleotide polymorphisms (SNPs) that were previously associated with oral phenotypes were analyzed (5, 6). In our previous work, we found significant associations between periapical lesions due to deep caries lesions in dentin and RHEB and dental caries and RPTOR. When combining patients that had concomitant dental caries, periodontitis, and periapical pathology, several markers in RHEB showed association (5). Further, a second study we conducted, in which a PheWAS (phenome-wide association study) was performed in individuals diagnosed with cancer unveiled that tooth loss/edentulism was associated with SNPs in AXIN2, and leukoplakia of oral mucosa was associated with both AXIN2 and RHEB (6). The goal of the present study was to test if the same genes associate with distinct oral health outcomes depending on the ethnic group of individuals who did not have cancer.
Methods
From our previous research (6), data of 6,100 subjects from the Dental Registry and DNA Repository project were available and used in this present study. From the total pool, 1,042 White and 266 Black patients were included in this analysis. These individuals were selected because they did not show any signs or history of cancer, however they were matched by sex, age, and geographic origin to the individuals that survived cancer in the sample in our previous study (6). Individuals that survived cancer have specific dental treatment needs in our sample and were not included in these analyses. Educational status was not available for the participants studied. Self-reported smoking and health history were assessed and followed the expected distributions of the population. Demographic data and risk factors associated with oral health outcomes can be found in Table 1. These data were originally obtained by the dentist when planning the treatment of the subjects using standard forms. The University of Pittsburgh Institutional Review Board (IRB # 0606091) approved this project. Written informed consent documents were obtained from all subjects. Age-appropriate assent documents were used for children younger than 14 years of age and signed informed consent documents were obtained from the parents.
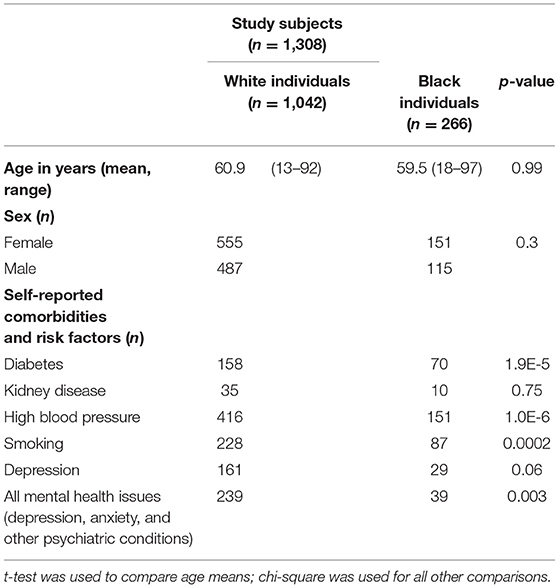
Table 1. Description of study participant demographic characteristics and risk factors associated with oral health outcomes.
Genotyping data from participants' DNA previously extracted from all subjects were used. Established protocols for DNA extraction and genotypic analysis have been previously described (12). Six SNPS that were selected in our previous studies (5, 6) and showed association with orofacial phenotypes were tested here; IRE1—rs196929, RHEB—rs2374261 and rs1109089, AXIN2—rs2240308 and rs11867417, and RPTOR—rs4396582. We ran separate analyses in self-reported White and Black groups to reduce possible confounding effects of population stratification. Table 2 lists the genes, the selected SNPs and their minor allele frequencies (MAF) in each group.
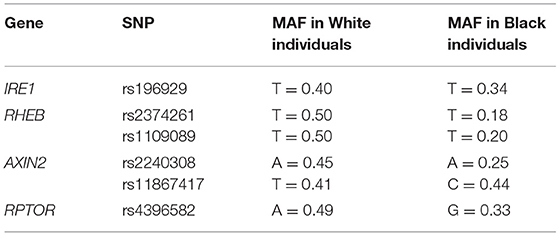
Table 2. Selected genes, SNPs and minor allele frequencies (MAF) of each ethnic group in our population.
Phenotypes tested included dental caries, diseases of the dental pulp and periapical tissues, diseases of the jaw, missing teeth or edentulism, ulcer, gingivitis, periodontitis, disorders of tooth development or eruption, tooth fracture, sleep related movement disorders (e.g., bruxism), diseases of salivary glands, fordyce granules, malocclusion, stomatitis, mucositis, lingual varicose veins, diseases of the tongue, torus, attrition, erosion and abrasion, temporomandibular joint disorder, and candidiasis.
Internal diagnostic codes from the dental registry were converted into “Phecodes” (13) in order to run the analysis using the PheWAS package, installed in R Studio software (14). The software finds specific comparison individuals for each affected patient according to the range of phenotypes entered in the PheWAS file. The additive genetic model was the test of choice whereas allele frequencies are calculated and compared between the formed groups.
Statistical power of our study to detect associations was estimated based on the disease prevalence reported by the Center of Disease Control (https://www.cdc.gov), when available for the specific oral phenotypes tested here, the number of cases and controls and the frequency of the associated SNP risk allele in our population. The calculation of statistical power for periodontitis, considering a prevalence of 47%, a marker B in linkage disequilibrium with our test locus A, the high-risk allele frequency for the allele A (set at 0.1), and the genotype risks for the Aa and AA genotypes relative to the baseline aa genotype risk was estimated for the smallest sample size of 42 cases and a case-control ratio of 1:4 as 72%, whereas the power consistently increases with larger sample sizes such as the ones available for the White group. We used a genetic power calculator for the statistical power analysis (15).
Results
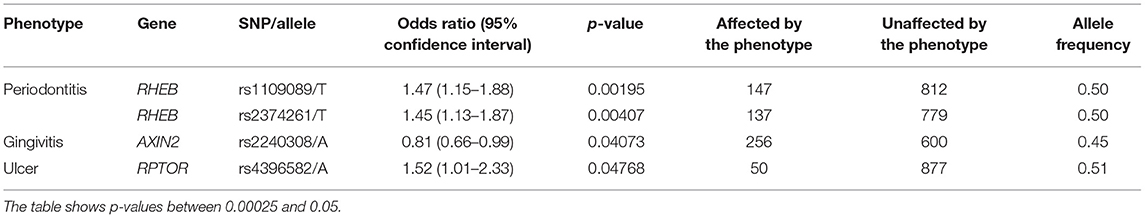
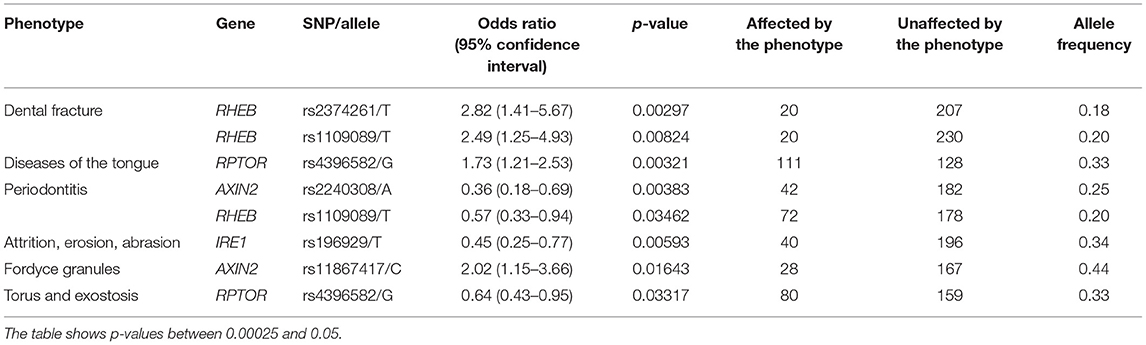
Our results showed that some phenotype associations differ by ethnic groups. The presence of ulcers and gingivitis were associated with RPTOR and AXIN2, respectively, in the self-reported White group (Table 3), and that was not the case for the self-reported Black group studied. On the other hand, phenotypes such as dental fracture, diseases of the tongue, attrition, erosion, abrasion, fordyce granules, and torus and exostosis were unique to the individuals who identify as Black (Table 4). RHEB was associated with periodontitis in both groups; however, the minor allele increases the likelihood of developing the disease in the White group, whereas it seems to yield a protective effect in individuals who identify as Black.
Discussion
Population stratification is a concern in genetic studies (16), and for that reason we decided to run analyses separately in self-identified Black and White groups, in particular to ask the question if the same phenotypes associate with the same genetic markers in the groups. The selection of individuals who did not have a history of cancer may have allowed for improved homogeneity, which may have increased statistical power. Only periodontitis associated with RHEB rs1109089 in both groups. With this approach, we were able to detect differences between conditions that those two groups might be susceptible to. Minor allele frequencies usually vary between different ethnic groups (17, 18) and creating more homogeneous study populations by separating the two main groups available in our registry allowed us to identify specific orofacial phenotypes related to them. One can argue that the approach of using self-reported geographic origin is a limitation. However, it has been reported that an individual's self-reported ancestry is sufficient to account for potential population stratification (19). Additionally, we have suggested before that there is an adequate consistency between self-reported and genetically driven geographic origin definitions in our Dental Registry and DNA Repository project (20).
The results showed that three genes (RHEB, RPTOR, and AXIN2) showed specific associations depending on the geographic origin. Black patients were more affected by diseases of the hard tissues such as dental fracture, attrition, erosion, abrasion, and torus or exostosis. Conversely, White patients seemed to be more frequently affected by diseases of the soft tissues such as gingivitis and ulcers. These associations with RHEB, RPTOR, and AXIN2 are not easily explained based on the function of these genes, therefore these differences may be due to socioeconomic disparities. Over the course of the project, government-assistance programs in Pennsylvania have been declining, but still at least ~50% of patients in our analyses benefit from this kind of support. These groups are over-represented by Black individuals. Expected higher frequencies of cardiovascular risks (21), diabetes (22), smoking (23) were seen among Black patients in our sample (Table 1). It was also expected to see less Black individuals self-reporting mental health issues despite concerns with food insecurity, unemployment, education, stress, and depressive symptoms, since they tend to be less served by mental health services (24). Dental fractures, attrition, and abrasion can be all related to higher levels of anxiety and poverty (25). We have also previously detected in our data from the Dental Registry and DNA Repository project potential bias on the types of treatments offered to Black patients in comparison to the White patients, with one being offered the less expensive and esthetic amalgam restorations more often than the other (26).
Although concerned with multiple testing, we did not apply the stringent Bonferroni correction and instead compared the results of the analyses between Black and White individuals. We reported before that this might be too strict of a correction which may lead to missing true biological associations (6, 27, 28). Another limitation present in our study was the low power to identify significant associations since there was a limited number of occurrences for a couple of the oral phenotypes. However, we were able to identify associations for the most common oral conditions available in the database. Particularly in the analysis of Black individuals, we found several associations even with a smaller sample size. Table 1 shows several expected differences between the two groups and the analyses presented here were not adjusted by these risk factors and having those concomitant risks may have impacted the distinct result found. Finally, we were not able to include additional groups of different geographic origins because of the lack of diversity in the database that would meet an adequate sample size for the analysis.
In summary, periodontitis associated with RHEB both in Black and White individuals with the same allele protecting Black patients and associating with increased likelihood in White patients. Additional associations found in Black individuals were not identified in White patients even with a larger sample size. Considering overall health characteristics and risk factors such as smoking habits imbalances between the groups, our results confirm a multifactorial aspect of the oral phenotypes studied. While this study focused on differences in genetic associations, social disparities that lead to higher levels of stress likely influence some of these observed differences and require further study.
Data Availability Statement
Requests to access these datasets should be directed to Alexandre R. Vieira, YXJ2MTFAcGl0dC5lZHU=.
Ethics Statement
The studies involving human participants were reviewed and approved by University of Pittsburgh Institutional Review Board. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
MB performed the analysis, interpreted data, and wrote the first draft of the manuscript. AM obtained support, helped with data collection, and critically reviewed the manuscript. AV created the concept, designed the study, collected data, helped with analysis, interpreted data, and wrote final draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
MB was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award number TL1TR001858 (Kraemer).
Disclaimer
The content is solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The University of Pittsburgh Dental Registry and DNA Repository provided data and DNA samples for this study.
References
1. Capurro DA, Iafolla T, Kingman A, Chattopadhyay A, Garcia I. Trends in income-related inequality in untreated caries among children in the United States: findings from NHANES I, NHANES III, and NHANES 1999-2004. Community Dent Oral Epidemiol. (2015) 43:500–10. doi: 10.1111/cdoe.12174
2. Griffin SO, Jones JA, Brunson D, Griffin PM, Bailey WD. Burden of oral disease among older adults and implications for public health priorities. Am J Public Health. (2012) 102:411–8. doi: 10.2105/AJPH.2011.300362
3. Huang DL, Park M. Socioeconomic and racial/ethnic oral health disparities among US older adults: oral health quality of life and dentition. J Public Health Dent. (2015) 75:85–92. doi: 10.1111/jphd.12072
4. Vieira AR, Hilands KM, Braun TW. Saving more teeth – a case for personalized care. J Pers Med. (2015) 5:30–5. doi: 10.3390/jpm5010030
5. Bezamat M, Deeley K, Khaliq S, Letra A, Scariot R, Silva RM, et al. Are mTOR and endoplasmic reticulum stress pathway genes associated with oral and bone diseases? Caries Res. (2019) 53:235–41. doi: 10.1159/000492675
6. Bezamat M, Harrison B, Zhou Y, Glickman KM, Telles V, Guirguis C, et al. Phenome-wide scan finds potential orofacial risk markers for cancer. Sci Rep. (2020) 10:4869. doi: 10.1038/s41598-020-61654-3
7. Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, et al. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. (2004) 74:1043–50. doi: 10.1086/386293
8. Callahan N, Modesto A, Meira R, Seymen F, Patir A, Vieira AR. Axis inhibition protein 2 (AXIN2) polymorphisms and tooth agenesis. Arch Oral Biol. (2009) 54:45–9. doi: 10.1016/j.archoralbio.2008.08.002
9. Letra A, Menezes R, Granjeiro JM, Vieira AR. AXIN2 and CDH1 polymorphisms, tooth agenesis, and oral clefts." Birth Defects Res A Clin Mol Teratol. (2009) 85:169–73. doi: 10.1002/bdra.20489
10. Wu X, Li Y, Wang F, Hu L, Li Y, Wang J, et al. Spatiotemporal expression of Wnt/beta-catenin signaling during morphogenesis and odontogenesis of deciduous molar in miniature pig. Int J Biol Sci. (2017) 13:1082–91. doi: 10.7150/ijbs.20905
11. Han Y, Zhou L, Ma L, Li D, Xu M, Yuan H, et al. The axis inhibition protein 2 polymorphisms and non-syndromic orofacial clefts susceptibility in a Chinese Han population. J Oral Pathol Med. (2014) 43:554–60. doi: 10.1111/jop.12162
12. Ranade K, Chang MS, Ting CT, Pei D, Hsiao CF, Olivier M, et al. High-throughput genotyping with single nucleotide polymorphisms. Genome Res. (2001) 11:1262–8. doi: 10.1101/gr.157801
13. Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. (2014) 30:2375–6. doi: 10.1093/bioinformatics/btu197
14. Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. (2010) 26:1205–10. doi: 10.1093/bioinformatics/btq126
15. Purcell S, Cherny SS, Sham PC. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. (2003) 19:149–50. doi: 10.1093/bioinformatics/19.1.149
16. Hellwege JN, Keaton JM, Giri A, Gao X, Edwards DRV, Edwards TL. Population stratification in genetic association studies. Curr Protoc Hum Genet. (2017) 95:1.22.1–23. doi: 10.1002/cphg.48
17. Cross DS, Ivacic LC, Stefanski EL, McCarty CA. Population based allele frequencies of disease associated polymorphisms in the Personalized Medicine Research Project. BMC Genet. (2010) 11:51. doi: 10.1186/1471-2156-11-51
18. Huang T, Shu Y, Cai Y-D. Genetic differences among ethnic groups. BMC Genomics. (2015) 16:1093. doi: 10.1186/s12864-015-2328-0
19. Hinrichs AL, Larkin EK, Suarez BK. Population stratification and patterns of linkage disequilibrium. Genet Epidemiol. (2009) 33(Suppl. 1):S88–92. doi: 10.1002/gepi.20478
20. Feng P, Wang X, Casado PL, Küchler EC, Deeley K, Noel J, et al. Genome wide association scan for chronic periodontitis implicates novel locus. BMC Oral Health. (2014) 14:84. doi: 10.1186/1472-6831-14-84
21. Erqou S, Kip KE, Mulukutla SR, Aiyer AN, Reis SE. Racial differences in the burden of coronary artery calcium and carotid intima media thickness between Blacks and Whites. Neth Heart J. (2015) 23:44–51. doi: 10.1007/s12471-014-0610-4
22. Buscemi J, Saiyed N, Silva A, Ghahramani F, Benjamins MR. Diabetes mortality across the 30 biggest U.S. cities: assessing overal trends and racial inequalities. Diabetes Res Cli Pract. (2021) 173:108652. doi: 10.1016/j.diabres.2021.108652
23. O'Reilly NL, Hager ER, Harrington D, Black MM. Assessment of risk for food insecurity among African American urban households: utilizing cumulative risk indices and latent class analysis to examine accumulation of risk factors. Transl Behav Med. (2020) 10:1322–9. doi: 10.1093/tbm/ibaa027
24. Morris RM, Sellwood W, Edge D, Colling C, Stewart R, Cupitt C, et al. Ethnicity and impact on the receipt of cognitive-behavioural therapy in people with psychosis or bipolar disorder: an English cohort study. BMJ Open. (2020) 10:e034913. doi: 10.1136/bmjopen-2019-034913
25. Ridley M, Rao G, Schilbach F, Patel V. Poverty, depression, and anxiety: causal evidence and mechanisms. Science. (2020) 370:eaay0214. doi: 10.1126/science.aay0214
26. Vieira AR, Mantravadi S. Recurrent dental caries in posterior teeth and tooth loss in Black versus White-adults in a setting that minimizes the effects of socioeconomic status. Clin Press. (2017) 1:7–10. doi: 10.28964/ClinPress-1-103
27. Vieira AR, McHenry TG, Daack-Hirsch S, Murray JC, Marazita ML. Candidate gene/loci studies in cleft lip/palate and dental anomalies finds novel suscptibility genes for clefts. Genet Med. (2008) 10:668–74. doi: 10.1097/GIM.0b013e3181833793
Keywords: periodontitis, health disparities, genetic association, racism, population substructure
Citation: Bezamat M, Modesto A and Vieira AR (2021) Phenome-Wide Association Study With Focus on Oral Health Disparities and Individuals Who Did Not Have Cancer. Front. Dent. Med. 2:641246. doi: 10.3389/fdmed.2021.641246
Received: 13 December 2020; Accepted: 06 April 2021;
Published: 28 April 2021.
Edited by:
Moritz Kebschull, University of Birmingham, United KingdomReviewed by:
Thomas Kocher, University of Greifswald, GermanyRyan Demmer, University of Minnesota Twin Cities, United States
Copyright © 2021 Bezamat, Modesto and Vieira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandre R. Vieira, YXJ2MTFAcGl0dC5lZHU=
 Mariana Bezamat
Mariana Bezamat Adriana Modesto
Adriana Modesto Alexandre R. Vieira
Alexandre R. Vieira