- Centre for Craniofacial and Regenerative Biology, Faculty of Dentistry, Oral & Craniofacial Sciences, Kings College London, London, United Kingdom
Pericytes represent a population of mesenchymal cells that are found in virtually all vascularised organs where they share a range of similar features, mostly associated with vascular maintenance roles. It is becoming increasingly apparent that organ to organ differences exist between pericytes that directly relate to tissue-specific functions of these cells. Here, we present novel data indicating that in dental structures, pericytes have an immunomodulatory role. We show evidence supporting the concept that pericytes are a source of multiple cytokines and chemokines within the dental pulp, capable of regulating the extravasation of circulating lymphocytes. Using the SmartSeq-2 method we performed single cell RNA-seq and show that this is attributed to a defined pericyte sub-population that exists in vivo. In addition, as a consequence of our transcriptomic screen, we detect and validate a novel marker that can identify mature pericytes capable of giving rise to odontoblasts in vivo. Using transcriptomic screening, this marker is also identified to be expressed in pericytes residing in multiple other tissues including bone marrow, lung, and pancreas.
Introduction
Terms such as mesenchymal stem cells, stromal stem cells, skeletal stem cells, bone marrow (or dental pulp) stem cells have all been used to refer to multipotent populations of cells in the mesenchymal/stromal compartment of organs (1–3). These stromal stem cells reside in the majority of vascularised tissues and some have a perivascular localization, commonly referred to as pericytes (formerly known as “rouget” cells) (4–6). Pericytes are present on the abluminal side of endothelial cells where they play a well-documented role in stabilizing endothelial networks both by mechanical support and by secretion of trophic factors (7). What has attracted the attention of the stem cell and regenerative biology field is evidence supporting the notion that pericytes are a source of mesenchymal stem cell precursors that generate mature mesenchymal cells that terminally differentiate into osteoblasts, odontoblasts, myoblasts, and adipocytes in vitro (8–12). It is also now established that the ability of pericytes to differentiate in such a way is not an artifact of tailored culture conditions but that this can be observed in vivo. To date, in vivo lineage tracing evidence exists showing pericytes give rise to multiple mesodermal derivatives in response to injury, such as osteoblasts (13), odontoblasts (14), and myoblasts (15). These observations pose the question of how pericytes in different organs control their differentiation into the appropriate cell type. Epigenetic and transcriptomic programs are active in dental pericytes that bias them into making an odontogenic fate decision if they are called to act as stem cells such as in a regeneration/repair context (16). This epigenetic programming of mesenchymal stem cell precursors has been shown to occur in a number of organs (17, 18). Using tooth pulp as a model we compared the properties of pericytes of the same origin (neural crest) in the same tissue (pulp) in a continuously growing vs. a non-growing tooth. Our data prompt reconsideration of the role of pericytes from that of being restricted to blood vessel maintenance. We show that pericytes in molars are a possible source of multiple cytokines and chemokines that regulate the local immune microenvironment. As shown by single cell RNAseq, these immunomodulatory factors originate from a discreet subpopulation of pericytes that do not correspond to the traditionally identified immunophenotype (CD146+/Endoglin+/Pdgfr-β+). In addition, we validate a novel marker from our transcriptomic screens that can be used to lineage trace pericytes that give rise to cells of mature mesenchymal phenotypes, including odontoblasts.
Materials and Methods
Mouse Strains
XlacZ4 [JAX:018625, B6.FVB-Tg(Fabp4-lacZ)4Mosh/J] was used to identify and isolate pericytes (19). Tagln-Cre [JAX: 017491, B6.Cg-Tg(Tagln-cre)1Her/J] (20) mice were crossed with a dual fluorescence reporter mouse line [JAX: 007576, Gt(ROSA)26Sortm4(ACTB−tdTomato, −EGFP)Luo/J] (21). The offspring will be referred to as Tagln-Cre;mTmG.
Cell Isolation
Homozygous XlacZ4 [JAX:018625, B6.FVB-Tg(Fabp4-lacZ)4Mosh/J] (19) pups were collected at postnatal days 5–7 and sacrificed. Incisors and first molars were extracted from the mandible and maxilla and placed into ice cold sterile phosphate buffered saline (PBS). The epithelium was removed from incisors using a fine needle and the remaining dental pulp placed in freshly made SB buffer (1% FBS, 10 mM HEPES buffer pH7, PBS). The tissue was then isolated by centrifugation (1 min × 12,000 g), cut into small pieces using fine scissors and re-suspended in 5 ml of Collagenase D (Roche, 11088866001) and Dispase (Stem cell technologies, 07923). The tissue was allowed to dissociate by incubating the suspension in a cell culture incubator at 37° in 5% CO2.
Fluorescence Activated Cell Sorting
The dissociated tissue suspension was stained for β-galactosidase activity using the Fluorescein Di-β-D-Galactopyranoside system (Invitrogen, D2920) as per the manufacturer's instructions. Cells were then run through a BD FACS Aria III fusion machine, and flow cytometry analysis performed on a BD LSR Fortessa machine. Data analysis was with FlowJo v10 software.
Tissue Processing
Postnatal day 5–7 (p 5–7) homozygous XlacZ4 or CD1 pups were sacrificed and teeth extracted into phosphate buffered saline (PBS). Tissues were fixed in 4%, 0.2% glutaraldehyde for 12 h in preparation for X-Gal staining. Following PBS washes, samples were decalcified in 19% ethylenediaminetetraacetic acid (Sigma, E3889) at 4°C for 2 days and then washed in PBS before an initial sucrose dehydration (15% Sucrose, 2 mM MgCl2, PBS) overnight at 4°C, followed by a second dehydration (50% Sucrose, 50% OCT, 2 mM MgCl2, PBS) for 6 h at room temperature. Finally, teeth were embedded in optimum cutting temperature compound (OCT, Thermo Fisher, UK) and stored at −80°C. Samples were sectioned at 8 μm on the Bright OTF5000 Cryostat and mounted on superfrost slides (4951Plus, Thermo Fisher, UK), then frozen at −80°C or used immediately for immunohistochemistry.
X-Gal Staining and DAB Immunohistochemistry
Slides were post-fixed in 0.2% glutaraldehyde for 10 min on ice, and then washed in ice cold PBS containing 2 mM MgCl2, prior to incubation with detergent rinse solution (10 mM MgCl2, 0.005% Sodium deoxycholate, 0.01% IGEPAL CA-630, PBS) for 10 min on ice. Slides were then placed in X-Gal staining solution [2 mM MgCl2, 0.005% Sodium deoxycholate, 0.01% IGEPAL CA-630, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 10 mM Tris-HCL pH7.3, 0.8 mg/ml X-Gal, PBS], at 37°C for 16 h. Slides were washed in PBS, and fixed in 4% PFA for 30 min at room temperature. Following 3 × 10 min PBS washes, endogenous peroxidase activity was blocked with 3% H2O2 (Acros, 20246500) for 15 min at room temperature. Following 3 × 10 min PBS washes, slides were incubated with blocking buffer (10% sheep serum, 0.1% Triton-X, 0.15% glycine, 2 mg/ml bovine serum albumin, PBS), for 1 h at room temperature. Subsequently slides were treated with primary antibody, diluted at a ratio of 1:300 in blocking buffer, overnight at 4°C. Unbound primary antibody was cleared by processing slides through 3 × 10 min washes of PBT2 (1% Tween-20, PBS), prior to incubation with biotinylated secondary antibody for an hour at room temperature. Unbound secondary antibody was subsequently cleared with 3 × 10 min PBT2 washes. Slides were incubated for 30 min at room temperature with avidin and biotinylated horse radish peroxidase (Vectastain Elite ABC HRP Kit, Universal, PK-6200) and washed 3 × 10 min in PBT2. Positive staining was visualized following addition of chromogen (Vectastain, Impact DAB peroxidase HRP kit) for 15–20 s. This reaction was stopped by placing in H2O and counterstained with haematoxylin for 30 s. For slides X-Gal stained, counterstaining was completed with exposure to Nuclear Fast Red (Sigma, N3020) for 3–5 s. Slides were dehydrated with increasing ethanol concentrations (70%, 90%, 95%, and 2 × 100%) before final xylene treatment (Sigma, 214736) 2 × 10 min. Sections were mounted with DPX (Merck, 100579), and images acquired on the Nikon Eclipse Ci. Antibodies used: Rabbit anti CD146 (Abcam, 75769), Rabbit anti CD105 (Abcam, 169545), Rabbit anti- Pdgfrb (Abcam, 32570), Biotinylated goat anti rabbit IgG (Vector laboratories, BA-100), Biotinylated rabbit anti-goat IgG (Vector laboratories, BA-500). All primary antibodies were used at a concentration of 1:300, and all secondary antibodies at a concentration of 1:500, in blocking buffer.
Transcriptomic Profiling
Dental pericytes (Figure 1A) were isolated as described above (illustrated in Figure 1C). Total RNA was obtained using the “Quick- RNA MicroPrep” kit (Zymo Research, R1051) according to manufacturer's instructions. Following poly-A selection, cDNA libraries were generated using SMARTer (Clontech, 634925). Barcoded libraries were then pooled and sequenced on an Illumina HiSeq 4000 (75 bp, paired end) at the Wellcome Trust Centre for Human Genetics (Oxford, United Kingdom). The paired-end libraries underwent quality control checks and trimming to filter out unreliably called nucleotides and to trim them from Illumina primers that were used during the library synthesis step. These fastq reads where then aligned to the mouse genome (GRCm38/mm10) using an RNAseq specific aligner. To quantify gene expression levels, count tables were generated using the mouse mm10 transcriptome as reference. The count tables of each sample group were then compared and subjected to expression testing to identify differentially expressed genes between the two conditions (Figure 1D). Raw reads were mapped to GRCm38/mm10 using Hisat2, and EdgeR together with the Cufflinks pipeline were used to identify genes with differential gene expression. Feature counts was also used to generate count tables for every sample. A gene was classified as being differentially expressed if it had a q < 0.05. The following algorithms were used for RNA-seq analysis: Hisat v2.0.3 (22), DESeq2 v2.11.38 (23), FeatureCounts v1.4.6p5 (24), the Cufflinks suite (25, 26), and EdgeR (27). Single cells were isolated as mentioned using FACS and RNA transcripts captured using the Smart-Seq2 protocol (28).
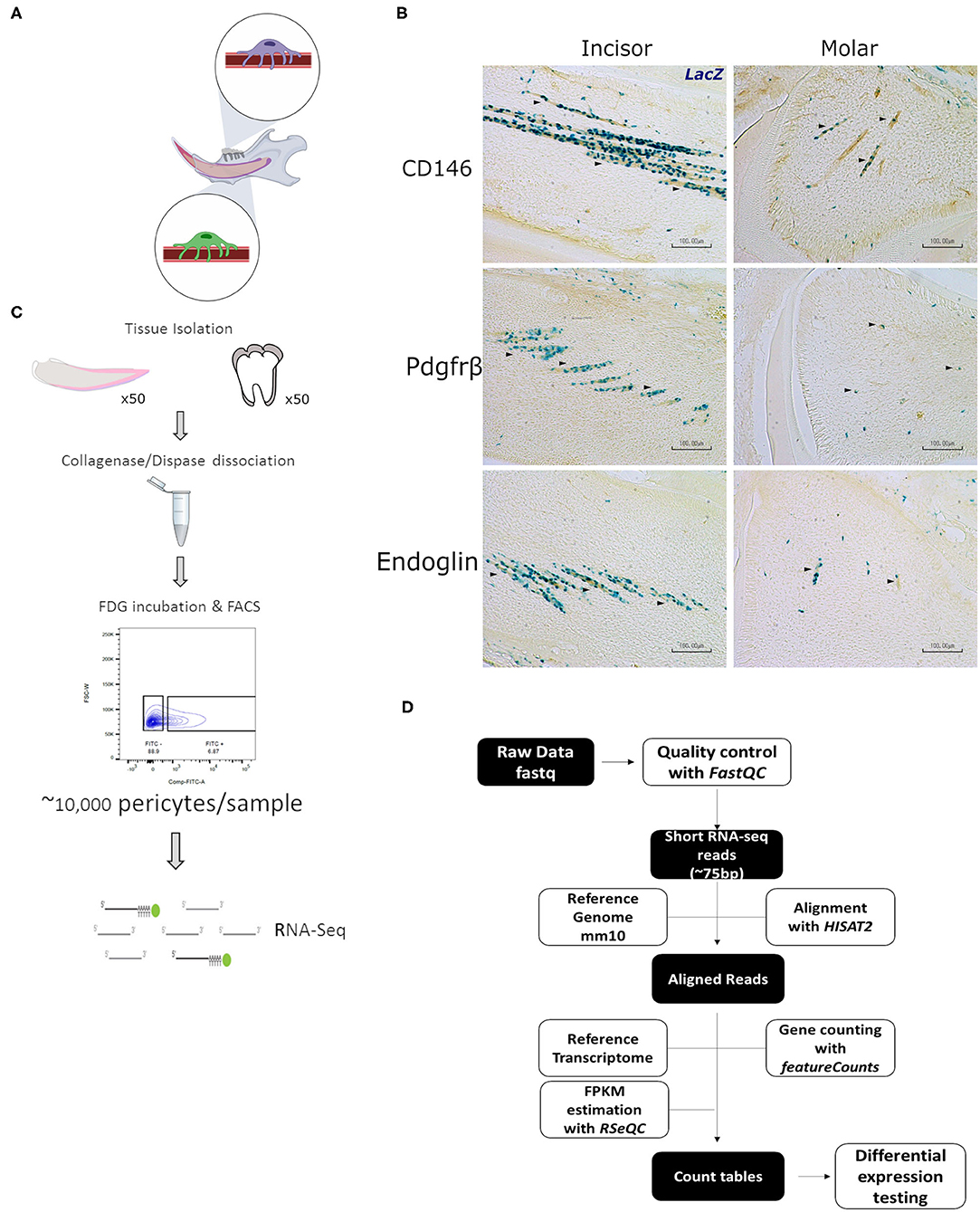
Figure 1. Characterization and isolation of LacZ+ cells. Schematic representation of mandibular area that is dissected for experiments indicating pericytes in incisor and molar (A). LacZ+ are seen to be perivascular to blood vessels in both incisor and molar pulp as shown by traditional staining using X-Gal. Immunohistochemical staining indicates that LacZ+ cells co-express CD146, platelet derived growth factor receptor β, and Endoglin as indicated by the arrowheads (B). Diagram indicative of the fluorescence activated cell sorting (FACS) strategy to isolate pericytes from incisor and molar pulp of XLacZ4 mice (C). Overview of RNA-seq bioinformatic analysis pipeline (D). This included, quality check, filtering of low quality reads, and align raw sequencing reads obtained from the sequencing experiment. These reads were then processed as shown to map to the mouse reference genome (mm10) and the constructed transcriptomes compared to find statistically significant differences in gene expression between incisor and molar pericytes (D).
Data Sharing
Molar RNA-seq data and single cell RNA-seq data are deposited in GEO (NCBI) under GSE150953. Incisor RNA-seq data are deposited under bioproject number: PRJNA420442 (16).
Results
XlacZ4+ Pericytes in Incisors and Molars Express Gene Markers Used to Identify Pericytes in vivo
Using post-natal pups homozygous for the XlacZ4 transgene we sought to isolate lacZ positive cells that have previously been validated to be pericytes. As suggested by the name, pericytes in all organs are found on the abluminal side of blood vessels (29). This is true for the majority of the mesenchymal compartments of multiple organs (4, 30). In mouse incisors, pericytes can be found intimately associated with blood vessels running the length of the central portion of the tooth. In molars, due to the more disorganized orientation of blood vessels, pericytes appear to be dotted around the mesenchyme (Figure 1B). Isolation of tissue from XLacZ4 animals, and subsequent histological processing showed, as expected, that in mouse incisors and molars, β-galactosidase expression, assayed using X-gal staining identifies cells intimately associated with blood vessels. In addition to their perivascular localization, these cells also express markers known to be expressed by pericytes in vivo. These include CD146, PDGF-Rβ, and Endoglin (Figure 1B). LacZ positive cells isolated from the XLacZ4 mouse have also been previously shown to be negative for a number of endothelial markers including Pecam1 (CD31), Nos3, Kdr, and Cdh5 (16). Having confirmed by both positive and negative selection that these cells are pericytes, we used a fluorescence activated cell sorting strategy to isolate the cells. The cell isolation process has been previously described (16) and detailed in the methods section. A number of documented bioinformatic tools were used to analyse the resulting RNA-seq libraries (Figure 1D). In brief, sequencing reads were quality checked and the integrity of the sequencing libraries was evaluated (Supplementary Figure 1). Reads identified as being of high quality were processed to generate count tables which were then subjected to differential expression testing to identify genes that were significantly dissimilar between the two pericyte populations (Figure 1D).
Molar and Incisor Pericytes Are Similar but Not Identical in Function
To confirm the identity of the FACS isolated (FDG+ve) cells as pericytes we firstly examined the gene transcription profiles of known marker genes that are reported as being expressed. Genome browser views confirmed high expression of poly-A captured sequencing reads mapping to the gene locus and exons of Acta2 (encoding αSMA), Mcam (CD146), Nes (nestin) (Figure 2A). All three of these are extensively described in the literature as being expressed in pericytes in multiple organs (4, 31). Isolated cells also expressed genes consistent with pericytes present in dental structures being of cranial neural crest origin (32, 33). This was demonstrated by their high expression of neural crest markers “Snail family transcriptional repressor 1” and 2 (Snai1 and Snai2) (Figure 2A). Following processing of sequencing reads and gene counting of each of the RNA-seq libraries, normalized FPKM tables were generated for each sample group. This allowed the identification of all genes expressed by pericytes. Comparing the expression of detected genes in the two groups it was evident that the overwhelming majority of genes expressed (n = 14,524) were shared by pericytes isolated from both molars and incisors (Figure 2B). Molar pericytes expressed a further 3,980 genes that were not detected in incisor pericytes. Respectively, incisor pericytes expressed 267 genes that could not be detected in molar pericytes (Figure 2B). The shared 14,524 genes were taken and categorized into protein families. Breakdown of this showed that the majority of shared genes were nucleic acid binding, such as transcription factors or co-factors (Figure 2C). This is consistent with what was expected as it would be assumed that pericytes irrespective of tissue of isolation, have similar transcription factor profiles. Three highly expressed genes encoding transcription factors, Twist1, Hmgb2, and Bzw1 are shown as illustrative examples (Figure 2D). Both Twist-1 and Hmgb2 have previously been shown to be important for maintenance of mesenchymal (stem) cells (34–37).
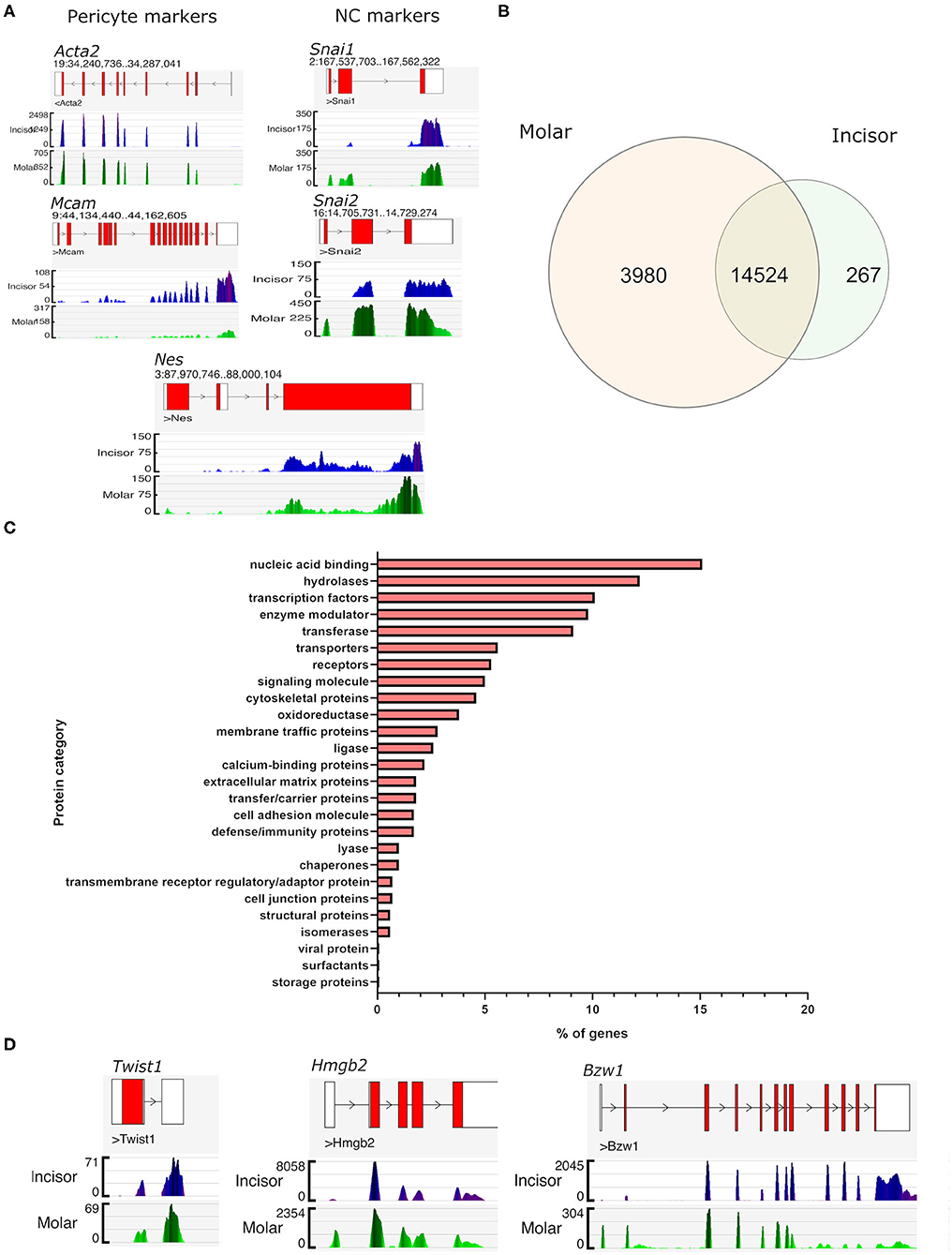
Figure 2. Gene expression similarities between incisor and molar pericytes. Genome browser views of the aligned RNA-seq libraries confirming expression of pericyte markers in both populations, including Acta2 (alpha smooth muscle actin), and Mcam (CD146). Genome browser views also confirm that both populations express neural crest cell markers as expected. These included Snai1, Snai2, and also Nes which has also been described as a pericyte marker (A). Using FPKM tables derived from the aligned RNA-seq data we identified all genes expressed by pericytes in each location and overlapped them to identify genes expressed by both populations (prior to differential expression testing). This revealed that an overwhelming majority of genes (n = 14,524) was shared by pericytes irrespective of their isolation from molar or incisor (B). Molar pericytes expressed a further 3,980 genes that were not detected in incisor pericytes. Respectively, incisor pericytes expressed 267 genes that could not be detected in molar pericytes (B). Breakdown of protein families that genes shared between the two pericyte populations contribute to (C). Illustrative Genome browser views of transcription factors expressed by both pericyte populations (D).
Differential Expression Testing Suggests Pericytes Have an Immunomodulatory Role in Molar Pulp Cells
Differential expression testing was performed to identify genes whose expression level is statistically different between the molar and incisor pericyte populations. The analysis revealed that 1,104 genes were significantly upregulated in molar pericytes. Contrastingly, 939 genes were significantly upregulated in the incisor pericyte population (Figure 3A). Genes upregulated in the incisor pericyte population were previously described and these have been shown to relate to their odontogenic capabilities (16). Enrichment analysis on the 1,104 genes upregulated in molar pericytes, revealed that they do not carry the same odontogenic gene expression signature as their incisor counterparts. Surprisingly, these genes are driving enrichment of a number of immunologically flavored pathways that regulate the immune system (Figure 3B). This highlights a previously unreported possible function of pericytes in molars (38). Enrichment analysis showed that cytokine and chemokine signaling makes a strong component of molar pericyte transcriptomic identity. The top enrichment pathways identified relating to an immune function are “chemokine signaling,” “lymphocyte chemotaxis,” “cytokine signaling,” and “response to tumor necrosis factor” (Figure 3B). Investigating the datasets revealed that molar pericytes express a number of cytokines and chemokines that are driving the enrichment of these pathways including “Ccl2,” “Ccl3,” “Ccl4,” “Ccl7,” and “Ccl22” (Figure 3C) From the chemokine family, we detect expression of “Cxcl1,” “Cxcl2,” “Cxcl9,” “Cxcl10,” and “Cxcl12.” A number of these are known potent chemo-attracters such as Cxcl1 for neutrophils (39) Cxcl2 for polymorphonuclear leukocytes (40) and Cxcl9 & 10 shown to promote extravasation of multiple white blood cells (41).
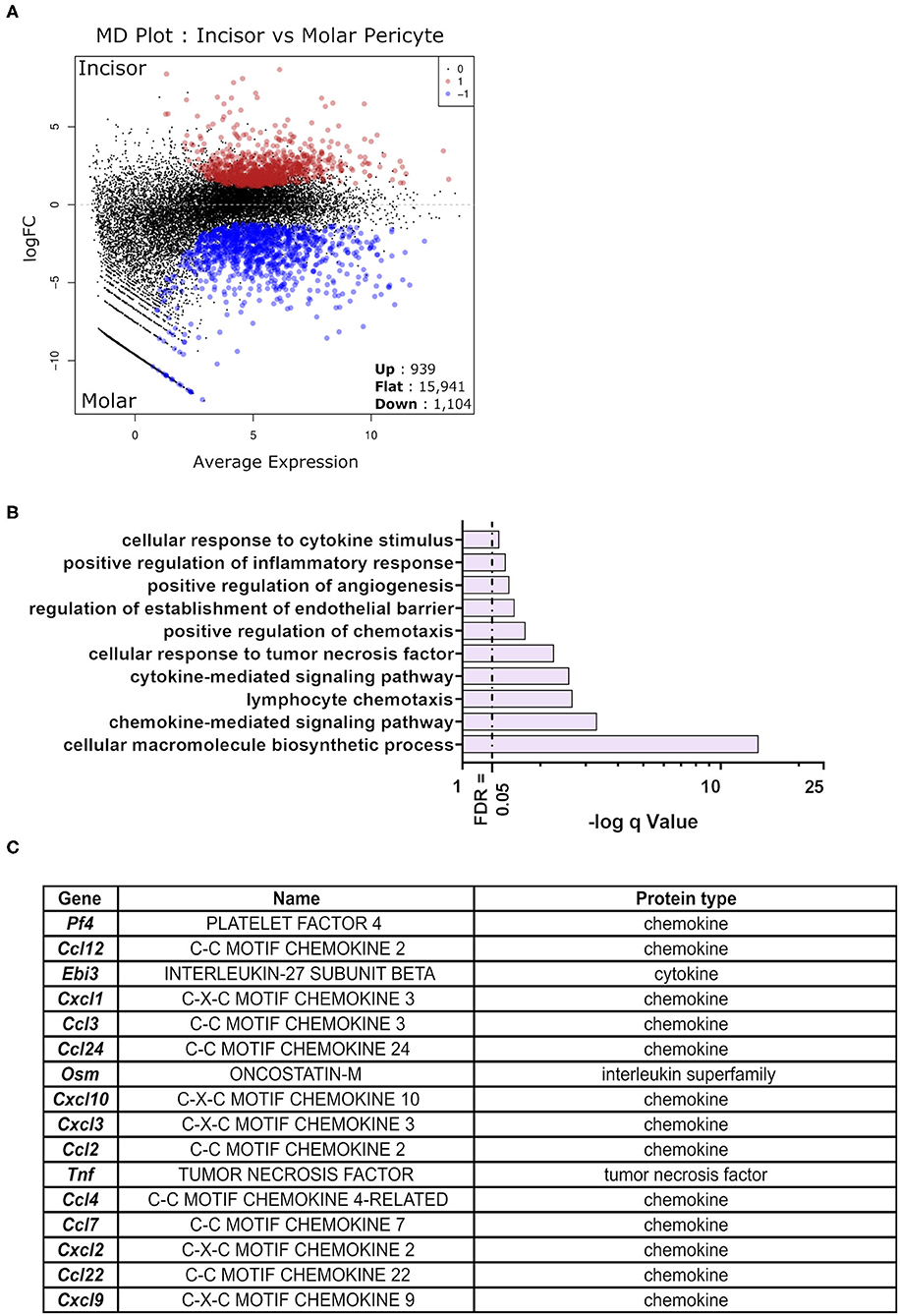
Figure 3. Differential expression testing on detected gene expression differences. MD plot showing the results post-differential expression testing on the sequenced RNA-seq libraries. Identifying 939 being significantly upregulated in incisor pericytes as opposed to 1,104 genes significantly upregulated in molar pericytes. Majority of genes detected was not significantly differentially expressed between the two populations (n = 15,941) (A). Pathway enrichment analysis on genes significantly upregulated in molar pericytes (n = 1,104) identifies key pathways that are active in these cells. An ongoing theme appears to be the involvement of these cells in cytokine production and chemotactic signaling (B). Table showing cytokines and chemokines identified to be transcribed by molar pericytes (C).
Single Cell RNA-Seq Using SmartSeq-2 Reveals Perivascular Sub-populations
It has previously been shown that the XLacZ4 transgene is expressed in populations of pericytes but no other marker is co-expressed with all lacZ+ cells in vivo (16, 19). Therefore, we were interested in identifying pericyte subpopulations and pericytes that were labeled by lacZ+ but not captured using traditional immunolabeling. Single cell RNA-seq was performed utilizing the SmartSeq-2 method (28) on molar and incisor XLacZ4 pericytes. Pericytes were captured from each organ, and bioinformatically analyzed using the Seurat pipeline (42). This revealed 3 clusters (Figure 4A) that did not show any distinction in clustering based on organ of isolation (Figure 4A′). We then performed an in silico analysis external to “Seurat” to determine if we could identify immature vs. mature pericytes. This was done using “CytoTrace” (43) which revealed two groups. Clusters “0” and “2” had a high “cytoTrace” signal indicating more immature cells while cluster “1” showed a low “cytoTrace” signal, indicating a more mature cluster (Figure 4B). Genes representing the transition from high to low cytoTrace (i.e., less to more mature) are also shown (Figure 4B′). When these genes were investigated using the original clustering pipeline to identify their localization in the dataset, high cytoTrace genes were clearly expressed in clusters “0” and “2” (Figure 4C). Conversely, low cytoTrace genes indicating more mature populations, were expressed in cluster “1” (Figure 4C′). These genes provide novel information regarding segregation of pericytes in vivo, and new markers that can be utilized to lineage-trace these cells. Expression of chemokines and cytokines previously shown as upregulated in the bulk-RNA-seq were also investigated. These were highly expressed in clusters “0” and “2” of both organs. Single cell-RNAseq also corroborated the previously shown bulk RNA-seq data as these are expressed highly in molar sub-populations (Figure 4D). Investigating these clusters for expression of commonly used pericyte markers such as CD146 (Mcam), and alpha smooth muscle actin (Acta2) showed these to be expressed ubiquitously (Figure 4D′). It is key to note the lower expression of these markers in clusters “0” and “2” which harbor the immune signature. This was not unexpected as evidence shows that pericytes with an immunomodulatory phenotype downregulate expression of classical pericyte markers such as alpha smooth muscle actin (44).
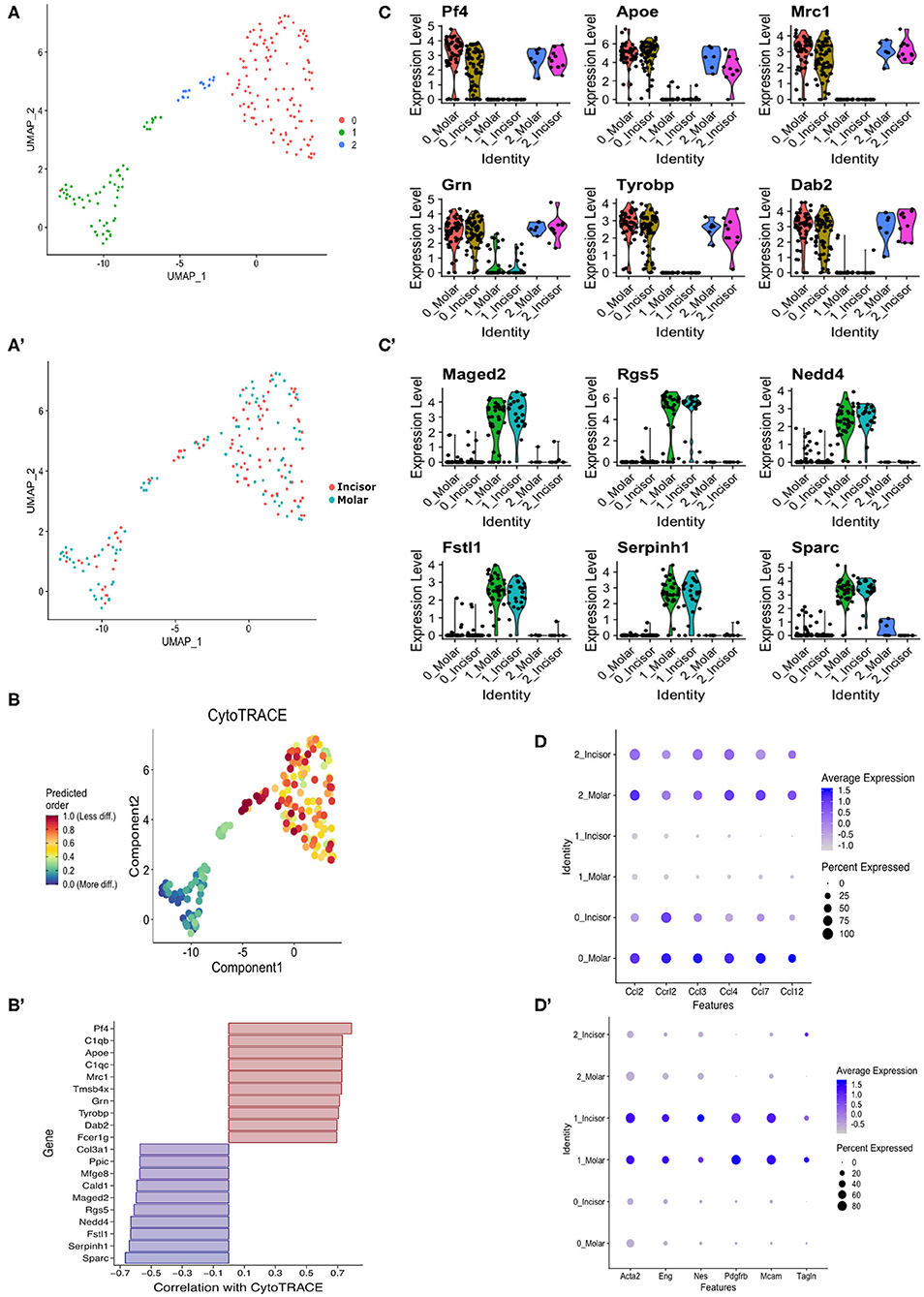
Figure 4. In silico stem cell prediction. UMAP clustering of incisor and molar pericytes identifies 3 transcriptionally distinct populations (A). There is no segregation of cells based on tissue of isolation (A′). CytoTRACE in silico analysis indicates that sub-cluster “0” is predicted as being less differentiated than the sub-cluster “1” and “2” (B). Genes correlated with CytoTRACE are shown with genes of high CytoTRACE correlation indicating expression in less differentiated cells (more stem) and low correlation indicating correlation with more differentiated cells (B′). Investigating CytoTRACE genes in sub-clusters identified tight correlation of positive CytoTRACE genes with cluster “0” and to a lesser extent “1” (C) and genes of negative CytoTRACE correlation with sub-cluster “1” (C′). Dotplot showing expression of previously identified cytokines in pericyte subpopulations (D). Dotplot showing expression of commonly used pericyte markers across pericyte subpopulations (D′).
Characterization of a Novel Marker Identifying Pericytes With Odontogenic Potential in vivo
During the RNA-seq screening a gene candidate emerged (Tagln) that was highly expressed in both molar and incisor datasets (Figure 5A). This was further investigated in the sc-RNAseq derived datasets where it showed high expression in the mature pericyte clusters (“1”), but also in a minority of cells in the immature clusters (“0” and “2”) (Figure 4D′). The expression signature of Tagln is thus similar to that of other commonly used markers for mature pericytes such as Endoglin, CD146, Platelet derived growth factor receptor beta and Nestin (Figure 4D′).
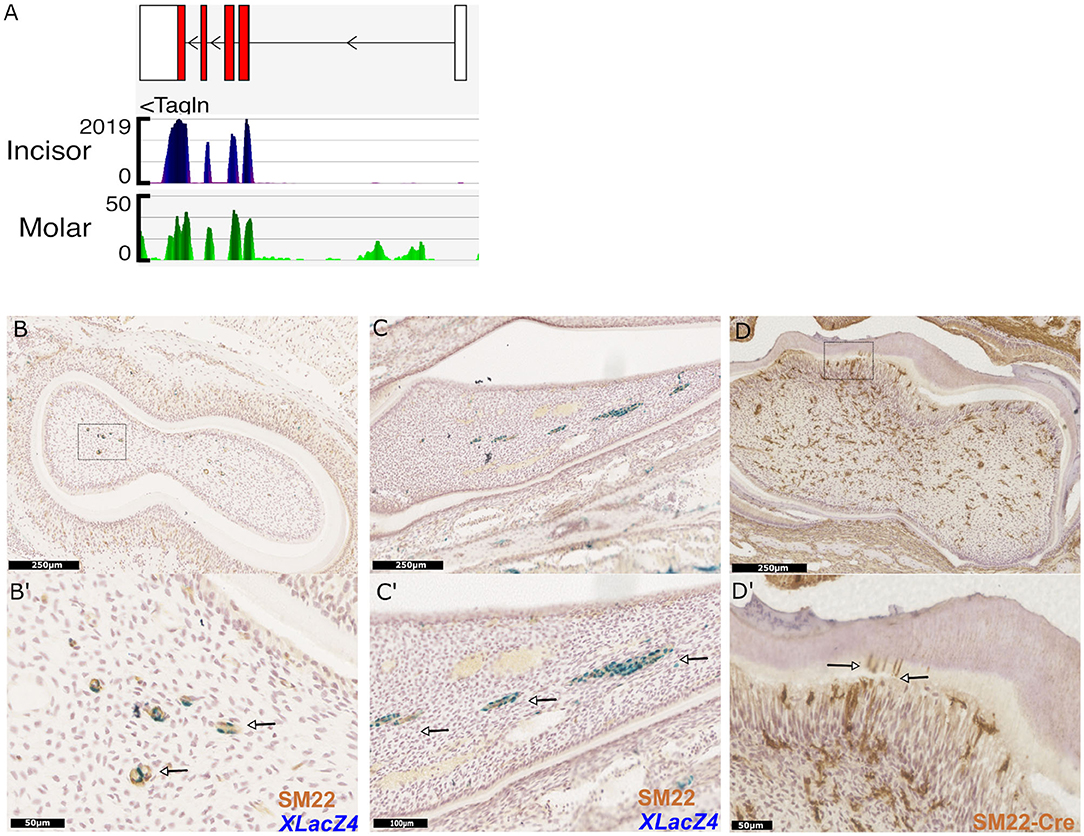
Figure 5. Identification and validation of a novel marker for dental pericytes. Genome browser views of Tagln (SM22), highly expressed in RNA-seq libraries constructed from isolated incisor and molar pericytes (A). Immunhistological staining of molar and incisor cryosections confirming co-expression of LacZ and SM22 (B,C respectively; magnification shown in B′,C′ with arrows indicating co-expression.) Immunohistological staining following genetic lineage tracing in TaglnCre-mTmG identifies odontoblasts positive for GFP expression in molars. Dentin tubules protruding into the dentine are also identified as shown by the arrows (D,D′).
Due to its high expression in pericytes in silico, we sought to investigate expression of the mature protein encoded by the Tagln gene, Transgelin (Sm22) in vivo. X-Gal staining was performed on cryosections of teeth isolated from XLacZ4 mice, followed by immunostaining using an Sm22 specific primary antibody. This validated expression of Sm22 in both molar and incisor pericytes positive for expression of lacZ (Figures 5B,B′,C,C′). Expression of Sm22 could not be detected in cells that were negative for expression of lacZ. Using a transgenic mouse line expressing Cre-recombinase under the control of the Tagln promoter we utilized genetic lineage tracing to identify the progeny of Sm22 expressing cells. Isolating tissue and performing immunohistological analysis on TaglnCre;mTmG molars indicated that cells expressing Sm22 gave rise to mature odontoblasts (Figure 5D). Their identity as odontoblasts being evident from their positioning under the dentine matrix, and also the apparent processes of labeled cells that extend into the dentine matrix (Figure 5D′). Sm22 is thus a marker that can be used to identify dental pericytes in vivo that have been shown to have odontogenic capabilities as they give rise to odontoblasts. Sm22+ pericytes are not only found in tooth pulp since data mining single cell datasets obtained from other organs revealed Sm22+ perivascular cells exist in the mesenchyme of lung (45), kidney (46), pancreatic islets (47), and bone marrow (48).
Discussion
Mesenchymal stem cell precursors, in the form of pericytes, reside in virtually all vascularised tissues (4). Their localization on the abluminal side of blood vessels, supports trophic, and mechanical support functions in maintaining endothelial cell networks (7). Expansion of pericytes in vitro shows they can give rise to MSC-like cells that irrespective of their tissue of origin can be stimulated to differentiate into mature mesenchymal lineage cells (49, 50). A number of studies have shown that pericytes isolated from mouse or human tissues are not equivalent in their transcriptomes or epigenomes and this is reflected in their differentiation potential in vivo and in vitro (13, 15, 16). Here we expand on this theme and investigated differences in pericyte transcriptomes isolated from the same tissue in the same organ that differ only in their adult growth. This in depth transcriptomic analysis revealed that pericytes in molars play a previously unidentified role in regulating the immune system, something which they do not appear to be doing to such an extent in incisors. These pathways implicate molar pericytes as being involved in chemokine signaling, cytokine signaling, and leukocyte activation. We also highlight a number of molecules as illustrative examples secreted by these pericytes that are driving enrichment of the above mentioned pathways. Pericytes have previously been reported to secrete immunomodulatory factors in many organs (51) or when seeded in specialized scaffolds in vitro (52). Pericytes have been shown to play a role in neutrophil trans-migration into the brain indicating that they are major contributors to immune defense systems (44, 53). Similarly, NG2 positive pericytes have been shown to regulate the movement of neutrophils across the basement membrane of mesenchymal tissues such as in muscle (53) in addition to secreting additional chemo-attractants to recruit more macrophages (54). This process of adopting an immunoregulatory phenotype has been attributed in part to a response to inflammatory factors such as interferon gamma (IFNγ) and Tumor necrosis factor alpha (TNFα) when secreted in the niche from supporting cells (55). Indeed in vivo studies have shown pericyte populations can constitutively release interleukins that are playing a role in regulating the local immune response in response to sterile inflammation (56). This immunoregulatory shift in pericyte behavior has been reported to interfere with expression of commonly used pericyte markers such as alpha smooth muscle actin (αSMA) (44). This would indicate that isolation strategies such as fluorescence activated cell sorting, relying on commonly used pericyte markers, may have previously missed these immunomodulatory pericytes. In the current study we show using single cell RNA sequencing that pericyte sub-populations exist in incisors and molars. A previously unidentified immune modulating sub-population exists in vivo that could potentially be playing a role in the context of sterile inflammation. Targeting this sub-population could help understand how pulp mesenchymal stem cells responds in cases of acute inflammation such as in damage (dental caries). A key question is why these “immune” pericytes are present in molar pulp but not incisor pulp? Both tooth types are subject to damage, odontoblast destruction, and pulp infection, however the key distinction is that in continuously growing incisors, pulp cells are completely replaced every 4 days and any infected cells would thus be rapidly cleared from the tooth tips. In non-growing molars however, little proliferation of pulp cells is observed and thus whereas infection in the incisors can be considered a transient event, molar infection needs to be controlled and an “immune” pericyte sub-population may be an important part of this control.
The other identified population has a more defined “mature pericyte” immunophenotype corresponding to cells co-expressing a number of commonly used molecules such as Endoglin, CD146 and alpha smooth muscle actin. An as of yet unreported gene Tagln was identified as being highly expressed in dental pericytes. This was confirmed be true in vivo using histological staining.
Remarkably, when lineage tracing Tagln expressing cells using an non-inducible Cre mouse, we identified that these pericytes differentiated into mature odontoblasts. Together, RNA-seq data presented here offer a valuable resource for the dental research community in addition to describing previously unappreciated cellular sub-populations that can act as additional targets for clinical intervention. Although there is a wealth of evidence showing that cultured stromal stem cells from many different tissues, including dental pulp, have effects on immune cell behavior in vitro, very little is known about the extent any of these immune modulatory processes occur in vivo (57, 58). Indeed, the most compelling in vivo evidence suggests the immune modulation observed when cells are injected systemically is indirect, occurring as a result of apoptosis of cells trapped in the lungs (59, 60). Since mobilization of endogenous (resident) stem cells is an intensive area of investigation in clinical regenerative approaches, including dentistry, an understanding of tissue-specific properties of the cells and their precursors is important to develop approaches that can be individually tailored to each tissue. Our results presented here showing that stem cell precursors in tooth pulp have transcriptome profiles suggestive of distinct organ-specific immune modulatory functions, can provide a platform for understanding the extent to which these cells modulate key reparative processes such as inflammation.
Data Availability Statement
Molar RNA-seq data and single cell RNA-seq data are deposited in GEO (NCBI) under GSE150953. Incisor RNA-seq data are deposited under bioproject number: PRJNA420442.
Ethics Statement
This animal study was reviewed and approved by KCL BSU.
Author Contributions
VY: collected and analyzed data. PS: acquisition of funding. VY and PS: experimental design, wrote, and reviewed the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
Research in the authors' laboratory was funded/supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St. Thomas' NHS Foundation Trust and King's College London and/or the NIHR Clinical Research Facility.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors would like to thank Ms. Hessah Alrubaian for aid optimizing the IHC protocols, Ms. Fernanda Suzano for genotyping services, Ms. Dhivya Chandrasekaran for mouse maintenance, the BRC flow cytometry core at Guy's Hospital for their services and ongoing support with flow cytometry, and the Wellcome Trust Centre for Human Genetics at Oxford University for their sequencing services.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdmed.2020.00006/full#supplementary-material
Supplementary Figure 1. Quality statistics on aligned RNA-seq datasets. Standard quality control checks were performed on datasets following the alignment to the mouse reference genome (mm10) prior to differential expression analysis. This showed that all samples sequenced had both high nucleotide calling accuracy, and sequence quality. Moreover, the datasets showed very little degree of duplication. We sequenced two biological replicates for incisor pericytes (A,B) and two for molar pericytes (C,D).
References
1. Wislet-Gendebien S, Laudet E, Neirinckx V, Alix P, Leprince P, Glejzer A, et al. Mesenchymal stem cells and neural crest stem cells from adult bone marrow: characterization of their surprising similarities and differences. Cell Mol Life Sci. (2012) 69:2593–608. doi: 10.1007/s00018-012-0937-1
2. Wexler SA, Donaldson C, Denning-Kendall P, Rice C, Bradley B, and Hows JM. Adult bone marrow is a rich source of human mesenchymal 'stem' cells but umbilical cord and mobilized adult blood are not. Br J Haematol. (2003) 121:368–74. doi: 10.1046/j.1365-2141.2003.04284.x
3. Uccelli A, Moretta L, and Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. (2008) 8:726–36. doi: 10.1038/nri2395
4. Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. (2008) 3:301–13. doi: 10.1016/j.stem.2008.07.003
5. Caplan AI. All MSCs are pericytes? Cell Stem Cell. (2008) 3:229–30. doi: 10.1016/j.stem.2008.08.008
6. Rouget C. Mémoire sur le développement, la structure et les propriétés physiologique des cappilaires sanguins et lymphatiques. Arch. Physiol. Norm. Pathol. (1873) 5:603–63.
7. Traktuev DO, Merfeld-Clauss S, Li J, Kolonin M, Arap W, Pasqualini R, et al. A population of multipotent CD34-positive adipose stromal cells share pericyte and mesenchymal surface markers, reside in a periendothelial location, and stabilize endothelial networks. Circ Res. (2008) 102:77–85. doi: 10.1161/CIRCRESAHA.107.159475
8. Huang GT, Gronthos S, and Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res. (2009) 88:792–806. doi: 10.1177/0022034509340867
9. Sung JH, Yang HM, Park JB, Choi GS, Joh JW, and Kwon CH. Isolation and characterization of mouse mesenchymal stem cells. Transplant Proc. (2008) 40:2649–54. doi: 10.1016/j.transproceed.2008.08.009
10. Romanov YA, Svintsitskaya VA, and Smirnov VN. Searching for alternative sources of postnatal human mesenchymal stem cells: candidate MSC-like cells from umbilical cord. Stem Cells. (2003) 21:105–10. doi: 10.1634/stemcells.21-1-105
11. Gerlach JC, Over P, Turner ME, Thompson RL, Foka HG, Chen WC, Peault B, Gridelli B, and Schmelzer E. Perivascular mesenchymal progenitors in human fetal and adult liverStem Cells Dev. (2012) 21:3258–69. doi: 10.1089/scd.2012.0296
12. Covas DT, Panepucci RA, Fontes AM, Silva WA Jr, Orellana MD, Freitas MC, et al. Multipotent mesenchymal stromal cells obtained from diverse human tissues share functional properties and gene-expression profile with CD146+ perivascular cells and fibroblasts. Exp Hematol. (2008) 36:642–54. doi: 10.1016/j.exphem.2007.12.015
13. Supakul S, Yao K, Ochi H, Shimada T, Hashimoto K, Sunamura S, et al. Pericytes as a source of osteogenic cells in bone fracture healing. Int J Mol Sci. (2019) 20:1079. doi: 10.3390/ijms20051079
14. Feng J, Mantesso A, De Bari C, Nishiyama A, and Sharpe PT. Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proc Natl Acad Sci USA. (2011) 108:6503–8. doi: 10.1073/pnas.1015449108
15. Sacchetti B, Funari A, Remoli C, Giannicola G, Kogler G, Liedtke S, et al. No identical “mesenchymal stem cells” at different times and sites: human committed progenitors of distinct origin and differentiation potential are incorporated as adventitial cells in microvessels. Stem Cell Rep. (2016) 6:897–913. doi: 10.1016/j.stemcr.2016.05.011
16. Yianni V, and Sharpe PT. Molecular programming of perivascular stem cell precursors. Stem Cells. (2018) 36:1890–904. doi: 10.1002/stem.2895
17. Yianni V, and Sharpe PT. Perivascular-derived mesenchymal stem cells. J Dent Res. (2019) 98:1066–72. doi: 10.1177/0022034519862258
18. Yianni V, and Sharpe PT. Epigenetic mechanisms driving lineage commitment in mesenchymal stem cells. Bone. (2020) 134:115309. doi: 10.1016/j.bone.2020.115309
19. Tidhar A, Reichenstein M, Cohen D, Faerman A, Copeland NG, Gilbert DJ, et al. A novel transgenic marker for migrating limb muscle precursors and for vascular smooth muscle cells. Dev Dyn. (2001) 220:60–73. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1089>3.0.CO;2-X
20. Holtwick R, Gotthardt M, Skryabin B, Steinmetz M, Potthast R, Zetsche B, et al. Smooth muscle-selective deletion of guanylyl cyclase-A prevents the acute but not chronic effects of ANP on blood pressure. Proc Natl Acad Sci USA. (2002) 99:7142–7. doi: 10.1073/pnas.102650499
21. Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L. A global double-fluorescent Cre reporter mouse. Genesis. (2007) 45:593–605. doi: 10.1002/dvg.20335
22. Kim D, Langmead B, and Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. (2015) 12:357–60. doi: 10.1038/nmeth.3317
23. Love MI, Huber W, and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
24. Liao Y, Smyth GK, and Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. (2014) 30:923–30. doi: 10.1093/bioinformatics/btt656
25. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. (2010) 28:511–5. doi: 10.1038/nbt.1621
26. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. (2012) 7:562–78. doi: 10.1038/nprot.2012.016
27. Robinson MD, McCarthy DJ, and Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
28. Ramskold D Luo S Wang YC Li R Deng Q Faridani OR . Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. (2012) 30:777–82. doi: 10.1038/nbt.2282
29. Wang S, Cao C, Chen Z, Bankaitis V, Tzima E, Sheibani N, et al. Pericytes regulate vascular basement membrane remodeling and govern neutrophil extravasation during inflammation. PLoS ONE. (2012) 7:e45499. doi: 10.1371/journal.pone.0045499
30. Jiang Y, Vaessen B, Lenvik T, Blackstad M, Reyes M, and Verfaillie CM. Multipotent progenitor cells can be isolated from postnatal murine bone marrow, muscle, and brain. Exp Hematol. (2002) 30:896–904. doi: 10.1016/S0301-472X(02)00869-X
31. Chen CW, Montelatici E, Crisan M, Corselli M, Huard J, Lazzari L, et al. Perivascular multi-lineage progenitor cells in human organs: regenerative units, cytokine sources or both? Cytokine Growth Factor Rev. (2009) 20:429–34. doi: 10.1016/j.cytogfr.2009.10.014
32. Yamanishi E, Takahashi M, Saga Y, and Osumi N. Penetration and differentiation of cephalic neural crest-derived cells in the developing mouse telencephalon. Dev Growth Differ. (2012) 54:785–800. doi: 10.1111/dgd.12007
33. Etchevers HC, Vincent C, Le Douarin NM, and Couly GF. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. (2001) 128:1059–68.
34. Meng T, Huang Y, Wang S, Zhang H, Dechow PC, Wang X, et al. Twist1 is essential for tooth morphogenesis and odontoblast differentiation. J Biol Chem. (2015) 290:29593–602. doi: 10.1074/jbc.M115.680546
35. Li Y, Lu Y, Maciejewska I, Galler KM, Cavender A, and D'Souza RN. TWIST1 promotes the odontoblast-like differentiation of dental stem cells. Adv Dent Res. (2011) 23:280–4. doi: 10.1177/0022034511405387
36. Cakouros D, Isenmann S, Cooper L, Zannettino A, Anderson P, Glackin C, and Gronthos S. Twist-1 induces Ezh2 recruitment regulating histone methylation along the Ink4A/Arf locus in mesenchymal stem cells. Mol Cell Biol. (2012) 32:1433–41. doi: 10.1128/MCB.06315-11
37. Taniguchi N, Carames B, Hsu E, Cherqui S, Kawakami Y, and Lotz M. Expression patterns and function of chromatin protein HMGB2 during mesenchymal stem cell differentiation. J Biol Chem. (2011) 286:41489–98. doi: 10.1074/jbc.M111.236984
38. Chen GY, and Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. (2010) 10:826–37. doi: 10.1038/nri2873
39. Schumacher C, Clark-Lewis I, Baggiolini M, and Moser B. High- and low-affinity binding of GRO alpha and neutrophil-activating peptide 2 to interleukin 8 receptors on human neutrophils. Proc Natl Acad Sci USA. (1992) 89:10542–6. doi: 10.1073/pnas.89.21.10542
40. Wolpe SD, Sherry B, Juers D, Davatelis G, Yurt RW, and Cerami A. Identification and characterization of macrophage inflammatory protein 2, Proc Natl Acad Sci USA. (1989) 86:612–6. doi: 10.1073/pnas.86.2.612
41. Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer Treat Rev. (2018) 63:40–47. doi: 10.1016/j.ctrv.2017.11.007
42. Satija R, Farrell JA, Gennert D, Schier AF, and Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. (2015) 33:495–502. doi: 10.1038/nbt.3192
43. Gulati GS, Sikandar SS, Wesche DJ, Manjunath A, Bharadwaj A, Berger MJ, et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science. (2020) 367:405–11. doi: 10.1126/science.aax0249
44. Pieper C, Marek JJ, Unterberg M, Schwerdtle T, and Galla HJ. Brain capillary pericytes contribute to the immune defense in response to cytokines or LPS in vitro. Brain Res. (2014) 1550:1–8. doi: 10.1016/j.brainres.2014.01.004
45. Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. (2017) 170:1134–48.e10. doi: 10.1016/j.cell.2017.07.034
46. Karaiskos N, Rahmatollahi M, Boltengagen A, Liu H, Hoehne M, Rinschen M, et al. A single-cell transcriptome atlas of the mouse glomerulus. J Am Soc Nephrol. (2018) 29:2060–8. doi: 10.1681/ASN.2018030238
47. Thompson PJ, Shah A, Ntranos V, Van Gool F, Atkinson M, and Bhushan A. Targeted elimination of senescent beta cells prevents type 1 diabetes. Cell Metab. (2019) 29:1045–1060.e10. doi: 10.1016/j.cmet.2019.01.021
48. Tikhonova AN, Dolgalev I, Hu H, Sivaraj KK, Hoxha E, Cuesta-Dominguez A, et al. The bone marrow microenvironment at single-cell resolution. Nature. (2019) 569:222–8. doi: 10.1038/s41586-019-1104-8
49. Zimmerlin L, Park TS, Donnenberg VS, Zambidis ET, and Donnenberg AD. Pericytes: a Ubiquitous Source of Multipotent Adult Tissue Stem Cells. Stem Cells in Aesthetic Procedures. Berlin; Heidelberg: Springer (2014). p. 135–48. doi: 10.1007/978-3-642-45207-9_9
50. Sims DE. Diversity within pericytes. Clin Exp Pharmacol Physiol. (2000) 27:842–6. doi: 10.1046/j.1440-1681.2000.03343.x
51. Sala E, Genua M, Petti L, Anselmo A, Arena V, Cibella J, et al. Mesenchymal stem cells reduce colitis in mice via release of TSG6, independently of their localization to the intestine. Gastroenterology. (2015) 149:163–76.e20. doi: 10.1053/j.gastro.2015.03.013
52. Menale C, Campodoni E, Palagano E, Mantero S, Erreni M, Inforzato A, et al. mesenchymal stromal cell-seeded biomimetic scaffolds as a factory of soluble rankl in rankl-deficient osteopetrosis. Stem Cells Transl Med. (2019) 8:22–34. doi: 10.1002/sctm.18-0085
53. Proebstl D, Voisin MB, Woodfin A, Whiteford J, D'Acquisto F, Jones GE, et al. Pericytes support neutrophil subendothelial cell crawling and breaching of venular walls in vivo. J Exp Med. (2012) 209:1219–34. doi: 10.1084/jem.20111622
54. Stark K, Eckart A, Haidari S, Tirniceriu A, Lorenz M, von Bruhl ML, et al. Capillary and arteriolar pericytes attract innate leukocytes exiting through venules and “instruct” them with pattern-recognition and motility programs. Nat Immunol. (2013) 14:41–51. doi: 10.1038/ni.2477
55. Krampera M, Cosmi L, Angeli R, Pasini A, Liotta F, Andreini A, et al. Role for interferon-gamma in the immunomodulatory activity of human bone marrow mesenchymal stem cells. Stem Cells. (2006) 24:386–98. doi: 10.1634/stemcells.2005-0008
56. Kovac A, Erickson MA, and Banks WA. Brain microvascular pericytes are immunoactive in culture: cytokine, chemokine, nitric oxide, and LRP-1 expression in response to lipopolysaccharide. J Neuroinflammation. (2011) 8:139. doi: 10.1186/1742-2094-8-139
57. Zhao Y, Wang L, Jin Y, and Shi S. Fas ligand regulates the immunomodulatory properties of dental pulp stem cells. J Dent Res. (2012) 91:948–54. doi: 10.1177/0022034512458690
58. Yamaza T, Kentaro A, Chen C, Liu Y, Shi Y, Gronthos S, et al. Immunomodulatory properties of stem cells from human exfoliated deciduous teeth. Stem Cell Res Ther. (2010) 1:5. doi: 10.1186/scrt5
59. Laing AG, Riffo-Vasquez Y, Sharif-Paghaleh E, Lombardi G, and Sharpe PT. Immune modulation by apoptotic dental pulp stem cells in vivo. Immunotherapy. (2018) 10:210–1. doi: 10.2217/imt-2017-0117
Keywords: pericyte, stem cell, mesenchymal, single cell RNAseq, bioinformatics, RNAseq, odontoblast, dentistry
Citation: Yianni V and Sharpe PT (2020) Transcriptomic Profiling of Dental Pulp Pericytes: An RNAseq Approach. Front. Dent. Med. 1:6. doi: 10.3389/fdmed.2020.00006
Received: 03 June 2020; Accepted: 14 July 2020;
Published: 25 August 2020.
Edited by:
Amerigo Giudice, University Magna Graecia of Catanzaro, ItalyReviewed by:
Claire Bardet, Université de Paris, FranceKun Xuan, The Fourth Military Medical University, China
Alessandra Pisciotta, University of Modena and Reggio Emilia, Italy
Copyright © 2020 Yianni and Sharpe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul T. Sharpe, cGF1bC5zaGFycGVAa2NsLmFjLnVr
 Val Yianni
Val Yianni Paul T. Sharpe
Paul T. Sharpe