- 1Department of Biology, San Francisco State University, San Francisco, CA, United States
- 2Department of Herpetology, California Academy of Science, San Francisco, CA, United States
- 3International Bird Rescue, Fairfield, CA, United States
The common murre (Uria aalge) is a species of seabird particularly vulnerable to several environmental stressors, including parasitic infection and oil contamination. However, the molecular response to these stressors is severely understudied. This study investigated the common murre’s transcriptomic responses to these stressors. Blood samples were collected from common murres undergoing rehabilitation at International Bird Rescue in Fairfield, CA. Total RNA was extracted from these samples, followed by library preparation and Illumina sequencing to generate whole transcriptome data. Differential gene expression analysis was conducted using DeSeq2 to identify genes significantly altered in response to oil contamination and parasitic infection. Differential gene expression analysis revealed 194 genes shared between oil-contaminated and infected birds, including key immune-related genes, such as ANXA2, LY96, and LY86. These genes play vital roles in mediating the production of pro-inflammatory cytokines. Gene Set Enrichment Analysis indicated significant alterations in stress, immune, and inflammatory responses, with additional lipid metabolism changes in contaminated birds. Our findings highlight the detrimental effects that these stressors have on wild birds. These findings suggest a generalized stress response and specific metabolic adaptations to oil exposure, providing insights for seabird conservation.
1 Introduction
Seabirds are highly vulnerable to many major environmental stressors such as pollution, oil spills, habitat loss, and harmful algal blooms (Phillips et al., 2011; Orgeret et al., 2022). They are also increasingly susceptible to disease from parasitic infections, with host–parasite interactions expected to significantly shift as a direct result of climate change (Khan et al., 2019). A seabird’s chance of exposure to these conditions may be increased when compared to other taxa because they have long lifespans and can traverse various continents throughout a single annual cycle (Boulinier et al., 2016). Behaviors such as migration, foraging, and dispersal have been shown to be important factors in the spread of infectious diseases and parasite transmission (Boulinier et al., 2016). The common murre (Uria aalge) is a representative of the seabirds in their susceptibility to the effects of climate change and other stressors (Phillips et al., 2011; Piatt et al., 2020). At wildlife rehabilitation centers, common murres are regularly admitted exhibiting symptoms and injuries related to oil contamination, anemia, disease, and starvation (Hampton et al., 2003; Henkel et al., 2014; Duerr et al., 2016). These environmental stressors can have significant health consequences for seabirds, including increased susceptibility to diseases like babesiosis.
Babesia is a protozoan parasite that is transmitted by ticks of the genus Ixodes, and infects the erythrocytes of mammalian, reptile, and avian hosts, resulting in the disease babesiosis (Puri et al., 2021; Zaki et al., 2021). Infection can lead to malaria-like symptoms such as fever, anemia, jaundice, and death in severe cases (Ristic and Kreier, 1984; Peirce et al., 2003; Vannier et al., 2015). Historically, babesiosis has had devastating impacts on the cattle industry and domestic animals, and a majority of Babesia research has focused on humans, mice, cattle, and dogs (Igarashi et al., 1999; Brown et al., 2006; Miles et al., 2012; Iwasaki and Medzhitov, 2015; Torina et al., 2020; Kumar et al., 2023), leaving a large gap in knowledge on avian species’ response to Babesia or apicomplexan parasites in general. The few investigations into the host’s response to infections have presented evidence of the innate and adaptive immune pathways playing a major role, with differential expression of genes relating to IL-1β, TNF-α, and IFN-γ (Brown et al., 2006; Beletic et al., 2021; Smith et al., 2021). The species Babesia uriae was first identified in 2009 in common murres undergoing rehabilitation and has been identified by light microscopy of blood smears in approximately 30% of murres admitted to International Bird Rescue’s California Wildlife hospital annually since 2016 (International Bird Rescue, unpublished; Yabsley et al., 2009). This parasite is closely related to B. poelea which infects brown boobies (Sula leucogaster) (Yabsley et al., 2009; Quillfeldt et al., 2011). Currently, there are 15 known species of avian Babesia which infect seabirds, and B. uriae is the first to be identified in a seabird in the Alcidae family (Peirce and Feare, 1978; Earle et al., 1993; Work and Rameyer, 1997; Merino, 1998; Peirce et al., 2003; Criado et al., 2006; Yabsley et al., 2006, 2009; Jefferies et al., 2008). Like other haemosporidian parasites in birds, acute infection is short and, if the individual survives this stage, results in chronic infection with lower parasitemia, regulated by the host’s immune system (Valkiūnas, 2005). Understanding how common murres respond to Babesia infection is crucial, as this parasite is becoming increasingly prevalent in seabird populations. Existing research on avian babesiosis has primarily focused on other species, but studies like Videvall et al. (2015) on Plasmodium infection in siskins provide valuable insights into potential immune system responses.
Plasmodium species and Babesia are similar in their evolutionary history, infection dynamics, and disease outcomes (Clark and Jacobson, 1998; Djokic et al., 2021; Inoue et al., 2013), so a similar response to both parasites in birds would be expected. Videvall et al. (2015) investigated the avian transcriptome changes to P. ashfordi infection in Eurasian siskins (Spinus spinus) at different stages of parasitemia and found that differential gene expression correlated with infection time. Moreover, gene set enrichment analysis (GSEA) was performed and found that gene ontology (GO) terms such as “immune system process,” “response to stress,” and “cell death regulation” were significantly overrepresented during peak infection (Videvall et al., 2015). Studies on the immune response to Babesia in mice have found further evidence of the innate immune response to mediate disease pathogenesis (Hemmer et al., 2000). Coupled with additional stressors that can activate an immune response, disease and symptoms can be dramatically exacerbated (Clark and Jacobson, 1998; Ye et al., 2020). In a recent study on Hawaiian honeycreepers, Hawai’i ‘Amakihi (Chlorodrepanis virens) were inoculated with P. relictum, the species of Plasmodium that led to the extinction of several species of honeycreepers (Paxton et al., 2023). This study focused on the host’s immune response to avian malaria and found that differences in adaptive and innate immune responses led to differences in host outcome to infection (Paxton et al., 2023). Similar to Babesia infection, oil contamination also poses a significant health threat to common murres.
In addition to pathogens, common murres also face the effects of oil spills, either from anthropogenic sources or from natural seepages (Hampton et al., 2003; Henkel et al., 2014). Seabirds exposed to petroleum oil risk feather contamination and ingestion, leading to severe health problems with direct impacts to the animal’s kidneys, liver, leukocyte populations, and blood cell physiology (Leighton et al., 1983; Fry and Lowenstine, 1985; Briggs et al., 1996). Feather contamination can cause a disruption of the bird’s outer plumage, which is crucial to their survival in the cold marine environment and leaves the individual at risk of hypothermia and death (Jenssen, 1994). Previous clinical studies in common murres have shown that ingestion of oil led to consistent decreases in body weight despite normal food consumption, indicating metabolic and malabsorption issues (Khan and Ryan, 1991). Another study on the immunosuppressive effects of oil and duck plague herpesvirus found that mallard ducks dosed with cyclophosphamide (CY) exhibited an immune response like that of duck plague herpesvirus, and a combination of both CY ingestion and duck plague herpesvirus resulted in higher death tolls than those with either CY or herpesvirus (Goldberg et al., 1990). Investigations into the transcriptome changes in response to oil contamination have primarily been performed on marine microbial species, coral, mice, and plants (Rivers et al., 2013; Alvarez et al., 2018; Liu et al., 2020; DeLeo et al., 2021). However, one transcriptomic study on seaside sparrows (Ammospiza maritima) that were indirectly exposed to petroleum in the Deep-Water Horizon (DWH) oil spill found significant upregulation of genes relating to hepatocellular proliferation and liver regeneration, while genes relating to necrosis, liver steatosis, and apoptosis were inhibited (Bonisoli-Alquati et al., 2020).
These two stressors, oil contamination and disease, are anticipated to increase in prevalence and severity due to anthropogenic climate change. In the case of oil contamination, changing ocean temperatures can lead to the release of previously trapped oil reserves by way of melting permafrost. In the case of Babesia, rising temperatures can expand the geographic range and increase the abundance of the invertebrate vectors of protozoan parasites, including Ixodes ticks. Investigating how a marine organism responds to these stressors can provide valuable insight into the potential impacts of climate change on marine organisms and can lead to long-term studies that can help inform conservation efforts, as well as develop strategies to protect vulnerable populations. The goal of this study was to map the changes in gene expression in response to differing environmental conditions that common murres, as well as other marine organisms face in the wild. To accomplish this broader goal, we identified differentially expressed genes in common murres infected with Babesia, as well as those contaminated with petroleum oil. We then performed GSEA to explore which biological pathways were enriched by differentially expressed genes in each condition. We hypothesized that both Babesia infection and oil contamination would lead to distinct transcriptional responses in Common murres, with each condition inducing expression changes of genes associated with biological pathways relating to immune functions. This work represents the first step in understanding the commonalities, differences, and underlying biological processes that occur in response to parasitic infection and oil contamination in seabirds.
2 Materials and methods
2.1 Sample collection
Biological samples used in this study were selected from a larger set of 261 Common murre samples collected by the staff at International Bird Rescue (IBR) in Fairfield, CA between May 13, 2021, to August 30, 2022 (Supplementary Table S1). Because of the cost of RNA sequencing, the number of biological replicates chosen for each condition was limited to three, including three samples from Common murres either infected with Babesia or contaminated with oil, used as a control group. While more replicate numbers are recommended for this type of analysis (ideally 12 per condition), DeSeq2 has been shown to be suitable for 3 or fewer replicated when the fold-change threshold is set to 0.5, and limma when the fold-change threshold is set to 2.0 (Schurch et al., 2016).
While it is difficult to obtain samples of healthy adult seabirds, several factors were taken into consideration to ensure there was as much similarity between the samples as possible. Even though all samples were collected over the duration of roughly a year, samples chosen for our control group were collected during the summer months: June and July of 2021, and June of 2022. All birds that were sampled were found beached in Santa Cruz County, California, and did not exhibit any major visible bodily injuries. They were listed in the IBR database as emaciated and dehydrated. Further, although one sample was aged as a hatch-year and the other two were identified as after hatch-year, all three samples were roughly similar in weight (600–800 g). All samples had relatively similar packed cell volume (PCV) values (>30%), normal total protein (TP) values (>4.0 g/dL), and normal buffy coat (BC) values (0–1%) (Newman et al., 1997). All birds were hydrated with a 5% by body weight 0.9% NaCl solution by the IBR staff before blood samples were taken. Any medication, supplements, and anti-parasitic were administered after sampling had taken place. Birds chosen for the oil contamination group were visually diagnosed with petroleum oil feather contamination and were reported to the Oiled Wildlife Care Network (OWCN) by IBR staff. To limit the amount of variability of each sample, it was ensured that samples chosen were not also later diagnosed with other major ailments that would also affect gene expression (i.e. fractured bones, joint luxation, etc.). Each bird was sampled at the time of entry to the rehabilitation center before any treatment was given.
For each sample, whole blood was collected using a 25-gauge needle attached to a 1 mL syringe from the medial metatarsal vein. Approximately 0.1 mL of blood was drawn from each bird, enough to be stored in Queen’s lysis buffer for DNA extraction (Longmire et al., 1997), in Invitrogen TRIzolTM LS ReagentTM (Thermo Fisher Scientific, Waltham, MA) for RNA preservation, as well as to make two blood smears for microscopy. The two blood smears were immediately set in methanol for fixation, and stained using JorVet Dip Quick Stain (Jorgenson Labs, Loveland, CO). Samples were frozen at −20°C until long-term storage was possible at −80°C at San Francisco State University prior to processing by the Avian Parasitology Laboratory.
2.2 DNA extraction and parasite detection
The Promega Wizard SV Genomic DNA Purification System was used to extract DNA from blood samples stored in Queen’s Lysis buffer following manufacturer’s instructions. Extracted DNA was stored at −20°C while unextracted DNA samples were kept at −80°C for long-term storage. 5’ and 3’ oligonucleotide primers [forward primer ITS15c (3’-CGATCGAGTGATCCGGTGAATTA-5’) and reverse primer ITS13b (5’-GCTGCGTCCTTCATCGTTGTG-3’)] were used, per Bostrom et al. (2008), and provided by Elim Biopharm Inc., Hayward, CA to amplify the ITS1 region of Babesia DNA. The thermocycler time and temperature profiles used are as follows: an initial activation step at 94.8°C for 1 min was followed by 35 cycles of amplification (94°C for 30 seconds, 62°C for 20 seconds, and 72°C for 30 seconds). The final extension was set at 72°C for 5 minutes, and then cooled to 4°C. Positive controls were identified using thin film microscopy and PCR, and PCR-grade water was used as a negative control. 91 common murre DNA extracts were tested for parasitic infection.
2.3 RNA extraction and sequencing
RNA was extracted from blood samples stored in TRIzol following the protocol in the Qiagen RNeasy Kit with the following modifications. A phenol-chloroform extraction was performed following the Qiagen RNeasy Lipid Tissue Mini Handbook to separate homogenate into aqueous and organic phases. Prior to extraction, samples were removed from −80°C and thawed to room temperature (approximately 15 minutes at room temperature). 200 mL of chloroform (ThermoScientific Chloroform 99% Extra Dry over Molecular Sieve, Stabilized AcroSeal) was added to the homogenate and shaken vigorously for 15 seconds and incubated on the benchtop at room temperature for 3 minutes. The samples were then centrifuged at 12,000 × g at 4°C for 15 minutes using an OWM Hermle Labnet Z 252 Mk Centrifuge. Standard extraction procedure was then applied until the column wash step. The Qiagen RNase-free DNase Set was added prior to the column wash procedure to ensure purity of the RNA. A final elution of 70 uL of RNase-free water was incubated on the column for 3 minutes and centrifuged at 12,000 g for 1 minute.
Total RNA was assessed for quality control using the Agilent Bioanalyzer Nano RNA chip (Agilent Technologies Inc. Santa Clara, USA) at San Francisco State University’s Genomics/Transcriptomics Analysis Core (GTAC). Peaks were assessed by RNA Integrity Number (RIN) values, which ranged from 7.3 to 9.6. A baseline RIN value of 5.0 was set to ensure proper quality. Due to budget restrictions, 3 individuals with the highest RIN value were selected for sequencing as representatives of each condition as biological replicates: Babesia-infected, petroleum oil-contaminated, and a control group for a total of 9 samples. Library preparation and RNA sequencing was performed by Novogene Co., LTD (Sacramento, CA) using the NovaSeq PE150 sequencing platform and strategy. Paired-end reads were generated in fastq format and released onto a remote server at SFSU. The estimated average number of reads was 17.6 million reads.
2.4 Gene expression analysis
Files were released and downloaded from Novogene and uploaded on to servers at the California Academy of Sciences in San Francisco, CA, as well as to the galaxy bioinformatics platform (The Galaxy Community at usegalaxy.org). The common murre reference genome and gene annotation files (Accession Number: SAMN12253989) were also uploaded to the galaxy platform. Trimmomatic was used to trim adapter sequences using the default job resource parameters (Bolger et al., 2014) (Galaxy version 0.38.0). FastQC was used to check the quality of the reads using default parameters (Andrews, 2010) (Galaxy version 0.73 + galaxy0). HISAT2 was used to align the reads to the genome sequence using the default parameters, which resulted in binary versions of sequence alignment maps (BAM) (Kim et al., 2015) (Galaxy version 2.2.1 + galaxy1).
FeatureCounts was then used to quantify reads using the default parameters (Liao et al., 2014) (Galaxy version 2.0.1 + galaxy2). Differential gene expression was performed using Limma-voom using the Benjamini and Hochberg (1995) p-value adjustment method and the p-value adjusted threshold of 0.05 was used as a false-discovery rate control (Law et al., 2014; Liu et al., 2015) (Galaxy version 3.50.1 + galaxy0). The TMM normalization method and robust settings were used to protect against outliers. Results were validated by running the same tests on DeSeq2 using the default job parameters (Love et al., 2014) (Galaxy version 2.11.40.7 + galaxy2.
For this analysis, 9 (Table 1) were selected and divided into three representative groups comprised of 3 birds each: a control group, in which birds were not infected with Babesia or contaminated, a Babesia-infected group, and an oil-affected group. It was originally planned to include replicate samples from birds that were both infected with Babesia and contaminated, however, we did not obtain RNA samples that were of sufficient quality for sequencing and analysis (RIN<6.5).
Two separate analyses were performed using three groups of biological replicates. To test how parasitic infection affects the Common murre, the 3 control group samples were set against the 3 Babesia infected samples. The second analysis determined how oil-contamination affected gene expression. The 3 control group samples were tested against the 3 oil-contaminated samples. Expression table data was input into R and volcano plots with labeled genes of interest were created for each test using the ggplot2 and dplyr packages (R Core Team, 2023; Wickham, 2016) (Supplementary Tables S2, S3). R and Microsoft Excel were used to filter through the gene expression data to determine the amount of overlap between the two analyses.
2.5 Gene set enrichment analysis (GSEA)
Statistically significant differentially expressed genes from the limma-voom analysis, were input into the g:Profiler database to convert gene names to ENSEMBL ID’s for GSEA (Raudvere et al., 2019). GSEA was performed in R using the ClusterProfiler package (Yu et al., 2012). Significantly expressed genes, in ENSEMBL format, and their associated fold change values were input into the package with the annotation file for Gallus gallus (org.Gg.eg.db) as a reference. While clusterProfiler offers curated gene sets for various organisms, avian annotations, excluding Gallus gallus, are currently unavailable. While using chicken gene sets might not perfectly capture common murre-specific biology, the enriched pathways still provided valuable insights into general stress response mechanisms potentially affected by the chosen stressors. The ontology parameter was set to test for biological function (BP) and set the p-value cutoff at 0.05. The Bonferroni-Hotchberg (BH) method for p-adjusted value to test for false positives (q-value). This method was performed for differentially expressed genes from each analysis. Pathways were identified and annotated using the Disease Ontology Semantic and Enrichment analysis (DOSE) (Yu et al., 2015).
3 Results
3.1 Transcriptome response
667 genes were differentially expressed (p < 0.05) in response to Babesia infection (Figures 1A, B). 380 (57%) genes were upregulated and 287 (43%) were downregulated (Table 2). Of these genes, 62 were unlabeled and had no known homologues. 39 of these unlabeled genes had a positive change in expression and 23 had a negative change in expression. The top significantly differentially expressed gene was Annexin A2 (ANXA2), which had a negative magnitude change in expression by the Babesia-infected group compared to the control group (p < 0.05).
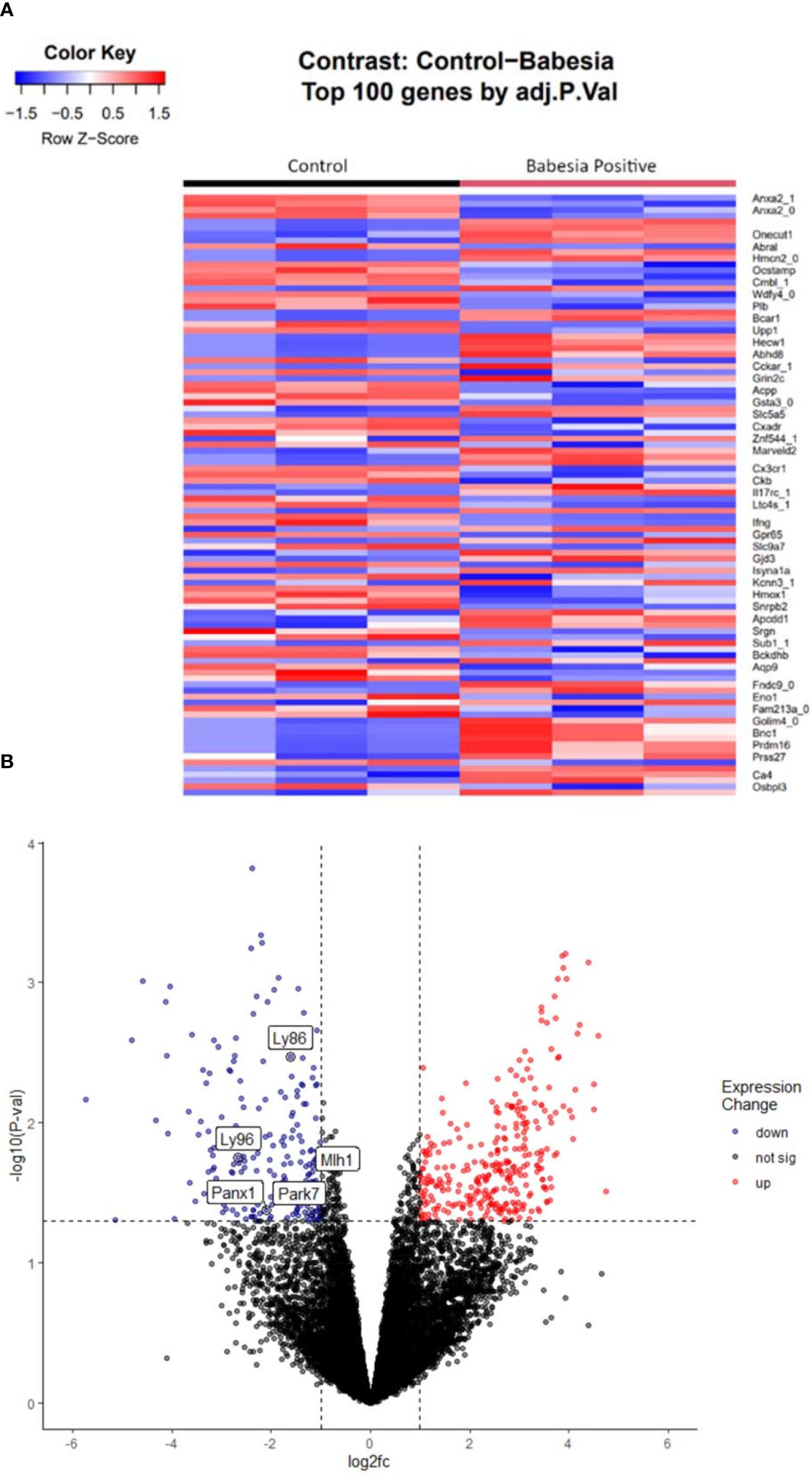
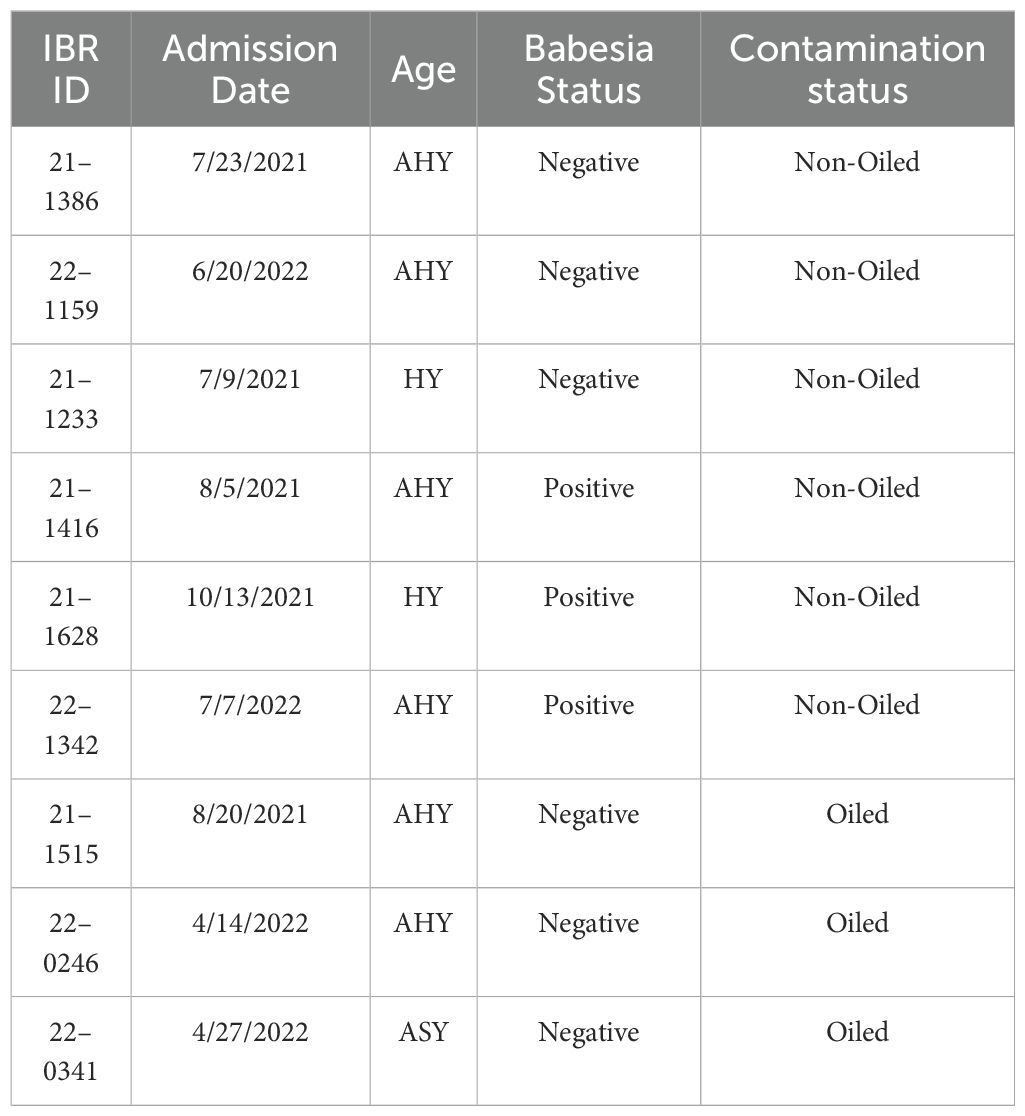
Figure 1. Heatmap (A) displaying the top 100 differentially expressed genes by adjusted p-value (q-value). Each column represents a sample, the left being the control group, and the right being the infected group. Each row represents a gene, with the magnitude of change in expression represented by a change in color (red being more upregulated, and blue being more downregulated). The volcano plot (B) displays the differentially expressed genes when analyzing the control group versus the Babesia-infected group. The log2(fold change) in gene expression is plotted on the x-axis, while the −log10 of the adjusted p-value is plotted on the y-axis. Genes that are significantly upregulated (p < 0.05 and log2foldchange ≥ 1) are shown in red, while genes that are significantly downregulated (p < 0.05 and log2foldchange ≤ −1) are shown in blue. Genes that are not significantly differentially expressed are shown in gray. Labeled genes, LY96 LY96, MLH1, PANX1, and PARK7, drive down GO terms related to immune response, defense response, and positive regulation of response to stimuli.
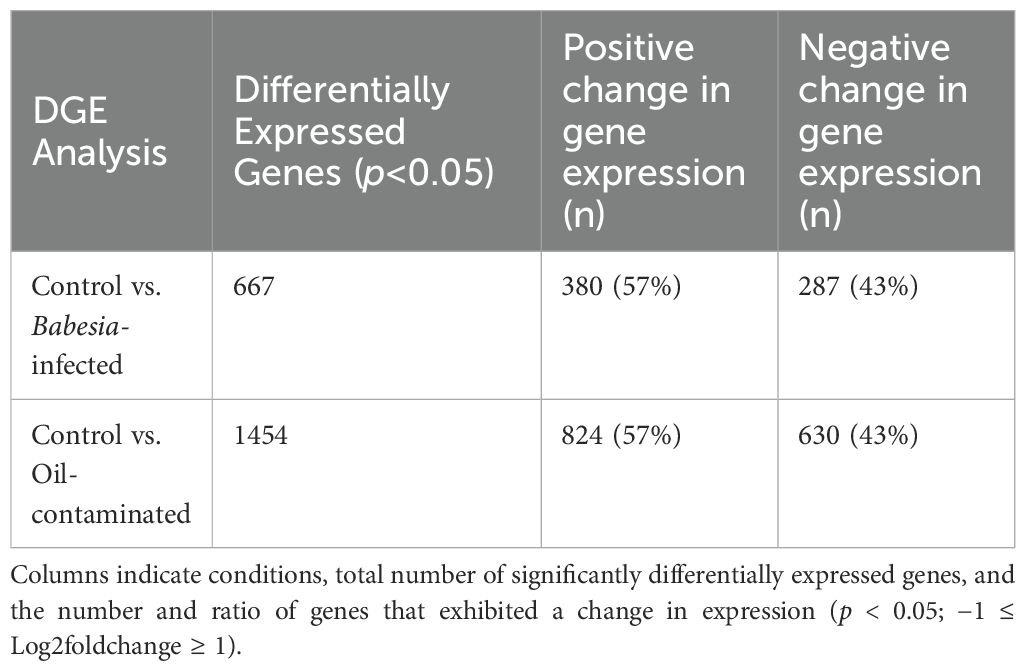
Table 2. Number of differentially expressed genes in response to Babesia infection and oil contamination in common murres.
Oil contamination elicited an even greater response, with 1454 total genes being differentially expressed (Figures 2A, B). Of those genes, 824 (57%) were upregulated and 631 (43%) were downregulated. 142 of these genes were unlabeled and had no known homologues. Of these uncharacterized genes, 72 were upregulated, and 70 were downregulated. The top significantly expressed gene was Interleukin-17 receptor protein E (IL17RE), which had a positive magnitude change in expression in the contaminated group. The proportion of significantly upregulated and downregulated genes (57% versus 43%, respectively) were the same in both analyses.
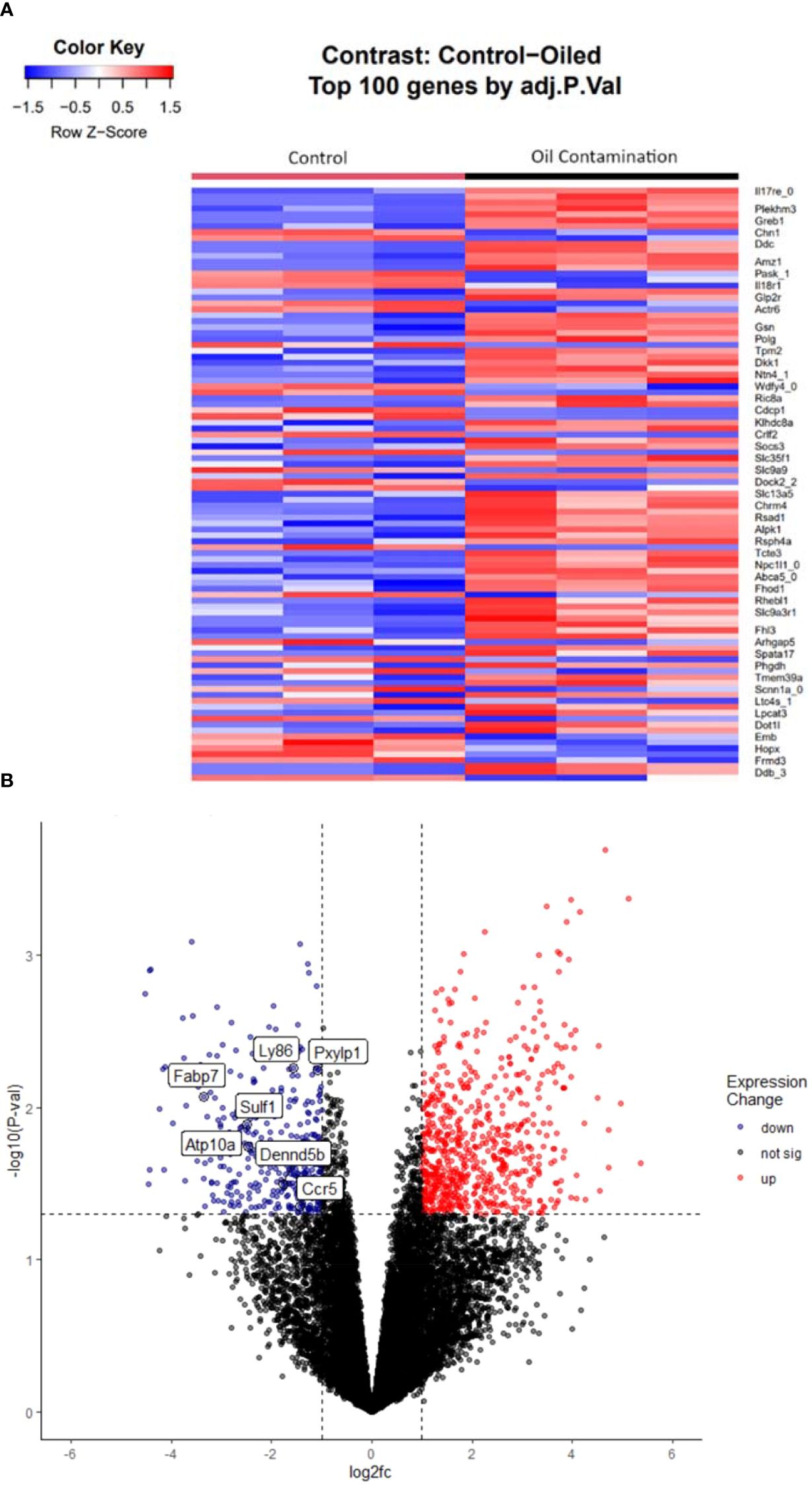
Figure 2. Heatmap (A) and volcano plot (B) displaying the top differentially expressed genes in response to oil contamination. Both figures show a much larger magnitude of differentially expressed genes, as described in Figure 1.
3.2 Similarities in gene expression and GSEA
We found that between the response to infection and the response to contamination, 194 of the same genes were differentially expressed by both groups (Supplementary Figure S1). This accounts for 32% of the genes that were expressed in response to infection and 17% that were expressed in response to contamination. This amount of overlap is much higher than what would be expected at random in response to different conditions (p < 0.001, Fisher’s Exact Test). Out of the shared 194 genes, 188 were expressed in the same direction by both groups. This accounts for 97% of the shared genes, indicating most of the expression change to both conditions represent a generalized response to stress, also indicating that the stress response to Babesia and oil contamination are highly similar. The remaining 6 shared genes had opposite directional changes in expression (Table 3).
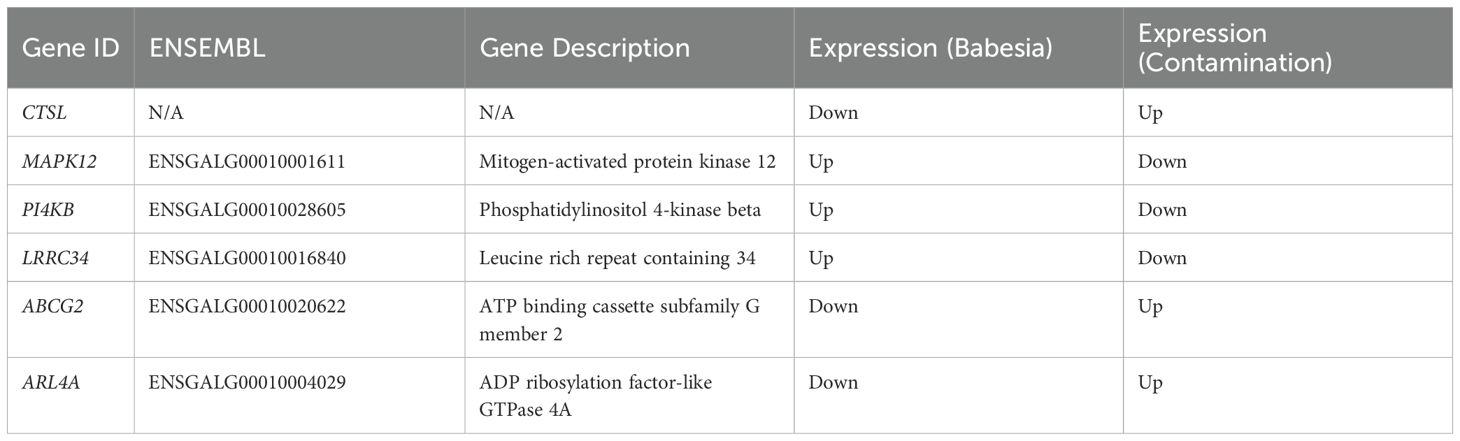
Table 3. Summary of the 6 genes that were differentially expressed in both the Babesia-infected group and the oil-contaminated group that were expressed in opposite directions.
Genes differentially expressed in response to Babesia infection led to suppression of biological pathways related to immune response (Figure 3). The top two enriched Gene Ontology terms related to biological processes were ‘defense response’ and ‘immune response.’ Similarly, terms associated with the immune system were suppressed in response to contamination (Figure 4). “Immune response” and “inflammatory response” were highly significantly suppressed. In each of these instances, there are only two genes significantly driving these patterns. In the infection analysis, “defense response” suppression is driven by Lymphocyte antigen 96 (LY96) and Parkinsonism associated deglycase (PARK7). LY96, as well as Lyphocyte antigen 86 (LY96), are also driving the GO “Positive regulation of response to stimuli,” and “immune response” is driven down by Pannexin 1 (PANX1) and MutL homolog 1 (MLH1). In the oil contamination analysis, both GO terms “immune response”, and “inflammatory response” are driven by LY96 and C-C Chemokine receptor type 5 (CCR5). Both analyses saw differential expressions of cytokines IL-17 and IL-18, as well as TNFAIP3. Contamination saw more DEG involved in cytokine pathways including SKIL, IL5RA, and TNFAIP8.
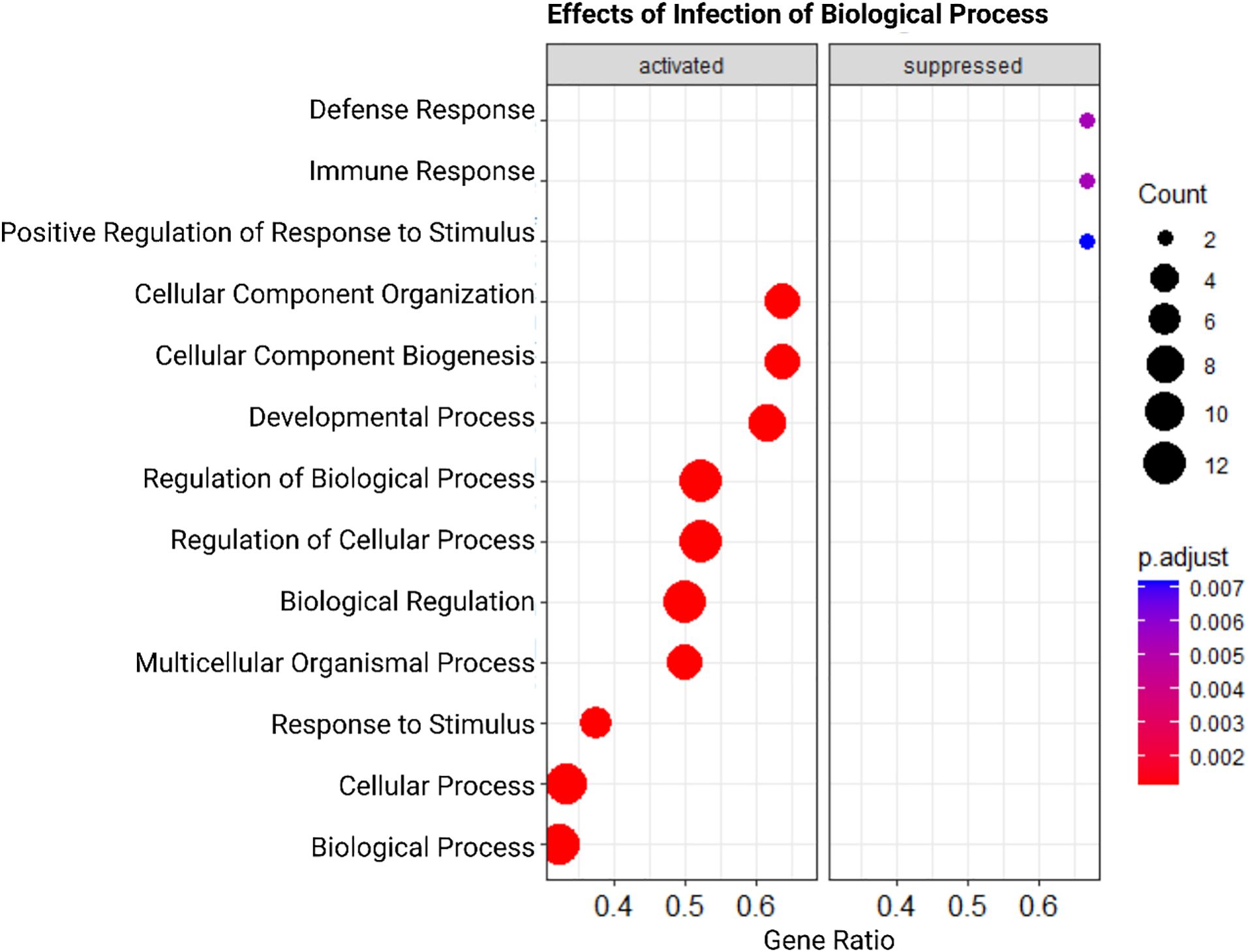
Figure 3. Dot-plot representation of Gene Set Enrichment Analysis (GSEA) results highlighting activated and suppressed biological functions in response to Babesia infection. Each dot represents a distinct biological function, positioned along the x-axis according to its enrichment score. Activated functions are shown on the left side of the plot, while suppressed functions are displayed on the right side. The color of each dot corresponds to the significance level of the enrichment score, with red dots indicating higher significance. The size of each dot represents the number of genes associated with a biological function. The x-axis represents the gene ratio, or the proportion of genes from the gene set that are present in the analyzed dataset. We can see that terms relating to immune response, inflammatory response, and response to stimulus are all suppressed.
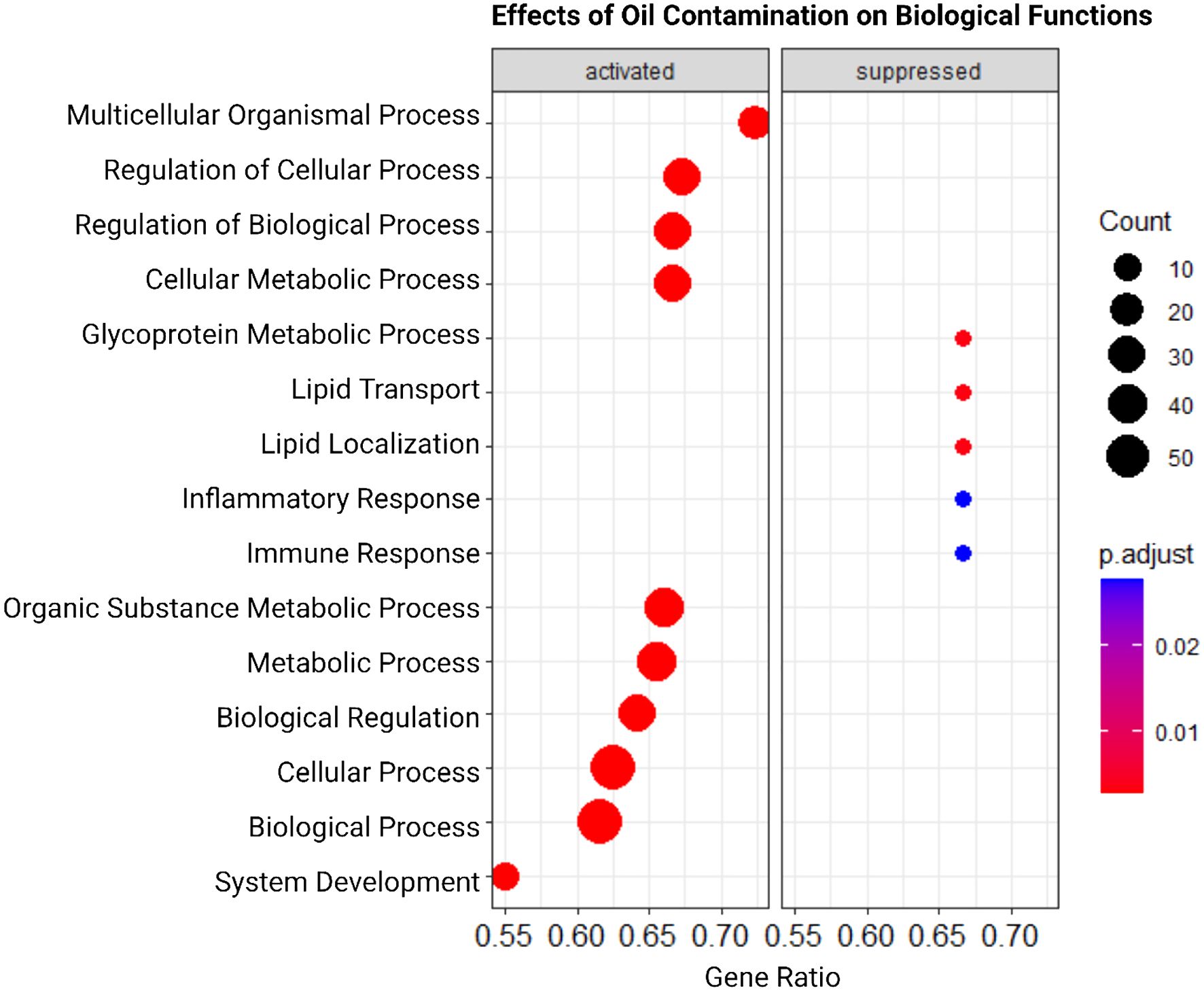
Figure 4. Dot-plot representation of Gene Set Enrichment Analysis (GSEA) results highlighting activated and suppressed biological functions in response to exposure to oil contamination, as described in Figure 3.
Also in the contamination group, ATPase phospholipid transporting 10A (ATP10A) and DENN Domain Containing 5B (DENND5B) drove “lipid transport.” “Lipid localization was also driven by ATP10A, as well as Fatty acid binding protein 7 (FABP7). “Glycoprotein metabolic process” was suppressed by Sulfatase 1 (SULF1) and 2-Phospoxylose Phospatase 1 (PXYLP1). Some other DEGs relating to lipid metabolism were PLA2G2E, ECHDC3, FADS1, FABP4, ACADL, LPIN2, CD36, and GATA2. PLA2GE secretes inflammatory lipid mediators (Gubern et al., 2008; Silverstein and Febbraio, 2009; Qin et al., 2018), ECHDC3 is involved in the biosynthesis of fatty acids (Sharma et al., 2019), FADS1 encodes a fatty acid desaturase (He et al., 2018), FABP4 is a gene related to the transport of fatty acids (Furuhashi et al., 2015), ACADL aids in fatty acid oxidation (Zhao et al., 2020), LPIN2 is involved in lipid synthesis (Dwyer et al., 2012).
4 Discussion
In this comparative analysis of gene expression, we investigated the molecular impacts of both oil contamination and infection from an apicomplexan parasite, Babesia, in common murres, using three biological replicates for each condition. Despite the small number of biological replicates, the results generated from this analysis are consistent with previous studies exploring similar questions (Videvall et al., 2015; Bonisoli-Alquati et al., 2020). The programs employed in this analysis, DeSeq2 and Limma-voom, while not specifically designed for small sample sizes, utilize various techniques that are designed to handle data with various levels of biological variability (Love et al., 2014). Both programs use shrinkage estimators, such as the Wald statistic or the moderated t-statistic. This reduces the variance of the estimated log2 fold changes, especially for genes with low counts, which in turn helps to differentiate true biological signals from random noise that is more prevalent in datasets with fewer replicates (Love et al., 2014). Both tools also employ normalization procedures that account for technical variability between samples, ensuring that differences in gene expression are more likely due to the biological conditions that are being compared, rather than technical artifacts (Costa-Silva et al., 2017). While there is, of course, room (and a need) for an improved study involving more biological replicates, we believe our results are significant and represent a major first step in uncovering the effects of environmental stress on a severely understudied species.
4.1 Avian host response to Babesia
GSEA analysis determined that biological functions relating to the immune response were the top two enriched pathways from this analysis. Further, these two processes were driven to suppression by two genes each; Defense response was driven by LY96 and PARK7, and inflammatory response was driven by LY96 and LY96. While these processes are suppressed, it is likely that the host is not suppressing the entire immune system but dampening certain aspects of the immune response. Lymphocyte antigen 96 (LY96) is a transmembrane protein that has been known to play a role in inflammation, immune suppression, and T cell activation, however, the role of this gene in the context of Babesia infection and its impact on the immune system is not fully understood (Li et al., 2023). The role of PARK7 in parasitic infection is also not well understood. The role of this gene is in antioxidative response and maintaining mitochondrial quality control (Wang et al., 2016). It is possible that LY96 acts as a negative regulator of T cell activation, potentially contributing to the dampened inflammatory and defense responses observed in this analysis. This process is regulatory that eukaryotes possess to suppress overly aggressive immune responses to prevent tissue damage. For example, a study on the role of LY96 in cancer progression found that suppression, or downregulation of LY96 can inhibit cancer development (Nie et al., 2022). PARK7 has been linked to inflammatory processes and may be a part of this suppression process.
While there is currently no information on the avian transcriptome response to Babesia, Videvall et al. (2015) analyzed the avian transcriptome response to Plasmodium ashfordi (2015), as well as the transcriptome of P. ashfordi while infecting an avian host (2017). Although Plasmodium and Babesia may elicit different overall transcriptome responses, we expect some level of overlap in the host response because both parasites undergo a life cycle stage where they infect, and destroy the red blood cells, leading to an inflammatory response (Clark and Jacobson, 1998; Hunt and Grau, 2003). Videvall et al. (2015) found differential expression of several genes that have also been found to be expressed by mice and humans in response to malaria infection. In the present study, two of these genes, Annexin A2 (ANXA2) and Lymphocyte antigen-86 (LY96), were downregulated in our Babesia-infected group. Both of these genes have also been widely found to be implicated in participating in the inflammatory response, however their role in Babesia infection is massively understudied.
ANXA2 has both anti- and pro-inflammatory roles in the innate immune response. ANXA2 aids in macro autophagy by interacting with Atg16, a protein that is related to autophagy (Dallacasagrande and Hajjar, 2020). In the context of pathogen-caused infection, ANXA2 plays crucial roles in the mediation of pro-inflammatory cytokines and promotion of anti-inflammatory signals (Dallacasagrande and Hajjar, 2020; Ma et al., 2021). There is also evidence that ANXA2 may be used to facilitate infection by bacteria such as Pseudomonas aeruginosa and Escheria coli (Dallacasagrande and Hajjar, 2020). LY96 is a protein-coding gene, that codes for the protein MD1, and has been shown to be produced by macrophages in the inflammatory response (Su et al., 2014; Thomas et al., 2016). The downregulation of these genes in Babesia-infected murres suggests potential mechanisms employed by the parasite to evade the host immune response. For instance, ANXA2 deficiency might hinder the ability of immune cells to infiltrate Babesia-infected tissues, allowing the parasite to establish itself. Similarly, downregulation of LY96 could suppress T-lymphocyte activation, thereby dampening the adaptive immune response. While these are speculative scenarios, they warrant further investigation to understand Babesia’s immunomodulatory strategies in common murres.
The presence of shared genes responding to multiple stresses points toward the possibility of a highly conserved host immune response to both babesiosis and malaria across species. Further research is required to interpret the precise mechanisms and implications of these shared genes in stress and immune responses.
Videvall et al. (2015) also analyzed the transcriptome of P. ashfordi after experimentally inoculating Eurasian siskins (Carduelis spinus). We compared the results of our Babesia-infected analysis to determine if any overlap in gene expression existed that may suggest expression from the Babesia parasite itself, rather than from the host. We found only one homologous gene that matched from this study, Subtilisin proteases 1 (SUB1; log fold change = 2.9088, p-value = 0.00618). This gene encodes proteins with essential roles in the maturation and proteolytic processing of merozoite antigens (Beeson et al., 2016). Although only one gene was found to be similarly differentially expressed, this is not surprising because, despite both Plasmodium and Babesia being apicomplexan type parasites, it is likely that they have developed distinct needs within erythrocytes, which can lead to differences in gene expression. Many of the genes, however, were found within our dataset, but fell well beneath our significance cutoff. This is most likely because, while comparing data from Plasmodium and Babesia may provide useful context, it is important to consider the limitations of comparing two different species. While both species are Apicomplexan type parasites, they belong to two distinctly different Apicomplexan orders that have diverged significantly, Plasmodium belongs to the order Haemosporidia, and Babesia belongs to Piroplasmida (Martinez-Ocampo, 2018). Furthermore, these two species have established unique life cycles and host-parasite interactions, the most distinct difference being that Plasmodium has a replication stage within the host’s liver, and Babesia does not (Hakimi et al., 2022). Further exploration comparing the similarities and differences between these two genera would be greatly beneficial in the optimization of these types of wildlife genomic studies.
4.2 Avian host response to oil contamination
Functions relating to lipid transport and localization were affected in response to petroleum oil contamination (Figure 4). Lipid transport and localization involves the coordination of various pathways to move insoluble lipolytic products throughout the body (Feingold et al., 2020). This process is essential to organisms because lipids play major roles as the building blocks of cellular membranes, aid in energy storage, act as signaling molecules between and within cells, and are required for proper protein functions. Lipid processes related to metabolism are centralized in the liver, and damage to the liver can affect the lipid amounts circulating throughout the body (Arvind et al., 2000). Further, damage to the kidneys has also been associated with disturbing lipid metabolism (Trevisan et al., 2006). Studies of the effects of petroleum oil on seabirds have reported problems with the kidneys and liver, with damage to these organs being the leading cause of mortality among patients (Briggs et al., 1996). Common murres and Cassin’s auklets (Ptychoramuphus aleuticus) exposed to petroleum oil in a controlled study exhibited liver problems such as hepatocellular dissociation (Fry and Lowenstine, 1985).
Petroleum oil may also disrupt lipid processes by directly damaging the lipid bilayer of cellular membranes, interfere with proteins that are responsible for transporting lipids throughout the body, and altering lipid environments locally by bioaccumulating in tissues. Studies on Polycyclic Aromatic Hydrocarbons (PAHs) bioaccumulation in marine life are limited, however, a study on the effects of PAHs in gelatinous zooplankton found that bioaccumulation occurred either dermally or through ingestion, and that the lipophilic nature of PAHs are the main cause of accumulation in an organism’s lipid content (Almeda et al., 2013). It is highly likely that the Common murres used for this study had significant levels of PAHs bioaccumulation because attempting to preen oiled feathers can likely lead to direct ingestion of PAHs. Dermal absorption of PAHs is also highly likely because contamination disrupts the barrier created by seabirds to protect them from the cold marine environment, leaving their skin exposed and vulnerable to further contamination.
Bonisoli-Alquati et al. (2020) studied the transcriptomes of Seaside sparrows (A. maritima) that were exposed, but not physically contaminated, to the Deepwater Horizon oil spill that occurred in the Gulf of Mexico in 2010. This was the only other published transcriptomic analysis of seabirds exposed to petroleum oil to our knowledge, and it was expected that our results would have many similarities, although the birds in that analysis were not physically contaminated by oil. Their results found significant upregulation of cyp1a2, a known marker of PAHs in birds and cytochrome p450 subunit member, as well as activation of the PAH-responsive aryl hydrocarbon receptor (AhR) pathway. While our results did not show upregulation of the cyp1a2 gene, we found significant downregulation of other p450 subunit members: cyp4b1_1 (log fold change = −2.536, p-value = 0.018), cyp4b1_0 (log fold change = −3.3510, p-value = 0.029), and cyp2r1 (log fold change = −1.0408, p-value = 0.0316).
The p450 family of enzymes (CYP), in humans, are membrane-bound proteins involved in drug detoxification and cellular metabolism (Zhao et al., 2021). In humans, the CYP 1 family is involved in drug metabolism, the CYP 2 family is primarily involved in drug and steroid metabolism, and CYP 4 is involved in fatty acid metabolism (Zhao et al., 2021). In the context of oil exposure, several studies have found expression of cyp1a in response to the deepwater horizon oil spill, mediated by AhR (Varanasi, 1989; Whitehead et al., 2012; Brewton et al., 2013; Dubansky et al., 2013; Crowe et al., 2014; Brown-Peterson et al., 2015; Xu et al., 2016, 2017). In a study of double-crested cormorants (Phalacrocorax auratus) experimentally dosed with synthetic (DWH Mississippi Canyon 252 oil), cyp1a was also found to have elevated expression levels in the liver (Alexander et al., 2017).
In the current study, cyp1a2 is not expressed, however, AhR is. This gene has been shown to be a regulator of cyp1a2 (Bonisoli-Alquati et al., 2020). This gene was significantly downregulated in our data, which may have led to the downregulation of cyp1a2, as well as other CYP enzymes such as cyp4b1 and cyp2r1. A possible explanation for the downregulation of AhR, as opposed to the activation of this gene, is related to resource allocation by the birds. It is highly likely that the oiled common murres admitted to IBR that were selected for this analysis were in such poor condition, that AhR and CYP enzymes were turned off as a survival mechanism. This method of allocating resources would prioritize essential functions such as maintaining body core temperature and organ function. Further research is needed to understand how the p450 CYP enzymes behave once the birds have had enough supportive care to return to a state of homeostasis.
Bonisoli-Alquati et al. (2020) also detected upregulation of the insulin-like growth factor binding protein (IGFBP1) gene, which was assisted by expression of the serine/threonine-protein kinase Pim3 (PIM3). Our results show a similar upregulation of the IGFBP1 gene in the oil contamination group (log fold change = 2.9766, p-value = 0.0128). This gene plays a major role in liver regeneration, giving further evidence that birds from our sample group experienced significant liver damage from PAH bioaccumulation. While PIM3 was not significantly upregulated, PIM1 was (log fold change = 1.4614, p-value = 0.0327), which is highly homologous to PIM3 (Julson et al., 2022). An investigation of PIM1 in knockout mice found expression of this gene to be associated with pancreatic, prostate, gastric, and colorectal cancers.
Finally, the Seaside sparrow study found ELOVL5 and ELOVL2, two elongases involved in polyunsaturated fatty acid synthesis, significantly downregulated. These elongases have downstream control in the adipose tissue, and downregulation of these genes may function in preventing lipid peroxidation from oxidative insults. These two elongases were also found in Great tits (Parus major) in a study on the effects of urban environments (Watson et al., 2017). While ELOVL5 and ELOVL2 were not significantly downregulated, ELOVL4 was (elovl4_0 log fold change = −2.9362, p-value = 0.020002). The elongase family consists of enzymes involved in the elongation of very-long-chain fatty acids and may have overlapping functions (Wang et al., 2023). It is possible that in Common murres, ELOV4 is the primary elongase for very-long-chain fatty acid synthesis, and in both Seaside sparrows and Great tits, ELOVL5 and ELOVL2 are the primary elongases.
4.3 Similarities in gene expression
As expected, Babesia and oil contamination saw DEGs relating to the innate immune system. NF-kB is a transcription factor that regulates expression pro-inflammatory cytokines, chemokines, innate immune cells, and T-cells (Liu et al., 2017). TNFAIP8, found to be induced by NF-kB (Niture et al., 2019), and FABP7, a promoter of NF-kB-driven inflammatory response (Killoy et al., 2020), were expressed by both groups. Birds that were contaminated with oil saw differential expression of genes associated with the regulation of Interleukin-1 beta (IL-1β), including CASP4, which regulates IL-1β synthesis in macrophages (Cheung et al., 2018), and CASP8, a gene that plays a role in modulating IL-1β and inflammation (Gurung and Kanneganti, 2015). IL-1β is a pro-inflammatory cytokine that is essential for the host’s ability to defend against infection (Lopez-Castejon and Brough, 2011).
Both analyses saw differential expressions of cytokines IL-17 and IL-18, as well as TNFAIP3. Contamination saw more DEG involved in cytokine pathways including SKIL, IL5RA, and TNFAIP8. SKIL has been shown to play a role in immune escape by upregulating autophagy (Ma et al., 2020). IL5RA is an Interleukin 5-receptor subunit that has been implicated in the regulation of white blood cells (Cheong et al., 2005). TNFAIP3 is a Tumor Necrosis Factor α-Induced Protein that mitigates the response to inflammation (Das et al., 2018). TNFAIP8 is another Tumor Necrosis Factor α-Induced Protein that helps maintain homeostasis during an immune response (Niture et al., 2019).
4.4 Cellular processes
Both infection and contamination lead to the activation of various cellular processes in common murres (Figures 3, 4). Notably, terms such as “regulation of cellular process,” “cellular metabolic process,” and “cellular component organization” were found to be among the activated processes in both groups. The activation of cellular response pathways may be a direct consequence of cell invasion by parasites, as shown in previous studies (Sumbria et al., 2021). Additionally, cell stress caused by the presence of these stressors might also contribute to the regulation of cellular processes. Iron plays a crucial role in numerous cellular activities (Rockfield et al., 2018), and its deficiency is a leading cause of anemia (Warner and Kamran, 2023). Specifically, hemolytic anemia, a type of anemia where red blood cells are destroyed, can result from the destruction of red blood cells (Hill and Hill, 2018). Babesia directly invades and replicates in the host’s red blood cells, ultimately leading to their destruction (Fry and Lowenstine, 1985; Niu et al., 2015; Wu et al., 2017). Similarly, ingestion of petroleum oil has been shown to cause hemolytic anemia in seabirds and other organisms (Leighton et al., 1983; Ostlere et al., 1988). The activation of cellular metabolic process and regulation of cellular process may be associated with the host’s defense mechanism. Both parasitic infection and oil contamination can impose significant metabolic burdens on the host, and upregulation of genes associated with this process may reflect enhanced energy production needs by the increase of immune cell recruitment and tissue repair. It may also be involved in maintaining cellular stability, or homeostasis in response to disruptions or damage caused by inflammation (Chovatiya and Medzhitov, 2014). Therefore, the activation of cellular processes observed in this study might serve as a protective mechanism by the host to mitigate excessive destruction of red blood cells caused by these stressors.
Conversely, there exists the potential for the activation of cellular processes to possess negative consequences in the context of the inflammatory response. The initial increase of metabolic activity in response to cellular invasion is beneficial, however, prolonged increase of metabolic activity can become detrimental (Blanco and Kaplan, 2023). Dysregulation of immune response can lead to tissue damage, inflammation, and autoimmunity (Blanco and Kaplan, 2023). In the context of the current study, while unlikely, it is possible that the activation of cellular processes may be attributed to dysregulation of immune cells and metabolism. Further experimental research is required to elucidate the function of cellular processes in the context of these conditions.
5 Conclusion
The findings of this study highlight the detrimental effects of these environmental stressors on the immune system of common murres. Both apicomplexan infection and oil contamination appear to suppress immune response mechanisms, potentially leaving the birds more susceptible to infections and compromising their overall health. Moreover, this analysis reveals specific impacts of oil contamination on the lipid metabolism of common murres. Genes associated with lipid transport and lipid localization were significantly overrepresented in response to oil contamination. This suggests a disruption in the birds’ ability to regulate lipid levels and perform essential functions related to lipid metabolism and glycometabolism. These results also highlight a need for more research in this field, ideally with more biological replicates to increase evidence of the genes observed in this study.
It is important to acknowledge that this study did not include technical replicates. This decision was made due to the author’s opinion that the high cost does not justify the added value, given that most DE studies do not include technical replicates. The authors are confident that the results reported are robust despite the lack of technical replicates. It also is important to emphasize that the results generated in this study should be taken with caution, as the sequencing and analysis of the transcriptomes of wildlife is notoriously difficult. One major concern in the present study that should be considered is the method implemented in choosing a control group. The samples collected are all from seabirds admitted into a wildlife hospital after being found beached for an unknown duration of time. Regardless of if the individual displays physical injuries upon admission, there may be underlying physiological or internal ailments that display themselves after intensive supportive care and the bird is returned to homeostasis. However, given the trends found in this study when compared to other studies, the results yielded are significant. Because of the concerns with forming a control group, it should be stated that these results should be regarded as baseline data, emphasizing the need for further experiments. However, this study highlights the potential benefit of using wildlife admitted to rehabilitation centers to study how anthropogenic changes affect their genomic profiles. Longitudinal studies can be implemented to collect blood samples at different time intervals, as opposed to at one single point. This would allow the researcher to track changes in expression before and after oil contamination has been washed off, parasite burden before and after treatment, and overall health over time. Such research could also investigate the effects of oil contamination across different vulnerable species, such as Western grebes (Aechmophorus occidentalis), with the purpose of comparing the results. Experimental validation of the results generated from this study would also be useful in informing future projects implementing similar methods.
Overall, this research sheds light on the intricate molecular responses of common murres to two distinct stressors—apicomplexan infection and oil contamination. Understanding the shared and unique pathways affected by these stressors is crucial for developing effective conservation strategies and mitigating the detrimental impacts on the health and survival of these iconic seabirds. Further investigations into the underlying mechanisms and long-term consequences of these gene expression changes will be valuable in enhancing our knowledge of the impacts of environmental stressors on avian populations, and ecosystems.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by Oiled Wildlife Care Network/International Bird Rescue IACUC. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CE: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. RQ: Data curation, Writing – review & editing. RD: Resources, Supervision, Validation, Writing – review & editing. SR: Formal analysis, Methodology, Software, Supervision, Validation, Writing – review & editing. RS: Funding acquisition, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was funded by the Genentech Foundation Scholars (G-7874540) and the NIH SFSU/UCSF Bridges to the Doctorate Fellowship (T32-GM142515). This project was supported by the California Department of Fish and Wildlife’s Oil Spill Prevention and Administration Fund through the Oiled Wildlife Care Network at the Karen C. Drayer Wildlife Health Center, School of Veterinary Medicine, University of California, Davis, CA.
Acknowledgments
We would like to thank the staff and volunteers at IBR for caring for these birds and for providing us with blood samples to make this work possible. We would also like to thank the members of the Avian Disease laboratory at SFSU for their support and contribution to this research. Sample shipment, processing, and sequencing was performed at the Novogene Corporation Inc. in Sacramento, CA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcosc.2024.1425484/full#supplementary-material
References
Alexander C. R., Hooper M. J., Cacela D., Smelker K. D., Caleshia S. C., Dean K. M., et al. (2017). Reprint of: CYP1A protein expression and catalytic activity in double-crested cormorants experimentally exposed to deepwater horizon mississippi canyon 252 oil. Ecotoxicol. Environ. Saf. 146, 68–755. doi: 10.1016/j.ecoenv.2017.05.015
Almeda R., Wambaugh Z., Wang Z., Hyatt C., Liu Z., Buskey E. J. (2013). Interactions between zooplankton and crude oil: toxic effects and bioaccumulation of polycyclic aromatic hydrocarbons. PLoS ONE 8 (6), e67212.
Alvarez M., de Carvalho J. F., Salmon A., Armand Cave-Radet M. A., El Amrani A., Foster T. E., et al. (2018). Transcriptome response of the foundation plant Spartina alterniflora to the Deepwater Horizon oil spill. Mol. Ecol. 27, 2986–3000. doi: 10.1111/mec.14736
Andrews S. (2010). FastQC: A quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Arvind A., Osganian S. A., Cohen D. E., Corey K. E., Feingold K.R., Anawalt B., Blackman M.R., et al (2000). “Lipid and lipoprotein metabolism in liver disease,” in Endotext. Eds. Feingold K. R., Anawalt B., Blackman M. R., Boyce A., Chrousos G., Corpas E., de Herder W. W., et al (MDText.com, Inc., South Dartmouth, MA).
Beeson J. G., Drew D. R., Boyle M. J., Feng G., Fowkes F. J. I., Richards J. S. (2016). Merozoite surface proteins in red blood cell invasion, immunity and vaccines against malaria. FEMS Microbiol. Rev. 40 (3), 343–372.
Beletić A., Janjić F., Radaković M., Spariosu K., Andrić J. F., Chandrashekar R., et al. (2021). Systemic inflammatory response syndrome in dogs naturally infected with babesia canis: association with the parasite load and host factors. Veterinary Parasitol. 291. doi: 10.1016/j.vetpar.2021.109366
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Society: Ser. B (Methodological) 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Blanco L. P., Kaplan M. J. (2023). Metabolic alterations of the immune system in the pathogenesis of autoimmune diseases. PloS Biol. 21, e3002084. doi: 10.1371/journal.pbio.3002084
Bolger A. M., Lohse M., Bjoern U. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bonisoli-Alquati A., Xu W., Stouffer P. C., Taylor S. S. (2020). Transcriptome analysis indicates a broad range of toxic effects of deepwater horizon oil on seaside sparrows. Sci. Total Environ. 720, 1375835. doi: 10.1016/j.scitotenv.2020.137583
Bostrom B., Wolf C., Greene C., Peterson D. S. (2008). Sequence conservation in the rRNA first internal transcribed spacer region of Babesia gibsoni genotype Asia isolates. Vet Parasitol. 152 (1-2), 152–157.
Boulinier T., Kada S., Ponchon A., Dupraz M., Dietrich M., Gamble A., et al. (2016). Migration, prospecting, dispersal? What host movement matters for infectious agent circulation? Integr. Comp. Biol. 56, 330–342. doi: 10.1093/icb/icw015
Brewton R. A., Fulford R., Griffitt R. J. (2013). Gene expression and growth as indicators of effects of the BP deepwater horizon oil spill on spotted seatrout (Cynoscion nebulosus). J. Toxicol. Environ. Health Part A 76, 1198–1209. doi: 10.1080/15287394.2013.848394
Briggs K. T., Yoshida S. H., Gershwin M. E. (1996). The influence of petrochemicals and stress on the immune system of seabirds. Regul. Toxicol. Pharmacol.: RTP 23, 145–155. doi: 10.1006/rtph.1996.0036
Brown W. C., Norimine J., Knowles D. P., Goff W. L. (2006). Immune control of babesia bovis infection. Veterinary Parasitol. Babesiosis 138, 75–87. doi: 10.1016/j.vetpar.2006.01.041
Brown-Peterson N. J., Krasnec M., Takeshita R., Ryan C. N., Griffitt K. J., Lay C., et al. (2015). A multiple endpoint analysis of the effects of chronic exposure to sediment contaminated with deepwater horizon oil on juvenile southern flounder and their associated microbiomes. Aquat. Toxicol. 165, 197–209. doi: 10.1016/j.aquatox.2015.06.001
Cheong H. S., Kim L. H., Park B. L., Choi Y. H., Park H., Hong S., et al. (2005). Association analysis of interleukin 5 receptor alpha subunit (IL5RA) polymorphisms and asthma. Hum. Genet. 50, 628–634. doi: 10.1007/s10038-005-0304-2
Cheung K. T., Sze D. M.-Y., Chan K. H., Leung P. H.-M. (2018). Involvement of caspase-4 in IL-1 beta production and pyroptosis in human macrophages during dengue virus infection. Immunobiology 223, 356–364. doi: 10.1016/j.imbio.2017.10.044
Chovatiya R., Medzhitov R. (2014). Stress, inflammation, and defense of homeostasis. Mol. Cell 54, 281–288. doi: 10.1016/j.molcel.2014.03.030
Clark I. A., Jacobson L. S. (1998). Do babesiosis and malaria share a common disease process? Ann. Trop. Med. Parasitol. 92, 483–488. doi: 10.1080/00034983.1998.11813306
Costa-Silva J., Domingues D., Lopes F. M. (2017). RNA-Seq differential expression analysis: An extended review and a software tool. PloS One 12, e0190152. doi: 10.1371/journal.pone.0190152
Criado A., Martinez J., Buling A., Barba J. C., Merino S., Jefferies R., et al. (2006). New data on epizootiology and genetics of piroplasms based on sequences of small ribosomal subunit and cytochrome b genes. Veterinary Parasitol. 142, 238–247. doi: 10.1016/j.vetpar.2006.07.004
Crowe K. M., Newton J. C., Kaltenboeck B., Johnson C. (2014). Oxidative stress responses of gulf killifish exposed to hydrocarbons from the deepwater horizon oil spill: potential implications for aquatic food resources. Environ. Toxicol. Chem. 33, 370–374. doi: 10.1002/etc.2427
Dallacasagrande V., Hajjar K. A. (2020). Annexin A2 in inflammation and host defense. Cells 9, 1499. doi: 10.3390/cells9061499
Das T., Chen Z., Hendriks R. W., Kool M. (2018). A20/tumor necrosis factor α-induced protein 3 in immune cells controls development of autoinflammation and autoimmunity: lessons from mouse models. Front. Immunol. 9. doi: 10.3389/fimmu.2018.00104
DeLeo D. M., Glazier A., Herrera S., Barkman A., Cordes E. E. (2021). Transcriptomic responses of deep-sea corals experimentally exposed to crude oil and dispersant. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.649909
Djokic V., Rocha S. C., Parveen N. (2021). Lessons learned for pathogenesis, immunology, and disease of erythrocytic parasites: plasmodium and babesia. Front. Cell. Infection Microbiol. 11, 685239. doi: 10.3389/fcimb.2021.685239
Dubansky B., Whitehead A., Miller J. T., Rice C. D., Galvez F. (2013). Multitissue molecular, genomic, and developmental effects of the deepwater horizon oil spill on resident gulf killifish (Fundulus grandis). Environ. Sci. Technol. 47, 5074–5082. doi: 10.1021/es400458p
Duerr R. S., Ziccardi M. H., Massey J.G. (2016). Mortality during treatment: factors affecting the survival of oiled, rehabilitated comm murres (Uria aalge). J. Wildlife Dis. 52, 495–505. doi: 10.7589/2015-03-054
Dwyer J. R., Donkor J., Zhang P., Csaki L. S., Vergnes L., Lee J. M., et al. (2012). Mouse lipin-1 and lipin-2 cooperate to maintain glycerolipid homeostasis in liver and aging cerebellum. Proc. Natl. Acad. Sci. 109, E2486–E2495. doi: 10.1073/pnas.1205221109
Earle R. A., Huchzermeyer F. W., Bennett G. F., Brossy J. J. (1993). Babesia pieircei sp. nov. from te jackass penguin. South African J. Zoo. 28, 88–90.
Feingold K. R., Anawalt B., Blackman M. R., Boyce A., Chrousos G., Corpas E., et al. (2020). “Introduction to Lipids and Lipoproteins,” in Endotext. Eds. Feingold K. R., Anawalt B., Blackman M. R., Boyce A., Chrousos G., Corpas E., de Herder W. W., et al (MDText.com, Inc., South Dartmouth (MA)).
Fry D.M., Lowenstine L. J. (1985). Pathology of common murres and cassin’s auklets exposed to oil. Arch. Environ. Contamination Toxicol. 14, 725–737. doi: 10.1007/BF01055780
Furuhashi M., Saitoh S., Shimamoto K., Miura T. (2015). Fatty acid-binding protein 4 (FABP4): pathophysiological insights and potent clinical biomarker of metabolic and cardiovascular diseases. Clin. Med. Insights Cardiol. 8, 23–33. doi: 10.4137/CMC.S17067
Goldberg D. R., Yuill T. M., Burgess E. C. (1990). Mortality from duck plague virus in immunosuppressed adult mallard ducks. J. Wildlife Dis. 26, 299–306. doi: 10.7589/0090-3558-26.3.299
Gubern A., Casas J., Barceló-Torns M., Barneda D., Rosa X. de la, Masgrau R., et al. (2008). Group IVA phospholipase A2 is necessary for the biogenesis of lipid droplets. J. Biol. Chem. 283, 27369–27382. doi: 10.1074/jbc.M800696200
Gurung P., Kanneganti T. D. (2015). Novel roles for caspase-8 in IL-1β and inflammasome regulation. Am. J. Pathol. 185, 17–25. doi: 10.1016/j.ajpath.2014.08.025
Hakimi H., Yamagishi J., Kawazu S. I., Asada M. (2022). Advances in understanding red blood cell modifications by Babesia. PloS Pathog. 18, e1010770. doi: 10.1371/journal.ppat.1010770
Hampton S., Ford R.G., Carter H. R., Abraham C., Humple D. (2003). Chronic oiling and seabird mortality from the sunken vessel S.S. Jacob luckenbach in central California. Mar. Ornithol. 31, 35–41.
He Z., Zhang R., Jiang F., Zhang H., Zhao A., Xu Bo, et al. (2018). FADS1-FADS2 genetic polymorphisms are associated with fatty acid metabolism through changes in DNA methylation and gene expression. Clin. Epigenet. 10, 113. doi: 10.1186/s13148-018-0545-5
Hemmer R. M., Ferrick D. A., Conrad P. A. (2000). Role of T cells and cytokines in fatal and resolving experimental babesiosis: protection in TNFRp55-/- mice infected with the human babesia WA1 parasite. J. Parasitol. 86, 736–742. doi: 10.1645/0022-3395(2000)086[0736:ROTCAC]2.0.CO;2
Henkel J. R., Sigel B. J., Taylor C. M. (2014). Oiling rates and condition indices of shorebirds on the northern Gulf of Mexico following the Deepwater Horizon oil spill. J. Field Ornithol. 85, 408–420. doi: 10.1111/jofo.12080
Hill A., Hill Q. A. (2018). Autoimmune hemolytic anemia. Hematol. Am. Soc. Hematol. Educ. Program 2018, 382–389. doi: 10.1182/asheducation-2018.1.382
Hunt N. H., Grau G. E. (2003). Cytokines: accelerators and brakes in the pathogenesis of cerebral malaria. Trends Immunol. 24, 491–499. doi: 10.1016/S1471-4906(03)00229-1
Igarashi I., Suzuki R., Waki S., Tagawa Y.-I., Seng S., Tum S., et al. (1999). Roles of CD4+ T cells and gamma interferon in protective immunity against babesia microtiInfection in mice. Infection Immun. 67, 4143–4148. doi: 10.1128/IAI.67.8.4143-4148.1999
Inoue S.-I., Niikura M., Mineo S., Kobayashi F. (2013). Roles of IFN-γ and Γδ T cells in protective immunity against blood-stage malaria. Front. Immunol. 4. doi: 10.3389/fimmu.2013.00258
Iwasaki A., Medzhitov R. (2015). Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353. doi: 10.1038/ni.3123
Jefferies R., Down J., McInnes L., Ryan U., Robertson H., Jakob-Hoff R., et al. (2008). Molecular characterization of babesia kiwiensis from the brown kiwi (Apteryx mantelli). J. Parasitol. 94, 557–560. doi: 10.1645/GE-1344.1
Jenssen B. M. (1994). Review article: effects of oil pollution, chemically treated oil, and cleaning on thermal balance of birds. Environ. pollut. (Barking Essex: 1987) 86, 207–215. doi: 10.1016/0269-7491(94)90192-9
Julson J. R., Marayati R., Beierle E. A., Stafman L. L. (2022). The role of PIM kinases in pediatric solid tumors. Cancers (Basel) 14, 3565. doi: 10.3390/cancers14153565
Khan J. S., Provencher J. F., Forbes M. R., Mallory M. L., Lebarbenchon C., McCoy K. D. (2019). “Chapter One - Parasites of Seabirds: A Survey of Effects and Ecological Implications,” in Advances in Marine Biology, vol. 82 . Ed. Sheppard C. (Amsterdam, Netherlands: Academic Press), 1–50.
Khan R. A., Ryan P. (1991). Long term effects of crude oil on common murres (Uria aalge) following rehabilitation. Bull. Environ. Contamination Toxicol. 46, 216–222. doi: 10.1007/BF01691940
Killoy K. M., Harlan B. A., Pehar M., Vargas M. R. (2020). FABP7 upregulation induces a neurotoxic phenotype in astrocytes. Glia 68, 2693–2704. doi: 10.1002/glia.23879
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kumar A., Kabra A., Igarashi I., Krause P. J. (2023). Animal models of the immunology and pathogenesis of human babesiosis. Trends Parasitol. 39, 38–52. doi: 10.1016/j.pt.2022.11.003
Law C. W., Chen Y., Shi W., Smyth GK. (2014). Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15 (2), R29. doi: 10.1186/gb-2014-15-2-r29
Leighton F. A., Peakall D. B., Butler R. G. (1983). Heinz-body hemolytic anemia from the ingestion of crude oil: A primary toxic effect in marine birds. Sci. (New York N.Y.) 220, 871–873. doi: 10.1126/science.6844918
Li W., Meng X., Yu T., Shi X., Dong W., Ruan H., et al. (2023). Gene LY96 is an M2 macrophage-related biomarker and is associated with immunosuppression in renal cell carcinoma. MedComm – Oncol. 2, e52. doi: 10.1002/mog2.52
Liao Y., Smyth G. K., Shi W. (2014). FeatureCounts: an efficient general-purpose program for assigning sequence reads to genomic features. Bioinf. (Oxford England) 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu R., Holik A. Z., Su S., Jansz N., Chen K., Leong H. S., et al. (2015). Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 43, e97. doi: 10.1093/nar/gkv412
Liu Y.-Z., Miller C. A., Zhuang Y., Mukhopadhyay S. S., Saito S., Overton E. B., et al. (2020). The impact of the deepwater horizon oil spill upon lung health—Mouse model-based RNA-seq analyses. Int. J. Environ. Res. Public Health 17, 5466. doi: 10.3390/ijerph17155466
Liu T., Zhang L., Joo D., Sun S. C. (2017). NF-κB signaling in inflammation. Signal Transduct Target 2, 17023. doi: 10.1038/sigtrans.2017.23
Longmire J., Maltbie M., Baker R. J. (1997). Use of ‘lysis buffer’ in DNA isolation and its implication for museum collections (Lubbock, TX: Museum of Texas Tech University), vol 163. doi: 10.5962/bhl.title.143318
Lopez-Castejon G., Brough D. (2011). Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 24, 189–195. doi: 10.1016/j.cytogfr.2011.10.001
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Ma K., Chen X., Liu W., Yang Y., Chen S., Sun J., et al. (2021). ANXA2 is correlated with the molecular features and clinical prognosis of glioma, and acts as a potential marker of immunosuppression. Sci. Rep. 11, 20839. doi: 10.1038/s41598-021-00366-8
Ma F., Ding M. G., Lei Y. Y., Luo L.H., Jiang S., Feng Y.H., et al. (2020). SKIL facilitates tumorigenesis and immune escape of NSCLC via upregulating TAZ/autophagy axis. Cell Death Dis. 11, 1028. doi: 10.1038/s41419-020-03200-7
Martínez-Ocampo F. (2018). “Genomics of Apicomplexa,” in Farm Animals Diseases, Recent Omic Trends and New Strategies of Treatment (London, United Kingdom: InTech).
Merino S. (1998). Babesia bennetti sp. from the Yellow-Legged Gull (Larus cachinnans, Aves, Laridae) on Benidorm Island, Mediterranean Sea. J. Parasitol. 84, 422–424. doi: 10.2307/3284504
Miles A.K., Bowen L., Ballachey B. E., Bodkin J. L., Murray M., Estes J. L., et al. (2012). Variations of transcript profiles between sea otters enhydra lutris from Prince William sound, Alaska, and clinically normal reference otters. Mar. Ecol. Prog. Ser. 451, 201–212. doi: 10.3354/meps09572
Newman S. H., Piatt J. F., White J. (1997). Hematological and plasma biochemical reference ranges of Alaskan seabirds: their ecological significance and clinical importance. Colonial Waterbirds 20, 492–504. doi: 10.2307/1521600
Nie K., Li J., Peng L., Zhang M., Huang W. (2022). Pan-cancer analysis of the characteristics of LY96 in prognosis and immunotherapy across human cancer. Front. Mol. Biosci. 9, 837393. doi: 10.3389/fmolb.2022.837393
Niture S., Moore J., Kumar D. (2019). TNFAIP8: inflammation, immunity and human diseases. J. Cell. Immunol. 1, 29–34.
Niu Q., Liu Z., Yu P., Yang J., Abdallah M. O., Guan G., et al. (2015). Genetic Characterization and Molecular Survey of Babesia bovis, Babesia bigemina, and Babesia ovata in Cattle, Dairy Cattle and Yaks in China. Parasites Vectors 8, 518. doi: 10.1186/s13071-015-1110-0
Orgeret F., Thiebault A., Kovacs K. M., Lydersen C., Hindell M. A., Thompson S. A., et al. (2022). Climate change impacts on seabirds and marine mammals: the importance of study duration, thermal tolerance and generation time. Ecol. Lett. 25, 218–239. doi: 10.1111/ele.13920
Ostlere L., Amos R., Wass J. A. (1988). Haemolytic anaemia associated with ingestion of naphthalene-containing anointing oil. Postgraduate Med. J. 64, 444–446. doi: 10.1136/pgmj.64.752.444
Paxton K. L., Cassin-Sackett L., Atkinson C. T., Videvall E., Campana M. G., Fleischer R. C. (2023). Gene expression reveals immune response strategies of naïve Hawaiian honeycreepers experimentally infected with introduced avian malaria. J. Heredity 114, 326–340. doi: 10.1093/jhered/esad017
Peirce M. A., Feare C. J. (1978). Piroplasmosis in the masked booby Sula dactylatra melanops in the Amirantes, Indian Ocean Vol. 98 (London, United Kingdom: British Ornithologists’ Club (1978).
Peirce M. A., Jakob-Hoff R. M., Twentyman C. (2003). New species of haematozoa from apterygidae in New Zealand. J. Natural History 37, 1797–1804. doi: 10.1080/00222930110109082
Phillips E. M., Zamon J. E., Nevins H. M., Gibble C. M., Duerr R. S., Kerr L. H. (2011). Summary of birds killed by a harmful algal bloom along the south Washington and North Oregon coasts during October 2009. Northwestern Nat. 92, 120–126. doi: 10.1898/10-32.1
Piatt J. F., Parrish J. K., Renner H. M., Schoen S. K., Jones T. T., Arimitsu M. L., et al. (2020). Extreme mortality and reproductive failure of common murres resulting from the Northeast Pacific marine heatwave of 2014-2016. PloS One 15, e0226087. doi: 10.1371/journal.pone.0226087
Puri A., Bajpai S., Meredith S., Aravind L., Krause P. J., Kumar S. (2021). Babesia microti: pathogen genomics, genetic variability, immunodominant antigens, and pathogenesis. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.697669
Qin D.-d., Yang Y.-f., Pu Z.-q., Liu D., Yu C., Gao P., et al. (2018). NR4A1 retards adipocyte differentiation or maturation via enhancing GATA2 and P53 expression. J. Cell. Mol. Med. 22, 4709–4720. doi: 10.1111/jcmm.13715
Quillfeldt P., Arriero E., Martínez J., Masello J. F., Merino S. (2011). Prevalence of blood parasites in seabirds - a review. Front. Zool. 8, 26. doi: 10.1186/1742-9994-8-26
R Core Team. (2023). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing.
Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., et al. (2019). G:Profiler: A web server for functional enrichment analysis and conversions of gene lists, (2019 Update). Nucleic Acids Res. 47, W191–W198. doi: 10.1093/nar/gkz369
Ristic M., Kreier J. P. (1984). “Malaria and Babesiosis: Similarities and Differences,” in Malaria and Babesiosis: Research Findings and Control Measures. Eds. Ristic M., Ambroise-Thomas P., Kreier J. (Springer Netherlands, Dordrecht), 3–33.
Rivers A. R., Sharma S., Tringe S. G., Martin J., Joye S. B., Moran M. A. (2013). Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 7, 2315–2329. doi: 10.1038/ismej.2013.129
Rockfield S., Chhabra R., Robertson M., Rehman N., Bisht R., Nanjundan M. (2018). Links between iron and lipids: implications in some major human diseases. Pharm. (Basel Switzerland) 11, 113. doi: 10.3390/ph11040113
Schurch N. J., Schofield P., Gierliński M., Cole C., Sherstnev A., Singh V., et al. (2016). How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 22, 839–851. doi: 10.1261/rna.053959.115
Sharma N. K., Key C.-C. C., Civelek M., Wabitsch M., Comeau M. E., Langefeld C. D., et al. (2019). Genetic regulation of enoyl-coA hydratase domain-containing 3 in adipose tissue determines insulin sensitivity in African Americans and Europeans. Diabetes 68, 1508–1522. doi: 10.2337/db18-1229
Silverstein R. L., Febbraio M. (2009). CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signaling 2, re3. doi: 10.1126/scisignal.272re3
Smith R. L., Goddard A., Boddapati A., Brooks S., Schoeman J. P., Lack J., et al. (2021). Experimental Babesia rossi infection induces hemolytic, metabolic, and viral response pathways in the canine host. BMC Genomics 22, 619. doi: 10.1186/s12864-021-07889-4
Su S., Zhu H., Xu X., Wang X., Dong Y., Kapuku G., et al. (2014). DNA methylation of the LY96 gene is associated with obesity, insulin resistance, and inflammation. Twin Res. Hum. Genet. 17, 183–191. doi: 10.1017/thg.2014.22
Sumbria D., Berber E., Mathayan M., Rouse B. T. (2021). Virus infections and host metabolism—Can we manage the interactions? Front. Immunol. 11. doi: 10.3389/fimmu.2020.594963
Thomas J., Odkhuu R. E., Nakashima A., Morita N., Kobayashi T., Yamai I., et al. (2016). Inflammatory responses increase secretion of MD-1 protein. Int. Immunol. 28, 503–512. doi: 10.1093/intimm/dxw031
Torina A., Blanda V., Villari S., Piazza A., Russa F. L., Grippi F., et al. (2020). Immune response to tick-borne hemoparasites: host adaptive immune response mechanisms as potential targets for therapies and vaccines. Int. J. Mol. Sci. 21, 8813. doi: 10.3390/ijms21228813
Trevisan R., Dodesini A. R., Lepore G. (2006). Lipids and renal disease. J. Am. Soc. Nephrol.: JASN 17, S145–S147. doi: 10.1681/ASN.2005121320
Valkiunas G. (2005). Avian malaria parasites and other haemosporidia (Boca Raton, FL, USA: CRC Press). doi: 10.1201/9780203643792
Vannier E. G., Diuk-Wasser M. A., Mamoun C. B., Krause P. J. (2015). Babesiosis. Infect. Dis. Clinics North America 29, 357–370. doi: 10.1016/j.idc.2015.02.008
Varanasi U., Ed. (1989). Metabolism of polycyclic aromatic hydrocarbons in the aquatic environment. Boca Raton, FL: CRC Press, Inc. 341p.
Videvall E., Cornwallis C. K., Palinauskas V., Valkiūnas G., Hellgren O. (2015). The avian transcriptome response to malaria infection. Mol. Biol. Evol. 32, 1255–1267. doi: 10.1093/molbev/msv016
Wang B., Cai Z., Tao K., Zeng W., Lu F., Yang R., et al. (2016). Essential control of mitochondrial morphology and function by chaperone-mediated autophagy through degradation of PARK7. Autophagy 12, 1215–1228. doi: 10.1080/15548627.2016.1179401
Wang X., Yu H., Gao R., Liu M., Xie W. (2023). A comprehensive review of the family of very-long-chain fatty acid elongases: structure, function, and implications in physiology and pathology. Eur. J. Med. Res. 28, 532. doi: 10.1186/s40001-023-01523-7
Warner M. J., Kamran M. T. (2023). “Iron Deficiency Anemia,” in StatPearls (StatPearls Publishing, Treasure Island (FL).
Watson H., Videvall E., Andersson M., Isaksson C. (2017). Transcriptome analysis of a wild bird reveals physiological responses to the urban environment. Sci. Rep. 7, 44180. doi: 10.1038/srep44180
Whitehead A., Dubansky B., Bodinier C., Garcia T. I., Miles S., Pilley C., et al. (2012). Genomic and physiological footprint of the deepwater horizon oil spill on resident marsh fishes. Proc. Natl. Acad. Sci. 109, 20298–20302. doi: 10.1073/pnas.1109545108
Wickham H. (2016). ggplot2: Elegant graphics for data analysis, 2nd Edn. New York: Springer. doi: 10.1007/978-3-319-24277-4_5
Work T. M., Rameyer R. A. (1997). Description and epizootiology of babesia poelea n. Sp. in brown boobies (Sula leucogaster (Boddaert)) on sand Island, Johnston Atoll, Central Pacific. J. Parasitol. 83, 734–738. doi: 10.2307/3284253
Wu J., Cao J., Zhou Y., Zhang H., Gong H., Zhou J. (2017). Evaluation on infectivity of babesia microti to domestic animals and ticks outside the ixodes genus. Front. Microbiol. 8, 1915. doi: 10.3389/fmicb.2017.01915
Xu E. G., Khursigara A. J., Magnuson J., Starr Hazard E., Hardiman G., Esbaugh A. J., et al. (2017). Larval red drum (Sciaenops ocellatus) sublethal exposure to weathered deepwater horizon crude oil: developmental and transcriptomic consequences. Environ. Sci. Technol. 51, 10162–10172. doi: 10.1021/acs.est.7b02037
Xu E. G., Mager E. M., Grosell M., Pasparakis C., Schlenker L. S., Stieglitz J. D., et al. (2016). Time- and oil-dependent transcriptomic and physiological responses to deepwater horizon oil in Mahi-Mahi (Coryphaena hippurus) embryos and larvae. Environ. Sci. Technol. 50, 7842–7851. doi: 10.1021/acs.est.6b02205
Yabsley M. J., Greiner E., Tseng F. S., Garner M. M., Nordhausen R. W., Ziccardi M. H., et al. (2009). Description of novel babesia species and associated lesions from common murres (Uria aalge) from California. J. Parasitol. 95, 1183–1188. doi: 10.1645/GE-1955.1
Yabsley M. J., Work T. M., Rameyer R. A. (2006). Molecular Phylogeny of Babesia poelea from Brown Boobies (Sula leucogaster) From Johnston Atoll, Central Pacific. J. Parasitol. 92, 423–425. doi: 10.1645/GE-617R.1
Ye Q., Wang B., Mao J. (2020). The pathogenesis and treatment of the ‘Cytokine storm’ in COVID-19. J. Infection 80, 607–613. doi: 10.1016/j.jinf.2020.03.037
Yu G., Wang L. G., Han Y., He Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A J. Integr. Biol. 16 (5), 284–287. doi: 10.1089/omi.2011.0118
Yu G., Wang L.-G., Yan G.-R., He Q.-Y. (2015). DOSE: an R/bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31, 608–609. doi: 10.1093/bioinformatics/btu684
Zaki A. A., Attia M. M., Ismael E., Mahdy O. A. (2021). Prevalence, genetic, and biochemical evaluation of immune response of police dogs infected with babesia vogeli. Veterinary World 14, 903–912. doi: 10.14202/vetworld.
Zhao M., Ma J., Li M., Zhang Y., Jiang B., Zhao X., et al. (2021). Cytochrome P450 enzymes and drug metabolism in humans. Int. J. Mol. Sci. 22, 12808. doi: 10.3390/ijms222312808
Keywords: seabird, RNA sequencing, differential expression analysis, oil contamination, Babesia, gene expression, environmental stressors
Citation: Esperanza CW, Quock RC, Duerr RS, Roy SW and Sehgal RNM (2024) Comparative gene expression responses to Babesia infection and oil contamination in a seabird. Front. Conserv. Sci. 5:1425484. doi: 10.3389/fcosc.2024.1425484
Received: 29 April 2024; Accepted: 26 July 2024;
Published: 22 August 2024.
Edited by:
Çağan H. Şekercioğlu, The University of Utah, United StatesReviewed by:
M. Çisel Kemahlı Aytekin, Koç University, TürkiyeMorteza Naderi, Koç University, Türkiye
Copyright © 2024 Esperanza, Quock, Duerr, Roy and Sehgal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlos W. Esperanza, Y2VzcGVyYW56YUBzZnN1LmVkdQ==
 Carlos W. Esperanza
Carlos W. Esperanza Rachel C. Quock1,2
Rachel C. Quock1,2 Ravinder N. M. Sehgal
Ravinder N. M. Sehgal