- 1Plant Conservation Genetics Laboratory, Center for Conservation and Sustainable Development, Missouri Botanical Garden, St. Louis, MO, United States
- 2Department of Biology, Saint Louis University, St. Louis, MO, United States
Leavenworthia (Brassicaceae) has served as a model group for investigating the evolution of mating systems in plants, yet several Leavenworthia species remain understudied. One such taxon is Leavenworthia exigua var. laciniata, one of three varieties of L. exigua, a winter-annual plant endemic to the central United States. Because L. exigua var. laciniata occupies a narrow geographic range and is experiencing major habitat loss, it was recently listed as threatened; however, little is known about its genetic diversity and implications for conservation. We conducted a range-wide population genetic study of L. exigua var. laciniata and L. exigua var. exigua to understand: (1) levels of genetic diversity within and among populations, (2) whether L. exigua var. laciniata is genetically distinct from L. exigua var. exigua, and (3) implications for conservation. L. exigua var. laciniata showed identical genotypes at all 16 microsatellite loci across most of its range, fixed heterozygosity at some loci, and significant heterozygote excesses, consistent with a lack of recombination associated with an asexual mating system, which has not been documented previously in Leavenworthia. Because L. exigua var. laciniata is an annual and the same genotype occurs across multiple populations, asexuality may be caused by apomixis, asexual reproduction via seed. In contrast, most populations of L. exigua var. exigua demonstrated population genetic patterns consistent with a self-compatible mating system. Because L. exigua var. laciniata is morphologically, geographically, and genetically distinct, it should be recognized as an evolutionarily significant unit for conservation. We recommend maintaining large population sizes to conserve evolutionary potential in L. exigua var. laciniata, as the likelihood that facultative sexual reproduction may occur may be greater in larger populations. Additional research in L. exigua var. laciniata is needed to confirm the occurrence of asexuality and apomixis, clarify its reproductive isolation from other taxa, and to understand whether it exhibits residual sexual reproduction, epigenetic variation, or phenotypic plasticity to help it persist in response to environmental variation. In the future, L. exigua var. laciniata may serve as an important model in which to investigate the conservation of threatened plant species with little genetic variation in a changing climate.
Introduction
The genus Leavenworthia Torr. (Brassicaceae) has long served as an important model system for investigating the evolution of plant mating systems (Busch and Urban, 2011). Leavenworthia comprises eight species of small, North American, winter-annual plants that primarily occupy limestone glades and glade-like open habitats with rocky, shallow soils. Several early studies of the genus reported variation in mating system among species, with some species exhibiting self-compatibility [L. aurea Torr., L. exigua Rollins, L. torulosa A. Gray, L. texana Mahler, and L. uniflora (Michx.) Britton], some species exhibiting self-incompatibility (L. stylosa A. Gray), and some showing variation among populations in self-incompatibility (L. alabamica Rollins and L. crassa Rollins) (Rollins, 1963; Lloyd, 1965; Solbrig, 1972; Solbrig and Rollins, 1977). This led to a large number of subsequent studies investigating the evolution of mating system in Leavenworthia and its consequences for population genetic variation (Lyons and Antonovics, 1991; Charlesworth and Yang, 1998; Liu et al., 1998; Busch, 2005a,b, 2006; Anderson and Busch, 2006; Busch et al., 2010, 2011; Busch and Urban, 2011; Koelling et al., 2011; Busch and Werner, 2012; Chantha et al., 2013; Baldwin and Schoen, 2017, 2019).
Despite the large number of studies focusing on Leavenworthia, the vast majority of research has focused on just a few well-studied species, whereas other species remain understudied and very poorly known. For example, because both L. alabamica and L. crassa show variation among populations in whether they are selfing or self-incompatible, a large number of studies have focused on these species (Busch et al., 2010, 2011; Koelling et al., 2011; Chantha et al., 2013; Baldwin and Schoen, 2017). Indeed, the molecular basis of self-incompatibility (Busch et al., 2008, 2011; Joly and Schoen, 2011) and a novel genetic locus controlling self-incompatibility unique to Leavenworthia has now been characterized in L. alabamica (Chantha et al., 2013; Baldwin and Schoen, 2017, 2019). Because other species exhibit contrasting mating systems, such as the self-incompatible L. stylosa and the predominantly self-fertilizing L. torulosa and L. uniflora, they have also been used extensively to compare how mating systems affect genetic diversity and other aspects of species biology (Charlesworth and Yang, 1998; Filatov and Charlesworth, 1999; Liu et al., 1999; Busch and Werner, 2012). In contrast, apart from descriptions of their taxonomy, natural history, or ecology (Rollins, 1956; Baskin and Baskin, 1972, 1981; Busch et al., 2008; Frings et al., 2019), comparatively little previous research has focused on L. exigua, L. aurea, and L. texana, all of which are listed as endangered or threatened in at least a portion of their ranges.
Leavenworthia exigua is a comparatively poorly known species distributed in Kentucky, Tennessee, Alabama, and northern Georgia. The species was divided into the following three intraspecific varieties based on morphology and geography: (1) Leavenworthia exigua var. lutea Rollins, which has yellow petals and is found only in Alabama, (2) Leavenworthia exigua var. exigua Rollins, which has white petals, lavender sepals, and is found in Tennessee and north Georgia, and (3) Leavenworthia exigua var. laciniata Rollins, which has white petals, green sepals, and is endemic to Kentucky (Rollins, 1963). All varieties were reported to be diploid, with a consistent chromosome number of n = 11, the lowest chromosome number in the genus. Chromosome numbers vary widely in Leavenworthia, with L. exigua, L. crassa, and L. alabamica exhibiting a chromosome count of n = 11, L. stylosa, L. torulosa, and L. uniflora exhibiting n = 15, and L. texana and L. aurea exhibiting n = 24. Based on self-pollination experiments conducted in L. exigua var. exigua and L. exigua var. lutea, L. exigua was reported to be self-compatible (Rollins, 1963), although to our knowledge, self-incompatibility in L. exigua var. laciniata has not been tested.
One of the three varieties of L. exigua is Kentucky glade cress (L. exigua var. laciniata), which has a naturally small distribution restricted to a small portion of Bullitt and Jefferson Counties in Kentucky. Because the geographic range of L. exigua var. laciniata is located just south of the Louisville, KY metropolitan area, which is rapidly expanding southward, its habitat is progressively being lost or degraded through residential and commercial development. Over half of the populations of L. exigua var. laciniata have already experienced declines, and many populations have been extirpated (USFWS, 2014). Because of these declines in both habitat and population size, L. exigua var. laciniata was recently listed as threatened under the U.S. endangered species act (USFWS, 2014).
Despite the fact that L. exigua var. laciniata is threatened, many aspects of its basic biology, mating system, and genetic diversity are unknown; additional research is needed to understand more about the species to facilitate conservation efforts. No previous population genetic study has focused on L. exigua var. laciniata, even though this information is critical for devising practical conservation strategies for threatened species (Frankham, 1995; Rossetto et al., 2021). For example, the amount of genetic diversity contained in populations, patterns of genetic structure, and the best strategy for protecting the genetic variation in L. exigua var. laciniata are currently unknown. Also unknown are whether the varieties of L. exigua are genetically distinct and whether they deserve recognition as distinct evolutionary significant units (ESU; Moritz, 1994).
In this study, we sampled multiple populations of two of the three varieties of L. exigua (L. exigua var. laciniata and L. exigua var. exigua), genotyped them at 16 polymorphic microsatellite loci, and analyzed the resulting genetic data to understand: (1) genetic diversity and structure within and among populations of L. exigua var. laciniata, (2) whether L. exigua var. laciniata is distinct genetically from L. exigua var. exigua, and (3) the implications for prioritization of in-situ and ex-situ conservation efforts. Based on the assumption that L. exigua var. laciniata is self-compatible (as this mating system was found in the other two varieties of L. exigua), we expected L. exigua var. laciniata to show patterns of population genetic diversity and structure similar to those found previously in other self-compatible species (e.g., Liu et al., 1998; Koelling et al., 2011; Swift et al., 2016; Edwards et al., 2019), namely, low within-population genetic diversity and high among-population diversity, low observed heterozygosity, and large positive inbreeding coefficients. Based on their allopatric geographic distributions and distinctive morphological features, we also expected that the varieties of L. exigua would be genetically distinct from one another and worthy of recognition as ESUs for conservation purposes.
Methods
Sample Collection
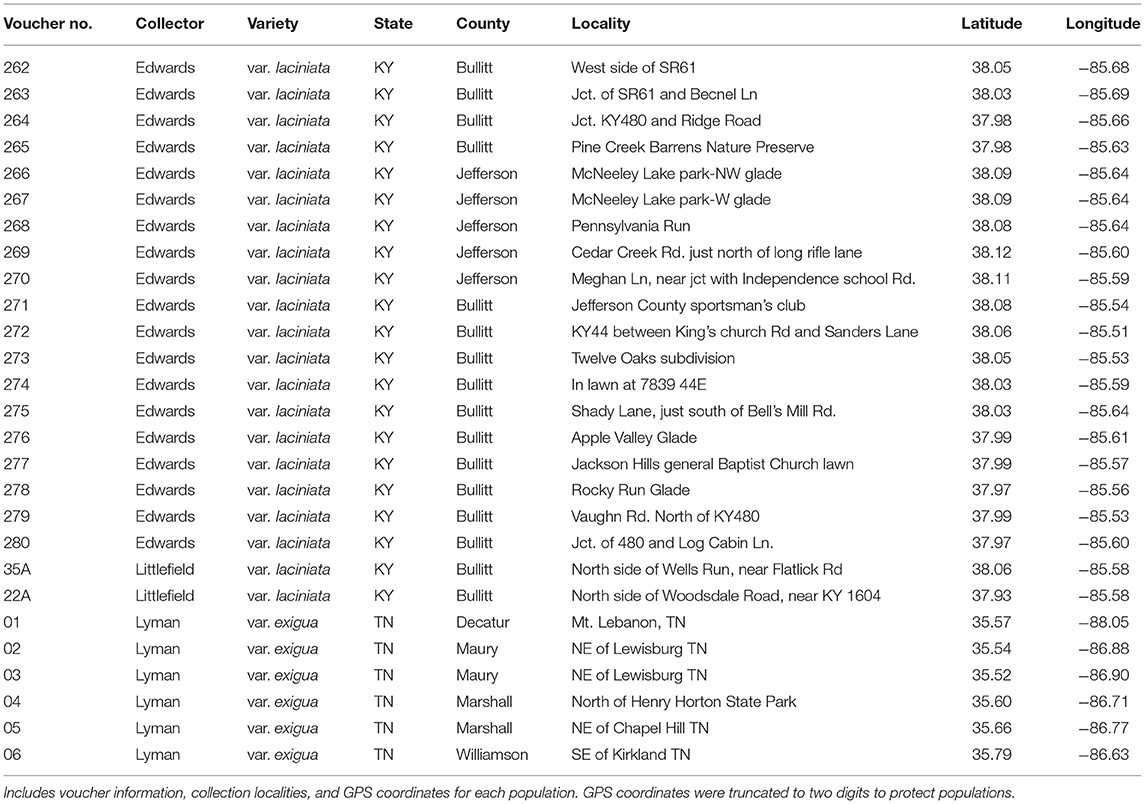
We conducted field work to sample populations of Leavenworthia exigua in April of 2016. We collected leaf tissue of between 16 and 25 individuals per population from 21 populations from throughout the geographic range of L. exigua var. laciniata and six populations of L. exigua var. exigua (Table 1; Figure 1A), resulting in 440 individuals of L. exigua var. laciniata and 136 individuals of L. exigua var. exigua, or 576 individuals in total. Individuals were collected as evenly as possible from throughout each site to capture the entire geographic extent of the population in each sampling location. Although L. exigua is not known to be clonal, we sampled from plants at least 1 meter apart to ensure that we sampled distinct individuals. Samples were dried in silica gel and brought back to the Conservation Genetics laboratory at the Missouri Botanical Garden for subsequent DNA analysis. We collected a voucher specimen in each population, which was deposited in the herbarium at the Missouri Botanical Garden (MO; Table 1). Although we made two attempts to locate and collect plants of L. exigua var. lutea, which at the time was known only from eight glades in six counties in Alabama, we were unable to locate these populations and therefore did not include L. exigua var. lutea in the study. A recent study reported that most historical populations of L. exigua var. lutea were extirpated, but also reported the discovery of several new populations (Frings et al., 2019).
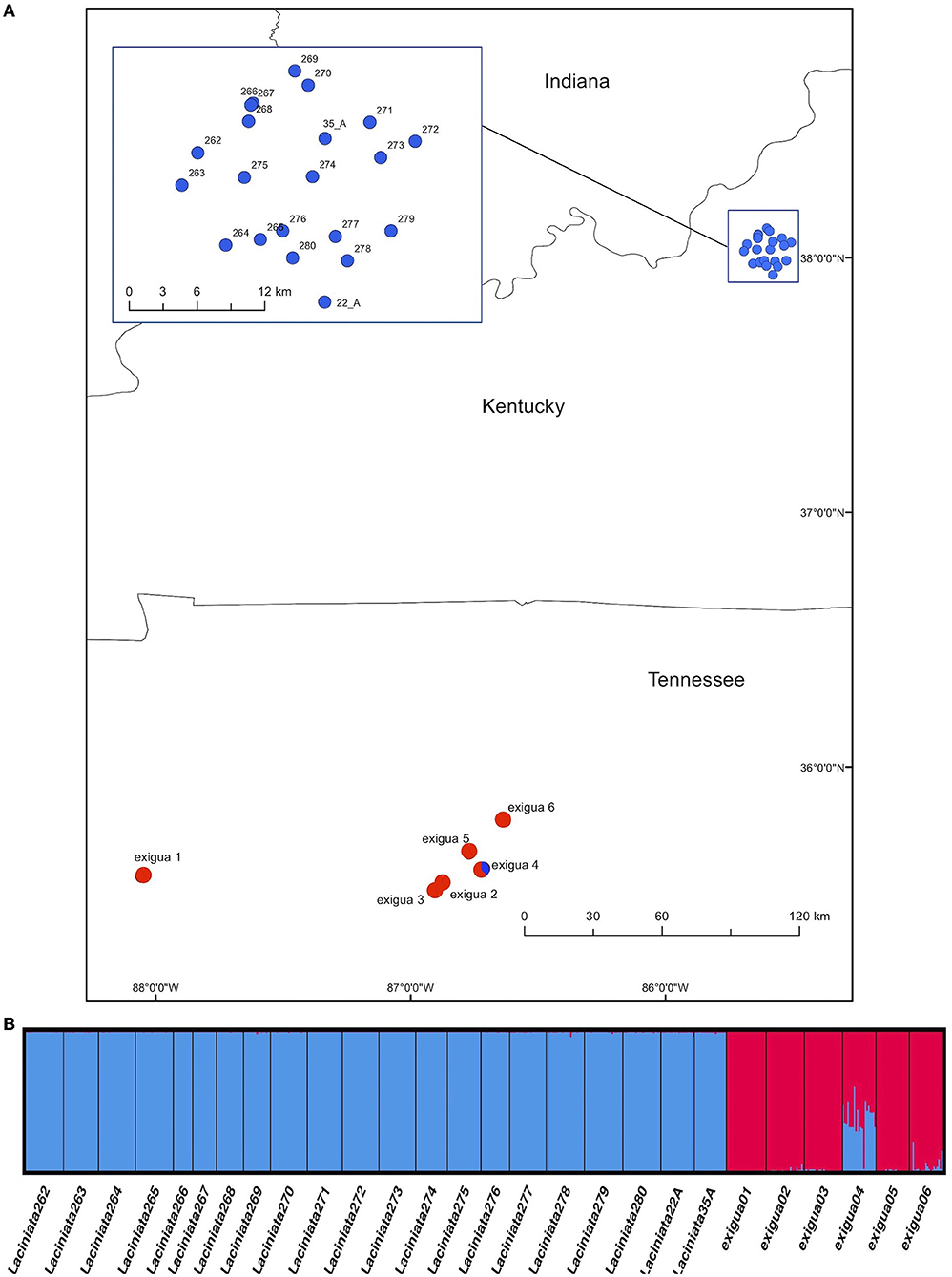
Figure 1. Map of sampling locations for Leavenworthia exigua and results of STRUCTURE analysis at K = 2. (A) Map showing sampling locations for Leavenworthia exigua, including 21 populations of Leavenworthia exigua var. laciniata in Kentucky and 6 populations of Leavenworthia exigua var. exigua in Tennessee. The pie charts indicate the proportional assignment of individuals in each population to the STRUCTURE clusters shown in (B). (B) STRUCTURE diagram, with each of the labeled blocks representing a group of individuals sampled at a location, each line representing an individual, and the colors representing the genetic clusters to which individuals are assigned.
Genetic Analysis
We extracted DNA from each individual using a CTAB approach (Doyle and Doyle, 1987). To identify microsatellites, we followed the protocol described in Swift et al. (2016). We carried out shotgun sequencing of genomic DNA extracted from two DNA samples of L. exigua var. laciniata. The source material for the DNA samples was bulked seed from the MBG seedbank (accession 2015-1048, a population that occurs within the range of L. exigua var. laciniata in Bullitt county, KY that was identified morphologically as L. exigua var. laciniata). Each sample contained several seeds of L. exigua var. laciniata derived from multiple maternal individuals. Library preparation was conducted using NEBnext Ultra DNA sample prep kits and NEBnext multiplex oligos (NEB). The indexed samples of L. exigua var. laciniata were pooled with samples from other studies and sequenced on an Illumina Miseq using 2 × 150 bp paired-end reads. We trimmed low-quality bases of the resulting reads and assembled the reads de novo into contigs using the Medium sensitivity/fast setting in Geneious (Biomatters Inc., Auckland, New Zealand). We searched contig consensus sequences for di-, tri-, and tetra-nucleotide microsatellite repeats using MSATCOMMANDER (Faircloth, 2008). Polymerase chain reaction (PCR) primers were designed from a subset of these sequences using the default settings in PRIMER3 (Rozen and Skaletsky, 2000; Faircloth, 2008).
We designed 144 primers and conducted an initial test for amplification using genomic DNA from two samples of L. exigua var. laciniata and one individual of L. exigua var. exigua. Microsatellite genotyping employed a universal dye-labeling approach, which involves adding an M13 tail (CACGACGTTGTAAAACGAC) to the 5' end of forward primers and use of a corresponding, fluorescently-labeled universal primer (Boutin-Ganache et al., 2001). PCR amplifications were performed in 10 μL reactions containing 0.5 U of GoTaq Flexi DNA polymerase (Promega corporation, Madison, Wisconsin, USA), 1× Promega Colorless GoTaq Flexi Buffer, 1.5 mM MgCl2, 4.5 pmol each of the reverse primer and one of four fluorescently labeled M13 primers (6-FAM, VIC, NED, or PET; Applied Biosystems), 0.18 pmol of the M13-tagged forward primer, and 0.5 mM of each dNTP. PCR temperature cycling conditions were as follows: (I) 3 min at 94°C, (II) denaturation for 30 s at 94°C, (III) annealing for 30 s at 52°C, (IV) extension for 45 s at 72°C, (V) 35 repetitions of steps 2–4, and (VI) a final elongation at 72°C for 20 min. We diluted PCR products 1:10 and sent them for genotyping on an ABI3730xl DNA Analyzer, with all genotyping carried out by loading 1 μL of the diluted PCR product, 9.0 μL of formamide, and 0.5 μL LIZ 500 size standard (Applied Biosystems, Waltham, Massachusetts, USA). All genotyping was conducted at the DNA analysis facility at Science Hill at Yale University.
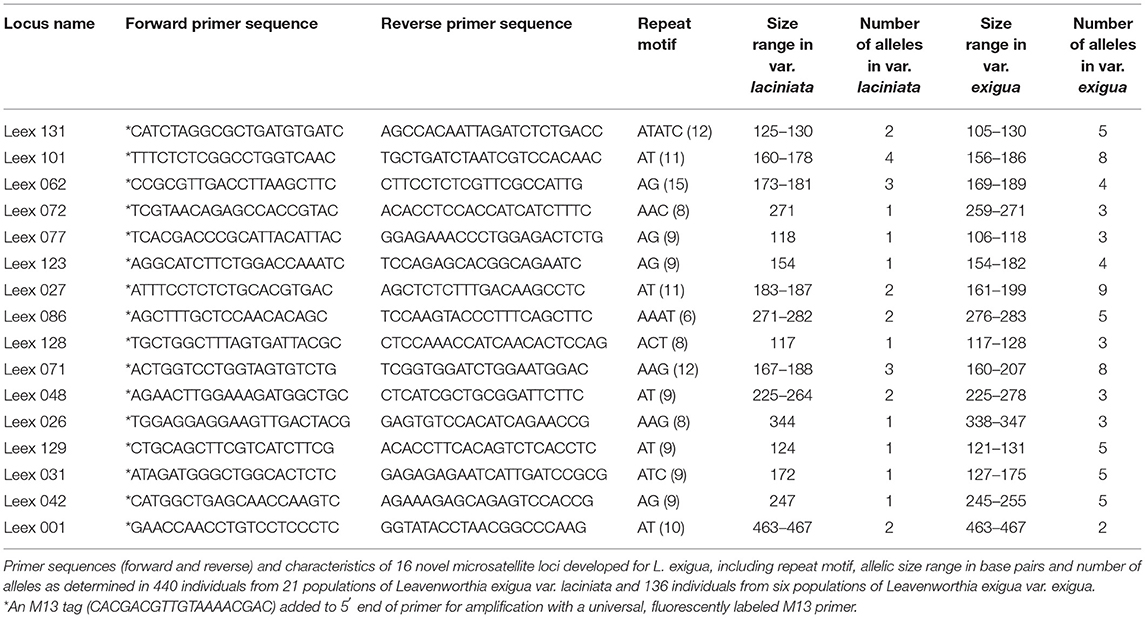
In the initial screening for amplification, we identified 67 loci that amplified in the two samples of L. exigua var. laciniata, of which 63 also amplified in L. exigua var. exigua. We selected 44 markers that amplified in all three samples for additional testing for amplification and polymorphism in 19 individuals of L. exigua, including 13 individuals of L. exigua var. laciniata and 6 individuals of L. exigua var. exigua. Although the 13 samples of L. exigua var. laciniata were sampled from throughout its geographic range, most loci exhibited little to no variation among these individuals. In contrast, we found that 21 of these loci were variable in the six individuals of L. exigua var. exigua. We therefore selected any marker that exhibited two or more alleles in L. exigua var. laciniata and those that were the most variable in L. exigua var. exigua. In total, we employed 16 markers to genotype all 576 individuals in this study (Table 2). For genotyping of all individuals, we used one of four different fluorescently labeled M13 primers to label each locus, and then we pooled the PCR products of the four uniquely labeled loci and ran them as described above. Fragment analysis and scoring were carried out using automated fragment scoring panels developed for each locus in GeneMarker version 1.6 (Soft Genetics LLC, State College, Pennsylvania, USA), and then the data were checked manually.
Data Analysis
We tested for deviations from Hardy-Weinberg equilibrium (HWE) at each locus/population combination using Fisher's exact tests in Genepop (Rousset, 2008). Population genetic diversity indices and summary statistics were estimated using Genodive (Meirmans, 2006) and GenAlEx (Peakall and Smouse, 2012); diversity indices included the average number of alleles per population, number of private alleles (AP), observed and expected heterozygosity (HO and HE, respectively), and the inbreeding coefficient (FIS). Because we observed large numbers of identical genotypes, we used Genodive to calculate Nei's clonal diversity (D) and to assign individuals to clones, with missing data not counted and a threshold of 0 differences to assign individuals to the same clone. We also analyzed the relatedness among individuals in the entire dataset using the software COANCESTRY v 1.0. Relatedness estimates were obtained using the triadic estimator (Wang, 2007) with 100 bootstraps while accounting for inbreeding. For relatedness estimates, values when individuals are unrelated, half sibs, full sibs, and parent-offspring are expected to be around 0, 0.25, 0.5, and 0.5, respectively (Wang, 2007). Values approaching 1 would indicate identical genotypes.
To understand the hierarchical partitioning of genetic variation within and among populations and individuals, we used analyses of molecular variance (AMOVA; Excoffier et al., 1992). Because patterns of genetic diversity were strikingly different between L. exigua var. exigua and L. exigua var. laciniata (see results), AMOVA was analyzed separately for the two taxa. We conducted AMOVA as implemented in Arlequin, using a locus-by-locus AMOVA with 1,000 permutations (Excoffier and Lischer, 2010).
Patterns of genetic structure were analyzed by calculating pairwise FST between all possible pairs of populations in Genodive. Because FST does not scale linearly with changes in diversity, we also calculated a standardized measure of pairwise population differentiation, Jost's D (Jost, 2008) in Genodive. We also used GenAlEx to calculate Nei's pairwise genetic distance among populations (Nei, 1972), which was visualized using a neighbor-joining phylogram using PAUP* (Swofford, 2002).
To investigate patterns of genetic structure in Leavenworthia exigua without a priori grouping of individuals into populations, we also analyzed our data using the Bayesian program STRUCTURE (Pritchard et al., 2000). We analyzed patterns of genetic structure in the entire data set using 10 independent runs at each K from 1 to 15, allowing for admixture, correlated allele frequencies, with population information used only to organize individuals in figures to understand how population designations corresponded to patterns of genetic structure. After preliminary analyses to determine the adequate burn-in and number of iterations, we used a burn-in of 250,000 iterations and a run length of 500,000 iterations for each chain. To ensure convergence and repeatability, we examined the groupings across all runs at each K in CLUMPAK (Kopelman et al., 2015) to ensure that the results of analyses were fully repeatable (Gilbert et al., 2012). To determine the optimal value of K, we used Structure Harvester (Earl and Vonholdt, 2012) to visualize ΔK (Evanno et al., 2005) and to plot the –ln likelihood values at each value of K. As recommended in the STRUCTURE documentation (Pritchard et al., 2000), we selected the K-value at which the –ln likelihood values began to plateau and that showed clear patterns of genetic structure.
Results
Patterns of Genetic Diversity in Leavenworthia exigua
In L. exigua var. laciniata, we found almost no genetic variation within and among populations. Eight of the 16 loci showed only a single invariant allele across all 440 individuals (Table 2), five loci showed only two alleles across all individuals of L. exigua var. laciniata, and only three loci had >2 alleles (Table 2). Of the loci that showed >1 allele, two loci demonstrated fixed heterozygosity with the two alleles invariant across all individuals of L. exigua var. laciniata, one locus showed fixed heterozygosity in all individuals of L. exigua var. laciniata except one alternative allele present in one population, and five loci showed a single invariant allele that was fixed across nearly all individuals of L. exigua var. laciniata except for one or two populations, where an alternative allele was present. Thus, the vast majority of individuals in L. exigua var. laciniata were all genetically identical with the exception of five populations that each differed from the dominant genotype by 1–2 private alleles (Table 3). We found a single genotype in each population and clonal diversity values were zero for all populations (Table 3). Further, assignment of individuals to clones identified only one single clone across all 440 individuals of L. exigua var. laciniata. Mean relatedness between individuals in each population was also extremely high, ranging from 0.83 to 1.0 (Table 3), and results largely showed high relatedness among individuals of L. exigua var. laciniata regardless of where the individuals were sampled (Figure 2). One exception to this is population 269, which showed two private alleles (Table 3) and showed lower relatedness values in comparison to individuals from different populations. Because several loci demonstrated fixed heterozygosity, the average number of alleles per locus was slightly greater than one for all populations of L. exigua var. laciniata (range 1.188–1.375; Table 3) and observed heterozygosity (range 0.106–0.188; Table 3) was greater than expected heterozygosity (range 0.061–0.115) in all populations, resulting in negative values of FIS in every population (range −0.464 to −1; Table 3). All but one population of L. exigua var. laciniata demonstrated significant deviations from Hardy-Weinberg equilibrium (i.e., significant heterozygote excess; Table 3) due to some loci exhibiting fixed heterozygosity across all individuals in a population.
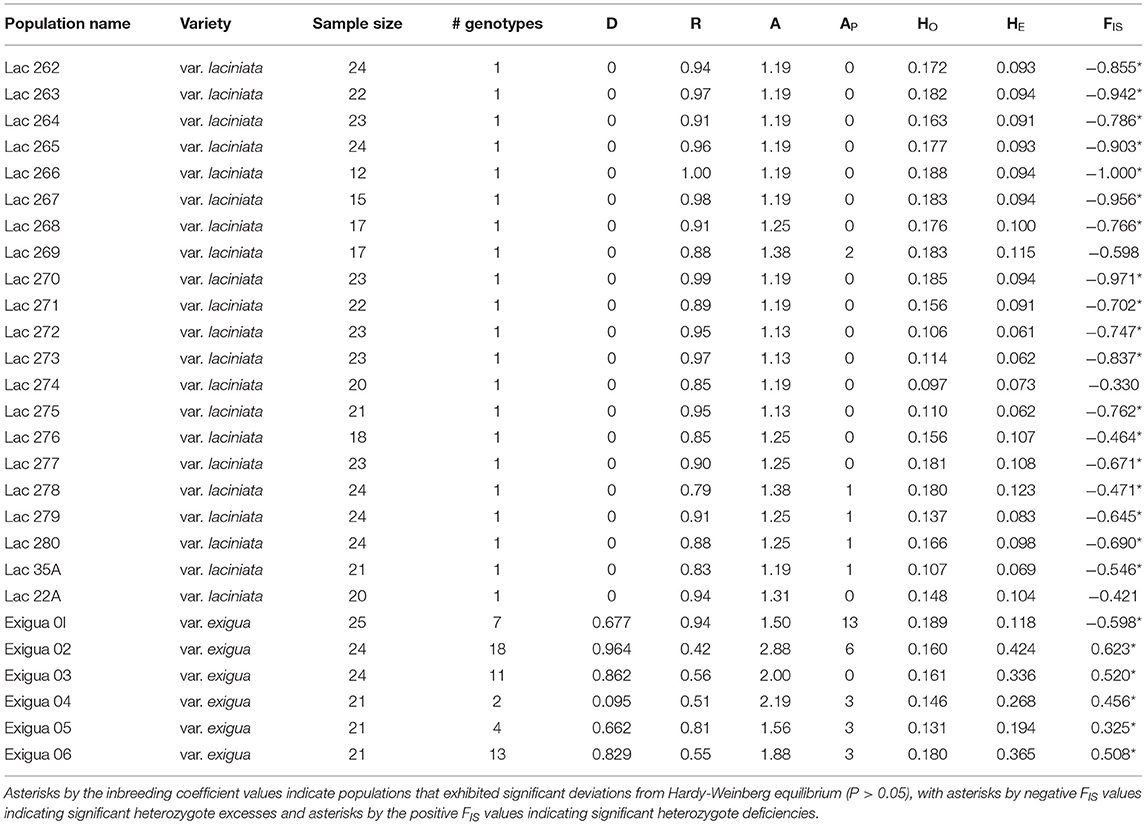
Table 3. Levels of genetic diversity in sampled populations of Leavenworthia exigua, including number of sampled individuals per population, number of genotypes found in each population, clonal diversity (D), mean relatedness among individuals within a population (R), average number of alleles (A), private alleles (AP) observed, and expected heterozygosity (HO and HE, respectively), the inbreeding coefficient (FIS).
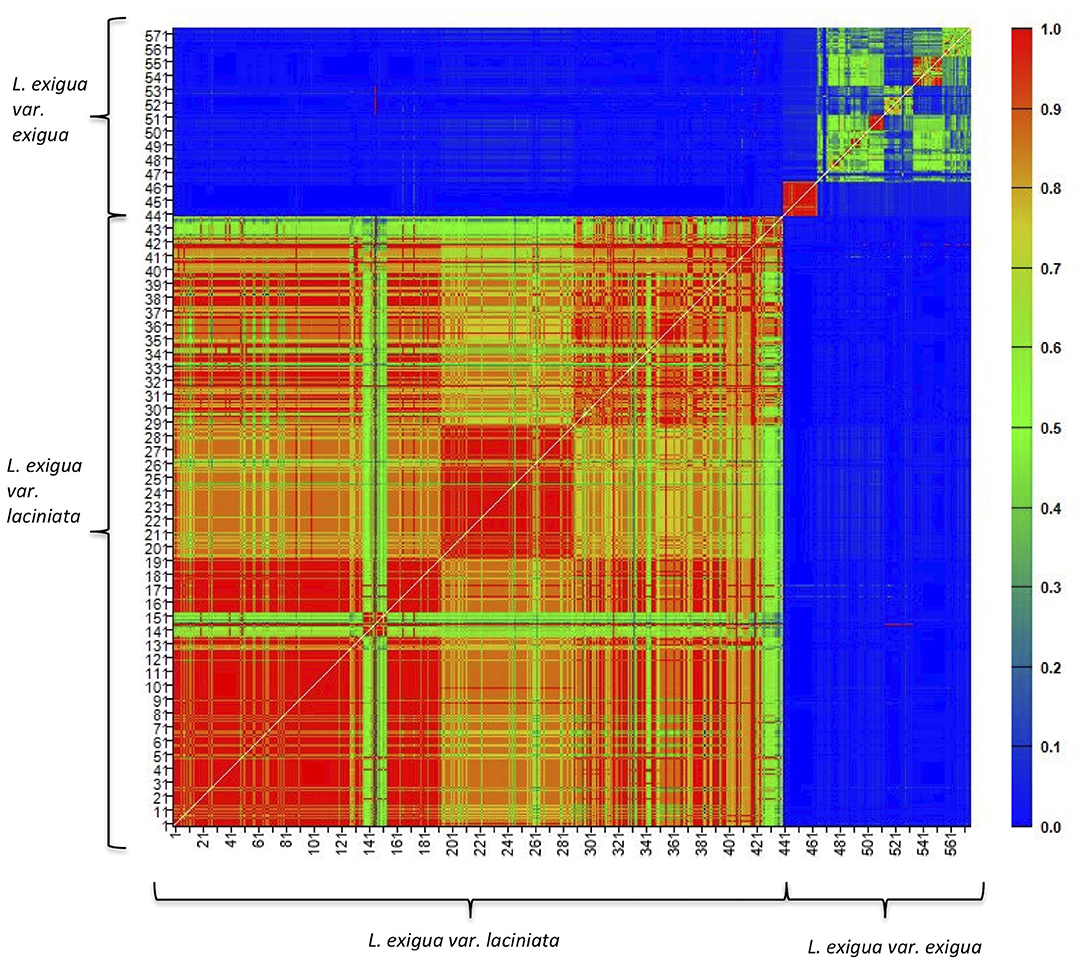
Figure 2. Heatmap of pairwise relatedness between all individuals of Leavenworthia exigua sampled in this study. Warm values indicate high levels of pairwise relatedness and cool values indicate lower relatedness. Individuals 1–440 were identified as L. exigua var. laciniata and 441–576 indicate individuals of L. exigua var. exigua.
In comparison to L. exigua var. laciniata, L. exigua var. exigua showed much greater genetic variation. Although a subset of individuals was genetically identical within each population, we found between 2 and 18 distinct genotypes in each population of L. exigua var. exigua and clonal diversity ranged from 0.095 to 0.964 (Table 3). For all populations of L. exigua var. laciniata except Exigua 01, mean relatedness values were lower than those found in L. exigua var. laciniata, ranging from 0.42 to 0.81 (Table 3), although they were still generally high, likely due to inbreeding in most populations (see below). Mean relatedness in population Exigua 01 was 0.95, similar to values found in L. exigua var. laciniata. Mean relatedness was also higher within populations than between populations (Figure 2). We found between 3 and 9 alleles at each locus across all 136 individuals of L. exigua var. exigua (Table 2). Within populations, the average number of alleles per locus ranged from 1.5 to 2.875 (Table 3). Populations of L. exigua var. exigua contained unique genetic variation, ranging from 3 to 13 private alleles in every population except population Exigua 3 (Table 3). For all populations except Exigua 01, expected heterozygosity ranged from 0.118 to 0.424, observed heterozygosity ranged from 0.131 to 0.189 (Table 3), and expected heterozygosity was greater than observed heterozygosity. Correspondingly, these populations demonstrated significant heterozygote deficiencies and FIS values that were <0.325 (Table 3). In contrast, population Exigua 01 exhibited greater observed heterozygosity than expected heterozygosity, a negative FIS value of −0.598, and a significant heterozygote deficiency, all due to three loci that exhibited fixed heterozygosity across the majority of individuals in the population.
Genetic Structure
AMOVA analyses to understand the hierarchical partitioning of genetic variation within and among populations and individuals revealed large differences in how genetic variation was partitioned in the two varieties of L. exigua. In L. exigua var. laciniata, populations were highly genetically similar to each other and individuals within populations were all genetically identical, such that 20% of the genetic variation was partitioned among populations, 0% was partitioned among individuals within populations, and 80% was found within individuals (Table 4). In L. exigua var. exigua, populations were genetically divergent from one another and at least some individuals within populations differed genetically, such that 56% of the variation was partitioned among populations, 13% of the variation was partitioned among individuals within populations, and 31% of the variation was found within individuals (Table 4).
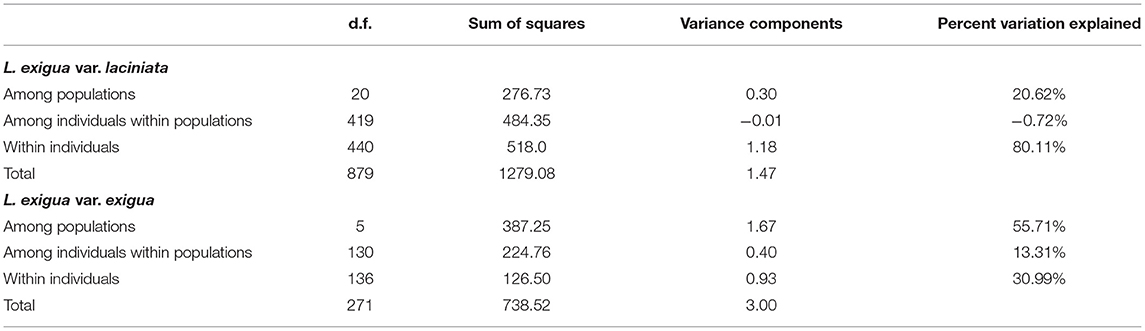
Table 4. Results of AMOVA analyses for Leavenworthia exigua, with L. exigua var. laciniata and L. exigua var. exigua analyzed separately.
Results of analyses of genetic structure as estimated by pairwise FST and Jost's D were similar to each other, with lower pairwise FST values between populations of L. exigua var. laciniata and greater values found both between populations of L. exigua var. exigua and between populations of L. exigua var. laciniata and L. exigua var. exigua (Supplementary Table 1). Most pairwise FST values between populations of L. exigua var. laciniata ranged from 0 to 0.15, but comparisons involving two specific populations (Lac269 and Lac35A) were all much greater, ranging between 0.06 and 0.49. Pairwise FST values ranged from 0.12 to 0.81 between populations of L. exigua var. exigua and from 0.50 to 0.89 between populations of L. exigua var. laciniata and L. exigua var. exigua (Supplementary Table 1). Jost's D, which is scaled according to the diversity present in a population, ranged from 0.00 to 0.08 between populations of L. exigua var. laciniata, from 0.08 to 0.81 between populations of L. exigua var. exigua, and from 0.34 to 0.81 between populations of L. exigua var. laciniata and L. exigua var. exigua (Supplementary Table 1).
Similar to the analyses of Pairwise FST and Jost's D, the phylogram based on Nei's genetic distance showed the populations of L. exigua var. laciniata forming a tight cluster, which was separated by a long branch from populations of L. exigua var. exigua, with populations of L. exigua var. exigua also separated by longer branches (Figure 3). Of the L. exigua var. exigua populations, the Exigua 04 population was placed as the most genetically similar to L. exigua var. laciniata, and the Exigua 01 population was the most genetically divergent from all other populations (Figure 3).
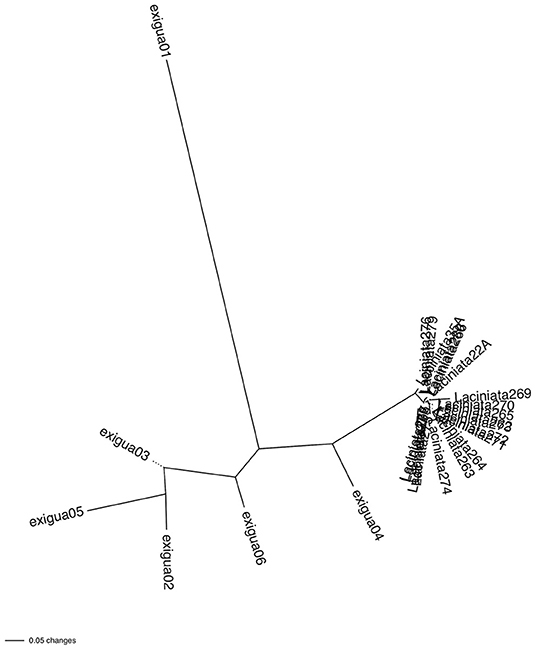
Figure 3. Unrooted neighbor-joining phylogram of Nei's genetic distance matrix of populations of Leavenworthia exigua, with branch lengths proportional to genetic distance among populations. The phylogram shows that Leavenworthia exigua var. laciniata populations are largely genetically identical or very similar to each other whereas Leavenworthia exigua var. exigua shows longer branch lengths, indicating genetic variation among populations.
For analyses with the Bayesian program STRUCURE, the Evanno method and plots of the –ln probability of the data both showed that the optimal number of clusters in the data was K = 2 (Supplementary Figure 1). At K = 2, STRUCTURE clustered all individuals of L. exigua var. laciniata into one cluster and the populations of L. exigua var. exigua into the other (Figure 1B) except the population Exigua 04 was shown to be admixed between the clusters containing the two varieties (Figure 1B). We also inspected results of analyses at greater values of K; K = 3 divided population Exigua 01 into its own group, whereas K = 4 did not show any additional clear patterns of genetic structure (results not shown).
Discussion
Genetic Diversity and Mating System in Leavenworthia exigua var. laciniata
Contrary to expectations, the results of this study suggest that L. exigua var. laciniata may exhibit an asexual mating system. Given that the other two varieties of L. exigua are self-compatible (Rollins, 1963), we expected that L. exigua var. laciniata would show population genetic patterns similar to other self-compatible Leavenworthia (Liu et al., 1998; Koelling et al., 2011), such as low within-population genetic variation, greater among-population variation, low observed heterozygosity, and high positive values of FIS. All of these were found in all populations of L exigua var. exigua except the Exigua 01 population. In contrast, L. exigua var. laciniata showed virtually no within- or among-population genetic variation at all. Assignment of individuals to clones assigned all 440 individuals of L. exigua var. laciniata to a single clone, and relatedness between individuals was high regardless of the population in which it was sampled. Results also showed that all individuals in 17 out of 22 populations were genetically identical to each other, while the only variation observed in L. exigua var. laciniata was unique fixed differences at 1–2 alleles per population in each of five populations, which may have arisen due to point mutations in the dominant genotype in each population that subsequently became fixed. We also observed fixed heterozygosity at several loci, significant excesses of heterozygotes relative to HWE, and high negative inbreeding coefficients, all of which are consistent with the lack of sexual recombination characteristic of asexuality (Balloux et al., 2003). Asexuality has not been documented previously in the genus Leavenworthia; the possibility of the existence of a taxon with an asexual mating system in Leavenwortha is particularly surprising given that it is such a well-studied genus used as a model for investigating the evolution of mating systems.
One alternative explanation for the lack of genetic variation found in L. exigua var. laciniata is that it is an artifact of the particular microsatellite markers developed for this study, possibly related in some way to how we isolated the microsatellite loci. However, we employed well-established marker-development protocols that we have used successfully to identify polymorphic microsatellites in a wide diversity of plant species (Swift et al., 2016; Edwards et al., 2019, 2021; Mohn et al., 2021). Furthermore, while developing the markers, we attempted to avoid ascertainment bias by incorporating material from multiple maternal individuals of L. exigua var. laciniata in the pool of seeds used for marker development, and we screened 44 loci for polymorphism in individuals sampled from across the range of L. exigua var. laciniata. Yet despite the efforts taken to isolate variable markers, we detected only a small amount of variation at six loci employed for genotyping, while the remaining 10 loci were invariant. These same markers were also variable within and among populations of L. exigua var. exigua despite being isolated from L. exigua var. laciniata. To verify the results of microsatellite analysis, we examined the results of a small preliminary AFLP analysis of 48 individuals grown from seeds sampled from five populations of L. exigua var. laciniata (B. Williams, unpublished data). Although asexuality is difficult to assess using dominant markers, this analysis also revealed low genetic diversity and extremely low genetic distances among populations (Nei's D = 0.01–0.06; B. Williams, unpublished data). However, additional comparative genome-wide genetic analyses of L. exigua var. laciniata and other related taxa are needed to explore the extent of genomic variation in L. exigua var. laciniata to test further the hypothesis of asexuality.
Although the microsatellite evidence generated in the present study may indicate an asexual mating system in L. exigua var. laciniata, how asexuality could be accomplished is an open question. In general, asexual reproduction can be achieved by plants in two ways: vegetative reproduction/clonality and apomixis. Vegetative reproduction/clonality is accomplished chiefly by budding from roots, rhizomes, stems, or storage organs such as tubers, resulting in the production of new ramets of the same genetic individual (Yang and Kim, 2016). Population genetic patterns for such species frequently reveal one or a few fixed heterozygous genotypes within a population, but these species usually exhibit differences in the genotypes present among populations (e.g., Gitzendanner et al., 2012; Fehlberg, 2017). Clonality also generally has the effect of increasing an individual's lifespan, such that clonal species are typically perennials with lifespans of >5 years, with some genets living up to tens of thousands of years (de Witte and Stöcklin, 2010). L. exigua var. laciniata has a winter-annual life history strategy, individuals are not thought to persist for more than 1 year, and plants do not develop rhizomes or stolons; it is therefore unlikely that vegetative reproduction is the means by which asexual reproduction is occurring in L. exigua var. laciniata. The fact that the same genotype of L. exigua var. laciniata is found across its range indicates that it dispersed among populations. This suggests that asexuality may instead be the result of apomixis, or asexual reproduction by seed. However, apomixis is extremely uncommon in the Brassicaceae, reported from only five genera (Boechera, Draba, Erysimum, Parrya, and Phoenicaulis) and one intergeneric hybrid (Raphanobrassica) (Mandáková et al., 2021) of the 349 accepted genera in the Brassicaceae (The Angiosperm Phylogeny Group et al., 2016). If asexuality in L. exigua var. laciniata is accomplished through apomixis, this would be an independent evolutionary origin of apomixis in the family. However, additional research is needed to investigate whether L. exigua var. laciniata is indeed apomictic.
Apomixis occurs through the formation of a diploid embryo without involving the fusion of haploid gametes or the act of fertilization (Hand and Koltunow, 2014). The mechanisms that cause apomixis are generally grouped into two categories: sporophytic or gametophytic (Hand and Koltunow, 2014). During sporophytic apomixis, the embryo sac develops following the typical angiosperm sexual pathway, but the embryo arises from diploid somatic ovule cells that surround the embryo sac (Hand and Koltunow, 2014). These embryo cells can develop to maturity only if the sexually derived embryo sac is fertilized, as the sexual and asexual embryos share the endosperm; a seed may therefore contain multiple embryos, although the sexual embryo does not always develop (Hand and Koltunow, 2014). In gametophytic apomixis, which is the more common type of apomixis in angiosperms, the embryo sac develops from either a diploid megaspore mother cell (diplospory) or from any diploid cell of the nucellus (apospory) (Hand and Koltunow, 2014).
Although extensive research has been conducted to understand apomixis in Boechera, presently, it is still unclear if L. exigua var. laciniata is apomictic and if so, whether similar mechanisms gave rise to apomixis in Leavenworthia. In Boechera, species vary in ploidy and sexuality; some species are diploid and sexual, some are diploid apomicts, and some are triploid apomicts. Although all apomixis in Boechera is gametophytic (Lovell et al., 2013), it can occur both through diplospory and apospory (Carman et al., 2019). Diploid apomicts are thought to have arisen either through hybridization (Beck et al., 2006; Carman et al., 2019) or spontaneously through genetic factors that cause apomixis (Lovell et al., 2013), whereas triploid apomicts are thought to have arisen through intraspecific hybridization between a diploid apomict and another, genetically divergent diploid species (Lovell et al., 2013). Although a previous phylogenetic study in Leavenworthia suggested that past hybridization occurred between L. exigua and L. torulosa (Beck et al., 2006), L. exigua var. laciniata was not included in this analysis, such that it is unclear whether hybridization may have led to its formation. Chromosome counts for all varieties of L. exigua were n = 11 (Rollins, 1963), the lowest number in the genus Leavenworthia, indicating that L. exigua var. laciniata is diploid. The diploid apomicts in Boechera produce seeds that have either with a 2C:5C or a 2C:6C embryo-to-endosperm ratio (Lovell et al., 2013). Preliminary flow cytometry of five seeds from one maternal individual of L. exigua var. laciniata showed that they may have the standard embryo-to-endosperm ratio of 2C:3C generally found in non-apomictic, sexual diploids (Tim Sharbel, pers. comm.). However, it is unclear whether this is consistent across maternal individuals and populations. Thus, flow cytometry of paired leaf and seed samples from multiple populations from all three varieties of Leavenworthia exigua and other Leavenworthia species with the same chromosome number is needed to further investigate the possibility of apomixis in L. exigua var. laciniata.
Genetic Distinctiveness of Varieties of L. exigua
The next goal of the study was to address whether L. exigua var. laciniata is genetically distinct from the other two varieties of L. exigua. In addition to the differences in morphology and distinct geographical ranges noted by Rollins (1963), the results of the present study show that the varieties may differ in mating system. In contrast to the putative asexual mating system exhibited by L. exigua var. laciniata, most populations of L. exigua var. exigua displayed population genetic patterns consistent with those found in other Leavenworthia species with a sexual, self-compatible mating system, including lower observed heterozygosity than expected heterozygosity, high positive inbreeding coefficients, and significant heterozygote deficiencies relative to Hardy-Weinberg equilibrium (Liu et al., 1998; Koelling et al., 2011). L. exigua var. exigua also showed a much greater number of distinct genotypes per population and overall greater genetic variation than L. exigua var. laciniata, with the majority of genetic variation partitioned among populations, as is common in self-compatible plant species. However, population Exigua 01 exhibited population genetic patterns more similar to those exhibited by L. exigua var. laciniata, potentially indicating that one population of L. exigua var. exigua may exhibit asexuality and that the mating system may vary among populations of L. exigua var. exigua. Additional sampling of populations across the range of L. exigua var. exigua is needed to investigate this possibility further. The two varieties analyzed in the present study are also significantly genetically differentiated, likely because gene flow, which acts to minimize differences among populations, may not be occurring between the two varieties because of geographical separation due to allopatry and potential asexuality in L. exigua var. laciniata.
Because we were unable to locate populations of L. exigua var. lutea, we were unable to analyze its mating system and its distinctiveness from the other two varieties of L. exigua. Previous analyses showed that L. exigua var. lutea is self-compatible, such that we expect that it would show population genetic patterns similar to those found in most populations of L. exigua var. exigua. L. exigua var. lutea is also allopatric, such that it may not have the opportunity to interbreed with the other two varieties; however, additional research is needed to test whether it is genetically distinct from the other two varieties of L. exigua. The extent of reproductive isolation among the three varieties of L. exigua is also unknown. If L. exigua var. laciniata is asexual, then it could be reproductively isolated from the other varieties of L. exigua, although many apomictic species retain viable pollen, serving as a mechanism facilitating sexual reproduction (Majeský et al., 2017). Additional analyses involving experimental crosses within and among the varieties paired with genetic and cytogenetic analyses of the resulting offspring are needed to further understand the extent to which the varieties exhibit reproductive isolation and whether L. exigua var. laciniata exhibits residual sexuality. If indeed L. exigua var. laciniata is asexual and reproductively isolated from the other two varieties of L. exigua, then it may warrant being recognized as a unique species. At the minimum, it should be considered as an evolutionary significant unit for conservation purposes.
Conservation Implications
The final goal of this study was to evaluate the implications for prioritization of in-situ and ex-situ conservation efforts for L. exigua var. laciniata. One of the main objectives of conservation genetics is to preserve the evolutionary potential of a species (Moritz, 2002). In sexual organisms, because recombination can produce different combinations of adaptive genes that can increase fitness, protecting the maximum amount of genetic variation in a species is thought to be important for preserving its adaptive capacity. In practice, researchers may identify unique genetic clusters and devise recommendations to protect these genetic groups to ensure the conservation of all unique subsets of genetic variation present in the species (e.g., Edwards et al., 2021). In L. exigua var. laciniata, the few observed genetic differences are probably the result of single mutations occurring in an individual that were then passed asexually to its offspring. Thus, to preserve the full range of genetic variation in L. exigua var. laciniata, it would therefore be necessary to ensure the protection of the five populations that exhibit these small mutations (i.e., those containing private alleles in Table 3), along with at least one population that has the dominant genotype.
Arguably, however, because the evolutionary process is somewhat different for asexual species, the strategy for protecting their evolutionary potential should be different than for sexual species. In asexual organisms, adaptation may occur if a mutation arises in an individual that increases its fitness relative to its competitors, in which case it will quickly spread through the population. This normally occurs infrequently (Jiang et al., 2010), as most mutations are neutral or have deleterious effects. Furthermore, in obligately asexual species, because no recombination occurs, when two beneficial mutations arise in a population, they cannot recombine and have to compete with each other (Gerrish and Lenski, 1998), such that one may be eliminated from the population. In the absence of recombination, deleterious mutations are also expected to accumulate in asexual species in a process known as Muller's ratchet (Muller, 1964). However, most apomictic plants exhibit some low levels of facultative sexual reproduction, in which case recombination can segregate multiple deleterious mutations into single linkage groups that can be eliminated through selection; this process was shown previously to reduce mutational load in an apomictic species with low rates of facultative sexual reproduction (Hodač et al., 2019). Although it is unknown whether facultative sexual reproduction occurs in L. exigua var. laciniata, because the likelihood of sexual reproduction likely increases in larger populations, we recommend protecting as many individuals as possible to maintain the evolutionary potential in L. exigua var. laciniata.
For ex-situ conservation, the normal strategy for seed banking for sexual organisms is to collect seed from a geographically representative set of populations to capture the full range of genetic diversity present in a species (Guerrant et al., 2014), which allows the ex-situ collection to serve as a backup in the case that unique genetic variation in the species is lost. Given that L. exigua var. laciniata exhibits virtually no genetic variation, this will be easily accomplished in L. exigua var. laciniata. However, even if they do not contain large amounts of genetic diversity, ex-situ collections could still serve as material for reintroductions to promote the persistence of the species. Reintroductions using a large number of individuals, regardless of source, would help increase population size, and concomitantly increase the evolutionary potential of the species.
Conclusions and Future Directions
Although the genus Leavenworthia has been used extensively as a model for investigating the evolution of mating systems in plants, the results of this study indicate that one of the most poorly known, understudied species in the genus may have a previously unrecognized, asexual mating system that is unique to the genus. The existence of an asexual species with virtually no genetic variation in such a well-studied genus offers important opportunities for future research. Given the interest in the evolution of mating systems in Leavenworthia, it is likely that the discovery of a new mating system in Leavenworthia will spark additional research into the origins and mechanisms underlying asexuality in Leavenworthia. Furthermore, the lack of genetic variation in L. exigua var. laciniata provides opportunities for understanding how genetic variation affects a species' response to its environment and its ability to persist; for example, we are currently investigating whether Leavenworthia species that differ in mating systems and levels of genetic variation exhibit differences in epigenomic and phenotypic plasticity in response to environmental stress. Similarly, we are also investigating whether epigenomic variation and phenotypic variation is present in the threatened L. exigua var. laciniata that may enable it to persist in response to environmental variation despite a lack of genetic variation. Indeed, L. exigua var. laciniata may be a useful model for understanding the importance (or lack thereof) of genetic variation in the persistence of rare plants and how to conserve a threatened species with virtually no genetic variation in a changing climate.
Data Availability Statement
The data presented in the study are deposited in the Figshare repository, http://dx.doi.org/10.6084/m9.figshare.19116224; and GenBank, accession number PRJNA808735.
Author Contributions
CE designed the study, acquired funding for the study, and conducted field work. BB and BW conducted lab work. BB made maps. CE and BW analyzed the data. CE wrote the manuscript. All authors revised the article and approved the final version of the manuscript.
Funding
Funding for this study was provided by the United States Fish and Wildlife Service and the Kentucky Natural Lands Trust. Financial support for the MBG conservation genetics program was provided in part by Philip and Sima Needleman, Stephen and Camilla Brauer, and the Bellwether Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Tara Littlefield, Joel Swift, Rachel Lyman, and Jennifer Garland for assistance with locality data, permitting, and field work; Matthew Albrecht for providing seed and access to research facilities; Rachel Lyman for assistance with laboratory work; Andrew Berry and the Bernheim Research forest for providing lodging during fieldwork; and Jennifer Garland for assistance with funding. We also thank the guest editor Roberta Gargiulo and three peer reviewers for their comments on a previous version of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcosc.2022.831085/full#supplementary-material
References
Anderson, I. A., and Busch, J. W. (2006). Relaxed pollinator-mediated selection weakens floral integration in self-compatible taxa of Leavenworthia (Brassicaceae). Am. J. Bot. 93, 860–867. doi: 10.3732/ajb.93.6.860
Baldwin, S. J., and Schoen, D. J. (2017). Genetic variation for pseudo-self-compatibility in self-incompatible populations of Leavenworthia alabamica (Brassicaceae). New Phytol. 213, 430–439. doi: 10.1111/nph.14109
Baldwin, S. J., and Schoen, D. J. (2019). Inbreeding depression is difficult to purge in self-incompatible populations of Leavenworthia alabamica. New Phytol. 224, 1330–1338. doi: 10.1111/nph.15963
Balloux, F., Lehmann, L., and De Meeus, T. (2003). The population genetics of clonal and partially clonal diploids. Genetics 164, 1635–1644. doi: 10.1093/genetics/164.4.1635
Baskin, J. M., and Baskin, C. C. (1972). The ecological life cycle of the cedar glade endemic Leavenworthia exigua var. exigua. Can. J. Bot. 50, 1711–1723. doi: 10.1139/b72-212
Baskin, J. M., and Baskin, C. C. (1981). Geographical distribution and notes on the ecology of the rare endemic Leavenworthia exigua var. laciniata. Castanea 46, 243–247.
Beck, J. B., Al-Shehbaz, I. A., and Schaal, B. A. (2006). Leavenworthia (Brassicaceae) revisited: testing classic systematic and mating system hypotheses. Syst. Bot. 31, 151–159. doi: 10.1600/036364406775971732
Boutin-Ganache, I., Raposo, M., Raymond, M., and Deschepper, C. F. (2001). M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. BioTechniques 31, 24–25. doi: 10.2144/01311bm02
Busch, J. W. (2005a). Inbreeding depression in self-incompatible and self-compatible populations of Leavenworthia alabamica. Heredity 94, 159–165. doi: 10.1038/sj.hdy.6800584
Busch, J. W. (2005b). The evolution of self-compatibility in geographically peripheral populations of Leavenworthia alabamica (Brassicaceae). Am. J. Bot. 92, 1503–1512. doi: 10.3732/ajb.92.9.1503
Busch, J. W. (2006). Heterosis in an isolated, effectively small, and self-fertilizing population of the flowering plant Leavenworthia alabamica. Evolution 60, 184–191. doi: 10.1111/j.0014-3820.2006.tb01092.x
Busch, J. W., Joly, S., and Schoen, D. J. (2010). Does mate limitation in self-incompatible species promote the evolution of selfing? The case of Leavenworthia alabamica. Evolution 64, 1657–1670. doi: 10.1111/j.1558-5646.2009.00925.x
Busch, J. W., Joly, S., and Schoen, D. J. (2011). Demographic signatures accompanying the evolution of selfing in Leavenworthia alabamica. Mol. Biol. Evol. 28, 1717–1729. doi: 10.1093/molbev/msq352
Busch, J. W., Sharma, J., and Schoen, D. J. (2008). Molecular characterization of Lal2, an SRK-like gene linked to the S-locus in the wild mustard Leavenworthia alabamica. Genetics 178, 2055–2067. doi: 10.1534/genetics.107.083204
Busch, J. W., and Urban, L. (2011). Insights gained from 50 years of studying the evolution of self-compatibility in Leavenworthia (Brassicaceae). Evol. Biol. 38, 15–27. doi: 10.1007/s11692-010-9104-5
Busch, J. W., and Werner, W. J. (2012). Population structure has limited fitness consequences in the highly selfing plant Leavenworthia uniflora (Brassicaceae). Int. J. Plant Sci. 173, 495–506. doi: 10.1086/665264
Carman, J. G., Mateo De Arias, M., Gao, L., Zhao, X., Kowallis, B. M., Sherwood, D. A., et al. (2019). Apospory and diplospory in diploid Boechera (brassicaceae) may facilitate speciation by recombination-driven apomixis-to-sex reversals. Front. Plant Sci. 10:724. doi: 10.3389/fpls.2019.00724
Chantha, S.-C., Herman, A. C., Platts, A. E., Vekemans, X., and Schoen, D. J. (2013). Secondary evolution of a self-incompatibility locus in the Brassicaceae genus Leavenworthia. PLoS Biol. 11:e1001560. doi: 10.1371/journal.pbio.1001560
Charlesworth, D., and Yang, Z. (1998). Allozyme diversity in Leavenworthia populations with different inbreeding levels. Heredity 81, 453–461. doi: 10.1046/j.1365-2540.1998.00415.x
de Witte, L. C., and Stöcklin, J. (2010). Longevity of clonal plants: why it matters and how to measure it. Ann. Bot. 106, 859–870. doi: 10.1093/aob/mcq191
Doyle, J. J., and Doyle, J. L. (1987). A rapid isolation procedure for small quantities of fresh tissue. Phytochem. Bull. 19, 11–15.
Earl, D. A., and Vonholdt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Edwards, C. E., Albrecht, M. A., Bassüner, B., and Yatskievych, G. A. (2019). Population genetic analysis reveals a predominantly selfing mating system and strong genetic structuring in a naturally fragmented, threatened plant. Conserv. Genet. 20, 1437–1448. doi: 10.1007/s10592-019-01226-9
Edwards, C. E., Tessier, B. C., Swift, J. F., Bassüner, B., Linan, A. G., Albrecht, M. A., et al. (2021). Conservation genetics of the threatened plant species Physaria filiformis (Missouri bladderpod) reveals strong genetic structure and a possible cryptic species. PLoS ONE 16:e0247586. doi: 10.1371/journal.pone.0247586
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes - application to human mitochondrial-DNA restriction data. Genetics 131, 479–491. doi: 10.1093/genetics/131.2.479
Faircloth, B. C. (2008). Msatcommander: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour. 8, 92–94. doi: 10.1111/j.1471-8286.2007.01884.x
Fehlberg, S. D. (2017). Knowledge of genetic diversity informs conservation of a rare, clonal wetland plant Lilaeopsis schaffneriana subsp. recurva (Apiaceae). J. Bot. Res. Instit. Texas 11, 99–510. doi: 10.17348/jbrit.v11.i2.1086
Filatov, D. A., and Charlesworth, D. (1999). DNA polymorphism, haplotype structure and balancing selection in the Leavenworthia pgic locus. Genetics 153, 1423–1434. doi: 10.1093/genetics/153.3.1423
Frankham, R. (1995). Conservation genetics. Annu. Rev. Genet. 29, 305–327. doi: 10.1146/annurev.ge.29.120195.001513
Frings, D. M., Davenport, L. J., Oberholster, C., and Hardig, T. M. (2019). Current status of Tennessee gladecress, Leavenworthia exigua var. lutea (Brassicaceae), in Alabama. Southeast. Nat. 18, 419–429. doi: 10.1656/058.018.0308
Gerrish, P. J., and Lenski, R. E. (1998). The fate of competing beneficial mutations in an asexual population. Genetica 102, 127–144. doi: 10.1023/A:1017067816551
Gilbert, K. J., Andrew, R. L., Bock, D. G., Franklin, M. T., Kane, N. C., Moore, J. S., et al. (2012). Recommendations for utilizing and reporting population genetic analyses: the reproducibility of genetic clustering using the program STRUCTURE. Mol. Ecol. 21, 4925–4930. doi: 10.1111/j.1365-294X.2012.05754.x
Gitzendanner, M. A., Weekley, C. W., Germain-Aubrey, C. C., Soltis, D. E., and Soltis, P. S. (2012). Microsatellite evidence for high clonality and limited genetic diversity in Ziziphus celata (Rhamnaceae), an endangered, self-incompatible shrub endemic to the Lake Wales Ridge, Florida, USA. Conserv. Genet. 13, 223–234. doi: 10.1007/s10592-011-0287-9
Guerrant, E. O., Havens, K., and Vitt, P. (2014). Sampling for effective ex situ plant conservation. Int. J. Plant Sci. 175, 11–20. doi: 10.1086/674131
Hand, M. L., and Koltunow, A. M. G. (2014). The genetic control of apomixis: asexual seed formation. Genetics 197, 441–450. doi: 10.1534/genetics.114.163105
Hodač, L., Klatt, S., Hojsgaard, D., Sharbel, T. F., and Hörandl, E. (2019). A little bit of sex prevents mutation accumulation even in apomictic polyploid plants. BMC Evol. Biol. 19:170. doi: 10.1186/s12862-019-1495-z
Jiang, X., Mu, B., Huang, Z., Zhang, M., Wang, X., and Tao, S. (2010). Impacts of mutation effects and population size on mutation rate in asexual populations: a simulation study. BMC Evol. Biol. 10:298. doi: 10.1186/1471-2148-10-298
Joly, S., and Schoen, D. J. (2011). Migration rates, frequency-dependent selection and the self-incompatibility locus in Leavenworthia (Brassicaceae). Evol. Int. J. Organic Evol. 65, 2357–2369. doi: 10.1111/j.1558-5646.2011.01300.x
Jost, L. (2008). G(ST) and its relatives do not measure differentiation. Mol. Ecol. 17, 4015–4026. doi: 10.1111/j.1365-294X.2008.03887.x
Koelling, V. A., Hamrick, J., and Mauricio, R. (2011). Genetic diversity and structure in two species of Leavenworthia with self-incompatible and self-compatible populations. Heredity 106, 310–318. doi: 10.1038/hdy.2010.59
Kopelman, N. M., Mayzel, J., Jakobsson, M., Rosenberg, N. A., and Mayrose, I. (2015). Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179–1191. doi: 10.1111/1755-0998.12387
Liu, F., Charlesworth, D., and Kreitman, M. (1999). The effect of mating system differences on nucleotide diversity at the phosphoglucose isomerase locus in the plant genus Leavenworthia. Genetics 151, 343–357. doi: 10.1093/genetics/151.1.343
Liu, F., Zhang, L., and Charlesworth, D. (1998). Genetic diversity in Leavenworthia populations with different inbreeding levels. Proc. R. Soc. Lond. Ser. B Biol. Sci. 265, 293–301. doi: 10.1098/rspb.1998.0295
Lloyd, D. G. (1965). Evolution of self-compatibility and racial differentiation in Leavenworthia (Cruciferae). Contrib. Gray Herb. Harvard Univ. 195, 3–134.
Lovell, J. T., Aliyu, O. M., Mau, M., Schranz, M. E., Koch, M., Kiefer, C., et al. (2013). On the origin and evolution of apomixis in Boechera. Plant Reprod. 26, 309–315. doi: 10.1007/s00497-013-0218-7
Lyons, E. E., and Antonovics, J. (1991). Breeding system evolution in Leavenworthia: breeding system variation and reproductive success in natural populations of Leavenworthia crassa (Cruciferae). Am. J. Bot. 78, 270–287. doi: 10.1002/j.1537-2197.1991.tb15754.x
Majeský, L., Krahulec, F., and Vašut, R. J. (2017). How apomictic taxa are treated in current taxonomy: a review. Taxon 66, 1017–1040. doi: 10.12705/665.3
Mandáková, T., Ashby, K., Price, B. J., Windham, M. D., Carman, J. G., and Lysak, M. A. (2021). Genome structure and apomixis in Phoenicaulis (Brassicaceae; Boechereae). J. Syst. Evol. 59, 83–92. doi: 10.1111/jse.12555
Meirmans, P. G. (2006). Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60, 2399–2402. doi: 10.1111/j.0014-3820.2006.tb01874.x
Mohn, R. A., Oleas, N. H., Smith, A. B., Swift, J. F., Yatskievych, G. A., and Edwards, C. E. (2021). The phylogeographic history of a range disjunction in eastern North America: the role of post-glacial expansion into newly suitable habitat. Am. J. Bot. 108, 1042–1057. doi: 10.1002/ajb2.1686
Moritz, C. (1994). Defining ‘evolutionarily significant units' for conservation. Trends Ecol. Evol. 9, 373–375. doi: 10.1016/0169-5347(94)90057-4
Moritz, C. (2002). Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst. Biol. 51, 238–254. doi: 10.1080/10635150252899752
Muller, H. J. (1964). The relation of recombination to mutational advance. Mutat. Res. Fundamental Mol. Mech. Mutag. 1, 2–9. doi: 10.1016/0027-5107(64)90047-8
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Rollins, R. C. (1963). The evolution and systematics of Leavenworthia (Cruciferae). Contrib. Gray Herb. Harvard Univ., 192, 3–98.
Rossetto, M., Yap, J. Y. S., Lemmon, J., Bain, D., Bragg, J., Hogbin, P., et al. (2021). A conservation genomics workflow to guide practical management actions. Glob. Ecol. Conserv. 26:e01492. doi: 10.1016/j.gecco.2021.e01492
Rousset, F. (2008). GENEPOP '007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 132, 365–386. doi: 10.1385/1-59259-192-2:365
Solbrig, O. T. (1972). Breeding system and genetic variation in Leavenworthia. Evolution 26, 155–160. doi: 10.1111/j.1558-5646.1972.tb00182.x
Solbrig, O. T., and Rollins, R. C. (1977). The evolution of autogamy in species of the mustard genus Leavenworthia. Evolution 31, 265–281. doi: 10.1111/j.1558-5646.1977.tb01007.x
Swift, J. F., Smith, S. A., Menges, E. S., Bassüner, B., and Edwards, C. E. (2016). Analysis of mating system and genetic structure in the endangered, amphicarpic plant, Lewton's polygala (Polygala lewtonii). Conserv. Genet. 17, 1269–1284. doi: 10.1007/s10592-016-0860-3
Swofford, D. L. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods), 4.0a169 Edn. Sunderland: Sinauer Associates.
The Angiosperm Phylogeny Group, Chase, M. W., Christenhusz, M. J. M., Fay, M. F., Byng, J. W., and Judd, W. S.. (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linnean Soc. 181, 1–20. doi: 10.1111/boj.12385
USFWS (2014). Determination of Threatened Status for Leavenworthia exigua var. laciniata (Kentucky Glade Cress). (79 FR 25683 25688).
Wang, J. (2007). Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet. Res. 89, 135–153. doi: 10.1017/S0016672307008798
Keywords: apomixis, asexuality, Brassicaceae, conservation genetics, genetic diversity, genetic structure, microsatellite, mating system
Citation: Edwards CE, Bassüner B and Williams BR (2022) Population Genetic Analysis of the Threatened Plant Leavenworthia exigua var. laciniata (Brassicaceae) Reveals Virtually No Genetic Diversity and a Unique Mating System. Front. Conserv. Sci. 3:831085. doi: 10.3389/fcosc.2022.831085
Received: 07 December 2021; Accepted: 11 March 2022;
Published: 30 March 2022.
Edited by:
Roberta Gargiulo, Royal Botanic Gardens, Kew, United KingdomReviewed by:
Jeremiah Busch, Washington State University, United StatesIsabel Marques, University of Lisbon, Portugal
Diego Hojsgaard, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Germany
Copyright © 2022 Edwards, Bassüner and Williams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christine E. Edwards, Y2hyaXN0aW5lLmVkd2FyZHNAbW9ib3Qub3Jn
 Christine E. Edwards
Christine E. Edwards Burgund Bassüner
Burgund Bassüner Brigette R. Williams
Brigette R. Williams