
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Conserv. Sci., 11 October 2022
Sec. Conservation Genetics and Genomics
Volume 3 - 2022 | https://doi.org/10.3389/fcosc.2022.1007100
 Tolulope I.N. Perrin-Stowe1,2
Tolulope I.N. Perrin-Stowe1,2 Yasuko Ishida3
Yasuko Ishida3 Dolores M. Reed4
Dolores M. Reed4 Emily E. Terrill2
Emily E. Terrill2 Oliver A. Ryder5,6
Oliver A. Ryder5,6 Jan E. Novakofski3,7
Jan E. Novakofski3,7 Nohra E. Mateus-Pinilla3,7,8,9
Nohra E. Mateus-Pinilla3,7,8,9 Budhan S. Pukazhenthi4
Budhan S. Pukazhenthi4 Alfred L. Roca1,3,7,9,10*
Alfred L. Roca1,3,7,9,10*Chronic wasting disease (CWD) is a prion disease of North American cervids. The transmission of CWD to endangered cervid species is of concern for captive breeding programs. Trans-species transmission could occur via direct contact with infected wild deer, or via prion contaminated fomites. Variation in the prion protein gene, PRNP, is associated with differences in CWD susceptibility among cervids. We therefore sequenced PRNP in 36 endangered Eld’s deer (Rucervus eldii thamin), detecting five synonymous and two non-synonymous SNPs. Three haplotypes were inferred, suggesting that genetic management in captive breeding programs has been effective at maintaining PRNP diversity. The haplotypes encoded two PrP protein variants. The more common Eld’s deer PrP variant encodes methionine at codon 208 and glutamine at codon 226. Because this protein variant is identical to a common PrP variant in white-tailed deer and mule deer and is especially common in white-tailed deer positive for CWD, we recommend reducing the frequency of this variant in the breeding stock, while implementing strict management practices to avoid exposure to wild North American cervids. The frequency of the other PrP variant, which differs from variants present in these North American cervids, was low. It has the potential to reduce susceptibility to CWD and thus could be increased in frequency. While PRNP haplotype frequencies should be shifted, genetic diversity should be maintained. Ultimately protein diversity may be protective should CWD infect the species, and trans-species polymorphisms are suggestive of past balancing selection and a potential fitness advantage for PRNP diversity.
The genetic management of endangered species in captive-breeding programs has been an important aspect of species conservation (Ballou, 1992). Maximum genetic diversity is maintained in the captive stock by equalizing founder contributions. This minimizes deleterious effects that can arise from inbreeding and genetic drift, which often impact small, isolated populations (Frankham, 2008). Genetic management can also be important when endangered populations are threatened by emerging diseases, which can be particularly devastating for populations that are small and genetically homogeneous (McKnight et al., 2017).
In captive breeding programs, genetic management is encouraged by the Association of Zoos and Aquariums (AZA), an independent accreditation organization that promotes the conservation, education, and animal welfare goals of zoos and aquariums in North America (AZA Board of Directors, 2020). In 2016, The AZA Species Survival Plan Yellow Program, which attempts to retain genetic diversity in endangered species among AZA institutions made a commitment to preserve the genetic diversity of the Eld’s deer (Reed et al., 2016).
Eld’s deer (Rucervus eldii, synonyms include Cervus eldii and Panolia eldii) are mid-sized deer that are endemic to the tropical forests and wetlands of Southeast Asia (Aung et al., 2001; Thu et al., 2019). The species originally ranged from Northeastern India to Myanmar, Thailand, Cambodia, Laos, and Hainan Island in China (Aung et al., 2001). Currently Eld’s deer are found in small remnants of their historic range due to overharvesting and habitat degradation (McShea et al., 1999), and are now listed as endangered by the International Union for the Conservation of Nature and Natural Resources (IUCN) (Gray et al., 2015).
Eld’s deer are in the subfamily Cervinae within the family Cervidae. The three subspecies of Eld’s deer are R. e. eldii, R. e. siamensis and R. e. thamin (Thu et al., 2019). The most common subspecies is R.e. thamin, which occurs in Myanmar in wild and captive populations (McShea et al., 1999). This subspecies is also managed in captive breeding programs in Eurasian and North American zoos (Thu et al., 2019), including the institutions accredited by the AZA that provided samples for the current study. In 2003, the Cervid Taxon Advisory Group of the AZA, which manages cervids including Eld’s deer, released recommendations designed to minimize the threat posed to the cervid stocks of AZA member institutions by the emergence of chronic wasting disease (CWD) (AZA Board of Directors, 2003).
Chronic wasting disease is a transmissible spongiform encephalopathy caused by misfolded prion proteins, which was first detected in mule deer in Colorado in the 1960s and has since spread across wild and captive cervid populations in at least 29 US states and two Canadian provinces, affecting white-tailed deer, mule deer, elk, and moose (Williams and Young, 1980; Williams and Young, 1982; Belay et al., 2004; Baeten et al., 2007; Rivera et al., 2019; Richards, 2021a; Richards, 2021b). CWD has spread to captive cervid populations in South Korea (CDC, 2022; Richards, 2021a; Richards, 2021b) and has also recently been detected in wild cervids in Finland, Norway, and Sweden (Pirisinu et al., 2018; Rivera et al., 2019; Richards, 2021b). Once CWD becomes endemic in a population, it can cause population declines (Haley et al., 2011; Saunders et al., 2012; Edmunds et al., 2016; Davenport et al., 2017).
CWD can be transmitted by direct contact of a susceptible animal with an infected cervid. Infectious prions can be shed by infected animals in semen (Kramm et al., 2020), urine (Haley et al., 2011), saliva, blood (Mathiason et al., 2006), and other bodily fluids (Miller et al., 2004; Gough and Maddison, 2010). CWD is a risk to the management of cervids in captive-breeding programs, even when direct contact with an infected animal is unlikely, because of the possibility of environmental transmission such as through dust inhalation of infectious particles (Miller et al., 2004; Gough and Maddison, 2010) or the infection risk posed by enclosures that previously housed CWD-positive deer (Mathiason et al., 2009). Prions can persist long term in the environment and can remain infectious in soil (depending on the composition of the soil) (Johnson et al., 2006; Kuznetsova et al., 2020) and have been detected in salt licks visited by wild deer (Plummer et al., 2018). Captive cervids such as Eld’s deer are at risk due to contaminated feed or bedding (Saunders et al., 2012; Henderson et al., 2015). Another concern is potential transmission during artificial insemination if the sire has infected semen (Kramm et al., 2020).
The prion protein is encoded by the gene PRNP. Variation in PRNP has been associated with differences in susceptibility to CWD in cervids. For example, in white-tailed deer (Odocoileus virginianus), two non-synonymous single nucleotide polymorphisms (SNPs) have been associated with reduced susceptibility to CWD: c.285A>C that encodes a histidine (H) instead of the more common glutamine (Q) at codon 95 and c.286G>A that encodes a serine (S) instead of the more common glycine (G) at codon 96 (Johnson et al., 2003; O'Rourke et al., 2004; Kelly et al., 2008; Brandt et al., 2015; Brandt et al., 2018). In mule deer (O. hemionus) two non-synonymous mutations, at codons 20 from aspartic acid (D) to glycine and 225 from serine to phenylalanine (F) have been identified (Jewell et al., 2005; Wilson et al., 2009; Zink et al., 2020; LaCava et al., 2021). In free-ranging mule deer in Wyoming and Colorado, a phenylalanine encoded by codon 225 is associated with a significantly lower CWD-positive rate compared to mule deer with a serine encoded by codon 225. In western Canada (Wilson et al., 2009) and Nebraska (Zink et al., 2020), a higher proportion of mule deer carrying one aspartic acid and one glycine at codon 20 was detected than mule deer carrying two aspartic acids at codon 20 in CWD-positive mule deer. In orally inoculated mule deer, PrPCWD was detected in the nervous system of deer carrying two serines at codon 225 deer after 189 days, but in deer carrying both a serine and a phenylalanine at codon 225 after 482 days, with the latter showing slower disease progression (Fox et al., 2006). An inoculation study of Rocky Mountain elk (Cervus canadensis nelsoni) with either leucine (L) or methionine (M) at codon 132 found that the incubation period of CWD was longest for LL individuals, intermediate for LM, and shortest for MM (Moore et al., 2020). When brain homogenate of CWD-infected elk of various genotypes at this codon was inoculated intracranially into transgenic mice, the incubation periods were found to similarly vary with elk genotype (Moore et al., 2020). In an oral inoculation study, caribou from North America encoding one asparagine (N) and one serine at codon 138 showed resistance to infection from CWD derived from elk and white-tailed deer (Mitchell et al., 2012). However, clinical CWD was detected in caribou with the same polymorphism in an intracranial inoculation study albeit with a reduction in symptomology as compared to the homozygotes (Moore et al., 2016). Fallow deer, in which 138N appears to be fixed (Robinson et al., 2019), showed no evidence of susceptibility to CWD under experimental exposure that mimicked natural transmission (Rhyan et al., 2011) but were found to be susceptible after intracerebral exposure with a prolonged incubation period (Hamir et al., 2011). Thus, when animals are intracerebrally or orally inoculated with variable doses of infectious material, this can lead to the clinical development of CWD even in animals with less susceptible genotypes. However, these experimental conditions may be considered extreme and not typical of susceptibility under natural conditions (Cullingham et al., 2020).
PRNP alleles that have been previously associated with CWD susceptibility can be examined in endangered cervids in captive breeding programs, as an initial step in extrapolating their potential susceptibility to CWD (Perrin-Stowe et al., 2021). In this study, we sequenced PRNP in Eld’s deer (R. e. thamin) housed in AZA-accredited facilities, to determine the degree of polymorphisms in PRNP, and to extrapolate the potential susceptibility of Eld’s deer to CWD based on the effects of PRNP polymorphisms on susceptibility to CWD in other cervid taxa.
The taxonomic classification of Eld’s deer has been a subject of discussion (Pitra et al., 2004; Heckeberg, 2020; Ghazi et al., 2021; Wong et al., 2021). Some sources assign Eld’s deer to the distinct genus Panolia following the nomenclature used by John Edward Gray in 1843 (Gray, 1843). In some publications, Eld’s deer are placed in the genus Rucervus, as they share morphological traits with the other species assigned to the genus, Rucervus duvaucelii and R. schomburgki (Geist, 1998; Wong et al., 2021). This designation is followed by the Species Survival Commission of the IUCN, as well as the AZA; thus, we use it here. However, some recent molecular studies have included Eld’s deer in the genus Cervus (Balakrishnan et al., 2003; Angom et al., 2017; Ghazi et al., 2021). The deer examined by the current study were exclusively from the subspecies R. e. thamin. When referring to Eld’s deer, we mean members of this subspecies, sometimes referred to as the thamin or Burmese brow-antlered deer.
Blood or tissue samples from 36 Eld’s deer individuals were used for this study. The AZA-accredited facilities that provided these samples were the Smithsonian’s National Zoo and Conservation Biology Institute (n = 21) in Front Royal, Virginia; the San Diego Zoo Wildlife Alliance (n = 6) in San Diego, California; the Wildlife Conservation Society at the Bronx Zoo (n = 6) in New York, New York; and the Sedgewick County Zoo (n = 3) in Wichita, Kansas (Supplementary Table 1). Samples were collected during routine veterinary care or came from stored blood or tissue collections. This research project was conducted under the Illinois Institutional Animal Care and Use Committee protocol 18212 and the Smithsonian Animal Care and Use Committee protocol #19-13.
DNA from tissue samples was extracted using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI). DNA from blood samples was extracted using the QIAamp DNA Blood Mini Kit (Qiagen, Germantown, MD). Blood sample lysing incubation time was extended to one hour, while tissue samples were lysed for 24 hours. The manufacturers’ instructions for both kits were followed for all other aspects of the protocols. PCR was conducted in 25 μl total volume, containing 1× PCR Buffer II (Applied Biosystems Inc.), final concentrations of 200 μM of each of the dNTPs, 1.5 mM MgCl2, 0.04 units/μl of AmpliTaq Gold DNA Polymerase (Applied Biosystems Inc.) and 0.4 μM of each oligonucleotide primer. The forward primer 223 (5’-acaccctctttattttgcag-3’) and the reverse primer 224 (5’-agaagataatgaaaacaggaag-3’) were used to amplify and sequence 830 bp encompassing the complete coding region within exon 3 of PRNP. Primer 223 is designed to amplify the functional PRNP gene by targeting introns, to avoid a processed pseudogene, which lacks introns, that has previously been detected in cervid taxa (O'Rourke et al., 2004).
The PCR cycling algorithm for PRNP amplification was as follows: initial denaturing at 95°C for 10 mins; 95°C for 30 s, 56°C for 30 s, and 72°C for 1 min (5 cycles); 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min (40 cycles); and a final extension at 72°C for 7 min (Perrin-Stowe et al., 2020). PCR amplification was confirmed on a 1.0% agarose gel with ethidium bromide using gel electrophoresis. The successful amplification products were enzyme-purified with Exonuclease I (New England Biolabs) and shrimp alkaline phosphatase (New England Biolabs) (Hanke and Wink, 1994). Purified PCR product (1 μl) and a primer (0.12 μM) were used for Sanger sequencing, in both directions, using the BigDye Terminator v3.1 Cycle Sequencing Kit (ABI).
In addition to each of the PCR primers, internal primers PRNP-IF 5’-atgctgggaagtgccatga-3’ and PRNP-IR 5’-catggcattcccagcat-3’ were also used to sequence the gene (Ishida et al., 2020). These sequences were then resolved on an ABI 3730XL DNA Sequencer at the Keck Center for Functional and Comparative Genomics at the University of Illinois at Urbana-Champaign. Sequences were then visually examined and assembled using the software Sequencher 5.4.6 (Gene Codes Corporation, Ann Arbor, MI).
The software package DnaSP utilizing the algorithm Phase was used to infer haplotypes (Stephens et al., 2001; Librado and Rozas, 2009); 10,000 iterations were run with 1000 burn-in iterations. Gene and haplotype identity was verified using NCBI Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Haplotype sequences were aligned using Sequencher, the open reading frames were confirmed, and the sequences were translated using MEGA X v.10.1 (Kumar et al., 2018). The distinct haplotype sequences were deposited in GenBank (accession numbers: OL961483-OL961485). The software PopART was used to generate and illustrate median-joining networks (under default parameters) (Bandelt et al., 1999; Leigh and Bryant, 2015). Haplotype and nucleotide diversity were calculated using DnaSP (Librado and Rozas, 2009). Confidence intervals for the haplotype frequencies (Hazra, 2017) were calculated using the following equation:
The complete coding region of PRNP was successfully sequenced in all 36 Eld’s deer individuals. Nucleotide diversity (π) was 0.00402. Out of the seven SNPs identified, five were synonymous: c.63G>C, c.114G>A, c.321G>A, c.516C>T and c.651T>C (Table 1). One of the two non-synonymous SNPs was c.624G>A, which encodes isoleucine (I) instead of methionine (M) at codon 208; 208I has been previously reported in two other cervid species (Table 2). The second non-synonymous SNP was c.676C>G, which encodes glutamic acid (E) instead of glutamine (Q) at codon 226; 226E has been previously reported in several cervid species (Jeong et al., 2007; Haley et al., 2017; Robinson et al., 2019) (Table 2). Each SNP identified (whether synonymous or nonsynonymous) was found in at least 22 chromosomes out of the 72 total chromosomes in the deer assessed.
Three haplotypes were inferred after the sequences were phased. Haplotypes were designated Ret1 through Ret3, in order of frequency. None of these three haplotype sequences has been previously reported among cervid sequences in Genbank. Haplotype diversity (Hd) in the Eld’s deer samples was 0.636. The haplotype with the highest frequency was used as the reference sequence for the Eld’s deer.
Haplotype Ret1 was detected in 35 of 72 (0.486 ± 0.115 [95% confidence interval: 95% CI]) phased Eld’s deer sequences and had the highest frequency among the samples (Table 1). The frequencies and 95% confidence intervals for each of the haplotypes are shown in Table 1, as are the SNPs present in each of the haplotypes. The three haplotypes encoded two different prion protein (PrP) variants. Haplotypes Ret1 and Ret3 encoded the same amino acid sequence (Table 2). An amino acid sequence identical to that encoded by Ret1 and Ret3 has been previously reported from at least one individual in a number of other cervid species: white-tailed deer (GenBank accession number: MG856905), Rocky Mountain mule deer (Odocoileus hemionus hemionus) (AAC33174), European roe deer (Capreolus capreolus) (MK103016), sika deer (MK103018), Reeves’s muntjac (Muntiacus reevesi) (MK103020), and Chinese water deer (MK103024) (Table 2). The other haplotype, Ret2, encodes an amino acid sequence identical to that encoded by haplotype Elad2 in Pere David’s deer (GenBank accession number: MW804583) (Perrin-Stowe et al., 2021). Haplotype Ret2 encodes an isoleucine (I) at codon 208 and glutamic acid (E) at codon 226. This variant will be referred to as PrP variant 208I;226E, while PrP variant encoded by Ret1 and Re3 will be referred to as PrP variant 208M;226Q.
A median-joining network of the three Eld’s deer haplotype sequences is shown in Figure 1. The differing amino acids encoded by the haplotypes are also shown. Ret1 and Ret3 are different at a single nucleotide, and thus more similar to each other than to Ret2 (separated from Ret1 and Ret3 by at least six nucleotides). The Eld’s deer haplotypes were also compared to PRNP sequences of other cervid taxa that had available sequences on GenBank and encoded variants of PrP (Figure 2, Table 2). Some Eld’s deer haplotypes are more similar to PRNP sequences in other cervid species than they are to each other. Haplotypes Ret1 and Ret3 are more similar to haplotypes of the Reeve’s muntjac and the Rocky Mountain mule deer than they are to haplotype Ret2 (Figure 2). Haplotype Ret2 is more similar to haplotypes carried by the Iberian red deer (C. elaphus hispanicus), the sika deer (with the 226E substitution), Rocky Mountain elk, and Pere David’s deer haplotype Elad2 (Figure 2).
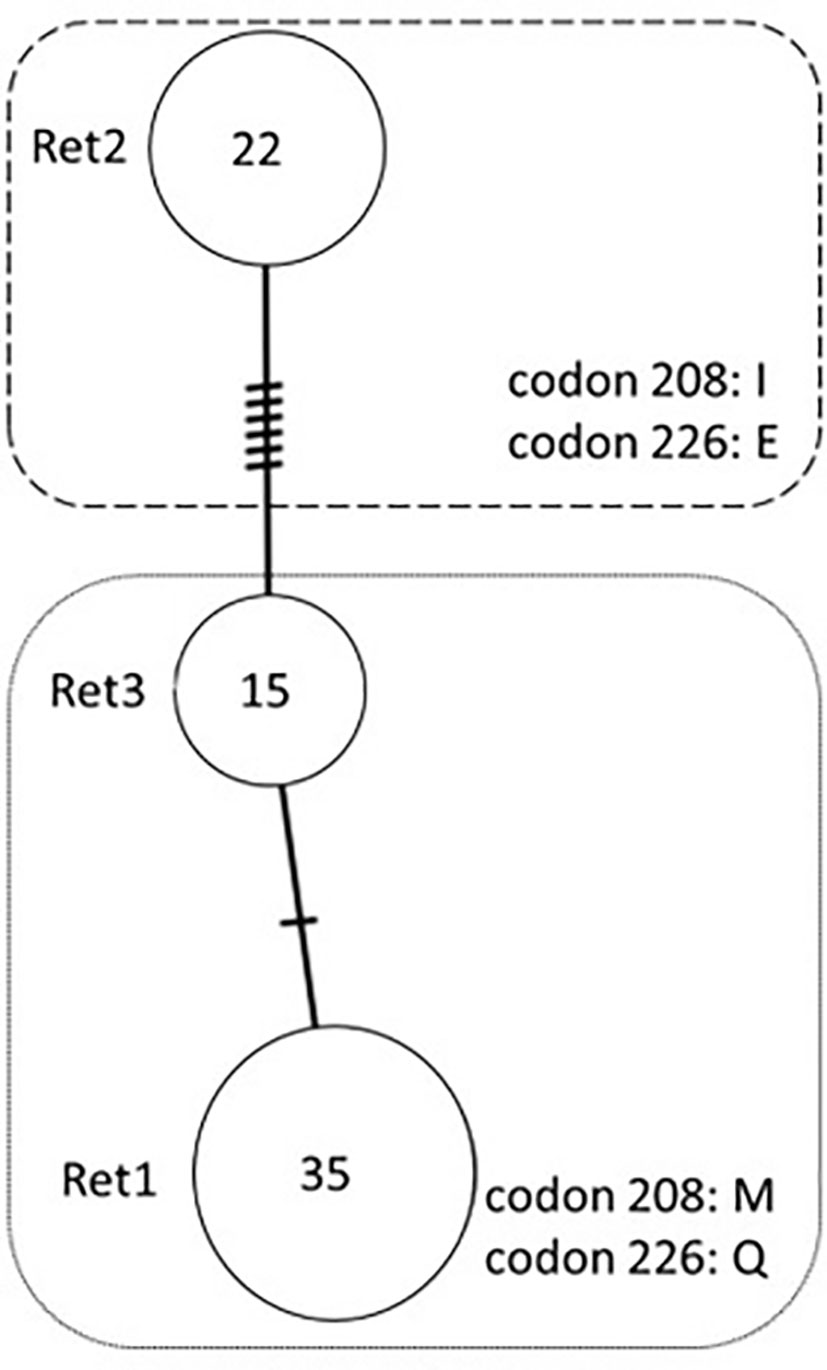
Figure 1 Median-joining network of PRNP haplotypes in Eld’s deer (Rucervus eldii thamin). Each circle represents a distinct haplotype; each hatch mark on the branches separating circles represents a mutation. The size of each circle is proportional to the number of chromosomes carrying the haplotype, which is also listed within the circle. The designation for each haplotype is listed beside the circle. There were two non-synonymous mutations, for which encoded amino acids are shown within the dotted boxes. The box outlined by small dots includes the two haplotypes that encode PrP variant 208M;226Q (Ret1 and Ret3). The box outlined by dashes includes the single haplotype that encodes PrP variant 208I;226E (Ret2).
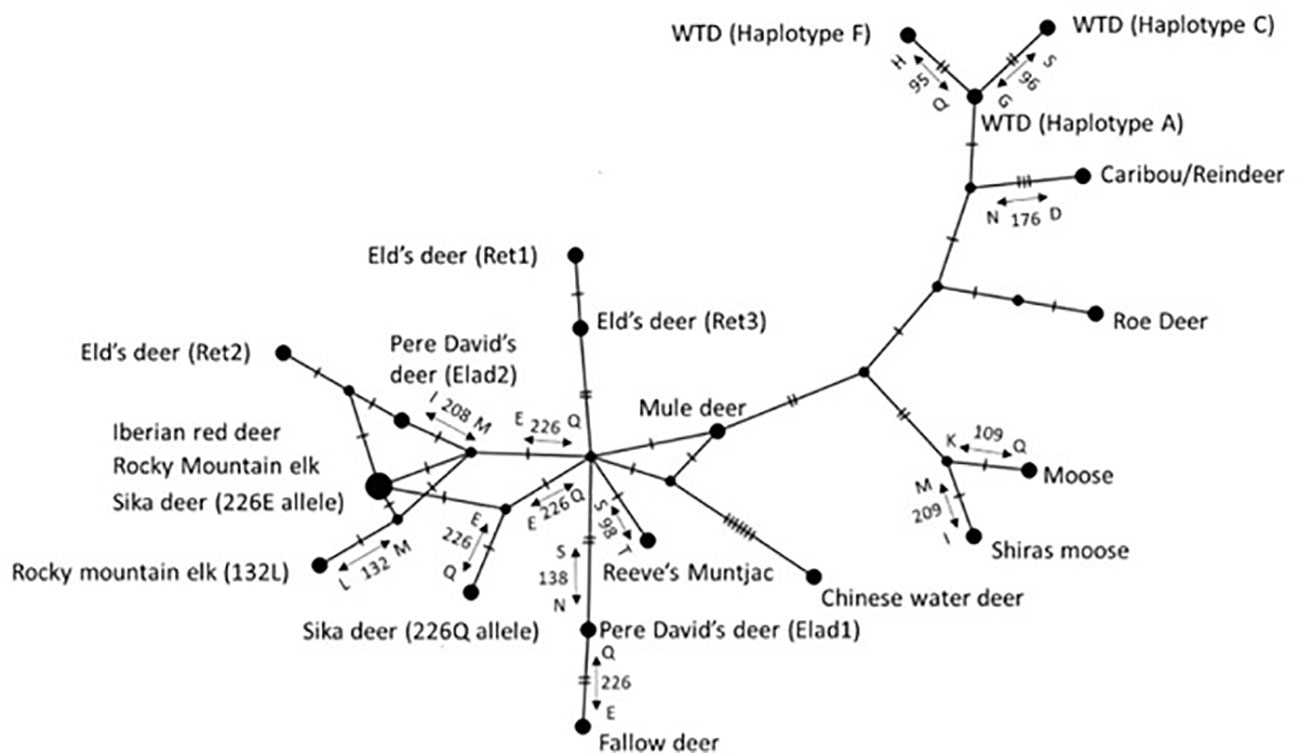
Figure 2 Median-joining network showing variation among PRNP sequences in cervid taxa. Each labeled circle represents a distinct haplotype and each hatch mark on the branches represents a single nucleotide substitution. For non-synonymous substitutions, the amino acid abbreviations are listed along the branches with their codon number. Eld’s deer haplotypes Ret1 and Ret3 group together, while haplotype Ret2 is more similar to sequences from other cervids than to haplotypes Ret1 and Ret3. WTD is the abbreviation for white-tailed deer. From three of the species shown, identical PRNP sequences have been reported: Iberian red deer (KT845864), sika deer (those with the 226E substitution; AY679695), and Rocky Mountain elk (EU082291). The node that represents the PRNP sequence from these three species is drawn larger than the nodes that represent the PRNP sequence from a single species.
In the PRNP coding region of 36 Eld’s deer individuals, seven polymorphisms were identified, comprising three haplotypes. The three haplotypes encoded two different PrP protein variants. One amino acid difference between the PrP variants was a methionine (M) to isoleucine (I) substitution at codon 208. This is a conservative amino acid substitution with both amino acids being relatively unreactive and hydrophobic (Betts and Russell, 2003). This substitution generally does not lead to considerable change in structure or function to a protein (Ohmura et al., 2001). However, in a recombinant protein misfolding cyclic amplification (PMCA) study, the M208I substitution has been reported to inhibit propagation of prions across the transmission barrier between deer and sheep (Harrathi et al., 2019). This finding may tentatively suggest that cervids that carry PrP with 208I may be less susceptible to CWD than cervids that carry PrP with 208M (Harrathi et al., 2019). However, that study was conducted in vitro using PMCA and thus this hypothesis would need to be tested, e.g., via additional inoculation experiments in vivo using brain homogenate from cervids with the relevant polymorphisms or in transgenic mice expressing cervid PrP.
The other polymorphism that distinguishes the Eld’s deer PrP variants is a glutamine (Q) to glutamic acid (E) amino acid substitution at codon 226. This is a conservative substitution; both amino acids are polar and have similar physiochemical characteristics (Betts and Russell, 2003). Transgenic mice expressing PrP with 226E (carried by Rocky Mountain elk and other cervid species) developed CWD through experimental inoculation more quickly than transgenic mice expressing PrP with 226Q (carried by mule deer and white-tailed deer) (Angers et al., 2009; Angers et al., 2010). The mice with the 226Q substitution showed a slower incubation rate but ultimately still experienced CWD infection (Angers et al., 2010; Bian et al., 2019). Red deer (Cervus elaphus elaphus) may encode either variant at codon 226 (Balachandran et al., 2010). A study of four inoculated red deer showed no difference in CWD incubation time between homozygotes for 226E or 226Q when compared to the heterozygote 226E;226Q (Balachandran et al., 2010). Depending on the CWD strains and cervid species, the residues at 226 may have a differential effect on CWD prion propagation efficiency, but neither of these residues seems to completely remove susceptibility to CWD. Further investigation into both non-synonymous substitutions within Eld’s deer specifically would be necessary to determine their potential role in CWD infection in this species.
Various other cervid species carry PRNP that encodes both 208M and 226Q, which are encoded by the Eld’s deer haplotypes Ret1 and Ret3 (PrP 208M;226Q) (Table 2). The Eld’s deer PrP 208M;226Q has the same amino acid sequence as the PrP variant found most frequently in white-tailed deer (Table 2); this PrP variant is disproportionately common among white-tailed deer positive for CWD (Ishida et al., 2020). Thus, management of Eld’s deer in captive breeding facilities may consider increasing the frequency of PrP variant 208I;226E, because Eld’s deer carrying PrP variant 208M;226Q may be at a greater risk for CWD transmission from free-ranging white-tailed deer, which are the cervids with the largest distribution among deer in North America (Hewitt, 2015). Free-ranging mule deer also carry this PrP variant (Table 2), and thus could also be a source of infection. This concern is notable given that interspecies transmission of CWD is common among cervids (Saunders et al., 2012) and that the AZA has already flagged transmission from free-ranging cervids as a potential threat to cervids in their facilities (AZA Board of Directors, 2020). Increasing the frequency of PrP 208I;226E would be preferred given that it differs in amino acid sequence from PrP in North American cervids. While other genetic management goals (such as equalizing founder contributions and avoidance of inbreeding) must be prioritized, the susceptibility of Eld’s deer to CWD infection could potentially be reduced by increasing the frequency of haplotypes encoding 208I;226E.
Trans-species polymorphisms, multiple alleles shared by more than one species, can be an indication of long-term balancing selection (Klein et al., 1998; Charlesworth, 2006; Koenig et al., 2019). Trans-species polymorphisms were evident when Eld’s deer PRNP sequences were compared to those of other cervids (Figure 2). In Eld’s deer, some PRNP haplotypes are less similar to each other than to PRNP sequences in other cervid taxa (Figure 2), providing support for this hypothesis. The two Eld’s deer PrP variants may have been present in a common ancestor of some species within the subfamily Cervinae (Klein et al., 1998; Charlesworth, 2006; Koenig et al., 2019), and persist within modern Eld’s deer. A similar pattern is shown when Pere David’s deer PRNP haplotypes are compared to the PRNP sequences of other cervid species (Perrin-Stowe et al., 2021). While there have been no reports of wild cervid taxa within Asia that have been exposed to the recent outbreak of CWD (we note that wider assessments are likely needed), historical exposure of ancestral populations to prion diseases driving balancing selection cannot be ruled out. There is evidence for balancing selection affecting PRNP in response to historical epidemics of transmissible spongiform encephalopathies caused by prions such as scrapie in sheep and kuru in humans (Mead et al., 2003; Slate, 2005; Nyström and Hammarström, 2014). Heterozygote advantage is believed to play a role in kuru in humans, with lower disease susceptibility for those encoding at codon 192 of the prion gene methionine in one chromosome and valine in the other chromosome (Nyström and Hammarström, 2014).
CWD susceptibility and progression can differ in incubation time and neuropathology due to variation both in prion strains and in PrP (Bruce et al., 1994; Bruce, 2003; Collinge and Clarke, 2007; Angers et al., 2014; Moore et al., 2020). This suggests that the presence of multiple PrP variants in a population could confer possible fitness benefits, as one variant may be more or less susceptible to a certain CWD strain than another. As noted, PrP variant 208M;226Q in Eld’s deer is identical to the most common PrP sequence carried by white-tailed deer and is also present in mule deer. This may potentially cause Eld’s deer that carry PRNP encoding the same PrP as wild North American cervids to be at greater risk of inter-species transmission of CWD than Eld’s deer with a different PrP variant. At the same time, deer that carry PrP variant 208I;226E might also be susceptible to certain prion strains because transgenic mice that carry 226E (which is carried by Rocky mountain elk among other species) develop disease faster than those that carry 226Q (carried by white-tailed deer, mule deer, and various other species) (Table 2) (Angers et al., 2010; Bian et al., 2019). Despite a potential risk, the potential advantage to deer that carry isoleucine (I) at position 208, and the benefits of maintaining more than one PrP variant in a population suggest that retention of both protein variants should be a goal in the management of Eld’s deer populations. In white-tailed deer, reduced susceptibility to CWD is provided to deer that are heterozygous and carry a single copy of a protective haplotype (Brandt et al., 2018; Ishida et al., 2020). This may suggest that the Ret1 and Ret3 haplotypes could be maintained in the stock in a heterozygous state with Ret2.
The interconnected population of Eld’s deer within AZA-accredited facilities are descended from 15 founders. The founder genome equivalent is 4.29, meaning that approximately four unrelated deer would have similar genetic diversity to the population managed in the captive breeding program (Reed et al., 2016). The AZA Species Survival Plan Yellow Program for Eld’s deer indicates that some founder lineages might be underrepresented within Eld’s deer (Reed et al., 2016). Despite this, the retention of three haplotypes of PRNP that encode two PrP variants in Eld’s deer would point to considerable success for the captive breeding program in maintaining the genetic diversity of the Eld’s deer stock. Furthermore, ex situ management of Eld’s deer within AZA facilities has included mandatory testing for CWD of all animals more than 6 months old that die (Spencer, 2008). Thanks to precautions (fencing, limiting interactions with wild cervids) taken by animal managers, none of the Eld’s deer to date have tested positive for CWD, demonstrating the importance of good animal husbandry practices in CWD prevention in ex situ managed Eld’s deer populations.
To our knowledge, this study is the first to report PRNP sequences and to identify PRNP polymorphisms in the Eld’s deer. Because Eld’s deer are endangered, and experimental methods can be invasive and costly, CWD inoculation experiments to directly determine their actual degree of susceptibility to CWD infection may be difficult to conduct or inappropriate. Therefore, studies that investigate the potential genetic susceptibility to CWD in Eld’s deer by comparing their PrP to that of other deer species that have the same PrP variants may provide information useful for managing the species to minimize the risk of CWD. Strict management practices that focus on avoiding contact between non-native cervid species and susceptible North American cervids (including contact with potentially contaminated feed, bedding and enclosures) are necessary to slow the spread of CWD. Additionally, an important management recommendation for Eld’s deer specifically is to lower the frequency of the Ret1 and Ret3 haplotypes encoding the PrP variant that is identical in amino acid sequence to a PrP variant that is common in North American deer and is disproportionately common among white-tailed deer with CWD.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. PRNP haplotypes of Eld’s deer generated from this study have been deposited in GenBank under the accession numbers OL961483-OL961485.
The animal study was reviewed and approved by Illinois Institutional Animal Care and Use Committee protocol 18212 and the Smithsonian Animal Care and Use Committee protocol #19-13.
TP-S generated data, conducted data analysis, and co-wrote the manuscript. YI assisted in data analysis and co-wrote the manuscript. DR and OR assisted in sample collection and contributed to interpretation and writing of the manuscript. ET assisted in sample collection and data generation. BP assisted in sample collection and the framing and writing of the manuscript. JN and NM-P contributed to the interpretation and writing of the manuscript. AR was responsible for the initiation of the study, helped manage the study and co-wrote the manuscript. All authors contributed to the article and approved the submitted version.
We thank the Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture, under project number ILLU 875-952 and ILLU 538-966 and the Francis M. and Harlie M. Clark Research Support Grant. Results in this project were compared to findings obtained from a study of CWD in Illinois WTD funded by The US Fish and Wildlife Service Federal Aid in Wildlife Restoration Project (W-146-R). Support was also provided by the Smithsonian’s National Zoo and Conservation Biology Institute via assistance from the Department of Conservation Medicine and animal management staff.
For collecting and providing Eld’s deer samples, we thank the Smithsonian’s National Zoo and Conservation Biology Institute in Front Royal, Virginia and the San Diego Zoo Institute for Conservation Research in San Diego, California. We also thank D. McAloose and J. Paré at the Wildlife Conservation Society at the Bronx Zoo in Bronx, New York; and S. Wilson and J. Kropf at the Sedgewick County Zoo in Wichita, Kansas.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcosc.2022.1007100/full#supplementary-material
Angers R., Christiansen J., Nalls A. V., Kang H. E., Hunter N., Hoover E. A., et al. (2014). Structural effects of PrP polymorphisms on intra- and interspecies prion transmission. Proc. Natl. Acad. Sci. 111 (30), 11169–11174. doi: 10.1073/pnas.1404739111
Angers R., Kang H. E., Napier D., Browning S., Seward T., Mathiason C., et al. (2010). Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science 328 (5982), 1154–1158. doi: 10.1126/science.1187107
Angers R., Seward T. S., Napier D., Green M., Hoover E. A., Spraker T. R., et al. (2009). Chronic wasting disease prions in elk antler velvet. Emerg. Infect. Dis. J. 15 (5), 696–703. doi: 10.3201/eid1505.081458
Angom S., Kumar A., Gupta S. K., Hussain S. A. (2017). Analysis of mtDNA control region of an isolated population of eld’s deer (Rucervus eldii) reveals its vulnerability to inbreeding. Mitochondrial DNA Part B 2 (1), 277–280. doi: 10.1080/23802359.2017.1325335
Aung M., McShea W. J., Htung S., Than A., Soe T. M., Monfort S., et al. (2001). Ecology and social organization of a tropical deer (Cervus eldi thamin). J. Mammalogy 82 (3), 836–847. doi: 10.1644/1545-1542(2001)082<0836:EASOOA>2.0.CO;2
AZA Board of Directors (2003) Guidelines for chronic wasting disease surveillance. Available at: https://www.aza.org/guidelines-for-chronic-wasting-disease-surveillance.
AZA Board of Directors (2020) Association of zoos and aquariums: about us. Available at: https://www.aza.org/about-us.
Baeten L. A., Powers B. E., Jewell J. E., Spraker T. R., Miller M. W. (2007). A natural case of chronic wasting disease in a free-ranging moose (Alces alces shirasi). J. Wildlife Dis. 43 (2), 309–314. doi: 10.7589/0090-3558-43.2.309
Balachandran A., Harrington N. P., Algire J., Soutyrine A., Spraker T. R., Jeffrey M., et al. (2010). Experimental oral transmission of chronic wasting disease to red deer (Cervus elaphus elaphus): early detection and late stage distribution of protease-resistant prion protein. Can. Vet. J. = La Rev. veterinaire Can. 51 (2), 169–178.
Balakrishnan C. N., Monfort S. L., Gaur A., Singh L., Sorenson M. D. (2003). Phylogeography and conservation genetics of eld's deer (Cervus eldi). Mol. Ecol. 12 (1), 1–10. doi: 10.1046/j.1365-294X.2003.01751.x
Ballou J. D. (1992). “Genetic and demographic considerations in endangered species captive breeding and reintroduction programs,” in Wildlife 2001: Populations. Eds. McCullough D. R., Barrett R. H. (Dordrecht: Springer Netherlands), 262–275.
Bandelt H. J., Forster P., Röhl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16 (1), 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Belay E. D., Maddox R. A., Williams E. S., Miller M. W., Gambetti P., Schonberger L. B. (2004). Chronic wasting disease and potential transmission to humans. Emerg. Infect. Dis. 10 (6), 977–984. doi: 10.3201/eid1006.031082
Betts M. J., Russell R. B. (2003). “Amino acid properties and consequences of substitutions,” in Bioinformatics for geneticists (Chichester, West Sussex, England: John Wiley and Sons, Ltd).
Bian J., Christiansen J. R., Moreno J. A., Kane S. J., Khaychuk V., Gallegos J., et al. (2019). Primary structural differences at residue 226 of deer and elk PrP dictate selection of distinct CWD prion strains in gene-targeted mice. Proc. Natl. Acad. Sci. 116 (25), 12478–12487. doi: 10.1073/pnas.1903947116
Brandt A. L., Green M. L., Ishida Y., Roca A. L., Novakofski J., Mateus-Pinilla N. E. (2018). Influence of the geographic distribution of prion protein gene sequence variation on patterns of chronic wasting disease spread in white-tailed deer (Odocoileus virginianus). Prion 12 (3-4), 204–215. doi: 10.1080/19336896.2018.1474671
Brandt A. L., Kelly A. C., Green M. L., Shelton P., Novakofski J., Mateus-Pinilla N. E. (2015). Prion protein gene sequence and chronic wasting disease susceptibility in white-tailed deer (Odocoileus virginianus). Prion 9 (6), 449–462. doi: 10.1080/19336896.2015.1115179
Bruce M. E. (2003). TSE strain variation: An investigation into prion disease diversity. Br. Med. Bull. 66 (1), 99–108. doi: 10.1093/bmb/66.1.99
Bruce M., Chree A., McConnell I., Foster J., Pearson G., Fraser H. (1994). Transmission of bovine spongiform encephalopathy and scrapie to mice: strain variation and the species barrier. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 343 (1306), 405–411. doi: 10.1098/rstb.1994.0036
CDC (2022) Chronic wasting disease (CWD): occurrence. Available at: https://www.cdc.gov/prions/cwd/occurrence.html.
Charlesworth D. (2006). Balancing selection and its effects on sequences in nearby genome regions. PloS Genet. 2 (4), e64. doi: 10.1371/journal.pgen.0020064
Cheng Y. C., Musiani M., Cavedon M., Gilch S. (2017). High prevalence of prion protein genotype associated with resistance to chronic wasting disease in one Alberta woodland caribou population. Prion 11 (2), 136–142. doi: 10.1080/19336896.2017.1300741
Collinge J., Clarke A. R. (2007). A general model of prion strains and their pathogenicity. Science 318 (5852), 930–936. doi: 10.1126/science.1138718
Cullingham C. I., Peery R. M., Dao A., McKenzie D. I., Coltman D. W. (2020). Predicting the spread-risk potential of chronic wasting disease to sympatric ungulate species. Prion 14 (1), 56–66. doi: 10.1080/19336896.2020.1720486
Davenport K. A., Mosher B. A., Brost B. M., Henderson D. M., Denkers N. D., Nalls A. V., et al. (2017). Assessment of chronic wasting disease prion shedding in deer saliva with occupancy modeling. J. Clin. Microbiol. 56 (1), e01243-01217. doi: 10.1128/JCM.01243-17
Edmunds D. R., Kauffman M. J., Schumaker B. A., Lindzey F. G., Cook W. E., Kreeger T. J., et al. (2016). Chronic wasting disease drives population decline of white-tailed deer. PloS One 11 (8), e0161127. doi: 10.1371/journal.pone.0161127
Fox K. A., Jewell J. E., Williams E. S., Miller M. W. (2006). Patterns of PrPCWD accumulation during the course of chronic wasting disease infection in orally inoculated mule deer (Odocoileus hemionus). J. Gen. Virol. 87(Pt 11), 3451–3461. doi: 10.1099/vir.0.81999-0
Frankham R. (2008). Genetic adaptation to captivity in species conservation programs. Mol. Ecol. 17 (1), 325–333. doi: 10.1111/j.1365-294X.2007.03399.x
Geist V. (1998). Deer of the world: Their evolution, behaviour, and ecology (Mechanicsburg, Pennsylvania: Stackpole books).
Ghazi M. G., Sharma S. P., Tuboi C., Angom S., Gurumayum T., Nigam P., et al. (2021). Population genetics and evolutionary history of the endangered eld’s deer (Rucervus eldii) with implications for planning species recovery. Sci. Rep. 11, 2564. doi: 10.1038/s41598-021-82183-7
Gough K. C., Maddison B. C. (2010). Prion transmission prion excretion and occurrence in the environment. Prion 4 (4), 275–282. doi: 10.4161/pri.4.4.13678
Gray J. E. (1843). List of the specimens of Mammalia in the collection of the British museum (London: The Trustees).
Gray T. N. E., Brook S. M., McShea W. J., Mahood S., Ranjitsingh M. K., Miyunt A., et al. (2015). “Rucervus eldii. the IUCN red list of threatened species 2015,” in International union for conservation of nature and natural resources. doi: 10.2305/IUCN.UK.2015-2.RLTS.T4265A22166803.en
Haley N. J., Mathiason C. K., Carver S., Zabel M., Telling G. C., Hoover E. A. (2011). Detection of chronic wasting disease prions in salivary, urinary, and intestinal tissues of deer: potential mechanisms of prion shedding and transmission. J. Virol. 85 (13), 6309–6318. doi: 10.1128/JVI.00425-11
Haley N. J., Rielinger R., Davenport K. A., Rourke K., Mitchell G., Richt J. A. (2017). Estimating chronic wasting disease susceptibility in cervids using real-time quaking-induced conversion. J. Gen. Virol. 98 (11), 2882–2892. doi: 10.1099/jgv.0.000952
Hamir A. N., Greenlee J. J., Nicholson E. M., Kunkle R. A., Richt J. A., Miller J. M., et al. (2011). Experimental transmission of chronic wasting disease (CWD) from elk and white-tailed deer to fallow deer by intracerebral route: final report. Can. J. Vet. Res. = Rev. Can. Recherche veterinaire 75 (2), 152–156.
Hanke M., Wink M. (1994). Direct DNA sequencing of PCR-amplified vector inserts following enzymatic degradation of primer and dNTPs. Biotechniques 17 (5), 858–860.
Harrathi C., Fernández-Borges N., Eraña H., Elezgarai S. R., Venegas V., Charco J. M., et al. (2019). Insights into the bidirectional properties of the sheep-deer prion transmission barrier. Mol. Neurobiol. 56 (8), 5287–5303. doi: 10.1007/s12035-018-1443-8
Hazra A. (2017). Using the confidence interval confidently. J. Thorac. Dis. 9 (10), 4125–4130. doi: 10.21037/jtd.2017.09.14
Heckeberg N. S. (2020). The systematics of the cervidae: a total evidence approach. PeerJ, 8, e8114. doi: 10.7717/peerj.8114
Henderson D. M., Denkers N. D., Hoover C. E., Garbino N., Mathiason C. K., Hoover E. A. (2015). Longitudinal detection of prion shedding in saliva and urine by chronic wasting disease-infected deer by real-time quaking-induced conversion. J. Virol. 89 (18), 9338–9347. doi: 10.1128/JVI.01118-15
Hewitt D. G. (2015). Hunters and the conservation and management of white-tailed deer (Odocoileus virginianus). Int. J. Environ. Stud. 72, 839 –849. doi: 10.1080/00207233.2015.1073473
Ishida Y., Tian T., Brandt A. L., Kelly A. C., Shelton P., Roca A. L., et al. (2020). Association of chronic wasting disease susceptibility with prion protein variation in white-tailed deer (Odocoileus virginianus). Prion 14 (1), 214–225. doi: 10.1080/19336896.2020.1805288
Jeong H. J., Lee J. B., Park S. Y., Song C. S., Kim B. S., Rho J. R., et al. (2007). Identification of single-nucleotide polymorphisms of the prion protein gene in sika deer (Cervus nippon laiouanus). J. Vet. Sci. 8 (3), 299–301. doi: 10.4142/jvs.2007.8.3.299
Jewell J. E., Conner M. M., Wolfe L. L., Miller M. W., Williams E. S. (2005). Low frequency of PrP genotype 225SF among free-ranging mule deer (Odocoileus hemionus) with chronic wasting disease. J. Gen. Virol. 86 (8), 2127–2134. doi: 10.1099/vir.0.81077-0
Johnson C., Johnson J., Clayton M., McKenzie D., Aiken J. (2003). Prion protein gene heterogeneity in free-ranging white-tailed deer within the chronic wasting disease affected region of Wisconsin. J. Wildlife Dis. 39 (3), 576–581. doi: 10.7589/0090-3558-39.3.576
Johnson C. J., Phillips K. E., Schramm P. T., McKenzie D., Aiken J. M., Pedersen J. A. (2006). Prions adhere to soil minerals and remain infectious. PloS Pathog. 2 (4), e32. doi: 10.1371/journal.ppat.0020032
Kaluz S., Kaluzova M., Flint A. P. (1997). Sequencing analysis of prion genes from red deer and camel. Prion 199 (1–2), 283–286. doi: 10.1016/s0378-1119(97)00382-x
Kelly A. C., Mateus-Pinilla N. E., Diffendorfer J., Jewell E., Ruiz M. O., Killefer J., et al. (2008). Prion sequence polymorphisms and chronic wasting disease resistance in Illinois white-tailed deer (Odocoileus virginianus). Prion 2 (1), 28–36. doi: 10.4161/pri.2.1.6321
Klein J., Sato A., Nagl S., O'hUigín C. (1998). Molecular trans-species polymorphism. Annu. Rev. Ecol. Systematics 29 (1), 1–21. doi: 10.1146/annurev.ecolsys.29.1.1
Koenig D., Hagmann J., Li R., Bemm F., Slotte T., Neuffer B., et al. (2019). Long-term balancing selection drives evolution of immunity genes in Capsella. eLife 8, e43606. doi: 10.7554/eLife.43606
Kramm C., Gomez-Gutierrez R., Soto C., Telling G., Nichols T., Morales R. (2020). In vitro detection of chronic wasting disease (CWD) prions in semen and reproductive tissues of white tailed deer bucks (Odocoileus virginianus). PloS One 14 (12), e0226560. doi: 10.1371/journal.pone.0226560
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Kuznetsova A., McKenzie D., Cullingham C., Aiken J. M. (2020). Long-term incubation PrPCWD with soils affects prion recovery but not iinfectivity. Pathogens 9 (4), 311. doi: 10.3390/pathogens9040311
LaCava M. E. F., Malmberg J. L., Edwards W. H., Johnson L. N. L., Allen S. E., Ernest H. B. (2021). Spatio-temporal analyses reveal infectious disease-driven selection in a free-ranging ungulate. R. Soc. Open Sci. 8 (8), 210802. doi: 10.1098/rsos.210802
Leigh J., Bryant D. (2015). PopART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6 (9), 1110–1116. doi: 10.1111/2041-210x.12410
Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 (11), 1451–1452. doi: 10.1093/bioinformatics/btp187
Mathiason C. K., Hays S. A., Powers J., Hayes-Klug J., Langenberg J., Dahmes S. J., et al. (2009). Infectious prions in pre-clinical deer and transmission of chronic wasting disease solely by environmental exposure. PloS One 4 (6), e5916–e5916. doi: 10.1371/journal.pone.0005916
Mathiason C. K., Powers J. G., Dahmes S. J., Osborn D. A., Miller K. V., Warren R. J., et al. (2006). Infectious prions in the saliva and blood of deer with chronic wasting disease. Science 314 (5796), 133–136. doi: 10.1126/science.1132661
McKnight D. T., Schwarzkopf L., Alford R. A., Bower D. S., Zenger K. R. (2017). Effects of emerging infectious diseases on host population genetics: a review. Conserv. Genet. 18 (6), 1235–1245. doi: 10.1007/s10592-017-0974-2
McShea W. J., Leimgruber P., Aung M., Monfort S. L., Wemmer C. (1999). Range collapse of a tropical cervid (Cervus eldi) and the extent of remaining habitat in central Myanmar. Anim. Conserv. 2 (3), 173–183. doi: 10.1111/j.1469-1795.1999.tb00063.x
Mead S., Stumpf M. P. H., Whitfield J., Beck J. A., Poulter M., Campbell T., et al. (2003). Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science 300 (5619), 640–643. doi: 10.1126/science.1083320
Miller M. W., Williams E. S., Hobbs N. T., Wolfe L. L. (2004). Environmental sources of prion transmission in mule deer. Emerg. Infect. Dis. 10 (6), 1003–1006. doi: 10.3201/eid1006.040010
Mitchell G. B., Sigurdson C. J., O’Rourke K. I., Algire J., Harrington N. P., Walther I., et al. (2012). Experimental oral transmission of chronic wasting disease to reindeer (Rangifer tarandus tarandus). PloS One 7 (6), e39055. doi: 10.1371/journal.pone.0039055
Moore S. J., Kunkle R., Greenlee M. H. W., Nicholson E., Richt J., Hamir A., et al. (2016). Horizontal transmission of chronic wasting disease in reindeer. Emerg. Infect. Dis. 22 (12), 2142–2145. doi: 10.3201/eid2212.160635
Moore S. J., Tatum T., Hwang S., Vrentas C., West Greenlee M. H., Kong Q., et al. (2020). Novel strain of the chronic wasting disease agent isolated from experimentally inoculated elk with LL132 prion protein. Sci. Rep. 10 (1), 3148. doi: 10.1038/s41598-020-59819-1
Nyström S., Hammarström P. (2014). Is the prevalent human prion protein 129M/V mutation a living fossil from a paleolithic panzootic superprion pandemic? Prion 8 (1), 2–10. doi: 10.4161/pri.27601
O'Rourke K. I., Spraker T. R., Hamburg L. K., Besser T. E., Brayton K. A., Knowles D. P. (2004). Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J. Gen. Virol. 85 (5), 1339–1346. doi: 10.1099/vir.0.79785-0
Ohmura T., Ueda T., Hashimoto Y., Imoto T. (2001). Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme. Protein Engineering Design Select. 14 (6), 421–425. doi: 10.1093/protein/14.6.421
Perrin-Stowe T. I. N., Ishida Y., Terrill E. E., Beetem D., Ryder O. A., Novakofski J. E., et al. (2021). Variation in the PRNP gene of pere david’s deer (Elaphurus davidianus) may impact genetic vulnerability to chronic wasting disease. Conserv. Genet. 23, 313–323. doi: 10.1007/s10592-021-01419-1
Perrin-Stowe T. I. N., Ishida Y., Terrill E. E., Hamlin B. C., Penfold L., Cusack L. M., et al. (2020). Prion protein gene (PRNP) sequences suggest differing vulnerability to chronic wasting disease for Florida key deer (Odocoileus virginianus clavium) and Columbian white-tailed deer (O. v. leucurus). J. Hered. 11 (6), 564–572. doi: 10.1093/jhered/esaa040
Pirisinu L., Tran L., Chiappini B., Vanni I., Di Bari M. A., Vaccari G., et al. (2018). Novel type of chronic wasting disease detected in moose (Alces alces), Norway. Emerg. Infect. Dis. 24 (12), 2210–2218. doi: 10.3201/eid2412.180702
Pitra C., Fickel J., Meijaard E., Groves C. (2004). Evolution and phylogeny of old world deer. Mol. Phylogenet. Evol. 33 (3), 880–895. doi: 10.1016/j.ympev.2004.07.013
Plummer I. H., Johnson C. J., Chesney A. R., Pedersen J. A., Samuel M. D. (2018). Mineral licks as environmental reservoirs of chronic wasting disease prions. PloS One 13 (5), e0196745–e0196745. doi: 10.1371/journal.pone.0196745
Reed D., Hatwood M., Lawless A., Andrews J. (2016). Population analysis & breeding and transfer plan: Eld’s deer (Rucervus eldii thamin) AZA species survival Plan®Yellow program.
Rhyan J., Miller M., Spraker T., McCollum M., Nol P., Wolfe L., et al. (2011). Failure of fallow deer (Dama dama) to develop chronic wasting disease when exposed to a contaminated environment and infected mule deer (Odocoileus hemionus). J. Wildlife Dis. 47 (3), 739–744. doi: 10.7589/0090-3558-47.3.739
Richards B. (2021a) Distribution of chronic wasting disease in north America. Available at: https://www.usgs.gov/media/images/distribution-chronic-wasting-disease-north-america-0.
Richards B. (2021b) Expanding distribution of chronic wasting disease. Available at: https://www.usgs.gov/centers/nwhc/science/expanding-distribution-chronic-wasting-disease?qt-science_center_objects=0#qt-science_center_objects.
Rivera N. A., Brandt A. L., Novakofski J. E., Mateus-Pinilla N. E. (2019). Chronic wasting disease in cervids: prevalence, impact and management strategies. Vet. Med. (Auckland N.Z.) 10, 123–139. doi: 10.2147/VMRR.S197404
Robinson A. L., Williamson H., Güere M. E., Tharaldsen H., Baker K., Smith S. L., et al. (2019). Variation in the prion protein gene (PRNP) sequence of wild deer in great Britain and mainland Europe. Vet. Res. 50 (1), 59. doi: 10.1186/s13567-019-0675-6
Saunders S. E., Bartelt-Hunt S. L., Bartz J. C. (2012). Occurrence, transmission, and zoonotic potential of chronic wasting disease. Emerg. Infect. Dis. 18 (3), 369–376. doi: 10.3201/eid1803.110685
Slate J. (2005). Molecular evolution of the sheep prion protein gene. Proc. Biol. Sci. 272 (1579), 2371–2377. doi: 10.1098/rspb.2005.3259
Spencer S. (2008) Chronic wasting disease surveillance and contingency plan Eastern Virginia rivers NWR complex. Available at: https://data.amerigeoss.org/pt_BR/dataset/chronic-wasting-disease-surveillance-and-contingency-plan-eastern-virginia-rivers-complex.
Stephens M., Smith N. J., Donnelly P. (2001). A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68 (4), 978–989. doi: 10.1086/319501
Thu A. M., Li G.-G., Zhang M., Thang T. H., Soe A. M., Naing W., et al. (2019). Group size and social organization of the endangered eld's deer (Rucervus eldii thamin): Results from a long-term study in Myanmar. Global Ecol. Conserv. 18, e00618. doi: 10.1016/j.gecco.2019.e00618
Williams E. S., Young S. (1980). Chronic wasting disease of captive mule deer: A spongiform encephalopathy. J. Wildlife Dis. 16 (1), 89–98. doi: 10.7589/0090-3558-16.1.89
Williams E. S., Young S. (1982). Spongiform encephalopathy of rocky mountain elk. J. Wildlife Dis. 18 (4), 465–471. doi: 10.7589/0090-3558-18.4.465
Wilson G. A., Nakada S. M., Bollinger T. K., Pybus M. J., Merrill E. H., Coltman D. W. (2009). Polymorphisms at the PRNP gene influence susceptibility to chronic wasting disease in two species of deer (Odocoileus spp.) in western Canada. J. Toxicol. Environ. Health Part a Current Issues 72 (17-18), 1025–1029. doi: 10.1080/15287390903084264
Wong M. H. G., Mo Y., Chan B. P. L. (2021). Past, present and future of the globally endangered eld’s deer (Rucervus eldii) on hainan island, China. Global Ecol. Conserv. 26, e01505. doi: 10.1016/j.gecco.2021.e01505
Keywords: brow-antlered deer, cervids, prion, thamin, transmissible spongiform encephalopathy
Citation: Perrin-Stowe TIN, Ishida Y, Reed DM, Terrill EE, Ryder OA, Novakofski JE, Mateus-Pinilla NE, Pukazhenthi BS and Roca AL (2022) Extrapolating the susceptibility of Eld’s deer (Rucervus eldii thamin) to chronic wasting disease from prion protein gene (PRNP) polymorphisms. Front. Conserv. Sci. 3:1007100. doi: 10.3389/fcosc.2022.1007100
Received: 29 July 2022; Accepted: 26 September 2022;
Published: 11 October 2022.
Edited by:
Hua Wu, Central China Normal University, ChinaReviewed by:
Rodrigo Morales, University of Texas Health Science Center at Houston, United StatesCopyright © 2022 Perrin-Stowe, Ishida, Reed, Terrill, Ryder, Novakofski, Mateus-Pinilla, Pukazhenthi and Roca. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alfred L. Roca, cm9jYUBpbGxpbm9pcy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.