- 1Molecular Invertebrate Systematics and Ecology Lab, Graduate School of Engineering and Science, University of the Ryukyus, Nishihara, Japan
- 2National Institute of Oceanography and Fisheries, Cairo, Egypt
- 3Sequencing Section, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan
- 4Department of Biochemistry, Genetics and Immunology, School of Biology, University of Vigo, Vigo, Spain
- 5Tropical Biosphere Research Center, University of the Ryukyus, Nishihara, Japan
Recently, sea cucumbers (Echinodermata: Holothuroidea) have been over-exploited in many areas of the world, including in the Ryukyu Islands, southern Japan, due to increases in their economic importance. Nevertheless, management and protection of sea cucumbers are insufficient worldwide. The black sea cucumber Holothuria (Halodeima) atra Jaeger, 1833, inhabits a large range across the Indo-West Pacific Ocean and is a widely harvested species. Here we conducted population genetic analyses on H. atra using partial mitochondrial DNA sequences of cytochrome c oxidase subunit I (COI) and 16S ribosomal RNA (16S) to examine 11 different populations around three island groups in the middle Ryukyus; Okinawajima Island, the Kerama Islands, and the Sakishima Islands, all within Okinawa Prefecture. We found 27 haplotypes for COI and 16 haplotypes for 16S. Locations within national and quasi-national parks (Zamami Island, Keramas, and Manza, Okinawajima; managed by the national Ministry of Environment and Okinawa Prefecture, respectively) had the highest number of haplotypes, whereas locations with less management and more anthropogenic pressure had lower numbers The mean of all samples' genetic diversity indices was moderate with regards to both haplotype and nucleotide diversity. According to our results, Zamami Ama was the most genetically diverse location based on both markers used, likely because it is located within Kerama-Shoto National Park with comparatively stricter regulations than most other locations. Based on our COI sequences, three-quarters of the locations with the highest haplotype diversity were found to be distant from Okinawajima Island, indicating that the genetic diversity of H. atra was reduced around Okinawajima Island. Our results possibly reflect negative impacts from anthropogenic pressures such as over-harvesting and coastal development, although future comprehensive research including sequences of nuclear loci is needed to confirm this hypothesis.
Introduction
Sea cucumbers (Echinodermata, Holothuroidea) are distributed across the world's oceans, from shallow coastal areas to the deep sea (Tyler et al., 1992; Purcell et al., 2016). Roughly 20 species of sea cucumbers are known to perform transverse fission (Dolmatov, 2014), and in some species, a large fraction of individuals produce asexual propagules (e.g., up to 76% of Holothuria atra experience fission each year at Fantome Island on the Great Barrier Reef; Uthicke, 1997). Among sea cucumbers, the order Holothuriida is widely distributed in tropical and subtropical waters (Purcell et al., 2012; Miller et al., 2017). Holothuriida species generally ingest small-sized organic matter and microalgae in sediment using their tentacles (Yingst, 1982; Mercier et al., 1999; Mfilinge and Tsuchiya, 2016; Purcell et al., 2016). It is well known that Holothuriida can enhance seagrass meadow productivity (Wolkenhauer et al., 2010), microphytobenthos productivity (Uthicke and Klumpp, 1998), calcium carbonate dissolution (Schneider et al., 2011, 2013), and can also buffer ocean acidification by discharging ammonia (Uthicke, 2001; Purcell et al., 2016). For all of these reasons Holothuriida species are considered ecosystem engineers that play crucial roles in diverse marine ecosystems, including coral reefs (Purcell et al., 2012).
Some Holothuriida species are prized as delicacies in Asia (Conand, 2004) and are therefore important commercially harvested species. However, the protection and management of these organisms have generally been insufficient. In recent years, overfishing of sea cucumbers has become a serious problem worldwide due to rising market demand (Uthicke and Conand, 2005; Toral-Granda et al., 2008; Purcell et al., 2013). Sea cucumbers in Japanese coastal waters have not been exempt from such problems, and some species have been over-harvested along the Japanese coastline for consumption, both domestically and for export to other Asian countries (Akamine, 2005).
Holothuria (Halodeima) atra (Jaeger, 1833), is one of the most common sea cucumber species along Indo-West Pacific coastlines and inhabits coral rubble, sandy tidal flats, and reef slopes (Purcell et al., 2012). The pelagic larval duration (PLD) of H. atra is estimated to be around 20 days (Ramofafia et al., 1995), and this time span is thought to be long enough for larvae to cross geographical regions. This species is known to have small and large morphotypes, but it is not yet clear whether they correspond with genetic variation (Uthicke et al., 2010). Although this species is not as commercially important as some other species such as Holothuria (Metriatyla) scabra Jaeger, 1833 or Thelenota ananas Jaeger, 1833, demand for H. atra has increased due to the depletion of other, more highly-valued species in the western central Pacific region (Kinch et al., 2008). Currently, H. atra is harvested in Micronesia, Polynesia, Melanesia, Australia, and New Zealand (Kinch et al., 2008), as well as in many Asian countries (Choo, 2008). Commercial harvesting of this species has also been reported in Okinawa Prefecture, southern Japan, although information on the local stock conditions is sparse (Purcell et al., 2014). Since H. atra is common in Okinawa and the Ryukyu Islands, investigating this species' population health is crucial for sustainable fishery management as well as for better preservation of local ecosystems.
In order to assess whether changes occur in the population's genetic composition of ecologically important organisms, molecular methodologies have been applied to marine organisms. Such methods have also been applied to holothurian species, and some studies, in various regions, have been performed examining their genetic diversity and population structure, mostly based on COI sequences. In Fiji, despite the number of specimens examined being low at some locations (three of the four populations considered had no more than 10 specimens each), only eight haplotypes were observed among 40 specimens. Such low genetic diversity has been proposed to be due to asexual recruitment processes such as fission (Eastwood et al., 2016). In contrast, Skillings et al. (2014) found that the haplotype diversity of 252 individuals of H. atra in Hawai‘i was relatively high compared to marine population genetic surveys of other species in the same area.
The Kuroshio Current, which is a warm tropical current that runs north along the Ryukyu Islands in southern Japan, is known as one of the most important oceanographic features across this region (Barkley, 1970), and is thought to be important in structuring the population of coral reef species in this region (Yasuda et al., 2014: Zayasu et al., 2016). In the central Ryukyus around Okinawajima Island, population genetic research has been conducted on local scale on the sea cucumber species Holothuria (Halodeima) edulis Lesson, 1830 (Soliman et al., 2016a) and Stichopus chloronotus Brandt, 1835 (Soliman et al., 2016b). These studies have indicated that even within small geographic areas around Okinawajima Island, genetically different populations can be distinguished in these species. Regarding H. atra, a previous study explored the genetic diversity of 44 specimens from two locations in southern Japan. However, only one population has been investigated from Okinawa (Skillings et al., 2011), and the local population connectivity of H. atra in Okinawa has still not been well studied. As well, research on sea cucumber population genetics on a larger scale to examine the effects of the Kuroshio Current have not been conducted. The knowledge of local genetic population structures is critical to ensure effective conservation measures (e.g., Fiji, López et al., 2017), such as in establishing conservation areas, such as Management Units (MUs). Since the global demand of holothurians has increased rapidly, deposition of such information is becoming increasingly important for better management. Thus, using partial sequences of cytochrome c oxidase subunit I (COI) and 16S ribosomal DNA (16S), we investigated the population genetic diversity and structure of H. atra around Okinawajima Island and nearby outlying islands in the central and southern Ryukyus. Finally, by investigating the genetic population diversity among locations and comparing population haplotype diversity of each island and site in this research, we discuss MUs. MUs are defined as “populations with significant divergence of allele frequencies at nuclear or mitochondrial loci, regardless of the phylogenetic distinctiveness of the alleles” (Moritz, 1994). Such populations are connected with weak gene flow, but may be possibly independent of each other, and thus the consideration of such units is quite important in the maintenance of potential Evolutionally Significant Units (ESU) (Moritz, 1994). Here, we define potential MUs for H. atra inhabiting Okinawa.
Materials and Methods
Sampling
Holothuria atra specimens were collected by reef walking and/or snorkeling between April to September 2019 at 11 locations across the central and southern Ryukyu Islands, from three island groups, Okinawajima Island (Chatan, Kayou, Kin, Manza, Odo, Sesoko and Uruma), the Kerama Islands (Zamami Ama and zamami Port), and the Sakishima Islands (Ishigaki St. 20 and Ishigaki St. 27) (Figures 1–3; Table 1). Zamami Island and Ishigaki Island are included in Kerama-Shoto and Iriomote-Ishigaki National Parks, respectively, whereas Manza is in Okinawa-Kaigan Quasi-National Park (Figure 1). Individual H. atra specimens were collected by hand, and a small amount of tissue was cut from around the mouth of each individual. All collections were non-lethal, and the individuals were returned to the original location from where they were taken. This non-lethal sampling method was confirmed to not require permits with the relevant prefectural administrator. In between sampling individuals, scissors were cleaned with 99% EtOH in order to avoid contamination. Tissues obtained were preserved in 99% EtOH until further investigation.
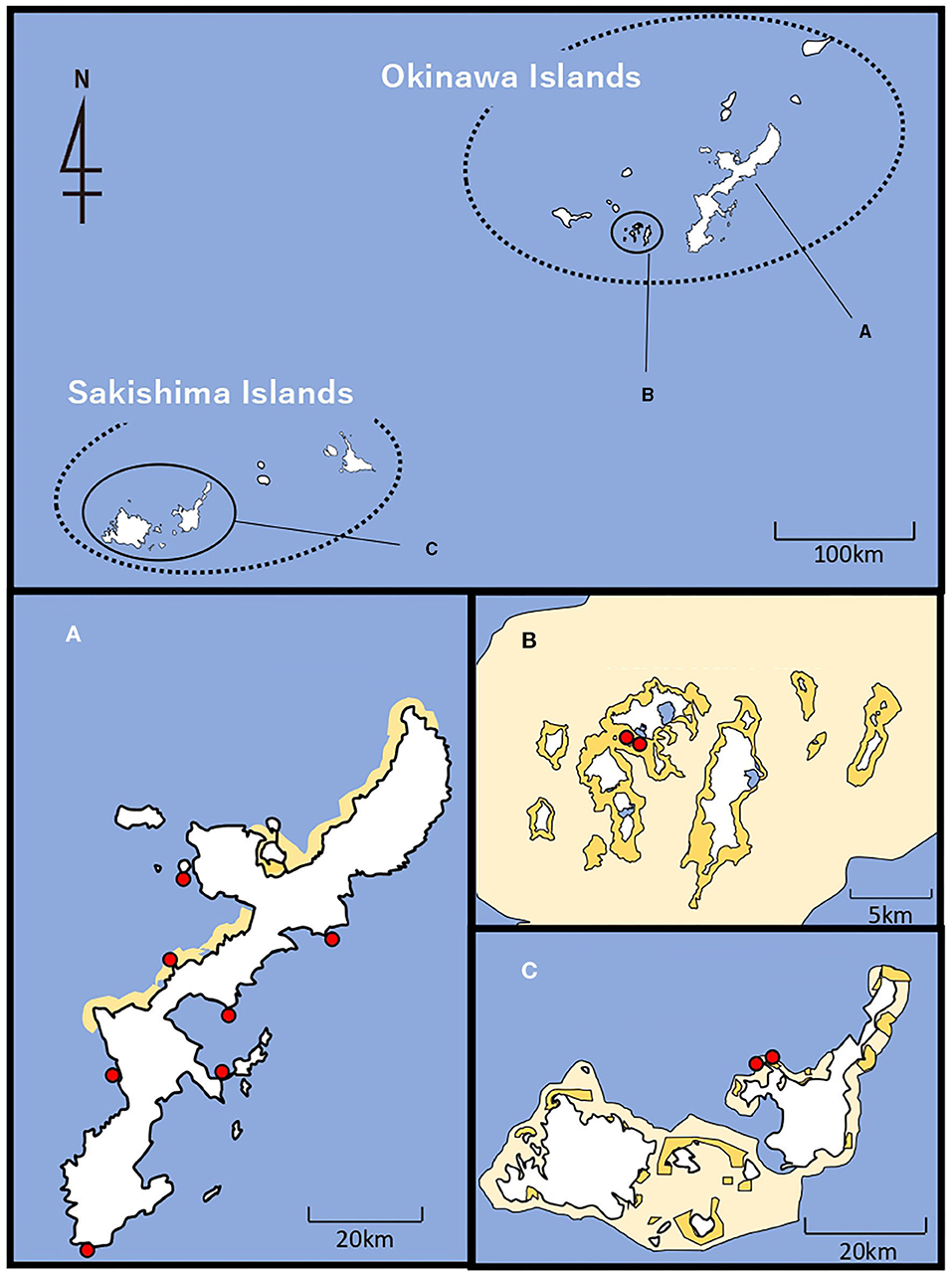
Figure 1. Map of sampling locations of H. atra in central and southern Ryukyu Islands. (A) Okinawajima Island, (B) Kerama Islands including Zamami Island and (C) Sakishima Islands including Ishigaki Island. National and quasi-national parks are shown with light yellow (=range of parks) and dark yellow (=marine protected areas). Red dots indicate sampling locations.
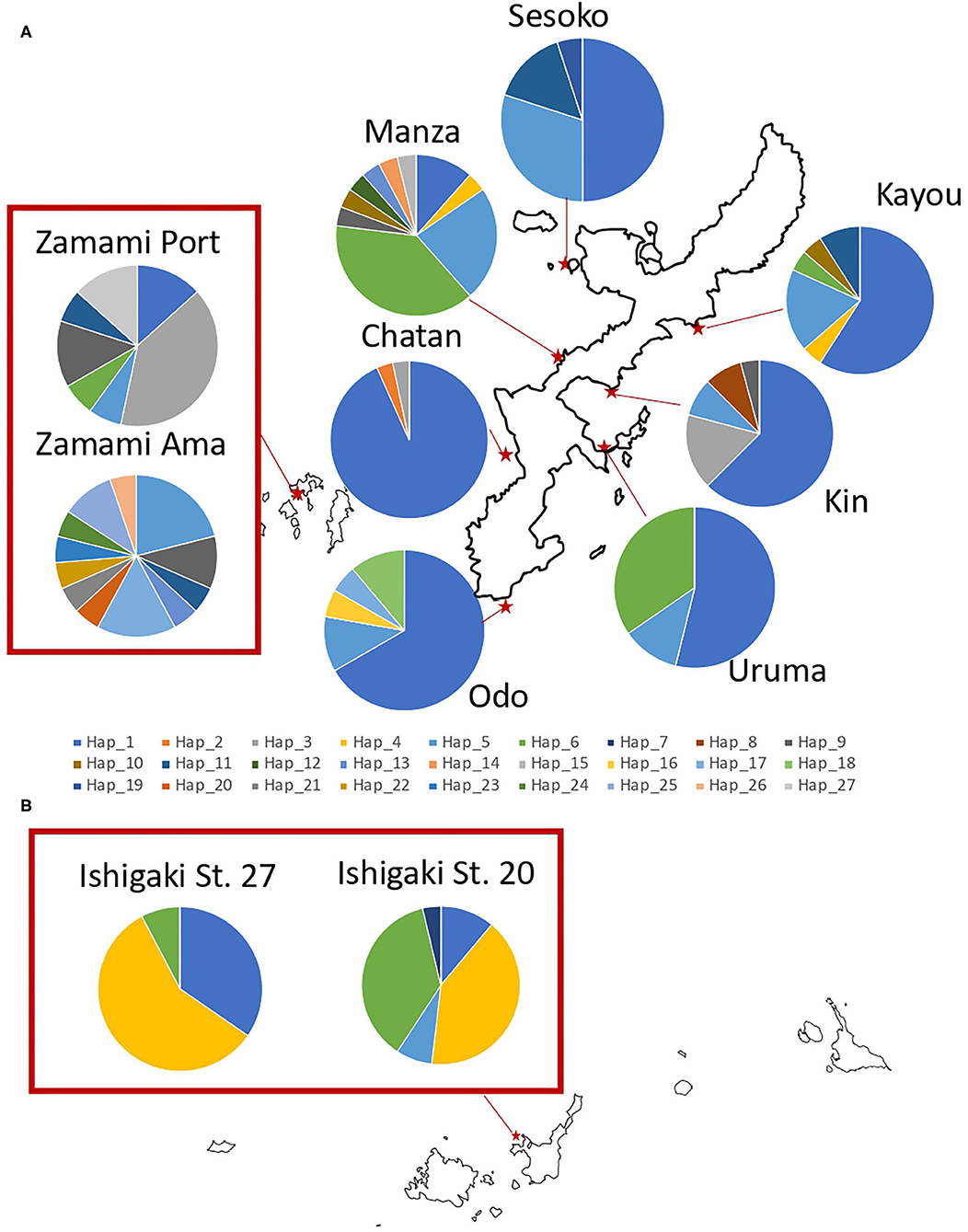
Figure 2. Sampling locations of H. atra with the proportion of each COI haplotype in (A) Okinawajima Island and the Kerama Islands, and (B) Ishigaki Island. Each color indicates a different haplotype.
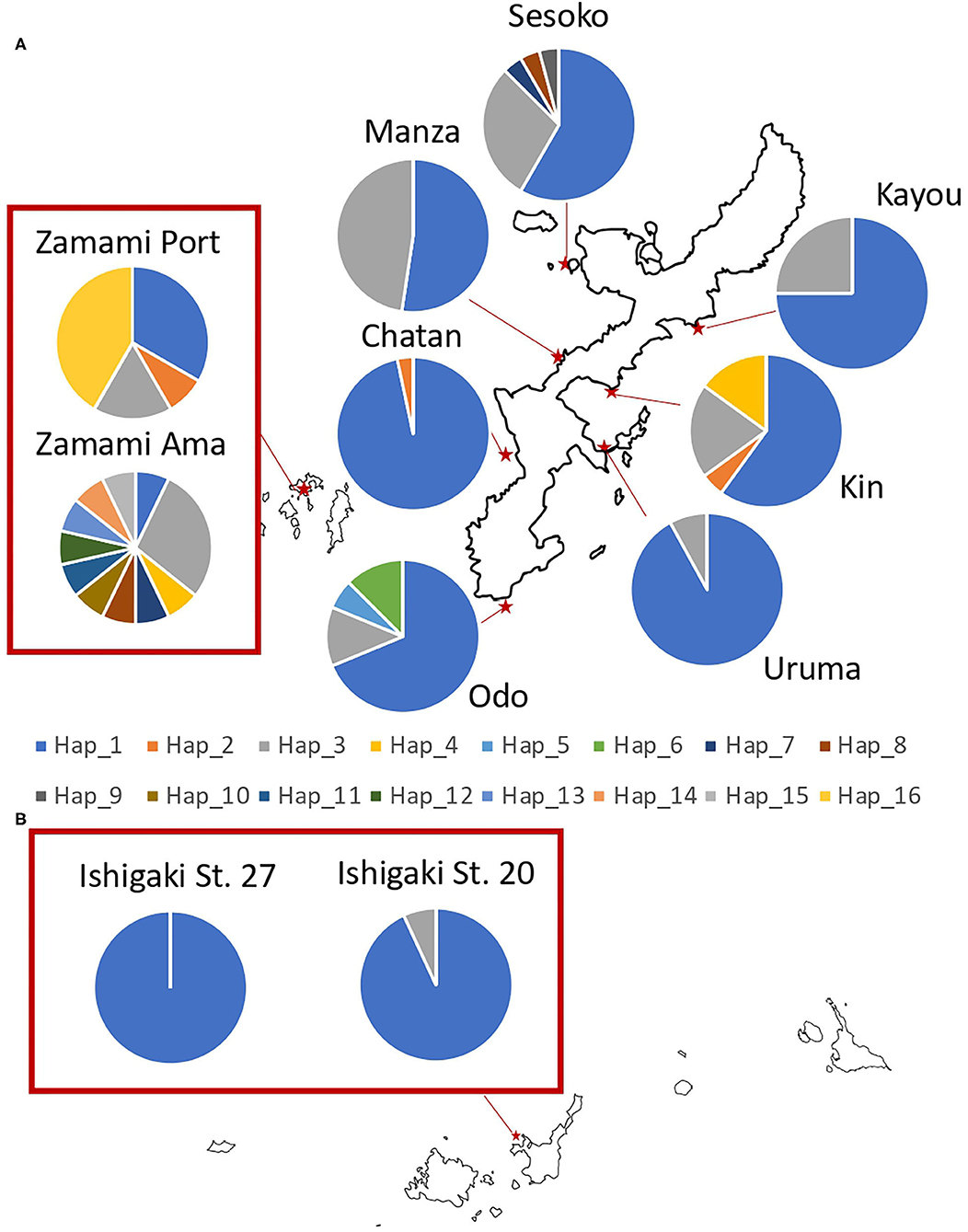
Figure 3. Sampling locations of H. atra with the proportion of each 16S haplotype in (A) Okinawajima Island and the Kerama Islands, and (B) Ishigaki Island. Each color indicates a different haplotype.
DNA Extraction, Amplification and Sequencing
Genomic DNA was extracted using a Qiagen DNeasy Blood & Tissue kit (Qiagen, Tokyo) following the manufacturer's protocol. After extraction, a partial region of cytochrome c oxidase ubunit I (COI) was amplified using the forward and reverse primers COIceF (5′-ACTGCCCACGCCCTAGTAATGATATTTTTTATGGTNATGCC-3′) and COIceR (5′-TCGTGTGTCTACGTCCATTCCTACTTRAACATRTG-3′) (Hoareau and Boissin, 2010) targeting ~700 bp long amplicons. For 16S ribosomal RNA (16S) a fragment was amplified using the forward and reverse primers 16S A-R (5′-CGCCTGTTTATCAAAAACAT-3′) and 16S B-R (5′-GCCGGTCTGAACTCAGATCACGT-3′) (Palumbi et al., 1991) targeting ~600 bp long amplicons.
PCR was conducted in a 20 μl reaction volume, with 7 μl of purified water, 10 μl of HotStarTaq® Plus Master Mix (Qiagen, Tokyo), 1 μl each of forward and reverse primers, and 1 μl of DNA. Amplification was performed under the following conditions: 95°C for 15 min, followed by 35 cycles of 30 s at 94°C, 45 s at 50°C and 1 min at 72°C, with a final extension of 10 min at 72°C. PCR clean-up was done with the Shrimp Alkaloid Phosphate (SAP) method following the manufacturer's protocol. For SAP, 0.3 μl of SAP solution, 0.15 μl of Exonuclease I solution and 2.55 μl of purified water were added, and then samples were incubated for 20 min at 37°C followed by 30 min at 83°C. Sequencing reactions were performed using a BigDyeTM Terminator v3.1 Cycle Sequencing Kit (ThermoFisher). The reaction mixtures contained a final volume of 10 μl and included 2.5 μl of BigDye 3.1 Master Mix and buffer, 0.5 μl of 10 μM primer, 1.5 μl of PCR product, and 5.5 μl of water. The following reaction conditions were applied: initial denaturation at 96°C for 1 min followed by 25 cycles of 96°C for 10 s, annealing at 50°C for 5 s and extension at 60°C for 4 min. Sequencing products were purified using Mag-Bind SeqDTR magnetic beads (Omega Bio-tek) following the manufacturer's instructions. The resulting purified products were analyzed on an ABI3130 Genetic Analyzer (Applied Biosystems, ThermoFisher) at the Genomic Unit, Scientific and Technological Support Center for Research (CACTI), University of Vigo, Spain.
Sequences Alignment and Phylogenetic Analyses
From each location, slightly different numbers of sequences were obtained from COI and 16S: Chatan (30 COI and 16S sequences), Uruma (26 of COI and 25 of 16S sequences), Odo (18 of COI and 16 of 16S sequences), Kin (24 of COI and 20 of 16S sequences), Manza (26 of COI and 21 of 16S sequences), Ishigaki Island (St. 20, 27 of COI and 29 of 16S sequences; St. 27, 26 of COI and 27 of 16S sequences), Zamami Island (Ama, 19 of COI and 14 of 16S sequences and Port, 15 of COI and 12 of 16S sequences), Kayou (22 of COI and 16 of 16S sequences) and Sesoko (20 of COI and 24 of 16S sequences) The obtained sequences were visually checked, assembled and aligned with Geneious v.8.1.9 (Kearse et al., 2012). Subsequently, COI and 16S sequences were compared with previously reported H. atra sequences using the Basic Local Alignment Search Tool (BLAST) on GenBank (www.ncbi.nlm.nih.gov/GenBank). The obtained sequences were then analyzed with those available from GenBank (clade 1—EU848217, EU848265, LC217308; clade 2—EU848222, EU848283, LC217309; clade 3—EU848266; also see Supplementary Table 2). After including these reference sequences, a COI alignment consisting of 260 sequences and 559 nucleotide positions was constructed. The partial 16S sequence of H. edulis (MZ081847) was added to the 16S alignment. Thus, 16S alignment consisted of 235 sequences and 349 nucleotide positions. For phylogenetic analyses, the best-fit substitution models (K2P+G for COI, T92+G for 16S) were selected with MEGA v.7 (Kumar et al., 2016). The Maximum Likelihood (ML) phylogenetic trees were built for both datasets using MEGA. Support for branches was assessed using a rapid bootstrap analysis with 1,000 pseudo-replicates. The Bayesian tree was also inferred with MrBayes v.3.2.7 (Ronquist et al., 2012), under K2P+G model. The MCMC ran for 1,000,000 generations, sampling every 1,000 steps, with 25% of the generated trees discarded as burn-in. Bootstrap values >50 and posterior probabilities values >0.95 are shown in the ML tree (Figure 4).
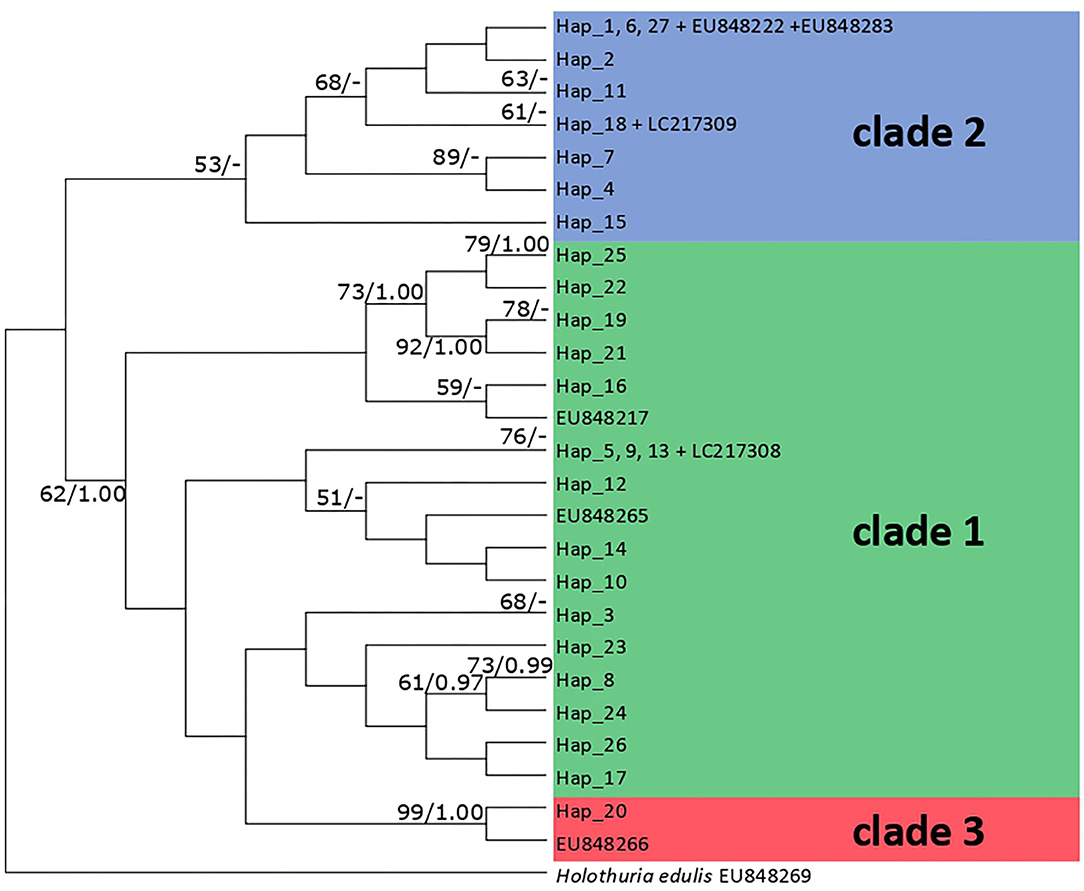
Figure 4. Maximum Likelihood phylogenetic tree based on COI sequences of H. atra. Only bootstrap values >50 are shown on the left, only >0.95 Bayesian support values are shown on the right. Each color indicates clades as reported in a previously published phylogeny (Uthicke et al., 2010). Note several haplotypes includes multiple samples. Please see also Supplementary Table 2 to match samples to each haplotype.
Genetic Diversity Indices and Neutrality Tests
Genetic diversity indices, including haplotypes (H), haplotype diversity (Hd), nucleotide diversity (π), and polymorphic sites (S), were calculated with DNAsp v.5.10.01 (Librado and Rozas, 2009) under default options for both datasets. Based on the results of the haplotype estimation, a median-joining network was generated for visualization of connectivity with PopART v.1.7 (Leigh and Bryant, 2015). Pairwise distance analysis was employed to investigate relationships between locations, and AMOVA was conducted in order to define genetic groups among different geographical areas. Both analyses were conducted with Arlequin v.3.5 (Excoffier and Lischer, 2010) with 10,000 replications. The same program was also used for Neutrality tests, Tajima's D test, and Fu's FS test. It is known that in Fu's FS test, negative values indicate recent population expansion or genetic hitchhiking, whereas positive values indicate recent bottleneck effect. Tajima's D score indicates population expansion or purification selection when positive and balancing selection, or population size decline when negative (Yamamichi and Innan, 2010).
Local Fishing Pressure Analyses
We calculated the number of fishers per total coast length at our locations by acquiring data on fisher populations, and numbers of fishing boats in publicly available data in the “Basic plan for coastal conservation of Ryukyu Islands” (Okinawa Prefecture Office, 2008), “Document of the Remote Islands No. 2, Industry” (Okinawa Prefecture Office, 2019) and the “47th Statistical Annual Report of Agriculture, Forestry and Fisheries of Okinawa Prefecture” (Okinawa General Bureau, 2019). The total population of fishers for each area was divided by the coastline length of each area (Zamami Village, Ishigaki City, Okinawajima Island).
We also calculated the amount of the catch of “others” per coastline length. The category “others” includes all collected marine animals aside from fishes, mammals, and mollusks for Zamami Village and Ishigaki City, and fishes, mammals, mollusks, cephalopods, crustaceans, and echinoids for Okinawajima Island (Okinawa General Bureau, 2019). The total “others” catch (Zamami Village and Ishigaki City−2016 report; Okinawajima−2017 report) of each area was divided by the coastline length of each area (Zamami Village, Ishigaki City and Okinawajima Island).
Management Units Investigation
To consider potential Management Units (MUs) of H. atra at the locations investigated, we explored the genetic diversity based on COI for each island, also considering the locations excluded from all other population genetic analyses due to small sample sizes (e.g., Miyako Island <10 specimens for each population; Supplementary Table 1). For this step, the same as in the separated locations' analyses, DNAsp v.5.10.01 (Librado and Rozas, 2009) under default options was employed to calculate genetic diversity indices including haplotypes (H), haplotype diversity (Hd), nucleotide diversity (π), and polymorphic sites (S). Based on the results obtained for haplotype estimation, a median-joining network was generated for visualization of connectivity with PopART v.1.7 (Leigh and Bryant, 2015).
Results
Sampling Locations and Numbers of Specimens
From a total of 295 specimens, 253 COI and 234 16S sequences were successfully sequenced and used in further analyses (Table 1, Supplementary Tables 1, 2). Although sampling was performed in other areas, including Oura Bay and several locations at Miyako Island, specimens from these locations were excluded from most analyses because of small sample sizes (<10).
Genetic Distances and Phylogenetic Tree Reconstruction
ML trees reconstructed based on both COI and 16S sequences did not show apparent correspondence with either location of specimens or body size of each specimen. The Bayesian tree reconstructed based on COI sequences showed a different topological structure compared to the ML tree, but with some nodes showing high posterior probabilities (Figure 4). Although the phylogenetic tree based on 16S showed no apparent clades (Supplementary Figures 2, 3), the phylogenetic tree based on COI sequences was composed of two major groups, one consisting of nine haplotypes (clade 2, bootstrap = 53%) and another including 17 haplotypes (clades 1, bootstrap = 62%) (Figure 4, also see Supplementary Figure 1). Haplotype 20 was sister to EU848266 into clade 3 (bootstrap = 99%). The greatest genetic distances for COI sequences were between haplotype 20 and haplotypes 6, 11, and 27 (4.1%; 23/559 bp), whereas for 16S, the greatest distances were between haplotype 8 and haplotypes 10 and 16 (2.3%; 8/349 bp). Haplotype 20 was found only at Zamami Ama, whereas the haplotypes in clade 2 appeared at all locations, and those within clade 1 at all locations except Ishigaki St. 27 (Figure 5).
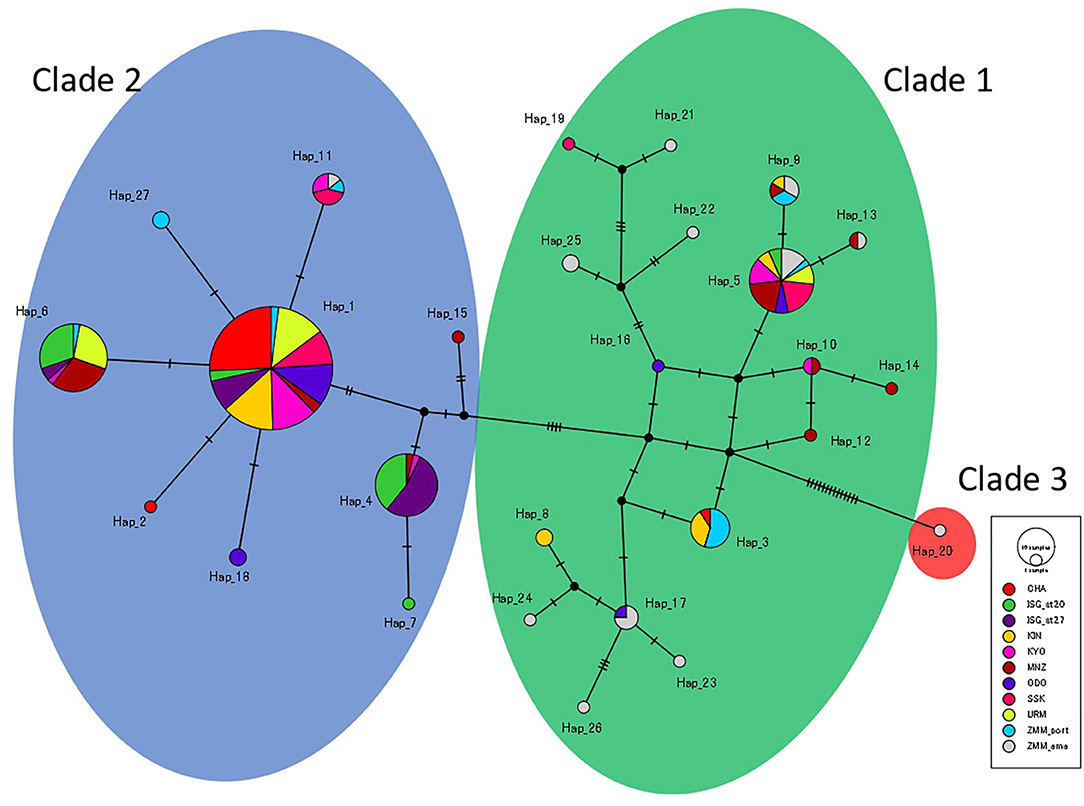
Figure 5. Median joining haplotype network based on COI sequences of H. atra. Each circle indicates haplotype with colors indicating the different locations. The size of each circle indicates the number of individuals included in each haplotype. Background colors show clades (green: clade 1, blue: clade 2, red: clade 3).
Genetic Diversity
Among the H. atra COI sequences (n = 253), 27 haplotypes were detected (Figures 2, 4; Table 1). The haplotype diversity (Hd = 0.77) and nucleotide diversity (π = 0.010) for all specimens were moderate. Chatan showed the lowest values of both haplotype and nucleotide diversities (Hd = 0.13 and π = 0.001, respectively). By contrast, among all locations, Zamami Ama had the highest values for both haplotype and nucleotide diversities (Hd = 0.94; π = 0.013), whereas Manza had the highest values (Hd = 0.81; π = 0.010) among Okinawajima populations. In terms of haplotype numbers, Zamami Ama had the greatest (= 12 haplotypes) followed by Manza (= 10 haplotypes) and Zamami Port (= 7 haplotypes), while Chatan, Uruma and Ishigaki St. 27 had the lowest (= 3 haplotypes each). Unique haplotypes were found at most locations except for Uruma, Kayou and Ishigaki St. 27. Zamami Ama had the greatest number of unique haplotypes (= 7 haplotypes) followed by Manza (= 3 haplotypes) and Odo (= 2 haplotypes). The most-shared haplotype was haplotype 1 followed by haplotype 5, which were found from 10 locations and nine locations, respectively (Figure 5).
Regarding 16S, 16 haplotypes were obtained in total with a relatively low haplotype diversity (Hd = 0.44). Ishigaki St. 27 had no haplotype variation, hence having the lowest haplotype diversity among our locations (Hd = 0.00). Likewise, Ishigaki St. 20 had the third-lowest haplotype diversity (Hd = 0.13). Apart from the Ishigaki populations, Chatan (Hd = 0.07) showed the lowest diversity followed by Uruma (Hd = 0.15). Populations in Zamami showed the highest haplotype diversity (Zamami Ama, Hd = 0.93 and Zamami Port, Hd = 0.74). The number of haplotypes was clearly the highest at Zamami Ama (N = 11), with Sesoko on Okinawajima with the second-most abundant number of haplotypes [= 5 haplotypes (Table 1)].
Population Genetic Structure
Pairwise Φst estimates based on COI sequences showed Chatan and both Ishigaki H. atra populations to be distinct from the other locations examined (Table 2). Zamami Ama was also significantly different from all other locations, including from Zamami Port despite being only ~1 km apart. Uruma showed significant differences for most comparisons except with Kayou and Odo. All Okinawajima Island populations, excluding Chatan and Uruma, did not show significant differences in any comparison.
When considering 16S sequences, Zamami Ama was significantly different from all other populations, including from the other population of Zamami Island (Zamami Port). Among Okinawajima populations, Chatan showed significant differences when compared to all other locations except with Uruma, and also the Uruma population had significant differences from Kin, Manza, and Sesoko.
AMOVA was conducted for COI and 16S sequences, manually dividing H. atra populations into groups based on pairwise Φst results. Most of the haplotype diversity was found within each population (COI = 72.76% and 16S = 69.88), but a considerable value was isolated by region (COI = 25.08% and 16S = 28.50%). The variations among populations within regions were small (COI = 2.16% and 16S = 1.62%). The results indicated that the central and southern Ryukyu Islands' populations could be divided into six groupings: Chatan, Uruma, Kin+ Kayou+ Manza+ Odo+ Sesoko, Ishigaki St. 20+ Ishigaki St. 27, Zamami Port, and Zamami Ama (Figure 6; Table 3).
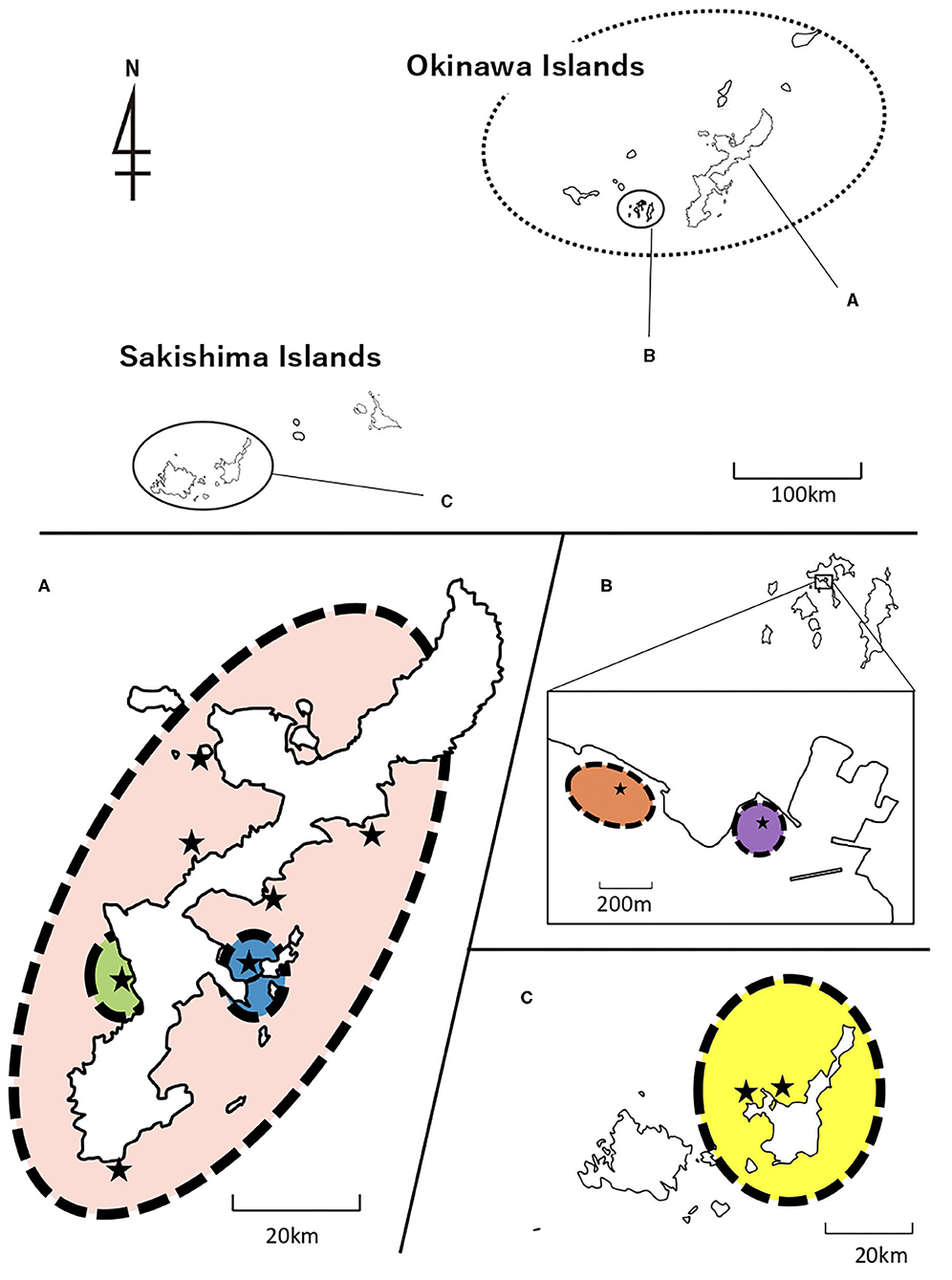
Figure 6. Population structure of H. atra supported with statistically significant values of AMOVA. (A) Okinawajima Island, (B) Kerama Islands including Zamami Island, and (C) Sakishima Islands including Ishigaki Island. Stars represent sampling locations. Colors show different population groups; Blue, Uruma; Green, Chatan; Pink, Rest of Okinawajima Island; Purple, Zamami Port; Orange, Zamami Ama; Yellow, Ishigaki Island. See Table 3 for statistical data.
Population Dynamics
Neutrality tests based on COI sequences indicated that only Chatan showed a significant negative p-value in its Tajima's D test (p < 0.001), while Zamami Ama had a marginally not significant value (p = 0.068) (Table 4). All Fu's FS values were positive except for that of the Zamami Ama population, yet none were statistically significant (Table 4).
The results based on 16S sequences were not fully consistent with the results obtained with COI sequences. Tajima's D of Sesoko and Chatan populations showed negative values (p = 0.044 and 0.032 respectively), whereas only Zamami Ama showed negative Fu's FS value (p = 0.001) (Table 4).
Local Fishing Pressure Analyses
Our results showed that the number of fishers per total coast length was less in Zamami Village (66/92; = 0.72 fishers/km) than for both Ishigaki City (275/184; = 1.49 fishers/km) and Okinawajima Island (2,272/650; = 3.50 fishers/km). Likewise, the numbers of fishing boats and annual catch of “others (tons)” (including sea cucumbers, utilized here because sea cucumber catches are not available) per total coastal length were also lower for Zamami Village (83 boats, 3 tons; 0.90 boats/km, 0.03 tons/km) than for both Ishigaki City (425 boats, 43 tons; 2.31 boats/km, 0.23 tons/km) and Okinawajima Island (1,755 boats, 4 tons; 2.7 boats/km, 0.01 tons/km).
Potential MUs Establishment
Among the islands investigated, Miyako Island showed the highest haplotype diversity (Hd = 0.98718) followed by Zamami Island (Hd = 0.94168), Ishigaki Island (Hd = 0.69737) and Okinawajima Island (Hd = 0.63980). Surprisingly, almost all the specimens from Miyako Island had different haplotypes (12 haplotypes within 13 individuals), and thus potential haplotype diversity at the area was assumed to be very high (Supplementary Table 3). Accordingly, each location possessed many unique haplotypes (Zamami Island: nine unique haplotypes, Okinawajima Island: eight unique haplotypes, and Miyako Island: five unique haplotypes), although Ishigaki had only one unique haplotype (Supplementary Figure 4).
Pairwise comparisons of genetic distance for all islands showed significant differences except for between Zamami and Miyako Islands, probably due to small sample sizes (Supplementary Table 4).
Discussion
Based on COI sequences, Uthicke et al. (2010) showed that H. atra includes three clades separated by a relatively large genetic differentiation. Following this, in Okinawa, Takano et al. (2017) confirmed the local presence of clades 1 and 2. In this study, the most common H. atra clade observed in Okinawa was clade 2 (Figure 5; Uthicke et al., 2010), and we confirmed in our study that this clade is common in the central and southern Ryukyus. On the other hand, in the current study, clade 3 was very rare, with only a single specimen observed from Zamami Ama. The maximum genetic distance between H. atra specimens for COI sequences obtained in this study (= 4.1%) was close to the maximum distance previously reported between New Caledonia and Great Barrier Reef individuals (4.5%, Uthicke et al., 2010). The phylogenetic tree based on 16S showed no obvious clades. Although 16S has been little used in genetic investigations of H. atra compared to COI, the maximum distance observed in our study (= 2.3%) does not support the existence of cryptic species within H. atra considering the minimum genetic distance (= 4.1%) between two different species in Holothuroidea as observed in a previous study (Kerr et al., 2005). However, further investigations are needed in order to establish if cryptic species truly exist within H. atra.
Even though there have been no previous focused studies on the genetic population structure of H. atra inhabiting the Ryukyus, our observed genetic diversities were relatively high compared to those of previous works conducted on the closely related species H. edulis and S. chloronotus across the same region (eight and three 16S haplotypes observed; Soliman et al., 2016a,b). Additionally, despite Skillings et al. (2011)'s results showed that the Okinawan H. atra population had the lowest genetic diversity in a study area spanning central to western Pacific Ocean with only three COI haplotypes, in the current study the actual diversity of this species seems to be much higher, comparable to the diversity observed in Hawai‘i (Hd = 0.88 in Hawai‘i in Skillings et al., 2011; Hd = 0.77 in Okinawa).
Potential reported causes of genetic diversity decline have included over-harvesting (González-Wangüemert et al., 2015), coastal development (Soliman et al., 2016a), and coastal alteration (Nehemia and Kochzius, 2017). Our study partially supports these previous observations as Chatan and Uruma, which have comparatively well-developed urban coastlines (Masucci and Reimer, 2019), had the smallest number of COI haplotypes. However, in the 16S dataset, Ishigaki St. 27 had the lowest number of haplotypes (one haplotype) followed by Ishigaki St. 20, Chatan, Uruma, Manza and Kayou (two haplotypes). Among these locations, only Chatan and Uruma have developed or artificial coastlines.
In the current study, Zamami Ama had the highest genetic diversity with many unique haplotypes in both the COI and 16S datasets. Similar results have been reported for the scleractinian coral Acropora digitifera Dana, 1846 in Okinawa, in which Zamami showed the highest number of unique haplotypes, particularly when compared with Okinawajima (Nakajima et al., 2010). The Kerama Islands, including Zamami Island, have been a quasi-national park from 1978 and have been included within Kerama-Shoto National Park since 2014. Similarly, Manza, where the highest COI genetic diversity around Okinawajima Island was observed in our study, has been within a quasi-national park since 1972. However, these results were not confirmed by the 16S analyses, in which Manza had only two haplotypes. In Japan, quasi-national and national parks have restrictions put on activities that may potentially disturb the natural environment such as coastal landfilling (land reclamation), sewage discharge, and construction (Ministry of Health Labor Welfare, 1957). High genetic diversity in protected areas have been reported around the world in other sea cucumber species, such as for example Holothuria polii Della Chiaje, 1824 and H. tubulosa Gmelin, 1791 in Turkey (González-Wangüemert et al., 2015). Therefore, we speculate that these environmental protections, as imperfect as they may be (Shinbo, 2016), could be crucial for the local conservation of H. atra's genetic diversity. Although Zamami Island waters are not a complete no-take zone, fisheries rules are much stricter than in completely non-protected areas.
To investigate this further, we compared the geographical regions in our study in terms of fishing pressure. Our results showed that the number of fishers and fishing boats per total coast length was much less in Zamami Village than in both Ishigaki City and Okinawajima Island. Likewise, the annual catch of “others” (including sea cucumbers) per total coastal length was also lower for Zamami Village than Ishigaki City, although the catch of “others” was at the same approximate level for Zamami Village and Okinawajima Island. It should be noted that the definition of “others” is different between Zamami Village and Okinawajima Island, and this may obscure the true amounts of sea cucumber harvests in each region. Another possibility is hand-to-hand trade (“hamauri” in Japanese), which has been reported in Okinawa (Okinawa General Bureau, 2017). Hamauri is an unregulated way of trading that makes it difficult to track the actual amount of harvested and traded organisms. Based on past research and the data available from Okinawa Prefecture regarding fishing pressure, we conclude that the high genetic diversity we observed in H. atra at Zamami is most likely due to the positive impacts of being within a national park, including benefits from reduced fishing pressures. To better confirm this, Okinawa Prefecture should work toward obtaining more accurate sea cucumber catch information.
In pairwise differentiation analyses based on COI sequences, Chatan and Zamami Ama were unique populations. As well, both Ishigaki populations were similar to each other but significantly different from all other populations. Chatan's COI results showed low genetic diversity with only three haplotypes, one of which was dominant (28/30 individuals). Surprisingly, the two Zamami populations (Ama and Port) showed a significant genetic break despite being only one km apart. Likewise, Kin and Uruma, separated by 13 km on Okinawajima Island's east coast, were significantly different. Similar results were previously observed for H. edulis in Okinawa (Soliman et al., 2016a), H. atra in Hawai‘i (Skillings et al., 2011), H. scabra in Australia, (Uthicke and Benzie, 2001), and also for H. mammata (Grube, 1840) and H. polii (Borrero-Pérez et al., 2011; Valente et al., 2015) in the Mediterranean Sea. These results indicate that even geographically close sea cucumber populations may retain unique genetic diversity, and therefore each population may be needed to be protected separately.
In addition, another explanation to the low gene flow between adjacent Zamami locations could be caused by high rates of sexual reproduction failure, as previously implied in other locations (Chao et al., 1994). This hypothesis is further supported by the fact that juveniles of Holothuriida species were not observed in previous studies (Chao et al., 1994; Uthicke and Benzie, 2001, 2002; Thorne et al., 2013). However, in the current study, the biological factor(s) determining genetic structure were not investigated in detail. Therefore, ecological surveys assessing the reproductive behavior and recruitment of H. atra at Zamami are needed in the future. At a same time, other DNA markers such as from the nuclear region may provide finer scale resolution of populations and should be investigated in the future.
From the AMOVA analyses, populations were categorized into six groups that were not island-dependent groupings, except for the grouping of the two Ishigaki locations together. Populations around Okinawajima Island were very disjunct, with three groupings (Chatan, Uruma, Sesoko + Kayou + Manza + Kin + Odo) that did not coincide to geographical groupings observed in other previous studies on other marine invertebrates (e.g., Nishikawa and Sakai, 2005). For example, previous studies conducted around Okinawajima Island on two different species of sea cucumbers (Soliman et al., 2016a,b), and on the amphipod Leucothoe vulgaris White and Reimer, 2012 (White et al., 2015), showed clear east and west coast genetic breaks; patterns that we did not observe in the current study. However, a complex disjunct pattern was previously observed in the tidal snail Batillaria flectosiphonata Ozawa, 1996 (Kojima et al., 2003) in Okinawajima Island and the authors speculated that the varied genetic composition may be caused by the complex geographical features of Okinawajima Island and/or different dispersal behavior.
Based on the results obtained here we found that H. atra populations from Okinawajima Island and those on more distant islands such as Zamami and Ishigaki Islands are genetically isolated from each other. This is consistent with many previous studies, which have confirmed isolation between Okinawajima—Kerama—Sakishima Islands (including Ishigaki) populations in the scleractinian corals Pocillopora damicornis Linnaeus, 1758, Goniastrea aspera Verrill, 1866, and A. digitifera, as well as between Okinawajima and Sakishima Islands in the tideland snail B. flectosiphonata (Adjeroud and Tsuchiya, 1999; Kojima et al., 2003; Nishikawa and Sakai, 2005; Shinzato et al., 2015). These results are somewhat surprising, given that it is often hypothesized that the Kuroshio Current should bring connectivity to coral reef organism populations around different islands, particularly those with planktonic larvae and comparatively high dispersal capabilities (Yorisue et al., 2020). It should be noted that it has been shown in a previous study that H. atra did not have significant genetic differentiation in pairwise comparisons between locations up to 2,000 km distant in Hawai‘i (Skillings et al., 2011). Therefore, the main factor(s) that determine H. atra population connectivity in Okinawa and the Kuroshio region remain unknown.
In the COI haplotype network, clade 2 had a star-like shape (dominant central haplotype connected to other haplotypes with few mutations), indicating recent population expansion (Ferreri et al., 2011). A star-like shape haplotype network is one of the common features of many echinoderm species (e.g., the blue sea star Linckia laevigata Linnaeus, 1758 in Otwoma and Kochzius, 2016; tropical sea urchins Acanthaster planci Linnaeus, 1758 and Tripneustes gratilla Linnaeus, 1758 in Liggins et al., 2014; sea cucumbers H. atra in Skillings et al., 2011; H. polii in Valente et al., 2015) and it is thought to show population expansion after the Last Glacial Maximum, when sea levels were much lower than present day (Ni et al., 2014). However, to better understand the historical fluctuation of echinoderm population sizes, more analyses on other species, combined with wider geographical surveys, should help confirm this theory in the future.
In this study, the Chatan population had a significantly negative Tajima's D value (Tajima's D = −2.23, P-value = 0.001) and Zamami Ama had a significantly negative Fu's FS value (Fu's FS = −5.74, P-value = 0.001). In the case of Zamami Ama, it is thought that this population has been expanding due to the presence of the highest number of unique haplotypes observed at any location, in both COI and 16S datasets. However, from previous studies, it has been inferred that smaller sea cucumber individuals tend to reproduce asexually rather than sexually (Bonham and Held, 1963; Chao et al., 1993, 1994; Uthicke, 1997). Another study indicated that nearshore reefs had more asexually reproduced individuals than mid-shelf reefs, likely induced by the high levels of environmental disturbances near shore (Uthicke et al., 1998). Chatan is also a location comparatively influenced by anthropogenic disturbances such as nutrient input from terrestrial runoff (Yang et al., 2013; Mukai et al., 2020). Therefore, Tajima's D test's significantly negative result in the Chatan population might reflect population expansion following a shift to asexual reproduction. However, to validate this hypothesis, further periodic investigations focusing on individual size fluctuations and seasonal changes of the occurrence of fission are needed.
From a conservation point of view, as each island examined here possessed many unique haplotypes as well as clear population genetic structure, we suggest considering each main islands' population as a different MU. Further investigations, including finer-scale genetic analyses, a larger number of loci, and long-term monitoring or observations are required in order to better manage and preserve ecologically important species such as H. atra.
Conclusions
In this study, we genetically assessed the diversity of H. atra populations at 11 different locations in the Ryukyu Islands, southern Japan. We found that some populations are isolated, despite being geographically very close to each other. From these results, H. atra population diversity in southern Japan needs to be considered at the local scale through the development, in conjunction with local fisheries communities, of effective management and conservations plans. Our results indicate that comparatively better conserved H. atra populations in national and quasi-national parks show higher genetic diversity compared to locations where coastlines have been altered and where less harvesting regulations exist. This implies that even partial restrictions on fishing may help even partially protect the genetic diversity of H. atra.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MZ026538—MZ026790, https://www.ncbi.nlm.nih.gov/genbank/, MZ081847, https://www.ncbi.nlm.nih.gov/genbank/, MZ090607—MZ090840, and https://www.ncbi.nlm.nih.gov/genbank/, MZ823508—MZ823528.
Author Contributions
Research plan was conceived by KH, TS, IF-S, and JDR. All field work performed by KH with help from AP. KH conducted all laboratory experiments and IF-S contributed to sequencing. Data analyses were performed by KH, TS, and JDR. Manuscript was written by KH, TS, AP, IF-S, and JDR. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Drs. Hin Boo Wee and Giovanni Masucci for advice on statistical analyses and Dr. Yee Wah Lau (all MISE, University of the Ryukyus) for checking English. As well, we thank Federico Clementoni (MISE), who helped with sampling. Zamami and Ishigaki Islands' expeditions were conducted with the Scripps Oceanographic Institute's 100 Island Challenge in summer 2019 and we thank Dr. Brian Zgliczynski, Lindsey Bonito, Christopher Sullivan, Sho Kodera, and Gabe Turner. We thank boat captains Takaya Naka at Ishigaki, Kazuya Hayashi at Okinawajima, Masaaki Isa at Miyako and Hisao Miyahira at Zamami. We thank Kazumi Inoha from the Okinawa Graduate School of Science and Technology (OIST) for logistical arrangements during the 100 Island Challenge expedition. We thank two reviewers for their constructive comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcosc.2021.736633/full#supplementary-material
References
Adjeroud, M., and Tsuchiya, M. (1999). Genetic variation and clonal structure in the scleractinian coral Pocillopora damicornis in the Ryukyu Archipelago, southern Japan. Mar. Biol. 134.4, 753–759. doi: 10.1007/s002270050592
Akamine, J. (2005). “The status of the sea cucumber fisheries and trade in Japan: past and present,” in FAO Fisheries Technical Paper, 39–48.
Bonham, K., and Held, E. E. (1963). Ecological observations on the sea cucumbers Holothuria atra and H. leucospilota at Rongelap Atoll, Marshall Islands. Pac. Sci. 17, 305–314.
Borrero-Pérez, G. H., González-Wangüemert, M., Marcos, C., and Pérez-Ruzafa, A. (2011). Phylogeography of the Atlanto-Mediterranean sea cucumber Holothuria (Holothuria) mammata: the combined effects of historical processes and current oceanographical pattern. Mol. Ecol. 20, 1964–1975. doi: 10.1111/j.1365-294X.2011.05068.x
Brandt, J. F. (1835). “Echinodermata ordo Holothurina,” in Prodromus Descriptionis Animalium ab H. Mertensio in Orbis Terrarum Circumnavigatione Observatorum. Fasc. I: 75 pps. Petropoli. 42–62. Available online at: https://books.google.com/books?id=9-KK6BsniXcC
Chao, S.-M., Chen, P., and Alexander, P. S. (1993). Fission and its effect on population structure of Holothuria atra (Echinodermata: Holothuroidea) in Taiwan. Mar. Biol. 116, 109–115. doi: 10.1007/BF00350738
Chao, S.-M., Chen, P., and Alexander, P. S. (1994). Reproduction and growth of Holothuria atra (Echinodermata: Holothuroidea) at two contrasting sites in southern Taiwan. Mar. Biol. 119, 565–570. doi: 10.1007/BF00354319
Choo, P. S. (2008). “Population status, fisheries and trade of sea cucumbers in Asia,” in Sea Cucumbers. A Global Review of Fisheries and Trade, eds V. Toral-Granda, A. Lovatelli, and M. Vasconcellos (FAO Fish. Aquacult. Tech. Paper No. 516), 81–118. Available online at: https://hdl.handle.net/20.500.12348/1511
Conand, C. (2004). “Present status of world sea cucumber resources and utilization: an international overview,” in Advances in Sea Cucumber Aquaculture and Management, eds A. Lovatelli, C. Conand, S. Purcell, S. Uthicke, J. F. Hamel, and A. Mercier, 13–23.
Dana, J. D. (1846). Zoophytes. United States Exploring Expedition during the years 1838-1842. Lea and Blanchard, Philadelphia. 7, 1–740. doi: 10.5962/bhl.title.70845
Della Chiaje, S. (1824). Descrizione zoologica ed anatomica di alcune specie di Oloturie. In: Memorie su la storia e anatomia degli animali senza vertebre del Regno di Napoli. Volume 1 Fratelli Fernandes, Napoli, 77–116.
Dolmatov, I. Y. (2014). Asexual reproduction in holothurians. Scien. World J. 27:234. doi: 10.1155/2014/527234
Eastwood, E. K., López, E. H., and Drew, J. A. (2016). Population connectivity measures of fishery-targeted coral reef species to inform marine reserve network design in Fiji. Sci. Rep. 6.1, 1–10. doi: 10.1038/srep19318
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10.3, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Ferreri, M., Qu, W., and Han, B. O. (2011). Phylogenetic networks: a tool to display character conflict and demographic history. Afr. J. Biotechnol. 10.60, 12799–12803. doi: 10.5897/AJB11.010
González-Wangüemert, M., Valente, S., and Aydin, M. (2015). Effects of fishery protection on biometry and genetic structure of two target sea cucumber species from the Mediterranean Sea. Hydrobiologia 743.1, 65–74. doi: 10.1007/s10750-014-2006-2
Grube, A. E. (1840). Actinien, Echinodermen und Würmer des Adriatischen- und Mittelmeers nach eigenen Sammlungen beschrieben. Königsberg: J.H. Bon. 92. Available online at: https://www.biodiversitylibrary.org/page/10662919
Hoareau, T. B., and Boissin, E. (2010). Design of phylum-specific hybrid primers for DNA barcoding: addressing the need for efficient COI amplification in the Echinodermata. Mol. Ecol. Resourc. 10.6, 960–967. doi: 10.1111/j.1755-0998.2010.02848.x
Jaeger, G. F. (1833). De Holothuriis. Gessnerianis, Turici. 40. Available online at: https://biodiversitylibrary.org/page/10588969
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28.12, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kerr, A. M., Janies, D. A., Clouse, R. M., Samyn, Y., Kuszak, J., and Kim, J. (2005). Molecular phylogeny of coral-reef sea cucumbers (Holothuriidae: Aspidochirotida) based on 16S mitochondrial ribosomal DNA sequence. Mar. Biotechnol. 7.1, 53–60. doi: 10.1007/s10126-004-0019-y
Kinch, J., Purcell, S., Uthicke, S., and Friedman, K. (2008). “Population status K. fisheries and trade of sea cucumbers in the Western Central Pacific,” in Sea Cucumbers: A Global Review of Fisheries and Trade, eds V. Toral-Granda, A. Lovatelli, and M. Vasconcellos (FAO Fish. Aquacult. Tech. Paper No. 516), 7–55.
Kojima, S., Kamimura, S., Kimura, T., Hayashi, I., Iijima, A., and Furota, T. (2003). Phylogenetic relationships between the tideland snails Batillaria flectosiphonata in the Ryukyu Islands and B. multiformis in the Japanese Islands. Zool. Sci. 20.11, 1423–1433. doi: 10.2108/zsj.20.1423
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33.7, 1870–1874 doi: 10.1093/molbev/msw054
Leigh, J. W., and Bryant, D. (2015). popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 6.9, 1110–1116. doi: 10.1111/2041-210X.12410
Lesson, R. P. (1830). Centurie zoologique, ou, Choix d'animaux rares, nouveaux ou imparfaitement connus. Paris & Strasbourg: F.G. Levrault and Bruxelles: Librairie parisienne. x + 11–244. Available online at: http://www.biodiversitylibrary.org/bibliography/41400#/summary
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25.11, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liggins, L., Gleeson, L., and Riginos, C. (2014). Evaluating edge-of-range genetic patterns for tropical echinoderms, Acanthaster planci and Tripneustes gratilla, of the Kermadec Islands, southwest Pacific. Bull. Mar. Sci. 90.1, 379–397. doi: 10.5343/bms.2013.1015
Linnaeus, C. (1758). Systema Naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis. Editio decima, reformata [10th revised edition], Vol. 1. Laurentius Salvius: Holmiae, 824. Available online at: https://biodiversitylibrary.org/page/726886
López, E. H., Eastwood, E., and Drew, J. (2017). “Genetic connectivity among populations of lollyfish (Holothuria atra),” in Fiji's Sea Cucumber Fishery: Advances in Science for Improved Management, eds S. Mangubhai, W. Lalavanua, and S. W. Purcell (Suva: Wildlife Conservation Society), 62–70.
Masucci, G. D., and Reimer, J. D. (2019). Expanding walls and shrinking beaches: loss of natural coastline in Okinawa Island, Japan. PeerJ 7:e7520. doi: 10.7717/peerj.7520
Mercier, A., Battaglene, S. C., and Hamel, J. F. (1999). Daily burrowing cycle and feeding activity of juvenile sea cucumbers Holothuria scabra in response to environmental factors. J. Exp. Mar. Biol. Ecol. 239.1, 125–156. doi: 10.1016/S0022-0981(99)00034-9
Mfilinge, P. L., and Tsuchiya, M. (2016). Changes in sediment fatty acid composition during passage through the gut of deposit feeding holothurians: Holothuria atra (Jaeger, 1883) and Holothuria leucospilota (Brandt, 1835). J. Lipids 457:9794. doi: 10.1155/2016/4579794
Miller, A. K., Kerr, A. M., Paulay, G., Reich, M., Wilson, N. G., Carvajal, J. I., et al. (2017). Molecular phylogeny of extant Holothuroidea (Echinodermata). Mol. Phylogen. Evol. 111, 110–131. doi: 10.1016/j.ympev.2017.02.014
Ministry of Health Labor and Welfare (1957). The ordinance for enforcement of the National Parks Act Ordinance of the Ministry of Health Labor, and Welfare, No. 41 (1957): Article 11.
Moritz, C. (1994). Defining ‘evolutionarily significant units' for conservation. Trends Ecol. Evol. 9.10, 373–375. doi: 10.1016/0169-5347(94)90057-4
Mukai, Y., Goto, A., Tashiro, Y., Tanabe, S., and Kunisue, T. (2020). Coastal biomonitoring survey on persistent organic pollutants using oysters (Saccostrea mordax) from Okinawa, Japan: Geographical distribution and polystyrene foam as a potential source of hexabromocyclododecanes. Sci. Total Environ. 739:140049. doi: 10.1016/j.scitotenv.2020.140049
Nakajima, Y., Nishikawa, A., Iguchi, A., and Sakai, K. (2010). Gene flow and genetic diversity of a broadcast-spawning coral in northern peripheral populations. PLoS ONE 5.6:49. doi: 10.1371/journal.pone.0011149
Nehemia, A., and Kochzius, M. (2017). Reduced genetic diversity and alteration of gene flow in a fiddler crab due to mangrove degradation. PLoS ONE 12:8. doi: 10.1371/journal.pone.0182987
Ni, G., Li, Q. I., Kong, L., and Yu, H. (2014). Comparative phylogeography in marginal seas of the northwestern Pacific. Mol. Ecol. 23.3, 534–548. doi: 10.1111/mec.12620
Nishikawa, A., and Sakai, K. (2005). Genetic connectivity of the scleractinian coral Goniastrea aspera around the Okinawa Islands. Coral Reefs 24.2, 318–323. doi: 10.1007/s00338-005-0484-4
Okinawa General Bureau (2017). Cabinet Office, General Affairs Department Research, and Planning Division. Exports and Industrial Use of Okinawan Sea, Cucumbers, 5–8.
Okinawa General Bureau (2019). Cabinet Office, Agriculture Forestry, and Fisheries Department Statistical Survey Division, 47th Statistical Annual Report of Agriculture Forestry, and Fisheries of Okinawa, Prefecture. 149–201.
Okinawa Prefecture Office (2008). Civil Engineering and Construction Department Coastal Disaster Management Division. Basic plan for coastal conservation of Ryukyu Islands. 21.
Okinawa Prefecture Office (2019). Planning Department, Regional and Remote Islands Division. Document of the remote islands No. 2, Industry, 18–43.
Otwoma, L. M., and Kochzius, M. (2016). Genetic population structure of the coral reef sea star Linckia laevigata in the Western Indian Ocean and Indo-West Pacific. PLoS One 11:10. doi: 10.1371/journal.pone.0165552
Ozawa, T. (1996). A new species of Batillaria (Gastropoda: Batillariidae) from Japan. Venus (Jpn. J. Malacol.) 55, 189–197.
Palumbi, S. R., Martin, A. P., Romano, S., Mcmilla, W. O., Stice, L., and Grabowski, G. (1991). The simple fool's guide to PCR. Honolulu: Department of Zoology, University of Hawai‘i.
Purcell, S., Choo, P. S., Akamine, J., and Fabinyi, M. (2014). Alternative product forms, consumer packaging and extracted derivatives of tropical sea cucumbers. SPC Beche-de-mer Inform. 34, 47–52.
Purcell, S. W., Conand, C., Uthicke, S., and Byrne, M. (2016) “Ecological roles of exploited sea cucumbers,” in Oceanography Marine Biology An Annual Review, eds R. N. Hughes, D. J. Hughes, I. P. Smith, A.C. Dale (Boca Raton, FL: CRC Press), 375–394.
Purcell, S. W., Mercier, A., Conand, C., Hamel, J. F., Toral-Granda, M. V., Lovatelli, A., et al. (2013). Sea cucumber fisheries: global analysis of stocks, management measures and drivers of overfishing. Fish Fish. 14.1, 34–59. doi: 10.1111/j.1467-2979.2011.00443.x
Purcell, S. W., Samyn, Y., and Conand, C. (2012). “Commercially important sea cucumbers of the world,” in FAO Species Catalogue for Fishery Purposes No. 6, eds N. De Angelis and A. Lovatelli, 1–6, 42–43.
Ramofafia, C., Gervis, M., and Bell, J. (1995). Spawning and early larval rearing of Holothuria atra. SPC Beche-de-mer Inform. Bull. 7, 2–6.
Ronquist, F., Teslenko, M. P., van der Mark, A., yres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61.3, 539–542. doi: 10.1093/sysbio/sys029
Schneider, K., Silverman, J., Kravitz, B., Rivlin, T., Schneider-Mor, A., Barbosa, S., et al. (2013). Inorganic carbon turnover caused by digestion of carbonate sands and metabolic activity of holothurians. Estuar. Coast. Shelf Sci. 133, 217–223. doi: 10.1016/j.ecss.2013.08.029
Schneider, K., Silverman, J., Woolsey, E., Eriksson, H., Byrne, M., and Caldeira, K. (2011). Potential influence of sea cucumbers on coral reef CaCO3 budget: A case study at One Tree Reef. J. Geophys. Res. Biogeosci. 116:G04032. doi: 10.1029/2011JG001755
Shinbo, T. (2016). Policy instruments for conserving hermatypic coral ecosystems and marine protected areas in Japan. (in Japanese) J. Rural Probl. 52, 76–82 doi: 10.7310/arfe.52.76
Shinzato, C., Mungpakdee, S., Arakaki, N., and Satoh, N. (2015). Genome-wide SNP analysis explains coral diversity and recovery in the Ryukyu Archipelago. Sci. Rep. 5, 1–8. doi: 10.1038/srep18211
Skillings, D. J., Bird, C. E., and Toonen, R. J. (2011). Gateways to Hawai‘i: genetic population structure of the tropical sea cucumber Holothuria atra. J. Mar. Biol. Article ID 783030. doi: 10.1155/2011/783030
Skillings, D. J., Bird, C. E., and Toonen, R. J. (2014). Comparative population structure of two edible Indo-Pacific coral reef sea cucumbers (Echinodermata: Holothuroidea). Bull. Mar. Sci. 90.1, 359–378. doi: 10.5343/bms.2013.1001
Soliman, T., Fernandez-Silva, I., and Reimer, J. D. (2016a). Genetic population structure and low genetic diversity in the over-exploited sea cucumber Holothuria edulis Lesson, 1830 (Echinodermata: Holothuroidea) in Okinawa Island. Conserv. Gene. 17.4, 811–821. doi: 10.1007/s10592-016-0823-8
Soliman, T., Takama, O., Fernandez-Silva, I., and Reimer, J. D. (2016b). Extremely low genetic variability within and among locations of the greenfish holothurian Stichopus chloronotus Brandt, 1835 in Okinawa Japan. Peer J. 4: E2410. doi: 10.7717/peerj.2410
Takano, T., Warén, A., and Kano, Y. (2017). Megadenus atrae n. sp., an endoparasitic eulimid gastropod (Mollusca) from the black sea cucumber Holothuria atra Jaeger (Aspidochirotida: Holothuriidae) in the Indo-West Pacific. System. Parasit. 94, 699–709. doi: 10.1007/s11230-017-9731-7
Thorne, B. V., Eriksson, H., and Byrne, M. (2013). Long term trends in population dynamics and reproduction in Holothuria atra (Aspidochirotida) in the southern Great Barrier Reef; the importance of asexual and sexual reproduction. J. Mar. Biol. Assoc. U K. 93.4, 1067–1072. doi: 10.1017/S0025315412000343
Toral-Granda, V., Lovatelli, A., and Vasconcellos, M. (eds.). (2008). Sea cucumbers. A global review of fisheries and trade. FAO Fish. Aquacult. Tech. Paper No. 516, 317.
Tyler, P. A., Young, C. M. D., Billett, S. M., and Giles, L. A. (1992). Pairing behaviour, reproduction and diet in the deep-sea holothurian genus Paroriza (Holothurioidea: Synallactidae). J. Mar. Biol. Assoc. UK. 72.2, 447–462. doi: 10.1017/S0025315400037814
Uthicke, S. (1997). Seasonality of asexual reproduction in Holothuria (Halodeima) atra, H.(H.) edulis and Stichopus chloronotus (Holothuroidea: Aspidochirotida) on the Great Barrier Reef. Mar. Biol. 129.3, 435–441. doi: 10.1007/s002270050184
Uthicke, S. (2001). Interactions between sediment-feeders and microalgae on coral reefs: grazing losses versus production enhancement. Mar. Ecol. Progr. Series 210, 125–138. doi: 10.3354/meps210125
Uthicke, S., and Benzie, J. A. H. (2001). Effect of bêche-de-mer fishing on densities and size structure of Holothuria nobilis (Echinodermata: Holothuroidea) populations on the Great Barrier Reef. Coral Reefs 19.3, 271–276. doi: 10.1007/s003380000118
Uthicke, S., and Benzie, J. A. H. (2002). A genetic fingerprint recapture technique for measuring growth in 'unmarkable' invertebrates: negative growth in commercially fished holothurians (Holothuria nobilis). Mar. Ecol. Progr. Series 241, 221–226. doi: 10.3354/meps241221
Uthicke, S., Benzie, J. A. H., and Ballment, E. (1998). Genetic structure of fissiparous populations of Holothuria (Halodeima) atra on the Great Barrier Reef. Mar. Biol. 132.1, 141–151. doi: 10.1007/s002270050380
Uthicke, S., Byrne, M., and Conand, C. (2010). Genetic barcoding of commercial Bêche-de-mer species (Echinodermata: Holothuroidea). Mol. Ecol. Resourc. 10.4, 634–646. doi: 10.1111/j.1755-0998.2009.02826.x
Uthicke, S., and Conand, C. (2005). Local examples of beche-de-mer overfishing: An initial summary and request for information. SPC Beche-de-mer Inform. Bull. 21, 9–14.
Uthicke, S., and Klumpp, D. W. (1998). Microphytobenthos community production at a near-shore coral reef: seasonal variation and response to ammonium recycled by holothurians. Mar. Ecol. Progr. Ser. 169, 1–11. doi: 10.3354/meps169001
Valente, S., Serrão, E. A., and González-Wangüemert, M. (2015). West versus East Mediterranean Sea: origin and genetic differentiation of the sea cucumber Holothuria polii. Mar. Ecol. 36.3, 485–495. doi: 10.1111/maec.12156
Verrill, A. E. (1866). Synopsis of the polyps and corals of the North Pacific Exploring Expedition, under Commodore C. Ringgold and Capt. John Rodgers, U.S.N., from 1853 to 1856. Collected by Dr. Wm. Stimpson, Naturalist to the Expedition. With descriptions of some additional species from the west coast of North America. Part III. Madreporaria. Communi. Essex Inst. 5, 17–50. Available online at: https://www.biodiversitylibrary.org/page/33204514
White, K. N., and Reimer, J. D. (2012). Commensal Leucothoidae (Crustacea, Amphipoda) of the Ryukyu Archipelago, Japan. Part I: ascidian-dwellers. ZooKeys 163:13. doi: 10.3897/zookeys.163.2003
White, K. N., Reimer, J. D., and Lorion, J. (2015). Preliminary analyses reveal strong genetic structure in populations of Leucothoe vulgaris (Crustacea: Amphipoda: Leucothoidae) from Okinawa, Japan. System. Biodiv. 14.1, 55–62. doi: 10.1080/14772000.2015.1078856
Wolkenhauer, S. M., Uthicke, S., Burridge, C., Skewes, T., and Pitcher, R. (2010). The ecological role of Holothuria scabra (Echinodermata: Holothuroidea) within subtropical seagrass beds. J. Mar. Biol. Assoc. U K. 90.2, 215–223. doi: 10.1017/S0025315409990518
Yamamichi, M., and Innan, H. (2010). Let's start! Ecogenomics (5) Detection of 1783 natural selection (vol. 1). Ecol. Soc. Japan Bull. 60, 293–302. doi: 10.18960/seitai.61.2_237
Yang, S. Y., Bourgeois, C., Ashworth, C. D., and Reimer, J. D. (2013). Palythoa zoanthid ‘barrens' in Okinawa: examination of possible environmental causes. Zool. Stud. 52.1:39. doi: 10.1186/1810-522X-52-39
Yasuda, N., Taquet, C., Nagai, S., Fortes, M., Fan, T. Y., Phongsuwan, N., et al. (2014). Genetic structure and cryptic speciation in the threatened reef-building coral Heliopora coerulea along Kuroshio Current. Bull. Mar. Sci. 90.1, 233–255. doi: 10.5343/bms.2012.1105
Yingst, J. Y. (1982). Factors influencing rates of sediment ingestion by Parastichopus parvimensis (Clark), an epibenthic deposit-feeding holothurian. Estuar. Coast. Shelf Sci. 14.2, 119–134. doi: 10.1016/S0302-3524(82)80040-6
Yorisue, T., Iguchi, A., Yasuda, N., Mizuyama, M., Yoshioka, Y., Miyagi, A., et al. (2020). Extensive gene flow among populations of the cavernicolous shrimp at the northernmost distribution margin in the Ryukyu Islands, Japan. R. Soc. Open Sci. 7.10:191731. doi: 10.1098/rsos.191731
Keywords: marine parks, mtDNA, genetics, Okinawa, Japan, benthos, lollyfish
Citation: Hamamoto K, Soliman T, Poliseno A, Fernandez-Silva I and Reimer JD (2021) Higher Genetic Diversity of the Common Sea Cucumber Holothuria (Halodeima) atra in Marine Protected Areas of the Central and Southern Ryukyu Islands. Front. Conserv. Sci. 2:736633. doi: 10.3389/fcosc.2021.736633
Received: 05 July 2021; Accepted: 23 August 2021;
Published: 27 September 2021.
Edited by:
Yoshiaki Tsuda, University of Tsukuba, JapanReviewed by:
Elena Buzan, University of Primorska, SloveniaKatherine Andrea Solari, Stanford University, United States
Copyright © 2021 Hamamoto, Soliman, Poliseno, Fernandez-Silva and Reimer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kohei Hamamoto, a29oZWloYW1hbW90bzk2QGdtYWlsLmNvbQ==
 Kohei Hamamoto
Kohei Hamamoto Taha Soliman
Taha Soliman Angelo Poliseno
Angelo Poliseno Iria Fernandez-Silva
Iria Fernandez-Silva James Davis Reimer
James Davis Reimer