
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 28 March 2025
Sec. Medicinal and Pharmaceutical Chemistry
Volume 13 - 2025 | https://doi.org/10.3389/fchem.2025.1557275
This article is part of the Research Topic Recent Advances in Synthetic Organic Chemistry at the Biomedical Interface: Honoring Professor Iwao Ojima on the Occasion of his 80th Birthday View all 8 articles
The majority of patients with androgen-dependent prostate cancer (PCa) develop resistance to hormone therapy after approximately 18–24 months of androgen deprivation therapy treatment. During this process, PCa cells progressively lose their sensitivity to androgens and evolve into castration-resistant prostate cancer leading to uncontrolled tumor growth and ultimately the failure of endocrine therapy. To develop potential anti-prostate cancer agents, in this study, we identified a novel ether-type arylpiperazine derivative as a potent androgen receptor (AR) antagonist, uncovering a series of effective antiproliferative compounds. The derivatives (7, 11, 17, 19, 20, 21, 22, 23, and 24) demonstrated strong cytotoxicity against cancer cells, with 17, 19, 20, and 23 showing significant androgen receptor antagonistic activity (Inhibition% >60) and robust AR binding affinities. The structure-activity relationship (SAR) of these developed derivatives was discussed based on data. Docking study suggested that the compound 19 mainly bind to AR ligand binding pocket site through Van der Waals’ force interactions. This research presents a promising lead compound for developing anticancer agents targeting prostate cancer therapy.
• A series of arylpiperazine derivative was synthesized.
• The anti-prostate cancer activities of derivatives were investigated.
• Binding affinity and antagonistic potency of derivatives were also investigated against AR.
• Some derivatives exhibited strong activities against AR and cancer cells.
• Molecular docking and SAR of derivatives were also studied.
According to the National Cancer Center of China’s 2024 National Cancer Report, there has been an increasing trend in both the incidence and mortality rates of prostate cancer in China in recent years, ranking sixth among the top ten malignant tumors in men (Zheng et al., 2024). The 2022 American Cancer Statistics Report estimated that prostate cancer would be the most common newly diagnosed cancer, accounting for 27%, and the second leading cause of cancer death, accounting for 11% (Siegel et al., 2022). The growth of prostate cancer cells depends on androgens, which exert their biological functions through the AR signaling pathway. Abnormal activation of the AR signaling pathway is the fundamental reason for the occurrence and development of prostate cancer (Dai et al., 2023; He et al., 2022; Jamroze et al., 2021).
Early-stage localized prostate cancer can be completely cured through surgical treatment and radiation therapy. For non-localized, inoperable prostate cancer patients, the first-line therapy is ADT (Choi et al., 2022). Endocrine therapy, while effective at controlling the progression of prostate cancer during the initial stages of treatment, leads to almost all initially hormone-sensitive tumors transforming into CRPC after 18–24 months of therapy (Vellky and Ricke, 2020). This presents significant clinical challenges, as there is currently no effective treatment regimen available for CRPC. Currently, there is no effective treatment for CRPC, although its molecular mechanisms of occurrence and development have not been fully elucidated, extensive research has found that 80% of advanced CRPC overexpress AR (Formaggio et al., 2021; Visakorpi et al., 1995), and the expression of AR in bone metastases is higher than in primary tumors (Fontana et al., 2022; Lu et al., 2020; Obinata et al., 2020). The application of next-generation ADTs (such as enzalutamide and abiraterone) can suppress the progression of CRPC by inhibiting AR in CRPC cells, and the absence of AR in CRPC cells can lead to cell death. This phenomenon indicates that the survival and growth of CRPC still depend on the AR signaling pathway, and the reactivation of AR is the fundamental cause of CRPC. Patients with CRPC constitute the main population at risk of dying from prostate cancer. Therefore, AR has become an important target for the treatment of prostate cancer. However, the development of resistance is a common issue in current endocrine therapies for prostate cancer (Schmidt et al., 2021).
Thus, finding and developing highly effective AR-targeted antagonists that combat resistance for the endocrine treatment of prostate cancer is an urgent need. Naftopidil (NAF, Figure 1), a class of arylpiperazine derivatives, selectively blocks the α1a/1d receptor subtypes, reduces the levels of dihydrotestosterone within the prostate tissue and cells, promotes apoptosis, and is currently used in the treatment of benign prostatic hyperplasia (Zhan et al., 2022). Furthermore, studies have found that NAF can effectively inhibit the proliferation of prostate cancer cells PC-3 and LNCaP, inducing apoptosis (Ishii et al., 2018; Iwamoto et al., 2017; Maesaka et al., 2021). Kinoyama et al. (2005), Kinoyama et al. (2004) reported that arylpiperazine derivatives exhibit significant AR antagonistic activity, with an IC50 of 0.11 μmol/L, compared to bicalutamide’s IC50 of 50 μmol/L. They can inhibit prostate hyperplasia without affecting serum testosterone levels. In recent years, our research team has conducted extensive preliminary basic research on the anti-prostate cancer activity of arylpiperazine derivatives, discovering that some arylpiperazine derivatives show good cytotoxic activity (Chen et al., 2018a; Chen et al., 2018b; Chen et al., 2017; Chen et al., 2015; Chen et al., 2019a; Qi et al., 2022a) and exhibit better antagonistic activity and affinity for AR (Chen et al., 2019a; Chen et al., 2019b).
Although the reported arylpiperazine derivatives possess significant AR antagonistic activity, there is less research on their resistance evaluation and antitumor molecular mechanisms. Based on the aforementioned studies, the drug design strategy of this project is to design and synthesize a new class of arylpiperazine derivatives based on naftopidil (Scheme 1) on the foundation of previous research. The aim is to investigate their biological activity, resistance, and antitumor molecular mechanisms, thereby obtaining new drugs with stronger antagonistic activity and resistance to treat prostate cancer.
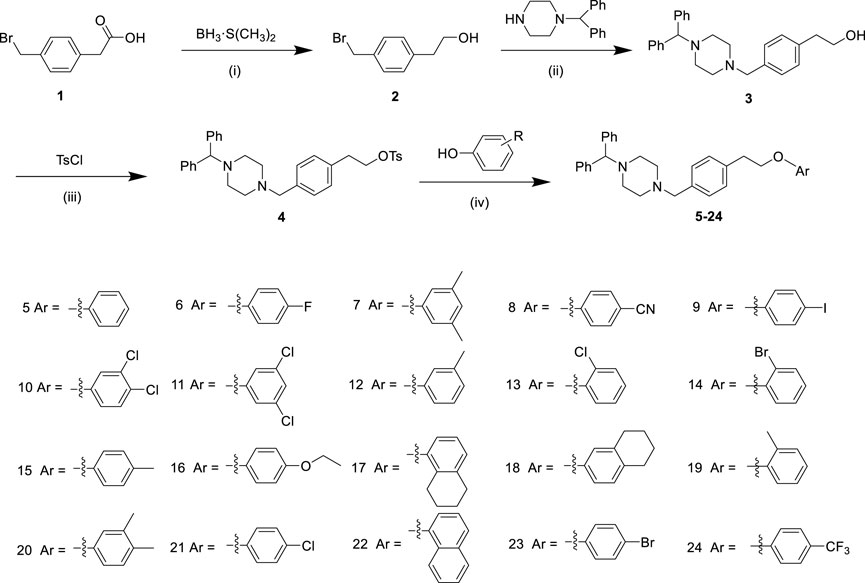
Scheme 1. The synthesis route of derivatives 5–24. Reagents and conditions: (i) BH3·S(CH3)2, THF, 0°C for 1 h, and then room temperature for 12 h; (ii) 1-(Diphenylmethyl) piperazine, K2CO3, CH3CN, reflux, 12 h; (iii) TsCl, Et3N and 4-dimethylaminopyridine, Cl2CH2, 0°C, 16 h; (iv) Phenol, K2CO3, CH3CN, reflux, 12 h.
Reagents and solvents were procured via commercial channels. Organic solvents underwent distillation before use. Melting points were determined using an uncalibrated SGW X-4 micro melting point apparatus. NMR spectra were acquired on a Bruker AVANCE-400 spectrometer in CDCl3, employing TMS as an internal standard, with chemical shifts reported in δ (ppm) and coupling constants in Hertz. HRMS spectra were documented on an AB Sciex X500R QTOF mass spectrometer (Foster City, CA, United States). HPLC chromatogram was performed on UltiMate 3000 with H2O and CH3CN as the mobile phase. The completion of all reactions was monitored by thin-layer chromatography (TLC) performed on pre-coated silica gel 60 F254 TLC plates (VWR), with observations made under ultraviolet light at wavelengths of 254 and/or 365 nm (Sun et al., 2022; Sun et al., 2025).
The cell lines PC-3, LNCaP, DU145 and WPMY-1 were purchased from the Cell Bank of the Chinese Academy of Sciences.
Compound 2 was synthesized using methods reported previously in the literature (Chen et al., 2018a; Chen et al., 2014).
Compound 3 was synthesized using methods reported previously in the literature (Chen et al., 2018b), and sesamol was substituted with 1-(diphenylmethyl)-piperazin. White solid (ethyl acetate). Yield: 70% from compound 1; M.p. 101.4°C–101.8°C; 1H NMR (400 MHz, CDCl3): δ 7.38 (d, J = 7.4 Hz, 4H), 7.25–7.20 (m, 6H), 7.13 (t, J = 8.6 Hz, 4H), 4.21 (s, 1H), 3.77 (t, J = 6.7 Hz, 2H), 3.46 (s, 2H), 2.80 (t, J = 6.6 Hz, 2H), 2.45 (s, 8H). HRMS (ESI) m/z [M + H]+: calcd for C26H31N2O: 387.2431, found: 387.2448.
Compound 4 was synthesized using methods reported previously in the literature (Chen et al., 2018b). White solid (ethyl acetate). M.p. 106.3°C–106.7°C; Yield, 87%. 1H NMR (400 MHz, CDCl3): δ 7.68 (d, J = 8.2 Hz, 2H), 7.39 (d, J = 7.4 Hz, 4H), 7.24 (t, J = 7.8 Hz, 6H), 7.19–7.12 (m, 4H), 7.02 (d, J = 7.8 Hz, 2H), 4.22 (s, 1H), 4.17 (t, J = 7.1 Hz, 2H), 3.48 (s, 2H), 2.90 (t, J = 7.1 Hz, 2H), 2.46 (s, 8H), 2.38 (s, 3H). HRMS (ESI) m/z [M + H]+: calcd for C33H37N2O3S: 541.2519, found: 541.2601.
Phenol (0.27 mmol, 1.5 equiv) and potassium carbonate (1.08 mmol, 6.0 equiv) were added to a solution of 4-((4-benzhydrylpiperazin-1-yl)methyl)phenethyl 4-methylbenzenesulfonate 4 (0.18 mmol, 1.0 equiv) in acetonitrile (CH3CN, 15 mL). The reaction mixture was heated to 85°C and stirred for 12 h. Afterward, the mixture was cooled down to room temperature. The reaction mixture was then filtered, and the filtrate was concentrated under vacuum. The residue was purified by silica gel column chromatography using a petroleum ether:ethyl acetate ratio of 25:1 (v/v) to obtain the corresponding product (5–24).
White solid (ethyl acetate); M.p. 96.3°C–96.8°C; Yield, 82%. The purity = 98.5%. 1H NMR (400 MHz, CDCl3): δ 7.39 (d, J = 7.6 Hz, 4H), 7.27 (s, 1H), 7.25 (d, J = 2.3 Hz, 2H), 7.23 (d, J = 7.0 Hz, 5H), 7.19 (d, J = 8.0 Hz, 2H), 7.14 (t, J = 7.2 Hz, 2H), 6.92 (d, J = 7.3 Hz, 1H), 6.88 (d, J = 8.2 Hz, 2H), 4.22 (s, 1H), 4.13 (t, J = 7.1 Hz, 2H), 3.48 (s, 2H), 3.05 (t, J = 7.1 Hz, 2H), 2.46 (s, 4H), 2.40 (s, 4H). 13C NMR (100 MHz, CDCl3): δ 158.8, 142.8, 137.0, 136.2, 129.5, 128.8, 128.4, 128.0, 126.9, 120.7, 114.6, 76.2, 68.6, 62.8, 53.4, 51.9, 35.5. HRMS (ESI) m/z [M + H]+: calcd for C32H35N2O: 463.2744, found: 463.2745.
Light yellow solid (ethyl acetate); M.p. 90.1°C–90.4°C; Yield, 78%. The purity = 98.3%. 1H NMR (400 MHz, CDCl3): δ 7.30 (d, J = 7.4 Hz, 4H), 7.15 (t, J = 7.2 Hz, 6H), 7.10 (d, J = 7.7 Hz, 2H), 7.06 (t, J = 7.3 Hz, 2H), 6.84 (t, J = 8.3 Hz, 2H), 6.72–6.69 (m, 2H), 4.14 (s, 1H), 3.99 (t, J = 7.0 Hz, 2H), 3.41 (s, 2H), 2.94 (t, J = 6.9 Hz, 2H), 2.39 (s, 4H), 2.32 (s, 4H). 13C NMR (100 MHz, CDCl3): δ 158.5, 155.5 (d, J = 112.5 Hz), 142.8, 137.0, 136.0, 129.5, 128.8, 128.5, 128.0, 126.9, 115.8 (d, J = 22.9 Hz), 115.6 (d, J = 8.0 Hz), 76.2, 69.4, 62.7, 53.3, 51.8, 35.5. HRMS (ESI) m/z [M + H]+: calcd for C32H34FN2O: 481.2650, found: 481.2647.
White solid (ethyl acetate); M.p. 123.2°C–123.5°C; Yield, 78%. The purity = 98%. 1H NMR (400 MHz, CDCl3): δ 7.39–7.37 (m, 4H), 7.21–7.18 (m, 8H), 7.12 (d, J = 7.0 Hz, 2H), 6.65–6.50 (m, 3H), 4.20 (s, 1H), 4.07 (t, J = 7.0 Hz, 2H), 3.46 (s, 2H), 3.01 (d, J = 6.6 Hz, 2H), 2.44 (s, 4H), 2.38 (s, 4H), 2.20 (s, 3H), 2.23 (s, 3H). 13C NMR (100 MHz, CDCl3): δ 159.0, 142.9, 139.2, 137.2, 136.2, 129.5, 128.5, 128.1, 127.0, 122.6, 122.5, 76.3, 68.6, 62.9, 53.4, 52.0, 35.7, 21.6. HRMS (ESI) m/z [M + H]+: calcd for C34H39N2O: 491.3057, found: 491.3059.
White solid (ethyl acetate); M.p. 112.3°C–112.7°C; Yield, 51%. The purity = 98.7%. 1H NMR (400 MHz, CDCl3): δ 7.53 (d, J = 8.6 Hz, 2H), 7.39 (d, J = 7.4 Hz, 4H), 7.24 (t, J = 7.3 Hz, 6H), 7.19–7.13 (m, 4H),6.90 (d, J = 8.6 Hz, 2H), 4.22 (s, 1H), 4.16 (t, J = 7.0 Hz, 2H), 3.49 (s, 2H), 3.07 (t, J = 6.9 Hz, 2H), 2.46 (s, 4H), 2.40 (s, 4H). 13C NMR (100 MHz, CDCl3): δ 162.1, 142.8, 136.6, 136.3, 134.0, 129.5, 128.8, 128.4, 128.0, 126.9, 119.2, 115.2, 104.0, 76.2, 69.0, 62.7, 53.4, 51.9, 35.2. HRMS (ESI) m/z [M + H]+: calcd for C33H34N3O: 488.2696, found: 488.2714.
White solid (ethyl acetate); M.p. 113.4°C–113.9°C; Yield, 76%. The purity = 98.7%. 1H NMR (400 MHz, CDCl3): δ 7.51 (d, J = 8.8 Hz, 2H), 7.39 (d, J = 7.5 Hz, 4H), 7.26 (s, 1H), 7.23 (d, J = 7.0 Hz, 5H), 7.18–7.13 (m, 4H), 6.64 (d, J = 8.8 Hz, 2H), 4.22 (s, 1H), 4.08 (t, J = 7.1 Hz, 2H), 3.49 (s, 2H), 3.03 (t, J = 7.0 Hz, 2H), 2.47 (s, 4H), 2.42 (s, 4H). 13C NMR (100 MHz, CDCl3): δ 158.7, 142.8, 138.2, 136.7, 136.2, 129.5, 128.8, 128.4, 128.0, 126.9, 117.0, 82.7, 76.2, 68.8, 62.7, 53.3, 51.8, 35.3. HRMS (ESI) m/z [M + H]+: calcd for C32H34IN2O: 589.1710, found: 589.1740.
Light yellow solid (ethyl acetate); M.p. 130.7°C–131.2°C; Yield, 68%. The purity = 98.8%. 1H NMR (400 MHz, CDCl3): δ 7.31 (d, J = 7.1 Hz, 4H), 7.19–7.13 (m, 7H), 7.09–7.04 (m, 4H), 6.86 (d, J = 2.7 Hz, 1H), 6.61 (dd, J = 8.8 Hz, J = 2.6 Hz, 1H), 4.13 (s, 1H), 3.99 (t, J = 7.0 Hz, 2H), 3.40 (s, 2H), 2.94 (t, J = 6.9 Hz, 2H), 2.37 (s, 4H), 2.31 (s, 4H). 13C NMR (100 MHz, CDCl3): δ 156.8, 141.7, 135.5, 135.4, 131.8, 129.6, 128.4, 127.7, 127.4, 126.9, 125.8, 122.8, 115.3, 113.5, 75.2, 68.2, 61.7, 52.3, 50.8, 34.2. HRMS (ESI) m/z [M + H]+: calcd for C32H33Cl2N2O: 531.1965, found: 531.1991.
Light yellow solid (ethyl acetate); M.p. 129.5°C–130.2°C; Yield, 77%. The purity = 97.9%. 1H NMR (400 MHz, CDCl3): δ 7.30 (d, J = 7.5 Hz, 4H), 7.14 (d, J = 6.6 Hz, 6H), 7.08–7.04 (m, 4H), 6.83 (s, 1H), 6.67 (s, 2H), 4.14 (s, 1H), 3.99 (t, J = 6.8 Hz, 2H), 3.40 (s, 2H), 2.93 (t, J = 6.8 Hz, 2H), 2.37 (s, 4H), 2.31 (s, 4H). 13C NMR (100 MHz, CDCl3): δ 159.9, 142.8, 136.6, 136.3, 135.4, 129.5, 128.8, 128.4, 128.0, 126.9, 121.0, 113.7, 76.3, 69.3, 62.8, 53.4, 51.9, 35.2. HRMS (ESI) m/z [M + H]+: calcd for C32H33Cl2N2O: 531.1965, found: 531.1985.
White solid (ethyl acetate); M.p. 125.1°C–125.6°C; Yield, 80%. The purity = 98.8%. 1H NMR (500 MHz, CDCl3): δ 7.45 (d, J = 7.5 Hz, 4H), 7.31 (s, 1H), 7.30 (d, J = 7.7 Hz, 5H), 7.26 (d, J = 8.0 Hz, 2H), 7.20 (dd, J = 15.0 Hz, J = 7.3 Hz, 3H), 6.79 (d, J = 7.5 Hz, 1H), 6.76–6.74 (m, 2H), 4.28 (s, 1H), 4.18 (t, J = 7.2 Hz, 2H), 3.55 (s, 2H), 3.11 (t, J = 7.1 Hz, 2H), 2.53 (s, 4H), 2.47 (s, 4H), 2.36 (s, 3H). 13C NMR (125 MHz, CDCl3): δ 158.9, 142.8, 139.5, 137.1, 136.1, 129.5, 129.2, 128.9, 128.5, 128.0, 126.9, 121.6, 115.5, 111.5, 76.2, 68.6, 62.7, 53.3, 51.9, 35.5, 21.6. HRMS (ESI) m/z [M + H]+: calcd for C33H37N2O: 477.2900, found: 477.2972.
White solid (ethyl acetate); M.p. 128.2°C–128.8°C; Yield, 74%. The purity = 98.2%. 1H NMR (400 MHz, CDCl3): δ 7.39 (d, J = 7.5 Hz, 4H), 7.33 (d, J = 7.3 Hz, 1H), 7.24 (t, J = 8.0 Hz, 8H), 7.15 (t, J = 6.9 Hz, 3H), 6.86 (d, J = 7.8 Hz, 2H), 4.22 (s, 1H), 4.18 (t, J = 7.0 Hz, 2H), 3.49 (s, 2H), 3.11 (t, J = 7.0 Hz, 2H), 2.46 (s, 4H), 2.39 (s, 4H). 13C NMR (100 MHz, CDCl3): δ 154.4, 142.8, 136.7, 136.3, 130.3, 129.4, 129.0, 128.4, 128.0, 127.6, 126.9, 123.0, 121.3, 113.4, 76.2, 69.8, 62.8, 53.3, 51.9, 35.4. HRMS (ESI) m/z [M + H]+: calcd for C32H34ClN2O: 497.2354, found: 497.2382.
White solid (ethyl acetate); M.p. 132.1°C–132.6°C; Yield, 81%. The purity = 98.5%. 1H NMR (400 MHz, CDCl3): δ 7.49 (d, J = 7.7 Hz, 1H), 7.38 (d, J = 7.4 Hz, 4H), 7.22 (d, J = 9.7 Hz, 8H), 7.18–7.12 (m, 3H), 6.81–6.75 (m, 2H), 4.21 (s, 1H), 4.14 (t, J = 6.8 Hz, 2H), 3.48 (s, 2H), 3.10 (t, J = 6.8 Hz, 2H), 2.46 (s, 4H), 2.40 (s, 4H). 13C NMR (100 MHz, CDCl3): δ 155.3, 142.8, 136.8, 136.2, 133.4, 129.5, 129.1, 128.5, 128.4, 128.0, 126.9, 121.9, 113.2, 112.3, 76.3, 69.9, 62.8, 53.4, 51.9, 35.5. HRMS (ESI) m/z [M + H]+: calcd for C32H34BrN2O: 541.1849, found: 541.1878.
White solid (ethyl acetate); M.p. 126.4°C–126.9°C; Yield, 72%. The purity = 98.3%. 1H NMR (400 MHz, CDCl3): δ 7.39 (d, J = 7.4 Hz, 4H), 7.26–7.20 (m, 8H), 7.17 (t, J = 5.9 Hz, 2H), 7.05 (d, J = 8.3 Hz, 2H), 6.78 (d, J = 8.4 Hz, 2H), 4.22 (s, 1H), 4.11 (t, J = 7.2 Hz, 2H), 3.48 (s, 2H), 3.04 (t, J = 7.1 Hz, 2H), 2.46 (s, 4H), 2.40 (s, 4H), 2.26 (s, 3H). 13C NMR (100 MHz, CDCl3): δ 156.7, 142.8, 137.0, 136.1, 129.9, 129.4, 128.8, 128.4, 128.0, 126.9, 117.9, 114.5, 76.2, 68.8, 62.8, 53.3, 51.9, 35.5, 20.5. HRMS (ESI) m/z [M + H]+: calcd for C33H37N2O: 477.2900, found: 477.2915.
White solid (ethyl acetate); M.p. 124.7°C–125.2°C; Yield, 67%. The purity = 98%. 1H NMR (400 MHz, CDCl3): δ 7.39 (d, J = 7.4 Hz, 4H), 7.26 (s, 1H), 7.23 (d, J = 7.9 Hz, 5H), 7.19 (d, J = 8.0 Hz, 2H), 7.14 (t, J = 7.4 Hz, 2H), 6.80 (s, 4H), 4.22 (s, 1H), 4.08 (t, J = 7.0 Hz, 2H), 3.95 (q, J = 13.8 Hz, J = 6.9 Hz, 2H), 3.50 (s, 2H), 3.03 (t, J = 7.0 Hz, 2H), 2.47 (s, 4H), 2.40 (s, 4H), 1.37 (t, J = 6.9 Hz, 3H). 13C NMR (100 MHz, CDCl3): δ 153.2, 152.9, 142.8, 137.1, 135.9, 129.5, 128.8, 128.4, 128.0, 126.9, 115.6, 115.4, 76.2, 69.4, 64.0, 62.7, 53.3, 51.8, 35.6, 15.0. HRMS (ESI) m/z [M + H]+: calcd for C34H39N2O2: 507.3006, found: 507.3019.
White solid (ethyl acetate); M.p. 115.8°C–116.3°C; Yield, 66%. The purity = 98.8%. 1H NMR (400 MHz, CDCl3): δ 7.68 (d, J = 8.0 Hz, 2H), 7.39 (d, J = 7.6 Hz, 4H), 7.27–7.22 (m, 5H), 7.17 (t, J = 7.6 Hz, 4H), 7.03 (d, J = 7.7 Hz, 2H), 4.22 (s, 1H), 4.17 (t, J = 7.1 Hz, 2H), 3.47 (s, 2H), 2.91 (t, J = 7.1 Hz, 2H), 2.45–2.40 (m, 12H), 1.42–1.28 (m, 4H). 13C NMR (100 MHz, CDCl3): δ 144.6, 142.8, 136.7, 134.9, 133.1, 129.8, 129.5, 128.7, 128.4, 128.0, 127.8, 126.9, 76.2, 70.6, 62.6, 53.3, 51.9, 35.0, 21.6. HRMS (ESI) m/z [M + H]+: calcd for C36H41N2O: 517.3214, found: 517.3219.
White solid (ethyl acetate); M.p. 116.0°C–116.7°C; Yield, 68%. The purity = 98.3%. 1H NMR (400 MHz, CDCl3): δ 7.39 (d, J = 7.6 Hz, 4H), 7.23 (dd, J = 15.2 Hz, J = 7.4 Hz, 8H), 7.17 (t, J = 6.6 Hz, 2H), 6.94 (d, J = 8.4 Hz, 1H), 6.64 (d, J = 8.4 Hz, 1H), 6.59 (s, 1H), 4.22 (s, 1H), 4.10 (t, J = 7.1 Hz, 2H), 3.51 (s, 2H), 3.04 (t, J = 7.1 Hz, 2H), 2.69 (t, J = 5.1 Hz, 4H), 2.49 (s, 4H), 2.43 (s, 4H), 1.75 (d, J = 2.6 Hz, 4H). 13C NMR (100 MHz, CDCl3): δ 156.6, 142.7, 138.1, 129.9, 129.5, 129.3, 128.8, 128.4, 128.0, 126.9, 114.5, 112.4, 76.2, 68.7, 62.7, 53.2, 51.7, 35.5, 28.6, 23.4, 23.2. HRMS (ESI) m/z [M + H]+: calcd for C36H41N2O: 517.3214, found: 517.3230.
White solid (ethyl acetate); M.p. 118.7°C–119.4°C; Yield, 83%. The purity = 98.9%. 1H NMR (400 MHz, CDCl3): δ 7.39 (d, J = 7.5 Hz, 4H), 7.25 (s, 2H), 7.22 (d, J = 7.2 Hz, 6H), 7.15 (d, J = 7.2 Hz, 2H), 7.10 (t, J = 7.6 Hz, 2H), 6.82 (t, J = 7.4 Hz, 1H), 6.76 (d, J = 8.5 Hz, 1H), 4.21 (s, 1H), 4.12 (t, J = 6.7 Hz, 2H), 3.50 (s, 2H), 3.06 (t, J = 6.7 Hz, 2H), 2.48 (s, 4H), 2.42 (s, 4H), 2.17 (s, 3H). 13C NMR (100 MHz, CDCl3): δ 157.0, 142.8, 137.5, 135.8, 130.7, 129.5, 129.0, 128.5, 128.0, 127.7, 126.9, 126.7, 120.3, 110.9, 76.2, 68.7, 62.7, 53.3, 51.8, 35.7, 16.3. HRMS (ESI) m/z [M + H]+: calcd for C33H37N2O: 477.2900, found: 477.2911.
White solid (ethyl acetate); M.p. 126.5°C–127.0°C; Yield, 81%. The purity = 98.4%. 1H NMR (400 MHz, CDCl3): δ 7.37 (d, J = 7.1 Hz, 4H), 7.24–7.19 (m, 8H), 7.14 (d, J = 7.2 Hz, 2H), 6.99 (d, J = 7.8 Hz, 1H), 6.69 (s, 1H), 6.61 (d, J = 6.0 Hz, 1H), 4.19 (s, 1H), 4.09 (t, J = 7.0 Hz, 2H), 3.52 (s, 2H), 3.02 (t, J = 6.9 Hz, 2H), 2.51 (s, 4H), 2.43 (s, 4H), 2.19 (s, 3H), 2.16 (s, 3H). 13C NMR (100 MHz, CDCl3): δ 157.0, 142.8, 137.7, 137.5, 130.4, 130.3, 129.8, 128.9, 128.7, 128.5, 128.0, 126.9, 116.3, 111.6, 76.2, 68.7, 62.6, 53.2, 51.6, 35.6, 20.0, 18.8. HRMS (ESI) m/z [M + H]+: calcd for C34H39N2O: 491.3057, found: 491.3067.
White solid (ethyl acetate); M.p. 129.6°C–130.1°C; Yield, 72%. The purity = 99%. 1H NMR (500 MHz, CDCl3): δ 7.45 (d, J = 7.2 Hz, 4H), 7.32 (s, 1H), 7.30 (d, J = 6.2 Hz, 5H), 7.26–7.24 (m, 4H), 7.21 (t, J = 7.4 Hz, 2H), 6.85 (d, J = 9.0 Hz, 2H), 4.28 (s, 1H), 4.15 (t, J = 7.1 Hz, 2H), 3.57 (s, 2H), 3.10 (t, J = 7.1 Hz, 2H), 2.54 (s, 4H), 2.47 (s, 4H). 13C NMR (125 MHz, CDCl3): δ 157.5, 142.8, 136.8, 136.0, 129.6, 129.3, 128.8, 128.5, 128.0, 126.9, 125.6, 115.9, 76.2, 69.0, 62.6, 53.3, 51.8, 35.4. HRMS (ESI) m/z [M + H]+: calcd for C32H34ClN2O: 497.2354, found: 497.2376.
White solid (ethyl acetate); M.p. 110.3°C–110.9°C; Yield, 75%. The purity = 98.5%. 1H NMR (500 MHz, CDCl3): δ 8.31 (d, J = 7.2 Hz, 1H), 7.83 (d, J = 7.1 Hz, 1H), 7.52–7.49 (m, 2H), 7.46 (d, J = 7.1 Hz, 5H), 7.39 (t, J = 7.8 Hz, 1H), 7.38–7.29 (m, 8H), 7.22 (t, J = 7.4 Hz, 2H), 6.84 (d, J = 7.4 Hz, 1H), 4.38 (t, J = 6.9 Hz, 2H), 4.29 (s, 1H), 3.57 (s, 2H), 3.27 (t, J = 6.9 Hz, 2H), 2.54 (s, 4H), 2.48 (s, 4H). 13C NMR (125 MHz, CDCl3): δ 154.6, 142.8, 137.3, 136.1, 134.5, 129.5, 128.9, 128.5, 128.0, 127.4, 126.9, 126.4, 125.9, 125.7, 125.2, 122.1, 120.3, 104.7, 76.2, 68.9, 62.7, 53.3, 51.9, 35.6. HRMS (ESI) m/z [M + H]+: calcd for C36H37N2O: 513.2900, found: 513.2927.
White solid (ethyl acetate); M.p. 127.2°C–127.8°C; Yield, 79%. The purity = 98.6%. 1H NMR (500 MHz, CDCl3): δ 7.44 (d, J = 7.3 Hz, 4H), 7.38 (dd, J = 7.0 Hz, J = 2.1 Hz, 2H), 7.31 (s, 1H), 7.29 (d, J = 8.0 Hz, 5H), 7.24 (d, J = 8.1 Hz, 2H), 7.20 (t, J = 7.4 Hz, 2H), 6.80 (d, J = 9.0 Hz, 2H), 4.27 (s, 1H), 4.14 (t, J = 7.1 Hz, 2H), 3.55 (s, 2H), 3.09 (t, J = 7.1 Hz, 2H), 2.54 (s, 4H), 2.48 (s, 4H). 13C NMR (125 MHz, CDCl3): δ 158.0, 142.8, 136.8, 136.0, 132.3, 129.6, 128.9, 128.5, 128.0, 126.9, 116.4, 112.9, 76.2, 69.0, 62.7, 53.3, 51.8, 35.4. HRMS (ESI) m/z [M + H]+: calcd for C32H34BrN2O: 541.1849, found: 541.1884.
White solid (ethyl acetate); M.p. 109.8°C–110.5°C; Yield, 68%. The purity = 98.7%. 1H NMR (500 MHz, CDCl3): δ 7.57 (d, J = 8.7 Hz, 2H), 7.46 (d, J = 7.3 Hz, 4H), 7.31 (t, J = 7.6 Hz, 6H), 7.27 (d, J = 8.0 Hz, 2H), 7.22 (t, J = 7.4 Hz, 2H), 6.98 (d, J = 8.7 Hz, 2H), 4.30 (s, 1H), 4.22 (t, J = 7.1 Hz, 2H), 3.57 (s, 2H), 3.14 (t, J = 7.1 Hz, 2H), 2.55 (s, 4H), 2.48 (s, 4H). 13C NMR (125 MHz, CDCl3): δ 161.3, 142.8, 136.6, 136.4, 129.6, 128.9, 128.5, 128.0, 126.9 (t, J = 4.6 Hz), 124.5 (dd, J = 539.0 Hz, J = 269.6 Hz), 122.9 (dd, J = 65.0 Hz, J = 32.5 Hz), 120.7 (d, J = 141.4 Hz), 114.5, 76.3, 68.9, 62.7, 53.4, 51.9, 35.3. HRMS (ESI) m/z [M + H]+: calcd for C33H34F3N2O: 531.2618, found: 531.2648.
PC-3 and WPMY-1 cells were cultured in Dulbecco’s modiffcation Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, United States) supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, UT, United States), 100 U/mL penicillin and 0.1 mg/mL streptomycin (Invitrogen). DU145 cells were cultured in RPMI1640 media supplemented with 10% fetal bovine serum (FBS, Hyclone), 100 U/mL penicillin and 0.1 mg/mL streptomycin (Invitrogen). LNCaP cells were cultured in F12 media supplemented with 10% fetal bovine serum (FBS, Hyclone), 100 U/mL penicillin and 0.1 mg/mL streptomycin (Invitrogen). The cells were incubated at 37°C in a humidified atmosphere with 5% CO2(Chen et al., 2025; Jiang et al., 2024).
The proliferative capacity of cells was evaluated through the implementation of a Cell Counting Kit-8 (CCK-8) assay, supplied by Dojindo (Japan), to quantify cellular growth. Post-transfection, cells were dispensed into a 96-well microplate at a density of 3 × 103 cells per well and incubated for intervals of 0, 24, 48, 72, 96, and 120 h. Following this, 10 μL of the CCK-8 solution was introduced into each well and the plates were returned to the incubator for an additional 2-h period at 37°C under 5% CO2 conditions. Absorbance readings at 450 nm were subsequently taken using an ELISA reader manufactured by Bio Tek (United States) (Chen et al., 2022; Zhou et al., 2024).
The compound concentrations were set at 30, 15, 7.5, 3.75, 1.88, and 0.94 μmol/L. The absorbance (A) was measured at 450 nm using a microplate reader. A linear regression analysis was performed plotting the logarithm of compound concentration against the inhibition rate to obtain a straight-line equation from which the half-maximal inhibitory concentration (IC50) of the compound, capable of inhibiting 50% of cancer cells, was determined. All experiments were repeated three times under identical conditions, and the mean value was taken as the final result.
Fireffy and Renilla luciferase activities, which are indicated as RLUs, were determined using Dual-Glo luciferase assay kits (Promega) according to the manufacturer’s instructions (Chen et al., 2019a; Chen et al., 2019b). RLUs were measured using a luminometer (GloMaxTM 96-Microplate Luminometer, Promega) and are reported as the mean ± SEM of three individual experiments. For agonists, fold of induction = LUinduced/RLUuninduced. For antagonists, % of control = 100 × RLU (agonist + antagonist)/RLU (agonist alone). All RLUs were normalized against ffreffy RLUs/Renilla RLUs. Data are expressed as EC50/IC50 values in μM, and the IC50 of phenylephrine (μM) was calculated by plotting the data using nonlinear regression analysis in Graph-Pad Prism 5 software.
The fluorescence polarization technique was used to analyze the binding of 7, 11, 17, 19, 20, 21, 22, 23, 24 and enzalutamide to the AR using the PolarScreen™ AR Competitor Assay, Green (lifetechnologies, A15880) according to the manufacturer’s instructions (Chen et al., 2019a; Chen et al., 2019b). Brieffy, the assay entails titration of the test compound against a preformed complex of Fluormone™AL Green and the AR-LBD (GST). The assay mixture was allowed to equilibrate at room temperature in 384-well black plates for 4 h, after which the fluorescence polarization values were measured in a SpectraMax® Paradigm® Multi-Mode Detection Platform (Molecular Devices) using an excitation wavelength of 485 nm and an emission wavelength of 535 nm. Data analysis for the ligand binding assays was performed using Prism software (GraphPad Software, Inc.).
Until now, three binding sites of androgen receptor have been reported, including LBP, AF2 and BF3 (Chen et al., 2019a; Chen et al., 2019b). In order to explore the mechanism of androgen receptor antagonism by the target compound 17, a dockingsimulation was carried out using AutoDock Vina software. The crystal structure of androgen receptor downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) was taken as the template protein to engage in docking simulation. In prepare, the exogenous ligand was removed and the hydrogen atoms were added to the system. To ensure the reliability of docking simulation, a redock process for the exogenous ligand was performed before docking analysis. Finally, one compound target (i.e., compound 17) with high AR antagonistic activity was docked into three potential binding sites (including LBP, AF2 and BF3) with 10 configurations.
Scheme 1 illustrated the synthesis of arylpiperazine derivatives 5–24 via a four-step reaction starting from 2-(4-(bromomethyl)phenyl)acetic acid. First, 2-(4-(bromomethyl)phenyl)acetic acid one was reduced to alcohol 2 with the presence of a borane–methyl sulfide complex (2 M in tetrahydrofuran) at 0°C for 1 h and at room temperature for 12 h. After the nucleophilic substitution reaction was carried out between compound 2 and 1-(diphenylmethyl)-piperazin using CH3CN as solvent in the presence of potassium carbonate at 85°C for 12 h gave 3 (70% yield from 1). Subsequently, compound 4 (87% yield) was obtained by reacting 3 with 4-toluene-sulfonyl chloride using CH2Cl2 as solvent in the presence of trimethylamine and a catalytic amount of 4-dimethylaminopyridine at 0°C for 16 h. Finally, compound 4 was treated with various phenol (1.5 equiv) in the presence of K2CO3 (6 eq) to obtain derivatives 5–24 in moderate yields (51%–83%). All synthesized analogs were confirmed using 1H-NMR, 13C-NMR, and HRMS.
The cytotoxic activity of derivatives 5–24 against three human prostate cancer cell lines (PC-3, LNCaP, and DU145) and one type of human normal prostate epithelial cells was evaluated using the CCK-8 method. The results are shown in Table 1.
In this study, the cytotoxic activity of ether arylpiperazine derivatives ranging from 5 to 24 was assessed against 4 cell lines: PC-3, LNCaP, DU145 (all human prostate cancer cell lines), and WPMY-1 (human normal prostatic epithelial cells) using the CCK-8 assay. The outcomes are summarized in Table 1. The data presented in the table reveal that several of these compounds exhibit pronounced cytotoxic activity against the tested cancer cell lines, with some demonstrating significantly greater potency compared to naftopidil. Notably: Compounds 7, 11, 17, 19, 20, 21, 23, and 24 exhibit exceptionally potent activity against PC-3 cells, characterized by IC50 values below 5 μM. For LNCaP cells, compounds 17, 19, 20, and 23 show particularly strong activity with IC50 values less than 5 μM. Compounds 19, 20, 23 and 24 also demonstrate pronounced cytotoxicity against DU145 cells. Moreover, the majority of compounds exhibited low cytotoxic character toward normal human prostate epithelial cells (WPMY-1).
The SAR investigation was mainly focused on the variation of the substitute’s type on the phenyl group as a required group for antitumor activity. (1) For instance, compared to phenylpiperazine compound 5, compounds 7 (IC50 = 2.95 μM) and 11 (IC50 = 1.59 μM), which introduce two symmetrical groups onto the phenyl ring, exhibit potent cytotoxic activity against PC-3 cells. In contrast, compound 12 (IC50 > 50 μM), featuring a methyl group as an electron-donating substituent in the meta position of the phenyl ring, displays significantly weaker cytotoxic activity against PC-3 than 5. Additionally, compound 24 (IC50 = 5.76 μM), with a trifluoromethyl group as a strong electron-withdrawing substituent on the phenyl ring, exhibits strong cytotoxic activity against DU145 cells. These activity results indicate that variations in substituents on the phenyl ring do have a certain impact on cytotoxic activity. (2) A comparison between compounds 10 and 11 reveals that compound 11 (IC50 = 1.59 μM) demonstrates exceptionally strong activity against PC-3 cells, while compound 10 exhibits weaker cytotoxic activity against PC-3. These activity results suggest that the introduction of symmetrical, weakly electron-withdrawing substituents at the 3 and 5 positions of the phenyl ring favors anticancer activity. (3) When comparing compound 19 with compounds 13 and 14, compound 19 (IC50 = 0.87 μM) exhibits more pronounced cytotoxic activity against DU145 cells. The results indicate that the introduction of electron-donating groups in the ortho position of the phenyl ring is more favorable for anticancer activity than the introduction of electron-withdrawing groups. (4) A comparison between compounds 12, 15, and 19 reveals that compound 19 (IC50 > 50 μM) exhibits weak activity against human normal prostatic epithelial cells. These activity results suggest that when introducing a methyl group at different positions on the phenyl ring, the ortho position favors anticancer activity more than other positions. (5) Comparing compound 7 with compound 12, compound 7 (IC50 = 2.95 μM) demonstrates more pronounced cytotoxic activity against PC-3 cells. These activity results indicate that the simultaneous introduction of methyl groups at the three and five positions of the phenyl ring favors anticancer activity more than the introduction of a methyl group only at the three position. (6) When comparing compounds 6, 8, 9, 15 and 21, compounds 23 and 24 exhibits more versatile and superior cytotoxic activity. These activity results suggest that the introduction of a bromo group or a trifluoromethyl group at the para position of the phenyl ring favors anticancer activity. (7) A comparison between compounds 17 and 18, compound 17 (IC50 = 1.89 μM, 1.04 μM, 7.32 μM) demonstrates better activity against the tested cell lines than compound 18 (IC50 = 5.02 μM, >50 μM, 14.36 μM). The results showed that the introduction of cycloalkyl at the 5 and 6 position of the phenyl ring was beneficial to anticancer activity (Figure 2). Above results can lead to a tool which can further design arylpiperazine derivatives as AR antagonists for in vitro and in vivo studies.
To further investigate whether these derivatives possess antagonistic activity against AR, this study adopted the scientific method of luciferase assay (Qi et al., 2022a; Qi et al., 2022b) to more accurately evaluate the antagonistic effects of these derivatives on AR (Table 2). During the implementation of the AR luciferase assay experiments, we specifically added 1 nM of the AR agonist R1881 for co-treatment, and quantitatively assessed the strength of the antagonistic activity based on the degree of inhibition of luciferase expression induced by R1881. According to the data presented in Table 2, it is evident that compounds 7, 11, 21, 22, and 24 exhibit relatively weak antagonistic effects on AR. However, notably, compounds 17, 19, 20, and 23 demonstrate significant antagonistic efficacy, with inhibition rates exceeding 55%. It is worth noting that these results do not entirely align with previous tests on anti-proliferative activity against cancer cells. These findings suggest that the skillful introduction of certain small molecular groups on the piperazine ring may significantly enhance their antagonistic activity against AR. The conclusions drawn from this study undoubtedly provide us with a powerful tool, which will aid us in delving deeper into the interaction mechanisms between piperazine derivatives and AR, thus laying a more solid theoretical foundation for future drug development.
Recent studies have shown that, in addition to the AR signaling pathway, estrogen receptors (ER) also play an important role in the pathogenesis of PCa(Belluti et al., 2023; Souza et al., 2023). Research by Sarswat et al. (Sarswat et al., 2011) discovered that arylpiperazines, in addition to inhibiting the transmission of AR signals, can also promote the expression of ER-β in prostate cancer cells. ER-β may function as a protective receptor, exerting inhibitory effects on the development and malignant progression of PCa(Chaurasiya et al., 2020; Lombardi et al., 2020). By acting on prostate cancer cells through these two signaling pathways simultaneously, arylpiperazines inhibit their proliferation.
To delve into the specific binding characteristics of these analogs that exhibit significant inhibitory activity against the AR, we conducted detailed binding affinity studies using fluorescence polarization (FP) technology (Chen et al., 2023; He et al., 2021; Xue et al., 2023). The experimental design was based on the competitive binding mechanism between fluorescent tracers and non-fluorescent antagonists, aiming to assess the interaction strength between a series of compounds numbered 7, 11, 17, 19, 20, 21, 22, 23 and 24 with AR (Table 3). Through this method, we were able to precisely measure the binding efficiency of each compound at different concentrations and summarize the results in Table 3. The study revealed that all tested analogs demonstrated strong binding affinity to AR, with IC50 values below 4 μmol (μM), indicating high binding affinity. Particularly noteworthy, compounds 17 and 19 exhibited outstanding binding performance, with IC50 values of 1.14 μM and 1.01 μM, respectively, surpassing not only other test samples but also the clinically used standard drug enzalutamide (IC50 = 2.56 μM). This suggests that these two compounds may serve as more effective candidates for AR antagonists.
Further analysis revealed a clear trend among the tested arylpiperazine derivatives: a direct correlation between the binding affinity of compounds to AR and their antagonistic activity. For instance, the tightly bound compounds 17 and 19 were also the most effective antagonists, achieving maximum inhibition rates of 70.3% and 71.5%, respectively. Additionally, while not as prominent as the former two, compounds 20 and 23 also exhibited strong antagonistic effects, with inhibition rates of 65.7% and 62.2%, respectively. These observations support the hypothesis that enhanced binding affinity may be a key factor in improving AR antagonistic activity. Based on these findings, it can be inferred that certain ether-substituted arylpiperazine derivatives, due to their excellent binding ability and antagonistic efficacy, hold potential for development as novel AR antagonists, especially in the field of prostate cancer treatment. Considering the exceptional characteristics displayed by compound 19, we have decided to focus on it for the next phase of research to explore its specific binding sites with AR and potential mechanisms of action. This will contribute to understanding how these compounds effectively block the AR signaling pathway, providing new strategies and methods for the treatment of prostate cancer.
To decipher the binding mode of these compounds, as well as to explore the detailed information about their major binding interactions with AR (Zhang et al., 2014) docking simulation is performed. The optimal antagonist, compound 19 was taken as the template molecule in this process, and three binding sites of AR, including ligand binding pocket (LBP), activation function-2 (AF2) and binding function 3 (BF3) (Axerio-Cilies et al., 2011; Lack et al., 2011), were all used to explore the binding affinities of this compound. The lowest docked energy values were summarized in Table 3.
As shown in Table 4, LBP binding site had the highest binding force of −11.01 kcal/mol, which indicated that AR LBP was the major binding site for compound 19. To better discover these binding interactions, the binding mode of compound 19 with AR was analyzed, and the detailed information was displayed in Figure 3. As shown in Figure 3, compound 19 could fit into the AR LBP site by the formation of Van der Waals’ force with 14 amino acid residues, such as Val 746, Trp 741 and Phe 764, and so on. These results showed that the compound 19 mainly bind to AR LBP site through the interaction of Van der Waals’ force.
In conclusion, this study reported the synthesis and biological evaluation of a series of novel arylpiperazine derivatives against three human prostate cancer cells and human prostate epithelial cells and AR, respectively. The results showed that the derivatives 7, 11, 17, 19, 20, 21, 22, 23 and 24 displayed strong cytotoxic activities against the tested cancer cells, and derivatives 17, 19, 20, and 23 exhibited relatively strong antagonistic potency against AR (Inhibition% >60) and exhibited potent AR binding affinities. Structure-activity relationship (SAR) studies indicated that the introduction of cycloalkyl groups at the m,p-position on the phenyl ring and a methyl group at the o-position on the phenyl ring favored enhanced activity. Docking study suggested that the compounds 19 mainly bind to AR ligand binding pocket (LBP) site through the interaction of Van der Waals’ force. These piperazine derivatives may guide the structural modification of novel anti-prostate cancer drugs.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
HJ: Writing–original draft, Conceptualization, Funding acquisition, Formal Analysis. HaC: Conceptualization, Writing–original draft, Methodology, Software. YW: Writing–original draft, Validation. HX: Investigation, Writing–original draft. HoC: Investigation, Conceptualization, Data curation, Funding acquisition, Writing–review and editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Natural Science Foundation of Science and Technology Projects of Guizhou Province (Grant no. Qian Ke He Foundation-ZK [2022] General 633), the Scientific and Technological Support Project of Guizhou Province (Grant no. Qian Ke He Zhi Cheng [2023] Yi Ban 262), 2022 Guangdong Basic, Applied Basic Research Fund and Enterprises Joint Fund of Public Health and Medicine Area (Grant No. 2022A1515220218) and the Henan Province Science and Technology Attack Plan Foundation (242102231047).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2025.1557275/full#supplementary-material
Axerio-Cilies, P., Lack, N. A., Nayana, M. R., Chan, K. H., Yeung, A., Leblanc, E., et al. (2011). Inhibitors of androgen receptor activation function-2 (AF2) site identified through virtual screening. J. Med. Chem. 54 (18), 6197–6205. doi:10.1021/jm200532b
Belluti, S., Imbriano, C., and Casarini, L. (2023). Nuclear estrogen receptors in prostate cancer: from genes to function. Cancers (Basel) 15 (18), 4653. doi:10.3390/cancers15184653
Chaurasiya, S., Widmann, S., Botero, C., Lin, C. Y., Gustafsson, J., and Strom, A. M. (2020). Estrogen receptor β exerts tumor suppressive effects in prostate cancer through repression of androgen receptor activity. PLoS One 15 (5), e0226057. doi:10.1371/journal.pone.0226057
Chen, H., Guan, X., Liu, Q., Yang, L., Guo, J., Gao, F., et al. (2022). Co-Assembled Nanocarriers of de novo Thiol-Activated hydrogen sulfide Donors with an RGDFF Pentapeptide for targeted therapy of non-small-cell Lung cancer. ACS Appl. Mater Interfaces 14 (48), 53475–53490. doi:10.1021/acsami.2c14570
Chen, H., Guo, S., Liu, Y., Jiang, H., Liao, Y.-X., Shen, J., et al. (2023). A stable NIR fluorescent probe for imaging lipid droplets in triple-negative breast cancer. Sens. Actuat B-Chem 398 (1), 134740. doi:10.1016/j.snb.2023.134740
Chen, H., Liang, X., Sun, T., Qiao, X., Zhan, Z., Li, Z., et al. (2018). Synthesis and biological evaluation of estrone 3-O-ether derivatives containing the piperazine moiety. Steroids 134, 101–109. doi:10.1016/j.steroids.2018.02.002
Chen, H., Liang, X., Xu, F., Xu, B., He, X., Huang, B., et al. (2014). Synthesis and cytotoxic activity evaluation of novel arylpiperazine derivatives on human prostate cancer cell lines. Molecules 19 (8), 12048–12064. doi:10.3390/molecules190812048
Chen, H., Wang, C. L., Sun, T., Zhou, Z., Niu, J. X., Tian, X. M., et al. (2018). Synthesis, biological evaluation and SAR of naftopidil-based arylpiperazine derivatives. Bioorg Med. Chem. Lett. 28 (9), 1534–1539. doi:10.1016/j.bmcl.2018.03.070
Chen, H., Xu, B. B., Sun, T., Zhou, Z., Ya, H. Y., and Yuan, M. (2017). Synthesis and antitumor activity of novel arylpiperazine derivatives containing the Saccharin moiety. Molecules 22 (11), 1857. doi:10.3390/molecules22111857
Chen, H., Xu, F., Liang, X., Xu, B. B., Yang, Z. L., He, X. L., et al. (2015). Design, synthesis and biological evaluation of novel arylpiperazine derivatives on human prostate cancer cell lines. Bioorg Med. Chem. Lett. 25 (2), 285–287. doi:10.1016/j.bmcl.2014.11.049
Chen, H., Yu, Y. Z., Tian, X. M., Wang, C. L., Qian, Y. N., Deng, Z. A., et al. (2019). Synthesis and biological evaluation of arylpiperazine derivatives as potential anti-prostate cancer agents. Bioorg Med. Chem. 27 (1), 133–143. doi:10.1016/j.bmc.2018.11.029
Chen, H., Zhang, J., Hu, P., Qian, Y., Li, J., and Shen, J. (2019). Synthesis, biological evaluation and molecular docking of 4-Amino-2H-benzo[h]chromen-2-one (ABO) analogs containing the piperazine moiety. Bioorg Med. Chem. 27 (20), 115081. doi:10.1016/j.bmc.2019.115081
Chen, J., Wang, C., Huang, X., Wan, R., Zhu, Z., Sun, G., et al. (2025). Quantum dots-Engineered Flexible Hydrogel as Plant-Wearable sensor for on-site Profiling Dynamic Pesticide Degradation. Adv. Funct. Mater, 2423643. doi:10.1002/adfm.202423643
Choi, E., Buie, J., Camacho, J., Sharma, P., and de Riese, W. T. W. (2022). Evolution of androgen deprivation therapy (ADT) and its new Emerging Modalities in prostate cancer: an Update for Practicing Urologists, Clinicians and Medical Providers. Res. Rep. Urol. 14, 87–108. doi:10.2147/rru.s303215
Dai, C., Dehm, S. M., and Sharifi, N. (2023). Targeting the androgen signaling Axis in prostate cancer. J. Clin. Oncol. 41 (26), 4267–4278. doi:10.1200/jco.23.00433
Fontana, F., Anselmi, M., and Limonta, P. (2022). Molecular mechanisms and genetic alterations in prostate cancer: from diagnosis to targeted therapy. Cancer Lett. 534, 215619. doi:10.1016/j.canlet.2022.215619
Formaggio, N., Rubin, M. A., and Theurillat, J. P. (2021). Loss and revival of androgen receptor signaling in advanced prostate cancer. Oncogene 40 (7), 1205–1216. doi:10.1038/s41388-020-01598-0
He, X., Ding, F., Sun, X., Zheng, Y., Xu, W., Ye, L., et al. (2021). Renovated multifunctional colorimetric/fluorometric sensor for simultaneous detection, imaging of pH variance and antimicrobial therapies. Sens. Actuat B-Chem 332, 129496. doi:10.1016/j.snb.2021.129496
He, Y., Xu, W., Xiao, Y. T., Huang, H., Gu, D., and Ren, S. (2022). Targeting signaling pathways in prostate cancer: mechanisms and clinical trials. Signal Transduct. Target Ther. 7 (1), 198. doi:10.1038/s41392-022-01042-7
Ishii, K., Matsuoka, I., Kajiwara, S., Sasaki, T., Miki, M., Kato, M., et al. (2018). Additive naftopidil treatment synergizes docetaxel-induced apoptosis in human prostate cancer cells. J. Cancer Res. Clin. Oncol. 144 (1), 89–98. doi:10.1007/s00432-017-2536-x
Iwamoto, Y., Ishii, K., Kanda, H., Kato, M., Miki, M., Kajiwara, S., et al. (2017). Combination treatment with naftopidil increases the efficacy of radiotherapy in PC-3 human prostate cancer cells. J. Cancer Res. Clin. Oncol. 143 (6), 933–939. doi:10.1007/s00432-017-2367-9
Jamroze, A., Chatta, G., and Tang, D. G. (2021). Androgen receptor (AR) heterogeneity in prostate cancer and therapy resistance. Cancer Lett. 518, 1–9. doi:10.1016/j.canlet.2021.06.006
Jiang, H., Zhang, H., and Jiang, S. (2024). CircDUSP22 Attenuates the Ferroptosis of prostate cancer cells via miR-18a-5p/SLC7A11/GPX4 signaling. Comb. Chem. High. Throughput Screen 27. doi:10.2174/0113862073324077240624094140
Kinoyama, I., Taniguchi, N., Kawaminami, E., Nozawa, E., Koutoku, H., Furutani, T., et al. (2005). N-Arylpiperazine-1-carboxamide derivatives: a novel series of orally active nonsteroidal androgen receptor antagonists. Chem. Pharm. Bull. (Tokyo) 53 (4), 402–409. doi:10.1248/cpb.53.402
Kinoyama, I., Taniguchi, N., Yoden, T., Koutoku, H., Furutani, T., Kudoh, M., et al. (2004). Synthesis and pharmacological evaluation of novel arylpiperazine derivatives as nonsteroidal androgen receptor antagonists. Chem. Pharm. Bull. (Tokyo) 52 (11), 1330–1333. doi:10.1248/cpb.52.1330
Lack, N. A., Axerio-Cilies, P., Tavassoli, P., Han, F. Q., Chan, K. H., Feau, C., et al. (2011). Targeting the binding function 3 (BF3) site of the human androgen receptor through virtual screening. J. Med. Chem. 54 (24), 8563–8573. doi:10.1021/jm201098n
Lombardi, A. P. G., Vicente, C. M., and Porto, C. S. (2020). Estrogen receptors promote Migration, Invasion and Colony formation of the androgen-independent prostate cancer cells PC-3 through β-Catenin pathway. Front. Endocrinol. (Lausanne) 11, 184. doi:10.3389/fendo.2020.00184
Lu, C., Brown, L. C., Antonarakis, E. S., Armstrong, A. J., and Luo, J. (2020). Androgen receptor variant-driven prostate cancer II: advances in laboratory investigations. Prostate Cancer Prostatic Dis. 23 (3), 381–397. doi:10.1038/s41391-020-0217-3
Maesaka, F., Tanaka, N., Nakai, Y., Asakawa, I., Tomizawa, M., Owari, T., et al. (2021). Comparison of disease-specific quality of life in prostate cancer patients treated with low-dose-rate brachytherapy: a randomized controlled trial of silodosin versus naftopidil. Int. J. Urol. 28 (11), 1171–1176. doi:10.1111/iju.14667
Obinata, D., Lawrence, M. G., Takayama, K., Choo, N., Risbridger, G. P., Takahashi, S., et al. (2020). Recent Discoveries in the androgen receptor pathway in castration-resistant prostate cancer. Front. Oncol. 10, 581515. doi:10.3389/fonc.2020.581515
Qi, Y., Chen, H., Chen, S., Shen, J., and Li, J. (2022). Synthesis, bioactivity, and molecular docking of novel arylpiperazine derivatives as potential AR antagonists. Front. Chem. 10, 947065. doi:10.3389/fchem.2022.947065
Qi, Y., Xue, B., Chen, S., Wang, W., Zhou, H., and Chen, H. (2022). Synthesis, biological evaluation, and molecular docking of novel hydroxyzine derivatives as potential AR antagonists. Front. Chem. 10, 1053675. doi:10.3389/fchem.2022.1053675
Sarswat, A., Kumar, R., Kumar, L., Lal, N., Sharma, S., Prabhakar, Y. S., et al. (2011). Arylpiperazines for management of benign prostatic hyperplasia: design, synthesis, quantitative structure-activity relationships, and pharmacokinetic studies. J. Med. Chem. 54 (1), 302–311. doi:10.1021/jm101163m
Schmidt, K. T., Huitema, A. D. R., Chau, C. H., and Figg, W. D. (2021). Resistance to second-generation androgen receptor antagonists in prostate cancer. Nat. Rev. Urol. 18 (4), 209–226. doi:10.1038/s41585-021-00438-4
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72 (1), 7–33. doi:10.3322/caac.21708
Souza, D. S., Macheroni, C., Vicente, C. M., Cavalheiro, R. P., Campo, V. L., and Porto, C. S. (2023). Estrogen receptors regulate galectin-3 in androgen-independent DU-145 prostate cancer cells. Oncol. Rep. 49 (5), 93. doi:10.3892/or.2023.8530
Sun, T., Chen, R., Huang, Q., Ba, M., Cai, Z., Hu, S., et al. (2022). Chromatographic Separation of Aromatic amine Isomers: a solved issue by a new Amphiphilic pillar[6]arene stationary phase. ACS Appl. Mater Interfaces 14 (50), 56132–56142. doi:10.1021/acsami.2c17889
Sun, T., Zhang, Y., Liu, H., Xu, X., Cai, Z., Hu, S., et al. (2025). Separation performances of extended pillar[6]arenes, a new stationary phase for gas chromatography. Talanta 283, 127098. doi:10.1016/j.talanta.2024.127098
Vellky, J. E., and Ricke, W. A. (2020). Development and prevalence of castration-resistant prostate cancer subtypes. Neoplasia 22 (11), 566–575. doi:10.1016/j.neo.2020.09.002
Visakorpi, T., Hyytinen, E., Koivisto, P., Tanner, M., Keinänen, R., Palmberg, C., et al. (1995). In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat. Genet. 9 (4), 401–406. doi:10.1038/ng0495-401
Xue, B., Hou, A., Du, Y., Qi, Y., Jiang, H., Zhou, H., et al. (2023). AIE donor-dependent photosensitizer for enhance photodynamic antibacterial interface. Surf. Interfaces 39, 102996. doi:10.1016/j.surfin.2023.102996
Zhan, H., Zhang, S., Li, L., Chen, Z., Cai, Y., Huang, J., et al. (2022). Naftopidil enantiomers suppress androgen accumulation and induce cell apoptosis via the UDP-glucuronosyltransferase 2B15 in benign prostate hyperplasia. J. Steroid Biochem. Mol. Biol. 221, 106117. doi:10.1016/j.jsbmb.2022.106117
Zhang, J., Li, Y., Chen, X., Pan, Y., Zhang, S., and Wang, Y. (2014). Systems pharmacology dissection of multi-scale mechanisms of action for herbal medicines in stroke treatment and prevention. PLoS One 9 (8), e102506. doi:10.1371/journal.pone.0102506
Zheng, R. S., Chen, R., Han, B. F., Wang, S. M., Li, L., Sun, K. X., et al. (2024). Cancer incidence and mortality in China, 2022. Zhonghua Zhong Liu Za Zhi 46 (3), 47–53. doi:10.1016/j.jncc.2024.01.006
Keywords: prostate cancer, antagonistic activity, binding affinities, docking study, AR antagonists
Citation: Jiang H, Chen H, Wang Y, Xu H and Chen H (2025) Synthesis, bioactivity, and molecular docking studies: novel arylpiperazine derivatives as potential new-resistant AR antagonists. Front. Chem. 13:1557275. doi: 10.3389/fchem.2025.1557275
Received: 08 January 2025; Accepted: 17 March 2025;
Published: 28 March 2025.
Edited by:
Greta Varchi, Consiglio Nazionale delle Ricerche (Bologna), ItalyReviewed by:
Moataz Ahmed Shaldam, Kafrelsheikh University, EgyptCopyright © 2025 Jiang, Chen, Wang, Xu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Jiang, eW9uZ2hvbmdqaWFuZ0AxNjMuY29t; Hong Chen, Y2hlbndleHBvQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.