- Department of Dermatology, Maternity and Child Health Hospital of Qinhuangdao, Qinhuangdao, China
Tumours and inflammation are serious risks to human health and are importantly regulated by the gas signalling molecule hydrogen sulphide. In this work, we report the rational design and synthesis of H2S donor molecules based on phenylthiophosphoryl dichloride nuclei and assess their efficacy against tumours and inflammation. We predicted its potential anticancer targets based on network pharmacology and then verified the inhibitory effect of the active compound S11 on the pathway PI3K/AKT by enzyme inhibition and molecular docking assay. In addition, compound S11 exhibited a potent anti-inflammatory effect on macrophages, effectively reducing the levels of inflammatory mediators TNF-α, IL-10 and HO-1. Compound S11 can be used as a new chemical entity for the discovery of new anti-cancer drugs or anti-inflammatory drugs.
1 Introduction
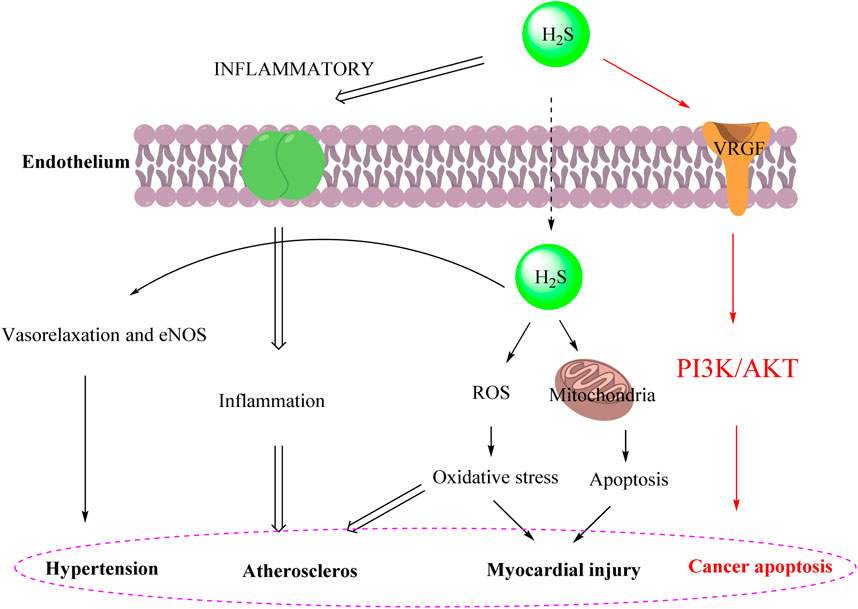
Hydrogen sulfide (H2S) is recognized as one of the three major gas signal transmitters along with nitric oxide (NO) and carbon monoxide (CO). In the past decade, H2S has been shown to have various biological activities, including positive regulatory effects in antioxidant mechanisms, promotion of angiogenesis, anti-inflammatory responses, and ion channel modulation (Hao et al., 2021; Li et al., 2021; Dilek et al., 2020). Furthermore, the cardioprotective effect of H₂S is associated with the inhibition of myocardial cell apoptosis following myocardial injury. The antioxidant effect of H2S is also reflected in preserving mitochondrial function by inhibiting mitochondrial respiration (Sanchez-Aranguren et al., 2020). Moreover, H2S plays a significant role in the hypertension (Goor et al., 2020), atheroscleros (Munteanu, 2023), myocardial injury (Kolluru et al., 2023) and cancer apoptosis (Gao et al., 2024) as illustrated in Figure 1.
Cancer can proliferate indefinitely by maintaining reproductive signalling or overexpressing growth factors to regenerate itself. It may also be initiated by aberrant activation of downstream signalling pathways, particularly the phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) pathway, or by aberrant inactivation of suppressor genes (Siegel et al., 2018; Moses et al., 2018). The critical role of the PI3K/AKT/mTOR (PAM) pathway in cell survival, proliferation, growth, and apoptosis influences make it a promising drug target in the war against cancer (Sanchez et al., 2019; Khan et al., 2019). Because H₂S gas is an important gas signalling transmitter, we can design a series of H₂S donors that can target and kill cancer cells by modulating the PI3K/AKT/mTOR pathway. Traditional drug discovery models can only verify the relationship between compounds and targets through a single experiment. In contrast, network pharmacology provides a “compound-protein/gene-disease” network approach, which operates on the principle of “network target, multi-component therapy” (Zhang et al., 2019). This makes it possible to analyse how small molecule drugs modulate disease at high throughput. Cyberpharmacology maps multi-drug ecological networks onto human and animal disease gene networks, providing a basis for identifying key disease-related drug targets (Nogales et al., 2022). In this study, a protein-protein interaction (PPI) network of compounds S11 associated with cancer was constructed using publicly available databases. Subsequently, potential targets of S11 for cancer treatment were identified. These targets were then analysed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment. Finally, it was validated using molecular docking methods with enzyme inhibition experiments. Similarly there is interest in the use of H2S donors for the treatment of inflammatory diseases. Therefore, it is of great interest to know as much as possible about H2S donors in vitro inflammatory systems. The anti-inflammatory effects of this series of hydrogen sulphide donors on macrophages in vitro were further tested in this context.
In a word, developing more effective organic H2S donors is of great significance for cancer and treating inflammation. This article designs a series of novel H2S donor molecules based on a sulfur-phosphorus core and the anti-tumor and anti-inflammatory mechanism is preliminarily explored, laying the foundation for the future design of H2S donor-based anti-inflammatory and anticancer drugs.
2 Results and discussion
2.1 Chemical synthesis
The main donor S1 of H2S was efficiently and concisely synthesized using benzene as the starting material. Compound S2-S16 were synthesized by reacting dichlorophenylthiophosphoryl with different amino alcohols, diols, or diamines in the presence of triethylamine (Scheme 1). In the 1H NMR spectra of compounds S1-S16, the chemical shifts of hydrogen atoms in benzene ring appeared in the range of 8.88–7.28 ppm. For most compounds, the signals of protons on OCH2 appeared in the range of 3.40–4.49 ppm. For compounds containing NH group, the chemical shifts of hydrogen in NH are in the range of 3.05–3.54 ppm. Our NMR data for compounds S2-S4 in this paper are consistent with the NMR data reported in Zhang’s article (Zhang et al., 2019).
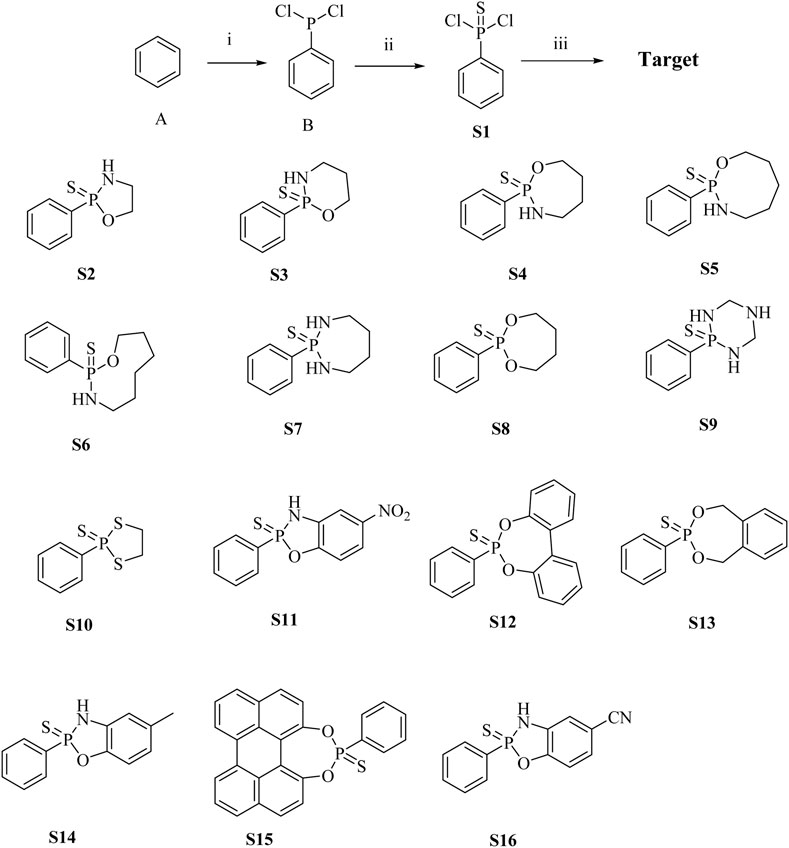
Scheme 1. Synthesis of H2S donor S1 - S16. Conditions and reagents: (i) AlCl3, PCl3, yield 78%; (ii) S, Benzene, yield 45%; (iii) Triethylamine, different amino alcohols, diols or diamines, yield 30%–45%.
2.2 H2S release ability of the compound and its influencing factors
The H2S-release capability of the donors was measured using the methylene blue (MB+) method (Sun et al., 2022). This method is based on the reaction of zinc acetate with H2S to form zinc sulfide, which then reacts with N, N-dimethyl-1,4-phenylenediamine sulfate to generate methylene blue in the presence of ferric ions under acid medium condition. The H2S is quantified by measuring the absorbance of the solution at 670 nm. Recently, literature has reported that TECP (a water-soluble phosphine reducing agent) or cysteine can promote the release of H2S. Therefore, in the presence of TECP or cysteine, the MB⁺ water titration method was used to evaluate the H₂S release ability (Nelmi et al., 2013; Hasegawa and Viles, 2014). The level of pH varies in various tissues and organs of the body; for example, the pH of the small intestine is about 8.0, whereas the pH of gastric juice is about 1.8. Therefore, we tested the release of H2S under different temperature and pH conditions. The results, as shown in Figure 2 and Supplementary Figure 1, showed that all compounds released H2S at room temperature. The release of H2S increased when the temperature was 37°C, indicating that temperature affects the amount of gas released. Different pH values have relatively little effect on such H2S donors. The release amount of compound S11 is the highest, indicating that the size of the cyclic structure and functional groups such as esters and amides have little effect on the release of H2S. However, the addition of a benzene ring can increase its release amount, which may be due to the electron transfer of the benzene ring increasing the release amount of H2S. The mechanism of H2S release was proposed, as shown in Figure 3.
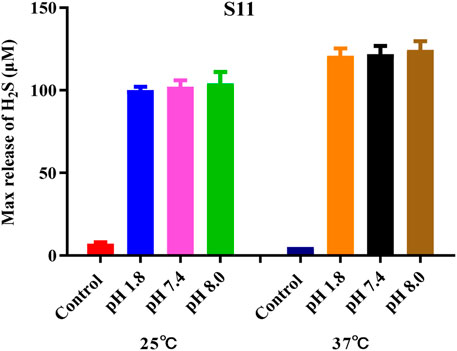
Figure 2. Amount of H2S released by compound S11. Each bar represents the mean ± SD (standard deviation) of three independent experiments.
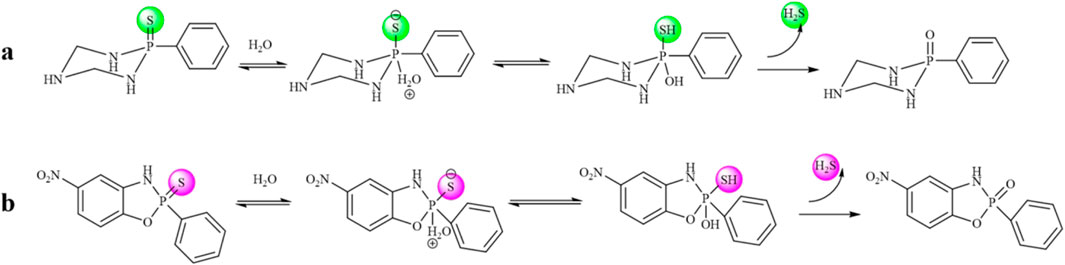
Figure 3. (A) Mechanism of compound S2-S10 releasing H2S. (B) Mechanism of compound S11-S16 releasing H2S.
2.3 The toxicity of the compounds
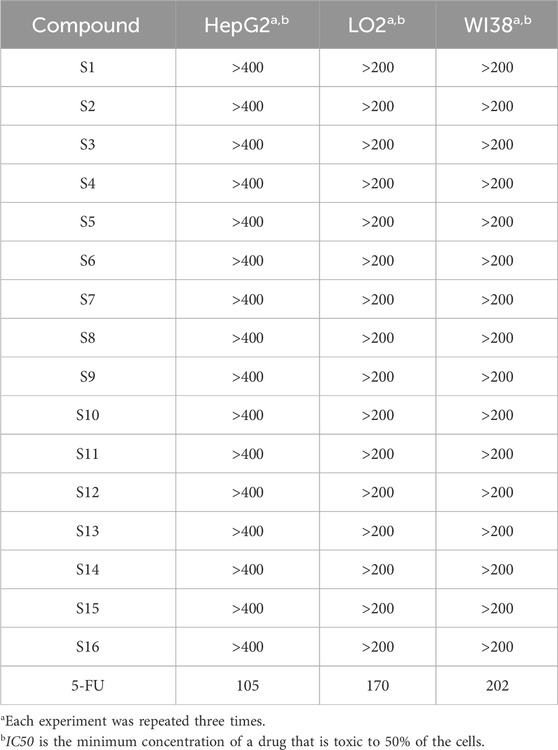
Compounds S1 - S16 were assessed for toxicities against LO2 cell lines (normal liver cell) and HepG2 cell lines (liver tumor cell) using MTT assay. Stock solutions of the tested compounds in DMSO (8,000 mM) were prepared freshly immediately prior to testing. Initially, cells were seeded in a 96-well plate at approximately 105 cells/well and allowed to adhere overnight. Cell survival relative to control was assessed after 24 h (Sanchez et al., 2019). The results are shown in Table 1. In summary, the toxicities of the series compounds are low to both normal cells LO2, WI38 and tumor cells HepG2, indicating that these compounds do not cause severe harm to the liver severely in the range of tested dose.
2.4 The antitumor activity of the compounds
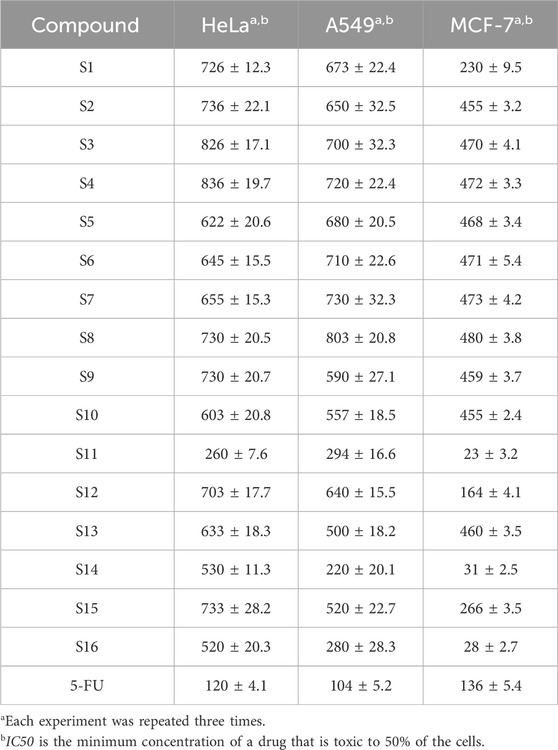
The anti-proliferative activity of H2S on tumor cells has been proven (Zhao et al., 2010; Faris et al., 2023; Liu et al., 2023). This experiment demonstrated that the P = S functional group is an effective functional group for releasing H2S and has good anti-tumour activity. The results, as shown in Table 2 and Figure 2, showed that S11 had high H2S release and good antiproliferative activity against MCF7. Based on the highest release of H2S with lower toxicity, we chose compound S11 for subsequent studies. We found that the amount of hydrogen sulfide released was positively correlated with the anticancer activity, and the highest amount of hydrogen sulfide was released from S11, S14 and S16, which showed the best activity against MCF7 cells. The activity of aminophenol derivatives was superior to that of 2,2′-dihydroxybiphenyl derivatives. The coupling of aminophenol derivatives to S1 with electron-withdrawing groups contributed to the activity, and nitro was preferred to cyano to methyl.
2.5 Assessment of anti-tumour mechanisms
2.5.1 Venn diagram construction and core PPI network screening
The Venn diagram in Figure 4A visualizes the overlap of S11 with tumour targets, revealing 26 common genes. It is suggested that S11 may regulate tumour cell death by interacting with these targets. We then constructed 2 networks using STRING for S11 (Figure 4B) and an intersecting network of S11 with cancer target (Figure 4C). Circles in Figure 4C indicate targets of drug-disease interactions; larger circles indicate more significant interactions between targets, suggesting that their respective functions are more important. Combined with Figure 4C, AKT is an important potential target for our analysed.
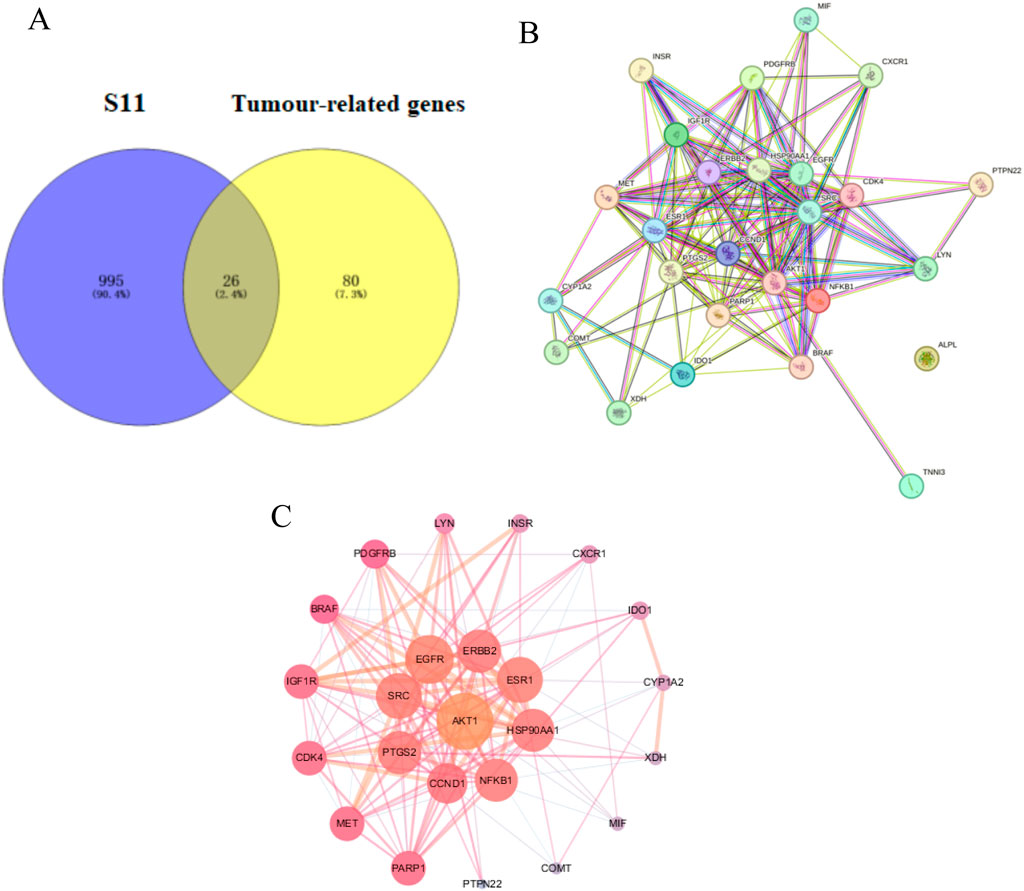
Figure 4. (A) Venn diagram reveals the relationship between S11 and Tumour-related genes. (B) Construction of a PPI network of the interactive PPI network of S11 and tumour targets by the STRING database. (C) The interactive PPI network of S11 and tumour targets by Cytoscape software.
2.5.2 GO and KEGG pathway enrichment analysis
The results of GO (BP, CC, MF) indicated that the top 10 targets are primarily membrane-associated and involved in lipid metabolism and biosynthesis processes (Figure 5A). KEGG enrichment bubble diagrams analysis identified two significant pathways relevant to S11’s therapeutic action, notably the PI3K-AKT signaling pathway and MAPK signaling pathway related to tumor disease (Figure 5B). These analyses underscore the important role of S11 in the treatment of tumor disease.
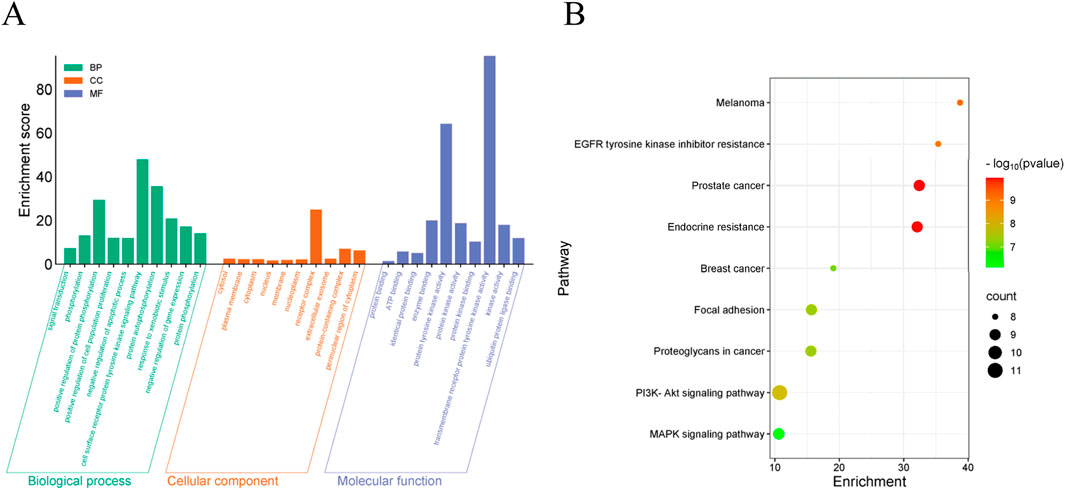
Figure 5. GO analysis and KEGG enrichment analysis of candidate target genes (A) Visualized enrichment bubble plots of biological processes (BP), cell components (CC) molecular function (MF). (B) Visualized enrichment bubble plots of Kyoto Encyclopedia of Genes and Genomes (KEGG). The term “RichFactor” refers to the proportion of differentially expressed transcripts found in a specific Gene Ontology (GO) entry compared to the total number of transcripts within that GO entry among all annotated transcripts. A higher RichFactor indicates a higher level of enrichment. The points on the graph are colored differently to indicate distinct P values, and the size of the points represents the number of target genes within the pathway.
2.5.3 Molecular docking
For this binding site (7F7W), both GlideScore and Model scores have a significant positive correlation with biological activity. This binding site has good tuberculosis with S11 and can form hydrogen bonding interactions (Figure 6). And the twist angle of S11 has changed, which may be caused by newly formed hydrophobic interactions and van der Waals forces between S11 and the ligand (Khan et al., 2022; Hu et al., 2022; Zhu et al., 2021). This indicates that our compound S11 may induce cell apoptosis by interacting with this protein, thereby inhibiting the PI3K/AKT signaling pathway mechanism.
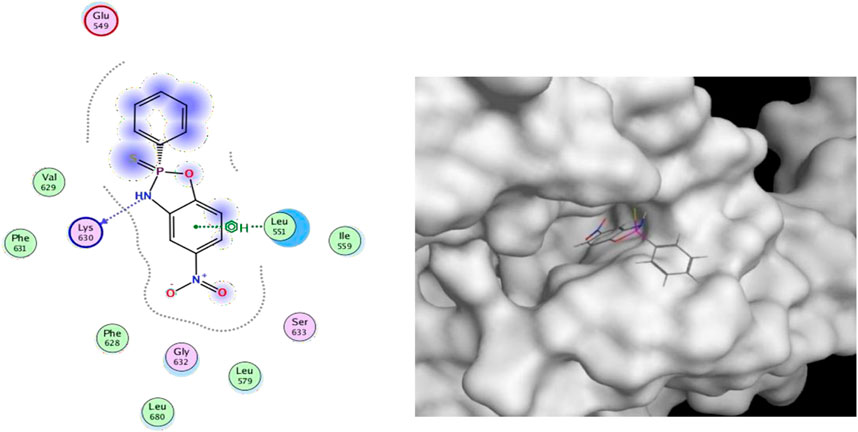
Figure 6. (Left) 2D ligands interaction diagram of the most active compounds (S11) with surrounding residues of the most probable binding site (Right) 3D diagram of compound S11 binding to this site.
2.5.4 Evaluation of anti-tumour targets
PI3K pathway aberrations are the most common in cancer and one of the most widely studied pathways in cancer therapy. To date, more than 30 PI3K inhibitors have entered clinical trials for various cancer types, and aberrant activation of PI3K and its downstream effectors, including Akt and mTOR, has been associated with a variety of cancers. AKT is a central node of the PI3K/AKT/mTOR signalling pathway, which is involved in the regulation of cell proliferation, differentiation and apoptosis and promotes tumourigenesis and metastasis, AKT is over-activated in more than 50% of tumours, including breast, lung, head and neck, endometrial, prostate and colorectal cancers (Hart et al., 2011). In conjunction with our previously predicted anticancer targets and potential mechanistic pathways, in order to further validate the antitumour mechanism of S11, we used a PI3K/AKT enzyme inhibition assay. The results, as shown in Table 3, showed that compound S11 had a better inhibitory effect on PI3Kα, PI3Kβ, PI3Kγ, and AKT using PI-103 as a control, validating our predicted mechanistic pathway. We tentatively suggest that S11 induces tumour cell death by regulating the PI3K/AKT/mTOR signalling pathway, which in turn induces tumour cell death.
2.6 The anti-inflammatory activity of the compounds
2.6.1 Effect of compounds on cell activity
To obtain more accurate anti-inflammatory results and minimize the impact of cell death caused by the test compounds on the outcomes, we first evaluated the effects of the compounds on RAW264.7 macrophage viability. The cells were divided into two groups: one treated with 1 mg/mL LPS and the other untreated. Macrophages were exposed to compounds at concentrations of 10, 50, and 100 μM for 24 h, followed by cell viability measurement using the CCK-8 assay (Huang et al., 2022; Flannigan and Wallace, 2015; Citi et al., 2020; Gemici and Wallace, 2015). The results showed that compound S11 had low toxicity to macrophages and had no significant effect on cell growth in the absence of LPS (Figure 7). In the presence of LPS stimulation, a slight reduction of 11% cell activity was observed.
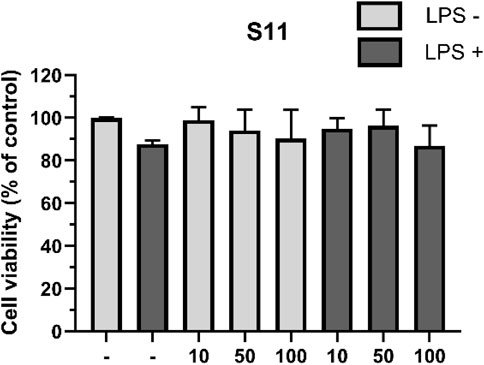
Figure 7. The effect of compounds on RAW264.7 cell viability. Mouse RAW264.7 macrophage cells were treated with compound S11 at concentrations of 10, 50, and 100 µM for 24 h in the presence or absence of 1 µg/L LPS. Each bar represents the average standard deviation of three independent experiments.
2.6.2 Impact of the compounds on nitrite production induced by LPS
Nitric oxide (NO) affects inflammation signaling pathways, regulating the intensity and timing of inflammatory responses, making it a crucial regulatory molecule in inflammation. LPS induces the production of nitric oxide (NO) by activating a series of signal transduction pathways. Upon NF-κB activation, transcription of the iNOS gene is promoted, leading to the synthesis of inducible nitric oxide synthase (iNOS). iNOS is an enzyme that produces large amounts of NO, activated under inflammatory conditions to release substantial nitric oxide (Kumar, 2023). In vivo, NO is easily oxidized by superoxide ions to generate nitrites. Studies have shown that H2S not only acts to scavenge peroxides but also undergoes chemical reactions with NO, forming novel nitrosylthiol compounds. We assess the anti-inflammatory activity of compounds by measuring nitrite production (Somensi et al., 2019).
Results indicate (Figure 8) that under conditions of 1 mg/mL LPS, compound S11 can reduce nitrite levels and showing significant inhibition of iNOS. S11 show clear dose-dependency. At 100 μM, compound S11 inhibits nitrite production by approximately 75%. Considering the H2S release results, the high level of H2S release in S11 compounds may be the reason for their significant anti-inflammatory activity.
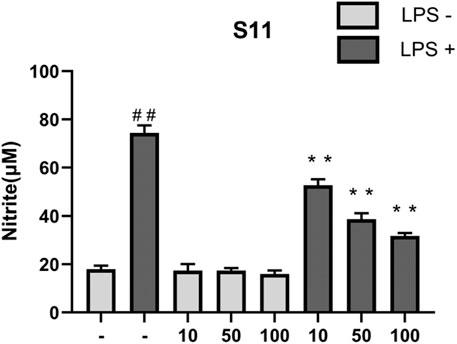
Figure 8. The effect of compounds on nitrite levels in RAW 264.7 cells. Mouse RAW264.7 macrophage cells were treated with compound S11 at concentrations of 10, 50, and 100 µM for 24 h in the presence or absence of 1 µg/mL LPS. Each bar represents the mean ± SD of three independent experiments. #p < 0.05, ##p < 0.01 vs untreated cells; *p < 0.05, **p < 0.01, vs LPS treated cells.
2.6.3 Effect of the compounds on LPS-mediated TNF-α and IL-10
Anti-inflammatory drugs typically exert their effects by inhibiting the NF-κB signaling pathway. Upon NF-κB activation, not only does it promote the transcription of the iNOS gene, but it also increases the expression of various inflammation-related genes such as TNF-α, IL-1β, etc. (Hou et al., 2006). Among these, TNF-α is a key pro-inflammatory cytokine in inflammation responses (You et al., 2022). It can activate immune cells, enhance inflammation signal transduction, and trigger inflammatory reactions, including leukocyte migration and cytokine release. Studies have shown that H2S can significantly reduce the number of neutrophils induced by LPS in the liver and lungs, and decrease TNF-α levels (Chiba et al., 2007; Pan et al., 2023; Huang et al., 2022).
Therefore, we assessed the anti-inflammatory ability of compounds by measuring their effect on TNF-α levels in RAW264.7 cells induced by LPS. Results showed (Figure 9A) that compared to the control group, S11 inhibited TNF-α expression (Figure 8). Compound S11 decreased TNF-α induction levels by 80%. H2S also promotes an increase in plasma IL-10 levels, thereby inhibiting inflammatory mediators. Comparatively (Figure 9B), compound S11 exhibited the highest induction level of IL-10, being three times that of the control group.
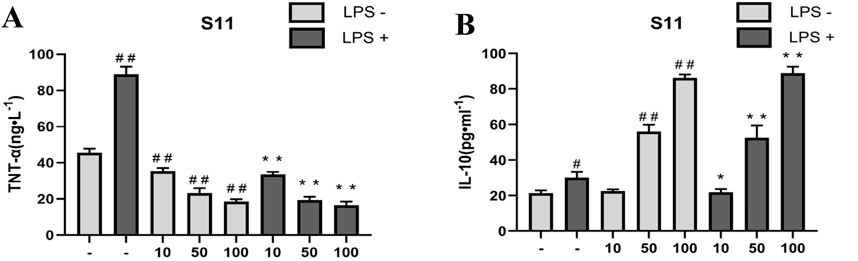
Figure 9. (A) The effect of compounds on TNT-α levels in RAW 264.7 cells. (B) The effect of compound on IL-10 level in RAW264.7 cells. Mouse RAW264.7 macrophage cells were treated with compound S11 at concentrations of 10, 50, and 100 µM for 24 h in the presence or absence of 1 µg/mL LPS. Each bar represents the mean ± SD of three independent experiments. #p < 0.05, ##p < 0.01 vs untreated cells; *p < 0.05, **p < 0.01, vs LPS treated cells.
2.6.4 Effect of the compounds on LPS-mediated HO-1
HO-1 is an important antioxidant enzyme involved in regulating intracellular redox balance. It reduces the generation of oxygen free radicals by degrading products such as carbon monoxide (CO) and biliverdin derived from heme, helping to inhibit oxidative stress and cell damage, and reducing the severity of inflammatory responses. Therefore, the expression level of HO-1 reflects the cell’s anti-inflammatory ability to some extent (Shen et al., 2022; Hou et al., 2021; Lee et al., 2022). Nrf2 is a major regulator of the antioxidant defense system in vivo, and is involved in signal transduction related to various intracellular defense mechanisms. HO-1 is a target protein downstream of Nrf2, and HO-1 and its products play beneficial roles in modulating inflammatory responses. It has been shown that active compounds can attenuate lipopolysaccharide-induced cellular inflammatory responses by increasing the amount of HO-1 and thereby activating the Nrf2/HO-1 pathway (Lee et al., 2022; Ali et al., 2018). In this experiment, compound S11 was used to treat RAW264.7 cells, and the expression levels of HO-1 were measured. The results showed that compound S11 promoted HO-1 expression to approximately twice the level of the control group (Figure 10).
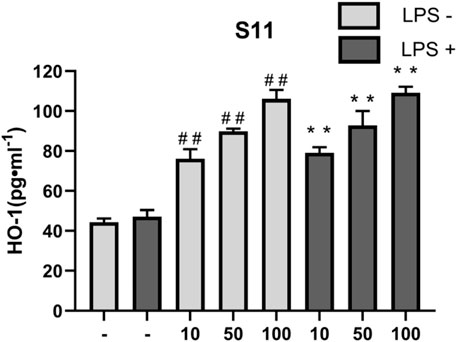
Figure 10. The effect of compounds on HO-1 levels in RAW 264.7 cells. Mouse RAW264.7 macrophage cells were treated with compound S11 at concentrations of 10, 50, and 100 µM for 24 h in the presence or absence of 1 µg mL-1 LPS. Each bar represents the mean ± SD of three independent experiments. #p < 0.05, ##p < 0.01 vs untreated cells; *p < 0.05, **p < 0.01, vs LPS treated cells.
3 Conclusion
This experiment aimed to investigate the multifaceted effects of H2S in anti-cancer, anti-inflammatory, and other aspects. To develop superior H2S donor drugs, we designed a series of H2S donor compounds based on thiol-phosphorus core and evaluated these compounds extensively. Specifically, evaluations included assessing the H2S releasing capability of the compounds, their toxicity and activity impact on cells, anti-tumour and anti-inflammatory effects. Through these assessments, we aimed to identify H2S donor drugs with excellent anti-tumour and anti-inflammatory activities, providing valuable references and guidance for drug development in related fields.
This experiment found that all compounds could release H2S, with compound S11 showing the most significant effect, possibly due to its benzene ring content. After treating LO2, HepG2, and WI38 cell lines with the compound S1-S13 showed low cytotoxicity. Compound S11 exhibited promising results in anticancer and anti-inflammatory assays. We performed target screening prediction by network pharmacology and validated it by molecular docking and enzyme inhibition assays, and found that compound S11 may cause cancer cell death by inhibiting the PI3K/AKT pathway. In vitro, S11 was found to have a better anti-inflammatory effect by the assay of some series of anti-inflammatory active factors TNF-α, IL-10 and HO-1.
4 Experimental section
All the key intermediates and final products were identified with 1H NMR and 13C NMR, recorded in a Bruker Avance 400 (1H at 400 MHz, 13C at 101 MHz), and chemical shifts were reported in parts per million using the residual solvent peaks as internal standards (CDCl3 = 7.26 ppm for 1H NMR and 77.16 ppm for 13C NMR).
4.1 Chemically synthetical experiments
4.1.1 Synthesis of compound dichloro (phenyl)phosphane (B)
We added 0.1 mmol of benzene, 0.3 mmol of phosphorus trichloride and 0.14 mmol of aluminium chloride to the reaction flask and refluxed with stirring for 5 h. Then add 0.13 mol of phosphorus trichloride and 45 mL of petroleum ether and stir the mixture at reflux for 30 min. After cooling to room temperature, filter the mixture under reduced pressure, distill the filtrate with petroleum ether at atmospheric pressure, and distill under reduced pressure. Collect fractions from 99°C–101°C with a yield of 78%. 1H NMR (400 MHz, Chloroform-d) δ 7.94 (t, J = 8.6 Hz, 2H), 7.60–7.50 (m, 3H). 13C NMR (101 MHz, Chloroform-d) δ 140.46 (d, J = 52.2 Hz), 132.79, 130.17 (d, J = 31.3 Hz), 129.00 (d, J = 7.9 Hz). 31P NMR (162 MHz, Chloroform-d) 169.55. TOF-MS, m/z [M + H+], calcd. for C6H6Cl2P+, 178.9506, found: 178.9531.
4.1.2 Synthesis of compound phenylphosphonothioic dichloride (S1)
Slowly add the settled sulfur powder in batches to the previous product (B), control the vulcanization process temperature to 30°C, most of the sulfur powder disappears, and the reaction solution becomes viscous. Then, raise the temperature to 80°C and stir for 1 h. Atmospheric and vacuum distillation removes unreacted phosphorus trichloride and benzene. Collect the 90°C–91°C fraction, which is the product phenyl thiophosphine dichloride. Yield 48%. 1H NMR (400 MHz, Chloroform-d) δ 8.13 (dd, J = 18.6, 7.9 Hz, 2H), 7.63 (dt, J = 6.0, 3.0 Hz, 1H), 7.60–7.50 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 138.07 (d, J = 118.2 Hz), 133.81 (d, J = 4.0 Hz), 129.99 (d, J = 15.2 Hz), 128.70 (d, J = 18.2 Hz). 31P NMR (162 MHz, Chloroform-d) 169.56. TOF-MS, m/z [M + H+], calcd. for C6H6Cl2SP+, 210.9227, found: 210.9237.
4.1.3 Synthesis of compound 2-phenyl-1,3,2-oxazaphospholidine 2-sulfide (S2)
Di-aminoethanol (2 mmol) and triethylamine (1 mL) were dissolved in dichloromethane. Phenylphosphine dichloride (2 mmol) was added slowly in an ice bath. The reaction is then carried out at room temperature for 8 h. The precipitate was filtered and the solution was collected and concentrated. The crude product was purified by column chromatography (PE/EA = 4:1) to give a white solid. Yield 43%. Separation and purification methods can also be found in the literature (zhang et al., 2019). 1H NMR (400 MHz, Chloroform-d) δ 8.00–7.75 (m, 2H), 7.54–7.29 (m, 3H), 4.53–4.40 (m, 1H), 4.38–4.23 (m, 1H), 3.80–3.60 (m, 1H), 3.54–3.34 (m, 1H), 3.16–2.99 (m, 1H). 13C NMR (101 MHz, Chloroform-d) δ 135.80 (d, J = 135.0 Hz), 132.14 (d, J = 3.2 Hz), 130.89 (d, J = 12.3 Hz), 128.42 (d, J = 14.8 Hz), 68.35, 43.97.31P NMR (162 MHz, DMSO) 169.50. TOF-MS, m/z [M + H+], calcd. for C8H11NOPS+, 199.0221, found: 200.0247.
4.1.4 Synthesis of compound 2-phenyl-1,3,2-oxazaphosphinane 2-sulfide (S3)
The method is the same as S2. Yield 47%. 1H NMR (400 MHz, Chloroform-d) δ 7.87 (ddd, J = 21.8, 13.8, 8.0 Hz, 2H), 7.48 (dt, J = 7.4, 4.8 Hz, 3H), 4.45 (td, J = 16.2, 7.1 Hz, 1H), 4.18–3.97 (m, 1H), 3.52–3.06 (m, 3H), 2.06 (ddt, J = 20.1, 10.1, 5.0 Hz, 1H), 1.62 (d, J = 14.3 Hz, 1H). 13C NMR (101 MHz, Chloroform-d) δ 133.67 (d, J = 133.7 Hz), 131.90 (d, J = 3.2 Hz), 130.77 (d, J = 11.3 Hz), 128.96 (d, J = 14.1 Hz), 67.84, 41.33, 26.70. 31P NMR (162 MHz, DMSO) 169.51. TOF-MS, m/z [M + H+], calcd. for C9H13NOPS+, 213.0377, found: 214.0355.
4.1.5 Synthesis of compound 2-phenyl-1,3,2-oxazaphosphepane 2-sulfide (S4)
The method is the same as S2. Yield 50%. 1H NMR (400 MHz, Chloroform-d) δ 7.87 (dd, J = 13.8, 7.2 Hz, 2H), 7.56–7.34 (m, 3H), 4.68–4.45 (m, 1H), 4.17 (dd, J = 22.9, 12.0 Hz, 1H), 3.45 (s, 1H), 3.09 (dd, J = 30.1, 14.8 Hz, 1H), 2.74 (d, J = 13.9 Hz, 1H), 1.97–1.67 (m, 3H), 1.63–1.45 (m, 1H). 13C NMR (101 MHz, Chloroform-d) δ 135.13 (d, J = 148.6 Hz), 131.41 (d, J = 3.2 Hz), 130.32 (d, J = 11.0 Hz), 128.37 (d, J = 14.5 Hz), 64.95, 42.65, 31.69, 29.70. 31P NMR (162 MHz, DMSO) 169.50. TOF-MS, m/z [M + H+], calcd. for C10H15NOPS+, 228.0534, found: 228.0599.
4.1.6 Synthesis of compound 2-phenyl-1,3,2-oxazaphosphocane 2-sulfide (S5)
The method is the same as S2. Yield 50%. 1H NMR (400 MHz, Chloroform-d) δ 7.75 (dd, J = 13.1, 8.2 Hz, 2H), 7.38–7.28 (m, 3H), 4.56 (qd, J = 9.7, 5.9 Hz, 1H), 4.07–3.85 (m, 1H), 3.49 (s, 1H), 3.02–2.84 (m, 1H), 2.78–2.62 (m, 1H), 1.98–1.82 (m, 1H), 1.73–1.33 (m, 5H). 13C NMR (101 MHz, Chloroform-d) δ 134.89 (d, J = 149.3 Hz), 130.81 (d, J = 3.1 Hz), 129.87 (d, J = 11.0 Hz), 127.93 (d, J = 14.5 Hz), 65.57, 53.39, 41.80, 29.57, 28.52, 23.62. 31P NMR (162 MHz, DMSO) 169.50. TOF-MS, m/z [M + H+], calcd. For C11H17NOPS+, 242.0690, found: 242.0677.
4.1.7 Synthesis of compound 2-phenyl-1,3,2-oxazaphosphonane 2-sulfide (S6)
The method is the same as S2. Yield 50%. 1H NMR (400 MHz, Chloroform-d) δ 7.82 (dd, J = 13.7, 8.0 Hz, 2H), 7.52–7.35 (m, 3H), 4.87–4.71 (m, 1H), 4.06–3.89 (m, 1H), 3.27 (s, 1H), 2.97 (t, J = 15.1 Hz, 1H), 2.84 (s, 1H), 1.86 (d, J = 14.8 Hz, 1H), 1.77–1.61 (m, 2H), 1.59–1.31 (m, 5H). 13C NMR (101 MHz, Chloroform-d) δ 134.51 (d, J = 149.7 Hz), 131.32 (d, J = 3.1 Hz), 130.66 (d, J = 10.8 Hz), 128.43 (d, J = 14.4 Hz), 62.82, 40.51, 28.53, 27.52, 20.60, 20.04.31P NMR (162 MHz, DMSO) 169.51. TOF-MS, m/z [M + H+], calcd. for C12H19NOPS+, 256.0847, found: 256.0866.
4.1.8 Synthesis of compound 2-phenyl-1,3,2-diazaphosphepane 2-sulfide (S7)
The method is the same as S2. Yield 50%. 1H NMR (400 MHz, Chloroform-d) δ 7.91 (dd, J = 13.6, 7.1 Hz, 2H), 7.47–7.31 (m, 3H), 3.34–3.11 (m, 2H), 3.09–2.84 (m, 4H), 1.73–1.52 (m, 4H). 13C NMR (101 MHz, Chloroform-d) δ 136.39 (d, J = 126.8 Hz), 131.29 (d, J = 3.0 Hz), 130.32 (d, J = 11.0 Hz), 128.28 (d, J = 13.6 Hz), 41.56, 41.53, 31.72. 31P NMR (162 MHz, DMSO) 169.51. TOF-MS, m/z [M + H+], calcd. for C10H16N2PS+, 227.0694, found: 227.0780.
4.1.9 Synthesis of compound 2-phenyl-1,3,2-dioxaphosphepane 2-sulfide (S8)
The method is the same as S2. Yield 50%. 1H NMR (400 MHz, Chloroform-d) δ 7.79 (dd, J = 13.5, 7.6 Hz, 2H), 7.46–7.29 (m, 3H), 4.24 (dd, J = 23.8, 5.8 Hz, 2H), 3.95 (td, J = 11.7, 5.9 Hz, 2H), 1.96–1.68 (m, 4H). 13C NMR (101 MHz, Chloroform-d) δ 132.34 (d, J = 149.7 Hz), 131.82 (d, J = 3.1 Hz), 130.10 (d, J = 11.3 Hz), 128.06 (d, J = 14.8 Hz), 67.07, 28.96. 31P NMR (162 MHz, DMSO) 169.51. TOF-MS, m/z [M + H+], calcd. for C10H14O2PS+, 229.0374, found: 229.0398.
4.1.10 Synthesis of compound 2-phenyl-1,3,5,2-triazaphosphinane 2-sulfide (S9)
The method is the same as S2. Yield 50%. 1H NMR (400 MHz, Chloroform-d) δ 7.90–7.65 (m, 2H), 7.42–7.27 (m, 3H), 3.50–2.69 (m, 7H). 13C NMR (101 MHz, Chloroform-d) δ 135.83 (d, J = 134.33 Hz), 132.78 (d, J = 13.1 Hz), 130.81 (d, J = 11.1 Hz), 128.11 (d, J = 14.1 Hz), 48.17, 42.12, 38.80. 31P NMR (162 MHz, DMSO) 169.51. TOF-MS, m/z [M + H+], calcd. for C8H13N3PS+, 214.0490, found: 214.0532.
4.1.11 Synthesis of compound 2-phenyl-1,3,2-dithiaphospholane 2-sulfide (S10)
The method is the same as S2. Yield 50%. 1H NMR (400 MHz, Chloroform-d) δ 8.12 (dd, J = 16.9, 7.4 Hz, 2H), 7.54–7.46 (m, 3H), 3.89–3.75 (m, 2H), 3.75–3.58 (m, 2H). 13C NMR (101 MHz, Chloroform-d) δ 137.18 (d, J = 83.5 Hz), 132.28 (d, J = 3.5 Hz), 131.34 (d, J = 12.8 Hz), 128.52 (d, J = 14.6 Hz), 43.00. 31P NMR (162 MHz, DMSO) 169.51. TOF-MS, m/z [M + H+], calcd. For C8H10PS+, 232.9604, found: 232.9677.
4.1.12 Synthesis of compound 5-nitro-2-phenyl-3H-benzo [d][1,3,2] oxazaphosphole 2-sulfide (S11)
The method is the same as S2. Yield 50%. 1H NMR (400 MHz, DMSO) δ 8.28–8.16 (m, 1H), 7.89 (d, J = 7.1 Hz, 2H), 7.82–7.73 (m, 1H), 7.72–7.61 (m, 1H), 6.85 (d, J = 9.7 Hz, 1H), 6.69 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 147.72, 134.81, 134.35 (d, J = 7.7 Hz), 134.11 (d, J = 3.1 Hz), 131.66 (d, J = 12.9 Hz), 131.28, 129.80, 128.99 (d, J = 15.6 Hz), 123.13, 117.16 (d, J = 4.0 Hz), 113.98. 31P NMR (162 MHz, DMSO) 169.51. TOF-MS, m/z [M + H+], calcd. For C12H10N2O3PS+, 293.0150, found: 293.0148.
4.1.13 Synthesis of compound 6-phenyldibenzo [d, f][1,3,2] dioxaphosphepine 6-sulfide (S12)
The method is the same as S2. Yield 50%. 1H NMR (400 MHz, Chloroform-d) δ 7.87 (dd, J = 14.3, 7.6 Hz, 2H), 7.59–7.47 (m, 3H), 7.42–7.24 (m, 6H), 7.10–7.01 (m, 2H). 13C NMR (101 MHz, Chloroform-d) δ 148.01 (d, J = 11.7 Hz), 133.05 (d, J = 3.1 Hz), 131.33 (d, J = 11.5 Hz), 129.70 (d, J = 35.0 Hz), 129.04 (d, J = 1.8 Hz), 128.04 (d, J = 15.0 Hz), 126.27 (d, J = 1.8 Hz), 121.98 (d, J = 3.7 Hz). 31P NMR (162 MHz, DMSO) 169.50. TOF-MS, m/z [M + H+], calcd. For C18H13PS+, 325.0374, found: 325.0332.
4.1.14 Synthesis of compound 3-phenyl-1,5-dihydrobenzo [e] [1,3,2] dioxaphosphepine 3-sulfide (S13)
The method is the same as S2. Yield 50%. 1H NMR (400 MHz, Chloroform-d) δ 8.04–7.86 (m, 2H), 7.55 (t, J = 7.3 Hz, 1H), 7.50–7.41 (m, 2H), 7.40–7.37 (m, 4H), 6.00 (t, J = 13.1 Hz, 2H), 4.94–4.80 (m, 2H). 13C NMR (101 MHz, Chloroform-d) δ 136.68, 133.02 (d, J = 3.1 Hz), 131.35 (d, J = 11.8 Hz), 129.19 (d, J = 53.5 Hz), 128.45 (d, J = 15.3 Hz). 67.47. 31P NMR (162 MHz, DMSO) 169.51. TOF-MS, m/z [M + H+], calcd. for C14H14O2PS+, 277.0374, found: 277.0331.
4.1.15 Synthesis of compound 5-methyl-2-phenyl-3H benzo [d][1,3,2] oxazaphosphole 2-sulfide (S14)
The method is the same as S2. Yield 30%. 1H NMR (400 MHz, Chloroform-d) δ 7.82 (dd, J = 14.7, 7.8 Hz, 2H), 7.51–7.44 (m, 1H), 7.42–7.34 (m, 2H), 6.66 (s, 2H), 6.46 (d, J = 7.9 Hz, 1H), 5.82 (d, J = 10.2 Hz, 1H), 2.17 (s, 3H). 13C NMR (101 MHz, Chloroform-d) δ 146.24 (d, J = 7.4 Hz), 134.05–133.51 (m), 132.15 (d, J = 3.2 Hz), 130.93 (d, J = 11.4 Hz), 128.71 (d, J = 14.8 Hz), 124.83, 121.53, 120.20 (d, J = 2.9 Hz), 116.40, 20.89. 31P NMR (162 MHz, DMSO) 169.52. TOF-MS, m/z [M + H+], calcd. for C13H13NOPS+, 262.0455, found: 262.0459.
4.1.16 Synthesis of 2-phenylperyleno [1,12-def][1,3,2] dioxaphosphepine 2-sulfide (S15)
The method is the same as S2. Yield 37%. 1H NMR (400 MHz, Chloroform-d) δ 8.09 (d, J = 8.8 Hz, 1H), 8.00 (d, J = 8.2 Hz, 1H), 7.92 (d, J = 8.1 Hz, 1H), 7.73 (ddd, J = 35.3, 19.9, 8.8 Hz, 3H), 7.59–7.46 (m, 4H), 7.42–7.29 (m, 4H), 6.91 (d, J = 8.8 Hz, 1H). 13C NMR (101 MHz, Chloroform-d) δ 151.05–144.86 (m), 133.39 (d, J = 3.1 Hz), 132.77 (d, J = 1.8 Hz), 132.19 (d, J = 1.5 Hz), 132.01 (d, J = 11.6 Hz), 131.73 (d, J = 1.3 Hz), 130.83 (dd, J = 42.0, 1.4 Hz), 128.51 (dd, J = 31.7, 14.0 Hz), 127.32 (d, J = 23.3 Hz), 126.76 (d, J = 6.5 Hz), 125.86, 122.13 (d, J = 2.4 Hz), 121.21 (d, J = 2.8 Hz). 31P NMR (162 MHz, DMSO) 169.52. TOF-MS, m/z [M + H+], calcd. for C26H16O2PS+, 423.0680, found: 423.0688.
4.1.17 Synthesis of compound 2-phenyl-3H-benzo [d][1,3,2] oxazaphosphole-5-carbonitrile 2-sulfide (S16)
The method is the same as S2. Yield 48%. 1H NMR (400 MHz, Chloroform-d) δ 7.83 (dd, J = 14.7, 7.8 Hz, 2H), 7.51–7.45 (m, 1H), 7.44–7.35 (m, 2H), 6.80 (dd, J = 8.3, 5.6 Hz, 3H), 6.65 (t, J = 7.3 Hz, 1H). 13C NMR (101 MHz, Chloroform-d) δ 145.65 (d, J = 7.8 Hz), 133.85, 132.18 (d, J = 3.2 Hz), 130.88 (d, J = 11.5 Hz), 128.76 (d, J = 14.8 Hz), 127.91 (d, J = 3.1 Hz), 123.23, 121.14, 119.40 (d, J = 3.2 Hz), 115.58. 31P NMR (162 MHz, DMSO) 169.50. TOF-MS, m/z [M + H+], calcd. for C26H16O2PS+, 423.0680, found: 423.0688.
4.2 Cell culture
RAW264.7 cells Cultivated in this laboratory, were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco) supplemented with 10% v/v heat inactivated fetal bovine serum (FBS) (Gibco), l-glutamine (4 mM), sodium pyruvate (1 mM), HEPES buffer (20 mM) (Hyclone), penicillin (100 U/mL) and streptomycin (100 μg/mL). Cells were seeded in non-tissue culture treated Petri dishes (Biomedia) and maintained at 37°C in a 5% CO2 humidified environment (Sanchez et al., 2019).
4.3 H2S measurement
The H2S-release capability of the donors was measured using the methylene blue (MB+) method. See Supplementary Material for more details (Sun et al., 2022; Hasegawa and Viles, 2014).
4.4 Venny map construction and core PPI network screening
We obtained SMILES of the active compound S11 from PubChem database, and then obtained S11 related information from TCSMP database, predicted and obtained related targets through PharmMapper database. Meanwhile, relevant targets were retrieved from Gene Cards database. Then five more databases were collected for cancer disease targets GenesCards database, CTD database, DisGeNET database, TTD database, OMIM database. Venn diagrams of S11 versus cancer targets were obtained and visualised using the online tool Venny 2.0 (https://bioinfogp.cnb.csic.es/tools/venn y/index. html). Subsequently, the identified cross-targets were uploaded to the STRING (https://cn.string-dbOrg/) database, which predicts protein interactions. The free nodes were hidden to obtain the PPI network of S11 with cancer. This part of the data was further analysed using Cytoscape 3.7.1 software (Zhang et al., 2019; Nogales et al., 2022).
4.5 GO and KEGG pathway enrichment analysis
To elucidate the biological significance of the gene list, we used Database Annotation, Visualisation and Integrated Discovery (DAVID), including GO and KEGG pathway enrichment analysis. We entered the cross-targets of S11 and cancer in the PPI network into DAVID, specifying the species as “Bos taurus”. We then performed functional analyses of graphene oxide, including biological processes (BP), cellular components (CC) and molecular functions (MF), as well as KEGG pathway enrichment analyses of the core targets, and concluded that a p-value of <0.05 was statistically significant. Finally, the OmicShare platform (https://www.omicshare.com/) was used to process the data (Zhang et al., 2019; Nogales et al., 2022).
4.6 Cell viability
The CCK-8 assay is a commonly used method for assessing cell viability, based on cellular metabolic activity. See Supplementary Material for more details (Huang et al., 2022; Flannigan and Wallace, 2015; Citi et al., 2020; Gemici and Wallace, 2015).
4.7 Nitrite level detection
The Griess assay is used to measure nitrite levels in biological samples, both in vivo and in vitro. See Supplementary Material for more details (Kumar, 2023).
4.8 Detection of cytokine levels
In this experiment, the levels of TNF-α, IL-10, and HO-1 cell factors were assessed using the ELISA method (Shen et al., 2022; Hou et al., 2021; Lee et al., 2022). See Supplementary Material for more details.
4.9 In vitro PI3K and ATK enzyme inhibitory assay
Enzyme activity assays were performed according to the reagent kit and references, and detailed procedures are in the Supplementary Material (Huang et al., 2022).
4.10 Docking and collecting data
Selection of previously described 7F7W targets for molecular docking with S11. See Supplementary Material for more details (Khan et al., 2022; Hu et al., 2022; Zhu et al., 2021).
4.11 Statistical analysis
The above experimental data are the mean ± SD of at least three independent experiments. SPSS 22.0 software was used to process the data, and one-way analysis of variance (ANOVA) was used to measure statistical differences between the two groups.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
CX: Conceptualization, Data curation, Writing–original draft, Writing–review and editing. NY: Supervision, Validation, Visualization, Writing–original draft. HY: Methodology, Project administration, Resources, Supervision, Validation, Writing–original draft. XW: Data curation, Writing–original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2024.1529211/full#supplementary-material
References
Ali, T., Kim, T., Rehman, S. U., Khan, M. S., Amin, F. U., Khan, M., et al. (2018). Natural dietary supplementation of anthocyanins via PI3K/Akt/Nrf2/HO-1 pathways mitigate oxidative stress, neurodegeneration, and memory impairment in a mouse model of alzheimer's disease. Mol. Neurobiol. 55 (7), 6076–6093. doi:10.1007/s12035-017-0798-6
Chiba, N., Masuda, A., Yoshikai, Y., and Matsuguchi, T. (2007). Ceramide inhibits LPS-induced production of IL-5, IL-10, and IL-13 from mast cells. J. Cell Physiol. 213 (1), 126–136. doi:10.1002/jcp.21101
Citi, V., Martelli, A., Brancaleone, V., Brogi, S., Gojon, G., Montanaro, R., et al. (2020). Anti-inflammatory and antiviral roles of hydrogen sulfide: rationale for considering H2S donors in COVID-19 therapy. Br. J. Pharmacol. 177 (21), 4931–4941. doi:10.1111/bph.15230
Dilek, N., Papapetropoulos, A., Toliver-Kinsky, T., and Szabo, C. (2020). Hydrogen sulfide: an endogenous regulator of the immune system. Pharm. Res. 161 (12), 105119–105132. doi:10.1016/j.phrs.2020.105119
Faris, P., Negri, S., Faris, D., Scolari, F., Montagna, D., and Moccia, F. (2023). Hydrogen sulfide (H2S): as a potent modulator and therapeutic prodrug in cancer. Curr. Med. Chem. 30 (40), 4506–4532. doi:10.2174/0929867330666230126100638
Flannigan, K. L., and Wallace, J. L. (2015). Hydrogen sulfide-based anti-inflammatory and chemopreventive therapies: an experimental approach. Curr. Pharm. Des. 21 (21), 3012–3022. doi:10.2174/1381612821666150514105413
Gao, W., Liu, Y. F., Zhang, Y. X., Wang, Y., Jin, Y. Q., Yuan, H., et al. (2024). The potential role of hydrogen sulfide in cancer cell apoptosis. Cell Death Discov. 10 (1), 114–119. doi:10.1038/s41420-024-01868-w
Gemici, B., and Wallace, J. L. (2015). Anti-inflammatory and cytoprotective properties of hydrogen sulfide. Meth. Enzymol. 555 (8), 169–193. doi:10.1016/bs.mie.2014.11.034
Goor, H., Born, J. C., Hillebrands, J. L., and Joles, J. A. (2020). Hydrogen sulfide in hypertension. Curr. Opin. Nephrol. Hypertens. 25 (2), 107–113. doi:10.1097/MNH.0000000000000206
Hao, Y., Wang, H., Fang, L., Bian, J., Gao, Y., and Li, C. (2021). H2S donor and bone metabolism. Front. Pharm. 12 (22), 661601–661625. doi:10.3389/fphar.2021.661601
Hart, S., Goh, K. C., Novotny-Diermayr, V., Tan, Y. C., Madan, B., Amalini, C., et al. (2011). Pacritinib (SB1518), a JAK2/FLT3 inhibitor for the treatment of acute myeloid leukemia. Blood Cancer J. 1 (11), e44–e408. doi:10.1038/bcj.2011.43
Hasegawa, U., and Vlies, A. J. (2014). Design and synthesis of polymeric hydrogen sulfide donors. Bio. Chem. 25 (7), 1290–1300. doi:10.1021/bc500150s
Hou, L., Zhang, J., Liu, Y., Fang, H., Liao, L., Wang, Z., et al. (2021). MitoQ alleviates LPS-mediated acute lung injury through regulating Nrf2/Drp1 pathway. Free. Radic. Biol. Med. 165 (31), 219–228. doi:10.1016/j.freeradbiomed.2021.01.045
Hou, R. C., Chen, Y. S., Chen, C. H., Chen, Y. H., and Jeng, K. C. (2006). Protective effect of 1,2,4-benzenetriol on LPS-induced NO production by BV2 microglial cells. J. Biomed. Sci. 13 (1), 89–99. doi:10.1007/s11373-005-9039-5
Hu, M., Yang, T., Yang, L., Niu, L., Zhu, J., Zhao, A., et al. (2022). Preclinical studies of Flonoltinib Maleate, a novel JAK2/FLT3 inhibitor, in treatment of JAK2V617F-induced myeloproliferative neoplasms. Blood. Cancer 12 (3), 37–45. doi:10.1038/s41408-022-00628-2
Huang, J., Chen, L., Wu, J., Ai, D., Zhang, J. Q., Chen, T. G., et al. (2022). Targeting the PI3K/AKT/mTOR signaling pathway in the treatment of human diseases: current status, trends, and solutions. J. Med. Chem. 65 (24), 16033–16061. doi:10.1021/acs.jmedchem.2c01070
Huang, W. R., Tu, J. X., Qiao, A. Q., and Chen, L. J. (2022). GW842166X alleviates osteoarthritis by repressing LPS-mediated chondrocyte catabolism in mice. Curr. Med. Sci. 42 (5), 1046–1054. doi:10.1007/s11596-022-2627-z
Khan, M. A., Jain, V. K., Rizwanullah, M., Ahmad, J., and Jain, K. (2019). PI3K/AKT/mTOR pathway inhibitors in triple-negative breast cancer: a review on drug discovery and future challenges. Drug. Discov. Today 24 (11), 2181–2191. doi:10.1016/j.drudis.2019.09.001
Khan, N. H., Wang, D., Wang, W., Shahid, M., Khattak, S., Ngowi, E. E., et al. (2022). Pharmacological inhibition of endogenous hydrogen sulfide attenuates breast cancer progression. Molecules 27 (13), 4049–49. doi:10.3390/molecules27134049
Kolluru, G. K., Shackelford, R. E., Shen, X., Dominic, P., and Kevil, C. G. (2023). Sulfide regulation of cardiovascular function in health and disease. Nat. Rev. Cardiol. 20 (2), 109–125. doi:10.1038/s41569-022-00741-6
Kumar, M. (2023). Hydrogen sulfide: from a toxic gas to a potential therapy for COVID-19 and inflammatory disorders. Nitric Oxide 140 (1), 8–15. doi:10.1016/j.niox.2023.08.002
Lee, M. H., Kang, C. H., Tae, L. K., Hyun, C. Y., Lee, S., and Kim, G. Y. (2022). Acertannin attenuates LPS-induced inflammation by interrupting the binding of LPS to the TLR4/MD2 complex and activating Nrf2-mediated HO-1 activation. Int. Immunopharmacol. 113 (PtA), 109–124. doi:10.1016/j.intimp.2022.109344
Li, Y., Chandra, T. P., Song, X., Nie, L., Liu, M., Yi, J., et al. (2021). H2S improves doxorubicin-induced myocardial fibrosis by inhibiting oxidative stress and apoptosis via Keap1-Nrf2. Techno Health Care 29 (1), 195–209. doi:10.3233/THC-218020
Liu, J., Tian, R., Sun, C., Guo, Y., Dong, L., Li, Y., et al. (2023). Microbial metabolites are involved in tumorigenesis and development by regulating immune responses. Front. Immunol. 19 (14), 1290414–1291134. doi:10.3389/fimmu.2023.1290414
Moses, C., Garcia, B., Harvey, A. R., and Blancafort, P. (2018). Hallmarks of cancer: the CRISPR generation. Eur. J. Cancer. 93 (12), 10–18. doi:10.1016/j.ejca.2018.01.002
Munteanu, C. (2023). Hydrogen sulfide and oxygen homeostasis in atherosclerosis: a systematic review from molecular biology to therapeutic perspectives. Int. J. Mol. Sci. 24 (9), 8376–8382. doi:10.3390/ijms24098376
Nelmi, O., Devarie-Baez, P. E. B., Bo, P., and Yu, Z. (2013). Chung-min, park. Light-induced hydrogen sulfide release from “caged” gem-dithiols. Acs. Chem. Biol. 15 (11), 2786–2789. doi:10.1021/ol401118k
Nogales, C., Mamdouh, Z. M., List, M., Kiel, C., Casas, A. I., and Schmidt, H. H. (2022). Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends. Pharmacol. Sci. 43 (2), 136–150. doi:10.1016/j.tips.2021.11.004
Pan, T., Xiao, Q., Fan, H. J., Xu, L., Qin, S. C., Yang, L. X., et al. (2023). Wuzi Yanzong Pill relieves MPTP-induced motor dysfunction and neuron loss by inhibiting NLRP3 inflammasome-mediated neuroinflammation. Metab. Brain. Dis. 38 (7), 2211–2222. doi:10.1007/s11011-023-01266-8
Sanchez, V. E., Nichols, C., Kim, H. N., Gang, E. J., and Kim, Y. M. (2019). Targeting PI3K signaling in acute lymphoblastic leukemia. Int. J. Mol. Sci. 20 (2), 412–418. doi:10.3390/ijms20020412
Sanchez-Aranguren, L. C., Ahmad, S., Dias, I. H. K., Alzahrani, F. A., Rezai, H., Wang, K., et al. (2020). Bioenergetic effects of hydrogen sulfide suppress soluble Flt-1 and soluble endoglin in cystathionine gamma-lyase compromised endothelial cells. Sci. Rep. 10 (1), 15810–15847. doi:10.1038/s41598-020-72371-2
Shen, J., Ma, X., He, Y., Wang, Y., Zhong, T., and Zhang, Y. (2022). Anti-inflammatory and anti-oxidant properties of Melianodiol on DSS-induced ulcerative colitis in mice. PeerJ 25 (2), 142099–e15109. doi:10.7717/peerj.14209
Siegel, R. L., Miller, K. D., and Jemal, A. (2018). Cancer statistics, 2018. Cancer. J. Clin. 68 (1), 7–30. doi:10.3322/caac.21442
Somensi, N., Rabelo, T. K., Guimarães, A. G., Quintans-Junior, L. J., Moreira, J. C. F., Gelain, D. P., et al. (2019). Carvacrol suppresses LPS-induced pro-inflammatory activation in RAW 264.7 macrophages through ERK1/2 and NF-kB pathway. Int. Immunopharmacol. 75 (23), 105743–106143. doi:10.1016/j.intimp.2019.105743
Sun, Y., Tang, X., Zhang, K., Liu, K., Li, Z., and Zhao, L. (2022). Hydrogen sulfide detection and zebrafish imaging by a designed sensitive and selective fluorescent probe based on resorufin. Spectrochim. Acta. Part. A. 264 (5), 120265–120340. doi:10.1016/j.saa.2021.120265
You, Z., Yang, Z., Cao, S., Deng, S., and Chen, Y. (2022). The novel KLF4/BIG1 regulates LPS-mediated neuro-inflammation and migration in BV2 cells via PI3K/Akt/NF-kB signaling pathway. Neuroscience 488 (15), 102–111. doi:10.1016/j.neuroscience.2022.01.014
Zhang, J., Zhang, Q., Wang, Y., Li, J., Bai, Z., Zhao, Q., et al. (2019). Toxicity, bioactivity, release of H2S in vivo and pharmaco-kinetics of H2S-donors with thiophosphamide structure. Eur. J. Med. Chem. 15 (176), 456–475. doi:10.1016/j.ejmech.2019.05.017
Zhang, R. X., Zhu, H., Bai, K., and Ning, K. (2019). Network pharmacology databases for traditional Chinese medicine: review and assessment. Front. Pharmacol. 10 (8), 123–128. doi:10.3389/fphar.2019.00123
Zhao, Y., Wang, H., and Xian, M. (2010). Cysteine-activated hydrogen sulfide (H2S) donors. J. Am. Chem. Soc. 133 (1), 15–17. doi:10.1021/ja1085723
Keywords: hydrogen sulfide, network pharmacology, anti-tumor, anti-inflammatory, phenylthiophosphoryl dichloride
Citation: Xu C, Yang N, Yu H and Wang X (2025) Design and synthesis of phenylthiophosphoryl dichloride derivatives and evaluation of their antitumour and anti-inflammatory activities. Front. Chem. 12:1529211. doi: 10.3389/fchem.2024.1529211
Received: 16 November 2024; Accepted: 24 December 2024;
Published: 21 January 2025.
Edited by:
Greta Varchi, Consiglio Nazionale delle Ricerche (Bologna), ItalyReviewed by:
Pawan Dhote, University of North Carolina at Chapel Hill, United StatesAnis Ur Rahman, Texas Tech University, United States
Copyright © 2025 Xu, Yang, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunyun Xu, Y2h1bnl1bnh1MDFAMTYzLmNvbQ==
 Chunyun Xu
Chunyun Xu Na Yang
Na Yang