- 1Beijing Center for Disease Prevention and Control, Beijing, China
- 2School of Public Health, Capital Medical University, Beijing, China
- 3School of Chinese Medicine, Yunnan University of Traditional Chinese Medicine, Kunming, China
Biotoxins are ranges of toxic substances produced by animals, plants, and microorganisms, which could contaminate foods during their production, processing, transportation, or storage, thus leading to foodborne illness, even food terrorism. Therefore, proposing simple, rapid, and effective detection methods for ensuring food free from biotoxin contamination shows a highly realistic demand. Aptamers are single-stranded oligonucleotides obtained from the systematic evolution of ligands by performing exponential enrichment (SELEX). They can specifically bind to wide ranges of targets with high affinity; thus, they have become important recognizing units in safety monitoring in food control and anti-terrorism. In this paper, we reviewed the technical points and difficulties of typical aptamer screening processes for biotoxins. For promoting the understanding of food control in the food supply chain, the latest progresses in rapid optical detection of biotoxins based on aptamers were summarized. In the end, we outlined some challenges and prospects in this field. We hope this paper could stimulate widespread interest in developing advanced sensing systems for ensuring food safety.
1 Introduction
Biotoxins, also known as natural toxins, are groups of inherently small molecules, peptides, or proteins, which are produced during the metabolism of plants, microorganisms, and animals (Montecucco, 2012). Food contamination by biotoxin usually occurs due to unsterilized processing, improper storage, as well as microbial spoilage, whereas others are inherent ingredients in plants, or might be produced by environmental stimuli, such as shellfish toxin (Rather et al., 2017). Biotoxins are prevalent in food-poisoning incidents around the world and are the main culprits of various acute and chronic human diseases. According to the World Health Organization (WHO), the consumption of mycotoxin-contaminated grains (Alshannaq and Yu, 2017), meats, and vegetables (especially fermented foods) with poor sanitation and bacterial contamination has been the first cause of foodborne illness (WHO, 2015). Aquatic and marine foods such as shellfish, fish, and even water are contaminated with algal biotoxins (Oliveira et al., 2011), as well as the normal metabolites produced by some plants for natural defenses and resistance; all of these become toxins in other organisms (Fletcher and Netzel, 2020). According to the classification of toxin-producing organisms, they are classified as animal toxins (snake venom and bee venom), plant toxins (lectins and alkaloids), microbial toxins (bacteria and molds), and marine biotoxins (algae and shellfish) (Picardo et al., 2020). Specifically, some highly toxic biotoxins, such as ricin, acacia toxin, and tetrodotoxin, have been listed in the Organization for the Prohibition of Chemical Weapons (OPCW) and have a risk of being used by terrorist organizations to threaten public security (Anderson, 2012). Biotoxins act on the cell membrane or ribosomal protein with high specificity, resulting in varying degrees of toxicity, which are not only harmful to the health of consumers (Rather et al., 2017) but also cause huge economic losses to the planting and aquaculture industries. Although biotoxins cannot replicate themselves, they are generally chemically stable and highly toxic. They can persist for a long time after an organism dies, further entering food chains, and finally triggering food safety and public health crises (Banach et al., 2020). In the past, due to the lack in knowledge and reliable detecting methods for biotoxins, food safety verification had been neglected, thus leading to thousands of foodborne poisoning events. With the continuous improvement of the economic level, people’s health awareness also improved significantly. Therefore, highly sensitive and rapid methods for detecting biotoxins are urgently required to ensure foods free from biotoxin contamination (Bacha et al., 2023).
Until now, various analytical tools have been proposed to determine biotoxin presence in foods, which can be mainly divided into laboratory precision detection based on high-performance liquid chromatography/mass spectrometry (HPLC–MS) (Ahuja et al., 2023) and on-site rapid screening represented by affinity-recognition sensors (Song et al., 2019; Sarkar et al., 2023). Between them, LC–MS-based methods allow precious, high throughput, and ultra-sensitive detection of biotoxins. Benefited by the development of high-resolution mass spectrometry (HRMS) and tandem mass spectrometry (MS/MS), the sensitivity of biotoxin detection could further reach to ultra-low levels, making them the gold standard for the identification and quantification of toxins in food by research workers, regulatory agencies, and the food industry. However, LC–MS-based methods generally involve complicated sample preparation, costly instruments, and well-trained personnel; these features confine their applications in daily monitoring. In contrast, on-site rapid screening biosensors, which are typically composed of highly specific recognition ligands (e.g., antibodies and aptamers) with advanced signal reporting materials, could well meet the need of the modern society for daily safety monitoring of food samples. With its rapid response, ease of use, and affordable advantages, it has become an indispensable and powerful tool to ensure the safety and quality of the food supply chain (Howes et al., 2014).
Aptamers are single-stranded DNA or RNA fragments of between 10 and 100 bases obtained from artistically synthesized libraries (Scheme 1). They show unique three-dimensional structures, which could form helices and single-stranded loop-like structures to match specific targets (small molecules, proteins, cells, etc.) through the non-covalent bonds, including hydrogen bonding, van der Waals forces, electrostatic interactions, and π–π* stacking interactions. Aptamers have multiple advantages of good stability, easy chemical modification, high affinity and specificity, as well as low immunogenicity. Until now, dozens of aptamers of biotoxins have been obtained by the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) technology, as listed in Table 1. Aptamers have also widely been used for constructing various biotoxin-specific biosensors (He et al., 2023). In this field, optical biosensors based on aptamer recognition (optical aptasensors) have gained more scientific attention and offer promising applications in the field of food safety and detection. They could transfer biorecognition events into quantifiable optical signals without being restricted by the environment and equipment, and they are rapid, accurate, cheap, and portable. Specifically, through integrating aptamers as signal reporting units with excellent optical materials (Loyez et al., 2022), such as upconversion luminescent NPs, colloidal gold NPs, and time-resolved fluorescent NPs, the detecting performance of aptasensors could further be improved (Liu B. et al., 2022).
In this paper, a concise description of aptamer screening processes for typical biotoxins and the key technical challenges were briefly introduced. Then, the source and characteristics of different biotoxins were summarized, and the general design for optical aptasensors using advanced materials and the nucleic acid isothermal amplification strategy were introduced in each chapter comprehensively. Finally, the challenges and directions of future research of the existing rapid detection methods were discussed. We hoped that this review would inspire research workers to develop more optical aptasensors with superior performance for ensuring food safety.
2 Screening aptamers for biotoxins and their challenges
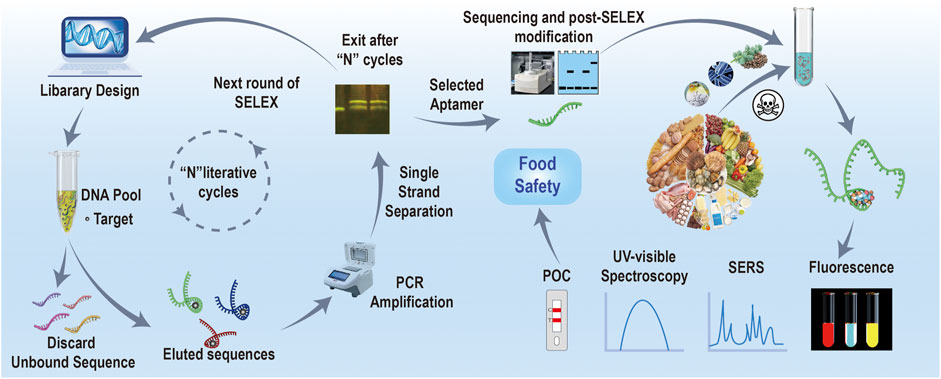
Nucleic acid aptamers are isolated by the Systematic Evolution of Ligands by EXponential enrichment (SELEX) screening technique, which was first proposed by Tuerk and Gold (1990). Using this technique, aptamers can be screened from a library of random single-stranded nucleic acid with high target affinity (Radom et al., 2013). Generally, SELEX for discovering biotoxin aptamers mainly involves four steps: 1. Oligonucleotide library design and optimization. Random nucleic acid libraries are usually obtained by chemical synthesis but can also be constructed by genomic DNA design and in vitro transcription (Pan and Clawson, 2010). 2. Improvement of screening methods: currently, dozens of SELEX methods and their derived strategies have been reported by different groups worldwide, including immobilized-based, non-immobilized-based, and assisted SELEX strategy for small-molecule targets (Abdelsayed et al., 2017; Yang et al., 2019). 3. Complex separation: the complex between the targets and their oligonucleotides on the surface of solid interfaces can be separated from the unbound ones, and then they directly undergo PCR amplified without elution (Stoltenburg et al., 2005). 4. Sub-library enrichment: after acquiring aptamers from the oligonucleotide library, single-strand splitting or truncation for structure optimization would be conducted subsequently for improving their affinities and specificities (Bawazer et al., 2014; Gu et al., 2018).
Developing a rational screening strategy is the footstone to obtaining well-performed aptamers (Yu et al., 2021) (Figure 1). Although standard screening processes for biotoxins have not been proposed at the present stage, some basic evaluating rules for enhancing the screening success rate are widely accepted currently (McKeague et al., 2015). First, biotoxins of larger molecular weights, fewer rotational bonds, and more aromatic moieties could be more suitable for SELEX because they contain more potential binding interfaces and sites, a lower entropic binding retardation, and potential π–π* stacking effect with aptamer bases. Second, the low polarity and poor water solubility of some biotoxins make them incompatible with traditional water phase conducted screenings. Third, biotoxins require specialized conjugation chemistry or complicated chemical synthesis steps to achieve immobilization, which causes challenges and complexities for SELEX. In the process of aptamer screening, it is usually necessary to immobilize the targets on certain solid phase carriers, which could be magnetic beads, graphene, gold nanoparticles (AuNPs), microcolumns, chips, etc., to separate nonbinding sequences with the complex (Lyu et al., 2021), which inevitably involve molecular structure derivation and chemical coupling processes. However, biotoxins have far fewer functional groups and simple chemical structures. Any alterations may result in fundamental molecular changes, changing the physicochemical properties inherently, which induce the failure of SELEX screening (Ellington and Szostak, 1992). Above all, aptamer screening is the foundation of building aptasensors for biotoxin analysis. Currently, only dozens of aptamers have been successfully screened, which is far from enough for biotoxins. To obtain more outstanding aptamers, investigating more trustworthy aptamer screening and optimization approaches is highly demanded (Wei et al., 2023). Molecular recognition units are the core component of optical sensors, which could fundamentally determine the specificity of the method. Generally, as a kind of biological recognition molecule, an aptamer is often compared with an antibody to highlight its superior functions. Although antibodies are commonly used as recognition ligands, obtaining biotoxin antibodies based on animals is technically challenging due to their high toxicity and poor immunogenicity. Compared with antibodies, aptamers have advantages as follows: being non-immunogenic, having good chemical stability, and not involving animal ethical issues. Moreover, as aptamers are chemically synthesized, they are easy to be structurally modified for improving the application performance (McKenzie et al., 2021).
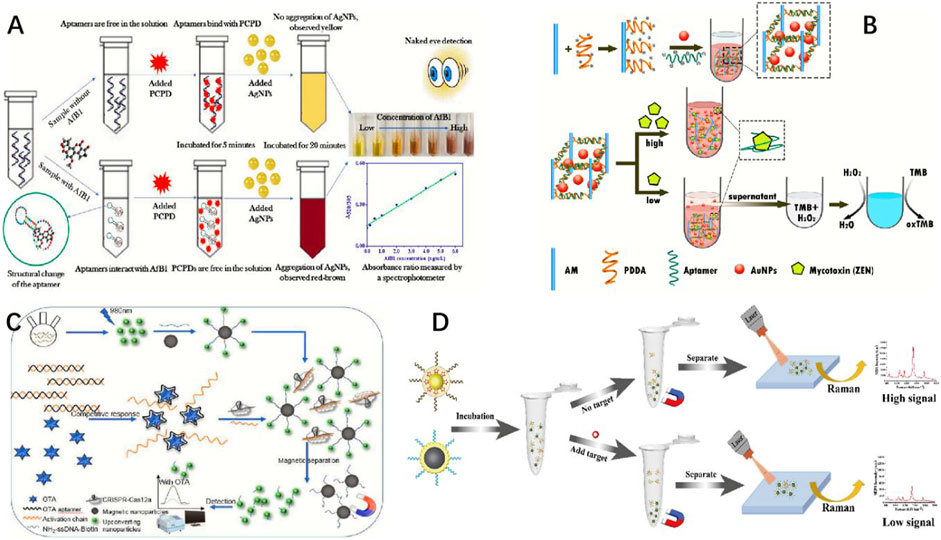
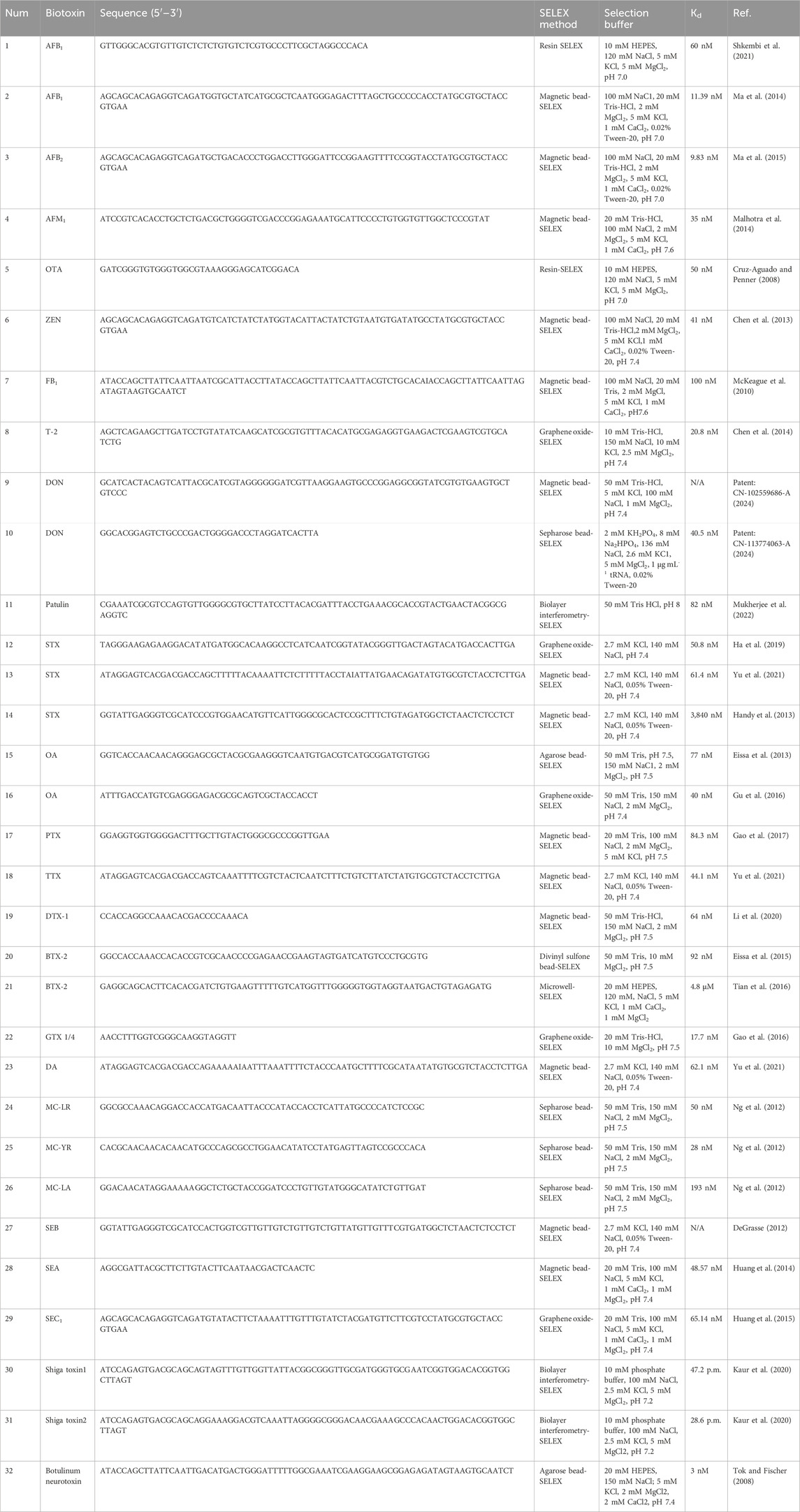
Figure 1. (A) Schematic demonstration of AfB1 detection based on colorimetric aptasensor utilizing the specific aptamer, PCPD, and AgNPs (Yu et al., 2021). (B) Schematic diagram of the AuNPs encapsulated PDDA-aptamer hydrogel for ultrasensitive colorimetry of ZEN (Lyu et al., 2021). (C) Schematic showing upconversion-mediated CRISPR-Cas12a biosensor that sensitively detects OTA (Niazi et al., 2022). (D) Schematic representation of the universal surface-enhanced Raman scattering (SERS) aptasensor platform for trace detection (Li et al., 2019).
3 Optical aptasensors for biotoxins
Optical aptasensors are always designed as portable analysis devices which use aptamers as recognition components to convert specific binding into measurable optical signals. Furthermore, benefiting from the recent advancement of nanomaterials (Niazi et al., 2022) and room-temperature nucleic acid signal amplification, these progresses push optical aptasensors of biotoxins to ultrasensitive detection levels (Li et al., 2019; Lei and Guo, 2022). Compared with existing detection methods, aptasensors do not require complex sample purifications and can complete sample detection within 30 min, which are more suitable for in situ detection, batch screening, and rapid diagnosis (Balbinot et al., 2021) (Figure 2).
Figure 2. Schematic illustration of (A) the label-free, rapid, and sensitive detection of ochratoxin A in colorimetric and fluorescent modes by engineering DNA G-quadruplex (Balbinot et al., 2021). (B) FL-SERS dual-mode detection of OTA (Zhang N. et al., 2022). (C) Engineered bifunctional aptamer and the MST assay (Tang and Li, 2017). (D) Detection mechanism based on a toehold-mediated cascade catalytic assembly and supramolecular DNAzyme nanostructures ([B/C/DNA1/DNA2]n) for mycotoxin detection (Tang et al., 2024).
The exploration of biotoxin aptamers as sensing elements has made unprecedented progress in the past 3 decades. Several advanced nanomaterials have been applied for constructing various optical aptasensors for rapidly detecting biotoxins (Zhang N. et al., 2022). The colorimetric aptasensor is an instrumental independent visual sensing manner (Hizir et al., 2016; Tang and Li, 2017; Tang et al., 2024). The fluorescent aptasensor is mainly utilized for sensing biotoxins by rapidly converting target-induced aptamer conformation changes to highly sensitive fluorescent signals (Chen et al., 2017; Zhang Q. et al., 2022; Khan et al., 2023a). The SERS aptasensor, recognized as a promising analytical technique of biotoxin detection (Muhammad and Huang, 2021), possesses multiple advantages of high sensitivity, rapid reports, and non-destructiveness (Zhao X. et al., 2021). Developing outstanding SERS substrates is highly necessary for building aptasensors (Zhao P. et al., 2021). In addition to pushing the development on nanomaterial-based signal reporting units, room temperature isothermal signal amplification strategies based on complementary hybridization of nucleic acids have also been regarded as important strategies to fulfill sensing signal amplification (Gu et al., 2021; Tan et al., 2021) (Figure 3). In this aspect, enzyme-assisted nucleic acid amplification (rolling circle amplification (RCA) with the assistance of DNA polymerases) (Ali et al., 2014), a kinetics-controlled enzyme-free nucleic acid amplification technique such as hybridization chain reaction (HCR) (Bi et al., 2017) and the catalytic hairpin assembly (CHA) (Liu et al., 2019) are all novel strategies used in optical aptasensors. Furthermore, the signal amplification methods assisted by nucleases, DNAzyme, such as exonuclease I (Exo I) (Lan et al., 2020), exonuclease III (Exo III) (Wang et al., 2021), deoxyribonuclease I (DNase I) (Gu et al., 2021), and ribonuclease H (RNase H) (Gu et al., 2021) have also shown great opportunities for constructing biotoxin-specific aptasensors. Beneficial from the biodegradable properties of both double-stranded ssDNA (dssDNA) and double-stranded DNA (dsDNA) in the presence of nucleases, the biotoxins can repeatedly participate in the biological cascade reaction (BCR), thus producing signal amplification (Ren et al., 2022).
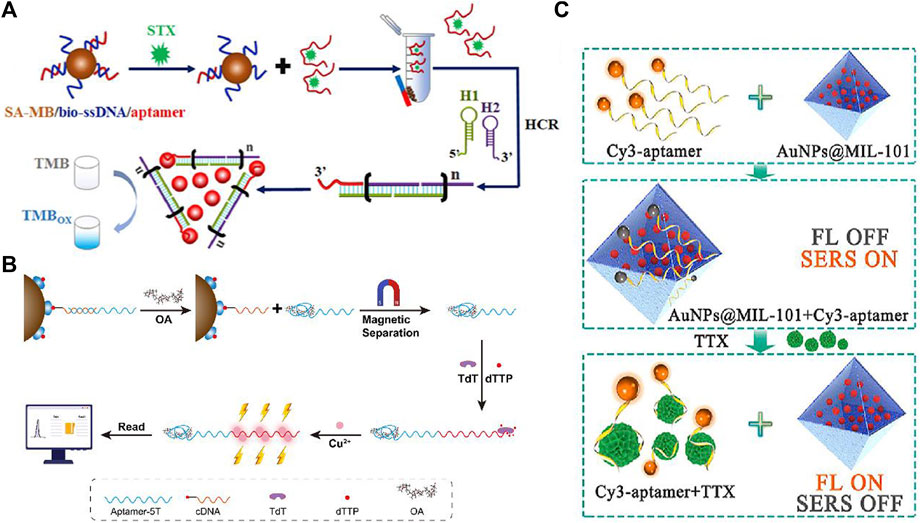
Figure 3. Schematic illustration of (A) the competitive colorimetric aptasensor transduced by hybridization chain reaction-facilitated catalysis of AuNP nanozyme for highly sensitive detection of saxitoxin (Gu et al., 2021). (B) Duplexed aptamer-isothermal amplification-based nucleic acid CuNP sensor for the detection of OA (Ali et al., 2014). (C) Fluorescence/SERS dual-mode aptasensor for analysis of TTX (Liu et al., 2019).
Moreover, smart synthetic DNA systems including DNA walker and the CRISPR-Cas system were also applied in fabricating aptasensors due to their programmatic behavior. Specifically, the repetitive cleavage reaction through highly directional mechanical movements was originally prelocked by the aptamer. The binding of biotoxins to the aptamer walker systems would activate this sensing system (Chen et al., 2022). For the CRISPR-Cas-based sensing system, the aptamer worked as a Cas protein activation chain. In the presence of biotoxins, the trans-cleavage activity of the Cas protein would be inhibited (Lin et al., 2022). In the nuclease-assisted signal amplification sensing systems, enzyme instability and rigorous reaction conditions might be adverse factors. Therefore, enzyme-free signal amplification aptasensors showed better durability in real applications. In this section, we described the current situation of high-performance optical aptasensors according to different types of biotoxins.
3.1 Optical aptasensors for mycotoxins
Mycotoxins are highly toxic secondary metabolites secreted by fungi. They are highly heat-resistant; therefore, they are hard to be removed through cooking and easily traverse into the food chain, affecting human and animal health (Eskola et al., 2020). According to the EU Rapid Alert System for Food and Feed (RASFF) statistic, mycotoxins might contaminate approximately a quarter of the world’s food and oil crops per year. Currently, agencies such as the World Health Organization and the Food Agriculture Organization have determined the permissible limit for mycotoxin contamination in food and feed to ensure food safety.
Colorimetric assay based on aggregation-induced Au/Ag NP color change, which are realized through precious regulating stability of NPs in a salt solution via an aptamer, has been widely used for designing visual mycotoxin aptasensors (Mirón-Mérida et al., 2021). In an AgNP-based colorimetric aptasensor, aflatoxin B1 (AFB1) achieved sensitive detection with an LOD of 0.09 ng/mL in food samples (Lerdsri et al., 2021). The signal amplification strategy was also widely integrated to the optical aptasensors for monitoring mycotoxins. The sensing platform was proposed with an LOD of 0.62 ng/mL, which could be utilized for AFB1 monitoring in complex sample matrices (Chen et al., 2016). Utilizing the peroxidase-like activity of AuNPs, a colorimetric aptasensor for zearalenone (ZEN) was designed using a chimeric aptamer. The aptasensor provided an LOD of 0.58 ng/mL, which could be well applied in real corn oil samples (Liu M. et al., 2022).
Fluorescent aptasensors have shown advantages of facile signal transduction, fast response, and high sensitivity, but they suffer from poor signal stability. In these designs, Förster resonance energy transfer (FRET) and the time-resolved fluorescent manner could overcome these shortcomings; thus, they have been widely utilized in the analysis of mycotoxins. Wang et al. designed a label-free aptasensor for FRET detection of trichothecenes A (T-2) toxin as low as 0.93 pg/mL. In this assay, T-2-specific aptamer functionalized using silver nanoclusters (apt-AgNCs) was synthesized as the fluorescent probe, whose fluorescence was initially quenched by MoS2 nanosheets (NSs). The presence of T-2 biotoxin led to the desorption of Apt-AgNCs from MoS2 NSs, which caused the fluorescence recovery in a target amount-dependent manner. This method showed good utility in risk assessment of T-2 toxin (Khan et al., 2018). Using a similar design, Wang et al. further synthesized a green-emitting gold nanocluster (Arg@ATT-AuNCs) as signal reporting element for the fluorescent sensing of T-2. The bioassay showed an LOD of 0.57 pg/mL with a linear range of 0.001–100 ng/mL (Khan et al., 2020).
Moreover, advanced nanomaterials have gained much attention for constructing sensors with enhanced performance that detect multiple mycotoxins simultaneously due to their creditable optical properties. Niazi et al. proposed a rapid time-resolved fluorescence (TRF)-based aptasensor for simultaneous recognition of ochratoxin A (OTA) and fumonisin B1 (FB1) using a multi-color, Ln-doped NPs (TRF-NPs) group. After method optimization, LODs of 0.019 pg/mL and 0.015 pg/mL for FB1 and OTA were achieved, respectively (Niazi et al., 2019a). In another case, a turn-on time-resolved fluorometric aptasensor is described for the simultaneous detection of ZEN, T-2, and AFB-1 in maize samples based on the multi-color, TRF-NP group, and tungsten disulfide nanosheets (WS2 NSs) are used as a quencher of time-resolved fluorescence. These methods showed great potentials in food safety fields (Niazi et al., 2019b). Based on a similar design, Khan et al. (2019) proposed a multicolored nanomaterial-based FRET sensing platform for the simultaneous detection of mycotoxins. The assay combined the dual-color gold nanoclusters (AuNCs) as fluorescence donors with WS2 NSs as a fluorescence quencher, which achieved simultaneous recognition of AFB1 and ZEN with a detection limit of 0.34 pg/mL and 0.53 pg/mL, respectively.
Hitabatuma et al. (2021) developed an aptasensor for OTA with an LOD of 0.247 pg mL−1. The aptamer binding to OTA induced the cDNA to hybridize with molecular beacon (MB). Following the MB stem unwinding, the FAM labeled on the MB would be far from the DABCYL. The fluorescence “turn-on” sensing mechanism is simple and fast, which exhibited specificity and sensitivity (LOD of 0.247 pg/mL) to OTA in a wheat sample. Fan et al. (2021) reported a dual-emission aptasensor based on the two fluorescent dyes: thioflavin T (ThT) and trans-2-[4’-(dimethylamino)styryl]-3-ethyl-1,3-benzothiazole (DMASEBT). The fluorescence of ThT was quenched after being inserted into DNA strands. In the working state, AFB1 would displace the bound ThT to the solution. The aptasensor was applied to the analysis of AFB1 with an LOD of 0.01 ng/mL in food samples successfully. Attributing to the development of novel nanomaterials, cobalt oxyhydroxide (CoOOH) nanosheets and graphitic carbon nitride quantum dots (gCNQDs) were used to fabricate a FRET-based aptasensor for OTA detection. This method was featured with ultra-sensitivity with an LOD as low as 201.9 pg/mL and was successfully applied to corn and barley flour (Bi et al., 2020). A sensitive fluorescent aptasensor for AFM-1 was proposed based on the time-resolved fluorescent NP as a signal probe and RCA to improve the sensitivity of the assay. The assay showed a lower detection limit (0.0194 pg/mL) than the previously reported assays (Niazi et al., 2020). In addition, CRISPR-Cas-assisted fluorescent aptasensors were constructed to analyze mycotoxins. Mao et al. (2022) applied CRISPR/Cas12a in designing an aptasensor for OTA detection. In the sensing process, an activated cDNA strand was first released from the sensors in the presence of OTA. The cDNA was subsequently hybridized with crRNA to cut UCNP-DNA linked on the Fe3O4 NPs, and then the fluorescence signal was turned on. This method was very sensitive, with an LOD of 0.83 ng/mL. The CRISPR/Cas12a-based sensing strategy had a great practical application prospect for various targets (Mao et al., 2022). SERS are well known as an attractive analytical tool with advantages of rapid and on-site ultrasensitive detection. SERS-based aptasensors are well developed as a promising tool for biotoxins. Constructing outstanding SERS substrates and SERS tags with superior properties is the key technology in this field. Chen et al. (2021) proposed a universal aptasensor for the analysis of ZEN based on Fe3O4@Au NPs and Au@Ag core-shell NPs, which could perform well in food samples with an LOD of 1.0 ng/L. A ratiometric SERS aptasensor for AFB1 was constructed based on hybrid nanomaterial, that is, Ti3C2Tx MXene-loaded AuNP dimers. Assembled AuNP dimers contained a rich SERS “hot spot,” which provided a strong SERS signal. MXenes nanosheet functioned as a support to the aptamer-modified AuNP dimers for achieving steady Raman signal. The presence of AFB1 could competitively bind to the aptamer, thus pushing AuNP dimers to be separated from MXenes NSs. This recognition processing causes the SERS intensity to decay with the increase of AFB1. The aptasensor showed an LOD as low as 0.6 pg/mL and could be well used in peanut samples (Wu et al., 2022).
To further improve the sensing reliability, dual-mode optical aptasensors have been developed for mycotoxin detection. Based on the feature of the G-quadruplex structure, He et al. (2022) developed a label-free and dual-mode aptasensor for OT, which contained both G-quadruplex/hemin DNAzym as the catalytic colorimetric unit and G-quadruplex ThT as the fluorescence reporting unit. Following the colorimetric manner, a DNA triplehelix switch was composed of aptamer and G-rich sequences. The binding of OTA resulted in the separation of the triple-helix, which subsequently released the ssDNA to bind hemin and stimulate color change. Under the fluorescent mode, the aptamer would combine with ThT to produce a strong signal. Beneficially from the structure flexibility of ssDNA, the G-quadruplex DNA assembly was rationally engineered to achieve dual-mode sensing for biotoxins. For instance, two kinds of NPs including AuNSs and AuNPs were modified with the aptamer and Cy3-modified cDNA, respectively. In the presence of OTA, the hybridization of aptamer and cDNA was broken, causing the disassembly of AuNSs and AuNPs. This process turned on the fluorescenc Cy3, but the SERS signal from Cy3 was decreased, achieving a dual-mode OTA ratio analysis. The aptasensor had good practicality in the analysis of coffee and wine samples (Wang et al., 2022).
Moreover, simultaneously detecting aptasensors have also been developed due to their outstanding time- and cost-saving features as well as fast and high-throughput testing. Optical aptasensors can also be constructed to simultaneously monitor multiple mycotoxins. Yang et al. (2021) designed a dual-targeted aptamer, which is the tandem of ZEN aptamer and OTA aptamer via a poly-T linker. Using this novel aptamer, simultaneous determination of ZEN and OTA as low as 0.12 nM was achieved based on the microscale thermophoresis in corn oil samples. Xiong et al. (2021) designed a dual DNA tweezer nanomachine that was originally inhibited by the anti-AFB1 aptamer and anti-OTA aptamer; both of them were labeled FAM and Cy5 fluorophores, respectively. The fluorescence was turned on in the presence of targets. The methods also showed good performance in cereals with LODs of 0.035 ng/mL for AFB1 and 0.1 ng/mL for OTA. Signal amplification based on CHA- and DNAzyme-cascaded hydrolysis reaction has also been applied for simultaneously sensing multiple mycotoxins. Several concatenated logic gates were used in the biosensors due to the versatility of these methods (Pan et al., 2022), which demonstrated that the multifunctional logic system had great potential for constructing biosensors for multiple mycotoxins. SERS aptasensors can also be designed to analyze multiple mycotoxins in one test. Song et al. (2022) developed an aptasensor based on diverse SERS tags and AuNP-modified 3D silica photonic crystal microsphere (SPCM) array with low LODs of 0.36 pg/mL for AFB1 and 0.034 pg/mL for OTA. Dual-mode aptasensors can also be developed for the simultaneous detection of multiple mycotoxins. For instance, based on the FRET and SERS manners, Wu et al. (2020) proposed an aptasensor for the simultaneous analysis of ZEN, FB1, and OTA. A long cDNA-labeled AuNP was designed to hybridize with the ZEN aptamer-labeled UCNP, the FB1 aptamer-labeled AuNP, and the OTA aptamer-labeled Cy5 simultaneously. In the presence of targets, the fluorescence signal of the UCNP or Cy5 increased, whereas the SERS signal of the AuNP decreased. Au nano-hybrid structures have emerged as important sensing materials, which could be synthesized to improve sensitivity and specificity. In Khan’s work, a fluorescence-labeled silica shell with Au NPs as the core was synthesized as a FL/SERS dual-mode nanoprobe for T-2 toxin. When exposed to T-2 toxin, the sensing system showed the concentration-dependent restoration of FL with the reduction of the SERS signal with the LOD of 85 p.m. Compared with ELISA, this method presented superior performance in wheat and maize (Khan et al., 2023b).
3.2 Optical aptasensor for marine toxins
Marine toxins, mainly generated by algae or phytoplankton during harmful algal blooms, are generally highly toxic. As they are continuously released in the environment, marine toxins are easily accumulated in aquatic and marine organisms such as mollusks and fishes through the food chain, finally posing a serious health threat to humans via consuming toxin-contaminated seafood (Wang et al., 2024). Thousands of marine toxins poisoning cases have been reported in the 21st century (Qiang et al., 2020). Marine toxin-contaminated food could lead to foodborne diseases such as food poisoning, diarrhea, indigestion, and neurotoxicity, even at low doses. In the United States, more than 3,000 people die owing to foodborne diseases annually. Therefore, it is highly necessary to design rapid, high-throughput, and cost-effective methods for the detection of multiple food contaminants. Thus, rapid detection of marine toxins in food is also an urgent task for health and safety. In this part, we summed up the optical aptasensors for typical marine toxins such as STX, OA, TTX, PTX, BTX, and DA (Nicolas et al., 2017).
Qiang et al. (2020) reported a colorimetric aptasensor based on salt-induced AuNP aggregation for quantitative analysis of STX at a concentration as low as 10 fM. By combining Au nanozymes and aptamer-triggered HCR, Li et al. proposed a colorimetric aptasensor for STX. By catalyzing the oxidation reaction of TMB-H2O2, an aptasensor with an LOD of 42.46 p.m. was achieved in real scallop samples (Zhao Y. et al., 2021). Fluorescent aptasensors have also been well developed for recognizing highly sensitive marine toxins. Xie et al. (2021) created an aptasensor for MC-LR analysis using FAM-labeled aptamers/AuNPs and DNase I, thus realizing fluorescence signal amplification. Shan et al. (2022) proposed a highly sensitive aptasensor for OA in seafood. In the presence of OA, they could bind with aptamers and produce long sequences with a poly(thymine) tail. The poly(thymine) tail becomes the copper nucleation sit. The fluorescence intensity derived from Cu nanoclusters could be in line with the OA concentrations. The aptasensor could achieve ultra-high sensitive detection of OA with an LOD of 1.1 pg/mL. Cheng reported an SERS-based aptasensor for STX for the first time. In the presence of STX, the STX would bind with aptamer, thus inducing tag molecules far from SERS substrate and the consequent SERS signal attenuation. This SERS aptasensor could be applied in shellfish samples with an LOD of 3.51 ng/mL (Cheng et al., 2019).
Introducing dual-mode sensing could improve the sensing performance of marine toxins in foods. Liu S. et al. (2022) demonstrated a fluorescence/SERS dual-mode aptasensor for the analysis of TTX. The Cy3 labeled aptamers were bound on the surface of novel AuNPs/MOF nanohybrids (MIL-101), which demonstrated fluorescence quenching and SERS enhancement. The presence of TTX would result in the fluorescence signal “turn on” and the SERS signal damping. This method showed distinguished sensitivity with an LOD of 6 pg/mL and demonstrated excellent practicability for screening TTX in puffer fish and clam. There were also novel aptasensors fabricated for sensing multiple typical marine toxins in one test. For example, Li et al. (2022) constructed a novel aptasensor for achieving simultaneous detection of three diarrheic shellfish toxins, including OA, DTX-1, and DTX-2. By integrating the TF-DSP aptamer with AuNPs@Fe2+ nanozyme activity, the assay showed good performance in real seafood samples.
3.3 Optical aptasensor of phytotoxins and bacterial toxins
Phytotoxins are naturally toxic plant-derived chemicals or proteins including siteloids, pyrrolizidine alkaloids, and lectins, and they could internalize into human cells and cause serious harm by inhibiting protein synthesis. Some of them are highly toxic to animals and humans if they contaminate food. Recent advancements in optical aptasensors have shown the availability for detection and monitoring (Günthardt et al., 2018). The reported aptamers for phytotoxins are mainly in ricin and abrin. Based on the unique peroxidase-like activity of aptamer-modified AuNPs, Hu et al. (2015) developed a simple colorimetric aptasensor for abrin (Figure 4). This method showed an LOD as low as 0.05 nM. Li et al. (2017) developed a novel strategy for analysis of ricin B-chain (RTB) based on isothermal strand-displacement polymerase reaction (ISDPR). In this design, a short blocker ssDNA was originally hybridized with the aptamer. In the presence of ricin, the blocker ssDNA was released and then hybridized with florescence-labeled hairpin probes to activate strong fluorescence. This technique could be applied to detect the RTB as well as the entire ricin toxin in the juice with an LOD of 0.6 mg/mL. To improve the performance of the “kinetic competition” aptasensor in complex matrices, Qi et al. proposed a ratiometric “kinetic competition” aptasensor using a dual fluorescence-labeled probe for RTA detection (Qi et al., 2020). The method was verified as a feasible method for RTA in sucrose, yeast, and baking soda powder samples.
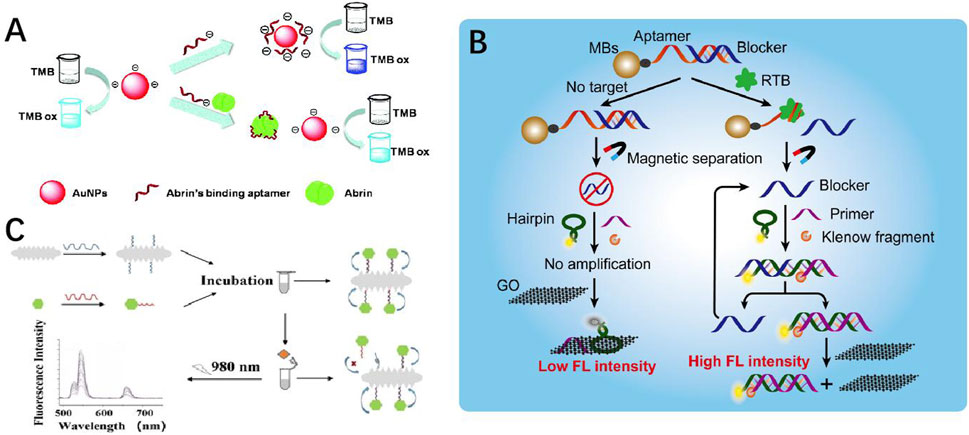
Figure 4. (A) Mechanism of the aptamer-based colorimetric biosensor for abrin (Ren et al., 2022). (B) Proposed ricin B-chain detection strategy (Chen et al., 2022). (C) Aptasensor based on AuNR@Pt-UCNPs for sensitive detection of SEB (Chen et al., 2016).
Bacterial toxins are a type of chemical derived from Clostridium, Salmonella, Staphylococcus, and Listeria pathogens that can invade host cells to cause foodborne infections in humans (Braun et al., 1994). Bacterial contamination has shown diverse causes, such as through unwashed hands, being present in raw milk or meat, and through contaminated water. By utilizing a similar aptamer-mediated gold nanoparticle aggregation mechanism, a rapid and easy colorimetric sensing was achieved for SEB using its aptamer and AuNPs in milk samples with LODs of 50 ng/mL visually and 0.5 ng/mL (Mondal et al., 2018). In addition, Wu et al. developed a fluorescent aptasensor for the analysis of SEB based on the aptamer-functionalized AuNR@Pt module and the cDNA-immobilized UCNP module. In another case, a fluorescent aptasensor for the analysis of SEA was designed by combining the aptamer functionalized AgNCs unit with the polypyrrole nanoparticles (PPyNPs). Aptamers could non-covalently bind onto the PPyNP, making the fluorescence of AgNCs turn on. The binding of SEA with the aptamer resulted in the release of AgNCs from the PPyNPs; therefore, the fluorescence was turned on under the stimulus of the target. In this method, an LOD of 0.3393 ng/mL was achieved in milk samples (Zhang et al., 2020).
3.4 Current challenges of optical aptasensors for biotoxins
In the past 3 decades, we have witnessed significant advancements in analytical techniques to manage the food safety problem of biotoxin contamination. UPLC–MS-based biotoxin analysis, including facilities, sophisticated analytical instruments, reagents, and logistics, made them unsuitable for daily monitoring. Currently, rich kinds of optical aptasensors were designed and they further demonstrated their real applications for biotoxins, including a variety of advanced analytical techniques and ideas, which could reduce the time and cost requirements. However, there are still some challenges that should be considered, such as authority, stability, and reliability, which make on-field rapid detection difficult. Here, we listed some significant challenges for rapid screening of biotoxins that are commonly encountered:
First, sample preparation in optical sensing is the inescapable critical step in real application. It was revealed that nearly 30% of the method variability originated from sample preparation. By paying attention to the key factors of sample preparation (size reduction, sampling size, and uniformity), the quality of rapid sensing results could be improved significantly. Considering the highly harmful nature of the biotoxins, the acceptable concentration range of different toxins varied in the same samples. Therefore, the sensing method should fully consider real needs. Third, food matrices are highly complex, which go through different processing processes, thus posing huge interferences for precise detection.
Rapid screening of biotoxins in foods shows huge market prospects in the future. Taking the mycotoxin testing market as an example, the testing market is set to reach a cumulative annual growth rate (CAGR) of 7.8%, which corresponds to a market of $1.4 billion by 2026 (EMR, 2022). As it indicates, food safety testing is an accelerated social need. In the light of this situation, the optical aptasensor should be further modified for higher precision while achieving the detection of multiple toxins simultaneously with high sensitivity and cost efficiency.
Author contributions
JS: conceptualization, investigation, writing–original draft, and writing–review and editing. MZ: writing–original draft and writing–review and editing. QG: investigation and writing–review and editing. BS: supervision and writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors are particularly grateful to Chunzheng Li for providing biotoxins for this research. They also thank Dingshuai Xue for help in writing the paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelsayed, M. M., Ho, B. T., Vu, M. M. K., Polanco, J., Spitale, R. C., and Lupták, A. (2017). Multiplex aptamer discovery through apta-seq and its application to ATP aptamers derived from human-genomic SELEX. ACS Chem. Biol. 12, 2149–2156. doi:10.1021/acschembio.7b00001
Ahuja, V., Singh, A., Paul, D., Dasgupta, D., Urajová, P., Ghosh, S., et al. (2023). Recent advances in the detection of food toxins using mass spectrometry. Chem. Res. Toxicol. 36, 1834–1863. doi:10.1021/acs.chemrestox.3c00241
Ali, M. M., Li, F., Zhang, Z., Zhang, K., Kang, D. K., Ankrum, J. A., et al. (2014). Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 43, 3324. doi:10.1039/c3cs60439j
Alshannaq, A., and Yu, J. H. (2017). Occurrence, toxicity, and analysis of major mycotoxins in food. Int. J. Environ. Res. Public Health 14, 632. doi:10.3390/ijerph14060632
Anderson, P. D. (2012). Bioterrorism: toxins as weapons. J. Pharm. Pract. 25, 121–129. doi:10.1177/0897190012442351
Bacha, S. A. S., Li, Y., Nie, J., Xu, G., Han, L., and Farooq, S. (2023). Comprehensive review on patulin and Alternaria toxins in fruit and derived products. Front. Plant Sci. 14, 1139757. doi:10.3389/fpls.2023.1139757
Balbinot, S., Srivastav, A. M., Vidic, J., Abdulhalim, I., and Manzano, M. (2021). Plasmonic biosensors for food control. Trends Food Sci. Technol. 111, 128–140. doi:10.1016/j.tifs.2021.02.057
Banach, J. L., Hoek-van den Hil, E. F., and van der Fels-Klerx, H. J. (2020). Food safety hazards in the European seaweed chain. Compr. Rev. Food Sci. Food Saf. 19, 332–364. doi:10.1111/1541-4337.12523
Bawazer, L. A., Newman, A. M., Gu, Q., Ibish, A., Arcila, M., Cooper, J. B., et al. (2014). Efficient selection of biomineralizing DNA aptamers using deep sequencing and population clustering. ACS Nano 8, 387–395. doi:10.1021/nn404448s
Bi, S., Yue, S., and Zhang, S. (2017). Hybridization chain reaction: a versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 46, 4281–4298. doi:10.1039/c7cs00055c
Bi, X., Luo, L., Li, L., Liu, X., Chen, B., and You, T. (2020). A FRET-based aptasensor for ochratoxin A detection using graphitic carbon nitride quantum dots and CoOOH nanosheets as donor-acceptor pair. Talanta 218, 121159. doi:10.1016/j.talanta.2020.121159
Braun, V., Pilsl, H., and Gross, P. (1994). Colicins: structures, modes of action, transfer through membranes, and evolution. Arch. Microbiol. 161, 199–206. doi:10.1007/BF00248693
Chen, J., Wen, J., Zhuang, L., and Zhou, S. (2016). An enzyme-free catalytic DNA circuit for amplified detection of aflatoxin B1 using gold nanoparticles as colorimetric indicators. Nanoscale 8, 9791–9797. doi:10.1039/c6nr01381c
Chen, L., Wen, F., Li, M., Guo, X., Li, S., Zheng, N., et al. (2017). A simple aptamer-based fluorescent assay for the detection of Aflatoxin B1 in infant rice cereal. Food Chem. 215, 377–382. doi:10.1016/j.foodchem.2016.07.148
Chen, R., Sun, Y., Huo, B., Mao, Z., Wang, X., Li, S., et al. (2021). Development of Fe3O4@Au nanoparticles coupled to Au@Ag core-shell nanoparticles for the sensitive detection of zearalenone. Anal. Chim. Acta 1180, 338888. doi:10.1016/j.aca.2021.338888
Chen, X. J., Huang, Y. K., Duan, N., Wu, S. J., Ma, X. Y., Xia, Y., et al. (2013). Selection and identification of ssDNA aptamers recognizing zearalenone. Anal. Bioanal. Chem. 405, 6573–6581. doi:10.1007/s00216-013-7085-9
Chen, X. J., Huang, Y. K., Duan, N., Wu, S. J., Xia, Y., Ma, X. Y., et al. (2014). Screening and identification of DNA aptamers against T-2 toxin assisted by graphene oxide. J. Agric. food Chem. 62, 10368–10374. doi:10.1021/jf5032058
Chen, Y., Meng, X., Lu, H., and Dong, H. (2022). Engineering DNA walkers for bioanalysis: a review. Anal. Chim. Acta 1209, 339339. doi:10.1016/j.aca.2021.339339
Cheng, S., Zheng, B., Yao, D., Wang, Y., Tian, J., Liu, L., et al. (2019). Determination of saxitoxin by aptamer-based surface-enhanced Raman scattering. Anal. Lett. 52, 902–918. doi:10.1080/00032719.2018.1505900
Cruz-Aguado, J. A., and Penner, G. (2008). Determination of ochratoxin A with a DNA aptamer. J. Agric. food Chem. 56, 10456–10461. doi:10.1021/jf801957h
DeGrasse, J. A. (2012). A single-stranded DNA aptamer that selectively binds to Staphylococcus aureus enterotoxin B. PloS one 7, e33410. doi:10.1371/journal.pone.0033410
Eissa, S., Ng, A., Siaj, M., Tavares, A. C., and Zourob, M. (2013). Selection and identification of DNA aptamers against okadaic acid for biosensing application. Anal. Chem. 85, 11794–11801. doi:10.1021/ac402220k
Eissa, S., Siaj, M., and Zourob, M. (2015). Aptamer-based competitive electrochemical biosensor for brevetoxin-2. Biosens. Bioelectron. 69, 148–154. doi:10.1016/j.bios.2015.01.055
Ellington, A. D., and Szostak, J. W. (1992). Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355, 850–852. doi:10.1038/355850a0
EMR (2022). Global mycotoxin testing market report and Forecast2023−2028; EMR. Available at: https://www.expertmarketresearch.com/reports/mycotoxin-testing-market (Accessed March 07, 2023).
Eskola, M., Kos, G., Elliott, C. T., Hajslova, J., Mayar, S., and Krska, R. (2020). Worldwide contamination of food-crops with mycotoxins: validity of the widely cited ‘FAO estimate’ of 25%. Crit. Rev. Food Sci. Nutr. 60, 2773–2789. doi:10.1080/10408398.2019.1658570
Fan, Y. Y., Li, J., Fan, L., Wen, J., Zhang, J., and Zhang, Z. Q. (2021). A label-free aptasensor based on a dual-emission fluorescent strategy for aflatoxin B1 detection. Sensors Actuators B-Chemical 346, 130561. doi:10.1016/j.snb.2021.130561
Fletcher, M. T., and Netzel, G. (2020). Food safety and natural toxins. Toxins (Basel) 12, 236. doi:10.3390/toxins12040236
Gao, S., Hu, B., Zheng, X., Cao, Y., Liu, D., Sun, M., et al. (2016). Gonyautoxin 1/4 aptamers with high-affinity and high-specificity: from efficient selection to aptasensor application. Biosens. Bioelectron. 79, 938–944. doi:10.1016/j.bios.2016.01.032
Gao, S., Zheng, X., Hu, B., Sun, M., Wu, J., Jiao, B., et al. (2017). Enzyme-linked, aptamer-based, competitive biolayer interferometry biosensor for palytoxin. Biosens. Bioelectron. 89, 952–958. doi:10.1016/j.bios.2016.09.085
Gu, H., Duan, N., Wu, S., Hao, L., Xia, Y., Ma, X., et al. (2016). Graphene oxide-assisted non-immobilized SELEX of okdaic acid aptamer and the analytical application of aptasensor. Sci. Rep. 6, 21665. doi:10.1038/srep21665
Gu, H., Duan, N., Xia, Y., Hun, X., Wang, H., and Wang, Z. (2018). Magnetic separation-based multiple SELEX for effectively selecting aptamers against saxitoxin, domoic acid, and tetrodotoxin. J. Agric. Food Chem. 66, 9801–9809. doi:10.1021/acs.jafc.8b02771
Gu, H., Hao, L., Ye, H., Ma, P., and Wang, Z. (2021). Nuclease-assisted target recycling signal amplification strategy for graphene quantum dot-based fluorescent detection of marine biotoxins. Mikrochim. Acta 188, 118. doi:10.1007/s00604-020-04684-y
Günthardt, B. F., Hollender, J., Hungerbühler, K., Scheringer, M., and Bucheli, T. D. (2018). Comprehensive toxic plants-phytotoxins database and its application in assessing aquatic micropollution potential. J. Agric. Food Chem. 66, 7577–7588. doi:10.1021/acs.jafc.8b01639
Ha, S. J., Park, J. H., Lee, B., and Kim, M. G. (2019). Label-free direct detection of saxitoxin based on a localized surface plasmon resonance aptasensor. Toxins 11, 274. doi:10.3390/toxins11050274
Handy, S. M., Yakes, B. J., DeGrasse, J. A., Campbell, K., Elliott, C. T., Kanyuck, K. M., et al. (2013). First report of the use of a saxitoxin-protein conjugate to develop a DNA aptamer to a small molecule toxin. Toxicon 61, 30–37. doi:10.1016/j.toxicon.2012.10.015
He, K., Sun, L., Wang, L., Li, W., Hu, G., Ji, X., et al. (2022). Engineering DNA G-quadruplex assembly for label-free detection of Ochratoxin A in colorimetric and fluorescent dual modes. J. Hazard. Mater. 423, 126962. doi:10.1016/j.jhazmat.2021.126962
He, Y., Yuan, J., Khan, I. M., Zhang, L., Ma, P., and Wang, Z. (2023). Research progress of aptasensor technology in the detection of foodborne pathogens. Food control. 153, 109891. doi:10.1016/j.foodcont.2023.109891
Hitabatuma, A., Pang, Y. H., Yu, L. H., and Shen, X. F. (2021). A competitive fluorescence assay based on free-complementary DNA for ochratoxin A detection. Food Chem. 342, 128303. doi:10.1016/j.foodchem.2020.128303
Hizir, M. S., Top, M., Balcioglu, M., Rana, M., Robertson, N. M., Shen, F., et al. (2016). Multiplexed activity of perAuxidase: DNA-capped AuNPs act as adjustable peroxidase. Anal. Chem. 88, 600–605. doi:10.1021/acs.analchem.5b03926
Howes, P. D., Chandrawati, R., and Stevens, M. M. (2014). Bionanotechnology. Colloidal nanoparticles as advanced biological sensors. Science 34, 1247390. doi:10.1126/science.1247390
Hu, J., Ni, P., Dai, H., Sun, Y., Wang, Y., Jiang, S., et al. (2015). Aptamer-based colorimetric biosensing of abrin using catalytic gold nanoparticles. Analyst 140, 3581–3586. doi:10.1039/c5an00107b
Huang, Y. K., Chen, X. J., Duan, N., Wu, S. J., Wang, Z. P., Wei, X. L., et al. (2015). Selection and characterization of DNA aptamers against Staphylococcus aureu enterotoxin C1. Food Chem. 166, 623–629. doi:10.1016/j.foodchem.2014.06.039
Huang, Y. K., Chen, X. J., Xia, Y., Wu, S. J., Duan, N., Ma, X. Y., et al. (2014). Selection, identification and application of a DNA aptamer against Staphylococcus aureu enterotoxin A. Anal. Methods 6, 690–697. doi:10.1039/c3ay41576g
Kaur, H., Shorie, M., and Sabherwal, P. (2020). Biolayer interferometry-SELEX for Shiga toxin antigenic-peptide aptamers and detection via chitosan-WSe2 aptasensor. Biosens. Bioelectron. 167, 112498. doi:10.1016/j.bios.2020.112498
Khan, I. M., Niazi, S., Akhtar, W., Yue, L., Pasha, I., Khan, M. K. I., et al. (2023a). Surface functionalized AuNCs optical biosensor as an emerging food safety indicator: fundamental mechanism to future prospects. Coord. Chem. Rev. 474, 214842. doi:10.1016/j.ccr.2022.214842
Khan, I. M., Niazi, S., Pasha, I., Khan, M. K. I., Yue, L., Ye, H., et al. (2023b). Novel metal enhanced dual-mode fluorometric and SERS aptasensor incorporating a heterostructure nanoassembly for ultrasensitive T-2 toxin detection. J. Mater. Chem. B 11, 441–451. doi:10.1039/d2tb01701f
Khan, I. M., Niazi, S., Yu, Y., Mohsin, A., Mushtaq, B. S., Iqbal, M. W., et al. (2019). Aptamer induced multicolored AuNCs-WS2 "turn on" fret nano platform for dual-color simultaneous detection of aflatoxinb1 and zearalenone. Anal. Chem. 91, 14085–14092. doi:10.1021/acs.analchem.9b03880
Khan, I. M., Niazi, S., Yu, Y., Pasha, I., Yue, L., Mohsin, A., et al. (2020). Fabrication of PAA coated green-emitting AuNCs for construction of label -free FRET assembly for specific recognition of T-2 toxin. Sensors Actuators B-Chemical 321, 128470. doi:10.1016/j.snb.2020.128470
Khan, I. M., Zhao, S., Niazi, S., Mohsin, A., Shoaib, M., Duan, N., et al. (2018). Silver nanoclusters based FRET aptasensor for sensitive and selective fluorescent detection of T-2 toxin. Sensors Actuators B-Chemical 277, 328–335. doi:10.1016/j.snb.2018.09.021
Lan, Y., Qin, G., Wei, Y., Wang, L., and Dong, C. (2020). Exonuclease I-assisted fluorescence aptasensor for tetrodotoxin. Ecotoxicol. Environ. Saf. 194, 110417. doi:10.1016/j.ecoenv.2020.110417
Lei, Z. L., and Guo, B. (2022). 2D material-based optical biosensor: status and prospect. Adv. Sci. (Weinh) 9, e2102924. doi:10.1002/advs.202102924
Lerdsri, J., Thunkhamrak, C., and Jakmunee, J. (2021). Development of a colorimetric aptasensor for aflatoxin B1 detection based on silver nanoparticle aggregation induced by positively charged perylene diimide. Food control. 130, 108323. doi:10.1016/j.foodcont.2021.108323
Li, C. H., Xiao, X., Tao, J., Wang, D. M., Huang, C. Z., and Zhen, S. J. (2017). A graphene oxide-based strand displacement amplification platform for ricin detection using aptamer as recognition element. Biosens. Bioelectron. 91, 149–154. doi:10.1016/j.bios.2016.12.010
Li, L., Ma, R., Zhao, Y., Wang, L., Wang, S., and Mao, X. (2022). Development of a colorimetric aptasensor fabricated with a group-specific aptamer and AuNPs@Fe2+ nanozyme for simultaneous detection of multiple diarrheic shellfish poisons. Talanta 246, 123534. doi:10.1016/j.talanta.2022.123534
Li, M., Chen, T., Gooding, J. J., and Liu, J. (2019). Review of carbon and graphene quantum dots for sensing. ACS Sens. 4, 1732–1748. doi:10.1021/acssensors.9b00514
Li, Z., Hu, B., Zhou, R., Zhang, X., Wang, R., Gao, Y., et al. (2020). Selection and application of aptamers with high-affinity and high-specificity against dinophysistoxin-1. RSC Adv. 10, 8181–8189. doi:10.1039/c9ra10600f
Lin, X., Li, C., Meng, X., Yu, W., Duan, N., Wang, Z., et al. (2022). CRISPR-Cas12a-mediated luminescence resonance energy transfer aptasensing platform for deoxynivalenol using gold nanoparticle-decorated Ti3C2Tx MXene as the enhanced quencher. J. Hazard Mater 433, 128750. doi:10.1016/j.jhazmat.2022.128750
Liu, B., Song, X., Lin, W. Y., Zhang, Y., Chen, B., and Zhang, B. Y. (2022a). Progress of optical biosensors for analyzing pathogens and organic pollutants in water since 2015. Environ. Rev. 30, 184–201. doi:10.1139/er-2021-0092
Liu, J., Zhang, Y., Xie, H., Zhao, L., Zheng, L., and Ye, H. (2019). Applications of catalytic hairpin assembly reaction in biosensing. Small 15, e1902989. doi:10.1002/smll.201902989
Liu, M., Zhang, J., Liu, S., and Li, B. (2022b). A label-free visual aptasensor for zearalenone detection based on target-responsive aptamer-cross-linked hydrogel and color change of gold nanoparticles. Food Chem. 389, 133078. doi:10.1016/j.foodchem.2022.133078
Liu, S., Huo, Y., Deng, S., Li, G., Li, S., Huang, L., et al. (2022c). A facile dual-mode aptasensor based on AuNPs@MIL-101 nanohybrids for ultrasensitive fluorescence and surface-enhanced Raman spectroscopy detection of tetrodotoxin. Biosens. Bioelectron. 201, 113891. doi:10.1016/j.bios.2021.113891
Loyez, M., DeRosa, M. C., Caucheteur, C., and Wattiez, R. (2022). Overview and emerging trends in optical fiber aptasensing. Biosens. Bioelectron. 196, 113694. doi:10.1016/j.bios.2021.113694
Lyu, C., Khan, I. M., and Wang, Z. (2021). Capture-SELEX for aptamer selection: a short review. Talanta 229, 122274. doi:10.1016/j.talanta.2021.122274
Ma, X. Y., Wang, W. F., Chen, X. J., Xia, Y., Duan, N., Wu, S. J., et al. (2015). Selection, characterization and application of aptamers targeted to Aflatoxin B2. Food control. 47, 545–551. doi:10.1016/j.foodcont.2014.07.037
Ma, X. Y., Wang, W. F., Chen, X. J., Xia, Y., Wu, S. J., Duan, N., et al. (2014). Selection, identification, and application of Aflatoxin B1 aptamer. Eur. Food Res. Technol. 238, 919–925. doi:10.1007/s00217-014-2176-1
Malhotra, S., Pandey, A. K., Rajput, Y. S., and Sharma, R. (2014). Selection of aptamers for aflatoxin M1 and their characterization. J. Mol. Recognit. 27, 493–500. doi:10.1002/jmr.2370
Mao, Z., Wang, X., Chen, R., Zhou, Z., Ren, S., Liang, J., et al. (2022). Upconversion-mediated CRISPR-Cas12a biosensing for sensitive detection of ochratoxin A. Talanta 242, 123232. doi:10.1016/j.talanta.2022.123232
McKeague, M., Bradley, C. R., De Girolamo, A., Visconti, A., Miller, J. D., and DeRosa, M. C. (2010). Screening and initial binding assessment of fumonisin B1 aptamers. Int. J. Mol. Sci. 11, 4864–4881. doi:10.3390/ijms11124864
McKeague, M., McConnell, E. M., Cruz-Toledo, J., Bernard, E. D., Pach, A., Mastronardi, E., et al. (2015). Analysis of in vitro aptamer selection parameters. J. Mol. Evol. 81, 150–161. doi:10.1007/s00239-015-9708-6
McKenzie, L. K., El-Khoury, R., Thorpe, J. D., Damha, M. J., and Hollenstein, M. (2021). Recent progress in non-native nucleic acid modifications. Chem. Soc. Rev. 50, 5126–5164. doi:10.1039/d0cs01430c
Mirón-Mérida, V. A., González-Espinosa, Y., Collado-González, M., Gong, Y. Y., Guo, Y., and Goycoolea, F. M. (2021). Aptamer-target-gold nanoparticle conjugates for the quantification of fumonisin B1. Biosens. (Basel) 11, 18. doi:10.3390/bios11010018
Mondal, B., Ramlal, S., Lavu, P. S., N, B., and Kingston, J. (2018). Highly sensitive colorimetric biosensor for staphylococcal enterotoxin B by a label-free aptamer and gold nanoparticles. Front. Microbiol. 9, 179. doi:10.3389/fmicb.2018.00179
Montecucco, C. (2012). 5. Bioterrorism and biological toxins. Toxicon 6, 99. doi:10.1016/j.toxicon.2012.04.006
Muhammad, M., and Huang, Q. (2021). A review of aptamer-based SERS biosensors: design strategies and applications. Talanta 227, 122188. doi:10.1016/j.talanta.2021.122188
Mukherjee, M., Appaiah, P., Sistla, S., Bk, B., and Bhatt, P. (2022). Bio-layer interferometry-based SELEX and label-free detection of patulin using generated aptamer. J. Agric. food Chem. 70, 6239–6246. doi:10.1021/acs.jafc.2c01591
Ng, A., Chinnappan, R., Eissa, S., Liu, H., Tlili, C., and Zourob, M. (2012). Selection, characterization, and biosensing application of high affinity congener-specific microcystin-targeting aptamers. Environ. Sci. Technol. 46, 10697–10703. doi:10.1021/es301686k
Niazi, S., Khan, I. M., Yan, L., Khan, M. I., Mohsin, A., Duan, N., et al. (2019a). Simultaneous detection of fumonisin1 and ochratoxin A using dual-color, time-resolved luminescent nanoparticles (NaY4:Ce, Tb and NH2-Eu/DPA@SiO2) as labels. Anal. Bioanal. Chem. 411, 1453–1465. doi:10.1007/s00216-019-01580-0
Niazi, S., Khan, I. M., Yu, Y., Pasha, I., Lv, Y., Mohsin, A., et al. (2020). A novel fluorescent aptasensor for aflatoxin M1 detection using rolling circle amplification and g-C3N4 as fluorescence quencher. Sensors Actuators B-Chemical 315, 128049. doi:10.1016/j.snb.2020.128049
Niazi, S., Khan, I. M., Yu, Y., Pasha, I., Shoaib, M., Mohsin, A., et al. (2019b). A "turn-on" aptasensor for simultaneous and time-resolved fluorometric determination of zearalenone, trichothecenes A and aflatoxin B1 using WS2 as a quencher. Microchim. Acta 186, 575. doi:10.1007/s00604-019-3570-y
Niazi, S., Khan, I. M., Yue, L., Ye, H., Lai, B., Korma, S. A., et al. (2022). Nanomaterial-based optical and electrochemical aptasensors: a reinforced approach for selective recognition of zearalenone. Food control. 142, 109252. doi:10.1016/j.foodcont.2022.109252
Nicolas, J., Hoogenboom, R. L. A. P., Hendriksen, P. J. M., Bodero, M., Bovee, T. F. H., Rietjens, I. M. C. M., et al. (2017). Marine biotoxins and associated outbreaks following seafood consumption: prevention and surveillance in the 21st century. Glob. Food Security-Agriculture Policy Econ. Environ. 15, 11–21. doi:10.1016/j.gfs.2017.03.002
Oliveira, J., Cunha, A., Castilho, F., Romalde, J. L., and Pereira, M. J. (2011). Microbial contamination and purification of bivalve shellfish: crucial aspects in monitoring and future perspectives – a mini-review. Food control. 22, 805–816. doi:10.1016/j.foodcont.2010.11.032
Pan, J., Deng, F., Liu, Z., Shi, G., and Chen, J. (2022). Toehold-mediated cascade catalytic assembly for mycotoxin detection and its logic applications. Anal. Chem. 94, 3693–3700. doi:10.1021/acs.analchem.1c05485
Pan, W., and Clawson, G. A. (2010). Primer-free aptamer selection using a random DNA library. Methods Mol. Biol. 629, 369–385. doi:10.1007/978-1-60761-657-3_24
Picardo, M., Núñez, O., and Farré, M. (2020). Suspect and target screening of natural toxins in the ter river catchment area in NE Spain and prioritisation by their toxicity. Toxins (Basel) 12, 752. doi:10.3390/toxins12120752
Qi, L., Han, X., and Du, Y. (2020). Improved sensitivity for ratiometric fluorescence detection of ricin based on "kinetic competition" aptasensing strategy. Sensors Actuators B-Chemical 314, 128073. doi:10.1016/j.snb.2020.128073
Qiang, L., Zhang, Y., Guo, X., Gao, Y., Han, Y., Sun, J., et al. (2020). A rapid and ultrasensitive colorimetric biosensor based on aptamer functionalized Au nanoparticles for detection of saxitoxin. Rsc Adv. 10, 15293–15298. doi:10.1039/d0ra01231a
Radom, F., Jurek, P. M., Mazurek, M. P., Otlewski, J., and Jeleń, F. (2013). Aptamers: molecules of great potential. Biotechnol. Adv. 31, 1260–1274. doi:10.1016/j.biotechadv.2013.04.007
Rather, I. A., Koh, W. Y., Paek, W. K., and Lim, J. (2017). The sources of chemical contaminants in food and their health implications. Front. Pharmacol. 8, 830. doi:10.3389/fphar.2017.00830
Ren, W., Pang, J., Ma, R., Liang, X., Wei, M., Suo, Z., et al. (2022). A signal on-off fluorescence sensor based on the self-assembly DNA tetrahedron for simultaneous detection of ochratoxin A and aflatoxin B1. Anal. Chim. Acta 1198, 339566. doi:10.1016/j.aca.2022.339566
Sarkar, D. J., Behera, B. K., Parida, P. K., Aralappanavar, V. K., Mondal, S., Dei, J., et al. (2023). Aptamer-based NanoBioSensors for seafood safety. Biosens. Bioelectron. 219, 114771. doi:10.1016/j.bios.2022.114771
Shan, W., Sun, J., Liu, R., Xu, W., and Shao, B. (2022). Duplexed aptamer-isothermal amplification-based nucleic acid-templated copper nanoparticles for fluorescent detection of okadaic acid. Sensors Actuators B-Chemical 352, 131035. doi:10.1016/j.snb.2021.131035
Shkembi, X., Svobodova, M., Skouridou, V., Bashammakh, A. S., Alyoubi, A. O., and O'Sullivan, C. K. (2021). Aptasensors for mycotoxin detection: a review. Anal. Biochem. 7, 644. doi:10.1016/j.ab.2021.114156
Song, L., Li, J., Li, H., Chang, Y., Dai, S., Xu, R., et al. (2022). Highly sensitive SERS detection for Aflatoxin B1 and Ochratoxin A based on aptamer-functionalized photonic crystal microsphere array. Sensors Actuators B-Chemical 364, 131778. doi:10.1016/j.snb.2022.131778
Song, S. H., Gao, Z. F., Guo, X., and Chen, G. H. (2019). Aptamer-based detection methodology studies in food safety. Food Anal. Methods 12, 966–990. doi:10.1007/s12161-019-01437-3
Stoltenburg, R., Reinemann, C., and Strehlitz, B. (2005). FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 383, 83–91. doi:10.1007/s00216-005-3388-9
Tan, X., Yu, W., Wang, Y., Song, P., Xu, Q., Ming, D., et al. (2021). A switchable and signal-amplified aptasensor based on metal organic frameworks as the quencher for turn-on detection of T-2 mycotoxin. Anal. Bioanal. Chem. 413, 6595–6603. doi:10.1007/s00216-021-03625-9
Tang, L., and Li, J. (2017). Plasmon-Based Colorimetric nanosensors for ultrasensitive molecular diagnostics. ACS Sens. 2, 857–875. doi:10.1021/acssensors.7b00282
Tang, Y., Yuan, J., Zhang, Y., Khan, I. M., Ma, P., and Wang, Z. (2024). Lateral flow assays based on aptamers for food safety applications. Food control. 155, 110051. doi:10.1016/j.foodcont.2023.110051
Tian, R. Y., Lin, C., Yu, S. Y., Gong, S., Hu, P., Li, Y. S., et al. (2016). Preparation of a specific ssDNA aptamer for brevetoxin-2 using SELEX. J. Anal. methods Chem. 2016, 1–8. doi:10.1155/2016/9241860
Tok, J. B., and Fischer, N. O. (2008). Single microbead SELEX for efficient ssDNA aptamer generation against botulinum neurotoxin. Chem. Commun. Camb. Engl. 16, 1883–1885. doi:10.1039/b717936g
Tuerk, C., and Gold, L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510. doi:10.1126/science.2200121
Wang, H., Zhao, B., Ye, Y., Qi, X., Zhang, Y., Xia, X., et al. (2022). A fluorescence and surface-enhanced Raman scattering dual-mode aptasensor for rapid and sensitive detection of ochratoxin A. Biosens. Bioelectron. 207, 114164. doi:10.1016/j.bios.2022.114164
Wang, K., He, B., Xie, L., Li, L., Yang, J., Liu, R., et al. (2021). Exonuclease III-assisted triple-amplified electrochemical aptasensor based on PtPd NPs/PEI-rGO for deoxynivalenol detection. Sensors Actuators B-Chemical 349, 130767. doi:10.1016/j.snb.2021.130767
Wang, Y. F., Javeed, A., Jian, C. Q., Zeng, Q. Y., and Han, B. N. (2024). Precautions for seafood consumers: an updated review of toxicity, bioaccumulation, and rapid detection methods of marine biotoxins. Ecotoxicol. Environ. Saf. 274, 116201. doi:10.1016/j.ecoenv.2024.116201
Wei, X., Ma, P., Mahmood, K. I., Zhang, Y., and Wang, Z. (2023). A review: construction of aptamer screening methods based on improving the screening rate of key steps. Talanta 253, 124003. doi:10.1016/j.talanta.2022.124003
WHO (2015). Estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007−2015. World Health Organization. Available at: https://www.who.int/publications/i/item/9789241565165
Wu, Z., He, D., Cui, B., Jin, Z., Xu, E., Yuan, C., et al. (2020). Trimer-based aptasensor for simultaneous determination of multiple mycotoxins using SERS and fluorimetry. Microchim. Acta 18, 495. doi:10.1007/s00604-020-04487-1
Wu, Z., Sun, D. W., Pu, H., Wei, Q., and Lin, X. (2022). Ti3C2Tx MXenes loaded with Au nanoparticle dimers as a surface-enhanced Raman scattering aptasensor for AFB1 detection. Food Chem. 372, 131293. doi:10.1016/j.foodchem.2021.131293
Xie, W., He, S., Fang, S., Liang, L., Shi, B., and Wang, D. (2021). Visualizing of AuNPs protection aptamer from DNase I enzyme digestion based on Nanopipette and its use for Microcystin-LR detection. Anal. Chim. Acta 1173, 338698. doi:10.1016/j.aca.2021.338698
Xiong, Z., Wang, Q., Xie, Y., Li, N., Yun, W., and Yang, L. (2021). Simultaneous detection of aflatoxin B1 and ochratoxin A in food samples by dual DNA tweezers nanomachine. Food Chem. 338, 128122. doi:10.1016/j.foodchem.2020.128122
Yang, W., Yu, H., Alkhamis, O., Liu, Y., Canoura, J., Fu, F., et al. (2019). In vitro isolation of class-specific oligonucleotide-based small-molecule receptors. Nucleic Acids Res. 47, e71. doi:10.1093/nar/gkz224
Yang, Y., Yin, Y., Wang, S., and Dong, Y. (2021). Simultaneous determination of zearalenone and ochratoxin A based on microscale thermophoresis assay with a bifunctional aptamer. Anal. Chim. Acta 1155, 338345. doi:10.1016/j.aca.2021.338345
Yu, H., Alkhamis, O., Canoura, J., Liu, Y., and Xiao, Y. (2021). Advances and challenges in small-molecule DNA aptamer isolation, characterization, and sensor development. Angew. Chem. Int. Ed. Engl. 60, 16800–16823. doi:10.1002/anie.202008663
Zhang, N., Li, J., Liu, B., Zhang, D., Zhang, C., Guo, Y., et al. (2022a). Signal enhancing strategies in aptasensors for the detection of small molecular contaminants by nanomaterials and nucleic acid amplification. Talanta 236, 122866. doi:10.1016/j.talanta.2021.122866
Zhang, Q., Kang, L., Yue, P., Shi, L., Wang, M., Zhou, L., et al. (2022b). Development of a graphene oxide nanosheet and double-stranded DNA structure based fluorescent "signal off" aptasensor for ochratoxin A detection in malt. Food Chem. 14, 100308. doi:10.1016/j.fochx.2022.100308
Zhang, X., Khan, I. M., Ji, H., Wang, Z., Tian, H., Cao, W., et al. (2020). A label-free fluorescent aptasensor for detection of staphylococcal enterotoxin A based on aptamer-functionalized silver nanoclusters. Polym. (Basel) 12, 152. doi:10.3390/polym12010152
Zhao, P., Liu, H., Zhu, P., Ge, S., Zhang, L., Zhang, Y., et al. (2021b). Multiple cooperative amplification paper SERS aptasensor based on AuNPs/3D succulent-like silver for okadaic acid quantization. Sensors Actuators B-Chemical 344, 130174. doi:10.1016/j.snb.2021.130174
Zhao, X., Wang, Y., Li, J., Huo, B., Huang, H., Bai, J., et al. (2021a). A fluorescence aptasensor for the sensitive detection of T-2 toxin based on FRET by adjusting the surface electric potentials of UCNPs and MIL-101. Anal. Chim. Acta 1160, 338450. doi:10.1016/j.aca.2021.338450
Keywords: biotoxins, aptamers, food safety, optical, biosensors
Citation: Sun J, Zhang M, Gao Q and Shao B (2024) Screening biotoxin aptamer and their application of optical aptasensor in food stuff: a review. Front. Chem. 12:1425774. doi: 10.3389/fchem.2024.1425774
Received: 30 April 2024; Accepted: 28 June 2024;
Published: 24 July 2024.
Edited by:
Shouyu Wang, Jiangnan University, ChinaReviewed by:
Ying Xiong, Central South University of Forestry and Technology, ChinaImran Mahmood Khan, The University of Nottingham Ningbo, China
Copyright © 2024 Sun, Zhang, Gao and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Shao, c2hhb2JpbmdjaEBzaW5hLmNvbQ==
 Jiefang Sun
Jiefang Sun Meng Zhang2
Meng Zhang2 Bing Shao
Bing Shao