- Department of Chemistry, Faculty of Science, Tokyo University of Science, Tokyo, Japan
We prepared L-amino acids (L-valine and L-serine, respectively) based on the Schiff base Cu2+ complexes CuSV and CuSS in the absence/presence of hydroxyl groups and their imidazole-bound compounds CuSV-Imi and CuSS-Imi to reveal the effects of hydroxyl groups on SOD activity. The structural and spectroscopic features of the Cu2+ complexes were evaluated using X-ray crystallography, UV-vis spectroscopy, and EPR spectroscopy. The spectroscopic behavior upon addition of lysozyme indicated that both CuSV and CuSS were coordinated by the imidazole group of His15 in lysozyme at their equatorial position, leading to the formation of hybrid proteins with lysozyme. CuSS-Imi showed a higher SOD activity than CuSV-Imi, indicating that the hydroxyl group of CuSS-Imi played an important role in the disproportionation of O2− ion. Hybridization of the Cu2+ complexes CuSV and CuSS with lysozyme resulted in higher SOD activity than that of CuSV-Imi and CuSS-Imi. The improvements in SOD activity suggest that there are cooperative effects between Cu2+ complexes and lysozyme.
1 Introduction
Reactive oxygen species (ROS), such as hydroxyl radicals (•OH), singlet oxygen (1O2), hydrogen peroxide (H2O2), and superoxide (O2−) are unavoidable byproducts of respiration in aerobic organisms. These ROS cause serious oxidative damage to biomolecules such as lipids, proteins, and nucleic acids. Among these, O2− is generated by the one-electron reduction of dioxygen (O2) upon enzymatic oxidation and oxygen delivery (Pryor, 1986). In addition to damaging biomolecules, O2− also induces the generation of other ROS, such as •OH and H2O2, under protic conditions (Hayyan et al., 2016). To remove O2− from their bodies, organisms have superoxide dismutase (SOD), which catalyzes the disproportionation of O2− to H2O2 and O2, as shown below.
As SODs play an important role in protecting biomolecules from oxidative damage, animals with higher SOD activity generally have a longer life expectancy (Tolmasoff et al., 1980).
SODs contain metal ion(s) in their active centers and catalyze the disproportionation of O2− as shown below.
At present, SODs typically contain Mn, Fe, Ni, Cu, and Zn as their central metals. SODs can be classified as MnSOD (McCord and Fridovich, 1969; Abreu et al., 2005; Kanematsu and Asada, 1979; Fréalle et al., 2005; del Rio et al., 2003; Sheng et al., 2014), FeSOD (Yost and Fridovich, 1973; Hatchikian and Henry, 1977; Opperdoes et al., 1977; Kirby et al., 1981; Pilon et al., 2011), NiSOD (Youn et al., 1996a; 1996b; Palenik et al., 2003; Barondeau et al., 2004; Wuerges et al., 2004), and CuZnSOD (Mann and Keilin, 1938; Goscin and Fridovich, 1972; Alscher et al., 2022), respectively. In this study, we focused on CuZnSOD, which has a dimetallic active center composed of Cu2+ and Zn2+ ions. While Zn2+ ions fix the secondary coordination sphere of the active center of CuZnSOD, Cu2+ ions catalyze the disproportionation of O2− ions, as shown below (Hart et al., 1999).
However, to apply native CuZnSOD as an O2− removing reagent, problems such as its poor stability and high cost must be addressed (O’Connor et al., 2012). To address these problems, hybridization of proteins and SOD-active metal complexes with small molecular weights has gained significant attention. In practice, ROS-removing ability is reported in Cu2+-bound albumin (Kato et al., 2014).
In this study, we focused on lysozyme because of its stability under ambient conditions. In addition, the His15 side chain of lysozyme is known as the binding site of metal ions such as Mn+ (Razavet et al., 2007), Ag+ (Panzner et al., 2011), Au+ (Ferraro et al., 2019), and Pt2+ (Casini et al., 2007).
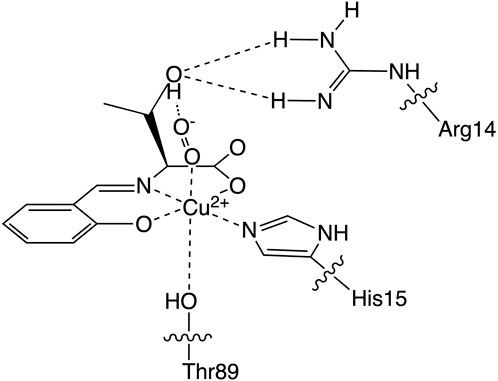
Based on these lysozyme properties, we have reported a hybrid protein composed of an SOD-active Cu2+ complex coordinated by a threonine derivative (CuST) and lysozyme (CuST@lysozyme), as shown in Figure 1 (Furuya et al., 2023). We determined its crystal structure, spectroscopic and electrochemical properties, and SOD activity. According to the crystallographic analysis, the Cu2+ center of CuST@lysozyme was coordinated by the imidazole group of His15 and the hydroxyl group of Thr89 residues of the lysozyme at the equatorial and apical positions, respectively. In addition, the hydroxyl group of CuST formed hydrogen bonds with the side chain of Arg14 of the lysozyme. The hybrid lysozyme exhibited SOD activity comparable to that of CuST.
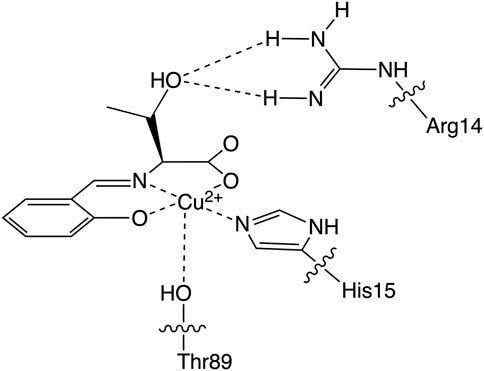
FIGURE 1. The Cu2+ center structure of the hybrid protein composed of the Cu2+ complex (CuST) and lysozyme.
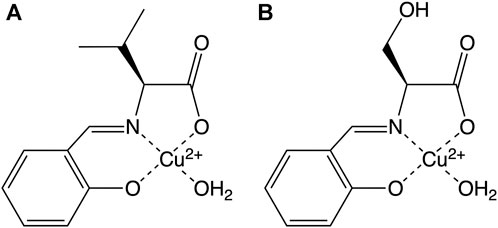
Since we speculated that the hydrogen bonds formed between the hydroxyl group of the Cu2+ complex and the Arg14 residue of lysozyme play an important role in the SOD activity, we prepared two Cu2+ complexes in the absence/presence of the hydroxyl group (CuSV and CuSS, respectively), as shown in Figure 2 to evaluate the hydrogen bond effects on their SOD activity. We revealed their crystal structure and spectroscopic and electrochemical properties and investigated their hybridization with lysozyme. In addition, we evaluated the O2− disproportionation activity of Cu2+ complex-bound hybrid lysozymes and discussed the effects of hydrogen bonds on SOD activity.
2 Materials and methods
2.1 Materials
Salicylaldehyde was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan), and the solvents were purchased from Kanto Chemical Co., Inc. (Tokyo, Japan). Other reagents were purchased from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan). All reagents were of the highest commercial grade and used without further purification.
2.2 Preparation of Cu2+ complexes
2.2.1 Preparation of CuSV
L-valine (118.0 mg, 1.01 mmol) and salicylaldehyde (121.5 mg, 1.00 mmol) were dissolved in 20 mL of methanol. The solution was subjected to microwave irradiation (Otani et al., 2021) at 358 K for 5 min to yield a yellow ligand solution. Copper (II) acetate monohydrate (200.9 mg, 1.01 mmol) was added to the solution and additionally treated with microwave irradiation at 358 K for 5 min to yield a green solution. The solution was placed under ambient conditions for 3–5 days to obtain green crystals suitable for X-ray analysis (yield: 135 mg, 43.8%).
The elemental analysis results calculated for C12H15NO4Cu are as follows: C: 47.92%, H: 5.03%, N: 4.66%, found, C: 47.78%, H: 5.12%, N: 4.61%. IR:1,468 cm−1 (νsym (C=O)), 1,600 cm−1 (νas (C=O)), 1,639 cm−1 (ν (C=N)), 3,232 cm−1 (ν (O−H)).
2.2.2 Preparation of CuSV-Imi
L-valine (117.1 mg, 1.00 mmol) and salicylaldehyde (122.2 mg, 1.00 mmol) were dissolved in 20 mL of methanol. The solution was subjected to microwave irradiation at 358 K for 5 min to yield a yellow ligand solution. Copper (II) acetate monohydrate (200.0 mg, 1.00 mmol) was added to the solution and additionally treated with microwave irradiation at 358 K for 5 min to yield a green solution. Imidazole (71.5 mg, 1.05 mmol) was added to the green solution, which was then microwave-irradiated at 358 K for 5 min to yield a dark green solution. The solution was placed under ambient conditions for 3–5 days, and dark-green crystals suitable for X-ray analysis were obtained (yield: 180.5 mg, 51.6%).
The elemental analysis results calculated for C15H17N3O3Cu are as follows: C: 51.35%, H: 4.88%, N: 11.98%, found, C: 51.27%, H: 4.92%, N: 12.03%. IR:1,465 cm−1 (νsym (C=O)), 1,586 cm−1 (νas (C=O)), 1,632 cm−1 (ν (C=N)).
2.2.3 Preparation of CuSS
L-serine (110.0 mg, 1.05 mmol) and salicylaldehyde (129.2 mg, 1.06 mmol) were dissolved in 20 mL of methanol. The solution was subjected to microwave irradiation at 358 K for 5 min to yield a yellow ligand solution. Copper (II) acetate monohydrate (203.0 mg, 1.02 mmol) was added to the solution and additionally treated with microwave irradiation at 358 K for 5 min to yield a green solution. The solution was placed under ambient conditions for 3–5 days to obtain green crystals (yield: 135 mg, 43.8%).
The elemental analysis results calculated for C10H11NO5Cu·H2O are as follows: C: 39.15%, H: 4.27%, N: 4.57%, found, C: 38.98%, H: 4.25%, N: 4.50%. IR:1,465 cm−1 (νsym (C=O)), 1,602 cm−1 (νas (C=O)), 1,628 cm−1 (ν (C=N)), 3,132, 3,236, 3,236 cm−1 (ν (O−H)).
2.2.4 Preparation of CuSS-Imi
L-serine (110.0 mg, 1.05 mmol) and salicylaldehyde (126.6 mg, 1.04 mmol) were dissolved in 20 mL of methanol. The solution was subjected to microwave irradiation at 358 K for 5 min to yield a yellow ligand solution. Copper (II) acetate monohydrate (201.6 mg, 1.01 mmol) was added to the solution and additionally treated with microwave irradiation at 358 K for 5 min to yield a green solution. Imidazole (68.9 mg, 1.01 mmol) was added to the green solution, which was then microwave-irradiated at 358 K for 5 min to yield a dark green solution. The solution was placed under ambient conditions for 3–5 days to obtain dark green crystals (yield: 150.7 mg, 44.6%).
The elemental analysis results calculated for C13H13N3O4Cu are as follows: C: 46.09%, H: 3.87%, N: 12.40%, found, C: 45.74%, H: 3.94%, N: 12.34%. IR: 1,462 cm−1 (νsym (C=O)), 1,600 cm−1 (νas (C=O)), and 1,626 cm−1 (ν (C=N)), 3,134 cm−1 (ν (O−H)).
2.3 Physical measurement
Microwave synthesis was performed using a Biotage initiator+. Elemental analyses (C, H, and N) were performed using a Vario EL cube analyzer at the Nagoya Institute of Technology. IR spectra were recorded on a JASCO FT-IR 4200 spectrophotometer in the range of 4,000–400 cm−1 at 298 K. UV–vis spectra were measured using a JASCO V-570 spectrophotometer in the range of 900–200 nm at room temperature. The solution concentrations ranged from 0.02 to 2.0 mM, and quartz cells (path length: 1.0 cm) were used. Electron paramagnetic resonance (EPR) spectra were recorded on a Bruker EMX-nano X-band EPR spectrometer at 77 K using 1 mM solutions of CuSV-Imi, CuSS-Imi, CuSV@lysozyme, and CuSS@lysozyme in quartz tubes as samples in quartz tubes. The cyclic voltammograms were measured by ALS/DY2323 BI-POTENTIOSTAT in a 0.1 M phosphate buffer solution (pH 7.0). Glassy carbon, Pt wire, and Ag/AgCl were used as the working, counter, and reference electrodes, respectively. The CV curves of CuSV-Imi (1.0 mM), CuSS-Imi (1.0 mM), CuSV@Lysozyme (1.0 mM CuSV +1.0 mM lysozyme), and CuSS@lysozyme (1.0 mM CuSS +1.0 mM lysozyme) were measured over four cycles under a nitrogen atmosphere at 298 K with a sweep rate of 100 mV/s. Fluorescence spectra were measured using a Jasco FP-6200 spectrofluorometer. The fluorescence wavelengths were in the range of 220–660 nm. The excitation wavelength was 260 nm. The concentrations of the solutions were in the range of 0–4.0 μM (CuSV, CuSS) + 0 or 4.0 μM (lysozyme).
2.4 X-ray crystallographic analysis
Single crystals of CuSV and CuSV-Imi were glued on top of the glass fibers and coated with a thin layer of epoxy resin to obtain the diffraction data. The intensity data were collected on a Bruker APEX2 CCD diffractometer with graphite-monochromated Mo Kα radiation (λ = 0.71073 Å). Data analysis was performed using the SAINT software package. The structures were solved by direct methods using SHELXS-97, expanded by Fourier techniques, and refined by full-matrix least-squares methods based on F2 using SHELXL-97 (Sheldrick, 2008). The empirical absorption correction was applied using the SADABS program. All non-hydrogen atoms were readily located and refined using anisotropic thermal parameters. All hydrogen atoms were located in geometrically calculated positions and refined using riding models.
2.5 Evaluation of SOD activities
The SOD activities of the Cu2+ complexes CuSV-Imi, CuSS-Imi, CuSV@lysozyme, and CuSS@lysozyme were evaluated using the WST SOD assay kit (Dojindo, Tokyo, Japan), which is based on the xanthine-xanthine oxidase method, according to the manufacturer’s instructions. The evaluation was performed in phosphate buffer (0.1 M, pH = 7.0) at 310 K with 2-(4-Iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt as indicator. IC50 values were determined by inhibition rates of 2000, 400, 80 and 16 μM solutions of the Cu2+ complexes. Absorbance was measured at 450 nm using a JASCO V-570 UV-vis spectrophotometer.
2.6 Computational details
All calculations were carried out with the Gaussian 09 package (Frisch et al., 2009) and all geometries of O2−-bound Cu2+ complexes in the triplet (S = 1) state were optimized at the density functional theory (DFT) level using B3LYP (Lee et al., 1988) functionals with 6-31+G(d) basis set (Clark et al., 1983) for all atoms. GaussView 5 (Dennington et al., 2009) was used to generate the starting structures and to visualize the optimized structures.
3 Results and discussion
3.1 Syntheses and crystal structures of the Cu2+ complexes
The ligands of the Cu2+ complexes were prepared by a reaction between salicylaldehyde and the corresponding L-amino acids (L-valine and L-serine). An equivalent amount of copper (II) acetate was added to the ligand solutions to form green CuSV and CuSS crystals. From the two Cu2+ complexes, a single CuSV crystal suitable for X-ray crystallographic analysis was obtained.
According to X-ray analysis, the crystal system of CuSV was orthorhombic, and judging from the systematic absence, the space group was P212121 (Akiyama et al., 2023). In the unit cell, we observed two Cu2+ complexes with a four- and five-coordinated structure, as shown in Figure 3. CuSV with a four-coordinated structure was bound by the phenolato O atom, carboxylato O atom, and imino N atom of the ligand and one water molecule on the equatorial plane. Based on the sum of the bond angles around the Cu2+ center (359.4°), the four-coordinated unit was concluded to have a square planar structure with an extremely small distortion. In contrast, the other Cu2+ complex with a five-coordinated structure was additionally coordinated with another water molecule at the apical position of the plane. Although the coordination of the apical water molecule was too weak (Cu2−O9 = 2.3663 (16), the Cu−N and Cu−O bonds were slightly longer than those of the four-coordination unit. The weak coordination of the apical water molecule is consistent to the elemental analysis of the powdered CuSV. According to the elemental analysis, powdered CuSV contains only one water molecule, indicating the apical water molecule is easily leave from the powdered CuSV. The small τ value (τ = 0.04) indicates that the structure of the five-coordinated unit is square pyramidal (Addison et al., 1984). These crystal structures are similar with previously reported Cu2+ complexes with the similar amino acid moiety (Katsuumi et al., 2020; Otani et al., 2021; Suzuki et al., 2023).
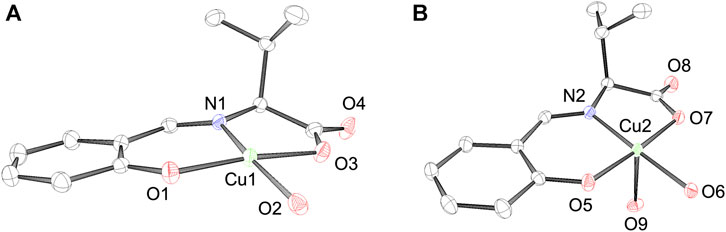
FIGURE 3. ORTEP drawings of Cu2+ complex CuSV with a (A) 4-coordinated and a (B) 5-coordinated structure. The thermal ellipsoids are drawn at 50% probability. Hydrogen atoms are omitted for clarity. Selected bond length (Å) and bond angles (degree): Cu1−N1 = 1.9205 (16), Cu1−O1 = 1.8955 (15), Cu1−O2 = 1.9339 (16), Cu1−O3 = 1.9629 (15), N1−Cu1−O1 = 94.40 (7), O1−Cu1−O2 = 94.04 (7), O2−Cu1−O3 = 88.01 (7), N1−Cu1−O3 = 82.90 (7), N1−Cu1−O2 = 170.16 (7), O1−Cu1−O3 = 171.32 (7), for structure (A), Cu2−N2 = 1.9243 (17), Cu2−O5 = 1.9432 (14), Cu2−O6 = 1.9411 (15), Cu2−O7 = 1.9956 (14), Cu2−O9 = 2.3663 (16), N2−Cu2−O5 = 92.86 (6), O5−Cu2−O6 = 89.41 (7), O6−Cu2−O7 = 95.00 (7), N2−Cu2−O7 = 82.74 (6), N2−Cu2−O9 = 93.05 (7), O5−Cu2−O9 = 97.98 (6), O6−Cu2−O9 = 86.56 (7), O7−Cu2−O9 = 82.34 (6), N2−Cu2−O6 = 177.73 (7), O5−Cu2−O7 = 175.60 (6), for structure (B), respectively.
Because we estimated the His15 side chain of lysozyme to be the Cu2+ complex-binding site, we also prepared imidazole-binding CuSV and CuSS (CuSV-Imi and CuSS-Imi) as lysozyme-bound structural models. CuSV-Imi and CuSS-Imi were synthesized by the reaction between CuSV/CuSS and an equivalent amount of imidazole. The reaction solutions were kept at 298 K, and dark green crystals were observed. Of the two crystals, CuSV-Imi yielded single crystals suitable for crystallographic analysis. Based on crystallographic analysis and the systematic absence, the crystal system and space group of CuSV-Imi were monoclinic and P21, respectively. The crystal structure of CuSV-Imi is shown in Figure 4. Upon reaction with imidazole, the water molecules of CuSV on the equatorial plane are replaced to form a square planar structure. Based on the bond angles around the Cu2+ center (359.3°), the square planar structure exhibits a small distortion. Through the coordination of imidazole on the plane, the other Cu−N and Cu−O bonds were slightly stretched. These bond elongations can be explained by the stronger electron donation of the imidazole compared to that of the water molecule. These structural features are similar to those of the previously reported Cu2+ complexes (Watanabe and Akitsu, 2012; Takeshita et al., 2015; Furuya et al., 2023), which has the same framework as CuSV-Imi.
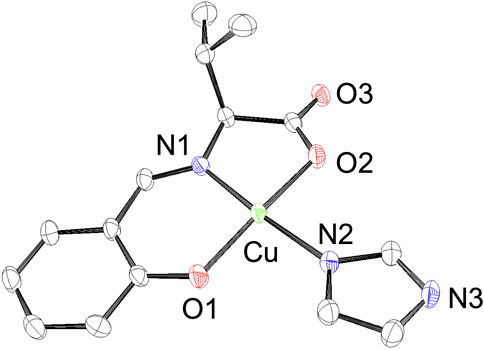
FIGURE 4. ORTEP drawings of Cu2+ complex CuSV-Imi with thermal ellipsoids drawn at 50% probability. Hydrogen atoms are omitted for clarity. Selected bond length (Å) and bond angles (degree): Cu−N1 = 1.927 (3), Cu−O1 = 1.909 (3), Cu−N2 = 1.961 (3), Cu−O2 = 1.971 (3), N1−Cu−O1 = 94.33 (12), O1−Cu−N2 = 91.54 (12), N2−Cu−O2 = 90.53 (11), N1−Cu−O2 = 82.91 (11), N1−Cu−N2 = 172.24 (13), O1−Cu−O2 = 171.11 (11).
3.2 Spectroscopic properties of the Cu2+ complexes
CuSV showed ν (C=N) vibration at 1,639 cm−1 and symmetric and asymmetric ν (COO−) vibrations at 1,468 cm−1 and 1,600 cm−1, respectively. The significant difference between symmetric and asymmetric ν(COO−) vibrations (≈140 cm−1) reflects the monodentate coordination of the carboxyl group (Reddy et al., 2011). CuSV also showed ν (O−H) vibration at 3,232 cm−1, which was due to the coordinating water molecule on the equatorial plane. In contrast, due to the replacement of water molecules and an imidazole on the equatorial plane, CuSV-Imi showed ν (C=N), symmetric and asymmetric ν (COO−) vibrations in slightly lower energy regions (1,632 cm−1, 1,465 cm−1, and 1,586 cm−1 respectively), while the ν (O−H) vibration was disappeared. The disappearance of the ν (O−H) vibration indicates that no water molecule is contained in the crystal of CuSV-Imi, which is consistent to the elemental analysis of CuSV. CuSS also showed ν (C=N), symmetric/asymmetric ν (COO−) vibrations at 1,628 cm−1, 1,465 cm−1, and 1,602 cm−1, respectively. In addition, CuSS showed ν (O−H) vibrations at 3,132 cm−1, 3,236 cm−1, and 3,320 cm−1. These ν (O−H) vibrations were assigned to hydroxyl groups of serine side chain, and coordinating/noncoordinating water molecules. Upon replacement of water molecule on the equatorial plane to imidazole, CuSS-Imi also showed ν (C=N), symmetric and asymmetric ν (COO−) vibrations in slightly lower energy regions (1,626 cm−1, 1,462 cm−1, 1,600 cm−1 respectively), while ν (O−H) vibrations assigned as coordinating and noncoordinating water molecules (3,236 cm−1, and 3,320 cm−1) were disappeared. These disappearances of ν (O−H) vibrations assigned as the water molecules are also consistent to the elemental analysis of CuSS-Imi.
The UV-vis spectra of the Cu2+ complexes are shown in Figure 5. UV-vis spectroscopic measurements revealed that CuSV showed absorption maxima at 266 nm (ε = 12,000 M−1 cm−1), 368 nm (ε = 5600 M−1 cm−1), and 664 nm (ε = 140 M−1 cm−1) assigned to π−π*, n−π*, and d-d transitions, whereas CuSS showed absorption maxima at 270 nm (ε = 19,000 M−1 cm−1), 367 nm (ε = 5400 M−1 cm−1), and 658 nm (ε = 110 M−1 cm−1). The similar UV-vis spectral features of CuSV and CuSS indicate that both Cu2+ complexes have similar structures in solution. In contrast, CuSV-Imi exhibited π−π*, n−π*, and d-d transitions at 269 nm (ε = 13,000 M−1 cm−1), 374 nm (ε = 4900 M−1 cm−1), and 622 nm (ε = 120 M−1 cm−1), whereas CuSS-Imi exhibited them at 269 nm (ε = 14,000 M−1 cm−1), 367 nm (ε = 5700 M−1 cm−1), 610 nm (ε = 130 M−1 cm−1), respectively. Upon replacing the water molecule on the equatorial plane with imidazole, both CuSV-Imi and CuSS-Imi exhibited d-d transitions in a higher energy region (622 and 610 nm) than CuSV and CuSS (664 and 658 nm). The higher energy shifts of the d-d transitions can be explained by the strong coordination of imidazole with the Cu2+ center. Higher energy shifts of their d-d transitions upon the replacement of water molecules with imidazole have been reported theoretically and experimentally (Furuya et al., 2023).
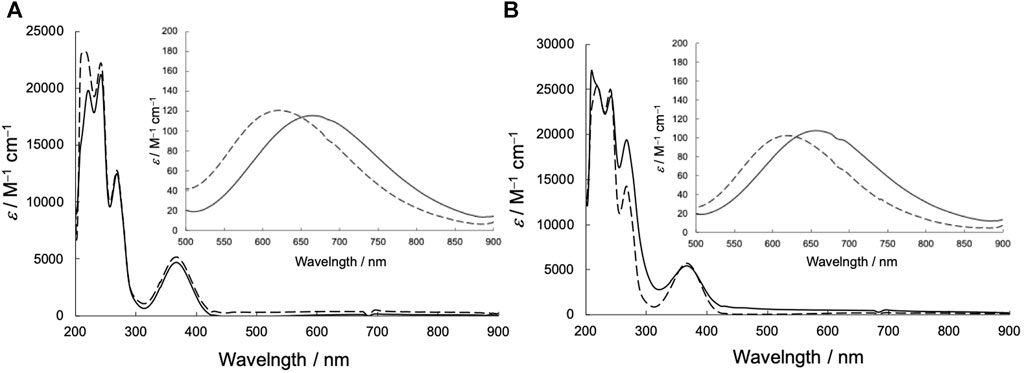
FIGURE 5. UV-vis spectra in methanol solution (0.02 mM) of (A) CuSV (solid line), CuSV-Imi (dashed line) and (B) CuSS (solid line), CuSS-Imi (dashed line). Insert: Expanded UV-vis spectra in 500–800 nm region (2 mM).
The EPR spectra of the Cu2+ complexes are shown in Figure 6. CuSV had g// and g⊥ values of 2.269 (|A//| = 173 G) and 2.065 (|A⊥| ≈ 20 G), respectively, whereas the g// and g⊥ values of CuSS were 2.265 (|A//| = 183 G) and 2.067 (|A⊥| ≈ 10 G), respectively. Both Cu2+ complexes show larger g// values than their g⊥ values, indicating that the Cu2+ centers have unpaired electrons in their dx2-y2 orbitals. These features are characteristic of Cu2+ complexes with square planar or pyramidal structures (Ramakrishnan et al., 2009). On the other hand, CuSV-Imi and CuSS-Imi showed g// and g⊥ values of 2.252 (|A//| = 173 G) and 2.063 (|A⊥| ≈ 10 G), and 2.253 (|A//| = 169 G) and 2.065 (|A⊥| ≈ 10 G), respectively. When the water molecule at the equatorial position was replaced with imidazole, both Cu2+ complexes exhibited smaller g// values. These smaller g// shifts can be explained by the strong coordination of imidazole at the equatorial position.
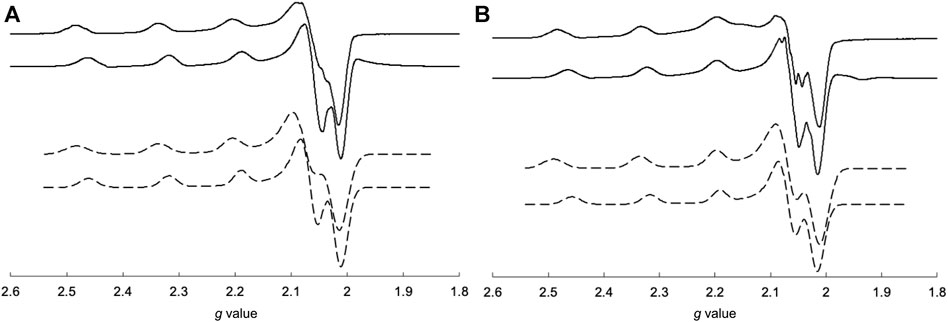
FIGURE 6. Comparisons of measured (solid lines) and simulated (dashed lines) EPR spectra of (A) CuSV (top), CuSV-Imi (bottom) and (B) CuSS (top), CuSS-Imi (bottom). All samples were prepared as 1 mM solutions in 0.1 M phosphate buffer (pH: 7.0) in quarts tubes and measured at 77 K.
3.3 Formation of hybrid proteins composed of the Cu2+ complexes and lysozyme
To confirm the formation of the hybrid proteins composed of Cu2+ complexes and lysozyme, the emission spectra of lysozyme solutions containing various concentrations of CuSV and CuSS were measured. As shown in Figures 7A, B, the lysozyme solutions produced emissions at 346 nm. These lysozyme solutions produced weaker emissions when the Cu2+ complex, CuSV or CuSS, was present at higher concentrations. In addition, based on the positive slopes obtained from the Stern–Volmer plots (Figures 7C, D), interactions and energy transitions between these Cu2+ complexes and lysozyme were suggested.
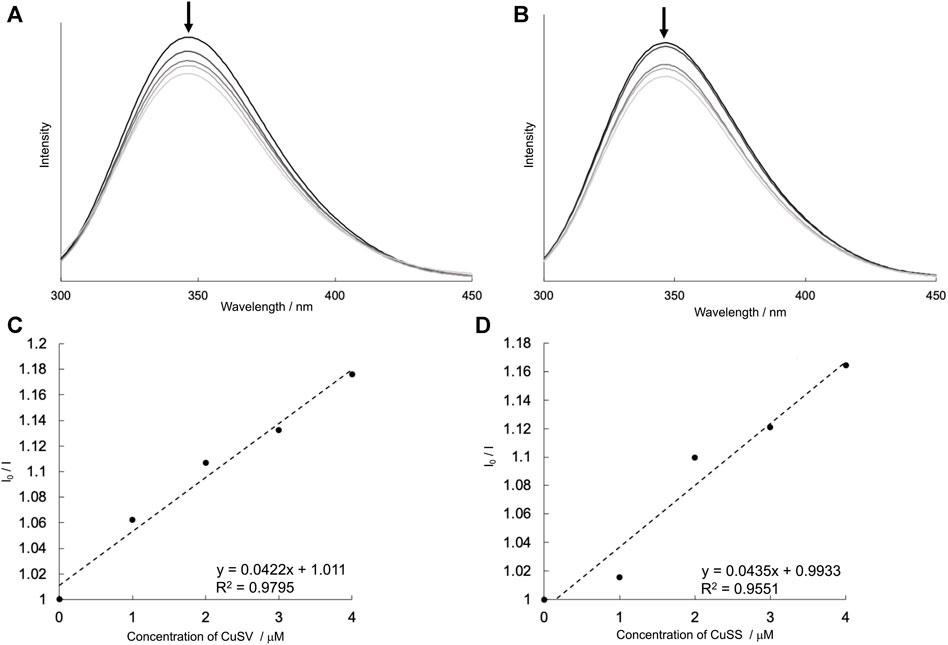
FIGURE 7. Emission spectra (λex = 260 nm) of lysozyme (4 μM) in presence of various concentrations of (A) CuSV (0–4 μM), (B) CuSS (0–4 μM), and their Stern–Volmer plots [(C,D), respectively]. These I0/I values were plotted by using their emission maxima (λem = 346 nm). All samples were prepared as 0.1 M phosphate buffer solution (pH = 7.0) and measured at 298 K.
UV-vis spectral investigations also indicated the coordination of lysozyme with Cu2+ complexes. The UV-vis spectra of CuSV and CuSS in the presence of various concentrations of lysozyme are shown in Figure 8. CuSV and CuSS exhibited their d-d transitions at 676 and 670 nm, respectively, in phosphate buffer. These absorptions shifted to a higher-energy region when lysozyme was present. These higher-energy shifts were also obtained by replacing water molecules on the equatorial planes of CuSV and CuSS with imidazole (Figure 5). Judging from these UV-vis spectral behaviors similar to those of previously reported CuST-bound lysozyme whose crystal structure are revealed (Furuya et al., 2023), coordination of the His15 side chain to the equatorial position of the Cu2+ centers of CuSV and CuSS are suggested.
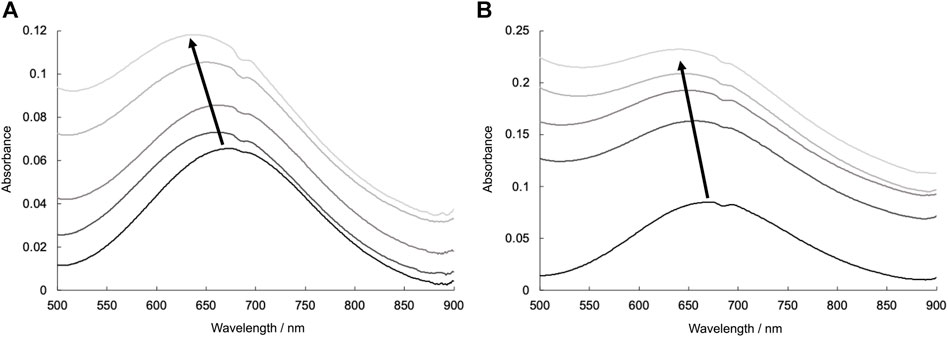
FIGURE 8. UV-vis spectra due to d-d transitions of (A) CuSV (0.8 mM) and (B) CuSS (0.8 mM) in presence of various concentrations of lysozyme (0–0.8 mM). All samples were prepared as 0.1 M phosphate buffer solution (pH = 7.0) and measured at 298 K.
EPR spectroscopy is useful for predicting the electronic structures around Cu2+ centers. The EPR spectral behavior of the Cu2+ complex in the presence of lysozyme is shown in Figure 9. As mentioned above, CuSV had g// and g⊥ values of 2.269 (|A//| = 173 G) and 2.065 (|A⊥| ≈ 20 G), respectively, whereas CuSS has values of 2.265 (|A//| = 183 G) and 2.067 (|A⊥| ≈ 10 G), respectively. They showed smaller g// values (2.251 (|A//| = 171 G) for CuSV and 2.255 (|A//| = 170 G) for CuSS) when the equivalent amounts of lysozyme were presented, while their g⊥ values showed only slight changes (2.065 (|A⊥| ≈ 10 G) for CuSV and 2.066 (|A⊥| ≈ 10 G) for CuSS). As mentioned, smaller shifts in the g// values were also observed for the imidazole coordination to CuSV and CuSS on the equatorial plane (Figure 6). Differential EPR spectra of CuSV and CuSS showed hyperfine splitting due to coordination of imino N atom of the ligands in their g⊥ regions. CuSV-Imi and CuSS-Imi also showed hyperfine splitting in their g⊥ regions with different splitting pattern from those of CuSV and CuSS, attributed to the coordination of imidazole on the equatorial plane. Furthermore, both CuSV and CuSS showed similar differential EPR spectra to CuSV-Imi and CuSS-Imi when an equivalent of lysozyme was presented. These EPR spectroscopic findings also indicated that CuSV and CuSS were bound to lysozyme by the coordination of the imidazole group of His15 on their equatorial plane.
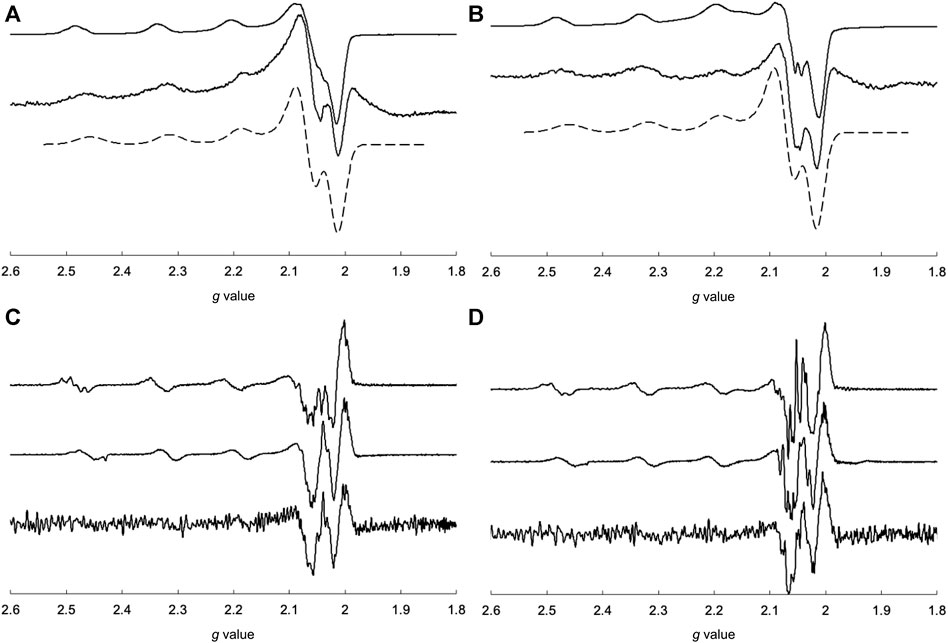
FIGURE 9. EPR spectra of (A) CuSV (top), measured and simulated 1:1 mixture of CuSV and lysozyme (middle, bottom), (B) CuSS (top), measured and simulated 1:1 mixture of CuSS and lysozyme (middle, bottom), and differential EPR spectra of (C) CuSV (top), CuSV-Imi (middle), 1:1 mixture of CuSV and lysozyme (bottom), (D) CuSS (top), CuSS-Imi (middle), 1:1 mixture of CuSS and lysozyme (bottom). All samples were prepared as 0.8 mM solution in 0.1 M phosphate buffer solution (pH = 7.0) in quarts tubes and measured at 77 K.
To investigate the electrochemical properties of the hybrid proteins, their cyclic voltammograms were measured and compared with those of CuSV-Imi and CuSS-Imi. A comparison of the voltammograms is presented in Figure 10. CuSV-Imi exhibits an irreversible redox pair. The reduction and oxidation waves were found at −0.51 V and +0.56 V (ΔE = 1.07 V), respectively. Based on the rest potential (+0.51 V) and the initial scan polarity (negative), the redox pair was identified as a Cu2+/Cu+. The significant peak separation (ΔE = 1.07 V) indicates a slow electron transfer caused by structural changes around the metal center during the redox process. CuSS-Imi also showed the reduction and oxidation waves due to the Cu2+/Cu+ process at −0.47 V and +0.57 V (ΔE = 1.04 V), respectively. Based on the comparison of the redox potentials and their separation, the hydroxyl group of CuSS-Imi has only a small effect on its electrochemical properties. On the other hand, the CuSV-bound lysozyme (CuSV@lysozyme) had Cu2+/Cu+ reduction and oxidation waves at −0.43 V and +0.16 V.
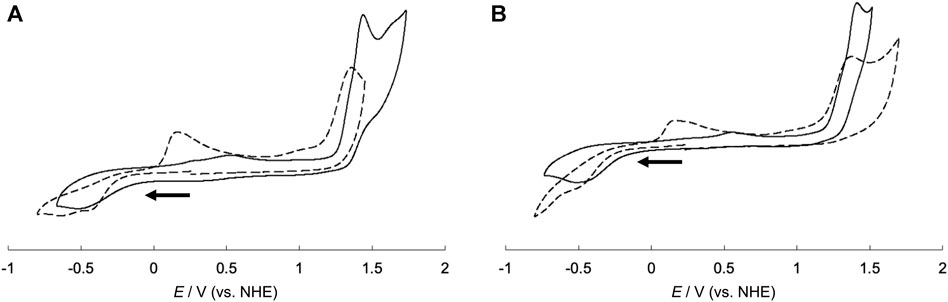
FIGURE 10. Cyclic voltammograms of (A) CuSV-Imi (solid line), CuSV@lysozyme (dashed line) and (B) CuSS-Imi (solid line), CuSS@lysozyme (dashed line). All samples were prepared as 1.0 mM solution in 0.1 M phosphate buffer solution (pH = 7.0). Glassy carbon, Pt wire, and Ag/AgCl electrodes were used as the working, counter, and reference electrodes, respectively and measured with a sweep rate of 100 mV/s. Potentials were converted from Ag/AgCl to NHE.
From the formation of the adduct, CuSV@lysozyme produced a smaller peak separation (ΔE = 0.59 V) than CuSV-Imi (ΔE = 1.07 V). CuSS-bound lysozyme (CuSS@lysozyme) also produced reduction and oxidation waves due to the Cu2+/Cu+ process at −0.48 V and +0.17 V and a smaller peak separation (ΔE = 0.65 V) than CuSS-Imi. These smaller peak separations suggest that the structural rearrangement around the metal center upon Cu2+/Cu+ processes were decreased by lysozyme hybridization. Our group previously reported the crystal structure of a lysozyme-bound Cu2+ complex (CuST@lysozyme) (Furuya et al., 2023). Crystallographic analysis revealed that the Cu2+ complex, CuST, was coordinated by the imidazole group of the His15 side chain. In addition, the Cu2+ center was fixed by weak coordination of the Thr89 side chain. Owing to the weak coordination, the Cu2+ centers of lysozyme-bound CuSV and CuSS can be fixed and decrease structural changes in the Cu2+/Cu+ process. The diminution of structural changes around the metal centers during the Cu2+/Cu+ process can favor the disproportionation activity of the O2− anion.
3.4 SOD activity evaluations of the hybrid proteins composed of the Cu2+ complexes and lysozyme
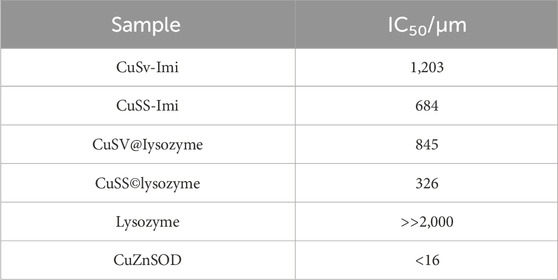
Spectroscopic measurements suggested that both CuSV and CuSS formed hybrid proteins with lysozymes. Therefore, we investigated the SOD activity of these hybrid proteins (CuSV@lysozyme and CuSS@lysozyme) to evaluate the effects of hybridization with lysozymes. SOD activity was evaluated as IC50 values obtained using the xanthine/xanthine oxidase method. The results of the SOD activity evaluation are summarized in Table 1. First, we compared the SOD activities of CuSV-Imi and CuSS-Imi. CuSS-Imi showed higher SOD activity (IC50 = 684 μM) than CuSV-Imi (IC50 = 1,203 μM). Based on the comparison of their structures, the SOD activity of CuSS-Imi can be affected by the hydroxyl group on the side chain of the serine moiety. In contrast, through hybridization with lysozyme, CuSV@lysozyme and CuSS@lysozyme had higher SOD activities (IC50 = 845 μM and 326 μM, respectively) than CuSV-Imi and CuSS-Imi. Based on the poor SOD activity of lysozyme (IC50 >> 2000 μM), the improvements in SOD activity were due to the cooperative effects of the Cu2+ complexes and lysozyme.
3.5 Theoretical investigations
According to comparisons of the SOD activities of the Cu2+ complexes and their lysozyme adducts, the presence of a hydroxyl group on the Cu2+ complex and hybridization with lysozyme improved SOD activity. To understand their SOD activity, theoretically optimized structures of O2−-bound Cu2+ complexes and their lysozyme adducts were compared. The optimized structures of O2−-bound CuSV-Imi and CuSS-Imi are shown in Figure 11. To the Cu2+ center of CuSV-Imi, a O2− anion coordinated on the equatorial position to form square pyramidal structure (τ = 0.10). By the coordination of O2− anion, the imidazole molecule was kicked out to the axial position and weakly coordinated (2.397 Å) to the Cu2+ center. The drastic structural change can prevent fast electron transfer between the metal center and superoxide. On the other hand, the Cu2+ center of CuSS-Imi was coordinated by a O2− anion to form distorted trigonal bipyramidal structure (τ = 0.62). Although the structure of the Cu2+ center was distorted by the coordination of a O2− anion, the imidazole molecule on the equatorial position of CuSS-Imi was not kicked out. In addition, the coordinated O2− anion formed hydrogen bond with hydroxyl group of the side chain of the serine moiety. Attributed to these structural features, the O2−-bound CuSS-Imi showed 128.9 kJ/mol lower energy than that of O2−-bound CuSV-Imi. These theoretical findings consistent to the higher SOD activity of CuSS-Imi than that of CuSV-Imi.
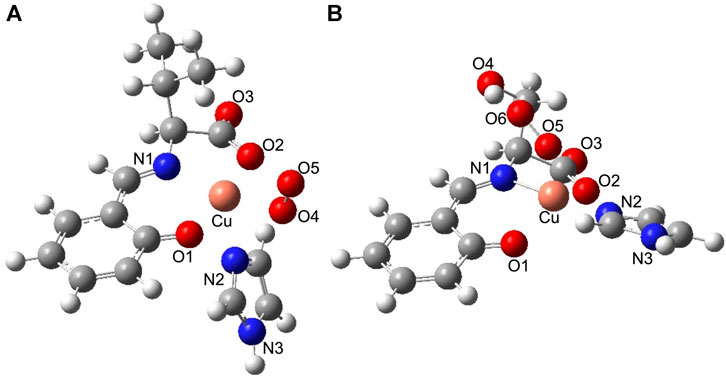
FIGURE 11. Theoretically proposed structures of (A) O2−-bound CuSV-Imi and (B) O2−-bound CuSS-Imi. Selected bond length (Å) and bond angles (degree): Cu−N1 = 2.022, Cu−O1 = 2.038, Cu−N2 = 2.397, Cu−O2 = 2.048, Cu−O4 = 1.975, N1−Cu−O1 = 90.156, O1−Cu−O4 = 92.369, O4−Cu−O2 = 96.667, O2−Cu−N1 = 81.359, N1−Cu−N2 = 106.462, O1−Cu−N2 = 90.077, O4−Cu−N2 = 88.433, O2−Cu−N2 = 89.232, N1−Cu−O4 = 164.894, O1−Cu−O2 = 170.915, for structure (A), and Cu−N1 = 1.940, Cu−O1 = 2.114, Cu−N2 = 1.993, Cu−O2 = 2.113, Cu−O5 = 2.169, N1−Cu−O1 = 91.113, O1−Cu−N2 = 93.168, N2−Cu−O2 = 92.353, O2−Cu−N1 = 80.806, N1−Cu−O5 = 98.791, O1−Cu−O5 = 95.293, N2−Cu−O5 = 86.225, O2−Cu−O5 = 128.491, N1−Cu−N2 = 173.086, O1−Cu−O2 = 136.136, for structure (B), respectively.
3.6 Discussion on the higher SOD activity of CuSS@lysozyme
Based on these experimental and theoretical investigations, the higher SOD activity of CuSV@lysozyme than that of CuSS@lysozyme can be explained as follows: i) Judging from the previously reported crystal structure of CuST@lysozyme (Furuya et al., 2023) and spectroscopic behaviors of CuSV and CuSS upon presence of lysozyme, the imidazole group of His15 residue and hydroxyl group of Thr89 bind to the Cu2+ center of CuSS on the equatorial and axial position, respectively. The theoretical investigation suggested that hydrogen bonds can be formed between the O2− anion and the hydroxyl group owing to the serine moiety of CuSS when the O2− anion coordinates with the metal center, as shown in Figure 12. The theoretical investigation also suggested that the coordination of the O2− anion to the metal center can be stabilized and promoted through the hydrogen bond. ii) By forming a hybrid protein with lysozyme, in addition to the coordination of the imidazole group of the His15 side chain with the metal center at the equatorial position, the hydroxyl group of Thr89 is weakly coordinated at the axial position. Electrochemical behaviors of CuSV@lysozyme and CuSS@lysozyme implied that the weakly coordinated Thr89 residue can contribute to reduce the structural changes during the Cu2+/Cu+ process. The reduced structural changes during the Cu2+/Cu+ process can improve electron transfer between O2− anions and enhance the SOD activity of the metal center.
4 Conclusion
In this study, we prepared Cu2+ complexes, CuSV and CuSS, in the absence/presence of hydroxyl groups, and their imidazole-bound compounds, CuSV-Imi and CuSS-Imi, to reveal the effects of hydroxyl groups on SOD activity. Crystallographic analysis and spectroscopic measurements suggested that the imidazole group coordinated with the Cu2+ ions at the equatorial positions of CuSV and CuSS, both in the solid state and in solution. Based on these properties and spectroscopic behaviors in the presence of lysozyme on CuSV and CuSS, the imidazole group of His15 in lysozyme can coordinate with the Cu2+ center in its equatorial position to form hybrid proteins with lysozyme. Electrochemical measurements indicated that the structural changes around the Cu2+ center of CuSV and CuSS during the Cu2+/Cu+ process were decreased by the formation of hybrid proteins with lysozyme.
A comparison of the SOD activities of CuSV-Imi and CuSS-Imi indicated that the hydroxyl group of CuSS-Imi played an important role in the disproportionation of the O2− ion. In addition, hybridization of the Cu2+ complexes CuSV and CuSS with lysozyme resulted in higher SOD activity than that of CuSV-Imi and CuSS-Imi. These improvements in SOD activity suggest that there are cooperative effects between Cu2+ complexes and lysozyme. These improvements in SOD activity can be explained by the stabilization of the O2−-bound metal center caused by the hydrogen bond between the coordinating O2− ion and the hydroxyl group. The diminution of the structural changes upon the redox of the metal center caused by the weak coordination of the hydroxyl group of the Thr89 side chain with the metal center also contributes to the improvement of the SOD activity.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: CCDC code of CuSV-Imi (2310763).
Author contributions
DN: Investigation, Writing–original draft, Writing–review and editing. YA: Investigation, Formal Analysis, Data curation, Writing–original draft. SS: Investigation, Formal Analysis, Data curation, Writing–original draft. RM: Writing - original draft, Data curation. TA: Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The elemental analyses were supported by the Equipment Sharing Division, Organization for Co-Creation Research and Social Contributions, Nagoya Institute of Technology, Japan. We would like to thank Editage (www.editage.jp) for English language editing. The elemental analyses were measured by the organization of Nagoya Institute of Technology.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abreu, I. A., Rodriguez, J. A., and Cabelli, D. E. (2005). Theoretical studies of manganese and iron superoxide dismutases: superoxide binding and superoxide oxidation. J. Phys. Chem. B 109, 24502–24509. doi:10.1021/jp052368u
Addison, A. W., Rao, T. N., Reedijk, J., Rijn, J., and Verschoor, G. C. (1984). Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen-sulphur donor ligands; the crystal and molecular structure of aqua[l,7-bis(N-methylbenzimidazol-2'-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 13, 1349–1356. doi:10.1039/dt9840001349
Akiyama, Y., Suzuki, S., Suda, S., Takiguchi, Y., Nakane, D., and Akitsu, T. (2023). Crystal structure and Hirshfeld surface analysis of mono/bis(aqua-κO)[N-(2-oxidobenzylidene)valinato-κ3O,N,O]copper(II): dimeric Schiff base copper(II) complexes having different numbers of coordinated water molecules. Acta Cryst. E79, 361–366. doi:10.1107/s2056989023002487
Alscher, R. G., Erturk, N., and Heath, L. S. (2022). Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53, 1331–1341. doi:10.1093/jexbot/53.372.1331
Barondeau, D. P., Kassmann, C. J., Bruns, C. K., Tainer, J. A., and Getzoff, E. D. (2004). Nickel superoxide dismutase structure and mechanism. Biochemistry 43, 8038–8047. doi:10.1021/bi0496081
Casini, A., Mastrobuoni, G., Temperini, C., Gabbiani, C., Francese, S., Moneti, G., et al. (2007). ESI mass spectrometry and X-ray diffraction studies of adducts between anticancer platinum drugs and hen egg white lysozyme. Chem. Commun. 43, 156–158. doi:10.1039/b611122j
Clark, T., Chandrasekhar, J., Spitznagel, G. W., and Schleyer, P. v. R. (1983). Efficient diffuse function-augmented basis sets for anion calculations. III. The 3-21+G basis set for first-row elements, Li–F. J. Comput. Chem. 4 4, 294–301. doi:10.1002/jcc.540040303
del Rio, L. A., Sandalio, L. M., Altomare, D. A., and Zilinskas, B. A., (2003). Mitochondrial and peroxisomal manganese superoxide dismutase: differential expression during leaf senescence. J. Exp. Bot. 54, 923–933. doi:10.1002/jcc.540040303
Dennington, R., Keith, T., and Millam, J. (2009). Gauss view. Shawnee Mission: Semichem Inc. Version 5.
Ferraro, G., Giorgio, A., Mansour, A. M., and Merlino, A. (2019). Protein-mediated disproportionation of Au(I): insights from the structures of adducts of Au(III) compounds bearing N,N-pyridylbenzimidazole derivatives with lysozyme. Dalton Trans. 48, 14027–14035. doi:10.1039/c9dt02729g
Fréalle, E., Noël, C., Viscogliosi, E., Camus, D., Dei-Cas, E., and Delhaes, L. (2005). Manganese superoxide dismutase in pathogenic fungi: an issue with pathophysiological and phylogenetic involvements. FEMS Immunol. Med. Microbiol. 45, 411–422. doi:10.1016/j.femsim.2005.06.003
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., et al. (2009). Gaussian 09, revision A.02. Wallingford: Gaussian Inc.
Furuya, T., Nakane, D., Kitanishi, K., Katsuumi, N., Tsaturyan, A., Shcherbakov, I. N., et al. (2023). A novel hybrid protein composed of superoxide-dismutase-active Cu(II) complex and lysozyme. Sci. Rep. 13, 6892. doi:10.1038/s41598-023-33926-1
Goscin, S. A., and Fridovich, I. (1972). The purification and properties of superoxide dismutase from Saccharomyces cerevisiae. Biochimica Biophysica Acta (BBA)-Enzymology 289, 276–283. doi:10.1016/0005-2744(72)90078-2
Hart, P. J., Balbirnie, M. M., Ogihara, N. L., Nersissian, A. M., Weiss, M. S., Valentine, J. S., et al. (1999). A structure-based mechanism for copper-zinc superoxide dismutase. Biochemistry 38, 2167–2178. doi:10.1021/bi982284u
Hatchikian, E. C., and Henry, Y. A. (1977). An iron-containing superoxide dismutase from the strict anaerobe Desulfovibrio desulfuricans (Norway 4). Biochimie 59, 153–161. doi:10.1016/s0300-9084(77)80286-1
Hayyan, M., Hashim, M. A., and AlNashef, I. M. (2016). Superoxide ion: generation and chemical implications. Chem. Rev. 116, 3029–3085. doi:10.1021/acs.chemrev.5b00407
Kanematsu, S., and Asada, K. (1979). Ferric and manganic superoxide dismutases in Euglena gracilis. Archives Biochem. Biophysics 195, 535–545. doi:10.1016/0003-9861(79)90380-1
Kato, R., Akiyama, M., Kawakami, H., and Komatsu, T. (2014). Superoxide dismutase activity of the naturally occurring human serum albumin-copper complex without hydroxyl radical formation. Chem. Asian J. 9, 83–86. doi:10.1002/asia.201301285
Katsuumi, N., Onami, Y., Pradhan, S., Haraguchi, T., and Akitsu, T. (2020). Crystal structure and Hirshfeld surface analysis of (aqua-κO)(methanol-κO)[N-(2-oxido-benzylidene)threoninato-κ3O,N,O’]copper(II). Acta Cryst. E76, 1539–1542. doi:10.1107/s2056989020011706
Kirby, T. W., Lancaster, J. R., and Fridovich, I. (1981). Isolation and characterization of the iron-containing superoxide dismutase of Methanobacterium bryantii. Archives Biochem. Biophysics 210, 140–148. doi:10.1016/0003-9861(81)90174-0
Lee, C., Yang, W., and Parr, R. G. (1988). Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter 37, 785–789. doi:10.1103/physrevb.37.785
Mann, T., and Keilin, D. (1938). Haemocuprein and hepatocuprein, copper-protein compounds of blood and liver in mammals. Proc. R. Soc. Lond. Ser. B Biol. Sci. 126, 303–315. doi:10.1098/rspb.1938.0058
McCord, J. M., and Fridovich, I. (1969). Superoxide dismutase. J. Biol. Chem. 244, 6049–6055. doi:10.1016/s0021-9258(18)63504-5
O Connor, M., Kellett, A., McCann, M., Rosair, G., McNamara, M., Howe, O., et al. (2012). Copper(II) complexes of salicylic acid combining superoxide dismutase mimetic properties with DNA binding and cleaving capabilities display promising chemotherapeutic potential with fast acting in vitro cytotoxicity against cisplatin sensitive and resistant cancer cell lines. J. Med. Chem. 55, 1957–1968. doi:10.1021/jm201041d
Opperdoes, F. R., Borst, P., and Spits, H. (1977). Particle-bound enzymes in the bloodstream form of Trypanosoma brucei. Eur. J. Biochem. 76, 21–28. doi:10.1111/j.1432-1033.1977.tb11566.x
Otani, N., Furuya, T., Katsuumi, N., Haraguchi, T., and Akitsu, T. (2021). Synthesis of amino acid derivative Schiff base copper(II) complexes by microwave and wet mechanochemical methods. J. Indian Chem. Soc. 98, 100004. doi:10.1016/j.jics.2021.100004
Palenik, B., Brahamsha, B., Larimer, F. W., Land, M., Hauser, L., Chain, P., et al. (2003). The genome of a motile marine Synechococcus. Nature 424, 1037–1042. doi:10.1038/nature01943
Panzner, M. J., Bilinovich, S. M., Youngs, W. J., and Leeper, T. C. (2011). Silver metallation of hen egg white lysozyme: X-ray crystal structure and NMR studies. Chem. Commun. 47, 12479–12481. doi:10.1039/c1cc15908a
Pilon, M., Ravet, K., and Tapken, W. (2011). The biogenesis and physiological function of chloroplast superoxide dismutases. Biochimica Biophysica Acta-Bioenergetics 1807, 989–998. doi:10.1016/j.bbabio.2010.11.002
Pryor, W. A. (1986). Oxy-radicals and related species: their formation, lifetimes, and reactions. Annu. Rev. Physiol. 48, 657–667. doi:10.1146/annurev.ph.48.030186.003301
Ramakrishnan, S., Rajendiran, V., Palaniandavar, M., Periasamy, V. S., Srinag, B. S., Krishnamurthy, H., et al. (2009). Induction of cell death by ternary copper(II) complexes of L-tyrosine and diimines: role of coligands on DNA binding and cleavage and anticancer activity. Inorg. Chem. 48, 1309–1322. doi:10.1021/ic801144x
Razavet, M., Artero, V., Cavazza, C., Oudart, Y., Lebrun, C., Fontecilla-Camps, J.-C., et al. (2007). Tricarbonylmanganese(I)–lysozyme complex: a structurally characterized organometallic protein. Chem. Commun. 43, 2805–2807. doi:10.1039/b703887a
Reddy, P. R., Shilpa, A., Raju, N., and Raghavaiah, P. (2011). Synthesis, structure, DNA binding and cleavage properties of ternary amino acid Schiff base-phen/bipy Cu(II) complexes. J. Inorg. Biochem. 105, 1603–1612. doi:10.1016/j.jinorgbio.2011.08.022
Sheldrick, G. M. (2008). A short history of SHELX. Acta Crystallogr. A Found. Crystallogr. 64, 112–122. doi:10.1107/s0108767307043930
Sheng, Y., Abreu, I. A., Cabelli, D. E., Maroney, M. J., Miller, A. F., Teixeira, M., et al. (2014). Superoxide dismutases and superoxide reductases. Chem. Rev. 114, 3854–3918. doi:10.1021/cr4005296
Suzuki, S., Akiyama, Y., Nakane, D., and Akitsu, T. (2023). Crystal structure and Hirshfeld surface analysis of (1H-imidazole-κN3) [N-(2-oxidobenzylidene) tyrosinato-κ3O,N,O’copper(II). Acta Cryst. E79, 596–599. doi:10.1107/s2056989023004735
Takeshita, Y., Takakura, K., and Akitsu, T. (2015). Multifunctional composites of chiral valine derivative Schiff base Cu(II) complexes and TiO2. Int. J. Mol. Sci. 16, 3955–3969. doi:10.3390/ijms16023955
Tolmasoff, J. M., Ono, T., and Cutler, R. G. (1980). Superoxide dismutase: correlation with life-span and specific metabolic rate in primate species. Proc. Natl. Acad. Sci. U.S.A. 77, 2777–2781. doi:10.1073/pnas.77.5.2777
Watanabe, Y., and Akitsu, T. (2012). Preparations of hybrid systems of L-amino acid derivatives of Schiff base Cu(II) and Zn(II) complexes and polyoxometalates. Asian Chem. Lett. 16, 9–18.
Wuerges, J., Lee, J.-W., Yim, Y.-I., Yim, H.-S., Kang, S.-O., and Carugo, K. D. (2004). Crystal structure of nickel-containing superoxide dismutase reveals another type of active site. Proc. Natl. Acad. Sci. U. S. A. 101, 8569–8574. doi:10.1073/pnas.0308514101
Yost, F. J., and Fridovich, I. (1973). An Iron-containing superoxide dismutase from Escherichia coli. J. Biol. Chem. 248, 4905–4908. doi:10.1016/s0021-9258(19)43649-1
Youn, H. D., Kim, E. J., Roe, J. H., Hah, Y. C., and Kang, S. O. (1996a). A novel nickel-containing superoxide dismutase from Streptomyces spp. Biochem. J. 318, 889–896. doi:10.1042/bj3180889
Keywords: Cu2+ complex, SOD activity, hybrid protein, lysozyme, hydrogen bond
Citation: Nakane D, Akiyama Y, Suzuki S, Miyazaki R and Akitsu T (2024) Improvement of the SOD activity of the Cu2+ complexes by hybridization with lysozyme and its hydrogen bond effect on the activity enhancement. Front. Chem. 11:1330833. doi: 10.3389/fchem.2023.1330833
Received: 31 October 2023; Accepted: 27 December 2023;
Published: 18 January 2024.
Edited by:
Mariana Carmen Chifiriuc, University of Bucharest, RomaniaReviewed by:
Sandra Rosanna Signorella, CONICET Instituto de Química Rosario (IQUIR)—Rosario, ArgentinaAmga Baldan, Rensselaer Polytechnic Institute, United States
Copyright © 2024 Nakane, Akiyama, Suzuki, Miyazaki and Akitsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daisuke Nakane, YTI5Nzc2QHJzLnR1cy5hYy5qcA==
 Daisuke Nakane
Daisuke Nakane Yukihito Akiyama
Yukihito Akiyama