- 1Department of Pharmacy, The First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology, Luoyang, China
- 2School of Nursing, College of Basic Medicine and Forensic Medicine, Henan University of Science and Technology, Luoyang, China
- 3School of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang, China
- 4State Key Laboratory of Quality Research in Chinese Medicine/Macau Institute for Applied Research in Medicine and Health, Macau University of Science and Technology, Macau, China
- 5Henan Wanliu Biotechnology Co., Ltd., Luoyang, China
- 6UNC Hospital, Chapel Hill, NC, United States
Indoleamine 2,3-dioxygenase 1 (IDO1) has attracted much attention in the field of cancer immunotherapy as an immunomodulatory enzyme. To identify potential IDO1 inhibitors, a novel series of compounds with N,N-diphenylurea and triazole structures were synthesized. The designed compounds underwent organic synthesis, and subsequent enzymatic activity experiments targeting IDO1 confirmed their activity at the molecular level. These experiments provided validation for the efficacy of the designed compounds in inhibiting IDO1, compound 3g exhibited an IC50 value of 1.73 ± 0.97 μM. Further molecular docking study further explained the binding mode and reaction potential of compound 3g with IDO1. Our research has resulted in a series of novel IDO1 inhibitors, which is beneficial to the development of drugs targeting IDO1 in numerous cancer diseases.
1 Introduction
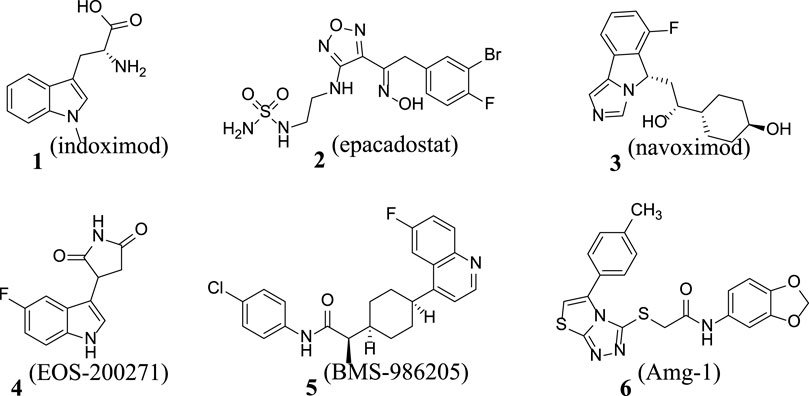
Cancer is one of the malignant diseases seriously threatening human health. According to the estimates by the World Health Organization, there are approximately 14 million new cancer cases diagnosed globally each year (Sun et al., 2022). The number of cases continues to increase, and the new cases are expected to increase to 21 million in 2030. The mortality rate of cancer is extremely high, and nearly 10 million cancer patients die every year. In both developing and developed countries, cancer is one of the leading causes of death. Currently, despite many achievements in cancer therapy, some challenges remain. As technology advances, immunotherapy has become a research hotspot for cancer treatment (Serafini et al., 2020). There are various forms of immunotherapy, which include approaches such as T cells activated in vitro, oncolytic virus, and natural killer cells and using antibodies or recombinant proteins to co-stimulate cells or block the so-called immune checkpoint pathways (Röhrig et al., 2012). Through investigating the tumor immune microenvironment, IDO1 was found to be a metabolic enzyme that could mediate tumor immune evasion and a potent immunomodulator (Pan et al., 2020). It plays a role in pathogenic inflammatory processes and generates immune tolerance to tumor antigens, mainly through facilitating tryptophan consumption and producing a series of toxic kynurenine metabolites, thereby achieving immune tolerance by activating the GCN2 pathway, inhibiting the mTOR pathway and the toxic effects of kynurenine, and promoting the differentiation of regulatory T cells (Nelp et al., 2018). Presently, IDO1 is overexpressed in a variety of tumor tissues, and IDO1 has emerged as a potential target for studying tumor immunosuppression (Muller et al., 2005). Since 2015, multiple IDO1 inhibitor drugs have been tested in clinical trials, such as indoximod, epacadostat, navoximod, EOS-200271, BMS-986205, and Amg-1 (Kumar et al., 2019; Hou X. X. et al., 2022; Kapron et al., 2023; Wang et al., 2023; Ye et al., 2023) (Figure 1; Figure 2).
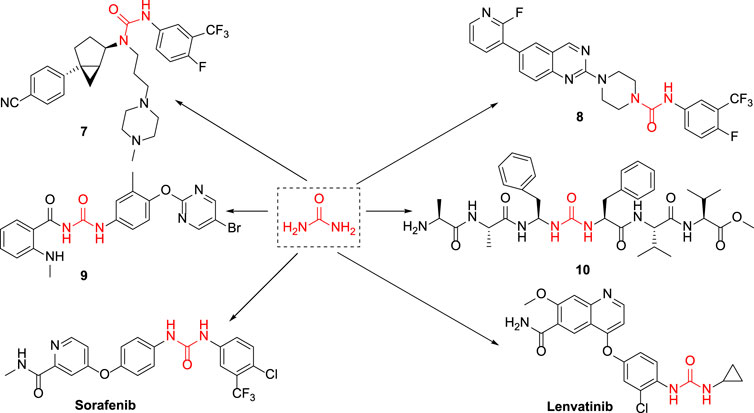
Urea is an organic compound that consists of carbon, nitrogen, oxygen, and hydrogen. In the 18th century, urea was first extracted from the urine of humans. In the 19th century, scientists found that the inorganic substance ammonium cyanate could be used as raw material to synthesize urea, which overturned the traditional vitalism theory and opened a new chapter in organic chemistry (Jr et al., 1993). Urea structural fragment is an important advantageous skeleton for drug development and is extensively used in the medical field (Nakagawa et al., 2009). For example, the melanin-concentrating hormone was associated with many physiological functions in the human body and played a pivotal role in especially the aspects of food intake and energy balance. Compound 7 with urea structure could be used as a melanin-concentrating hormone antagonist and is effective in reducing body weight (McBriar et al., 2006). Kotakadi et al. reported that compound 8 with urea structure had an excellent inhibitory effect on the proliferation of Staphylococcus aureus (Babu et al., 2015). Khanet al. discovered that compound 9 could effectively repress pancreatic cancer cells and laryngeal squamous carcinoma cells. The Lipton laboratory found that compound 10 exhibited a moderate inhibitory effect on HIV-1 protease. The currently marketed antineoplastic drugs containing urea structure include lenvatinib for the treatment of kidney or thyroid cancer, which is a tyrosine kinase inhibitor drug able to effectively slow down or terminate the growth of tumor cells (Zhang et al., 2020). Sorafenib is also included, which is a new multi-targeted oral drug for cancer therapy (Tang et al., 2020). It is used to treat gastrointestinal stromal tumors and metastatic renal cells that are not responding to standard therapy or are resistant, which could selectively target the receptors of certain proteins and is considered to act as a molecular switch during tumor growth (Yau et al., 2022). Its aforementioned indications have obtained the “fast track” designation granted by the FDA in the United States.
Considering the excellent biological activity performance of urea compounds, to find new and more efficient IDO1 inhibitor drugs, we aimed to link diphenylurea compounds with different substituents to triazole compounds, which were widely used in the modification of drug molecules (Rohrig et al., 2019; Qi et al., 2020; Zhang et al., 2020). Ten diphenylurea-linked 1,2,3-triazole derivatives were synthesized, these compounds were designed with the expectation of exhibiting specifict functions in inhibiting the activity of IDO1.
2 Chemistry
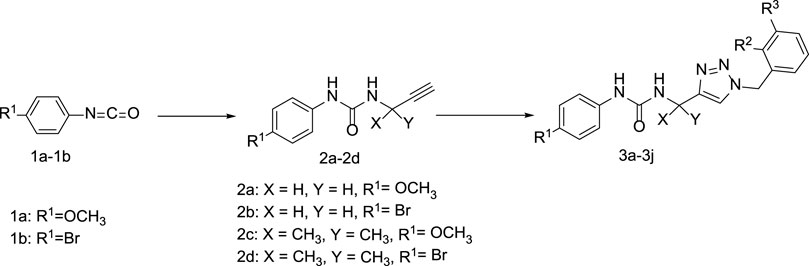
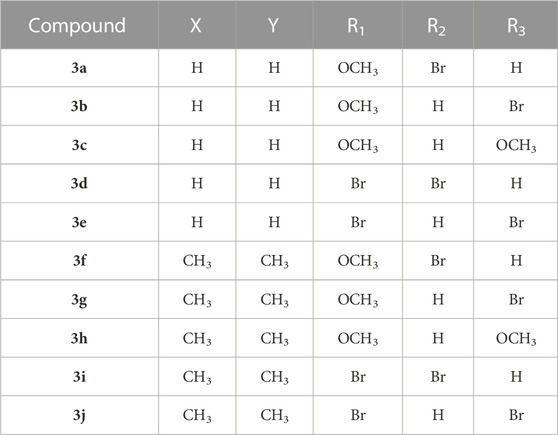
Detailed operation as follows: The phenyl isocyanate derivatives (1a-1b) reacted with prop-2-yn-1-amine (or 2-methylbut-3-yn-2-amine) and triethylamine in the dichloromethane to form compound 2a-2b. Compound 2a-2b reacted with some azide derivatives to give compound 3a-3j as shown in Figure 3 and Table 1. The molecular structures of these compounds were analyzed using 1H NMR and 13C NMR. Furthermore, we conducted additional testing to evaluate the anti-IDO1 activity of these ten compounds.
3 Results and discussion
3.1 IDO1 inhibition study
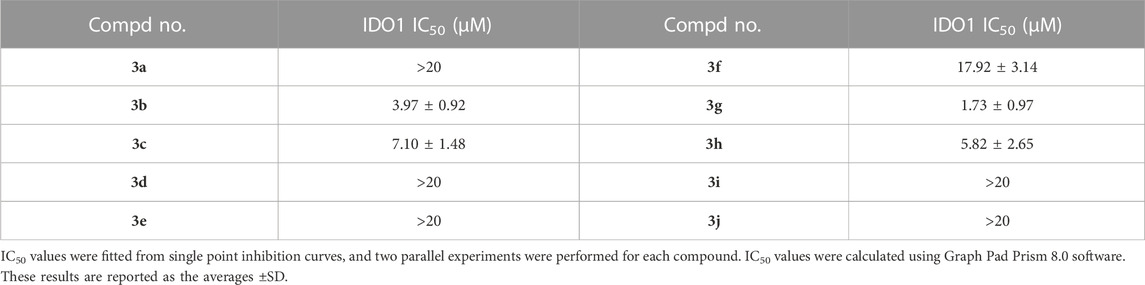
Based on a comprehensive review of the literature, we employed a Hela cell-based functional assay to investigate the IDO1 inhibition activities of the designed compounds. (Table 2). We employed Amg-1 as a positive control, whose IC50 value was measured to be 3.97 µM under our experimental conditions, which was close to the actual value (Hou X. et al., 2022). As shown in Table 2, the activities of compound 3a, 3c, 3g, and 3h against IDO1 were all below 10 μM, of which 3g reached 1.73 ± 0.97 μM. This indicates that some give synthesized compounds have good inhibitory activity.
3.2 Molecular docking studies of compound 3a, 3b, 3f, and 3g
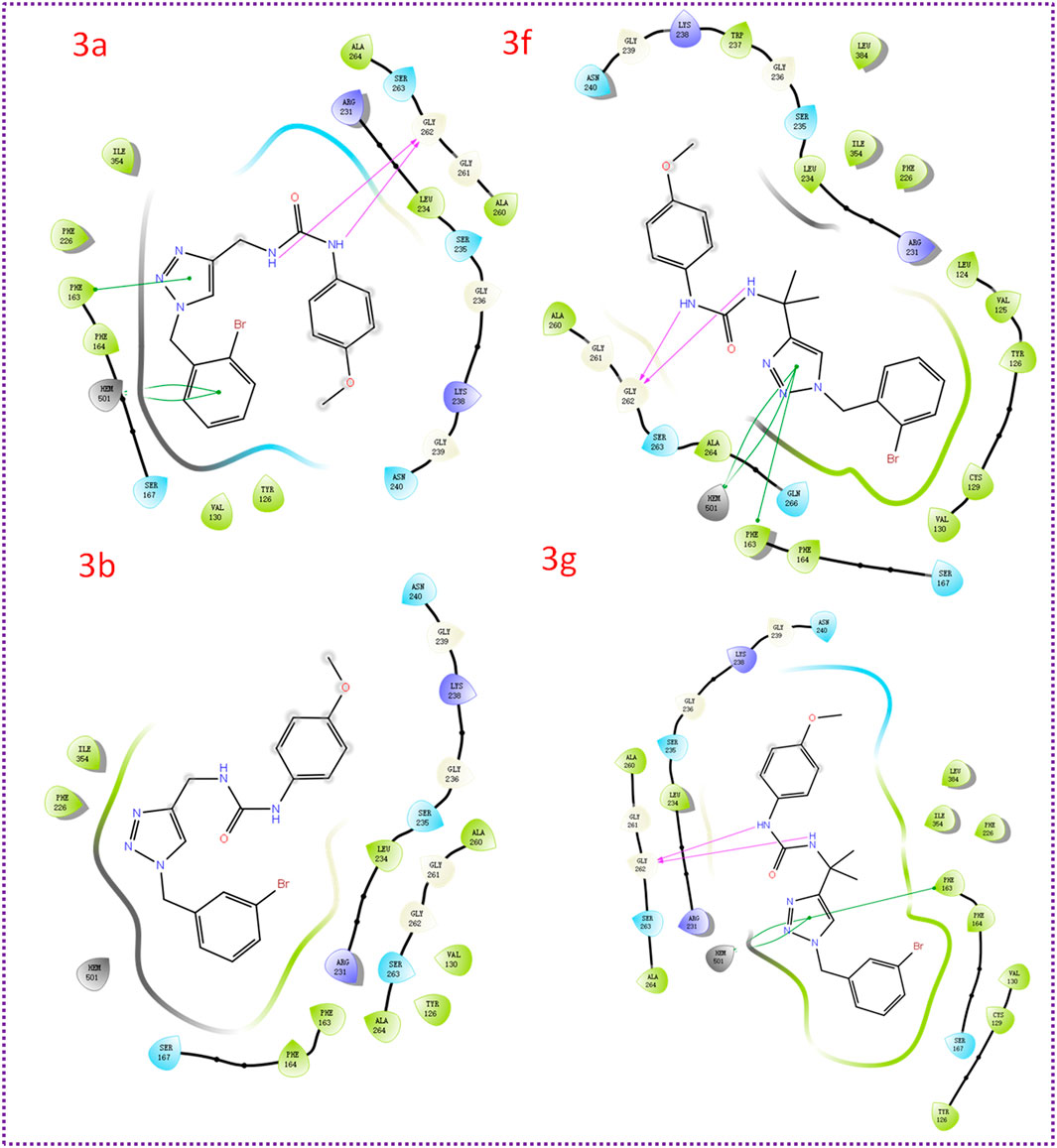
To gain insights into the binding mode of the active compounds to IDO1, we selected compounds 3a, 3b, 3f, and 3g as model compounds. These compounds were chosen due to their varying levels of activity, allowing us to examine the potential differences in their binding interactions with IDO1. As shown in Figure 4, the docking scores of compounds 3a, 3b, 3f and 3g with IDO1 were −6.424, −6.292, −7.665 and −6.876 kcal/mol, respectively. The 1,2,3-triazolyl group in the structures of compounds 3f and 3g were nearby the heme. And the binding mode of compounds 3f and 3g are similar to several reported IDO1 inhibitors. The 1,2,3-triazole group in the structure of compound 3a and 3b were far away from the heme iron and have weaker affinities for IDO1. The docking results indicated that compounds 3f and 3g were more capable of coordinating with ferrous ions than 3a and 3b.
4 Conclusion
Ten series of compounds that contain the N,N-diphenylurea structure and the 1,2,3-triazole structure were designed and synthesized. In vitro experiments were conducted to assess the compounds’ activity, and the results showed that the compounds with an isopropyl linker displayed better activity than those with a methyl linker. Notably, compound 3g exhibited the most potent inhibitory effect on IDO1, with an IC50 value of 1.73 ± 0.97 μM. Further molecular docking studies confirmed that compound 3g is the most promising of the synthesized compounds.
5 Experimental protocols
5.1 Materials and chemistry
The 1,2,3-triazole derivatives were synthesized in-house by our research group. All reagents and solvents obtained from commercially available source were used without further treatment. 1H NMR and 13C NMR spectra were acquired in DMSO-d6 solution with a Bruker 600 spectrometer. Chemical shifts (d) were given in parts per million with tetramethylsilane as internal reference and coupling constants were expressed in hertz. Hela cell line was purchased from ATCC (Virginia). Recombinant human IFN-γ was purchased from R&D systems (Emeryville, CA). The 3.05 Ntrichloroacetic acid, was purchased from Sigma Aldrich (St. Louis, MI).
5.2 General procedure for preparation of compound 2a (The method is suitable for 2b)
To a solution of 1-isocyanato-4-methoxybenzene (1.5 g, 0.01 mol) in CH2Cl2 (100 mL) was added prop-2-yn-1-amine (0.6 g, 0.011 mol) in one portion. After stirring at room temperature for 1.0 h, the mixture was concentrated, and the residue was purified by column chromatography on silica gel (eluent: PE: EA = 5:1) to give compound 2a as a solid.
5.3 General procedure for preparation of compound 3a (The method is suitable for 3b-3j)
The reaction reagents were compound 2a (0.2 g, 1.0 mmol) and 1-(azidomethyl)-2-bromobenzene (0.2 g, 1.2 mmol), which were added to 15 mL of mixed solvent (water: THF: tert-butanol = 1:1:1). The reaction was performed in copper sulfate pentahydrate (0.1 mmol) and sodium ascorbate (0.2 mmol) at 80°C. After completion of the reaction, the mixture was extracted through CH2Cl2 (10 mL×3). The organic phase was combined and concentrated in vacuo. The desired compound 3a was isolated by column chromatography (CH2Cl2/MeOH = 20:1).
1-((1-(2-bromobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-3-(4-methoxyphenyl)urea (3a). Gray solid. LC-MS(ESI): m/z 416.07 (M + H)+. Yield: 81.4%. 1H NMR (600 MHz, DMSO-d6) δ 8.34 (s, 1H, NH-H), 7.97 (s, 1H, NH-H), 7.69 (d, J = 6.0 Hz, 1H, CH-H), 7.40 (t, J1 = 6.0 Hz, J2 = 6.0 Hz, 1H, Ar-H), 7.32–7.27 (m, 3H, Ar-H), 7.14 (d, J = 6.0 Hz, 1H, Ar-H), 6.81 (d, J = 6.0 Hz, 2H, Ar-H), 6.47 (t, J1 = 6.0 Hz, J2 = 6.0 Hz, 1H, Ar-H), 5.66 (s, 2H, CH2-H), 4.32 (d, J = 6.0Hz, 2H, CH2-H), 3.69 (s, 3H, OCH3-H). 13C NMR (150 MHz, DMSO-d6) δ 155.7, 154.4, 135.5, 133.9, 133.3, 130.8, 130.8, 128.7, 123.6, 123.3, 119.9, 114.3, 55.6, 53.2, and 35.3.
1-((1-(3-bromobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-3-(4-methoxyphenyl)urea (3b). Gray solid. LC-MS(ESI): m/z 416.07 (M + H)+. Yield: 89.7%. 1H NMR (600 MHz, DMSO-d6) δ 8.34 (s, 1H, NH-H), 8.08 (s, 1H, NH-H), 7.53 (d, J = 6.0 Hz, 2H, CH-H, Ar-H), 7.35–7.28 (m, 4H, Ar-H), 6.81 (d, J = 12.0 Hz, 2H, Ar-H), 6.47 (s, 1H, Ar-H), 5.59 (s, 2H, CH2-H), 4.31 (s, 2H, CH2-H), 3.69 (s, 3H, OCH3-H). 13C NMR (150 MHz, DMSO-d6) δ 155.8, 154.4, 139.2, 133.9, 131.4, 131.4, 131.2, 127.5, 123.7, 122.2, 119.9, 114.3, 55.6, 52.3, and 35.4.
1-((1-(3-methoxybenzyl)-1H-1,2,3-triazol-4-yl)methyl)-3-(4-methoxyphenyl)urea (3c). White solid. LC-MS(ESI): m/z 368.17 (M + H)+. Yield: 82.8%. 1H NMR (600 MHz, DMSO-d6) δ 8.34 (s, 1H, NH-H), 8.03 (s, 1H, NH-H), 7.29–7.26 (m, 3H, CH-H, Ar-H), 6.89 (d, J = 6.0 Hz, 2H, Ar-H), 6.86 (d, J = 6.0 Hz, 1H, Ar-H), 6.81 (d, J = 6.0 Hz, 2H, Ar-H), 6.46 (s, 1H, Ar-H), 5.53 (s, 2H, CH2-H), 4.31 (s, 2H, CH2-H), 3.73 (s, 3H, OCH3-H), 3.69 (s, 3H, OCH3-H). 13C NMR (150 MHz, DMSO-d6) δ 159.8, 155.8, 154.4, 138.0, 133.9, 130.3, 120.5, 119.9, 114.3, 114.2, 113.8, 55.6, 55.5, 53.1, and 35.4.
1-((1-(2-bromobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-3-(4-bromophenyl)urea (3days). White solid. LC-MS(ESI): m/z 463.97 (M + H)+. Yield: 66.9%. 1H NMR (600 MHz, DMSO-d6) δ 8.71 (s, 1H, NH-H), 7.99 (s, 1H, NH-H), 7.68 (d, J = 6.0 Hz, 1H, CH-H), 7.40–7.36 (m, 5H, Ar-H), 7.31 (t, J1 = 6.0 Hz, J2 = 6.0 Hz, 1H, Ar-H), 7.15 (d, J = 6.0 Hz, 1H, Ar-H), 6.64 (s, 1H, Ar-H), 5.66 (s, 2H, CH2-H), 4.34 (d, J = 6.0 Hz, 2H, CH2-H). 13C NMR (151 MHz, DMSO-d6) δ 155.3, 140.2, 135.4, 133.3, 131.8, 130.8, 130.8, 128.7, 123.7, 123.3, 120.0, 112.9, 53.3, and 35.3.
1-((1-(3-bromobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-3-(4-bromophenyl)urea (3e). White solid. LC-MS(ESI): m/z 463.97 (M + H)+. Yield: 90.3%. 1H NMR (600 MHz, DMSO-d6) δ 8.71 (s, 1H, NH-H), 8.06 (s, 1H, NH-H), 7.53 (d, J = 6.0 Hz, 2H, CH-H, Ar-H), 7.39–7.35 (m, 4H, Ar-H), 7.33–7.30 (m, 1H, Ar-H), 6.64 (s, 1H, Ar-H), 5.59 (s, 2H, CH2-H), 4.33 (d, J = 6.0 Hz, 2H, CH2-H). 13C NMR (150 MHz, DMSO-d6) δ 155.3, 140.2, 139.2, 131.8, 131.4, 131.4, 131.1, 127.5, 123.5, 122.2, 120.0, 112.9, 52.3, and 35.3.
1-(2-(1-(2-bromobenzyl)-1H-1,2,3-triazol-4-yl)propan-2-yl)-3-(4-methoxyphenyl)urea (3f). White solid. LC-MS(ESI): m/z 444.10 (M + H)+. Yield: 77.2%. 1H NMR (600 MHz, DMSO-d6) δ 8.23 (s, 1H, NH-H), 7.99 (s, 1H, NH-H), 7.68 (d, J = 12.0Hz, 1H, CH-H), 7.38 (t, J1 = 12.0Hz, J2 = 12.0Hz, 1H, Ar-H), 7.30 (t, J1 = 12.0Hz, J2 = 12.0Hz, 1H, Ar-H), 7.23–7.20 (m, 2H, Ar-H), 7.06 (d, J = 6.0Hz, 1H, Ar-H), 6.78 (d, J = 12.0Hz, 2H, Ar-H), 6.40 (s, 1H, Ar-H), 5.65 (s, 2H, CH2-H), 3.68 (s, 3H, OCH3-H), 1.64 (s, 6H, 2CH3-H). 13C NMR (150 MHz, DMSO-d6) δ 154.8, 154.3, 154.2, 135.7, 134.0, 133.3, 130.7, 130.4, 128.7, 123.0, 122.3, 119.5, 114.3, 55.6, 53.2, 50.5, 40.5, and 28.9.
1-(2-(1-(3-bromobenzyl)-1H-1,2,3-triazol-4-yl)propan-2-yl)-3-(4-methoxyphenyl)urea (3 g). White solid. LC-MS(ESI): m/z 444.10 (M + H)+. Yield: 82.4%. 1H NMR (600 MHz, DMSO-d6) δ 8.22 (s, 1H, NH-H), 8.05 (s, 1H, NH-H), 7.53 (s, 2H, CH-H, Ar-H), 7.36–7.29 (m, 2H, Ar-H), 7.23–7.20 (m, 2H, Ar-H), 6.78 (d, J = 18.0Hz, 2H, Ar-H), 6.39 (s, 1H, Ar-H), 5.57 (s, 2H, CH2-H), 3.67 (s, 3H, OCH3-H), 1.62 (s, 6H, 2CH3-H). 13C NMR (150 MHz, DMSO-d6) δ 154.7, 154.5, 154.2, 139.3, 134.0, 131.4, 131.4, 131.1, 127.4, 122.3, 122.0, 119.5, 114.3, 55.6, 52.2, 50.4, and 28.9.
1-(2-(1-(3-methoxybenzyl)-1H-1,2,3-triazol-4-yl)propan-2-yl)-3-(4-methoxyphenyl)urea (3 h). White solid. LC-MS(ESI): m/z 396.20 (M + H)+. Yield: 87.7%. 1H NMR (600 MHz, DMSO-d6) δ 8.23 (s, 1H, NH-H), 8.01 (s, 1H, NH-H), 7.29–7.20 (m, 3H, CH-H, Ar-H), 6.89–6.77 (m, 5H, Ar-H), 6.39 (s, 1H, Ar-H), 5.52 (s, 2H, CH2-H), 3.72 (s, 3H, OCH3-H), 3.68 (s, 3H, OCH3-H), 1.63 (s, 6H, 2CH3-H). 13C NMR (150 MHz, DMSO-d6) δ 159.9, 154.7, 154.4, 154.2, 138.1, 134.0, 133.4, 130.3, 121.9, 120.3, 119.5, 114.4, 114.3, 113.9, 113.8, 55.5, 55.5, 53.0, 50.5, and 28.9.
1-(2-(1-(2-bromobenzyl)-1H-1,2,3-triazol-4-yl)propan-2-yl)-3-(4-bromophenyl)urea (3i). White solid. LC-MS(ESI): m/z 492.00 (M + H)+. Yield: 79.6%. 1H NMR (600 MHz, DMSO-d6) δ 8.58 (s, 1H, NH-H), 8.01 (s, 1H, NH-H), 7.68 (d, J = 12.0Hz, 1H, CH-H), 7.40–7.28 (m, 6H, Ar-H), 7.05 (d, J = 12.0Hz, 1H, Ar-H), 6.57 (s, 1H, Ar-H), 5.65 (s, 2H, CH2-H), 1.64 (s, 6H, 2CH3-H). 13C NMR (150 MHz, DMSO-d6) δ 154.3, 154.1, 140.3, 135.6, 133.3, 133.3, 131.8, 130.7, 130.4, 128.7, 123.0, 122.3, 119.8, 112.6, 87.7, 53.2, 50.6, 40.5, and 28.8.
1-(2-(1-(3-bromobenzyl)-1H-1,2,3-triazol-4-yl)propan-2-yl)-3-(4-bromophenyl)urea (3j). White solid. LC-MS(ESI): m/z 492.00 (M + H)+. Yield: 72.8%. 1H NMR (600 MHz, DMSO-d6) δ 8.58 (s, 1H, NH-H), 8.07 (s, 1H, NH-H), 7.54–7.52 (m, 2H, CH-H, Ar-H), 7.46–7.43 (m, 2H, Ar-H), 7.36–7.34 (m, 2H, Ar-H), 7.31–7.28 (m, 2H, Ar-H), 6.57 (s, 1H, Ar-H), 5.58 (s, 2H, CH2-H), 1.63 (s, 6H, 2CH3-H). 13C NMR (150 MHz, DMSO-d6) δ 154.3, 154.2, 140.3, 139.3, 132.0, 131.7, 131.4, 131.4, 131.1, 127.4, 122.3, 122.1, 120.7, 119.8, 112.6, 52.2, 50.6, and 28.8.
5.4 IDO1 enzymatic inhibition assay
To assess the inhibitory effect of the designed compounds against IDO1, Hela cells were seeded at 50,000–60,000 cells per well in 100 μL of complete Dulbecco’s modified eagle medium growth medium in a 96-well plate and incubated for 12–18 h. On the second day, diluted inhibitor at a final concentration of 100 ng/mL human IFN-γ was added (100 μL per well) and incubated at 37°C with 5% CO2 for 18 h. On the third day, 140 μL of medium was transferred into a new 96-well plate, and the protein was precipitated with 10 μL of 6.1 N trichloroacetic acid (CCl3COOH) at 50°C for 30 min. The plate was then centrifuged at 2,500 rpm for 10 min, and 100 μL of supernatant was transferred to another 96-well plate and mixed with 100 μL of 2% (w/v) p-dimethylaminobenzaldehyde in CH3COOH. The plate was incubated at 25°C for 10 min, and the yellow color derived from kynurenine was measured at 480 nm using a microplate reader (PE, United States). Inhibition curves with IC50 values were generated using Prism 6.0.
5.5 Molecular docking
Docking experiments were conducted using Schrödinger (Schrödinger LLC 2015; United States). The 4PK5 complex system with Amg-1 as the co-crystal ligand was chosen for the study. The protein was processed and optimized using the Protein Preparation Wizard, which involved hydrogenation of the protein, assignment of bond orders, creation of zero-order bonds with metals, creation of disulfide bonds, removal of water molecules larger than 5 Å from het groups, structural optimization, and energy minimization. The ligands were constructed using ChemDraw and optimized using the LigPrep module under the condition of pH 7.0 ± 2.0 for protonation and generation of stereoisomers. Molecular lattices centered on Amg-1 were generated, and metal coordination constraints centered on heme iron were applied. Docking was performed using SP precision.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Author contributions
X-XH, Z-YW and J-XY conceived the study, designed the experiments, and supervised all research. AZ prepared the draft of manuscript. EG and L-FM revised the manuscript. H-LW analyzed the data and performed molecular docking. All authors reviewed the manuscript.
Conflict of interest
Author L-FM was employed by the company Henan Wanliu Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2023.1222825/full#supplementary-material
References
Babu, D. S., Srinivasulu, D., and Kotakadi, V. S. (2015). Synthesis of novel N-aryl-4-[6-(2-fluoropyridin-3-yl)quinazolin-2-yl]-piperazine-1-carboxamide or -carbothioamide derivatives and their antimicrobial activity. Chem. Heterocycl. Compd. 51, 60–66. doi:10.1007/s10593-015-1660-2
Hou, X., Gong, X., Mao, L., Zhao, J., and Yang, J. (2022a). Discovery of novel 1,2,3-triazole derivatives as Ido1 inhibitors. Pharm. (Basel) 15, 1316. doi:10.3390/ph15111316
Hou, X. X., Gong, X. Q., Mao, L. F., Sun, G., and Yang, J. X. (2022b). Design, synthesis and biological evaluation of erlotinib-based Ido1 inhibitors. Front. Pharmacol. 13, 940704. doi:10.3389/fphar.2022.940704
Jr, O. W., Lew, N. L., Liu, Y., Lowrie, E. G., and Lazarus, J. M. (1993). The urea reduction ratio and serum albumin concentration as predictors of mortality in patients undergoing hemodialysis. N. Engl. J. Med. 329, 1001–1006. doi:10.1056/NEJM199309303291404
Kapron, B., Plazinska, A., Plazinski, W., and Plech, T. (2023). Identification of the first-in-class dual inhibitors of human DNA topoisomerase IIα and indoleamine-2,3-dioxygenase 1 (Ido 1) with strong anticancer properties. J. Enzyme Inhibition Med. Chem. 38, 192–202. doi:10.1080/14756366.2022.2140420
Kumar, S., Waldo, J. P., Jaipuri, F. A., Marcinowicz, A., Van Allen, C., Adams, J., et al. (2019). Discovery of clinical candidate (1r,4r)-4-((R)-2-((S)-6-Fluoro-5H-imidazo[5,1-a]isoindol-5-yl)-1-hydroxyethyl)cyclohexan-1-ol (navoximod), a potent and selective inhibitor of indoleamine 2,3-dioxygenase 1. J. Med. Chem. 62, 6705–6733. doi:10.1021/acs.jmedchem.9b00662
Mcbriar, M. D., Guzik, H., Shapiro, S., Xu, R., Paruchova, J., Clader, J. W., et al. (2006). Bicyclo[3.1.0]hexyl urea melanin concentrating hormone (MCH) receptor-1 antagonists: Impacting hERG liability via aryl modifications. Bioorg. Med. Chem. Lett. 16, 4262–4265. doi:10.1016/j.bmcl.2006.05.069
Muller, A. J., Duhadaway, J. B., Donover, P. S., Sutanto-Ward, E., and Prendergast, G. C. (2005). Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat. Med. 11, 312–319. doi:10.1038/nm1196
Nakagawa, T., Lomb, D. J., Haigis, M. C., and Guarente, L. P. (2009). SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137, 560–570. doi:10.1016/j.cell.2009.02.026
Nelp, M. T., Kates, P. A., Hunt, J. T., Newitt, J. A., Balog, A., Maley, D., et al. (2018). Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form. Proc. Natl. Acad. Sci. 115, 3249–3254. doi:10.1073/pnas.1719190115
Pan, S., Zhou, Y., Wang, Q., Wang, Y., Tian, C., Wang, T., et al. (2020). Discovery and structure-activity relationship studies of 1-aryl-1H-naphtho[2,3-d] [1,2,3]triazole-4,9-dione derivatives as potent dual inhibitors of indoleamine 2,3-dioxygenase 1 (Ido1) and trytophan 2,3-dioxygenase (TDO). Eur. J. Med. Chem. 207, 112703. doi:10.1016/j.ejmech.2020.112703
Qi, Z. Y., Hao, S. Y., Tian, H. Z., Bian, H. L., Hui, L., and Chen, S. W. (2020). Synthesis and biological evaluation of 1-(benzofuran-3-yl)-4-(3,4,5-trimethoxyphenyl)-1H-1,2,3-triazole derivatives as tubulin polymerization inhibitors. Bioorg. Chem. 94, 103392. doi:10.1016/j.bioorg.2019.103392
Röhrig, U. F., Majjigapu, S. R., Grosdidier, A., Bron, S., Stroobant, V., Pilotte, L., et al. (2012). Rational design of 4-aryl-1,2,3-triazoles for indoleamine 2,3-dioxygenase 1 inhibition. J. Med. Chem. 55, 5270–5290. doi:10.1021/jm300260v
Rohrig, U. F., Reynaud, A., Majjigapu, S. R., Vogel, P., Pojer, F., and Zoete, V. (2019). Inhibition mechanisms of indoleamine 2,3-dioxygenase 1 (Ido1). J. Med. Chem. 62, 8784–8795. doi:10.1021/acs.jmedchem.9b00942
Serafini, M., Torre, E., Aprile, S., Grosso, E. D., Gesù, A., Griglio, A., et al. (2020). Discovery of highly potent benzimidazole derivatives as indoleamine 2,3-dioxygenase-1 (Ido1) inhibitors: From structure-based virtual screening to in vivo pharmacodynamic activity. J. Med. Chem. 63, 3047–3065. doi:10.1021/acs.jmedchem.9b01809
Sun, G., Mao, L., Deng, W., Xu, S., Zhao, J., Yang, J., et al. (2022). Discovery of a series of 1,2,3-triazole-containing erlotinib derivatives with potent anti-tumor activities against non-small cell lung cancer. Front. Chem. 9, 789030. doi:10.3389/fchem.2021.789030
Tang, W., Chen, Z., Zhang, W., Cheng, Y., Zhang, B., Wu, F., et al. (2020). The mechanisms of sorafenib resistance in hepatocellular carcinoma: Theoretical basis and therapeutic aspects. Signal Transduct. Target Ther. 5, 87. doi:10.1038/s41392-020-0187-x
Wang, K., Song, L. H., Liang, Q. L., Zhang, Y., Ma, X. L., Wang, Q., et al. (2023). Discovery of novel sulfonamide chromone-oxime derivatives as potent indoleamine 2,3-dioxygenase 1 inhibitors. Eur. J. Med. Chem. 254, 115349. doi:10.1016/j.ejmech.2023.115349
Yau, T., Park, J. W., Finn, R. S., Cheng, A. L., Mathurin, P., Edeline, J., et al. (2022). Nivolumab versus sorafenib in advanced hepatocellular carcinoma (CheckMate 459): A randomised, multicentre, open-label, phase 3 trial. Lancet Oncol. 23, 77–90. doi:10.1016/S1470-2045(21)00604-5
Ye, K., Wang, K., Wang, T., Tang, H., Wang, L., Zhang, W., et al. (2023). Design, synthesis, biological evaluation of urea substituted 1,2,5-oxadiazole-3-carboximidamides as novel indoleamine 2,3-dioxygenase-1 (Ido1) inhibitors. Eur. J. Med. Chem. 250, 115217. doi:10.1016/j.ejmech.2023.115217
Keywords: indoleamine 2,3-dioxygenase 1, 1,2,3-triazole, N,N-diphenylurea, molecular docking, inhibitor
Citation: Hou X-X, Wu Z-Y, Zhan A, Gao E, Mao L-F, Wang H-L and Yang J-X (2023) Synthesis and activity study of novel N,N-diphenylurea derivatives as IDO1 inhibitors. Front. Chem. 11:1222825. doi: 10.3389/fchem.2023.1222825
Received: 15 May 2023; Accepted: 25 May 2023;
Published: 20 June 2023.
Edited by:
Hong Chen, Luoyang Normal University, ChinaCopyright © 2023 Hou, Wu, Zhan, Gao, Mao, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Xue Yang, bHVjeWZseTg4MTEwNEAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Xi-Xi Hou1†
Xi-Xi Hou1† An Zhan
An Zhan Long-Fei Mao
Long-Fei Mao