- 1Department Biotechnology and Bioinformatics, Yogi Vemana University, Kadapa, India
- 2Department of Medical Laboratories, College of Applied Medical Sciences, Qassim University, Buraidah, Saudi Arabia
- 3Department of Agriculture Science, Faculty of Agro-Based Industry, Universiti Malaysia Kelantan, Jeli, Malaysia
- 4Department of Biochemistry, Faculty of Medicine, Bioscience, and Nursing, MAHSA University, Jenjarom, Malaysia
- 5Department of Periodontics, Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical Sciences, Chennai, India
- 6Department of Foundation, RCSI & UCD Malaysia Campus, Georgetown, Malaysia
- 7Department of Pharmacognosy, Faculty of Pharmacy, Ain shams University, Cairo, Egypt
Abstract
Nanobiotechnology is a popular branch of science that is gaining interest among scientists and researchers as it allows for the green manufacturing of nanoparticles by employing plants as reducing agents. This method is safe, cheap, reproducible, and eco-friendly. In this study, the therapeutic property of Piper nigrum fruit was mixed with the antibacterial activity of metallic copper to produce copper nanoparticles. The synthesis of copper nanoparticles was indicated by a color change from brown to blue. Physical characterization of Piper nigrum copper nanoparticles (PN-CuNPs) was performed using UV-vis spectroscopy, FT-IR, SEM, EDX, XRD, and Zeta analyzer. PN-CuNPs exhibited potential antioxidant, antibacterial, and cytotoxic activities. PN-CuNPs have shown concentration-dependent, enhanced free radical scavenging activity, reaching maximum values of 92%, 90%, and 86% with DPPH, H2O2, and PMA tests, respectively. The antibacterial zone of inhibition of PN-CuNPs was the highest against Staphylococcus aureus (23 mm) and the lowest against Escherichia coli (10 mm). PN-CuNPs showed 80% in vitro cytotoxicity against MCF-7 breast cancer cell lines. Furthermore, more than 50 components of Piper nigrum extract were selected and subjected to in silico molecular docking using the C-Docker protocol in the binding pockets of glutathione reductase, E. coli DNA gyrase topoisomerase II, and epidermal growth factor receptor (EGFR) tyrosine to discover their druggability. Pipercyclobutanamide A (26), pipernigramide F (32), and pipernigramide G (33) scored the highest Gibbs free energy at 50.489, 51.9306, and 58.615 kcal/mol, respectively. The ADMET/TOPKAT analysis confirmed the favorable pharmacokinetics, pharmacodynamics, and toxicity profiles of the three promising compounds. The present in silico analysis helps us to understand the possible mechanisms behind the antioxidant, antibacterial, and cytotoxic activities of CuNPs and recommends them as implicit inhibitors of selected proteins.
Introduction
Nanotechnology emerged as an interdisciplinary approach with various applications. The emergence of nanoparticles (NPs) created a scientific revolution in terms of providing a sustainable environment for humans. NPs are promising due to their optical, catalytic, and surface-to-volume ratio (Singh et al., 2018; Sarma et al., 2019) and are currently employed to treat a variety of ailments, as well as for energy storage and drug/gene delivery systems (Pomerantseva et al., 2019; Fries et al., 2021).
Physical and chemical methods employed for the synthesis of NPs include lithography, ultrasonic fields, UV irradiation, and photochemical reduction (Mafuné et al., 2002; Naik et al., 2002). However, these methods require hazardous chemicals as reducing agents. Hence, green synthesis of nanoparticles was introduced as an alternative method that used either microbes or plant extract of medicinal plants as reducing agents. Green synthesis of nanoparticles by microbes is a time-consuming process (Thakkar et al., 2010) for maintenance, but a plant-extract-mediated process requires less time. Green synthesis of copper nanoparticles (CuNPs) using plants has been reported recently with Cocculus hirsutus (Ameena et al., 2022), Kigelia africana fruit, and Sesbania aculeata (Tamil Elakkiya et al., 2022). Natural compounds can act as an alternative for novel drug synthesis, and plant-derived compounds showed high efficiency and easy availability (Cragg and Newman, 2013). Medicinal herbs have various bioactive molecules exhibiting various biological properties, such as antioxidant, antimicrobial (Mseddi et al., 2020), antibiofilm (Gad-Elkareem et al., 2019), anti-quorum sensing (Snoussi et al., 2016), and antidiabetic (Moussa et al., 2013; Noumi et al., 2018) properties.
Gold, silver, platinum, ruthenium, zinc, and copper are various metals used for the synthesis of metal NPs (Akintelu et al., 2019; Akintelu and Folorunso, 2019; Folorunso et al., 2019; Nasrollahzadeh et al., 2019; Almatroudi et al., 2020; Anjum and Riazunnisa, 2022). Among these metal NPs, copper nanoparticles (CuNPs) are gaining popularity because of their high surface area, low cost, and eco-friendliness. Physical characteristics of CuNPs such as morphology, crystallinity, composition (Cuevas et al., 2015), a simple methodology for synthesis, and easy modification into the required shape and size make them exceptional (Balasooriya et al., 2017). The biological activities of CuNPs make them an ideal source for antibiotic production, as new antibiotics are urgently needed (Chung et al., 2017; Moussa et al., 2020). According to the WHO, 177 ± 16 ppb is the permissible limit of copper (Sudha et al., 2012) in stored food and drinking water. Copper vessels purify drinking water by killing Escherichia coli, rapidly and efficiently destroying bacteria through a contact-killing mode (Santo et al., 2011).
Applying in silico molecular docking to augment drug discovery research has gained much scientific interest in recent years. Not only does it save time and effort spent on inactive molecules, but also resources are spared to develop promising drug entities to clinical stages. Virtual screening allows the use of chemical structures of literature compounds compiled from eminent databases, filtered, and prepared to adopt a proper experimental design to predict the activity inside the binding pockets of certain enzymes (Altyar et al., 2022). Piper nigrum, also known as pepper, is a member of the Piperaceae family. It is known as the “king of spices” (Takooree et al., 2019). P. nigrum seed is used as an acceptable food additive (Meghwal and Goswami, 2013). Piperine, an alkaloid responsible for the pungent odor, is used in traditional medicine and as an insecticide; it also has anticonvulsant properties. Phytochemicals present in black pepper include phenols, flavonoids, alkaloids, amides, essential oils, lignans, neolignans, terpenes, and chalcones (Liu et al., 2010; Shityakov et al., 2019).
The water extract obtained from P. nigrum fruits was shown to be rich in pungent principles such as alkamides and piperine alkaloids; thus, these compounds were assembled from the literature to perform virtual screening. The same crude extract was utilized in all the bioassays described herein. The literature search revealed more than 55 compounds isolated from P. nigrum fruits along with volatile metabolites. Due to the presence of biologically active components, they are considered ideal natural therapeutic agents against bacterial, cancer, and fungal cells (Aldaly, 2010; Reshmi et al., 2010; Zhang et al., 2021). Various bio-activities were correlated with the alkaloid/alkamide components, particularly, piperine (Tyagi et al., 2021). While 1% of P. nigrum fruits comprised essential oils, dominated by α-pinene, gluulol, α-terpinene, and β-caryophyllene, 5%–9% represented pungent principle alkamides. In this study, approximately 60 alkamide/alkaloid compounds and their derivatives were selected to investigate their binding properties and potential as antioxidants, antibacterial, and antitumor agents. Furthermore, ADMET/TOPKAT analysis was conducted to reveal their pharmacokinetic properties, pharmacodynamics, and toxicity.
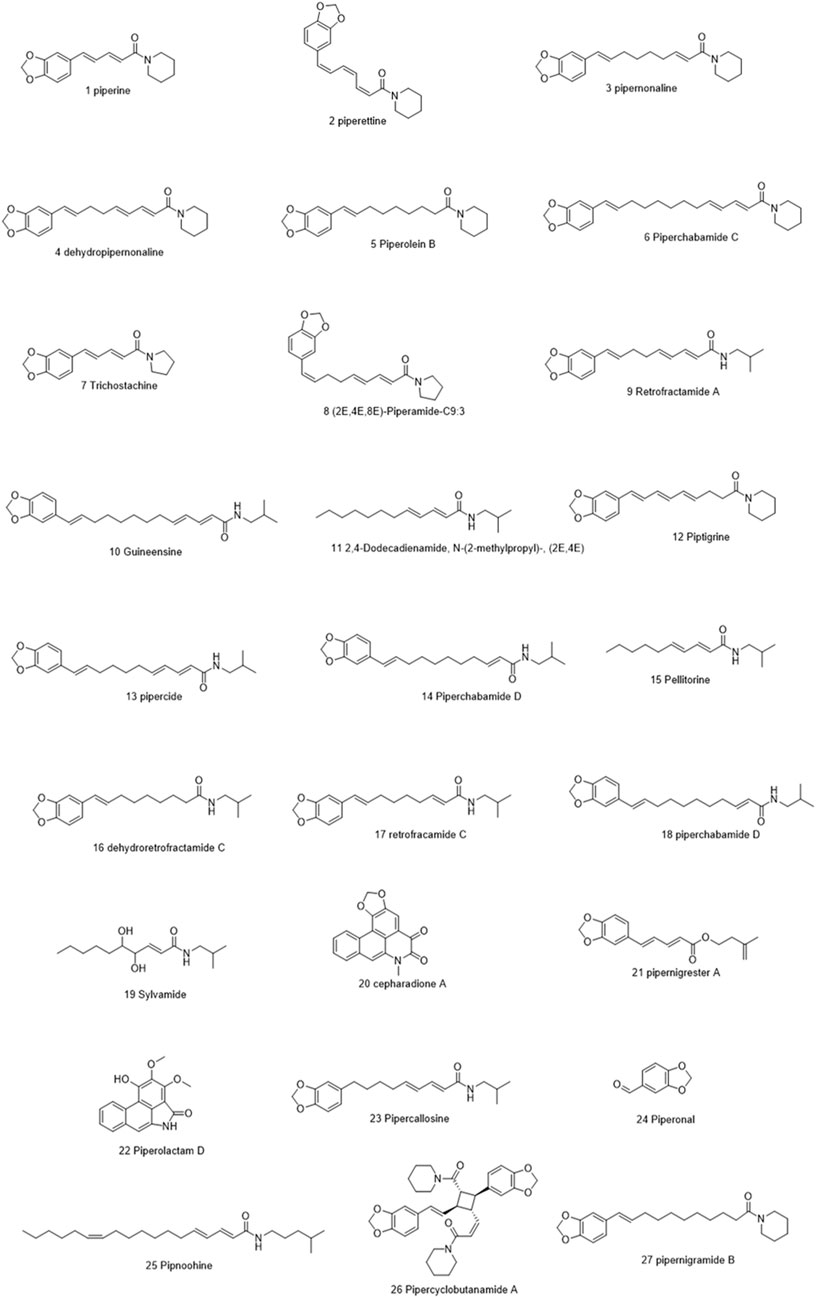
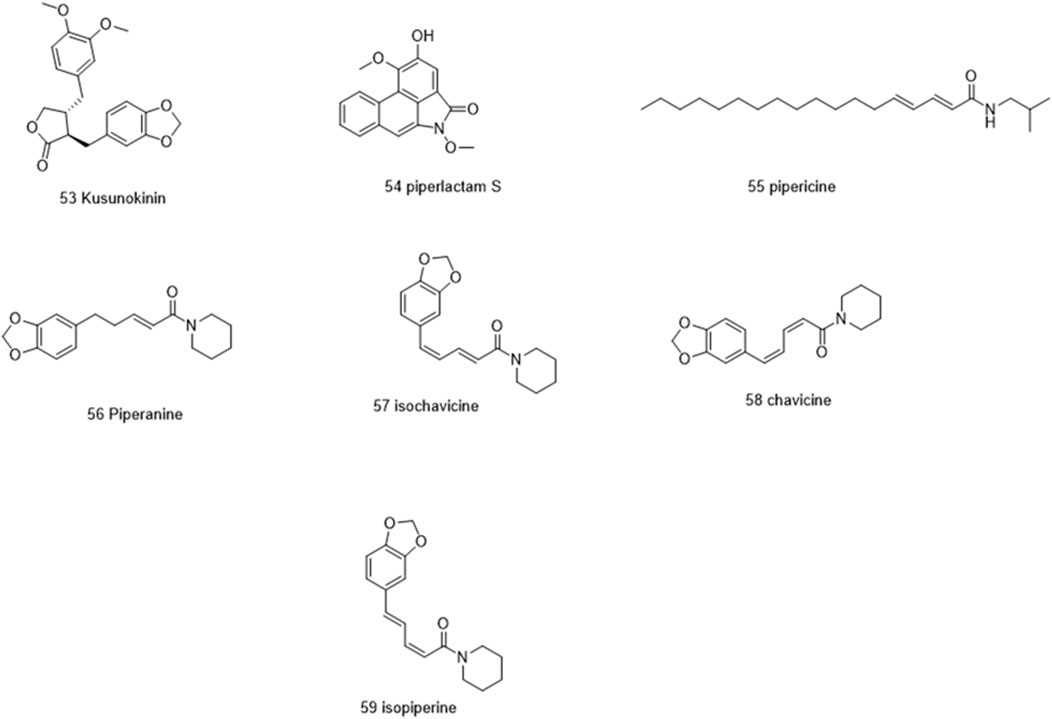
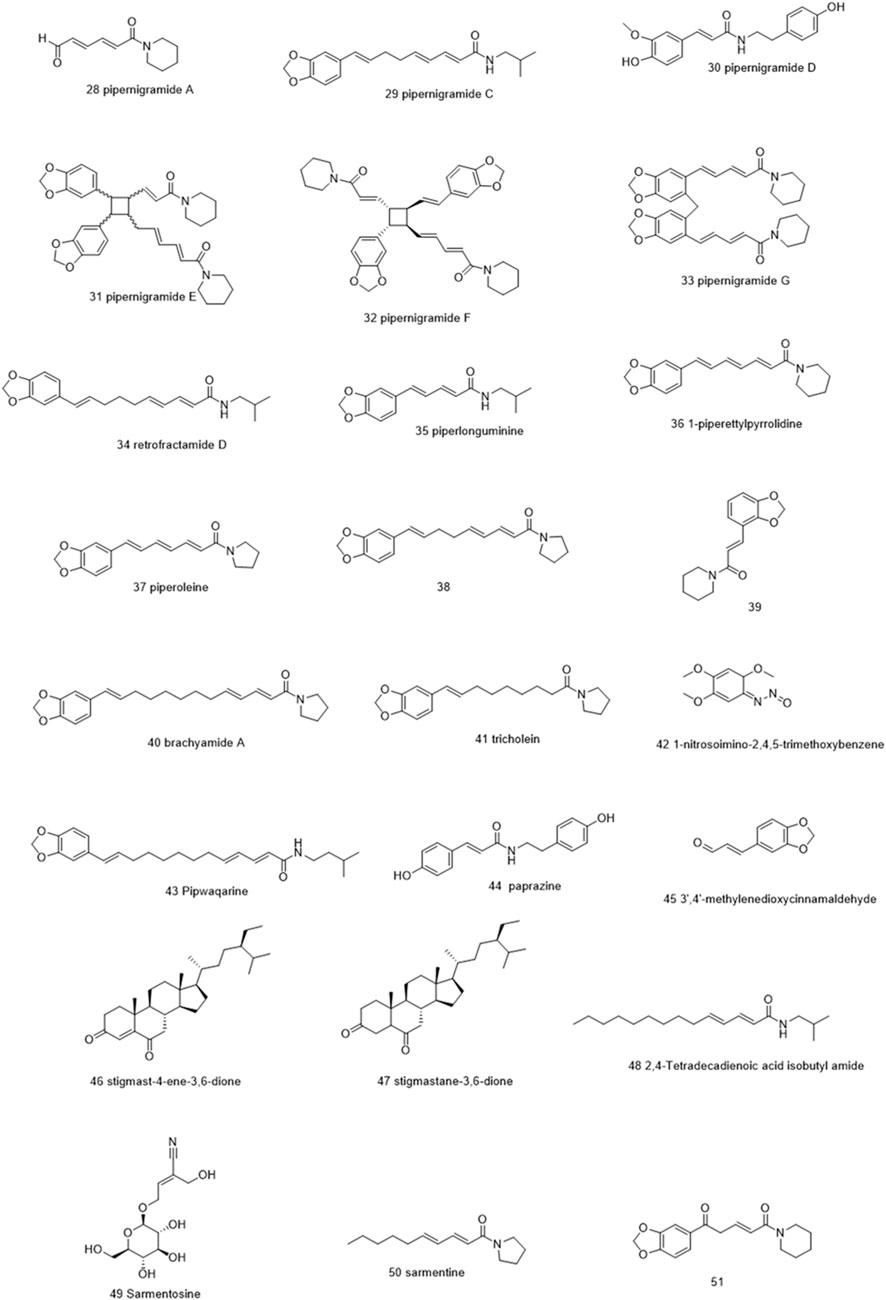
Molecules selected for the in silico study are as follows: the active chemosensate components in black pepper, sharing piperidine, pyrrolidine, and isobutyl amide group, and classified as two categories of amides: the piperonal moiety-associated amides and the long chain fatty acid unsaturated amides (Dawid et al., 2012). The anti-inflammatory alkamides, including pipernonaline, guineensine, pellitorine, retrofractamide C, piperolein B, (2E,4Z,8E)-N-[9-(3,4-methylenedi-oxyphenyl)-2,4,8-nonatrienoyl]piperidine, piperchabamide D, dehydropipernonaline, retrofractamides, and pipernigramides, are also investigated (Figures 1–3) (Scott et al., 2005; Rho et al., 2007; Yun et al., 2018; Shityakov et al., 2019; Barata et al., 2021).
Piperine, the well-known alkaloid isolated from P. nigrum hundreds of years ago, showed the lowest threshold of pungency followed by piperyline and piperettine. Indeed, the isobutyl amine- and pyrrolidine-containing compounds revealed a higher threshold than any of the piperidine analogs (Dawid et al., 2012) Some of these compounds, such as piperonylamine, pipericide, sarmentosine, sarmentine, and chavicine, are bioactive compounds with pharmacological effects (Figure 1) (Majdalawieh and Carr, 2010). In particular, piperine is believed to be the main bioactive chemical component with antimicrobial activities purified from P. nigrum (Mohammed et al., 2016).
Bacterial antibiotic resistance will result in serious human health problems and deaths. If this issue is not taken seriously, approximately 10 million people will die from antimicrobial-resistant bacterial infections by 2050. Recently, nanotechnology-based strategies have attracted attention to the development of new diagnostic technologies and devices in modern medicine. CuNPs have been extensively used in the medical field due to their superb antimicrobial nature against multidrug-resistant pathogens.
Phytochemicals present in P. nigrum fruit extract might be involved in the reduction and stabilization of CuNP synthesis. Therefore, the current study focuses on the green synthesis of P. nigrum fruit-based copper nanoparticles using aqueous P. nigrum fruit extract, as well as their physical and biological characterization (antioxidant, antibacterial, and in vitro cytotoxicity). The invention of the unique size, shape, and structure of CuNPs exhibits multispectrum activity and has the potential to obstruct bacterial, cancer growth, and infection prevalence in clinical applications. Furthermore, we evaluated these in vitro activities with in silico studies to determine the correlation between them. Molecular docking studies were performed on the key active compounds in black pepper to manifest their druggability in the active sites of epidermal growth factor receptor (EGFR), DNA gyrase topoisomerase II, and glutathione reductase.
Materials and methods
Piper nigrum fruit collection and preparation of fruit extract
P. nigrum fruits were purchased from the local market in Kadapa, Andhra Pradesh, India. India is the major producer of P. nigrum. A voucher specimen of P. nigrum fruit (from the sample collected from a local market in Kadapa, Andhra Pradesh) was deposited and authenticated by Dr. A. Madhusudhana Reddy, Head of Herbarium Division, Dept. of Botany, Yogi Vemana University, Kadapa, India (the voucher specimen number of Piper nigrum fruit is YVU/KR-5301-5302).
P. nigrum fruits were washed, shade-dried, and powdered before being used. A measure of 10 g of powdered P. nigrum was mixed with 100 mL of double-distilled water and boiled in a water bath at 70–80°C for 15–20 min. The solution was cooled and filtered using Whatman No: 1 filter paper. The filtrate solution was then stored at 4 C for further analysis (Tyagi et al., 2021).
Synthesis of P. nigrum fruit extract copper nanoparticles
P. nigrum copper nanoparticles (PN-CuNPs) were synthesized by adding 10 mL of fruit extract dropwise to 200 mL of copper acetate (3 mM) solution on a magnetic stirrer. The color change of the solution from brown to blue represents the formation of PN-CuNPs. The reduction of copper ions was monitored using a UV-visible spectrometer (Thermo Scientific Evolution 201) with a wavelength range of 200–800 nm. The reaction mixture was centrifuged at 10,000 rpm for 10 min, and the pellet was washed with distilled water and air-dried. The synthesized PN-CuNPs were stored at 4°C for further analysis.
Characterization of PN-CuNPs
UV-vis spectrophotometry analysis
The absorption maxima of 2 mL of freshly prepared reaction mixture of PN-CuNPs were obtained using a UV-Vis spectrophotometer (Thermo Scientific Evolution 401). The spectra were recorded with a wavelength range of 200–800 nm at regular time intervals (10, 15, 30, 45, 30, 45, 60, 120, and 180 min) using double-distilled water as a blank. We calculated the band gap energy using the UV spectrum data following Sabouri et al. [74].
X-ray diffraction analysis of PN-CuNPs
X-ray diffraction (XRD) analysis of PN-CuNPs was performed on a PANLytical ‘X’ Pert Pro diffractometer operated at 40 kV and 30 mA with Cu K-alpha radiation. The average size of PN-CuNPs can be calculated using the Debye–Scherer equation as follows:
where D is the crystallite size of the copper nanoparticles, λ is the wavelength, β denotes the full width at half maximum of the diffraction peak, k denotes the Scherer constant with a value from 0.9–1, and θ denotes the Bragg angle.
FT-IR analysis of PN-CuNPs
Dehydrated PN-CuNPs and P. nigrum fruit extract were used to analyze functional groups. A measure of 10 mg of PN-CuNPs and P. nigrum fruit extract was mixed with 100 mg of KBr, vigorously ground into a fine powder, and compressed into diaphanous pellet discs followed by FT-IR [PerkinElmer (Spectrum Two model), UK VERTEX 70 model, Bruker, Germany]. Chemical and functional groups of the samples were obtained in the range of 400–4000 cm−1 at room temperature. The probable biomolecules responsible for the reduction, capping, and effective stabilization of the PN-CuNPs were recorded using an FT-IR spectrophotometer (Kanniah et al., 2021).
SEM and EDAX analysis of PN-CuNPs
The morphology and chemical composition of PN-CuNPs were examined by scanning electron microscopy (Model: EVO 18; Carl Zeiss, Germany) equipped with an energy-dispersive X-ray spectrometer (EDX). EDX was performed at an acceleration voltage of 20–40 keV. Thin films of PN-CuNPs were placed on a gold-coated grid by sprinkling a very small amount of PN-CuNPs on the grid. Thin films on the SEM grid were dried for 5 min and examined at different angles.
Particle size and zeta potential determination
The zeta potential and size of PN-CuNPs were measured by a Zetasizer Nano series instrument between 0.1 and 10,000 nm (Malvern Nano-ZS90). The stability of the nanoparticle sample was measured by maintaining the temperature of 25°C dispersion, 0.894 mPa of dispersion medium viscosity, and 0.073 mS/cm conductivity with 3.8 V electrode voltages.
In vitro antioxidant assays
DPPH free radical scavenging assay of P. nigrum fruit extract and PN-CuNPs
P. nigrum fruit extract and biosynthesized PN-CuNPs were subjected to 2,2-diphenyl-1-picrylhydrazyl (DPPH) analysis. Different concentrations of PN-CuNPs (25–150 μg/mL) and fruit extract were mixed with 3 mL of methanolic solution containing DPPH radicals (0.1 mM). Ascorbic acid was taken as a positive control. After 30 min, the absorbance was determined at 517 nm and converted into % using the following formula:
where A0 = absorbance of the control and A1 = absorbance of the test.
Hydrogen peroxide scavenging assay of P. nigrum fruit extract and PN-CuNPs
A modified protocol (Pavithra and Vadivukkarasi, 2015) was used for the H2O2 scavenging assay. Different concentrations of PN-CuNPs (25–150 μg/mL) and P. nigrum fruit extract were mixed with 40 mM H2O2 solution and allowed to incubate for 10 min, and the absorbance was measured at 230 nm. A positive control of ascorbic acid was used.
where A0 = absorbance of the control, and A1 = absorbance of the test.
Phosphomolybdenum assay of P. nigrum fruit extract and PN-CuNPs
A modified protocol (Pavithra and Vadivukkarasi, 2015) was used to estimate total antioxidant activity. Molybdate reagent solution was prepared by mixing 0.6 M sulfuric acid, 28 mM sodium phosphate, and 4 mM ammonium molybdate. Different concentrations of PN-CuNPs (25–150 μg/mL) and P. nigrum fruit extract were added to each test tube, which individually contained 3 mL of distilled water and 1 mL of molybdate reagent solution, followed by incubation in a water bath at 95°C for 90 min. After incubation, these test tubes were cooled to room temperature for 20–30 min, and the absorbance was measured at 695 nm using ascorbic acid as a positive control.
Antibacterial activity of P. nigrum fruit extract and PN-CuNPs
The antibacterial activity of PN-CuNPs and P. nigrum fruit extract was examined using the agar well method. Staphylococcus aureus and Bacillus subtilis (Gram-positive) and Escherichia coli and Proteus vulgaris (Gram-negative) were chosen to investigate the antibacterial efficacy of PN-CuNPs and P. nigrum fruit extract. Different concentrations of PN-CuNPs (25, 50, 75, 100, and 150 μg/mL) and P. nigrum fruit extract were used along with positive control ampicillin. Plates were incubated at 37°C for 24 h, and the diameter of the antibacterial zone of inhibition was measured.
Cytotoxicity of P. nigrum fruit extract and PN-CuNPs
The cytotoxicity of synthesized PN-CuNPs and P. nigrum fruit extract against MCF-7 cells was measured by MTT assay (cell lines were purchased from the National Centre for Cell Science (NCCS), Pune, India). MCF-7 cells were seeded at a density of 5 × 104 cells/well in 96-well plates, then treated with various concentrations of PN-CuNPs (25–200 μg/mL) and P. nigrum fruit extract, and incubated at 37°C for 24 h at 5% CO2 and 95% humidity. The cells were treated with MTT (5 mg/mL), and the absorbance was recorded at 570 nm.
In silico molecular docking studies of P. nigrum bioactive compounds with EGFR, DNA gyrase, and glutathione reductase
A total of 58 compounds were selected from the literature as the most bioactive components of P. nigrum fruits, specifically, alkamide/alkaloids. The molecular docking was designed to examine the binding sites of three proteins: glutathione reductase (Pdb ID: 1Xan), DNA gyrase (Pdb ID: 1Kzn), and EGFR (Pdb ID: 1m17). The X-ray crystal structures were downloaded from the Protein Data Bank (PDB) website (https://www.rcsb.org/), with their binding native ligands, HXP xanthene, clorobiocin, and erlotinib, respectively. The 3D chemical structures of the tested molecules and native ligands were prepared, and their energy was minimized by the MMFF94x force field using our previous protocol (Moussa et al., 2020). The computational study was performed using Discovery Studio 4.5 software at Ain Shams University, Faculty of Pharmacy, where ligands and enzymes were prepared using the molecular docking C-Docker protocols (Abdel Razek et al., 2020; Moussa et al., 2020; Ibrahim and Moussa, 2021).
By employing the pH ionization mode to 7.5, the physiological state is best matched and simulated with respect to the types of conformers, isomers, or tautomers produced; accordingly, the ligand’s Lipinski filter is adjusted. The root mean square deviation (RMSD) value was calculated for the docking poses and found to be 1.0, which proved the accuracy of the reproduced poses and validated the docking process. This was accomplished by removing the co-crystallized ligand from the active site (experimental) and superimposing with the docked native ligand (calculated).
The in vitro standards ampicillin, ascorbic acid, and doxorubicin were docked in their respective binding pockets to better assess and compare the significance of the ligands’ binding interactions and free energy. After docking, visualization was conducted using Discovery Studio Visualizer 2022, and the top 10 poses were selected for each tested molecule according to the Gibbs free energy. Preferences were given between molecules according to their interaction similarity, covalent and non-covalent, with the binding ligands and standards. The binding Gibbs free energy was obtained from the following formula and as per an implicit solvation model to present the best poses:
Here, ∆Gbinding denotes the binding energy of the ligand–protein interaction.
Ecomplex denotes the potential energy for the complex of the ligand bound with the protein.
Eprotein denotes only the protein potential energy.
Eligand denotes only the ligand potential energy.
Molecular docking simulations
The Glide docking wizard embedded in Maestro was employed to filter ligands and measure their receptor-free binding energy according to the extra precision criteria (XP). The docking score generated (Glide XP docking score) is calculated via an “XP Pose Rank” property using the following equation that takes into account the E-model and the Glide score:
Docking score (Glide score) = a*vdW + b*Coul + Hbond + Lipo + Metal + RotB + site.
The default settings adjusted in the grid generation were the OPLS 2005 force field, the van der Waals factor (1.00), and a 0.25 control factor for the workload. The grid box dimensions utilized were 14 Å × 14 Å × 14 Å, which are sufficient for ligand fitting. The best poses were assigned according to the lowest Gibbs free energy, RMSD values, and favorable orientation in the binding pockets. System validation was carried out by re-docking the two co-crystallized ligands into their primary protein pockets to measure the RMSD values, which were revealed to be 0.0301, 0.0041, and 0.6626 for 1m17, 1Kzn, and 1Xan, respectively. The values of RMSD less than 2 between the reference ligands before and after the molecular docking indicated the accuracy and precision of the docking protocols used.
Results and discussion
Synthesis of PN-CuNPs
The color change from brown to blue validated the synthesis of PN-CuNPs. This was mediated by P. nigrum fruit extract, which is involved in the reduction of Cu+ to Cu0. UV-visible spectral analysis revealed a strong peak at 500 nm due to surface plasmon resonance (Supplementary Figure S1B) (Ghosh et al., 2020; Mali et al., 2020; Sreeja et al., 2020). Secondary metabolites such as alkaloids, essential oils, polyphenolic compounds, and terpenoids act as stabilizing and capping agents and may be responsible for the reduction of Cu+ to Cu0 (Ghosh et al., 2020).
Characterization of PN-CuNPs
UV-vis spectrophotometer analysis of PN-CuNPs
The copper solution turned from brown to blue color after the addition of P. nigrum fruit extract powder, which indicated the synthesis of PN-CuNPs (Supplementary Figure S1B). The color change occurred due to the surface plasmon resonance of CuNPs (Mali et al., 2020; Sreeja et al., 2020). Supplementary Figure S1C shows that the optical band gap was calculated using the Tauc relation by plotting (ahn)2 vs. hn and extrapolating the linear portion of the curve to (ahn)2 ¼ 0. Hence, the optical energy band gaps of the PN-CuNPs were 3.7 eV. The band gap energy reveals that the PN-CuNPs have semiconductor properties.
X-ray diffraction analysis of PN-CuNPs
The crystalline nature of the synthesized PN-CuNPs was characterized by XRD analysis and is presented in Supplementary Figure S2. The signals at different 2 theta values associated with the particular designs support the production of copper nanoparticles. For instance, the planes (11–1), (111), (20–2), (202), (−113), (31–1), (220), (221), and (22–2) are indexed for the peaks at 2 h values of 35.46, 38.62, 48.78, 53.58, 59.10, 61.42, 66.14, 67.86, and 74.93. These peaks match those in the ICDD PDF card no. 96-901-6327 standard database. The average size of PN-CuNPs crystal created through green synthesis was approximately 14 nm calculated using the Debye–Scherer equation. The aforementioned results are consistent with Amer and Awwad (2020) and Ghosh et al. (2020).
FT-IR analysis of PN-CuNPs
FT-IR data of P. nigrum extract and PN-CuNPs represent probable functional groups of phytochemicals involved in bio-reduction as well as stabilization of PN-CuNPs (Supplementary Figure S3; Supplementary Table S1). The 3801 cm−1 absorption band of PN-CuNPs corresponds to the O-H stretch of alcohol. The absorption band of PN-CuNPs at 3377 cm−1 is due to N-H stretching, which is due to aliphatic primary amine functional groups. The 1,522 cm−1 band corresponds to C-H and C=C functional groups. The peaks of both extract and NPs are represented in Supplementary Table S1. Some of the peaks decreased in intensity, and some of the peaks shifted in their position. The decrease in the intensities and absorbance frequencies of the peaks at 3377, 2929, and 2854 cm−1 indicate the involvement of amines and alkane groups (N-H and C-H stretch) in the bio-reduction of NPs. PN-CuNPs exhibit an asymmetric stretching absorption band of 1408 cm−1. Phytochemicals present in P. nigrum, such as alkaloids, essential oils, polyphenolic compounds, and terpenoids, were involved in the bio-reduction of Cu+ into PN-CuNPs (Kanniah et al., 2021; Bawazeer et al., 2022).
SEM and EDX spectral analysis of PN-CuNPs
The external morphology of PN-CuNPs, which form a spindle-shaped structure, was observed through SEM (Supplementary Figure S4A). Elemental analysis of PN-CuNPs was investigated by energy-dispersive X-ray analysis (Supplementary Figure S4B) at 1 and 8 keV. It displays the percent and composition of Cu metal as a significant peak, and other small peaks correspond to biomolecules capping the PN-CuNPs. Supplementary Figure S4B illustrates the percentages of Cu and other elements.
Particle size and zeta potential determination of PN-CuNPs
The hydrodynamic size and surface charge of PN-CuNPs were revealed by a particle size analyzer and zeta potential analyzer. The average diameter of PN-CuNPs was 30–32 nm with negative surface zeta potential (−50 mV), and it remained constant over a period confirming the stability and the strong repulsion among the particles (Supplementary Figures S5A, B). This observation can be attributed to the presence of biomolecules as stabilizing agents in the leaf extract. The zeta potential measures the surface charge of particles. As the zeta potential increases, the surface charge of the particles increases. The zeta potential greatly influences particle stability in suspension through electrostatic repulsion between particles. Our results are consistent with Majdalawieh and Carr (2010) and Mohammed et al. (2016).
In vitro antioxidant assays
Free radical scavenging assay of P. nigrum fruit extract and PN-CuNPs
PN-CuNPs and P. nigrum fruit extracts were subjected to DPPH assay. PN-CuNPs showed an elevated DPPH free radical scavenging activity compared to P. nigrum fruit extracts. A higher concentration of PN-CuNPs resulted in more free radical scavenging activity. PN-CuNPs showed the highest inhibition, approximately 92%, at a concentration of 150 μg/mL (Supplementary Figure S6) (Nahak and Sahu, 2011; Alminderej et al., 2021).
Hydrogen peroxide scavenging assay of P. nigrum fruit extract and PN-CuNPs
PN-CuNPs and P. nigrum fruit extract were examined for their H2O2 free radical scavenging activity. PN-CuNPs exhibited a higher rate of H2O2 free radical scavenging activity than P. nigrum fruit extracts. Increasing the concentration of PN-CuNPs yielded more free radical scavenging activity. PN-CuNPs showed the highest inhibition, approximately 90%, at a concentration of 150 μg/mL (Supplementary Figure S6B).
Total antioxidant activity of P. nigrum fruit extract and PN-CuNPs
The total antioxidant activity (TAA) of PN-CuNPs and P. nigrum fruit extracts was determined by the phosphomolybdenum method. PN-CuNPs exhibited an increased free radical scavenging activity compared to P. nigrum fruit extracts. Increased concentration of PN-CuNPs resulted in a higher TAA. The TAA of PN-CuNPs ranged from 68% to 86%. PN-CuNPs showed the highest amount of TAA, 86%, at a concentration of 150 μg/mL (Supplementary Figures S6C, S7). CuNPs synthesized from Cocculus hirsutus leaf extract also exhibited significant free radical scavenging activity (Ameena et al., 2022).
Antibacterial activity of P. nigrum fruit extract and PN-CuNPs
PN-CuNPs and P. nigrum fruit extracts were tested for antibacterial activity against human pathogenic microbes such as S. aureus (Gram-positive), B. subtilis (Gram-positive), E. coli (Gram-negative), and P. vulgaris (Gram-negative) by the agar well diffusion method. PN-CuNPs showed the highest antibacterial zone of inhibition with increasing concentration. The antibacterial zone of inhibition was in the range of 10 ± 0.17 mm to 23 ± 0.52 mm (Supplementary Figures S8A–D, S9). S. aureus exhibited the maximum antibacterial zone of inhibition of approximately 23 ± 0.52 mm (Supplementary Figure S8A) at a 100 μg/mL concentration, and the minimum zone of inhibition was found with E. coli (10 ± 0.17 mm) at 25 μg/ml, as depicted in Supplementary Figure S8C. Therefore, S. aureus was more susceptible to PN-CuNPs (Shah et al., 2019; Alminderej et al., 2021).
Proposed mechanism of antibacterial activity of PN-CuNPs
PN-CuNPs exhibited a larger antibacterial zone of inhibition against Gram-positive bacteria such as S. aureus than against Gram-negative bacteria. Copper is an essential metal for the growth of microbes at lower concentrations, but at higher concentrations, it will inhibit the growth of bacteria by penetrating the bacterial cell wall. PN-CuNPs penetrate the bacterial cell wall and stop the growth of bacteria. The size and shape of CuNPs influence their antibacterial activity. The free surface energy of the particles changes with size and morphology, as does the pH inside the cells (Karlsson et al., 2015). The proposed mechanism of action of antibacterial activity of PN-CuNPs is depicted in Supplementary Figure S10. The mechanism of action of PN-CuNPs occurs by enzyme interaction with –SH groups leading to DNA damage and, finally, generation of oxidative stress leading to ROS production (Santo et al., 2012; Zain et al., 2014), which has an antibacterial effect (Supplementary Figure S10).
Cytotoxicity assay of P. nigrum fruit extract and PN-CuNPs
CuNPs have numerous biomedical applications in medicine and drug delivery and have anti-angiogenic properties of cancer. PN-CuNPs and P. nigrum fruit extracts were subjected to cytotoxic effect investigation against the MCF-7 breast cancer cell line by MTT assay. PN-CuNPs exhibited cytotoxicity against cancer cells. At 200 μg/mL, PN-CuNPs showed cytotoxic activity of approximately 80% against the MCF-7 cell line (Supplementary Figure S11). The results shown in Supplementary Figure S11 indicated that PN-CuNPs caused a significant decrease in the viability of the MCF-7 cells when compared to the P. nigrum fruit extract (Chung et al., 2017; Lovika et al., 2021). The correlation graph between antioxidant and cytotoxic activity indicated that both assays are statistically linked with each other (Supplementary Figure S12).
The anticancer activity of CuNPs synthesized using a green process that involves medicinal plants was confirmed by previous studies (Jacob et al., 2012; Vivek et al., 2012; Suman et al., 2013). The study of Suman et al. (2013) clarified that the HeLa cell line was suppressed at high doses of Morinda citrifolia metallic NPs. Piper longum leaf extract NPs inhibited Hep-2 cell lines (Sankar et al., 2014). Annona squamosa leaf extract NPs suppressed MCF-7 cell lines (Greenwell and Rahman, 2015). The anticancer properties of CuNPs were influenced by the size, form, and surface coating of NPs. The size of CuNPs is the most influential of these parameters (Phaniendra et al., 2015). The photocatalytic and cytotoxic activity of nickel oxide nanoparticles and nickel nanosheets was evaluated using Salvia hispanica seed extract and tragacanth gum (Sabouri et al., 2020; Sabouri et al., 2021).
Proposed mechanism of cytotoxicity activity of PN-CuNPs
The significant anticancer potential of PN-CuNPs synthesized by P. nigrum fruit extracts against common MCF-7 breast cancer cell lines is linked to their antioxidant activities. Previous similar studies revealed that CuNPs and medicinal plants reduced tumors by suppressing free radicals (Greenwell and Rahman, 2015). Free radicals induced mutations in DNA and RNA, resulting in altered gene expression and, finally, leading to a proliferation of cancerous cells (Phaniendra et al., 2015). Elevated levels of free radicals in the various organs led to angiogenesis and tumorigenesis (Hielscher and Gerecht, 2015). The proposed mechanism of the cytotoxic activity of PN-CuNPs is depicted in Supplementary Figure S13. Medicinal plant-based PN-CuNPs inhibit cancer cell growth (Desai et al., 2008). In the current study, the viability of MCF-7 cells was reduced, suggesting anticancer activity of PN-CuNPs. Understanding the detailed mechanism requires further study.
Docking interactions
Glutathione reductase was used as a binding enzyme to virtually investigate the potential of the selected compounds as antioxidants. It is responsible for providing reduced glutathione to most cells and can control ROS.
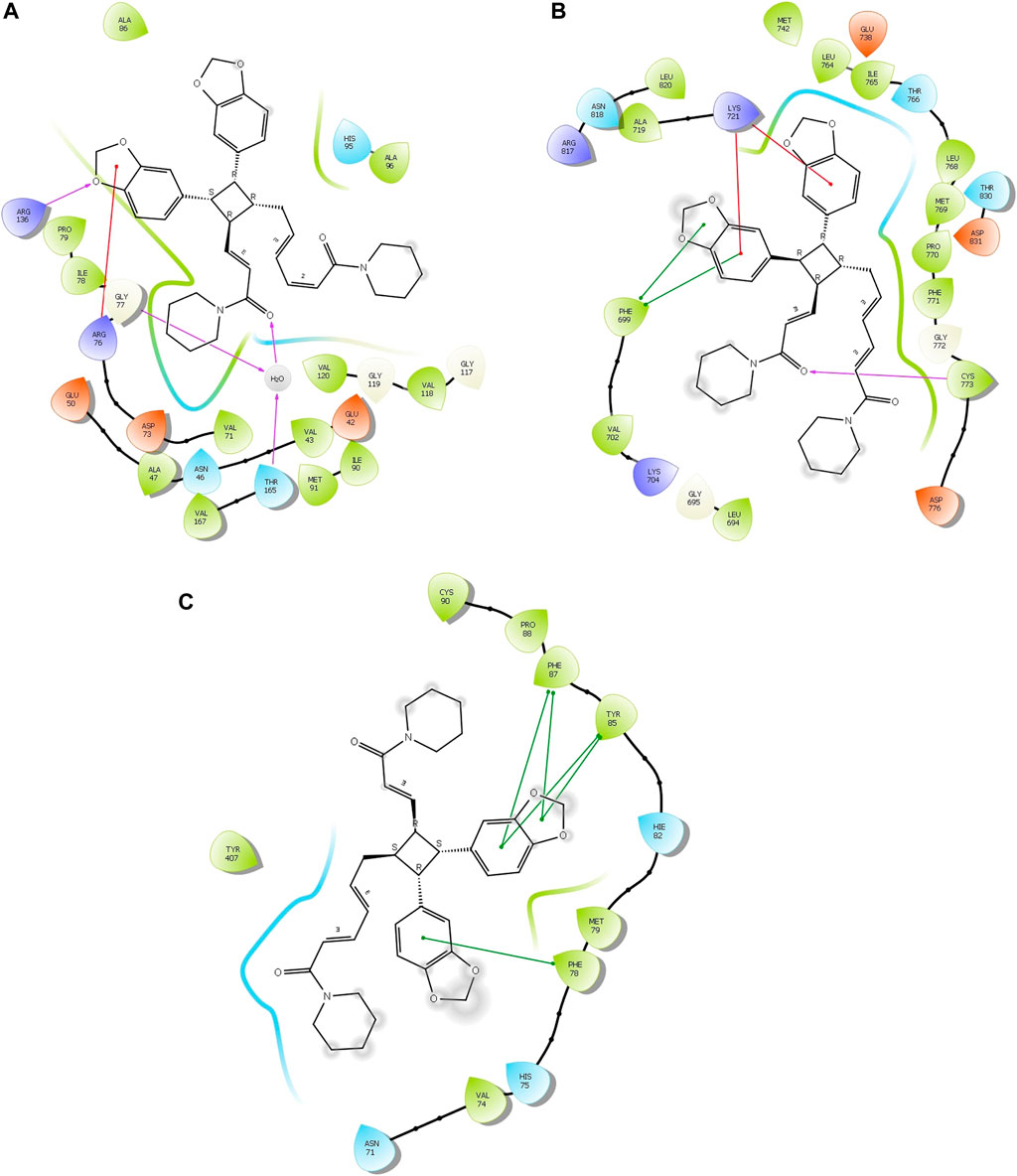
Four compounds, namely, pipernigramide F 32, pipercyclobutanamide A 26, guineensine 10, and brachyamide A 40, reported the best binding free energy in the glutathione reductase enzyme as non-competitive inhibitors by fitting allosterically to the NADPH/FAD binding pocket. The scored ΔG values were 40.18, 41.83, 38.58, and 42.03 kcal/mol, respectively, while the ascorbic acid control scored 25.31 kcal/mol. The key amino acid residues involved in the noncovalent interactions were Asn71, His75, and His82, with both the native ligand HXP xanthene and ascorbic acid. Pipernigramide F 32 manifested seven firm interactions with the vital amino acid residues in the active site through its cyclobutane and aromatic rings together with the proton acceptors as the dioxole and carbonyl moieties detailed as a π–π -stacked bond with His75, a C-H bond with His82, π–alkyl bonds with His82, Trp70, and Phe82, a conventional H bond with Asn71, and a π–π T-shaped bond with Tyr407 (Figure 4A). Pipercyclobutanamide A 26 revealed eight noncovalent binding modes, which were π–sigma bonds with Phe78, three π–alkyl bonds with Phe78, Leu438, Val74, and Tyr407, C-H bonds with His75, π–donor H bonds with His82, and conventional H bonds with His75. Guineensine 10 showed several desirable interactions as five π–alkyl bonds with Tyr407, His82, Leu438, Val74, and Trp70, one π–π --stacked bond with Phe78, two conventional H bonds with Asn71 and His75, and one π–sigma bond with Phe78.
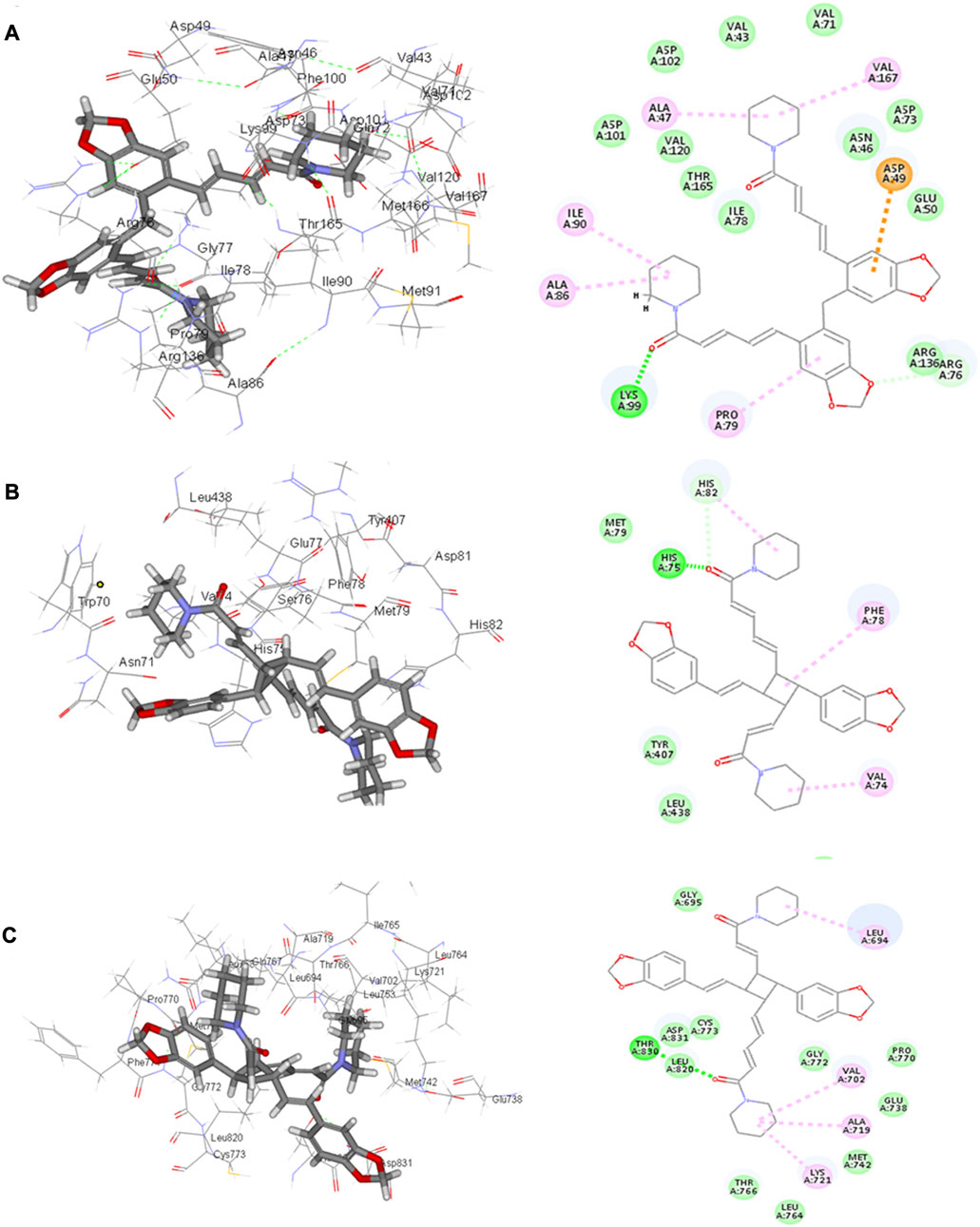
FIGURE 4. 2D and 3D binding of the most potent compound 32 in the binding pockets of (A) glutathione reductase, (B) DNA gyrase topoisomerase II, and (C) EGFR tyrosine kinase.
Brachyamide A 40 demonstrated π–π-stacked bonds with Phe78, C-H bonds with Glu77, His75, Tyr407, and His82, and alkyl bonds between the hydrophobic residues Leu438, Val74, Phe78, and the respective non-polar portions of the ligand. Additionally, Van der Waals forces were revealed with Tyr407.
The same interactions were revealed clearly using the Maestro module of the Schrodinger platform by applying the Glide XP docking protocol where the top 9 highest scoring compounds were 31 (pipernigramide E), 30 (pipernigramide D), 32 (pipernigramide F), 26 (pipercyclobutanamide A), 43 (pipwaqarine), 6 (piperchabamide C), 53 (kusunokinin), 41 (tricholein), and 19 (sylvamide). H bond interactions and π–π interactions were shown by Phe78, Phe87, Tyr85, His8, His75, and Tyr407 (Table 1).
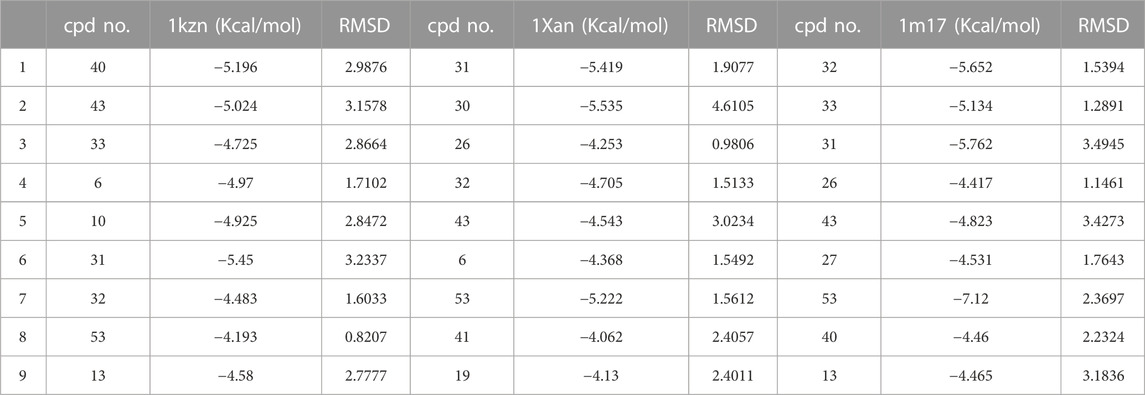
TABLE 1. A list of the top 9 scoring compounds with their ∆G values and RMSD values in the binding sites of 1Kzn, 1Xan, and 1m17.
The importance of the E. coli DNA gyrase topoisomerase II (1Kzn) has become clear as it is involved in maintaining the chromosomal super helicity in the microbial transcription and replication processes. While the B subunit of the 1Kzn molecule encompasses the ATPase active site in the N-terminal domain, the C-domain interacts with the DNA to alter the super-helical form of the DNA substrate. Due to the rise in the number of resistant microbial strains, new inhibitors of the gyrase enzyme are desperately needed. Herein, the docking of the selected P. nigrum fruit components was performed in the ATP hydrolysis binding site, and amino acid residues with the most influence, as detailed and described by Gross et al., (2003), were identified. D73 and N46 are reported to synchronize with Mg+2 in the ATP binding site. The best binding bioactive compounds in the DNA gyrase active site are pipernigramide G 33, pipnoohine 25, pipercyclobutanamide A 26, and pipernigramide G 33. The amide group in compound 33 interacted through nine non-covalent bonds; a conventional H bond with Lys99; the aromatic and saturated rings exhibited four alkyl interactions with Ala86, Pro79, Val167, and Ala47; Van der Waals (vdW) forces between Arg136 and the dioxole ring. C-H bonds with Arg76 and between proton donors and Asp101 were recorded in the DNA gyrase pocket as well as a π–anion interaction with Asp49 (Figure 4B). The ΔG binding energy was –58.615 kcal/mol, which is superior to the positive control ampicillin (−49.407 kcal/mol) (Supplementary Table S1).
Pipnoohine 25 revealed a conventional H bond between Arg136 and the amide group. Alkyl interactions between the hydrophobic side chain, Ala86, and Pro79 were detected. Pipercyclobutanamide A 26 showed a conventional H bond with Gly77, and a π–anion interaction between Glu50, Arg76, and the aromatic ring. Pro79, Ile78, Ile90, Ala86, and Arg76 were involved in alkyl-type interactions with unsaturated and saturated rings. A π–π T-shaped bond was identified between Phe50 and the benzo dioxole ring, and a C-H bond was identified between Asp73, Asn46, and Asp49 and proton donors.
The core interacting residues in the standard ampicillin within 3 Å were a conventional H bond between the amine group and residue Asn46, attractive charges between Lys99 and the acidic anions, and vdW forces and alkyl interactions with Ile78 and Asp49. The aforementioned amino acids comparably interacted with the selected compounds, especially pipernigramide G 33, pipnoohine 25, and pipercyclobutanamide A 26, confirming their potential potency as antimicrobial agents.
Using the Xtra precision protocol of the Maestro module of the Schrodinger platform, similar polar and non-polar interactions were detected and verified with their comparative binding energies. Residues such as Arg136, Gly77, Thr165, and Arg76 represented core H bond interactions, while vdW interactions were noted with Asp49, Arg136, Gly77, Thr165, Asp49, Asn46, Ile90, Met166, and Val167. The best poses were selected based on the lowest binding energies, lowest RMSD values, and favorable interactions with the native ligand.
As far as EGFR tyrosine kinase (1m17) is concerned, the docking study was carried out in the same active site, the ATP binding site, as the native co-crystallized ligand erlotinib, a major tyrosine kinase inhibitor approved by the FDA (Sharifi-Rad et al., 2022). After careful analysis, the binding modes of pipernigramide G 33 and pipernigramide F 32 (Figure 4C) proved to be promiscuous and scored the lowest Gibbs free energy, −54.1347 and −62.1958 kcal/mol, respectively, with a minus sign indicating the quick auspicious fitting in the EGFR binding pocket, which is even better than erlotinib, whose free energy was −47.3483 kcal/mol (Supplementary Table S1). Within a measured distance of 2 Å, compound 33 showed a conventional H bond formation between the residues Lys721, Asp831, Thr830, and Thr766 and its amide carbonyl group. This was also evident between Met769 and the oxygen atom of the dioxole rings.
Four alkyl interactions between the aromatic moiety and four amino acid residues, Val702, Leu820, Ala719, and Leu694, stabilized the binding of the molecule over the hydrophobic bed. vdW forces between the piperidine protons and Pro770 added more to the non-polar favorable interactions; additionally, a π–donor hydrogen bond between Cys773 and the hydrophobic aromaticity and a C-H bond detected between Glu738 and the proton donors of the piperidine ring were identified.
The lowest ΔG value was reserved to pipernigramide F 32 (Figure 4C) with interaction forces as follows: a π–sulfur bond between Cys773 and the aromatic rings; conventional H bonds between the benzo dioxole ring and Lys721, Met769, Thr830, and Thr766; three C-H bonds with Pro770 and Glu738; alkyl interactions between the piperidine ring and Val702, Ala719, and Gly772; and a C-H bond noticed between the Gly695 and the dioxole ring. Moreover, Asp831 interacted through a C-H bond with the protons of the piperidine ring.
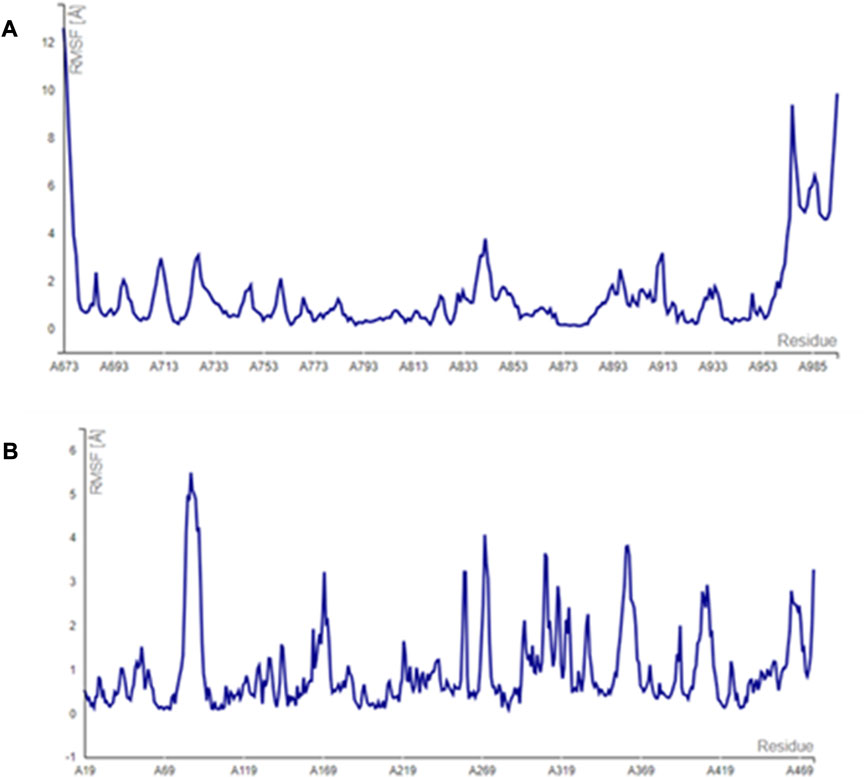
The main vital forces responsible for bioactivity were the number of different interactions, their types, bond distance measured, and strength as compared to the ligand erlotinib. Erlotinib’s good binding affinity comprised its unique interactions, namely, a salt bridge formation with Asp831, alkyl interactions with Lys721 and Met742, Leu820, and Val720, and an H bond with Cys773. vdW forces contributed to a firmer binding through Gly695, Glu738, and Leu694, all within a distance of less than 2 Å. Comparable interactions were identified in the bioactive compounds, particularly, pipernigramide F 32 and pipernigramide G 33, suggesting their promising potential as antitumor agents with tyrosine kinase inhibition activity (Figure 5). Protein stability was measured through flexibility studies manifesting 10 modules with RMSD values within the range of 2–4 Å, which suggested the stability of protein residues during interactions (Figure 6).
The vital factors that contributed to better binding forces and whose significance was most noted were the length of the molecule in terms of the number of carbons in the side chain; for instance, 26 was of firmer binding energy than pipernigramide F 32 owing to the only difference between them, which was the side chain length. Second, the presence of the piperidine ring improved fitting into the enzyme pocket as shown by comparing the ΔG value of brachyamide A 40 and guineensine 10. Even the unsaturation in a single bond contributed to a noticeable decrease in binding strength as is clear in the pairs of compounds 4/5, 16/17, and 13/14. Moreover, the size of the molecule and its orientation in the active site proved to be of considerable importance as is evident from the smallest three docked molecules, piperonal 24, 1-nitrosoimino-2,4,5-trimethoxybenzene 42, and 3′,4′-methylenedioxycinnamaldehyde 45, whose binding forces were the least compared to their congeners. It appeared that appropriate contact with the hydrophobic bed was essential for achieving the best possible binding interactions, which required either long chain molecule or a multicyclic structure; additionally, pyrrolidine was favored over the piperidine ring as observed from the two compounds, 1-piperettylpyrrolidine 36 and piperoleine 37 (Mafuné et al., 2002). The standard ascorbic acid proton acceptor groups showed conventional hydrogen bonds with His75, Asn71, and His82, which highlighted the core value of the number of proton acceptors in the binding ligand. It is worth mentioning that the best fitting compounds 32, 26, 10, and 40 manifested the same types of interactions with the amino acid residues. Similarly, the native ligand HXP xanthene revealed analogous types of interactions to the ascorbic acid control.
Binding interactions in the 1m17 were studied using Xtra precision protocol in the Maestro module of the Schrodinger platform, which depended on the intricately calculated “XP Pose Rank” property based on both the Glide score and the E model scores.
The key amino acids involved in interactions were Lys721, Asp831, Thr830, Met769, Cys773, and Pro770, confirming the poses obtained before with the C-Docker protocol (Table 1). The molecular modeling study was further validated using the Maestro module of the Schrodinger platform and indicated the top 9 compounds to be 40 (brachyamide A), 43 (pipwaqarine), 33 (pipernigramide G), 32 (pipernigramide F), 31 (pipernigramide E), 53 (kusunokinin), 6 (piperchabamide C), 10 (guineensine), and 26 (pipercyclobutanamide A) in the three binding pockets of the three target enzymes. Their binding free energy ranged between −7.12 and −4.13 kcal/mol (Tables 1, 2).
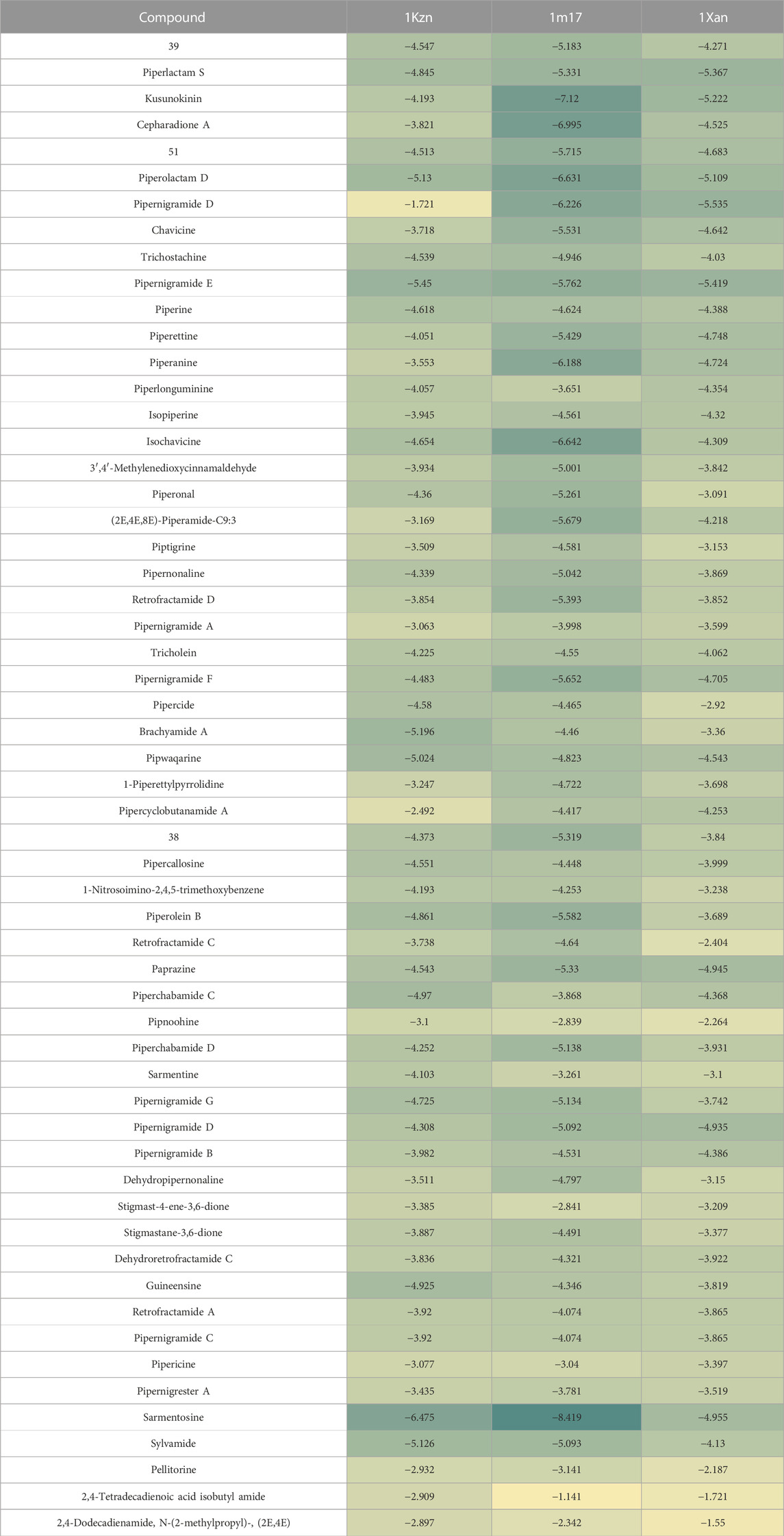
TABLE 2. Heatmap showing the free binding energy of selected pepper compounds in the binding pockets of target enzymes.
Validation by redocking the co-crystallized ligands was performed in the three enzymes 1m17, 1Kzn, and 1Xan, revealing RMSD values of 0.0301, 0.0041, and 0.6626, respectively. RMSD values were calculated with limits indicating that accurate docking protocols were measured. The selection of compounds depended not only on the Gibbs free energy but also on the lowest RMSD values and favorable binding interactions, as explained in the following paragraphs (Figure 7).
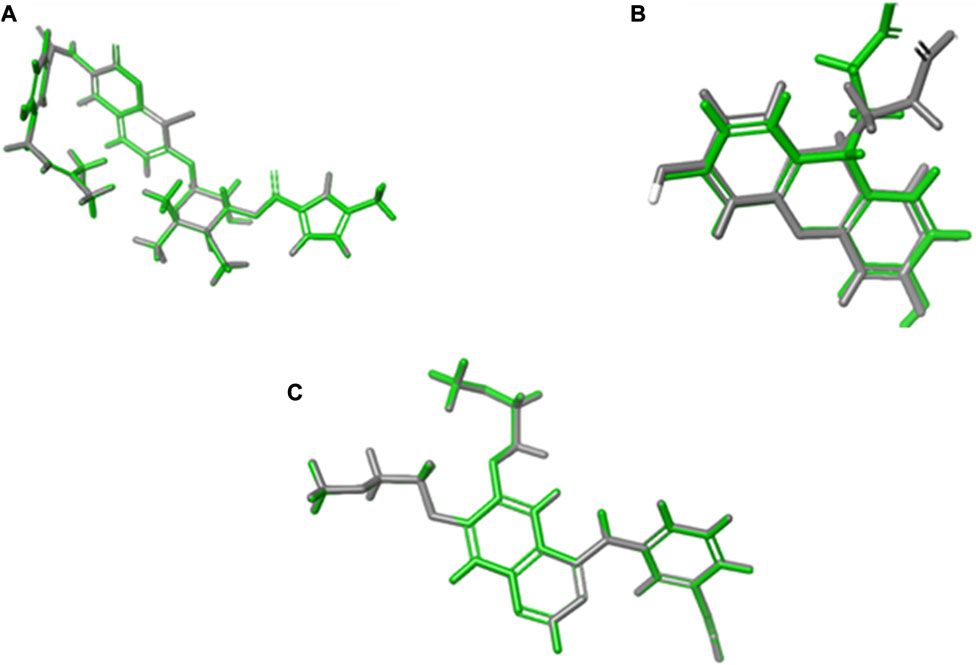
FIGURE 7. RMSD calculations through overlays between the native co-crystallized and redocked ligands. (A) 1Kzn, (B) 1Xan, and (C) 1m17 with the native ligand in green and docked ligand in gray.
The tremendous need to unveil new antibacterial molecules has become evident in the last 30 years when multiresistant organisms such as methicillin-resistant staphylococci (MRSA), vancomycin-resistant enterococci and staphylococci (VRSA), and erythromycin-resistant pneumonia began to spread widely. Natural products as a source of potent antimicrobial agents play a key role in the pursuit of new antibiotics. From the molecular modeling study and the proposed mechanisms, the antibacterial effect was regarded as largely based on the redox capacity of the PN-CuNPs (Cu+1 and Cu+2) through its interaction with SH groups, causing DNA damage and toxicity to bacteria. Furthermore, copper in high concentration can inhibit enzymes, block protein functional groups, and provide a source of hydroperoxide free radicals.
An antioxidant effect was evident through in vitro and in silico studies from the binding to the glutathione reductase human protein and the bacterial DNA gyrase. An anticancer effect was manifested by the favorable binding to the EGFR (PDB ID: 1m17) enzyme pocket manifested by H bonding and vdW force interactions through its Cys773 amino acid to a piperidyl amide group in most of the top-scoring compounds, which supported our proposed mechanism.
ADMET and TOPKAT analysis
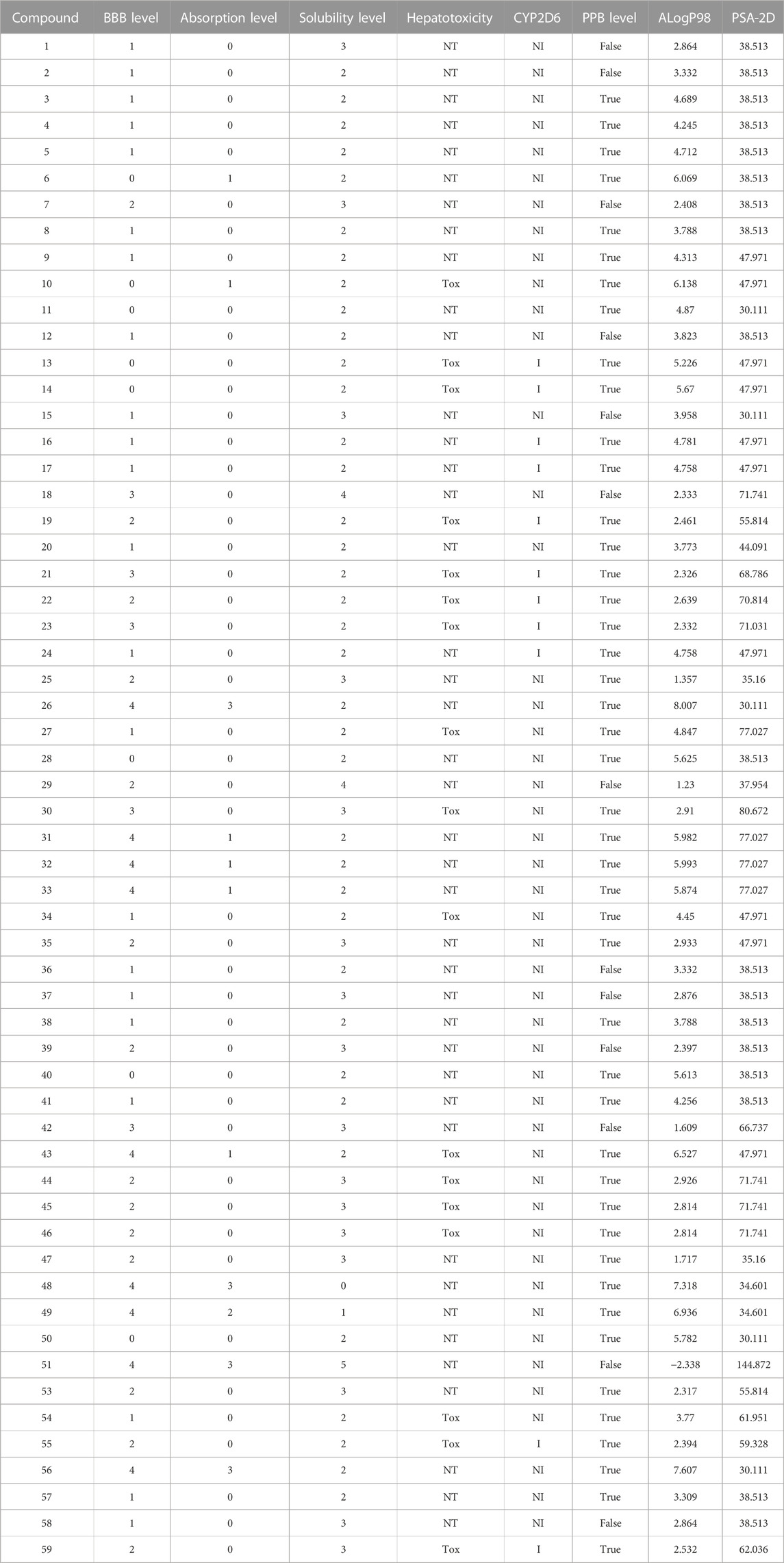
ADMET prediction of the selected alkamides indicated their pharmacodynamic, pharmacokinetic, and toxicity properties (Table 3). The cytochrome inhibition and hepatotoxicity profile were reported for only a few compounds. Penetration levels in the blood–brain barrier (BBB) were ranked from 0 to 4, indicating the very high penetration to the lowest levels. In the same way, absorption was expressed on the scale 0, 1, 2, and 3, indicating good, moderate, low, and very low absorption levels, respectively. The most potent compounds revealed a BBB penetration level of 4 with more than 90% binding to plasma proteins, indicating its promising drug likelihood (Table 3).
Pipernigramide E 31, F 32, and G 33 demonstrated moderate absorption, yet pipercyclobutanamide A 26 manifested a very low absorption score. The solubility score of 2 indicated low solubility for the compounds pipernigramide E 31, F 32, and G 33 and pipercyclobutanamide A 26, which possibly require a suitable formulation to assist delivery to the active site. Most of the alkamides tested revealed moderate intestinal absorption characteristics, as evidenced by the ADMET plot in Figure 8, where they were in the 99% absorption ellipse, with only compounds 52–54 located outside the 99% absorption range. BBB penetration with values of 0 and 1 was either very high penetration or high penetration, except for compounds 35–39, which fell outside the 99% confidence ellipse of BBB absorption (Table 3; Figure 8). Regarding plasma protein binding, most of the compounds showed more than 90% binding except for 1, 2, 7, 12, 15, 18, 29, 36, 37, 39, 42, 51, and 58. The solubility levels of the chosen compounds were of values 2 or 3, indicating low to moderate solubility, except for compounds 48 and 49, which were identified as being insoluble and of very low solubility levels. A few compounds manifested binding to CYP2D6, such as 13, 14, 16, 17, 19, 21–24, and 55.
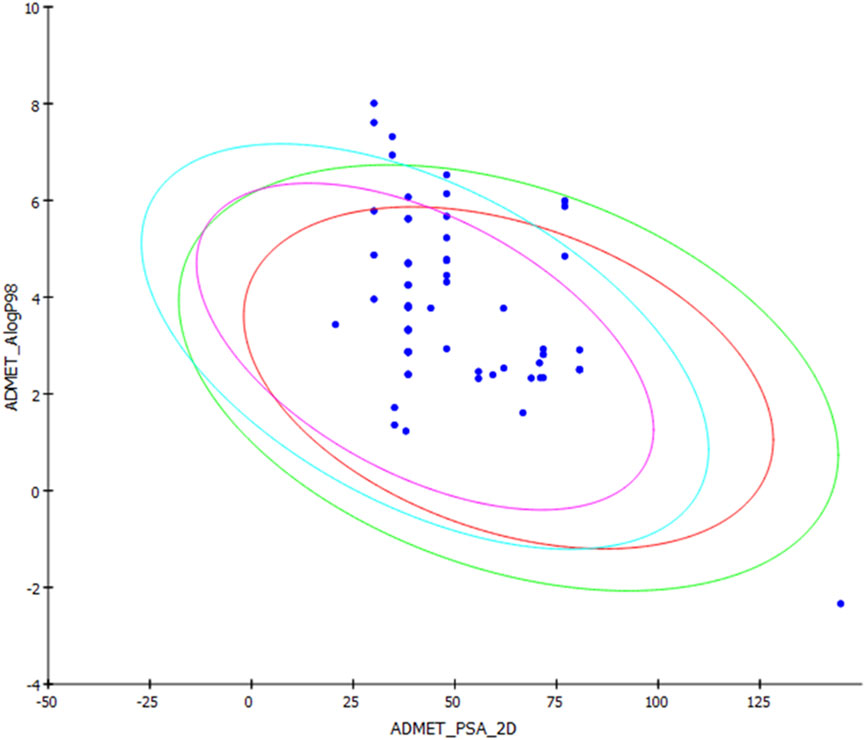
FIGURE 8. ADMET plot for chosen alkamides from Piper nigrum fruits, showing the blood–brain barrier (BBB) 99% and 95% confidence limit ellipses and the human intestinal absorption models in ADMET_AlogP98.
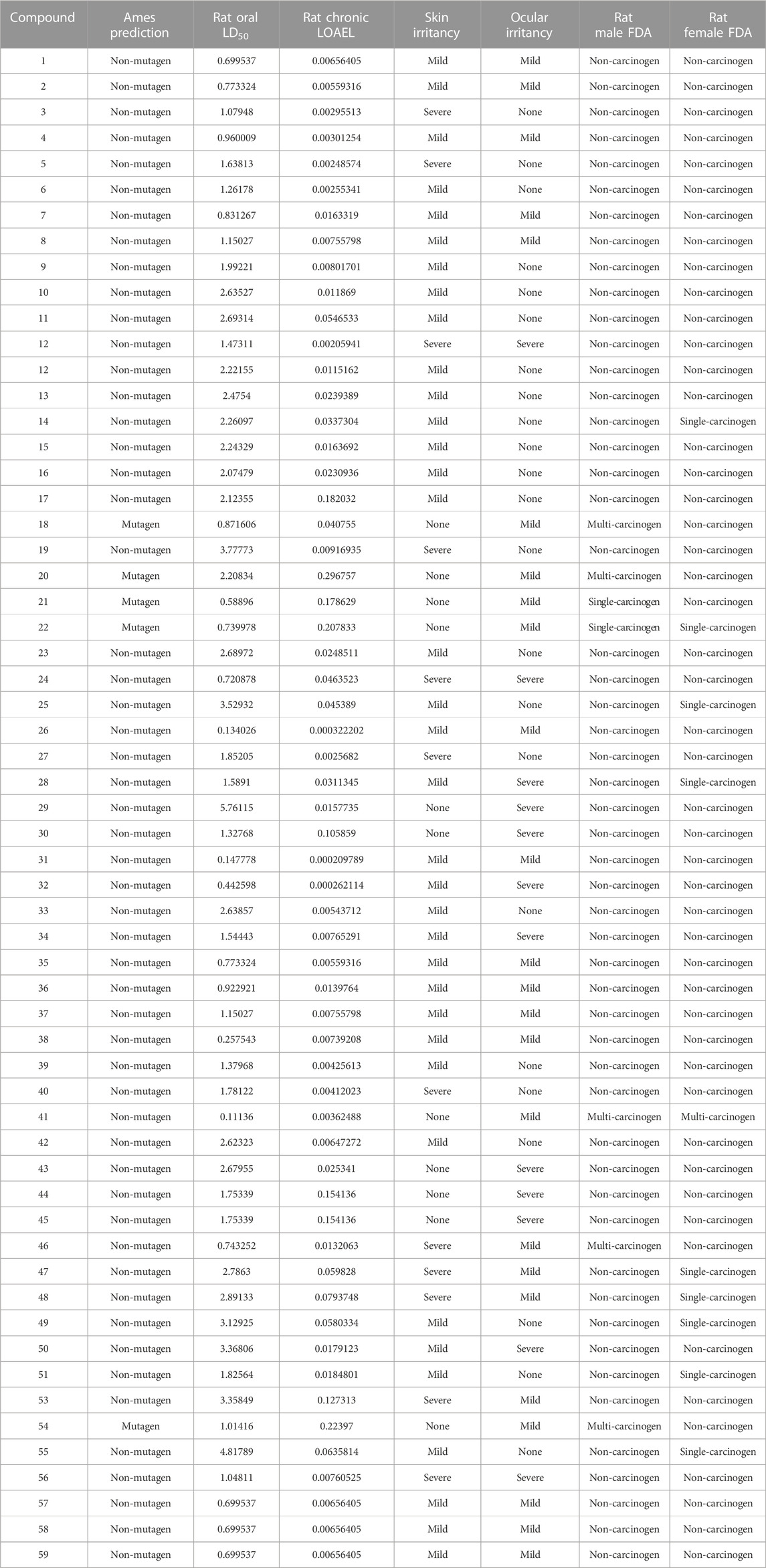
The TOPKAT analysis revealed that all the tested compounds were non-mutagenic and non-carcinogenic against male and female rats FDA except for 18, 20–22, and 54 (Table 4). The LD50 measured orally in rats ranged between 0.11 and 4.81 g/kg body wt., whereas the lowest observed adverse effect level (LOAEL) values ranged between 0.296 and 0.0002 g/kg body wt. Ocular irritancy and skin irritancy were mild for most of the compounds.
The antibiotic resistance problem has exceeded expectations with serious health decline and death rates [79]. Without significant intervention, more than 10 million people are estimated to die because of a lack of effective antibiotics against resistant microbial species. The use of adequately designed CuNPs in the medical field is a promising strategy to tackle multiresistant pathogens, especially if the use is supported with computational approaches to verify their molecular efficacy [80, 81].
Conclusion
The green synthesis of CuNPs using an aqueous extract of P. nigrum fruit extract was confirmed by UV-vis absorption, EDX analysis, FT-IR spectrometry, and XRD diffraction pattern. The particle size of the synthesized PN-CuNPs was 30–32 nm with a zeta potential of approximately −50 mV. This study demonstrated that both P. nigrum fruit extract and synthesized PN-CuNPs displayed considerable antioxidant, antibacterial, and antitumor activities. Virtual screening techniques were employed on more than 55 alkamide/alkaloid compounds using different molecular modeling software: Discovery Studio 4.5 software and the validated C-Docker protocol and Maestro Schrodinger platforms. This unveiled the promising free binding energies of most of the compounds, which were superior compared to their native ligands. Careful analysis of docking results manifested key amino acid interactions and binding forces as well as vital factors involved in structure binding relationships. An ADMET/TOPKAT study indicated that the most potent compounds were of low toxicity and favorable pharmacodynamic and pharmacokinetic profiles.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MK, experimental work. MV, biological assays. NR, experimental work and draft writing. HK, manuscript revision. AM, manuscript draft writing, revision, and editing. KR, conceptualization, supervision, and validation. SC, manuscript revision. GR, manuscript revision. AM, conceptualization, experimental design, methodology, molecular docking, validation, manuscript draft writing, revision, and editing. All authors contributed to the article and approved the submitted version.
Acknowledgments
This research article is a result of the collaboration between Ain Shams University, Yogi Vemana University Kadapa, Qassim University, and University Malaysia Kelantan. The authors thank the laboratory assistants involved in this project. AM is deeply grateful to the Department of Medicinal Chemistry in the Faculty of Pharmacy, Ain Shams University, for providing the software used in the virtual screening and molecular docking work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2023.1218588/full#supplementary-material
References
Abdel Razek, M. M., Moussa, A. Y., El-Shanawany, M. A., and Singab, A. N. B. (2020). A new phenolic alkaloid from halocnemum strobilaceum endophytes: Antimicrobial, antioxidant and biofilm inhibitory activities. Chem. Biodivers. 17 (10), e2000496. doi:10.1002/cbdv.202000496
Akintelu, S., and Folorunso, A. (2019). Characterization and antimicrobial investigation of synthesized silver nanoparticles from Annona muricata leaf extracts. J. Nanotechnol. 6, 1–5. doi:10.24966/ntmb-2044/100022
Akintelu, S. A., Folorunso, A. S., Oyebamiji, A. K., and Erazua, E. A. (2019). Antibacterial potency of silver nanoparticles synthesized using Boerhaavia diffusa leaf extract as reductive and stabilizing agent. Int. J. Pharma Sci. Res. 10 (12), 374–380. doi:10.24966/ntmb-2044/100022
Alao, I. F., Oyekunle, I. P., Iwuozor, K. O., and Emenike, E. C. (2022). Green synthesis of copper nanoparticles and investigation of its anti-microbial properties. J. Chem. Sect. B 4 (1), 39–52.
Aldaly, Z. T. (2010). Antimicrobial activity of piperine purified from Piper nigrum. J. Basrah Res. 36, 54–61.
Almatroudi, A., Khadri, H., Azam, M., Rahmani, A. H., Khaleefah, A., Khaleefah, F., et al. (2020). Antibacterial, Antibiofilm and anticancer activity of biologically synthesized silver nanoparticles using seed extract of Nigella sativa. Processes 8 (4), 388. doi:10.3390/pr8040388
Alminderej, F., Bakari, S., Almundarij, T. I., Snoussi, M., Aouadi, K., and Kadri, A. (2021). Antimicrobial and wound healing potential of a new chemotype from Piper cubeba L. essential oil and in silico study on S. aureus tyrosyl-tRNA synthetase protein. Plants 10 (2), 205. doi:10.3390/plants10020205
Altyar, A. E., Youssef, F. S., Kurdi, M. M., Bifari, R. J., and Ashour, M. L. (2022). The role of cannabis sativa L. As a source of cannabinoids against coronavirus 2 (SARS-CoV-2): An in silico study to evaluate their activities and ADMET properties. Molecules 27 (9), 2797. doi:10.3390/molecules27092797
Ameena, S., Rajesh, N., Anjum, S. M., Khadri, H., Riazunnisa, K., Mohammed, A., et al. (2022). Antioxidant, antibacterial, and anti-diabetic activity of green synthesized copper nanoparticles of Cocculus hirsutus (menispermaceae). Appl. Biochem. Biotechnol. 194, 4424–4438. doi:10.1007/s12010-022-03899-4
Amer, M., and Awwad, A. (2020). Green synthesis of copper nanoparticles by Citrus limon fruits extract, characterization and antibacterial activity.
Anjum, S., and Riazunnisa, K. (2022). Fine ultra-small ruthenium oxide nanoparticle synthesis by using catharanthus roseus and moringa oleifera leaf extracts and their efficacy towards in vitro assays, antimicrobial activity and catalytic: Adsorption kinetic studies using methylene blue dye. J. Clust. Sci. 33 (3), 1103–1117. doi:10.1007/s10876-021-02037-0
Balasooriya, E. R., Jayasinghe, C. D., Jayawardena, U. A., Ruwanthika, R. W. D., Mendis de Silva, R., and Udagama, P. V. (2017). Honey mediated green synthesis of nanoparticles: New era of safe nanotechnology. J. Nanomater 2017, 1–10. doi:10.1155/2017/5919836
Barata, L. M., Andrade, E. H., Ramos, A. R., de Lemos Of, , Setzer, W. N., Byler, K. G., et al. (2021). Secondary metabolic profile as a tool for distinction and characterization of cultivars of black pepper (Piper nigrum L) cultivated in Pará State, Brazil. Int. J. Mol. Sci. 22 (2), 890. doi:10.3390/ijms22020890
Bawazeer, S., Khan, I., Rauf, A., Aljohani, A. S., Alhumaydhi, F. A., Khalil, A. A., et al. (2022). Black pepper (Piper nigrum) fruit-based gold nanoparticles (BP-AuNPs): Synthesis, characterization, biological activities, and catalytic applications–A green approach. Green Process Synth. 11 (1), 11–28. doi:10.1515/gps-2022-0002
Chung, I. M., Abdul Rahuman, A., Marimuthu, S., Vishnu Kirthi, A., Anbarasan, K., Padmini, P., et al. (2017). Green synthesis of copper nanoparticles using Eclipta prostrata leaves extract and their antioxidant and cytotoxic activities. Exp. Ther. Med. 14 (1), 18–24. doi:10.3892/etm.2017.4466
Cragg, G. M., and Newman, D. J. (2013). Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 1830 (6), 3670–3695. doi:10.1016/j.bbagen.2013.02.008
Cuevas, R., Durán, N., Diez, M., Tortella, G., and Rubilar, O. (2015). Extracellular biosynthesis of copper and copper oxide nanoparticles by Stereum hirsutum, a native white-rot fungus from chilean forests. J. Nanomater 2015, 1–7. doi:10.1155/2015/789089
Dawid, C., Henze, A., Frank, O., Glabasnia, A., Rupp, M., Buning, K., et al. (2012). Structural and sensory characterization of key pungent and tingling compounds from black pepper (Piper nigrum L). J. Agric. food Chem. 60 (11), 2884–2895. doi:10.1021/jf300036a
Desai, A. G., Qazi, G. N., Ganju, R. K., El-Tamer, M., Singh, J., Saxena, A. K., et al. (2008). Medicinal plants and cancer chemoprevention. Curr. Drug Metab. 9 (7), 581–591. doi:10.2174/138920008785821657
Folorunso, A., Akintelu, S., Oyebamiji, A. K., Ajayi, S., Abiola, B., Abdusalam, I., et al. (2019). Biosynthesis, characterization and antimicrobial activity of gold nanoparticles from leaf extracts of Annona muricata. J. Nanostructure Chem. 9 (2), 111–117. doi:10.1007/s40097-019-0301-1
Fries, C. N., Curvino, E. J., Chen, J-L., Permar, S. R., Fouda, G. G., and Collier, J. H. (2021). Advances in nanomaterial vaccine strategies to address infectious diseases impacting global health. N. Nanotech 16 (4), 1–14. doi:10.1038/s41565-020-0739-9
Gad-Elkareem, M. A., Abdelgadir, E. H., Badawy, O. M., and Kadri, A. (2019). Potential antidiabetic effect of ethanolic and aqueous-ethanolic extracts of Ricinus communis leaves on streptozotocin-induced diabetes in rats. PeerJ 7, e6441. doi:10.7717/peerj.6441
Ghosh, M. K., Sahu, S., Gupta, I., and Ghorai, T. K. (2020). Green synthesis of copper nanoparticles from an extract of jatropha curcas leaves: Characterization, optical properties, CT-DNA binding and photocatalytic activity. RSC Adv. 10 (37), 22027–22035. doi:10.1039/d0ra03186k
Greenwell, M., and Rahman, P. (2015). Medicinal plants: Their use in anticancer treatment. Int. J. Pharm. Sci. Res. 6 (10), 4103–4112. doi:10.13040/IJPSR.0975-8232.6(10).4103-12
Gross, C. H., Parsons, J. D., Grossman, T. H., Charifson, P. S., Bellon, S., Jernee, J., et al. (2003). Active-site residues of Escherichia coli DNA gyrase required in coupling ATP hydrolysis to DNA supercoiling and amino acid substitutions leading to novobiocin resistance. Antimicrob. agents Chemother. 47 (3), 1037–1046. doi:10.1128/aac.47.3.1037-1046.2003
Hielscher, A., and Gerecht, S. (2015). Hypoxia and free radicals: Role in tumor progression and the use of engineering-based platforms to address these relationships. Free Radic. Biol. Med. 79, 281–291. doi:10.1016/j.freeradbiomed.2014.09.015
Ibrahim, N., and Moussa, A. Y. (2021). A comparative volatilomic characterization of Florence fennel from different locations: Antiviral prospects. Food & Funct. 12 (4), 1498–1515. doi:10.1039/d0fo02897e
Jacob, S. J. P., Finub, J., and Narayanan, A. (2012). Synthesis of silver nanoparticles using Piper longum leaf extracts and its cytotoxic activity against Hep-2 cell line. Colloids Surf. B 91, 212–214. doi:10.1016/j.colsurfb.2011.11.001
Kanniah, P., Chelliah, P., Thangapandi, J. R., Gnanadhas, G., Mahendran, V., and Robert, M. (2021). Green synthesis of antibacterial and cytotoxic silver nanoparticles by Piper nigrum seed extract and development of antibacterial silver based chitosan nanocomposite. Int. J. Biol. Macromol. 189, 18–33. doi:10.1016/j.ijbiomac.2021.08.056
Karlsson, H., Toprak, M., and Fadeel, B. (2015). Handbook on the toxicology of metals. Amster Netherlands: Elsevier.
Liu, Y., Yadev, V. R., Aggarwal, B. B., and Nair, M. G. (2010). Inhibitory effects of black pepper (Piper nigrum) extracts and compounds on human tumor cell proliferation, cyclooxygenase enzymes, lipid peroxidation and nuclear transcription factor-kappa-B. Nat. Product. Commun. 5 (8), 1934578X1000500. doi:10.1177/1934578x1000500822
Lovika, B., Fauziah, F., and Bellatasie, R. (2021). Cytotoxic activity of piper cubeba: A review. Int. Res. J. Pharm. 4 (3), 12–18.
Mafuné, F., Kohno, J-y., Takeda, Y., and Kondow, T. (2002). Full physical preparation of size-selected gold nanoparticles in solution: Laser ablation and laser-induced size control. J. Phys. Chem. B 106 (31), 7575–7577. doi:10.1021/jp020577y
Majdalawieh, A. F., and Carr, R. I. (2010). In vitro investigation of the potential immunomodulatory and anti-cancer activities of black pepper (Piper nigrum) and cardamom (Elettaria cardamomum). J. Med. Food 13 (2), 371–381. doi:10.1089/jmf.2009.1131
Mali, S. C., Dhaka, A., Githala, C. K., and Trivedi, R. (2020). Green synthesis of copper nanoparticles using Celastrus paniculatus Willd. leaf extract and their photocatalytic and antifungal properties. Biotechnol. Rep. 27, e00518. doi:10.1016/j.btre.2020.e00518
Meghwal, M., and Goswami, T. (2013). Piper nigrum and piperine: An update. Phytother. Res. 27 (8), 1121–1130. doi:10.1002/ptr.4972
Mohammed, G. J., Omran, A. M., and Hussein, H. M. (2016). Antibacterial and phytochemical analysis of Piper nigrum using gas chromatography-mass Spectrum and Fourier-transform infrared spectroscopy. Int. J. PharmacoPhytochem Res. 8 (6), 977–996.
Moussa, A., Labib, R., and Ayoub, N. (2013). Isolation of chemical constituents and protective effect of Pistacia khinjuk against CCl4–induced damage on HepG2 cells. Phytopharmacology 4 (4), 1–9.
Moussa, A. Y., Lambert, C., Stradal, T. E., Ashrafi, S., Maier, W., Stadler, M., et al. (2020). New peptaibiotics and a cyclodepsipeptide from Ijuhya vitellina: Isolation, identification, cytotoxic and nematicidal activities. Antibiotics 9 (3), 132. doi:10.3390/antibiotics9030132
Mseddi, K., Alimi, F., Noumi, E., Veettil, V. N., Deshpande, S., Adnan, M., et al. (2020). Thymus musilii velen. As a promising source of potent bioactive compounds with its pharmacological properties: In vitro and in silico analysis. Arab. J. Chem. 13 (8), 6782–6801. doi:10.1016/j.arabjc.2020.06.032
Nahak, G., and Sahu, R. (2011). Phytochemical evaluation and antioxidant activity of Piper cubeba and Piper nigrum. J. Appl. Pharma Sci. 1 (8), 153.
Naik, R. R., Stringer, S. J., Agarwal, G., Jones, S. E., and Stone, M. O. (2002). Biomimetic synthesis and patterning of silver nanoparticles. Nat. mater 1 (3), 169–172. doi:10.1038/nmat758
Nasrollahzadeh, M., Mahmoudi-Gom Yek, S., Motahharifar, N., and Ghafori Gorab, M. (2019). Recent developments in the plant-mediated green synthesis of Ag-based nanoparticles for environmental and catalytic applications. Chem. Rec. 19 (12), 2436–2479. doi:10.1002/tcr.201800202
Noumi, E., Merghni, A., Alreshidi, M. M., Haddad, O., Akmadar, G., De Martino, L., et al. (2018). Chromobacterium violaceum and Pseudomonas aeruginosa PAO1: Models for evaluating anti-quorum sensing activity of melaleuca alternifolia essential oil and its main component terpinen-4-ol. Molecules 23 (10), 2672. doi:10.3390/molecules23102672
Pavithra, K., and Vadivukkarasi, S. (2015). Evaluation of free radical scavenging activity of various extracts of leaves from Kedrostis foetidissima (Jacq) Cogn. Food Sci. Hum. Wellness 4 (1), 42–46. doi:10.1016/j.fshw.2015.02.001
Phaniendra, A., Jestadi, D. B., and Periyasamy, L. (2015). Free radicals: Properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 30 (1), 11–26. doi:10.1007/s12291-014-0446-0
Pomerantseva, E., Bonaccorso, F., Feng, X., Cui, Y., and Gogotsi, Y. (2019). Energy storage: The future enabled by nanomaterials. Science 366 (6468), eaan8285. doi:10.1126/science.aan8285
Reshmi, S., Sathya, E., and Devi, P. S. (2010). Isolation of piperdine from Piper nigrum and its antiproliferative activity. J. Med. Plant Res. 4 (15), 1535–1546.
Rho, M-C., Lee, S. W., Park, H. R., Choi, J-H., Kang, J. Y., Kim, K., et al. (2007). ACAT inhibition of alkamides identified in the fruits of Piper nigrum. Phytochemistry 68 (6), 899–903. doi:10.1016/j.phytochem.2006.11.025
Sabouri, Z., Akbari, A., Hosseini, H. A., Khatami, M., and Darroudi, M. (2020). Tragacanth-mediate synthesis of NiO nanosheets for cytotoxicity and photocatalytic degradation of organic dyes. Bioprocess Biosyst. Eng. 43 (7), 1209–1218. doi:10.1007/s00449-020-02315-7
Sabouri, Z., Rangrazi, A., Amiri, M. S., Khatami, M., and Darroudi, M. (2021). Green synthesis of nickel oxide nanoparticles using Salvia hispanica L(chia) seeds extract and studies of their photocatalytic activity and cytotoxicity effects. Bioprocess Biosyst. Eng. 44 (11), 2407–2415. doi:10.1007/s00449-021-02613-8
Sankar, R., Maheswari, R., Karthik, S., Shivashangari, K. S., and Ravikumar, V. (2014). Anticancer activity of Ficus religiosa engineered copper oxide nanoparticles. Mater Sci. Eng. C 44, 234–239. doi:10.1016/j.msec.2014.08.030
Santo, C. E., Lam, E. W., Elowsky, C. G., Quaranta, D., Domaille, D. W., Chang, C. J., et al. (2011). Grass G: Bacterial killing by dry metallic copper surfaces. Appl. Environ. Microbiol. 77 (3), 794–802. doi:10.1128/aem.01599-10
Santo, C. E., Quaranta, D., and Grass, G. (2012). Antimicrobial metallic copper surfaces kill Staphylococcus haemolyticus via membrane damage. Microbiologyopen 1 (1), 46–52. doi:10.1002/mbo3.2
Sarma, G. K., Sen Gupta, S., and Bhattacharyya, K. G. (2019). Nanomaterials as versatile adsorbents for heavy metal ions in water: A review. Environ. Sci. Pollut. Res. 26 (7), 6245–6278. doi:10.1007/s11356-018-04093-y
Scott, I. M., Puniani, E., Jensen, H., Livesey, J. F., Poveda, L., Sánchez-Vindas, P., et al. (2005). Analysis of Piperaceae germplasm by HPLC and LCMS: A method for isolating and identifying unsaturated amides from piper spp extracts. J. Agric. food Chem. 53 (6), 1907–1913. doi:10.1021/jf048305a
Shah, R., Pathan, A., Vaghela, H., Ameta, S. C., and Parmar, K. (2019). Green synthesis and characterization of copper nanoparticles using mixture (Zingiber officinale, Piper nigrum and Piper longum) extract and its antimicrobial activity. Chem. Sci. Trans. 8 (1), 63–69.
Sharifi-Rad, J., Quispe, C., Durazzo, A., Lucarini, M., Souto, E. B., Santini, A., et al. (2022). Resveratrol’biotechnological applications: Enlightening its antimicrobial and antioxidant properties. J. Herb. Med. 32, 100550. doi:10.1016/j.hermed.2022.100550
Shityakov, S., Bigdelian, E., Hussein, A. A., Hussain, M. B., Tripathi, Y. C., Khan, M. U., et al. (2019). Phytochemical and pharmacological attributes of piperine: A bioactive ingredient of black pepper. Eur. J. Med. Chem. 176, 149–161. doi:10.1016/j.ejmech.2019.04.002
Singh, J., Dutta, T., Kim, K-H., Rawat, M., Samddar, P., and Kumar, P. (2018). ‘Green’synthesis of metals and their oxide nanoparticles: Applications for environmental remediation. J. nanobiotechnology 16 (1), 84–24. doi:10.1186/s12951-018-0408-4
Snoussi, M., Dehmani, A., Noumi, E., Flamini, G., and Papetti, A. (2016). Chemical composition and antibiofilm activity of Petroselinum crispum and Ocimum basilicum essential oils against Vibrio spp. strains. Microb. Pathog. 90, 13–21. doi:10.1016/j.micpath.2015.11.004
Sreeja, C., Philip, K. A., Shamil, K., Asraj, O., and Sreeja, S. (2020). Biomediated green synthesis of copper nanoparticles using piper nigrum leaf extract. J. Nanosci. 6, 908–910. doi:10.30799/jnst.305.20060303
Sudha, V. P., Ganesan, S., Pazhani, G., Ramamurthy, T., Nair, G., and Venkatasubramanian, P. (2012). Storing drinking-water in copper pots kills contaminating diarrhoeagenic bacteria. J. Health Popul. Nutr. 30 (1), 17–21. doi:10.3329/jhpn.v30i1.11271
Suman, T., Rajasree, S. R., Kanchana, A., and Elizabeth, S. B. (2013). Biosynthesis, characterization and cytotoxic effect of plant mediated silver nanoparticles using Morinda citrifolia root extract. Colloids Surf. B Biointerfaces 106, 74–78. doi:10.1016/j.colsurfb.2013.01.037
Takooree, H., Aumeeruddy, M. Z., Rengasamy, K. R., Venugopala, K. N., Jeewon, R., Zengin, G., et al. (2019). A systematic review on black pepper (Piper nigrum L): from folk uses to pharmacological applications. Crit. Rev. Food Sci. Nutr. 59 (Suppl. 1), S210–S243. doi:10.1080/10408398.2019.1565489
Tamil Elakkiya, V., Meenakshi, R., Senthil Kumar, P., Karthik, V., Ravi Shankar, K., Sureshkumar, P., et al. (2022). Green synthesis of copper nanoparticles using Sesbania aculeata to enhance the plant growth and antimicrobial activities. Int. J. Environ. Sci. Technol. 19 (3), 1313–1322. doi:10.1007/s13762-021-03182-9
Thakkar, K. N., Mhatre, S. S., and Parikh, R. Y. (2010). Biological synthesis of metallic nanoparticles. Nanomed Nanotechnol. Biol. Med. 6 (2), 257–262. doi:10.1016/j.nano.2009.07.002
Tyagi, P. K., Tyagi, S., Gola, D., Arya, A., Ayatollahi, S. A., Alshehri, M. M., et al. (2021). Ascorbic acid and polyphenols mediated green synthesis of silver nanoparticles from Tagetes erecta L. aqueous leaf extract and studied their antioxidant properties. J. Nanomater. 2021, 2021–2029. doi:10.1155/2021/6515419
Vivek, R., Thangam, R., Muthuchelian, K., Gunasekaran, P., Kaveri, K., and Kannan, S. (2012). Green biosynthesis of silver nanoparticles from Annona squamosa leaf extract and its in vitro cytotoxic effect on MCF-7 cells. Process Bioche 47 (12), 2405–2410. doi:10.1016/j.procbio.2012.09.025
Yun, Y. S., Noda, S., Takahashi, S., Takahashi, Y., and Inoue, H. (2018). Piperine-like alkamides from Piper nigrum induce BDNF promoter and promote neurite outgrowth in Neuro-2a cells. J. Nat. Med. 72 (1), 238–245. doi:10.1007/s11418-017-1140-3
Zain, N. M., Stapley, A. G., and Shama, G. (2014). Green synthesis of silver and copper nanoparticles using ascorbic acid and chitosan for antimicrobial applications. Carbohydr. Polym. 112, 195–202. doi:10.1016/j.carbpol.2014.05.081
Keywords: Piper nigrum, Piper nigrum copper nanoparticles, cytotoxicity, molecular docking, alkamides, ADMET/TOPKAT, antibacterial activity, antioxidant activity
Citation: Kiranmayee M, Rajesh N, Vidya Vani M, Khadri H, Mohammed A, Chinni SV, Ramachawolran G, Riazunnisa K and Moussa AY (2023) Green synthesis of Piper nigrum copper-based nanoparticles: in silico study and ADMET analysis to assess their antioxidant, antibacterial, and cytotoxic effects. Front. Chem. 11:1218588. doi: 10.3389/fchem.2023.1218588
Received: 07 May 2023; Accepted: 14 August 2023;
Published: 05 September 2023.
Edited by:
Eslam B. Elkaeed, AlMaarefa University, Saudi ArabiaReviewed by:
Premlata Kumari, Sardar Vallabhbhai National Institute of Technology Surat, IndiaIbrahim Eissa, Al-Azhar University, Egypt
Nurul Iman Aminudin, International Islamic University Malaysia, Malaysia
Copyright © 2023 Kiranmayee, Rajesh, Vidya Vani, Khadri, Mohammed, Chinni, Ramachawolran, Riazunnisa and Moussa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashaimaa Y. Moussa, QXNoYWltYWFfeWVoaWFAcGhhcm1hLmFzdS5lZHUuZWc=; Khateef Riazunnisa, a2hhdGVlZnJpYXpAZ21haWwuY29t; Gobinath Ramachawolran, ci5nb2JpbmF0aEByY3NpdWNkLmVkdS5teQ==
 Modumudi Kiranmayee
Modumudi Kiranmayee Nambi Rajesh
Nambi Rajesh M. Vidya Vani1
M. Vidya Vani1 Arifullah Mohammed
Arifullah Mohammed Gobinath Ramachawolran
Gobinath Ramachawolran Khateef Riazunnisa
Khateef Riazunnisa Ashaimaa Y. Moussa
Ashaimaa Y. Moussa