- Guangxi Key Laboratory of Guangxi Key Laboratory of Electrochemical Energy Materials, School of Chemistry and Chemical Engineering, Institute of Optical Materials and Chemical Biology, Guangxi University, Nanning, China
As isothermal, enzyme-free signal amplification strategies, hybridization chain reaction (HCR) and catalytic hairpin assembly (CHA) possess the advantages such as high amplification efficiency, excellent biocompatibility, mild reactions, and easy operation. Therefore, they have been widely applied in DNA-based biosensors for detecting small molecules, nucleic acids, and proteins. In this review, we summarize the recent progress of DNA-based sensors employing typical and advanced HCR and CHA strategies, including branched HCR or CHA, localized HCR or CHA, and cascaded reactions. In addition, the bottlenecks of implementing HCR and CHA in biosensing applications are discussed, such as high background signals, lower amplification efficiency than enzyme-assisted techniques, slow kinetics, poor stability, and internalization of DNA probes in cellular applications.
1 Introduction
Due to their high programmability, stability, and biocompatibility, nucleic acids are popular in the development of dynamic circuits and nanostructures (Zhang et al., 2014; Sadowski et al., 2014; Wang et al., 2017). To date, various DNA-based biosensors have been reported for bioanalytical applications (Yuan et al., 2018; Mo et al., 2023; Mo et al., 2021). A DNA-based biosensor consists of three main components: a recognition unit, an amplification unit, and a transducer (Figure 1). The recognition unit is used to interact with analytes. In which target DNA/RNA could be recognized via Watson-Crick base pairing. Moreover, the development of aptamers has expanded the scope of analytes to include metal ions, small molecules, proteins, and living cells (Hansen-Bruhn et al., 2018; Qin et al., 2019; Song et al., 2022). Upon binding to the target, the recognition unit undergoes a conformational change and exposes the trigger domain for the amplification unit, which then initiates the amplification process. Various amplification techniques have been developed to enhance sensitivity, including HCR and CHA (Cheglakov et al., 2015; Chai and Miao, 2019; Wang et al., 2021). These amplification strategies could be divided into two categories: enzyme-assisted amplification and enzyme-free amplification. The first category relies on the catalytic capability of enzymes to realize nucleic acid amplification, such as polymerase chain reaction (PCR) (Saiki et al., 1988) and rolling circle amplification (RCA) (Zhao et al., 2008). While the sensitivity could be greatly improved, the use of enzymes has limited their application in complex environments. On the other hand, enzyme-free amplification, such as hybridization chain reaction (HCR) (Dirks and Pierce, 2004) and catalytic hairpin assembly (CHA) (Yin et al., 2008), could amplify nucleic acids through toehold-mediated strand displacement. Notably, these methods could operate under mild and isothermal conditions, without the involvement of enzymes or thermal cyclers. These features make them suitable for applications in biological environments, especially in living cells. The isothermal, enzyme-free amplification techniques, HCR and CHA, provide high amplification efficiency, high biocompatibility, ease of operation, and low cost. In addition, several advanced techniques have recently been reported to improve performance, such as branched HCR or CHA, and localized HCR or CHA. The last component of the biosensor was the transducer, which transformed the nucleic acid information into a measurable signal, such as fluorescent, colorimetric, and electrochemical signals (Huang et al., 2020; Sun et al., 2021). In this review, we will summarize the research progress on biosensors based on HCR and CHA, as well as related advanced amplification methods, with particular attention to sensing platforms based on pure DNA materials, such as nucleic acid circuits or DNA nanostructures. The challenges encountered in the development of sensors will also be discussed. We hope this paper will provide updated information for sensing strategies based on HCR and CHA, and facilitate their applications in bioanalysis, clinical diagnosis, and environmental monitoring.
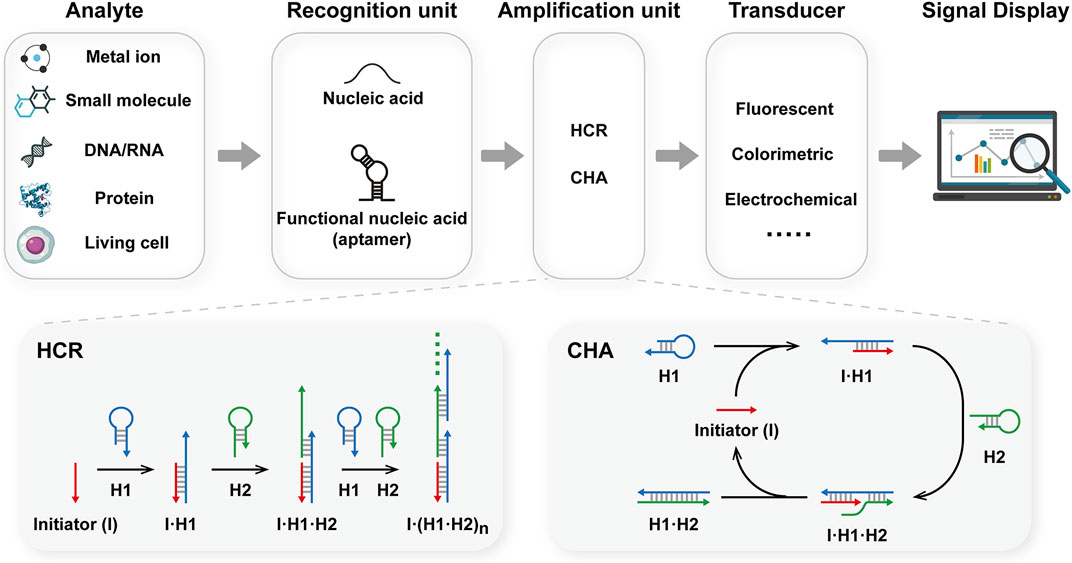
FIGURE 1. Schematic illustration of the construction of DNA-based biosensors integrated with HCR or CHA.
2 DNA-based biosensors integrated with HCR or CHA
2.1 Biosensors based on traditional or advanced HCR
2.1.1 Traditional HCR
Classical HCR was firstly developed by Dirks and Pierce (Dirks and Pierce, 2004), which involves an initiator and two metastable hairpins. The initiator could trigger the alternate opening of hairpins, forming nicked double-stranded DNA structures. As a simple and powerful amplification method, HCR has been incorporated into a number of sensing platforms. A high-fidelity amplified FISH system was reported by Marras et al. (2019), in which the target mRNA interacted with the binary probe to release the HCR initiator. This system could distinguish single nucleotide variations within a single mRNA. In another study, Yang et al. (2019) realized the simultaneous detection of miRNA and pre-miRNA in living cells through two pairs of HCR hairpin probes, showing detection limits of 680 pM and 820 pM, respectively (Figure 2A). Later, Tang et al. (2020) integrated HCR with the dual-split aptamer probe (DSAP) for activatable cancer cell detection. In this design, DSAP could specifically recognize cancer cells and release the trigger for HCR, leading to in-situ HCR on the cell membrane and amplified fluorescence. This strategy could reach a detection limit of 20 cells/200 uL. Overall, these studies proved the potential of HCR in biosensing and bioimaging.
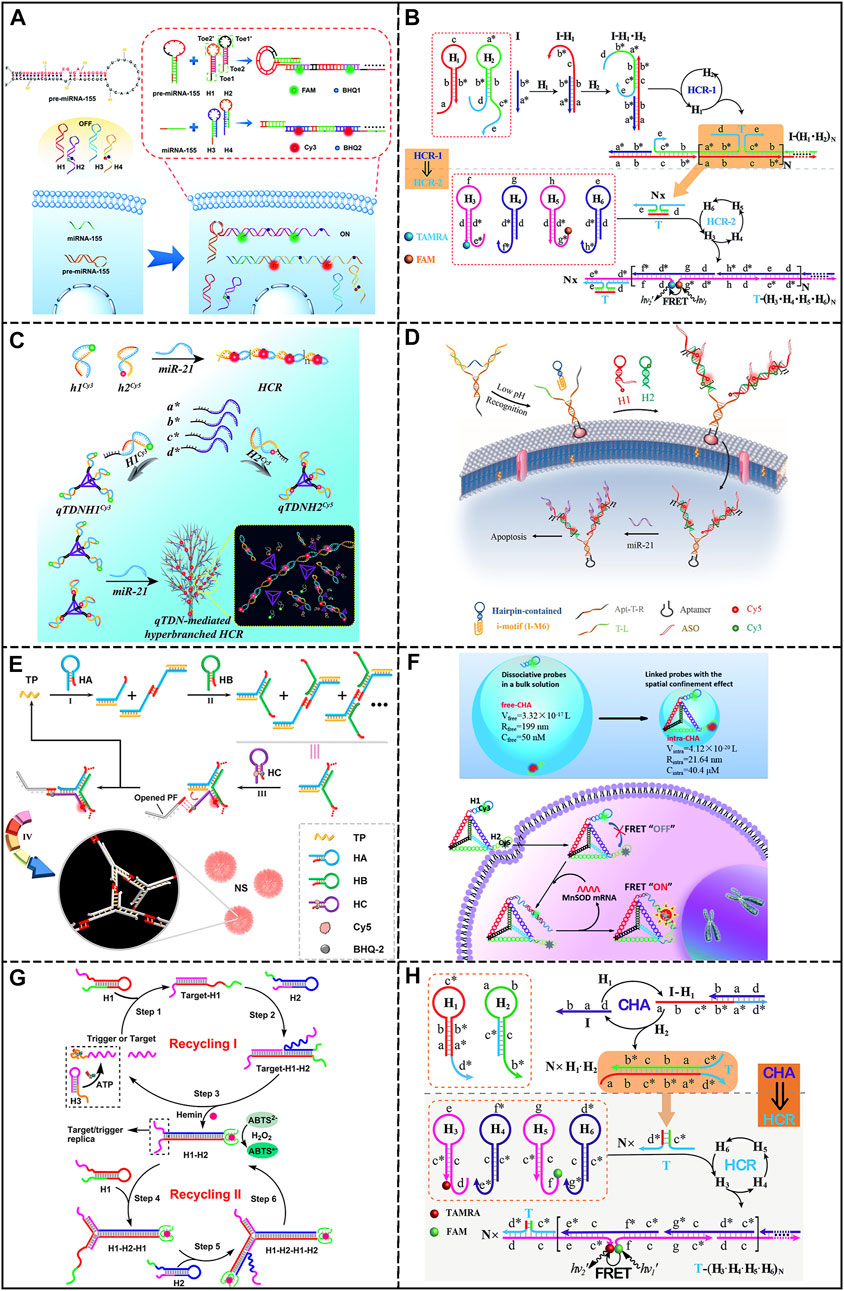
FIGURE 2. DNA-based biosensors integrated with HCR or CHA. (A) Biosensor based on traditional HCR. Reproduced from (Yang et al., 2019) with permission from the Royal Society of Chemistry. (B) Biosensor based on branched HCR. Reproduced from (Wei et al., 2018a) with permission from the Royal Society of Chemistry. (C) Biosensor based on localized HCR. Reproduced from (Wang et al., 2019a) with permission from the Royal Society of Chemistry. (D) Biosensor based on other HCR. Reproduced from (Ma et al., 2019) with permission from the American Chemical Society. (E) Biosensor based on branched CHA. Reproduced from (Liu et al., 2020) with permission from the American Chemical Society. (F) Biosensor based on localized CHA. Reproduced from (Qing et al., 2020) with permission from the Royal Society of Chemistry. (G) Biosensor based on self-replicating CHA. Reproduced from (Dai et al., 2017) with permission from the American Chemical Society. (H) Biosensor based on cascaded CHA-HCR strategy. Reproduced from (Wang et al., 2018) with permission from the Royal Society of Chemistry.
2.1.2 Branched HCR
Branched hybridization chain reaction (bHCR) involves hierarchical reactions of HCR, producing branched DNA polymers and higher levels of amplification efficiency. For example, Wang’ group (Wei et al., 2018a) developed a concatenated HCR system for miRNA detection. As shown in Figure 2B, target RNA could trigger the first layer of HCR, exposing the initiator for the second layer of HCR, resulting in a shortened distance between FRET donor and acceptor. The detection limit was calculated to be 3 pM. Soon after, the same group integrated the concept of cascade HCR with logic gate operation for intracellular miRNA imaging. In this study, the presence of miRNA was transduced to the initiator of cascading HCR through biocomputing operations. By rationally designing the sensing module, binary or advanced logic gates were achieved. (Gong et al., 2019). To obtain better sensitivity, electrochemical detection has also been implemented with branched HCR for miR-141 detection, achieving a LOD of 0.18 fM (Li et al., 2019). Additionally, by integrating aptamers into the branched HCR system, biomolecules, such as ATP, can be detected. In a study by Zhou et al. (2019), the binding of ATP aptamer to the target released the initiator for branched HCR, leading to FRET signals with a detection limit of 170 nM.
2.1.3 Localized HCR
The kinetics of typical HCR is limited since the working principle is based on random collision of hairpins. To accelerate the reaction, localized HCR (LHCR) was developed based on the spatial-confinement effect. The reaction time could be reduced by anchoring the reactants to DNA nanostructures in order to reach higher local concentration of hairpins. For instance, Bui et al. (2017) immobilized the HCR hairpins along a 1D linear DNA track, which enabled the hairpins to hybridize along the track. The kinetics of this strategy was six times faster than traditional HCR due to the locality effect. To further apply the concept of LHCR to sensing applications; Wu et al. (2020a) developed a tripartite Y-shaped DNA probe for mRNA imaging in living mice. Y-shaped scaffolds were functionalized with HCR hairpins and a folate (FA) ligand that could selectively enter tumor cells, leading to HCR assemblies on the scaffold in response to miR-21. Moreover, taking advantage of the stability of Y-shaped structure, the probe was successfully applied for high-contrast miRNA imaging in living mice. In addition, 3D DNA nanostructures have also been employed for LHCR. As illustrated in Figure 2C, Wang et al. (2019a) reported hyperbranched HCR mediated by tetrahedral DNA nanostructures (qTDN). By modifying HCR hairpins on the vertices of qTDN, the reaction orientation was extended, as well as the collision probability and reaction kinetics. The reaction rate is about 70 times faster than that of traditional HCR, and the detection limit was 2.14 pM. These reports confirmed the acceleration effect of LHCR.
2.1.4 Other HCR
In addition to the strategies mentioned above, other HCR technologies have been developed. A recent study by Cheng et al. (2021) used proximity-dependent surface hybridization chain reactions to design an electrochemical biosensor for miRNA detection. MiRNA triggered HCR and the formation of duplexes with single-stranded dangling tails, which were labeled with ferrocene (Fc) and could be used to identify the sequence fixed on the electrode. After identification, the Fc tags were brought proximal to the electrode, which induced amplified electrochemical signals with a LOD of 3 fM. Furthermore, Jiang et al. (2021) presented a dumbbell hybridization chain reaction (DHCR), which improved the amplification performance by a novel morphology of the DNA nanostructure. This system consisted of four probes (A, B, C, D), in which probes A and B contained PSA split aptamers that could recognize PSA. However, in the absence of PSA, the unhybridized parts of probes A and B could alternatively open dumbbell probes C and D, forming a stacked DNA nanostructure and enhancing electrochemical signals. The LOD for PSA was 0.5 pg/mL. Moreover, HCR can be regarded as an ideal choice for target cell imaging and gene delivery. Ma et al. (2019) reported a bipedal hybridization chain (BiHCR) for activable cancer cell imaging (Figure 2D). A Y-shaped nanoprobe was constructed containing an i-motif sequence, an aptamer sequence and two trigger strands for HCR. After specific binding between the aptamer and cancer cells, the acidic extracellular environment triggered a conformational change in the i-motif structure, exposing the HCR initiators. Which further induced in-situ bipedal HCR and produced FRET signals. In addition, Peng et al. (2022) developed a double loop-stem mediated HCR (D-HCR) for label-free biosensing. The presence of miR-21 not only triggered the formation of DNA nanowires, but also released previously locked G4 aptamers. The LOD of 4.60 pM was achieved by combining D-HCR with thioflavin T (ThT) for fluorescent detection, and 13.60 pM was achieved by incorporating Hemin for colorimetric detection.
2.2 Biosensors based on traditional or advanced CHA
2.2.1 Traditional CHA
The CHA was firstly introduced by Yin’s group (Yin et al., 2008), in which an initiator could catalyze the duplex formation between two metastable hairpins, resulting in the amplification of double-stranded DNA products. Owing to the feature of simple operation, mild conditions, and flexibility, CHA has been widely applied in the development of sensors. For example, a universal CHA system (uniCHA) was proposed by Zhang et al. (2022) to detect various piRNAs and miRNAs. An adaptable starting hairpin containing the CHA initiator was employed in this system. The targets would interact with the starting hairpin to expose the initiator, which could trigger CHA and generate FRET signals. It is noteworthy that the sequence of the starting sequence can be adjusted depending on the target. Therefore, this system has been applied to the detection of various piRNAs and miRNAs. In addition, CHA can be integrated with lateral flow immunoassay (LFIA) to detect SARS-CoV-2 RNA (Zou et al., 2021). In this study, target RNA catalyzed the formation of a H1-H2 duplex, which was then detected by LFIA via digoxin/biotin recognition. The detection limit was calculated to be 2000 copies/mL. These studies demonstrate the potential of CHA for clinical diagnosis.
2.2.2 Branched CHA
Branched CHA (bCHA) usually employ three or more hairpin probes to form three-way or four-way junctions upon the initiator. Due to the higher energy barriers, bCHA had a much lower background signal than traditional CHA, providing higher detection sensitivity. Yue et al. (2019) employed bCHA for miRNA imaging in living cells by anchoring three CHA hairpins on a Y-shaped DNA scaffold. After entering the cell, the probe could undergo bCHA through the catalysis of miR-155, leading to the formation of DNA dendrimers and in-situ imaging of miRNA. In another study, Liu et al. (2020) proposed a miRNA imaging method based on in-situ growth of DNA nanostructures by introducing palindromic sequences to bCHA hairpins. According to Figure 2E, upon miR-21, bCHA could be triggered to form Y-shaped structures, which could further assemble into 3D nanospheres owing to the palindromic fragments at the end of the hairpins. The process of bCHA also caused separation of fluorophore and quencher, leading to the recovery of fluorescence and reaching a LOD of 1.4 pM. Other than miRNA detection, bCHA has also found its application in antibiotic detection, Zhang et al. (2019) developed an aptasensor for the simultaneous detection of chloramphenicol (CAP) and kanamycin (Kana). In the presence of CAP or Kana, CHA hairpins can be catalyzed to form three-arm or four-arm branched junctions, which could be distinguished by microchip electrophoresis (MCE) based on migration time. And the detection limits for CAP and Kana were 0.49 pg/mL and 0.52 pg/mL, respectively.
2.2.3 Localized CHA
Localized CHA (LCHA) takes advantage of the spatial-confinement effect by limiting CHA hairpins in a compact area, leading to higher amplification efficiency and reaction rate. Similar to LHCR, the hairpins could be fixed on 1D, 2D, or 3D DNA nanostructures to realize LCHA, such as DNA origami (Dai et al., 2021), Y-shaped scaffolds (Wu et al., 2020b), DNA nanocubes (Liu et al., 2019; Feng et al., 2022), DNA triangular prisms (DTP) (Yang et al., 2020), and DNA nanolanterns (Wang et al., 2022a). Recently, a nanowire-based LCHA for miRNA imaging was reported by Wei et al. (2018b). Target RNA catalyzed the hybridization between adjacent hairpins on the nanowire, leading to the recovery of quenched fluorescence. The LOD of this method was 2.0 pM, which was about 11.8 times lower than typical CHA. In addition, Bai et al. (2022) developed a dispersion-to-localization of catalytic hairpin assembly (DL-CHA) strategy. In the presence of miR-21, CHA was induced between hairpins anchored to linker strands, which promoted the formation of DNA ladders. A high signal-to-background (S/B) ratio was reached at 37.8, with a wide linear range of 100 pM to 100 nM. Moreover, Qing et al. (2020) developed an intramolecular CHA strategy for mRNA imaging, where the distance between reactants were shortened by anchoring the CHA hairpins on a DNA tetrahedron (Figure 2F). The local concentration of hairpins was about 808 times higher than free CHA, which increased the initial reaction rate by 15.6 times. Later, an intermolecular CHA strategy for FRET imaging of miRNA-155 in cells was proposed by Wang et al. (2022b). By anchoring CHA hairpins on separate DNA tetrahedrons, the targets could trigger CHA between DNA nanostructures, resulting in polymerized products and amplified FRET signals.
2.3 Biosensors based on cascaded reaction
For the development of biosensors with higher sensitivity, multiple amplification strategies could be cascaded, whereby the product of the upstream reaction acts as a trigger for the downstream reaction. In this section, we will discuss recent advances in biosensors based on cascade reactions involving HCR and CHA.
2.3.1 Cascaded reaction of CHA and CHA
Cascaded CHA could achieve exponential signal growth by interconnecting multiple CHA circuits. For example, Guk et al. (2019) reported a self-circulation CHA system for miRNA detection. According to their design, miR-34a initiated the first CHA cycle and released the catalyst for the second CHA cycle, which in turn produced the initiator for the first cycle. Owing to its versatility, this strategy has been adapted for interferon-γ (IFN-γ) detection, reaching a LOD of 0.6 pM (Wang et al., 2019b). Besides, another promising approach is the self-replicating catalytic hairpin assembly system (SRCHA), in which only two hairpins were required for the cascade reaction. As illustrated in Figure 2G, the target could trigger the first CHA cycle and generate duplexes with exposed trigger sequences. Therefore, the CHA product may serve as a new CHA initiator. This system has been successfully applied to the detection of metal ions, biomolecules, and nucleic acids (Dai et al., 2017; Pan et al., 2019).
2.3.2 Cascaded reaction of CHA and HCR
The cascaded integration of CHA and HCR is expected to provide higher amplification efficiency than single amplification. As shown in Figure 2H, Wang et al. (2018) established a general CHA-HCR sensing platform, where the outputs of CHA induced by the target miRNA could serve as the initiators for HCR circuits, showing a LOD of 2 pM. Later, Li et al. (2022) integrated the concept of palindrome-based HCR (PHCR) with CHA for miR-21 detection. The presence of miR-21 catalyzed the formation of CHA products, which further initiated HCR between hairpins with palindromic domains, leading to the formation of crosslinking networks through the assembly of HCR products. With amplified FRET signals, target RNA could be detected down to 10 pM. The strategy of HCR-CHA has also been employed in sensors. Wei et al. (2016) integrated split CHA initiators into HCR hairpins. The presence of miR-144 could trigger HCR and co-localize the split sequences, which further initiated the CHA process and induced amplified fluorescence, reaching the LOD of 0.3 fM.
3 Conclusions and perspectives
The last decade has seen a rapid development in HCR and CHA for the biosensing applications. Based on their features of being isothermal and enzyme-free, these techniques have been employed for the detection of metal ions, small molecules, proteins, cells, tissues and living animals.
Despite significant progress, the applications of isothermal and enzyme-free amplification still face several challenges. 1) To improve detection accuracy, the problem of background leakage must be resolved. During the spontaneous breathing process of DNA duplexes, non-specific reactions between hairpins of HCR or CHA may occur in the absence of the initiator, leading to a high background signal. To minimize background signals, more effort should be directed toward rationally designing the sequence in order to avoid unexpected reactions. 2) In terms of sensitivity, traditional HCR or CHA showed limited amplification efficacy compared to enzyme-assisted amplification. To increase sensitivity, the development of cascaded amplification strategies would be beneficial (Wang et al., 2018; Wang et al., 2019b). 3) The kinetics are relatively slow since typical HCR and CHA rely on random collisions of hairpins. The strategy of localized HCR or CHA could provide a promising method to achieve a faster reaction time by exploiting the spatial-confinement effect (Wang et al., 2019a; Qing et al., 2020). 4) The majority of reported strategies focus on singlexed detection. However, it is necessary for clinical diagnosis of diseases to determine the level of multiple markers. Thus, constructing multiplexed sensing platform is more beneficial for biomedical applications. However, the sequence of hairpins needs to be designed for different targets, thereby limiting their applications in multi-target analysis. Fortunately, several universal sensing systems have been proposed, where the initiator was embedded in a starting hairpin. By adjusting the sequence of the starting hairpins according to the target, the sequence of the remaining hairpins could remain unchanged (Huang et al., 2016; Zhang et al., 2022). 5) The cellular delivery of nucleic acid probe remains a challenging task. Luckily, the development of DNA nanotechnology has offered a bunch of nanocarriers with controllable geometry, easy synthesis, and high biocompatibility. Integrating HCR or CHA hairpins on DNA nanostructures is a promising approach for achieving intracellular internalization (Liang et al., 2014; Raniolo et al., 2019; Peng et al., 2021). 6) The stability of nucleic acid probes is a major concern, especially in real samples, such as food or blood. There has been evidence that DNA nanocarriers can be effective at increasing stability (Li et al., 2021). Alternatively, there is the option of using artificial nucleic acids, such as L-DNA, peptide nucleic acids, and locked nucleic acids (Olson et al., 2017).
In spite of the challenges, continuous attention has been paid to the development of isothermal, enzyme-free amplification techniques. It is believed that more advanced amplification strategies based on HCR and CHA will be emerging, and more targets will be detected with the aid of aptamers or antibodies, and more signal transduction methods (e.g., mass spectrometry, ultrasound imaging, magnetic resonance imaging) may be integrated to develop multifunctional sensors. Eventually, these efforts will result in more useful tools for bioanalysis, clinical diagnosis, and environmental monitoring.
Author contributions
LM and WL directed and supervised the research. LM and WH contributed to the draft writing. ZL, DL, RQ, and MM contributed to literature collection, reviewing and editing. CY assisted with the editing and visualization.
Funding
This work was financially supported by the National Natural Science Foundation of China (Nos. 21877048, 22077048, and 22277014), Guangxi Natural Science Foundation (Nos. 2022GXNSFBA035533, AD21220061, 2021GXNSFDA075003, 2022GXNSFBA035482), the startup fund of Guangxi University (A3040051003, A3040051011).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bai, H., Yan, Y., Li, D., Fan, N., Cheng, W., Yang, W., et al. (2022). Dispersion-to-localization of catalytic hairpin assembly for sensitive sensing and imaging microRNAs in living cells from whole blood. Biosens. Bioelectron. 198, 113821. doi:10.1016/j.bios.2021.113821
Bui, H., Miao, V., Garg, S., Mokhtar, R., Song, T., and Reif, J. (2017). Design and analysis of localized DNA hybridization chain reactions. Small 13, 1602983. doi:10.1002/smll.201602983
Chai, H., and Miao, P. (2019). Bipedal DNA walker based electrochemical genosensing strategy. Anal. Chem. 91, 4953–4957. doi:10.1021/acs.analchem.9b01118
Cheglakov, Z., Cronin, T. M., He, C., and Weizmann, Y. (2015). Live cell MicroRNA imaging using cascade hybridization reaction. J. Am. Chem. Soc. 137, 6116–6119. doi:10.1021/jacs.5b01451
Cheng, Y. H., Liu, S. J., and Jiang, J. H. (2021). Enzyme-free electrochemical biosensor based on amplification of proximity-dependent surface hybridization chain reaction for ultrasensitive mRNA detection. Talanta 222, 121536. doi:10.1016/j.talanta.2020.121536
Dai, J., He, H., Duan, Z., Guo, Y., and Xiao, D. (2017). Self-replicating catalyzed hairpin assembly for rapid signal amplification. Anal. Chem. 89, 11971–11975. doi:10.1021/acs.analchem.7b01946
Dai, J., Xing, C., Lin, Y., Huang, Y., Yang, Y., Chen, Z., et al. (2021). Localized DNA catalytic hairpin assembly reaction on DNA origami for tumor-associated microRNA detection and imaging in live cells. Sens. Actuators B Chem. 344, 130195. doi:10.1016/j.snb.2021.130195
Dirks, R. M., and Pierce, N. A. (2004). Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. U. S. A. 101, 15275–15278. doi:10.1073/pnas.0407024101
Feng, Y., Liu, Q., Zhao, X., Chen, M., Sun, X., Li, H., et al. (2022). Framework nucleic acid-based spatial-confinement amplifier for miRNA imaging in living cells. Anal. Chem. 94, 2934–2941. doi:10.1021/acs.analchem.1c04866
Gong, X., Wei, J., Liu, J., Li, R., Liu, X., and Wang, F. (2019). Programmable intracellular DNA biocomputing circuits for reliable cell recognitions. Chem. Sci. 10, 2989–2997. doi:10.1039/C8SC05217D
Guk, K., Hwang, S. G., Lim, J., Son, H. Y., Choi, Y., Huh, Y. M., et al. (2019). Fluorescence amplified sensing platforms enabling miRNA detection by self-circulation of a molecular beacon circuit. Chem. Commun. 55, 3457–3460. doi:10.1039/C9CC00351G
Hansen-Bruhn, M., Nielsen, L. D. F., and Gothelf, K. V. (2018). Rapid detection of drugs in human plasma using a small-molecule-linked hybridization chain reaction. ACS Sens. 3, 1706–1711. doi:10.1021/acssensors.8b00439
Huang, J., Wang, H., Yang, X., Quan, K., Yang, Y., Ying, L., et al. (2016). Fluorescence resonance energy transfer-based hybridization chain reaction for in situ visualization of tumor-related mRNA. Chem. Sci. 7, 3829–3835. doi:10.1039/C6SC00377J
Huang, X., Jia, J., Lin, Y., Qiu, B., Lin, Z., and Chen, H. (2020). A highly sensitive electrochemiluminescence biosensor for pyrophosphatase detection based on click chemistry-triggered hybridization chain reaction in homogeneous solution. ACS Appl. Mat. Interfaces. 12, 34716–34722. doi:10.1021/acsami.0c10542
Jiang, Y., Chen, X., Feng, N., and Miao, P. (2021). A highly sensitive aptasensor for the detection of prostate specific antigen based on dumbbell hybridization chain reaction. Sens. Actuators B Chem. 340, 129952. doi:10.1016/j.snb.2021.129952
Li, C., Luo, S., Wang, J., Shen, Z., and Wu, Z. S. (2021). Nuclease-resistant signaling nanostructures made entirely of DNA oligonucleotides. Nanoscale 13, 7034–7051. doi:10.1039/D1NR00197C
Li, C., Zhang, J., Gao, Y., Luo, S., and Wu, Z. S. (2022). Nonenzymatic autonomous assembly of cross-linked network structures from only two palindromic DNA components for intracellular fluorescence imaging of miRNAs. ACS Sens. 7, 601–611. doi:10.1021/acssensors.1c02504
Li, Y., Huang, C., and Li, Y. (2019). Ultrasensitive electrochemiluminescence detection of MicroRNA via one-step introduction of a target-triggered branched hybridization chain reaction circuit. Anal. Chem. 91, 9308–9314. doi:10.1021/acs.analchem.9b02580
Liang, L., Li, J., Li, Q., Huang, Q., Shi, J., Yan, H., et al. (2014). Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew. Chem. Int. Ed. 53, 7745–7750. doi:10.1002/anie.201403236
Liu, L., Rong, Q., Ke, G., Zhang, M., Li, J., Li, Y., et al. (2019). Efficient and reliable MicroRNA imaging in living cells via a FRET-based localized hairpin-DNA cascade amplifier. Anal. Chem. 91, 3675–3680. doi:10.1021/acs.analchem.8b05778
Liu, R., Zhang, S., Zheng, T. T., Chen, Y. R., Wu, J. T., and Wu, Z. S. (2020). Intracellular nonenzymatic in situ growth of three-dimensional DNA nanostructures for imaging specific biomolecules in living cells. ACS Nano 14, 9572–9584. doi:10.1021/acsnano.9b09995
Ma, W., Chen, B., Zou, S., Jia, R., Cheng, H., Huang, J., et al. (2019). I-Motif-Based in situ bipedal hybridization chain reaction for specific activatable imaging and enhanced delivery of antisense oligonucleotides. Anal. Chem. 91, 12538–12545. doi:10.1021/acs.analchem.9b03420
Marras, S. A. E., Bushkin, Y., and Tyagi, S. (2019). High-fidelity amplified FISH for the detection and allelic discrimination of single mRNA molecules. Proc. Natl. Acad. Sci. U.S.A. 116, 13921–13926. doi:10.1073/pnas.1814463116
Mo, L., Liang, D., He, W., Yang, C., and Lin, W. (2021). Ratiometric and amplified fluorescence nanosensor based on a DNA tetrahedron for miRNA imaging in living cells. J. Mat. Chem. B 9, 8341–8347. doi:10.1039/D1TB01537K
Mo, L., Liang, D., Mo, M., Yang, C., and Lin, W. (2023). Dual-detection of miRNAs in living cells via hybridization chain reaction on DNA tetrahedron. Sens. Actuators B Chem. 375, 132955. doi:10.1016/j.snb.2022.132955
Olson, X., Kotani, S., Yurke, B., Graugnard, E., and Hughes, W. L. (2017). Kinetics of DNA strand displacement systems with locked nucleic acids. J. Phys. Chem. B 121, 2594–2602. doi:10.1021/acs.jpcb.7b01198
Pan, J., Zeng, L., and Chen, J. (2019). An enzyme-free DNA circuit for the amplified detection of Cd2+ based on hairpin probe-mediated toehold binding and branch migration. Chem. Commun. 55, 11932–11935. doi:10.1039/C9CC06311K
Peng, X., Fang, S., Ji, B., Li, M., Song, and J., Qiu, L., et al. (2021). DNA nanostructure-programmed cell entry via corner angle-mediated molecular interaction with membrane receptors. Nano. Lett. 21, 6946–6951. doi:10.1021/acs.nanolett.1c02191
Peng, X., Yang, J., Liang, W., Sun, Y., Mei, X., Zhang, G., et al. (2022). Double loop-stem hairpins mediated hybridization chain reaction: A multifunctional DNA molecular tool to produce the intact aptamer for label-free biosensing. Sens. Actuators B Chem. 369, 132327. doi:10.1016/j.snb.2022.132327
Qin, Y., Li, D., Yuan, R., and Xiang, Y. (2019). Netlike hybridization chain reaction assembly of DNA nanostructures enables exceptional signal amplification for sensing trace cytokines. Nanoscale 11, 16362–16367. doi:10.1039/C9NR04988F
Qing, Z., Hu, J., Xu, J., Zou, Z., Lei, Y., Qing, T., et al. (2020). An intramolecular catalytic hairpin assembly on a DNA tetrahedron for mRNA imaging in living cells: Improving reaction kinetics and signal stability. Chem. Sci. 11, 1985–1990. doi:10.1039/C9SC04916A
Raniolo, S., Croce, S., Thomsen, R. P., Okholm, A. H., Unida, V., Iacovelli, F., et al. (2019). Cellular uptake of covalent and non-covalent DNA nanostructures with different sizes and geometries. Nanoscale 11, 10808–10818. doi:10.1039/C9NR02006C
Sadowski, J. P., Calvert, C. R., Zhang, D. Y., Pierce, N. A., and Yin, Peng. (2014). Developmental self-assembly of a DNA tetrahedron. ACS Nano 8, 3251–3259. doi:10.1021/nn4038223
Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn, G. T., et al. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487–491. doi:10.1126/science.2448875
Song, Z., Zhou, Y., Shen, M., Zhao, D., Hu, H., Zeng, S., et al. (2022). MUC1 detection and in situ imaging method based on aptamer conformational switch and hybridization chain reaction. Talanta 239, 123129. doi:10.1016/j.talanta.2021.123129
Sun, W., Wang, Y., Zhu, Z., Wang, Y., Zhang, M., Jiang, L., et al. (2021). Accurate and nonpurified identification of extracellular vesicles using dual-binding recognition mode. Anal. Chem. 93, 12383–12390. doi:10.1021/acs.analchem.1c02259
Tang, J., Lei, Y., He, X., Liu, J., Shi, H., and Wang, K. (2020). Recognition-driven remodeling of dual-split aptamer triggering in situ hybridization chain reaction for activatable and autonomous identification of cancer cells. Anal. Chem. 92, 10839–10846. doi:10.1021/acs.analchem.0c02524
Wang, H., Li, C., Liu, X., Zhou, X., and Wang, F. (2018). Construction of an enzyme-free concatenated DNA circuit for signal amplification and intracellular imaging. Chem. Sci. 9, 5842–5849. doi:10.1039/C8SC01981A
Wang, J., Wang, D. X., Ma, J. Y., Wang, Y. X., and Kong, D. M. (2019a). Three-dimensional DNA nanostructures to improve the hyperbranched hybridization chain reaction. Chem. Sci. 10, 9758–9767. doi:10.1039/C9SC02281C
Wang, P., Meyer, T. A., Pan, V., Dutta, P. K., and Ke, Y. (2017). The beauty and utility of DNA origami. Chem 2, 359–382. doi:10.1016/j.chempr.2017.02.009
Wang, S., Li, X., Jiang, B., Yuan, R., and Xiang, Y. (2019b). Target-triggered programming of cascaded catalytic hairpin assemblies for enzyme-free and highly sensitive sensing of cytokines. Sens. Actuators B Chem. 298, 126929. doi:10.1016/j.snb.2019.126929
Wang, W., Peng, Y., Wu, J., Zhang, M., Li, Q., Zhao, Z., et al. (2021). Ultrasensitive detection of 17β-estradiol (E2) based on multistep isothermal amplification. Anal. Chem. 93, 4488–4496. doi:10.1021/acs.analchem.0c04681
Wang, Y., Bai, Y., Cao, L., Li, L., Zhan, L., Zuo, H., et al. (2022b). Catalytic hairpin assembled polymeric tetrahedral DNA frameworks for MicroRNA imaging in live cells. Biosens. Bioelectron. 197, 113783. doi:10.1016/j.bios.2021.113783
Wang, Y. X., Wang, D. X., Wang, J., Liu, B., Tang, A. N., and Kong, D. M. (2022a). DNA nanolantern-mediated catalytic hairpin assembly nanoamplifiers for simultaneous detection of multiple microRNAs. Talanta 236, 122846. doi:10.1016/j.talanta.2021.122846
Wei, J., Gong, X., Wang, Q., Pan, M., Liu, X., Liu, J., et al. (2018a). Construction of an autonomously concatenated hybridization chain reaction for signal amplification and intracellular imaging. Chem. Sci. 9, 52–61. doi:10.1039/C7SC03939E
Wei, Q., Huang, J., Li, J., Wang, J., Yang, X., Liu, J., et al. (2018b). A DNA nanowire based localized catalytic hairpin assembly reaction for microRNA imaging in live cells. Chem. Sci. 9, 7802–7808. doi:10.1039/C8SC02943A
Wei, Y., Zhou, W., Li, X., Chai, Y., Yuan, R., and Xiang, Y. (2016). Coupling hybridization chain reaction with catalytic hairpin assembly enables non-enzymatic and sensitive fluorescent detection of microRNA cancer biomarkers. Biosens. Bioelectron. 77, 416–420. doi:10.1016/j.bios.2015.09.053
Wu, H., Chen, T. T., Wang, X. N., Ke, Y., and Jiang, J. H. (2020a). RNA imaging in living mice enabled by an in vivo hybridization chain reaction circuit with a tripartite DNA probe. Chem. Sci. 11, 62–69. doi:10.1039/C9SC03469B
Wu, H., Zhou, W. J., Liu, L., Fan, Z., Tang, H., Yu, R. Q., et al. (2020b). In vivo mRNA imaging based on tripartite DNA probe mediated catalyzed hairpin assembly. Chem. Commun. 56, 8782–8785. doi:10.1039/D0CC03596C
Yang, F., Cheng, Y., Cao, Y., Dong, H., Lu, H., Zhang, K., et al. (2019). Sensitively distinguishing intracellular precursor and mature microRNA abundance. Chem. Sci. 10, 1709–1715. doi:10.1039/C8SC03305F
Yang, F., Cheng, Y., Zhang, Y., Wei, W., Dong, H., Lu, H., et al. (2020). Bioinspired framework nucleic acid capture sensitively and rapidly resolving MicroRNAs biomarkers in living cells. Anal. Chem. 92, 4411–4418. doi:10.1021/acs.analchem.9b05304
Yin, P., Choi, H. M. T., Calvert, C. R., and Pierce, N. A. (2008). Programming biomolecular self-assembly pathways. Nature 451, 318–322. doi:10.1038/nature06451
Yuan, B., Chen, Y., Sun, Y., Guo, Q., Huang, J., Liu, J., et al. (2018). Enhanced imaging of specific cell-surface glycosylation based on multi-FRET. Anal. Chem. 90, 6131–6137. doi:10.1021/acs.analchem.8b00424
Yue, S., Song, X., Song, W., and Bi, S. (2019). An enzyme-free molecular catalytic device: Dynamically self-assembled DNA dendrimers for in situ imaging of microRNAs in live cells. Chem. Sci. 10, 1651–1658. doi:10.1039/C8SC04756A
Zhang, F., Nangreave, J., Liu, Y., and Yan, H. (2014). Structural DNA nanotechnology: State of the art and future perspective. J. Am. Chem. Soc. 136, 11198–11211. doi:10.1021/ja505101a
Zhang, K., Gan, N., Shen, Z., Cao, J., Hu, F., and Li, T. (2019). Microchip electrophoresis based aptasensor for multiplexed detection of antibiotics in foods via a stir-bar assisted multi-arm junctions recycling for signal amplification. Biosens. Bioelectron. 130, 139–146. doi:10.1016/j.bios.2019.01.044
Zhang, L. M., Gao, Q. X., Chen, J., Li, B., Li, M. M., Zheng, L., et al. (2022). A universal catalytic hairpin assembly system for direct plasma biopsy of exosomal PIWI-interacting RNAs and microRNAs. Anal. Chim. Acta. 1192, 339382. doi:10.1016/j.aca.2021.339382
Zhao, W., Ali, M. M., Brook, M. A., and Li, Y. (2008). Rolling circle amplification: Applications in nanotechnology and biodetection with functional nucleic acids. Angew. Chem. Int. Ed. 47, 6330–6337. doi:10.1002/anie.200705982
Zhou, Y., Yang, L., Wei, J., Ma, K., Gong, X., Shang, J., et al. (2019). An autonomous nonenzymatic concatenated DNA circuit for amplified imaging of intracellular ATP. Anal. Chem. 91, 15229–15234. doi:10.1021/acs.analchem.9b04126
Zou, M., Su, F., Zhang, R., Jiang, X., Xiao, H., Yan, X., et al. (2021). Rapid point-of-care testing for SARS-CoV-2 virus nucleic acid detection by an isothermal and nonenzymatic Signal amplification system coupled with a lateral flow immunoassay strip. Sens. Actuators B Chem. 342, 129899. doi:10.1016/j.snb.2021.129899
Keywords: hybridization chain reaction, catalytic hairpin assembly, biosensor, disease diagnosis, cell imaging
Citation: Mo L, He W, Li Z, Liang D, Qin R, Mo M, Yang C and Lin W (2023) Recent progress in the development of DNA-based biosensors integrated with hybridization chain reaction or catalytic hairpin assembly. Front. Chem. 11:1134863. doi: 10.3389/fchem.2023.1134863
Received: 31 December 2022; Accepted: 10 February 2023;
Published: 16 February 2023.
Edited by:
Yifan Lyu, Hunan University, ChinaCopyright © 2023 Mo, He, Li, Liang, Qin, Mo, Yang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiying Lin, d2VpeWluZ2xpbjIwMTNAMTYzLmNvbQ==
 Liuting Mo
Liuting Mo Wanqi He
Wanqi He Weiying Lin
Weiying Lin