- 1School of Chemistry, University of Birmingham, Birmingham, United Kingdom
- 2School of Biosciences, University of Birmingham, Birmingham, United Kingdom
- 3Linear Diagnostics Ltd., Birmingham, United Kingdom
A plug-and-play sandwich assay platform for the aptamer-based detection of molecular targets using linear dichroism (LD) spectroscopy as a read-out method has been demonstrated. A 21-mer DNA strand comprising the plug-and-play linker was bioconjugated onto the backbone of the filamentous bacteriophage M13, which gives a strong LD signal due to its ready alignment in linear flow. Extended DNA strands containing aptamer sequences that bind the protein thrombin, TBA and HD22, were then bound to the plug-and-play linker strand via complementary base pairing to generate aptamer-functionalised M13 bacteriophages. The secondary structure of the extended aptameric sequences required to bind to thrombin was checked using circular dichroism spectroscopy, with the binding confirmed using fluorescence anisotropy measurements. LD studies revealed that this sandwich sensor design is very effective at detecting thrombin down to pM levels, indicating the potential of this plug-and-play assay system as a new label-free homogenous detection system based on aptamer recognition.
1 Introduction
The SARS-CoV-2 pandemic has starkly highlighted how molecular diagnostics can make a very significant impact on healthcare outcomes. It is clear that both the antibody-based lateral flow device (LFD) test and DNA-based tests like PCR have saved countless lives and reduced the global severity of the pandemic. But nevertheless the diagnostic response to the outbreak has also shown that there is still much room for improvement in modern healthcare diagnostics. (Alafeef and Pan, 2022). Even established tests that use PCR and LFDs can suffer from a combination of various issues such as time and sensitivity, as well as limitations in throughput. In addition, nucleic acid tests such as PCR are largely restricted to the laboratory, slowing down the testing process further and in some instances negating their use. These observations make it clear that efforts need to continue in generating new testing modalities to resolve these issues. This paper details the next stage of development of our bacteriophage/linear dichroism (LD) assay system that offers an alternative target read-out method to more established techniques. (Michel et al., 2020; Trotter et al., 2020; Sun et al., 2019; Chen et al., 2019). This system has been previously developed for detection using antibodies (Pacheco-Gómez et al., 2012) and DNA (Ali et al., 2020) and offers high levels of sensitivity alongside speed and simplicity. Here we extend our work on this signal read-out methodology to encompass DNA aptamer-based detection.
The LD/bacteriophage assay is based on the observation that filamentous bacteriophage (equivalent to a filament that is 1 μm long by 10 nm wide) can be aligned in fluid flow. This in turn aligns the chromophores within the bacteriophage, which can then be detected using LD spectroscopy. Crucially any other material present in solution that does not align has no LD signal, meaning that impurities do not perturb the spectrum. In previous work, we exploited this observation for an assay by derivatising a bacteriophage (M13 in this case) with moieties that could bind to an assay target at physiological pH. Upon target binding, the hydrodynamic behaviour of the particle changed, altering alignment and hence the LD signal. We accordingly demonstrated that this novel LD sensing methodology could be used to detect pathogenic E. coli using antibodies (Pacheco-Gómez et al., 2012) as well as DNA from pathogens. (Ali et al., 2020).
In the case of DNA sensing using LD spectroscopy, we generated a “sandwich assay” where the DNA target bridges between two M13 bacteriophages in order to disrupt alignment. (Ali et al., 2020). We found it sufficient to covalently attach DNA strands to the M13 bionanoparticles that were complementary to different sequence regions of the target to bring about a change in LD signal. However, in the case of detecting a non-nucleic acid target using similar DNA-functionalised particles, a slightly more intricate design was required. Here we show the proof-of-concept of a so-called plug-and-play method for target recognition using LD spectroscopy (Scheme 1). It consists of a highly modular M13 sensing platform that in principle can be used for interaction with any combination of aptameric sequence for which target binding brings about a change in LD signal.
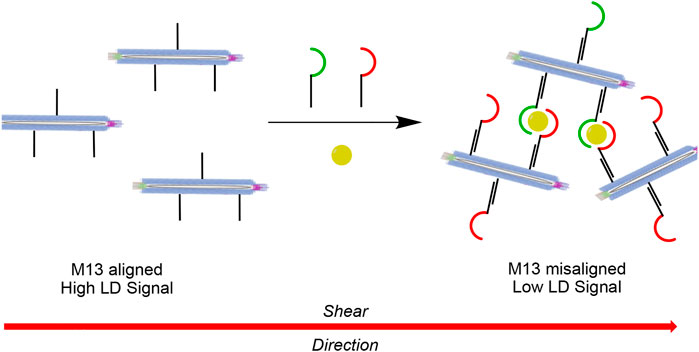
SCHEME 1. Schematic representation of the plug-and-play method for target recognition using LD spectroscopy. Each M13 bacteriophage is covalently attached to copies of the same DNA strand, which are then bound to complementary DNA strands containing one-half of a recognition element for the target of interest. The presence of the target (shown in yellow) causes aggregation of the particles in solution through the formation of multiple sandwich complexes, which results in misalignment of the particles in shear flow and hence a reduction in the LD signal.
2 Results and discussion
2.1 Design
In order to demonstrate the desired change in M13 alignment for LD sensing of non-DNA targets using aptamers, it was decided to continue to explore our sandwich assay design that had already been shown to be highly effective for DNA sensing. (Ali et al., 2020). For the target, we chose the protein thrombin, the main regulatory enzyme in the blood coagulation cascade, for two reasons. Firstly its aptamer binding properties are well characterized and understood, making it a popular and highly effective target exemplar for a number of other sensor platforms. (Ikebukuro et al., 2004; Xiao et al., 2005; Wang et al., 2008; Cho et al., 2008; Wang and Zhao, 2012; Deng et al., 2014; Kim et al., 2018; Malecka and Ferapontova, 2021; Neupane and Stine, 2021). Secondly two different thrombin aptamers are known (TBA and HD22), enabling a relatively straightforward design for a sandwich assay, which involves the simultaneous binding of different regions of the thrombin by two separate DNA strands. Indeed others have already demonstrated the effectiveness of this approach in various sandwich assays for thrombin (Neupane and Stine, 2021; Ikebukuro et al., 2004; Wang and Zhao, 2012).
As we had previously demonstrated an effective bioconjugation technique involving thiol-ene and amide coupling chemistries to append DNA strands to lysine residues (pVIII protein) of the M13 bacteriophage, (Carr-Smith et al., 2015; Ali et al., 2020), we decided to adopt the same approach here. However instead of directly attaching the two different aptamers to the M13 chassis, we used exactly the same linker strand as the one employed previously for DNA sensing (Ali et al., 2020). Using this approach, the same functionalised chassis can be connected via DNA base-pairing to any aptamer of choice by attaching a sequence to the aptamer strand that is complementary to the linker strand (Scheme 1). Such a plug-and-play methodology enables a highly flexible and modular approach to the sensor platform, in which various binding motifs may be connected to the M13 chassis via DNA-mediated base-paring, without the need to undertake new bioconjugation reactions for different assays.
Scheme 2 illustrates the method used for the construction of the molecular components of the thrombin assay system. First the M13 bacteriophages are prepared in bulk and covalently attached to multiple copies of the universal plug-and-play linker strand PPL (here only one strand is shown for clarity) to form the functionalised M13-PPL bionanoparticles. Next the two DNA aptamers for thrombin, TBA and HD22, with extended sequences complementary to M13-PPL, are annealed via DNA duplex formation to complete the assembly of the recognition system. Our hypothesis was that the resulting aggregation of M13 bionanoparticles brought about by simultaneous sandwich complexation of thrombin by the two aptamers would result in a change in linear alignment in flow and hence a change in the LD signal, as shown schematically in Scheme 1.
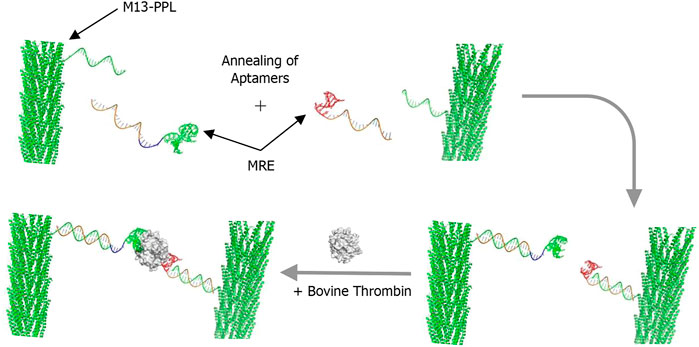
SCHEME 2. The thrombin sandwich assay design. DNA-modified M13 bacteriophage (M13-PPL) is annealed to two different extended aptamers (molecular recognition elements or MREs), to which the target, the protein thrombin, is added for detection. Note: In this study, an average of 15 PPL strands were conjugated to each M13 chassis, although only one is shown here for clarity.
2.2 Synthesis and characterisation
The sequences of the oligonucleotides used in this study are shown in Table 1. It lists the two aptamers as extended forms (TBA15 and HD22T5), the plug-and-play linker (PPL) and a control strand with a scrambled sequence (NS). The PPL sequence is a 21-mer oligonucleotide containing a thiol moiety at its 3’ end (prepared as a disulfide) and a chromophore tag, 6-carboxyfluorescein (6-FAM), for quantification purposes. This PPL strand was conjugated to the bacteriophage via a short linker molecule to form M13-PPL, one of four DNA-functionalised phage systems that we reported previously. (Ali et al., 2020). Its preparation, purification using size exclusion chromatography (SEC) and quantification using UV/vis absorbance spectroscopy is outlined in the Supplementary Material S1. A batch of M13-PPL from our previous study (Ali et al., 2020) was used again here, which contained an average of fifteen PPL strands conjugated onto its coat protein.

TABLE 1. The DNA oligonucleotides used in this study. PPL, prepared previously, (Ali et al., 2020) is shown in its thiol form to indicate its attachment point to the M13 chassis. The two extended aptamers TBA15 and HD22T5 are listed with their thrombin binding (aptameric) regions in red and green respectively and their complementary regions to the PPL strand in orange.
As described above, the two thrombin aptamers TBA and HD22 can bind at different exosites (secondary binding sites) on the protein surface, making them ideal candidates for a sandwich assay. The interaction of these aptamers with thrombin has been studied in detail since their SELEX screening for TBA in 1992 (Bock et al., 1992) and HD22 in 1997. (Tasset et al., 1997). The G-quadruplex forming TBA was extended from its 5’ end with the complementary sequence to that of PPL to give the 36-mer TBA15. It has previously been shown (De Rache et al., 2012) that elongation of this aptamer sequence does not require a spacer sequence to retain formation of the G-quadruplex. However HD22 forms a short folded internal duplex in addition to a G-quadruplex. Therefore, to mitigate any adverse effect on this more intricate folded conformation, a short spacer sequence of five thymine bases was inserted between the aptamer sequence and the sequence complementary to PPL to give some additional flexibility to the resulting extended 55-mer strand HD22T5. CD studies (see Supplementary Material S1) confirmed that sequence elongation of both aptamers had no detrimental effect on their secondary structures.
2.3 Binding studies
Gel electrophoretic mobility shift assays (EMSAs) were undertaken to establish the thrombin binding properties of the extended DNA aptamer strands, with these tested in tandem to establish whether thrombin could be bound by both strands. Accordingly, equimolar amounts of TBA15 and HD22T5 were combined and then thrombin was added to each sample in increasing amounts. These mixtures were incubated at 37°C for 30 min to ensure equilibration before each experiment. As shown in an experiment that stained for DNA (Figure 1), upon increasing the concentration of thrombin across the gel (lanes 1–6), TBA15 became almost completely retained at the top of the gel once a thrombin concentration of 2 µM was reached. Interestingly this change was less dramatic for HD22T5, indicating that a higher proportion of this aptamer remained unbound by thrombin at the same concentration. These results can not be explained by differences in binding affinity as the aptamer HD22 is the stronger binder of thrombin. (Zhao et al., 2021). Instead they are more likely to be ascribed to differences in binding stoichiometry, given that it has previously been shown (Tasset et al., 1997) that two TBA aptamers can bind one thrombin molecule simultaneously, whereas this is not the case for HD22, which can only bind at exosite II. As for two distinct bands corresponding to thrombin-DNA complexes being observed higher up the gel, it is possible that the lower band, which first appears at sub-stoichiometric concentrations of thrombin, corresponds to the 2/1 sandwich complex, with the higher band (highlighted in an orange box) being the 1/1 complex because of its lower negative charge density. However, an alternative explanation is that different forms or aggregates of thrombin may be bound by these aptamers under these gel conditions, which is supported by other bands for the target being revealed by a protein stain of this gel (see Supplementary Material S1).
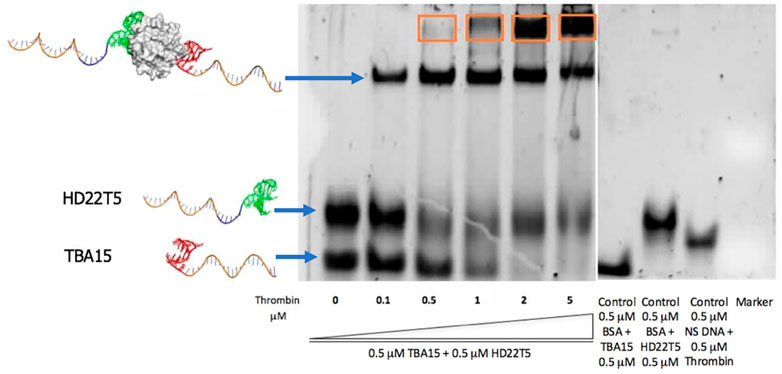
FIGURE 1. EMSA of TBA15 and HD22T5 (both at 0.5 μM) with increasing concentrations of thrombin (0, 0.1, 0.5, 1, 2, 5 μM) (lanes 1–6). Control bovine serum albumin, BSA (0.5 μM) + TBA15 (0.5 μM), BSA (0.5 μM) + HD22T5 (0.5 μM), non-specific DNA, NS (0.5 μM) + thrombin (0.5 μM) (lanes 7–9). SYBR Gold® nucleic acid stain used to visualize gel. Electrophoresis buffer used: 1.0 x TBE +10 mM KCl. The band in an orange box is ascribed to other stoichiometries or aggregates (see text for details). For more experimental details, see Supplementary Material S1.
With the binding of thrombin by both extended aptamers confirmed by EMSAs, their duplex formation in solution with strand PPL needed to be confirmed before undertaking the LD sensing studies. Fluorescence anisotropy (FA) was used for this purpose, which involved titrating in TBA15 and HD22T5 respectively to solutions of the PPL strand. As expected, an increase in the FA signal was observed for both extended aptamers as their molecular tumbling time decreased upon forming a larger complex. (Vafaei et al., 2021). These data indicate that saturation of binding is reached with the addition of one molar equivalent of extended aptamer, corresponding to the formation of a 1/1 duplex in each case (Figure 2). The HD22T5-PPL duplex had a slightly larger change in FA, which can be explained by HD22T5 being the larger aptamer. As expected, the addition of a scrambled non-specific strand NS (Table 1) brought about no change in FA.
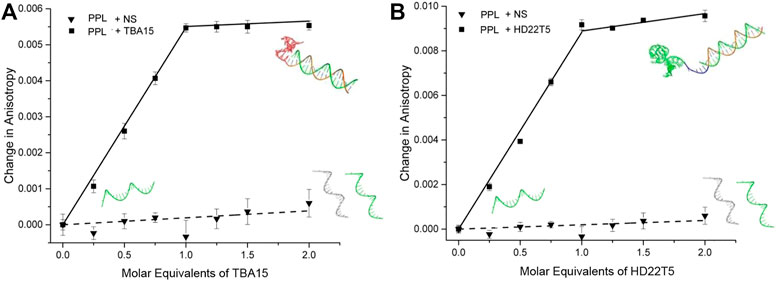
FIGURE 2. Fluorescence anisotropy changes of PPL strand upon the addition of (A) TBA15 (squares) and NS non-specific DNA (triangles), and (B) HD22T5 (squares) and NS non-specific DNA (triangles). [PPL] as disulfide = 40 nM in the following buffer: 20 mM Tris-HCl, 1 mM MgCl2, 120 mM NaCl, 10 mM KCl, 2 mM CaCl2, pH 7.4. Measurements performed in triplicate, with error bars presented as a standard error of the mean (SEM).
2.4 LD sensing studies
With it confirmed that these extended aptamers could bind thrombin as well as form stable duplexes with the individual PPL strand, the PPL-conjugated M13 bacteriophage bionanoparticles M13-PPL were then studied using linear dichroism spectroscopy to determine whether the aptamer-mediated detection of thrombin could be achieved. First of all, the extended aptamers were individually incubated with M13-PPL stocks at physiological pH before being combined at 20.5 nM each to make the M13-PPL-TBA15/M13-PPL-HD22T5 sandwich assay sensor platform. The assay solution was then subjected to linear flow in a Couette flow cell, with the LD signal monitored at 225 nm as described previously. (Pacheco-Gómez et al., 2012; Carr-Smith et al., 2015; Ali et al., 2020).
Upon the addition of thrombin in the picomolar to nanomolar range, a significant decrease in LD signal at 225 nm was observed, resulting from misalignment of the bacteriophage aptamer probes in flow (Figure 3). This result is similar to what was previously observed in the detection of DNA strands, (Ali et al., 2020), with a series of control studies strongly indicating that the sensing system responds well to aggregation of M13 particles caused by formation of multiple sandwich complexes involving both extended aptamers. These control studies included studies in the presence of one extended aptamer alone, in which only a very small change in signal was observed for HD22T5, with a slightly larger change for TBA15 (an average of 30% of the signal for the combined aptamer set up). This difference is most likely caused by some simultaneous binding by TBA15 to exosites I and II of thrombin in a 1:2 sandwich complex, as discussed earlier. These results indicate that any 1:1 complex formation with these aptamers does not appear to cause a significant change in LD signal, as this would not trigger M13 particle aggregation and therefore a change in alignment. Further control experiments with bovine serum albumin (BSA), which is not bound by these aptamers, also showed no significant change in LD signal (red), with an average LD signal decrease of only 8%. Finally, the PPL-functionalised M13 bionanoparticle system was also tested for thrombin in the absence of both aptamers (blue). As expected, no significant change in LD signal was observed (ca. 4% decrease in LD signal), which confirms that no thrombin binding occurs in the absence of the extended aptamers. The data shown in Figure 3B indicates that thrombin can be detected using LD spectroscopy down to the low pM range, which is competitive for this particular target compared to some other more established read-out methods. (Ikebukuro et al., 2004; Xiao et al., 2005; Wang et al., 2008; Cho et al., 2008; Wang and Zhao, 2012; Deng et al., 2014; Kim et al., 2018; Malecka and Ferapontova, 2021; Neupane and Stine, 2021).
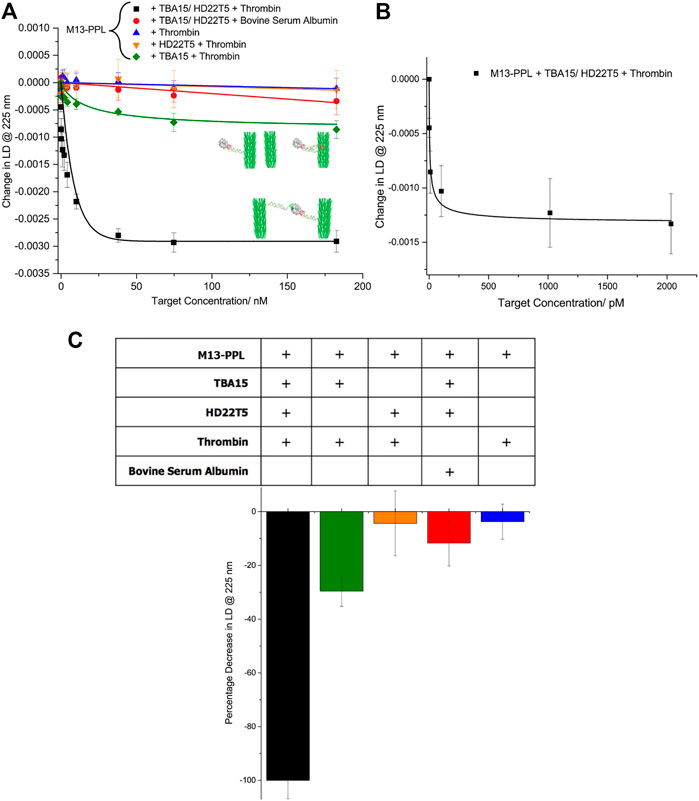
FIGURE 3. (A) Change in LD signal at 225 nm upon addition of targets at increasing concentrations (0–183 nM): M13-PPL-TBA15 (20.5 nM) + M13-PPL-HD22T5 (20.5 nM) + thrombin (black squares), M13-PPL-TBA15 (20.5 nM) + thrombin (green diamonds), M13-PPL-HD22T5 (20.5 nM) + thrombin (orange triangles), M13-PPL-TBA15 (20.5 nM) + M13-PPL-HD22T5 (20.5 nM) + BSE (red circles), M13-PPL + thrombin; (B) Change in LD signal at 225 nm for M13-PPL-TBA15 (20.5 nM) + M13-PPL-HD22T5 (20.5 nM) + thrombin at increasing concentrations (0, 1.0, 10, 100, 1,000, 2000 p.m.) (C) Percentage decrease in LD signal at 225 nm observed at a final target concentration of 183 nM for different conditions used, compared to the full assay system. Experiments performed in the following buffer: 20 mM Tris-HCl, 1 mM MgCl2, 120 mM NaCl, 10 mM KCl, 2 mM CaCl2, pH 7.4. M13-PPL was incubated with aptamer (TBA15 and HD22T5) for 30 min before the addition of targets. Measurements performed in triplicate, with error bars presented as a standard error of the mean (SEM). For more details on the LD technique, see Supplementary Material S1.
3 Conclusion
In conclusion, this study reports the first example of an aptamer-functionalised bacteriophage system, enabling the design of a label-free homogenous assay system for non-nucleic acid targets using an LD read-out. The use of LD spectroscopy for sensing offers the potential to test contaminated samples with ease and at speed as no washing or separation steps are required before quantification and the background signal is low. The main advantage of this new plug-and-play LD sensor platform is its modularity in that different aptamer sequences can be connected to a DNA-linked M13 chassis without performing any new bioconjugation steps to the bacteriophage. If this sandwich assay system were to be taken forward for targets other than thrombin, the same DNA-functionalised chassis could be made as a batch stock of the viral scaffold, with other aptamers functionalised in the same way with sequences complementary to the same PPL strand. This would greatly reduce the time taken to develop an assay from aptamer generation to assay testing.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
HL performed the experimental studies with additional contributions from AA and JC. MH, JT, and TD supervised the work. JT oversaw the preparation and writing of the manuscript.
Funding
This work was supported by the BBSRC, Midlands Integrative Biosciences Training Partnership (MIBTP).
Acknowledgments
The authors wish to thank the Midlands Integrative Biosciences Training Partnership (MIBTP) and the School of Chemistry for PhD studentship support to HL, AA, and JC. The Centre for Chemical and Materials Analysis at the University of Birmingham is acknowledged for technical support. We acknowledge the EPSRC for the award of a Leadership Fellowship to JT (EP/G007578/1).
Conflict of interest
Author MH was employed by the company Linear Diagnostics Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2023.1040873/full#supplementary-material
References
Alafeef, M., and Pan, D. (2022). Diagnostic approaches for COVID-19: lessons learned and the path forward. ACS Nano 16, 11545–11576. doi:10.1021/acsnano.2c01697
Ali, A., Little, H. A., Carter, J. G., Douglas, C., Hicks, M. R., Kenyon, D. M., et al. (2020). Combining bacteriophage engineering and linear dichroism spectroscopy to produce a DNA hybridisation assay. RSC Chem. Biol. 1, 449–454. doi:10.1039/d0cb00135j
Bock, L. C., Griffin, L. C., Latham, J. A., Vermaas, E. H., and Toole, J. J. (1992). Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355, 564–566. doi:10.1038/355564a0
Carr-Smith, J., Pacheco-Gómez, R., Little, H. A., Hicks, M. R., Sandhu, S., Steinke, N., et al. (2015). Polymerase chain reaction on a viral nanoparticle. ACS Synth. Biol. 4, 1316–1325. doi:10.1021/acssynbio.5b00034
Chen, C., Liu, W., Tian, S., and Hong, T. (2019). Novel surface-enhanced Raman spectroscopy techniques for DNA, protein and drug detection. Sensors 19, 1712. doi:10.3390/s19071712
Cho, H., Baker, B. R., Wachsmann-Hogiu, S., Pagba, C. V., Laurence, T. A., Lane, S. M., et al. (2008). Aptamer-based SERRS sensor for thrombin detection. Nano Lett. 8, 4386–4390. doi:10.1021/nl802245w
De Rache, A., Kejnovská, I., Vorlíčková, M., and Buess-Herman, C. (2012). Elongated thrombin binding aptamer: A G-quadruplex cation-sensitive conformational switch. Chem. Eur. J. 18, 4392–4400. doi:10.1002/chem.201103381
Deng, B., Lin, Y., Wang, C., Li, F., Wang, Z., Zhang, H., et al. (2014). Aptamer binding assays for proteins: The thrombin example—a review. Anal. Chim. Acta 837, 1–15. doi:10.1016/j.aca.2014.04.055
Ikebukuro, K., Kiyohara, C., and Sode, K. (2004). Electrochemical detection of protein using a double aptamer sandwich. Anal. Lett. 37, 2901–2909. doi:10.1081/al-200035778
Kim, H., An, Z., and Jang, C.-H. (2018). Label-free optical detection of thrombin using a liquid crystal-based aptasensor. Microchem. J. 141, 71–79. doi:10.1016/j.microc.2018.05.010
Malecka, K., and Ferapontova, E. E. (2021). Femtomolar detection of thrombin in serum and cerebrospinal fluid via direct electrocatalysis of oxygen reduction by the covalent G4-hemin-aptamer complex. ACS Appl. Mat. Interfaces 13, 37979–37988. doi:10.1021/acsami.1c03784
Michel, B. Y., Dziuba, D., Benhida, R., Demchenko, A. P., and Burger, A. (2020). Probing of nucleic acid structures, dynamics, and interactions with environment-sensitive fluorescent labels. Front. Chem. 8, 112. doi:10.3389/fchem.2020.00112
Neupane, D., and Stine, K. J. (2021). Electrochemical sandwich assays for biomarkers incorporating aptamers, antibodies and nanomaterials for detection of specific protein biomarkers. Appl. Sci. 11, 7087. doi:10.3390/app11157087
Pacheco-Gómez, R., Kraemer, J., Stokoe, S., England, H. J., Penn, C. W., Stanley, E., et al. (2012). Detection of pathogenic bacteria using a homogeneous immunoassay based on shear alignment of virus particles and linear dichroism. Anal. Chem. 84 (1), 91–97. doi:10.1021/ac201544h
Sun, Y., Peng, Z., Li, H., Wang, Z., Mu, Y., Zhang, G., et al. (2019). Suspended CNT-Based FET sensor for ultrasensitive and label-free detection of DNA hybridization. Biosens. Bioelectron. 137, 255–262. doi:10.1016/j.bios.2019.04.054
Tasset, D. M., Kubik, M. F., and Steiner, W. (1997). Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 272, 688–698. doi:10.1006/jmbi.1997.1275
Trotter, M., Borst, N., Thewes, R., and von Stetten, F. (2020). Review: Electrochemical DNA sensing – principles, commercial systems, and applications. Biosens. Bioelectron. 154, 112069. doi:10.1016/j.bios.2020.112069
Vafaei, S., Allabush, F., Tabaei, S. R., Male, L., Dafforn, T. R., Tucker, J. H. R., et al. (2021). Förster resonance energy transfer nanoplatform based on recognition-induced fusion/fission of DNA mixed micelles for nucleic acid sensing. ACS Nano 15, 8517–8524. doi:10.1021/acsnano.1c00156
Wang, W., Chen, C., Qian, M. X., and Zhao, X. S. (2008). Aptamer biosensor for protein detection based on guanine-quenching. Sens. Actuators, B Chem. 129, 211–217. doi:10.1016/j.snb.2007.07.125
Wang, X., and Zhao, Q. (2012). A fluorescent sandwich assay for thrombin using aptamer modified magnetic beads and quantum dots. Microchim. Acta 178, 349–355. doi:10.1007/s00604-012-0850-1
Xiao, Y., Lubin, A. A., Heeger, A. J., and Plaxco, K. W. (2005). Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew. Chem. Int. Ed. 44, 5456–5459. doi:10.1002/anie.200500989
Keywords: aptamer, diagnostics, DNA, bionanoparticle, M13, sandwich assay
Citation: Little HA, Ali A, Carter JG, Hicks MR, Dafforn TR and Tucker JHR (2023) A plug-and-play aptamer diagnostic platform based on linear dichroism spectroscopy. Front. Chem. 11:1040873. doi: 10.3389/fchem.2023.1040873
Received: 09 September 2022; Accepted: 11 April 2023;
Published: 09 May 2023.
Edited by:
Carme Fabrega, Spanish National Research Council (CSIC), SpainReviewed by:
Surajit Chatterjee, Case Western Reserve University, United StatesAnnemieke Madder, Ghent University, Belgium
Copyright © 2023 Little, Ali, Carter, Hicks, Dafforn and Tucker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James H. R. Tucker, ai50dWNrZXJAYmhhbS5hYy51aw==; Timothy R. Dafforn, dC5yLmRhZmZvcm5AYmhhbS5hYy51aw==
 Haydn A. Little
Haydn A. Little Aysha Ali
Aysha Ali Jake G. Carter1,2
Jake G. Carter1,2 James H. R. Tucker
James H. R. Tucker