- 1School of Chemistry and Astbury Centre of Structural Biology, University of Leeds, Leeds, United Kingdom
- 2Iceni Glycoscience Ltd., Norwich, United Kingdom
- 3Department of Chemistry and Manchester Institute of Biotechnology, University of Manchester, Manchester, United Kingdom
The chemoenzymatic synthesis of a series of dual N- and C-terminal–functionalized cholera toxin B subunit (CTB) glycoconjugates is described. Mucin 1 peptides bearing different levels of Tn antigen glycosylation [MUC1(Tn)] were prepared via solid-phase peptide synthesis. Using sortase-mediated ligation, the MUC1(Tn) epitopes were conjugated to the C-terminus of CTB in a well-defined manner allowing for high-density display of the MUC1(Tn) epitopes. This work explores the challenges of using sortase-mediated ligation in combination with glycopeptides and the practical considerations to obtain high levels of conjugation. Furthermore, we describe methods to combine two orthogonal labeling methodologies, oxime- and sortase-mediated ligation, to expand the biochemical toolkit and produce dual N- and C-terminal–labeled conjugates.
Introduction
The development of a methodology for the site-specific conjugation of glycopeptides and carbohydrate epitopes to multimeric scaffolds is important because of their potential as vaccines, therapeutics, or in glycan/glycopeptide arrays (Doores et al., 2006; Godula and Bertozzi, 2012; Branson et al., 2014; Ulrich et al., 2014; Pifferi et al., 2020). In many cases it is advantageous to be able to attach more than one distinct group to a scaffold in a controlled manner, which requires the use of orthogonal ligation strategies; a variety of approaches for protein ligations, such as cysteine derivatization (Dadová et al., 2018; Tessier et al., 2019), azide–alkyne cycloaddition (Wang et al., 2003; Agard et al., 2004; Bruins et al., 2018), and oxime ligation (Geoghegan and Stroh, 1992; Chen et al., 2003) have been combined to enable orthogonal labeling (Smith et al., 2009; Yi et al., 2011; Rashidian et al., 2013; Matos et al., 2020). Another popular ligation strategy employs transpeptidases (Harmand et al., 2018; Fottner et al., 2021; Zhang et al., 2021); for example, the sortases that are responsible for reversibly catalyzing the attachment of virulence factors to the cell walls of Gram-positive bacteria (Mazmanian et al., 1999; Popp et al., 2007). Sortase A (SrtA) from Staphylococcus aureus ligates proteins carrying an LPXTGX recognition motif to substrates carrying an N-terminal glycine residue (Ton-That et al., 2000; Spirig et al., 2011). This class of reactions has been extensively optimized both by engineering the substrate and the enzymes involved (Morgan et al., 2022). In particular, directed evolution has yielded sortases with enhanced activity (Chen et al., 2011, 2016; Hirakawa et al., 2012), perhaps most notably Sortase 7M which exhibits enhanced kinetics and Ca2+-independent activity (Wuethrich et al., 2014). Sortase ligation has been used for the cyclisation of glycopeptides (Wu et al., 2011), conjugation of two mucin glycopeptides (Matsushita et al., 2009), synthesis of GPI analogues (Guo et al., 2009), and generation of glycoform-defined VHH–MUC1 vaccine constructs (Fang et al., 2017), but we are not aware of reports of sortase-mediated ligation of glycopeptides to multimeric proteins to generate multivalent neoglycoproteins.
Here, we report synthetic studies for the conjugation of MUC1 glycopeptides to the cholera toxin B subunit (CTB). Mucin 1 (MUC1) is an extensively O-glycosylated protein whose extracellular domain extends up to 200–500 nm beyond the surface of the cell, with a total molecular weight of 250–500 kDa (Brayman et al., 2004; Hattrup and Gendler, 2008; Nath and Mukherjee, 2014). The extracellular domain comprises a highly conserved variable number of tandem repeats (VNTR) of 20 amino acids, which is repeated between 20–125 times depending on the individual (Baldus et al., 2004; Hattrup and Gendler, 2008). This VNTR domain is rich in serine and threonine permitting heavy O-glycosylation and proline, which contributes to the rod-like structure of the extracellular domain (Baldus et al., 2004; Hollingsworth and Swanson, 2004). Dramatic changes to MUC1 glycosylation are associated with various carcinomas (Gendler, 2001; Awaya et al., 2004; Singh et al., 2006; Krishn et al., 2016). Abnormal truncated O-linked glycans are recognized as potential cancer markers and result in more of the peptide backbone being exposed to the environment (Burchell et al., 2001; Munkley, 2016). The Tn (core) antigen is the simplest of the tumor associated carbohydrate antigens comprising of α-O-GalNAc linked to either serine or threonine.
We report methods to attach MUC1 glycopeptides to the cholera toxin B subunit (CTB). The natural role of this pentameric glycan-binding protein is to deliver a toxic ADP-ribosyltransferase enzyme into intestinal cells (Sánchez and Holmgren, 2008). However, the non-toxic CTB subunit is also widely used as a neuronal cell tracer (Vercelli et al., 2000; Saleeba et al., 2019), and it has also been used as a scaffold for the preparation of multivalent neoglycoprotein inhibitors of bacterial toxin adhesion and as a mucosal adjuvant (Holmgren et al., 1993, 2005; Pizza et al., 2001; Branson et al., 2014). It is our interest in these applications of the CTB protein that has led us to develop a range of ligation methods for the site-specific modification of this protein at its N-terminus and also through site-specific incorporation of azidohomoalanine for bioorthogonal ligation reactions (Branson et al., 2014; Machin et al., 2019; Haigh et al., 2020; Balmforth et al., 2021). The work reported here extends our toolbox of methods to C-terminal modification of CTB by exploring the feasibility of using sortase to conjugate naturally occurring glycopeptide epitopes to produce a series of well-defined glycoconjugates. Furthermore, we also demonstrate the feasibility of orthogonal modification combining sortase and oxime ligation to add additional functionality to the N-terminus. The ability to expand the functionality of site-specific glycoconjugates for immobilization, add additional carbohydrate epitopes, or to increase immunogenicity is key to developing relevant and useful therapeutics or increasing the chances of detecting glycan-binding proteins.
Materials and methods
General procedure for solid-phase glycopeptide synthesis
Low loading Rink Amide MBHA resin (loading: 0.33–0.35 mmol/g, 100 µmol) was shaken in DMF (5 ml) for 2 h, filtered, and washed with DMF. Fmoc deprotection: resin-bound peptide intermediates were shaken in 20% piperidine in DMF (3 × 5 ml × 3 min) and washed with DMF, CH2Cl2, and DMF. Couplings: the resin was treated with a 5-fold excess of Fmoc-amino acid (except for the Fmoc-Ser/Thr (GalNAc)-OH which was employed in a 2-fold excess) in 2 ml DMF containing HCTU (5 equiv) and DIPEA (10 equiv). The mixture was shaken and coupling times were 40 min (except for the Fmoc-Ser/Thr (GalNAc)-OH and the subsequent coupling each of which were 12 h in duration). The excess reagents removed by filtration, and the resin was washed with DMF, CH2Cl2 and DMF. O-acetyl deprotection: following coupling of the final amino acid, resin was washed with methanol, shaken in 70% hydrazine hydrate in methanol (3 × 5 ml × 5 min), and washed with methanol. Glycopeptide cleavage: following Fmoc and O-acetyl deprotection, the resin was washed with CH2Cl2 and methanol, before drying under vacuum. The resin was suspended in TFA:H2O:TIS (95:2.5:2.5, 5 ml) and shaken for a maximum of 2 h. The mixture was filtered and washed with TFA. The solution was concentrated under a stream of N2 to <1 ml. Glycopeptide obtained by ether precipitation, isolated by centrifugation and pellet further washed with ether. The white precipitate was dissolved in H2O and lyophilized to give white foam. The yield and characterization data of all synthesized peptides/glycopeptides can be found in Supplementary Material.
Determination of sortase-mediated ligation efficiency
For a total volume of 100 µl, CTB–LPETGA (200 µM) and appropriate peptide/glycopeptide (3 mM, 15 equiv) were mixed, and the reaction mixture was made up to volume with the HEPES buffer {+DMSO [10% v/v (final)], if required} before the addition of Sortase 7M (30 µM, 15 mol%). The reaction mixture was incubated at 25°C, and timepoints for analysis by SDS-PAGE were taken at 2, 5, 10, 20, 30, 60, 90, and 120 min. For time point samples, 5 µl of the reaction was diluted in 15 µl H2O and mixed with 5 × SDS loading buffer (5 µl). Samples were heated to 95°C for 5 min before being frozen and stored at −20°C before analysis by SDS-PAGE. Samples were run on a 15% Tris-glycine SDS-PAGE gel, stained with Coomassie blue (12 h), destained, and imaged using a Bio-Rad Gel Doc XR system. The relative quantity of labeled product and unlabeled starting material was quantified by densitometry (Bio-Rad Image Lab 6.0.1, Bio-Rad Laboratories Ltd.). Data were analyzed in OriginPro 2019b (OriginLab Corporation, Northampton, MA, United States). Original SDS-PAGE gels are included within the Supplementary Material.
General procedure for preparative scale sortase ligation using CBD-Sortase 7M
For a total volume of 200 µl, the protein to be labeled (200 µM) and peptide/glycopeptide (3,000 µM, 15 eq.) were mixed, and the reaction mixture was made up to volume with the appropriate buffer before the addition of His6-CBD-Srt7M (30 µM, 15 mol%). The reaction mixture was incubated for 2 h at 25°C. Once the reaction has reached completion by mass spectrometry, 200 µl of chitin resin [50% slurry in TBS (pH 7.2)] was added, and the mixture incubated on ice for 15 min. The solution was filtered using a centrifugal cellulose acetate filter (4,000 × g, 1 min). The excess substrate was removed by either diafiltration, dialysis, or size exclusion chromatography [Superdex 75 10/300, Tris-buffered saline (pH 7.2)].
General procedure for Thr/Ser oxidation and oxime ligation
Protein to be labeled was buffer exchanged into sodium phosphate buffer (pH 7.4); it is very important that the buffer does not contain potassium. Performing reactions in the presence of K+ ions will result in oxidation failing to reach completion (Brabham et al., 2020). Oxidation: the protein to be labeled (500 µl, 200 µM) in sodium phosphate buffer was mixed with L-methionine (4 µl of 250 mM stock, final concentration 2.0 mM, 10 eq) and NaIO4 (2 µl of 250 mM stock, final concentration 1.0 mM, 5 eq). The reaction was followed by ES-MS, and it typically completed within 10 min. The protein was purified using a G-25 minitrap desalting column equilibrated and eluted with sodium phosphate buffer (pH 6.8). Oxime ligation: to the oxidized protein obtained from the G-25 minitrap desalting column (1 ml, ∼100 µM) was added aniline (18 µl, final concentration 200 mM, 1,000 equiv.) and the oxyamine substrate (8 µl of 250 mM stock in DMSO, final concentration 2.0 mM, 10 equiv.). The mixture was briefly mixed by vortexing and incubated at 37°C for 8–16 h. The oxime-labeled protein was purified using a PD-10 desalting column equilibrated and eluted with sodium phosphate buffer (pH 6.8). The reactions were followed by protein HRMS, which are included within the Supplementary Material.
Results and discussion
Preparation of glycopeptide and protein substrates
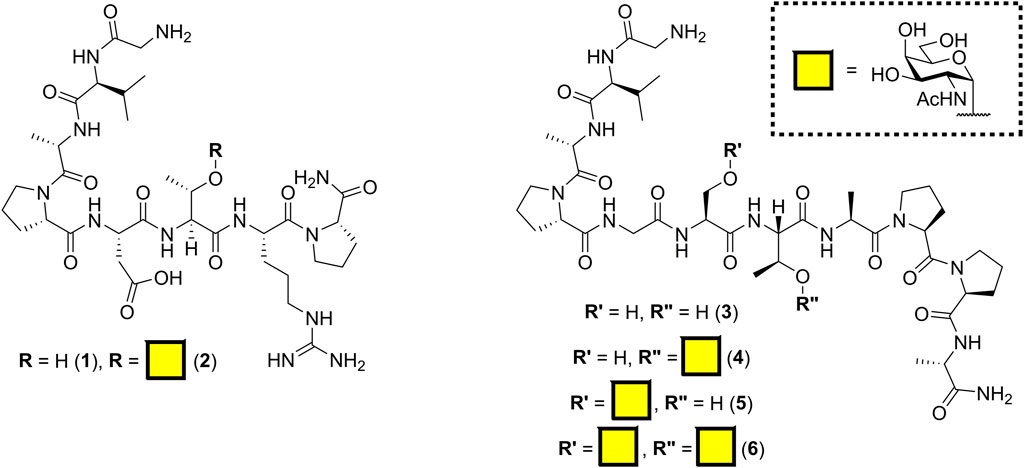
The antibody SM3 (identified from the sera of breast cancer patients) and the mouse class I MHC product H-2Db are known to bind regions of incompletely glycosylated MUC1, either APDTRP or APGSTAPPA, respectively (Apostolopoulos et al., 1998; Dokurno et al., 1998; Burchell et al., 2001; Martínez-Sáez et al., 2015). Previously, Fang et al. (2017) reported the C-terminal sortase conjugation of αMUC1(Tn) glycopeptide [GGGCTSAPDT (GalNAc)RPAP] to camelid-derived heavy chain–only antibody fragments (VHHs) using Sortase 5M. Our initial aim was to keep the linker length to a minimum; therefore, the SM3 and H-2Db sequences were synthesized by solid-phase peptide synthesis with a Gly–Val N-terminal tag for sortase-mediated ligation. Inclusion of valine provided both a minimal length spacer and a unique signal enabling quantification of peptide concentration by NMR spectroscopy. Peracetylated α-O-GalNAc-Ser/Thr amino acids, synthesized from D-galactosamine as outlined in Supplementary Scheme S1 (Xu et al., 2004; De Silva et al., 2009), were incorporated into the peptides to create all three potential GalNAc (Tn) glycosylation patterns (Figure 1). To minimize off-resin manipulation of the glycopeptides, O-acetyl protecting groups were removed on resin using 70% hydrazine hydrate in methanol. This deacetylation reaction reached completion within 15 min rather than 2 h as previously reported (Mitchell et al., 2001). Glycopeptides were cleaved from the resin using standard TFA cleavage conditions; no acid-catalyzed cleavage of GalNAc moiety was observed, provided cleavage was not allowed to proceed for more than 2 h.
Cholera toxin B subunit (CTB) was chosen as the model pentameric scaffold protein for glycopeptide conjugation. A synthetic gene for cholera toxin B subunit harboring a C-terminal NGGNLPETGA extension including a sortase recognition motif (CTB–LPETGA) was constructed by assembly PCR. The protein was expressed in E. coli and isolated from the growth media by ammonium sulfate precipitation, followed by nickel affinity and size exclusion chromatography steps. Characterization of CTB–LPETGA by SDS-PAGE and HRMS revealed a truncation present in approximately 10% of the CTB protomer units corresponding to 11 amino acids from the C-terminus being absent including the LPETGA sortase motif (Supplementary Figure S1). The cause of the truncation could potentially have been premature termination of translation or subsequent proteolysis; however, addition of protease inhibitors to the growth media (both prior to induction and ammonium sulfate precipitation) or to the buffer used to resuspend the ammonium sulfate precipitate did not have any effect. Having 10% of peptide truncation means that on average there are 4.5 sortase sites available for ligation per pentamer, rather than the expected 5, but this is unlikely to have any negative impact on any downstream application of the ligation methods reported here.
Effect of DMSO and Pro4 on ligation efficiency
Peptide 1 and glycopeptide 2, based on the SM3 sequence, were ligated to CTB–LPETGA using Sortase 7M (Wuethrich et al., 2014), and the reactions were followed by SDS-PAGE and quantified by densitometry. After 2 h conjugation with the SM3 peptide 1 had reached its maximum conversion of 78% (Figure 2A). Conjugation with the SM3 glycopeptide 2 reached a maximum conversion of 52%, approximately 25 percentage points lower than observed for the non-glycosylated peptide under the same conditions (Figure 2B). Considering that the GalNAc residue is attached to Thr-6 of the peptide substrate, it seemed unlikely that glycosylation was having a detrimental steric effect on the ligation reaction. We therefore postulated that the sugar residue might cause some conformational change in the peptide that reduced ligation efficiency. Therefore, each reaction was repeated in the presence of 10% DMSO to act as chaotropic agent and disrupt any putative hydrogen bonding within the peptide/glycopeptide (Jackson and Mantsch, 1991). No change in conjugation efficiency was observed for peptide 1 (Figure 2A); however, a significant increase in the level of conjugation was observed for glycopeptide 2. After 2 h, 73% of the available sortase sites were converted to the product, which was an increase approximately 20% compared to the reaction without DMSO present (Figure 2B). The rate and total level of conjugation for peptide 1 and glycopeptide 2 were comparable in the presence of 10% DMSO. No significant reduction in rate of labeling was observed, indicating that this concentration of DMSO was sufficient to interrupt any unfavorable intramolecular interactions within the glycopeptide, but insufficient to impair Sortase 7M activity, and unlikely to perturb the highly stable CTB structure (Goins and Friere, 1988).
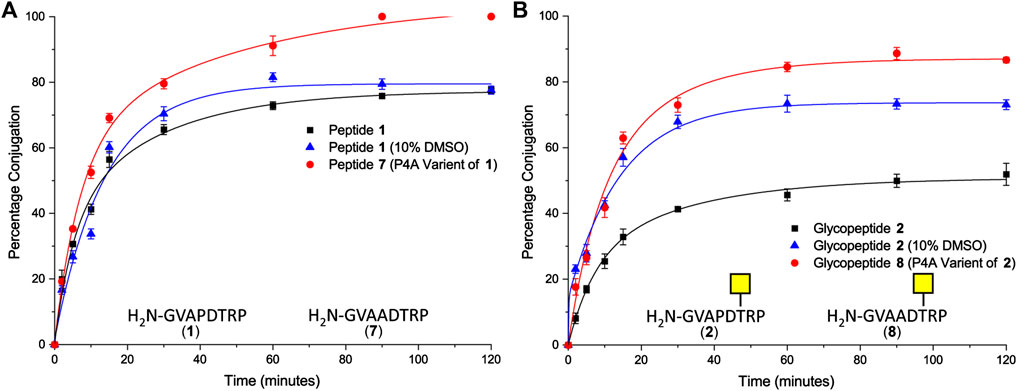
FIGURE 2. (A) Time-course of Sortase 7M–mediated ligation of CTB–LPETGA and peptide 1 based on the SM3 recognition sequence or peptide 7 (P4A variant of peptide 1). (B) Time-course of Sortase 7M–mediated ligation of CTB–LPETGA and glycopeptide 2 based on the SM3 recognition sequence or glycopeptide 8 (P4A variant of glycopeptide 2). Percentage conjugation an average of three densitometry measurements and error bars indicate 2 × standard deviation.
Next, the effect of the peptide sequence on conjugation efficiency was evaluated. The 20 amino acid MUC1 tandem repeat domain is known to be highly kinked, which in part is induced by its five proline residues (Brayman et al., 2004). The second set of experiments focused on the N-terminal motif of GVAPXS/T [where X is D (SM3) or G (H-2Db)] to evaluate if the proline residue closest to the nucleophilic glycine residue could be the cause of the poor conjugation efficiency for the glycopeptide. Variants of the SM3 peptide and glycopeptide substrates were synthesized in which Pro4 was changed to Ala and ligation was repeated in the absence of DMSO. After 1.5 h, quantitative ligation was achieved for the P4A peptide 7 (Figure 2A) and conjugation of P4A glycopeptide 8 reached a maximum level of 87% (Figure 2B). Ligation of both P4A substrates were a significant improvement over the natural substrates and confirmed the Pro4 had an influence on conjugation efficiency of both the peptide and glycopeptide substrates. It is possible that ligation of glycopeptide 8 could have improved further if the reaction were spiked with 10% DMSO; however, as this was not the native SM3 epitope, further experiments to confirm this hypothesis were not performed.
Effect of N-terminal flexibility on glycopeptide ligation efficiency
Although removal of the proline residue resulted in near quantitative ligation, it is unclear if the proline residue may induce important secondary structure within the glycopeptide. It was considered important not to perturb the conformation of the (glyco) peptides if the methodology were to be used to develop glycoconjugates, which could be used as therapeutics or detection of novel carbohydrate binding proteins. Therefore, the effect of introducing various length linkers between the sortase handle and the MUC1 sequence was investigated to determine if conjugation efficiency could be improved whilst maintaining amino acids that could be important for secondary structure. One, two, or three copies of 8-amino-3,6-dioxaoctanoic acid (AEEAc-OH), each equivalent to insertion of PEG3, were introduced between the GV sortase handle and the SM3 sequence. Furthermore, the effect of increasing N-terminal flexibility was investigated by changing the Gly–Val sortase handle to a triglycine motif (12) in analogy to the longer [GGGCTSAPDT (GalNAc)RPAP] sequence previously investigated (Fang et al., 2017). All four extended peptide sequences (9, 10, 11, 12) gave statistically significant improvements in conjugation (12–21 percentage points) relative to glycopeptide 2 under the same conditions (Figure 3). In the case of compound 12, the improvement in conjugation is likely due to the increased flexibility of the N-terminus rather than the additional length provided by the addition of a single amino acid (GGG vs. GV) (Figure 3). Only peptide 10 with the PEG6 equivalent linker showed a significant improvement in the level of conjugation relative to the glycopeptide bearing the N-terminal triglycine (12).
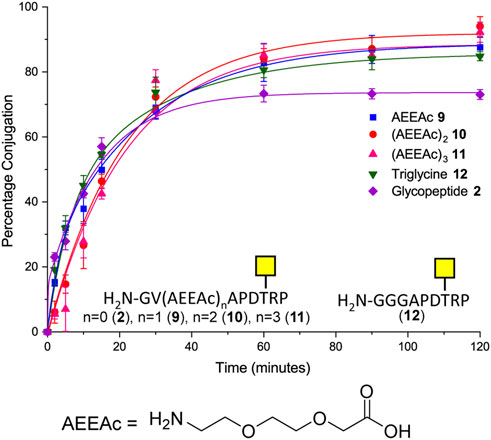
FIGURE 3. Time-course of Sortase 7M–mediated ligation of CTB–LPETGA and glycopeptide 2 and 9–12 based on the SM3 recognition sequence. The reaction performed in the presence of 10% DMSO. AEEAc, 8-amino-3,6-dioxaoctanoic acid. Percentage conjugation an average of three densitometry measurements and error bars indicate 2 × standard deviation.
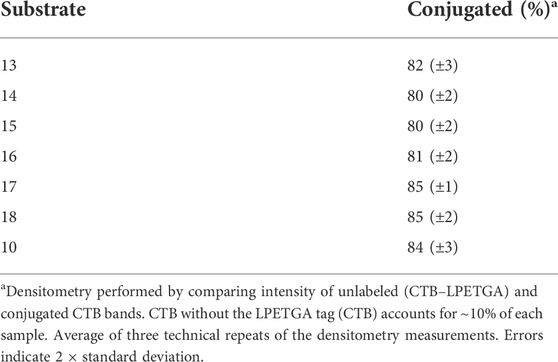
As the double AEEAc linker showed the highest levels of conjugation in the preliminary experiments, the peptides and glycopeptides based on the SM3 (APDTRP) and H-2Db (APGSTAPPA) motifs were resynthesized with the double AEEAc linker (n = 2). After 2 h the level of conjugation between CTB–LPETGA and the new peptides and glycopeptides containing a PEG6 equivalent linker (13–18) in the presence of 10% DMSO was between 80%–85% (by SDS-PAGE and densitometry) (Figure 4; Table 1). Compared to the original glycosylated SM3 substrate 2 in the absence of DMSO, the optimized linker length and conditions for glycopeptide 10 resulted in an increased conversion of 32 ± 4 percentage points.
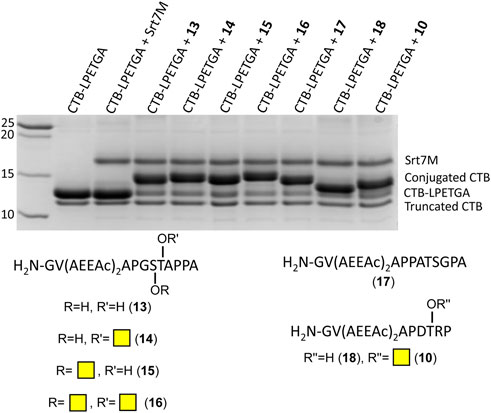
FIGURE 4. SDS-PAGE gel showing sortase ligation after 2 h using peptides (13, 17, and 18) and glycopeptides (10, 14–16) containing a double length AEEAc linker, equivalent to PEG6. Levels of conjugation listed in Table 1.
Development and testing of chitin-binding Sortase 7M variant
The ability to remove the sortase enzyme rapidly from the preparative scale ligation reactions efficiently would be vital as prolonged exposure to sortase will eventually lead to hydrolysis of the ligated product (Morgan et al., 2022). The standard methodology for removing sortase from ligation reactions by exploiting the affinity of its His-tag for Ni-NTA resin was not possible because of the natural affinity of CTB for Ni-NTA resin (Dertzbaugh and Cox, 1998). Attempts to exploit the difference in affinity of these two proteins for the Ni-NTA resin were unsuccessful, and further attempts to isolate the ligation product by size exclusion chromatography (SEC) led to increased levels of hydrolysis compared to the crude sample prior to being loaded on the SEC column. Although immobilization of sortase was a potential solution (Steinhagen et al., 2013; Witte et al., 2015), once immobilized the activity of the enzyme is reduced. As a result, we decided to investigate an alternative fusion construct of Sortase 7M which could be used in solution for the reaction and then selectively removed at the end of the reaction. An N-terminal chitin-binding domain (CBD) was selected because of its effectively irreversible binding to chitin resin and its small size which should not interfere with activity of the enzyme.
To ensure the CBD domain did not interfere with the transpeptidase activity, the conjugation of CTB to the SM3 glycopeptide with the double length AEEAc linker (10) was repeated using both the Sortase 7M and CBD-Sortase 7M variant. Although the chitin-binding variant of Sortase 7M was found to ligate slightly faster over the first 30 min compared to the non–chitin-binding variant, at timepoints beyond 1 h the difference in conjugation between the two enzymes was negligible with conjugation reaching 90% for both enzymes after 2 h (Figure 5). The efficiency of removing CBD-Sortase 7M from ligation reactions was evaluated by repeating the experiments with SM3 peptide 18 and glycopeptide 10. After 2 h reaction in the presence of CBD-Sortase 7M, chitin resin was added and the mixture was incubated on ice to minimize any further reaction past the deemed endpoint prior to removal of the resin by centrifugal filtration. An analysis of samples before and after use of the chitin resin showed complete removal of the CBD-Sortase 7M, and no significant difference in the level of conjugation before and after removal of chitin resin was observed (Figure 6).
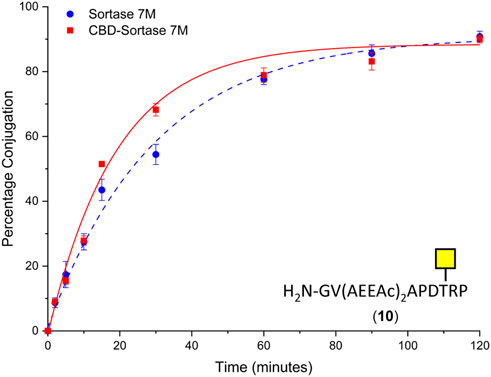
FIGURE 5. Time-course for ligation of CTB–LPETGA and glycopeptide 10 using Sortase 7M (■) and chitin-binding domain (CBD) Sortase 7M (•). Percentage conjugation an average of three densitometry measurements and error bars indicate 2 × standard deviation.
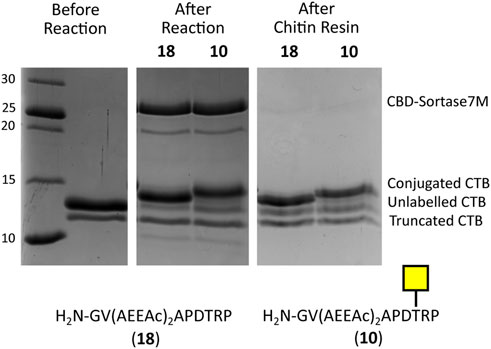
FIGURE 6. SDS-PAGE gel demonstrating removal of CBD-Sortase 7M from reaction mixture using 1.5 eq of chitin resin. No change in the level of conjugation is observed before and after removal of CBD-Sortase 7M.
Combining orthogonal N- and C-terminal ligation methodologies
Orthogonal modification of the C- and N-terminus of a protein scaffold allows the opportunity to introduce more than one new functionality at specific sites on a protein scaffold. Oxime ligation at the N-terminus of CTB was first described for the construction of well-defined CTB-viral peptide immunogens (Chen et al., 2003). We have previously applied oxime ligation of CTB for the preparation of multivalent neoglycoprotein bacterial toxin inhibitors and for N-terminal biotinylation of CTB for phage display (Branson et al., 2014; Balmforth et al., 2021). To explore if oxime and C-terminal sortase-mediated ligation could be used orthogonally, the N-terminus of CTB–LPETGA was first biotinylated before the peptides and glycopeptides (10, 13–18) were conjugation to the C-terminus using CBD-Sortase 7M. We chose to perform the ligation reactions in this order as periodate oxidation of a glycoprotein bearing terminal GalNAc residues risks oxidation of the sugar residues. An alternative strategy of N-terminal periodate oxidation followed by sortase ligation to attach the glycopeptides and then oxime ligation to introduce the biotin groups would not be practical as delays to the oxime step result in cyclisation of the N-terminal aldehyde group onto the peptide backbone (Rose et al., 1999).
The N-terminal threonine residue was oxidized using sodium periodate (5 eq) in sodium phosphate buffer. This reaction typically reaches completion within 5 min in sodium phosphate (Figures 7A,B), whereas the reaction does not reach completion in phosphate-buffered saline containing potassium ions, which are known to hinder periodate reactivity (Brabham et al., 2020). Previously oxime ligation of alkoxyamine–PEG4–biotin was used for the N-terminal biotinylation of wt CTB; this was sufficient to allow effective binding to streptavidin for phage display screening (Balmforth et al., 2021). Using the same alkoxyamine–PEG4–biotin (10 eq) in the presence of aniline (1% v/v) at 37°C, oxime biotinylation of the oxidized CTB reached completion within 16 h (Figure 7C). When the oxidized CTB was used below 200 µM, a mixture of labeled product (12,924.5 Da) and N-terminal cyclisation (12,508.3 Da) was observed (Rose et al., 1999). For optimal oxime ligation reactions, an oxidized CTB concentration above 400 µM was required to minimize N-terminal cyclisation and obtain quantitative N-terminal biotinylation of both CTB–LPETGA and the truncated CTB.
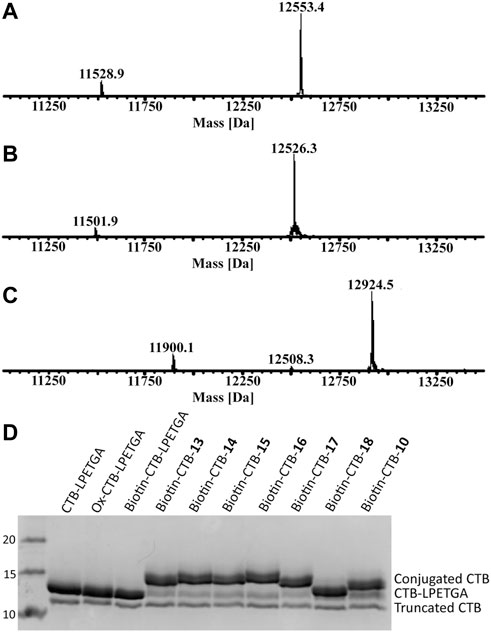
FIGURE 7. (A) ESI-MS of CTB–LPETGA (12,553.4 Da) and truncated CTB (11,528.9 Da) before treatment with NaIO4. (B) ESI-MS of CTB–LPETGA following oxidation with periodate. (C) ESI-MS following treatment of oxidised-CTB with alkoxyamine-PEG4-biotin and aniline, showing quantitative N-terminal biotinylation of both substrates. (D) SDS-PAGE gel of CTB–LPETGA, oxidized CTB–LPETGA, biotinylated CTB–LPETGA, and C-terminally conjugated and biotinylated CTB constructs. Levels of conjugation listed in Table 2.
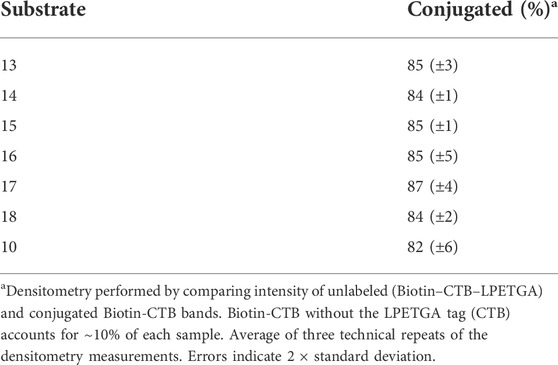
Having achieved quantitative N-terminal biotinylation, preparative scale sortase ligation of the biotin–CTB–LPETGA with the (AEEAc)2 peptides and glycopeptides (10, 13–18) was performed using CBD-Sortase 7M in the presence of 10% DMSO. After 2 h, each reaction was placed on ice and CBD-Sortase 7M removed using chitin resin. The level of conjugation was evaluated by SDS-PAGE and densitometry following purification by size exclusion chromatography to remove the excess peptide/glycopeptide (Figure 7D; Table 2). No significant difference in sortase ligation efficiency was observed between CTB–LPETGA with and without the N-terminal biotin tag.
Conclusion
In conclusion, we have described the optimization of C-terminal sortase ligation for a series of MUC1(Tn) glycopeptide epitopes and the orthogonal labeling of both the N- and C-terminus of the cholera toxin B subunit using two site-specific methods. The N-terminus of CTB–LPETGA was quantitatively labeled with an alkoxyamine-functionalized biotin via oxime ligation. Sortase-mediated ligation could then be used to ligate a series of MUC1(Tn) epitopes to the C-terminus with a conjugation efficiency of 82%–87%. Although demonstrated here with biotinylation, it should be straight forward to extend N-terminal functionalization with other peptides, glycopeptides, or immunogenic epitopes given the ease of incorporation of alkoxyamine functionalization to small molecules or to peptides during solid-phase peptide synthesis. Furthermore, we have demonstrated the addition of an N-terminal chitin-binding domain to Sortase 7M has no negative effect enzyme activity and can be used to remove the enzyme rapidly and efficiently once the deemed endpoint of the reaction has been reached.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; the raw data associated with this paper including SDS-PAGE images, NMR and mass spectra are openly available from the University of Leeds data repository. https://doi.org/10.5518/1194.
Author contributions
JPD, RAF, SD, MEW, and WBT developed the concepts and designed the experiments. JPD and DCM conducted the experiments and analyzed the data under the supervision of MEW, WBT, and SD. JPD wrote the initial draft of the manuscript. JPD, MEW, and WBT reviewed and edited the manuscript. WBT, MEW, SD, and RAF were responsible for funding acquisition. All authors listed have made a substantial, crucial, and direct contribution to the work, and approved it for publication.
Acknowledgments
The authors thank the Biotechnology and Biological Research Council (BB/M011151/1) and Iceni Glycoscience for PhD studentship for JD. The authors would like to acknowledge Jeanine Williams, University of Leeds, for technical support with analytical HPLC.
Conflict of interest
Author SD was employed by the company Iceni Glycoscience Ltd. RF is a director and shareholder of Iceni Glycoscience Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.958272/full#supplementary-material
References
Agard, N. J., Prescher, J. A., and Bertozzi, C. R. (2004). A strain-promoted [3 + 2] Azide−Alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 126, 15046–15047. doi:10.1021/ja044996f
Apostolopoulos, V., Chelvanayagam, G., Xing, P.-X., and McKenzie, I. F. C. (1998). Anti-MUC1 antibodies react directly with MUC1 peptides presented by class I H2 and HLA molecules. J. Immunol. 161, 767–775.
Awaya, H., Takeshima, Y., Yamasaki, M., and Inai, K. (2004). Expression of MUC1, MUC2, MUC5AC, and MUC6 in atypical adenomatous hyperplasia, bronchioloalveolar carcinoma, adenocarcinoma with mixed subtypes, and mucinous bronchioloalveolar carcinoma of the lung. Am. J. Clin. Pathol. 121, 644–653. doi:10.1309/U4WG-E9EB-FJN6-CM8R
Baldus, S. E., Engelmann, K., and Hanisch, F.-G. (2004). MUC1 and the MUCs: a family of human mucins with impact in cancer biology. Crit. Rev. Clin. Lab. Sci. 41, 189–231. doi:10.1080/10408360490452040
Balmforth, M. R., Haigh, J., Kumar, V., Dai, W., Tiede, C., Tomlinson, D. C., et al. (2021). Piggybacking on the cholera toxin: Identification of a CTB-binding protein as an approach for targeted delivery of proteins to motor neurons. Bioconjug. Chem. 32, 2205–2212. doi:10.1021/ACS.BIOCONJCHEM.1C00373
Brabham, R. L., Keenan, T., Husken, A., Bilsborrow, J., McBerney, R., Kumar, V., et al. (2020). Rapid sodium periodate cleavage of an unnatural amino acid enables unmasking of a highly reactive α-oxo aldehyde for protein bioconjugation. Org. Biomol. Chem. 18, 4000–4003. doi:10.1039/D0OB00972E
Branson, T. R., McAllister, T. E., Garcia-Hartjes, J., Fascione, M. A., Ross, J. F., Warriner, S. L., et al. (2014). A protein-based pentavalent inhibitor of the cholera toxin B-subunit. Angew. Chem. Int. Ed. 53, 8323–8327. doi:10.1002/anie.201404397
Brayman, M., Thathiah, A., and Carson, D. D. (2004). MUC1: a multifunctional cell surface component of reproductive tissue epithelia. Reprod. Biol. Endocrinol. 2, 4. doi:10.1186/1477-7827-2-4
Bruins, J. J., Blanco-Ania, D., van der Doef, V., van Delft, F. L., and Albada, B. (2018). Orthogonal, dual protein labelling by tandem cycloaddition of strained alkenes and alkynes to ortho -quinones and azides. Chem. Commun. 54, 7338–7341. doi:10.1039/C8CC02638F
Burchell, J. M., Mungul, A., and Taylor-Papadimitriou, J. (2001). O-Linked glycosylation in the mammary gland: changes that occur during malignancy. J. Mammary Gland. Biol. Neoplasia 6, 355–364. doi:10.1023/a:1011331809881
Chen, I., Dorr, B. M., and Liu, D. R. (2011). A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl. Acad. Sci. U. S. A. 108, 11399–11404. doi:10.1073/pnas.1101046108
Chen, J., Zeng, W., Offord, R., and Rose, K. (2003). A novel method for the rational construction of well-defined immunogens: the use of oximation to conjugate cholera toxin B subunit to a Peptide−Polyoxime complex. Bioconjug. Chem. 14, 614–618. doi:10.1021/bc025651u
Chen, L., Cohen, J., Song, X., Zhao, A., Ye, Z., Feulner, C. J., et al. (2016). Improved variants of SrtA for site-specific conjugation on antibodies and proteins with high efficiency. Sci. Rep. 6, 31899. doi:10.1038/srep31899
Dadová, J., Galan, S. R., and Davis, B. G. (2018). Synthesis of modified proteins via functionalization of dehydroalanine. Curr. Opin. Chem. Biol. 46, 71–81. doi:10.1016/j.cbpa.2018.05.022
De Silva, R. A., Wang, Q., Chidley, T., Appulage, D. K., and Andreana, P. R. (2009). Immunological response from an entirely carbohydrate antigen: design of synthetic vaccines based on Tn-PS A1 conjugates. J. Am. Chem. Soc. 131, 9622–9623. doi:10.1021/ja902607a
Dertzbaugh, M. T., and Cox, L. M. (1998). The affinity of cholera toxin for Ni2+ ion. Protein Eng. Des. Sel. 11, 577–581. doi:10.1093/protein/11.7.577
Dokurno, P., Bates, P. A., Band, H. A., Stewart, L. M. D., Lally, J. M., Burchell, J. M., et al. (1998). Crystal structure at 1.95 å resolution of the breast tumour-specific antibody SM3 complexed with its peptide epitope reveals novel hypervariable loop recognition. J. Mol. Biol. 284, 713–728. doi:10.1006/jmbi.1998.2209
Doores, K. J., Gamblin, David. P., and Davis, B. G. (2006). Exploring and exploiting the therapeutic potential of glycoconjugates. Chem. Eur. J. 12, 656–665. doi:10.1002/chem.200500557
Fang, T., Van Elssen, C. H. M. J., Duarte, J. N., Guzman, J. S., Chahal, J. S., Ling, J., et al. (2017). Targeted antigen delivery by an anti-class II MHC VHH elicits focused αMUC1(Tn) immunity. Chem. Sci. 8, 5591–5597. doi:10.1039/C7SC00446J
Fottner, M., Weyh, M., Gaussmann, S., Schwarz, D., Sattler, M., and Lang, K. (2021). A modular toolbox to generate complex polymeric ubiquitin architectures using orthogonal sortase enzymes. Nat. Commun. 12, 6515. doi:10.1038/s41467-021-26812-9
Gendler, S. J. (2001). MUC1, the renaissance molecule. J. Mammary Gland. Biol. Neoplasia 6, 339–353. doi:10.1023/A:1011379725811
Geoghegan, K. F., and Stroh, J. G. (1992). Site-directed conjugation of nonpeptide groups to peptides and proteins via periodate oxidation of a 2-amino alcohol. application to modification at N-terminal serine. Bioconjug. Chem. 3, 138–146. doi:10.1021/bc00014a008
Godula, K., and Bertozzi, C. R. (2012). Density variant glycan microarray for evaluating cross-linking of mucin-like glycoconjugates by lectins. J. Am. Chem. Soc. 134, 15732–15742. doi:10.1021/ja302193u
Goins, B., and Friere, E. (1988). Thermal stability and intersubunit interactions of cholera toxin in solution and in association with its cell-surface receptor ganglioside GM1. Biochemistry 27, 2046–2052. doi:10.1021/bi00406a035
Guo, X., Wang, Q., Swarts, B. M., and Guo, Z. (2009). Sortase-catalyzed peptide−Glycosylphosphatidylinositol analogue ligation. J. Am. Chem. Soc. 131, 9878–9879. doi:10.1021/ja903231v
Haigh, J. L., Williamson, D. J., Poole, E., Guo, Y., Zhou, D., Webb, M. E., et al. (2020). A versatile cholera toxin conjugate for neuronal targeting and tracing. Chem. Commun. 56, 6098–6101. doi:10.1039/D0CC01085E
Harmand, T. J., Bousbaine, D., Chan, A., Zhang, X., Liu, D. R., Tam, J. P., et al. (2018). One-pot dual labeling of IgG 1 and preparation of C-to-C fusion proteins through a combination of sortase A and butelase 1. Bioconjug. Chem. 29, 3245–3249. doi:10.1021/acs.bioconjchem.8b00563
Hattrup, C. L., and Gendler, S. J. (2008). Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol. 70, 431–457. doi:10.1146/annurev.physiol.70.113006.100659
Hirakawa, H., Ishikawa, S., and Nagamune, T. (2012). Design of Ca 2+ -independent Staphylococcus aureus sortase a mutants. Biotechnol. Bioeng. 109, 2955–2961. doi:10.1002/bit.24585
Hollingsworth, M. A., and Swanson, B. J. (2004). Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer 4, 45–60. doi:10.1038/nrc1251
Holmgren, J., Adamsson, J., Anjuère, F., Clemens, J., Czerkinsky, C., Eriksson, K., et al. (2005). Mucosal adjuvants and anti-infection and anti-immunopathology vaccines based on cholera toxin, cholera toxin B subunit and CpG DNA. Immunol. Lett. 97, 181–188. doi:10.1016/j.imlet.2004.11.009
Holmgren, J., Lycke, N., and Czerkinsky, C. (1993). Cholera toxin and cholera B subunit as oral-mucosal adjuvant and antigen vector systems. Vaccine 11, 1179–1184. doi:10.1016/0264-410x(93)90039-z
Jackson, M., and Mantsch, H. H. (1991). Beware of proteins in DMSO. Biochimica Biophysica Acta - Protein Struct. Mol. Enzym. 1078, 231–235. doi:10.1016/0167-4838(91)90563-F
Krishn, S. R., Kaur, S., Smith, L. M., Johansson, S. L., Jain, M., Patel, A., et al. (2016). Mucins and associated glycan signatures in colon adenoma–carcinoma sequence: prospective pathological implication(s) for early diagnosis of colon cancer. Cancer Lett. 374, 304–314. doi:10.1016/j.canlet.2016.02.016
Machin, D. C., Williamson, D. J., Fisher, P., Miller, V. J., Wildsmith, G. C., Ross, J. F., et al. (2019). Sortase-modified cholera toxoids show specific golgi localization. doi:10.26434/CHEMRXIV.9125201.V1
Martínez‐Sáez, N., Castro-López, J., Valero-González, J., Madariaga, D., Compañón, I., Somovilla, V. J., et al. (2015). Deciphering the non-equivalence of serine and threonine O -glycosylation points: implications for molecular recognition of the Tn antigen by an anti‐MUC1 antibody. Angew. Chem. Int. Ed. 54, 9830–9834. doi:10.1002/anie.201502813
Matos, M. J., Brown, L., Bernardim, B., Guerreiro, A., Jiménez-Osés, G., and Bernardes, G. J. L. (2020). Sequential dual site-selective protein labelling enabled by lysine modification. Bioorg. Med. Chem. 28, 115783. doi:10.1016/j.bmc.2020.115783
Matsushita, T., Sadamoto, R., Ohyabu, N., Nakata, H., Fumoto, M., Fujitani, N., et al. (2009). Functional neoglycopeptides: synthesis and characterization of a new class of MUC1 glycoprotein models having core 2-based O -glycan and complex-type N -glycan chains. Biochemistry 48, 11117–11133. doi:10.1021/bi901557a
Mazmanian, S. K., Liu, G., Ton-That, H., and Schneewind, O. (1999). Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763. doi:10.1126/science.285.5428.760
Mitchell, S. A., Pratt, M. R., Hruby, V. J., and Polt, R. (2001). Solid-phase synthesis of O-linked glycopeptide analogues of enkephalin. J. Org. Chem. 66, 2327–2342. doi:10.1021/jo005712m
Morgan, H. E., Turnbull, W. B., and Webb, M. E. (2022). Challenges in the use of sortase and other peptide ligases for site-specific protein modification. Chem. Soc. Rev. 51, 4121–4145. doi:10.1039/D0CS01148G
Munkley, J. (2016). The role of sialyl-Tn in cancer. Int. J. Mol. Sci. 17, 275–283. doi:10.3390/ijms17030275
Nath, S., and Mukherjee, P. (2014). MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 20, 332–342. doi:10.1016/j.molmed.2014.02.007
Pifferi, C., Ruiz-de-Angulo, A., Goyard, D., Tiertant, C., Sacristán, N., Barriales, D., et al. (2020). Chemical synthesis and immunological evaluation of new generation multivalent anticancer vaccines based on a Tn antigen analogue. Chem. Sci. 11, 4488–4498. doi:10.1039/D0SC00544D
Pizza, M., Giuliani, M. M., Fontana, M. R., Monaci, E., Douce, G., Dougan, G., et al. (2001). Mucosal vaccines: non toxic derivatives of LT and CT as mucosal adjuvants. Vaccine 19, 2534–2541. doi:10.1016/s0264-410x(00)00553-3
Popp, M. W., Antos, J. M., Grotenbreg, G. M., Spooner, E., and Ploegh, H. L. (2007). Sortagging: a versatile method for protein labeling. Nat. Chem. Biol. 3, 707–708. doi:10.1038/nchembio.2007.31
Rashidian, M., Kumarapperuma, S. C., Gabrielse, K., Fegan, A., Wagner, C. R., and Distefano, M. D. (2013). Simultaneous dual protein labeling using a triorthogonal reagent. J. Am. Chem. Soc. 135, 16388–16396. doi:10.1021/ja403813b
Rose, K., Chen, J., Dragovic, M., Zeng, W., Jeannerat, D., Kamalaprija, P., et al. (1999). New cyclization reaction at the amino terminus of peptides and proteins. Bioconjug. Chem. 10, 1038–1043. doi:10.1021/bc9900587
Saleeba, C., Dempsey, B., Le, S., Goodchild, A., and McMullan, S. (2019). A student’s guide to neural circuit tracing. Front. Neurosci. 13, 897. doi:10.3389/fnins.2019.00897
Sánchez, J., and Holmgren, J. (2008). Cholera toxin structure, gene regulation and pathophysiological and immunological aspects. Cell. Mol. Life Sci. 65, 1347–1360. doi:10.1007/s00018-008-7496-5
Singh, A. P., Chauhan, S. C., Bafna, S., Johansson, S. L., Smith, L. M., Moniaux, N., et al. (2006). Aberrant expression of transmembrane mucins, MUC1 and MUC4, in human prostate carcinomas. Prostate 66, 421–429. doi:10.1002/pros.20372
Smith, J. J., Conrad, D. W., Cuneo, M. J., and Hellinga, H. W. (2009). Orthogonal site-specific protein modification by engineering reversible thiol protection mechanisms. Protein Sci. 14, 64–73. doi:10.1110/ps.04965405
Spirig, T., Weiner, E. M., and Clubb, R. T. (2011). Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 82, 1044–1059. doi:10.1111/j.1365-2958.2011.07887.x
Steinhagen, M., Zunker, K., Nordsieck, K., and Beck-Sickinger, A. G. (2013). Large scale modification of biomolecules using immobilized sortase a from Staphylococcus aureus. Bioorg. Med. Chem. 21, 3504–3510. doi:10.1016/j.bmc.2013.03.039
Tessier, R., Ceballos, J., Guidotti, N., Simonet-Davin, R., Fierz, B., and Waser, J. (2019). Doubly orthogonal” labeling of peptides and proteins. Chem 5, 2243–2263. doi:10.1016/j.chempr.2019.06.022
Ton-That, H., Mazmanian, S. K., Faull, K. F., and Schneewind, O. (2000). Anchoring of surface proteins to the cell wall of Staphylococcus aureus. J. Biol. Chem. 275, 9876–9881. doi:10.1074/jbc.275.13.9876
Ulrich, S., Boturyn, D., Marra, A., Renaudet, O., and Dumy, P. (2014). Oxime ligation: a chemoselective click-type reaction for accessing multifunctional biomolecular constructs. Chem. Eur. J. 20, 34–41. doi:10.1002/chem.201302426
Vercelli, A., Repici, M., Garbossa, D., and Grimaldi, A. (2000). Recent techniques for tracing pathways in the central nervous system of developing and adult mammals. Brain Res. Bull. 51, 11–28. doi:10.1016/S0361-9230(99)00229-4
Wang, Q., Chan, T. R., Hilgraf, R., Fokin, V. v., Sharpless, K. B., and Finn, M. G. (2003). Bioconjugation by copper(I)-Catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 125, 3192–3193. doi:10.1021/ja021381e
Witte, M. D., Wu, T., Guimaraes, C. P., Theile, C. S., Blom, A. E. M., Ingram, J. R., et al. (2015). Site-specific protein modification using immobilized sortase in batch and continuous-flow systems. Nat. Protoc. 10, 508–516. doi:10.1038/nprot.2015.026
Wu, Z., Guo, X., and Guo, Z. (2011). Sortase a-catalyzed peptide cyclization for the synthesis of macrocyclic peptides and glycopeptides. Chem. Commun. 47, 9218. doi:10.1039/c1cc13322e
Wuethrich, I., Peeters, J. G. C., Blom, A. E. M., Theile, C. S., Li, Z., Spooner, E., et al. (2014). Site-specific chemoenzymatic labeling of aerolysin enables the identification of new aerolysin receptors. PLoS ONE 9, e109883. doi:10.1371/journal.pone.0109883
Xu, R., Hanson, S. R., Zhang, Z., Yang, Y.-Y., Schultz, P. G., and Wong, C.-H. (2004). Site-specific incorporation of the mucin-type N -acetylgalactosamine-α-O-threonine into protein in escherichia c oli. J. Am. Chem. Soc. 126, 15654–15655. doi:10.1021/ja044711z
Yi, L., Sun, H., Itzen, A., Triola, G., Waldmann, H., Goody, R. S., et al. (2011). One-pot dual-labeling of a protein by two chemoselective reactions. Angew. Chem. Int. Ed. 50, 8287–8290. doi:10.1002/anie.201100840
Keywords: sortase, glycopeptide, glycoconjugates, oxime ligation, protein modification, transpeptidase, neoglycoproteins
Citation: Dolan JP, Machin DC, Dedola S, Field RA, Webb ME and Turnbull WB (2022) Synthesis of cholera toxin B subunit glycoconjugates using site-specific orthogonal oxime and sortase ligation reactions. Front. Chem. 10:958272. doi: 10.3389/fchem.2022.958272
Received: 31 May 2022; Accepted: 26 July 2022;
Published: 14 September 2022.
Edited by:
M. Carmen Galan, University of Bristol, United KingdomReviewed by:
Xue-Long Sun, Cleveland State University, United StatesHidde Ploegh, Boston Children’s Hospital, Harvard Medical School, United States
Copyright © 2022 Dolan, Machin, Dedola, Field, Webb and Turnbull. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: W. Bruce Turnbull, dy5iLnR1cm5idWxsQGxlZWRzLmFjLnVr; Michael E. Webb, bS5lLndlYmJAbGVlZHMuYWMudWs=
†Present address: Jonathan P. Dolan, School of Chemical and Physical Sciences and Centre for Glycoscience Research and Training, Keele University, Keele, Staffordshire, United Kingdom
 Jonathan P. Dolan
Jonathan P. Dolan Darren C. Machin1
Darren C. Machin1 Michael E. Webb
Michael E. Webb W. Bruce Turnbull
W. Bruce Turnbull