- 1School of Pharmacy, College of Medicine, National Taiwan University, Taipei, Taiwan
- 2Genomics Research Center, Academia Sinica, Taipei, Taiwan
- 3Department of Applied Science, National Taitung University, Taitung, Taiwan
- 4Department of Chemistry, National Cheng Kung University, Tainan, Taiwan
The extracellular human endo-6-O-sulfatases (Sulf-1 and Sulf-2) are responsible for the endolytic cleavage of the 6-sulfate groups from the internal D-glucosamine residues in the highly sulfated subdomains of heparan sulfate proteoglycans. A trisaccharide sulfate, IdoA2OS-GlcNS6S-IdoA2OS, was identified as the minimal size of substrate for Sulf-1. In order to study the complex structure with Sulf-1 for developing potential drugs, two trisaccharide analogs, IdoA2OS-GlcNS6OSO2NH2-IdoA2OS-OMe and IdoA2OS-GlcNS6NS-IdoA2OS-OMe, were rationally designed and synthesized as the Sulf-1 inhibitors with IC50 values at 0.27 and 4.6 μM, respectively.
Introduction
Heparan sulfate (HS) proteoglycans, which are ubiquitously distributed on the cell surface and in the extracellular matrix and basement membrane, play significant roles in adhesion, recognition, and signal transduction events (Vives et al., 2014). HS is a polyanionic polysaccharide belonging to the glycosaminoglycan (GAG) families, covalently bound to a core protein with a tetrasaccharide linkage region (El Masri et al., 2020). The HS backbone is composed of an alternative disaccharide repeating unit with all 1→4-linkages of N-acetyl-α-D-glucosamine (GlcNAc) and β-D-glucuronic acid (GlcA)/α-L-iduronic acid (IdoA). The acetyl group of GlcNAc could possibly be hydrolyzed to yield the amino group followed by N-sulfation, and/or the 2-O position of the uronic acid and/or the 3-O and/or 6-O positions of GlcN could undergo sulfation through a series of enzymatic modifications (Kreuger and Kjellén, 2012). The highly sulfated regions of HS, which were characterized as S-domains, are involved in binding with various proteins, such as fibroblast growth factors, transforming growth factor-β, Wnt, and bone morphogenetic protein (Li and Kusche-Gullberg, 2016). Changes in the HS sulfation patterns may cause dissociations with these proteins, resulting in up or downregulation of the corresponding signal transduction factors. Human endo-O-sulfatases 1 and 2 (Sulf-1 and Sulf-2) are two isoforms of the extracellular endo-6-O-sulfatases responsible for the hydrolysis of the sulfate groups at the 6-O positions of the internal D-glucosamine residues (Justo et al., 2022). Both enzymes modulate the sulfation patterns and regulate the HS-protein interactions (Hanson et al., 2004; El Masri et al., 2017). Numerous diseases have been proved to be related to the overexpression of Sulf-1 and Sulf-2, including gastric and pancreatic cancers, invasive breast carcinoma, lung adenocarcinoma, and osteoarthritis (OA) (Lai et al., 2003; Viviano et al., 2004; Otsuki et al., 2008; Otsuki et al., 2010; Rosen and Lemjabbar-Alaoui, 2010; Hur et al., 2012; Chanalaris et al., 2019; Lin et al., 2020; Severmann et al., 2020; Shamdani et al., 2020; Chivu-Economescu et al., 2022). Thus, the development of effective inhibitors may provide detailed information on their complex structures with Sulfs at the molecular level and offer an opportunity for structure-activity relationship for new drug discovery.
In our previous study, a variety of HS oligosaccharides with different chain lengths and N- and O-sulfation patterns was screened for the substrate specificity of Sulf-1, and a trisaccharide sulfate, IdoA2OS-GlcNS6OS-IdoA2OS 1 (Figure 1), was identified as the minimal size of substrate for Sulf-1 (Chiu et al., 2020). A substrate analog 2 with the sulfonamide at the 6′-O position of GlcNS and a 5-amino-1-n-pentyl group at the 1-O position of IdoA had been developed as an inhibitor of Sulf-1. Although compound 2 could be immobilized on the chip of Surface Plasmon Resonance (SPR) for measurement of the dissociation constant (KD) with Sulf-1 or possibly be applied as a probe to identify new Sulfs via its attachment to the magnetic nanoparticles, the 5-amino-1-n-pentyl linker may cause the difficulty for crystallization of compound 2 with Sulf-1 for X-ray determination of the complex structure since its flexibility and the amino group might create an additional salt bridge with protein. Furthermore, the linker part may not be appropriate for the development of potential drug candidates. Inspired by the structure of Fondaparinux 3 (Chang et al., 2014), an anticoagulant drug used in the clinic, the methyl group is introduced to replace the 5-amino-1-n-pentyl group at the reducing end, and the substrate 4 and the 6′-O-sulfonamide inhibitor 5 are designed and prepared to test their activities for Sulf-1. Additionally, the 6′-N-sulfate 6 is synthesized for its interaction with Sulf-1 and its comparison with compounds 4 and 5.
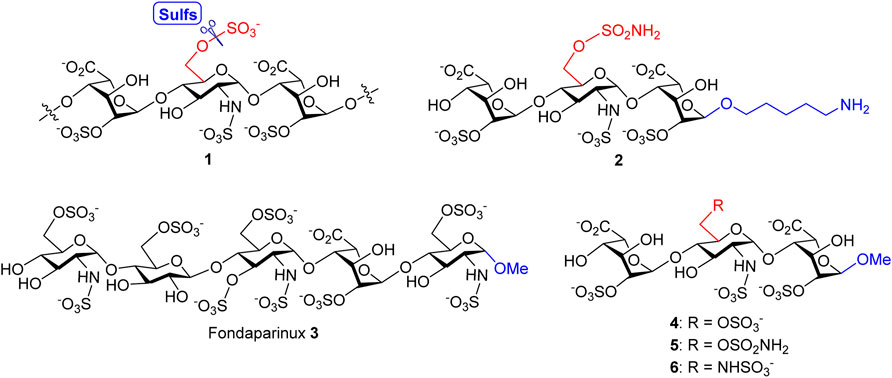
FIGURE 1. Structures of Sulf-1’s trisaccharide unit 1 and inhibitor 2 with a 5-amino-1-n-pentyl linker, the anticoagulant Fondaparinux 3, and the proposed Sulf-1’s substrate 4 and inhibitors of the 6′-O-sulfonamide 5 and the 6′-N-sulfate 6 with the methyl group at 1-O.
Results and Discussion
Synthesis of the Sulf-1 substrate 4. The backbone assembly of the target trisaccharide 4 is approached through a [1 + 2] strategy. The preparation of the disaccharide acceptor 13 is illustrated in Scheme 1. A coupling of the D-glucosamine-derived thioglycoside 7 with the 1,6-anhydro-β-L-idopyranosyl 4-alcohol 8 was carried out to yield the desired α-disaccharide 9 (71%), according to our previous report (Zulueta et al., 2012). Copper (II) trifluoromethanesulfonate-catalyzed acetolysis of compound 9 to open the 1,6-anhydro ring in acetic anhydride furnished the 1,6-diacetate 10 (88%), which underwent anomeric acetyloxy replacement via a combination of trimethylsilyl p-toluenyl thioether (TMSSTol) and zinc iodide (ZnI2) to give the corresponding thioglycoside 11 in 76% yield. Treatment of the disaccharide donor 11 with methanol in the presence of N-iodosuccinimide (NIS) and trifluoromethanesulfonic acid (TfOH) as the activators afforded the expected α-methyl disaccharide 12 (60%) because of neighboring group participation of the 2-O-benzoyl group. The stereochemistry was determined through a series of NMR spectral analyses, indicating the correlation of W-coupling between 1-H and 3-H in the 2D spectrum (Supplementary Material, Supplementary Pages S19–S22). Removal of the 2-naphthylmethyl (2-NAP) group at the 4′-O position of compound 12 using 2,3-dichloro-5,6-dicyano-1,4-benzoquinone (DDQ) provided the desired 4′-alcohol 13 in 79% yield.
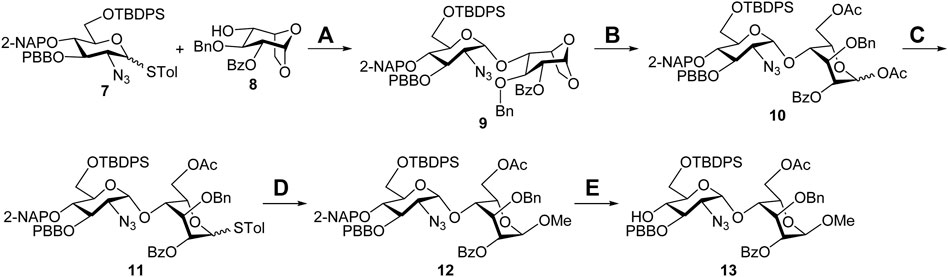
SCHEME 1. Synthesis of the disaccharide acceptor 13. Reagents and conditions: (A) NIS, TfOH, CH2Cl2, 3 Å MS, −78°C to −20°C, 4 h, 71%; (B) Cu(OTf)2, Ac2O, RT, 1 h, 88%; (C) TMSSTol, ZnI2, CH2Cl2, 0°C to RT, 3 h, 76%; (D) NIS, TfOH, MeOH, CH2Cl2, 3 Å MS, −50°C, 2 h, 60%; (E) DDQ, CH2Cl2/H2O = 19/1, RT, 3 h, 79%. Ac, acetyl; Bn, benzyl; Bz, benzoyl; DDQ, 2,3-dichloro-5,6-dicyano-1,4-benzoquinone; 2-NAP, 2-naphthylmethyl; NIS, N-iodosuccinimide; PBB, p-bromobenzyl; TBDPS, tert-butyldiphenylsilyl; Tf, trifluoromethanesulfonyl; TMS, trimethylsilyl; Tol, 4-methylphenyl.
With the glycosyl acceptor 13 in hand, the total synthesis of substrate 4 is depicted in Scheme 2. Benzylation of the 4-alcohol 8 (Ag2O, BnBr) led to the ether 14 (94%), which was opened under acetolysis conditions to generate the 1,6-diacetate 15 (87%). Anomeric substitution of compound 15 promoted by ZnI2 and TMSSTol was carried out, and the thioglycoside 16 was obtained in a 74% yield (Hu et al., 2011). Highly stereoselective α-glycosylation of the donor 16 with the acceptor 13 using NIS/TfOH delivered the desired trisaccharide 17 (71%), and all acyl groups in 17 were removed under Zemplén transesterification conditions to give the tetraol 18 (86%). Oxidation of 18 with (2,2,6,6-tetramethylpiperidin-1-yl)oxyl (TEMPO) and bis(acetoxy)iodobenzene (BAIB) furnished the dilactone 19 (76%), which was subjected upon cleavage of the tert-butyldiphenylsilyl (TBDPS) group with HF•pyridine complex at the 6′-O position yielding the 6′-alcohol 20 (78%). Opening of two lactone rings in compound 20 employing 1 M lithium hydroxide aqueous solution provided the 2,6′,2″-triol 21 (90%), which could be sulfonated with SO3•Et3N complex to afford the corresponding 2,6′,2″-tri-O-sulfate 22 (77%). Hydrogenolysis of compound 22 with Pd(OH)2/C and H2 in neutral buffer solution, allowing removal of the 1,3″,4″-tri-O-benzyl and 3′-O-p-bromobenzyl groups and reducing the 2′-C-azido group to the 2′-C-amino group, led to the desired product 23 (90%). N-Sulfonation of the amine 23 with SO3•pyridine complex at ca. pH 9.5, controlled by the addition of 1 M NaOH aqueous solution, furnished a crude mixture, which was purified through sequential Sephadex G10 size-exclusion column chromatography and Dowex 50WX8-Na+ ion-exchange column chromatography to yield target molecule 4 (Na+ salt, 77%). The structure of compound 4 was characterized through analyses of its 1D and 2D NMR spectra (Supplementary Material, Supplementary Pages S45–S49). The molecular weight of 4 (M+6Na++H+, calculated for C19H26NO29S4Na6+ 997.8854, found 997.8815) was further confirmed by the high-resolution electrospray ionization mass spectrum (Supplementary Material, Supplementary Page S50).
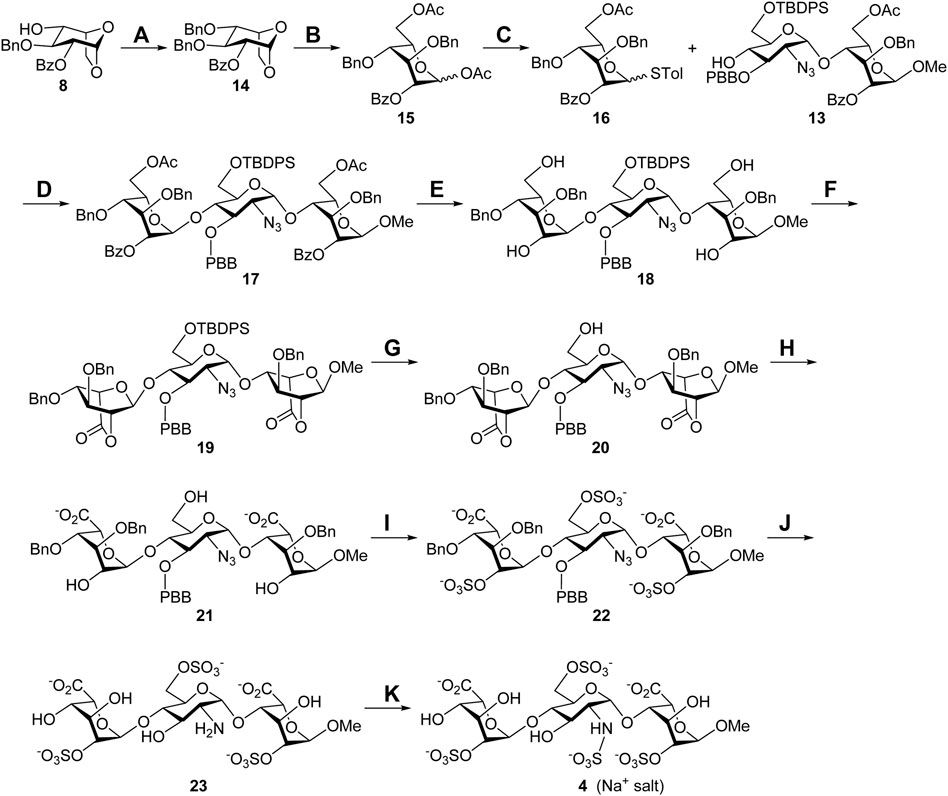
SCHEME 2. Synthesis of the Sulf-1 substrate 4. Reagents and conditions: (A) BnBr, Ag2O, CH2Cl2, 3 Å MS, RT, 48 h, 94%; (B) Cu(OTf)2, Ac2O, RT, 1 h, 87%; (C) TMSSTol, ZnI2, CH2Cl2, 0°C to RT, 3 h, 74%; (D) NIS, TfOH, CH2Cl2, 3 Å MS, −40°C, 2 h, 71%; (E) NaOMe, CH2Cl2/MeOH = 1/1, 0°C to RT, 24 h, 86%; (F) TEMPO, BAIB, CH2Cl2/H2O = 2/1, RT, 6 h, 76%; (G) HF•pyridine, THF, 0°C to rt, 24 h, 78%; (H) 1 M LiOH(aq), THF, RT, 1 h, 90%; (I) SO3•Et3N, DMF, 65°C, 24 h, 77%; (J) Pd(OH)2/C, H2(g), phosphate buffer (pH = 7.0), MeOH, RT, 48 h, 90%; (K) SO3•pyridine, 1 M NaOH(aq), H2O, RT, 48 h, 77%. BAIB, [bis(acetoxy)iodo]benzene; DMF, N,N-dimethylformide; TEMPO, (2,2,6,6-tetramethylpiperidin-1-yl)oxyl; THF, tetrahydrofuran.
Synthesis of the 6′-O-sulfonamide trisaccharide inhibitor 5. Scheme 3 describes the synthesis of HS trisaccharide analog 5 containing the 6′-O-sulfonamide as the Sulf-1 inhibitor. Starting from the common trisaccharide intermediate 20, the 6′-hydroxy group was treated with N-benzyl sulfamoyl chloride and pyridine to give the N-benzyl-6′-O-sulfonamide derivative 24 (78%). It should be noted that the reaction needed to be quenched in 15 min since the NH-proton could be removed by the base and further coupled with N-benzyl sulfamoyl chloride to yield the unwanted side product when the reaction time is prolonged. The lactone rings of compound 24 were opened under basic conditions to provide the 2,2″-diol 25 (90%), which was converted into the corresponding di-O-sulfate 26 (70%) via sulfonation at the 2-O and 2″-O positions. Global deprotection of 26 under hydrogenolysis conditions led to the product 27 (73%), which underwent N-sulfonation to furnish the desired 6′-O-sulfonamide inhibitor 5 (Na+ salt) in 75% yield after consecutive purification through the size-exclusion column and ion-exchange column. The structure of compound 5 was determined via the 1D and 2D NMR spectral analyses (Supplementary Material, Supplementary Pages S57–S61). By using the trisaccharide 5 as a representative example, the assignments of all protons, including the splitting patterns and coupling constants, were examined by the 1D-Total Correlation Spectroscopy (1D-TOCSY) experiments in detail. As indicated in Figure 2, three isolated spectroscopic patterns were generated by 1H connectivity through J-coupling after the excitation of selective 1H nuclei at a given frequency (MacKinnon et al., 2016). The complicated proton NMR patterns of compound 5 could be resolved in three individual pyranosyl rings (purple one: non-reducing end L-iduronic acid sugar unit; blue one: D-glucosamine unit; red one: reducing end L-iduronic acid sugar unit) and characterized by combination with basic 1D and 2D NMR spectra. Starting from the 2D-HMBC spectrum, the correlation of 3JC-H coupling between the anomeric carbon of the reducing end L-iduronic acid and the methyl protons at 1-O was identified. Two sets of 2JC-H coupling correlations between the anomeric carbon of the L-iduronic acid and H-5 were also recognized. All of the anomeric hydrogen atoms could be illustrated through the 1JC-H coupling between the corresponding anomeric carbon via the 2D-HSQC spectroscopy. In comparison with the 1D-TOCSY spectra, the red individual 1H spectrum was elucidated for the reducing end L-iduronic acid sugar unit via the corresponding H-1 and H-5. The purple individual 1H spectrum corresponded to the nonreducing end L-iduronic acid sugar unit through the H-5” correlation. The remaining blue individual 1H spectrum represented the D-glucosamine sugar unit. Finally, all proton peaks were assigned with the combination of 2D-COSY spectrum analysis. These techniques were applied to identify the structures of all compounds in this study. In addition, the high-resolution electrospray ionization mass spectrum further confirmed the correct molecular weight of 5 (M-6H++5Na+, calculated for C19H26N2O28S4Na5− 972.9049, found 972.9021) in comparison with the results of computational simulation (Supplementary Material, Supplementary Page S62).
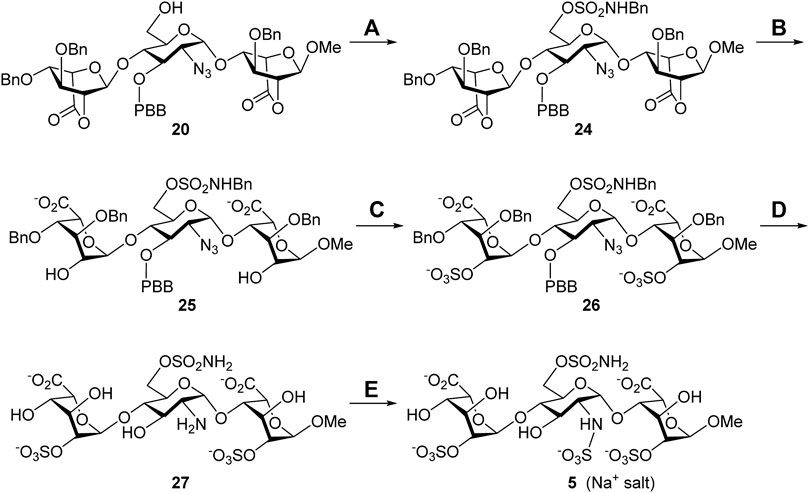
SCHEME 3. Synthesis of the 6′-O-sulfonamide trisaccharide inhibitor 5. Reagents and conditions: (A) ClSO2NHBn, pyridine, RT, 15 min, 78%; (B) 1 M LiOH(aq), THF, RT, 1 h, 90%; (C) SO3•Et3N, DMF, 65°C, 24 h, 70%; (D) Pd(OH)2/C, H2(g), phosphate buffer (pH = 7.0), MeOH, RT, 48 h, 73%; (E) SO3•pyridine, 1 M NaOH(aq), H2O, RT, 48 h, 75%.
Synthesis of the 6′-N-sulfate trisaccharide inhibitor 6. The aforementioned common building block 6′-alcohol 20 was utilized to synthesize the 6′-N-sulfate derivative 6 (Scheme 4). The reaction of 20 with diphenylphosphoryl azide (DPPA) and 1,8-diazabicyclo [5.4.0]undec-7-ene (DBU) in toluene successfully transformed the 6′-hydroxy group into the 6′-azido group, and the expected 2′,6′-diazide 28 was obtained in 60% yield. Such harsh conditions, at 100°C for 24 h, were needed for the counterattack of the azide anion to the 6′-phosphate intermediate through SN2 nucleophilic substitution. With compound 28 ready, a similar four-step conversion was carried out to generate the final target molecule 6, including the opening of two lactone rings (29, 86%), 2,2″-di-O-sulfonation (30, 86%), global deprotection by hydrogenolysis reduction, (31, 40%), and 2′,6′-di-N-sulfonation (6, 78%). Perhaps due to the chelation effect of the amino groups with the palladium catalyst causing difficult separation, the 2′,6′-diamine 31 was isolated in moderate yield. The structure of compound 6 was determined via the 1D and 2D NMR spectral analyses (Supplementary Material, Supplementary Pages S69–S73). The high-resolution electrospray ionization mass spectrum has confirmed the correct molecular weight of 6 (M + Na+, calculated for C19H28N2O28S4Na5+ 974.9194, found 974.9195) in comparison with the result of computational simulation (Supplementary Material, Supplementary Page S74).
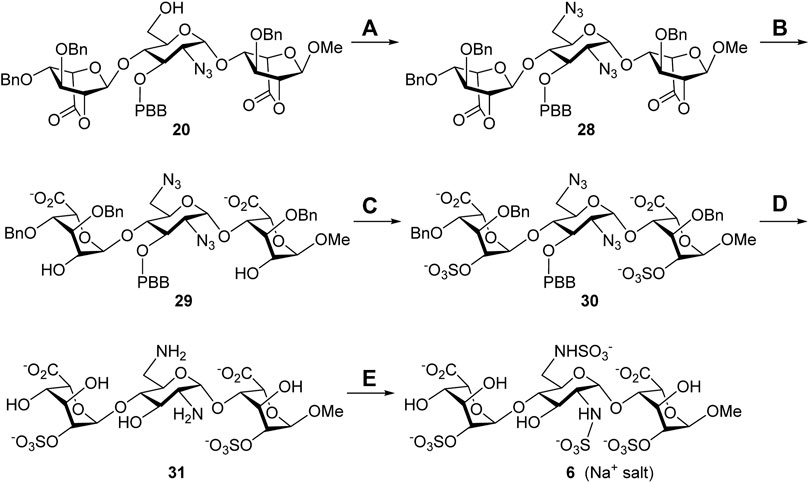
SCHEME 4. Synthesis of the 6′-N-sulfate trisaccharide inhibitor 5. Reagents and conditions: (A) DPPA, DBU, toluene, 100°C, 24 h, 60%; (B) 1 M LiOH(aq), THF, rt., 1 h, 86%; (C) SO3•Et3N, DMF, 65°C, 24 h, 86%; (D) Pd(OH)2/C, H2(g), phosphate buffer (pH = 7.0), MeOH, rt., 48 h, 40%; (E) SO3•pyridine, 1 M NaOH(aq), H2O, rt., 48 h, 78%. DBU, 1,8-diazabicyclo [5.4.0]undec-7-ene; DPPA, diphenylphosphoryl azide.
Sulf-1 inhibition assay. Human Sulf-1 was overexpressed and purified according to the procedure of the previous report (Chiu et al., 2020). The fluorescent assay was employed to measure the inhibitory activity by the released 4-methylumbelliferone (4-MU) from hydrolysis of 4-methylumbelliferyl sulfate (4-MUS) in the presence of human Sulf-1 and the inhibitor. Compounds 4, 5, and 6 were examined in these experiments. Since the methyl trisaccharide 4 exhibited similar substrate activity as 1 toward Sulf-1, it did not have any inhibition property (IC50 ˃ 100 μM). The IC50 value of the 6′-O-sulfonamide trisaccharide 5 was measured at 0.27 μM, and the kinetic studies of the inhibitory activity showed a competitive inhibition with the Ki value of 0.14 μM (Figure 3). This indicated that replacement of the methyl moiety from 5-amino-1-n-pentyl at the 1-O position maintained a similar level of inhibitory activity to Sulf-1, suggesting that the 5-amino-pentyl group of 2 did not contribute to additional interaction with Sulf-1. Interestingly, the 6′-N-sulfate trisaccharide 6 with the IC50 value of 4.6 μM confirmed that it was an inhibitor to Sulf-1. Although the structures of compounds 4 and 6 were only one bioisosteric modification from O-sulfate to N-sulfate at the 6′-position, the interactions with Sulf-1 were different. These results have revealed that the sulfonamide group could be an appropriate moiety to inhibit Sulf-1 activity (Hanson et al., 2004). The N-sulfate moiety, albeit with lower inhibition, provided alternative considerations for the design of new inhibitors.
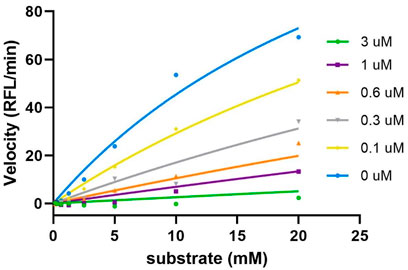
FIGURE 3. Kinetics studies of inhibition of compound 5. The enzyme activities were evaluated using various concentrations of substrate (0∼20 mM) and inhibitor (0∼3 uM). The results were then fitted into the competitive model using the equation Km, Obs = Km*(1+[I]/Ki), and Y=Vmax*X/(Km, Obs + X) (GraphPad). The Ki value was determined as 0.14 μM.
Conclusion
The total syntheses of the 1-O- and 6′-C-modified HS trisaccharides 4, 5, and 6 via the 6′-alcohol 20 as the common intermediate have been successfully developed. Both bicyclo[2.2.2]-lactone rings in 20 block the 2,2″-dihydroxy groups, eliminating two protecting groups and allowing the functional group transformation at the 6′-hydroxy group. A straightforward four-step transformation, including lactone opening, O-sulfonation, hydrogenolysis, and N-sulfonation, has efficiently yielded the desired products 4, 5, and 6. The 1-O-methyl-modified trisaccharide 4 has been identified as the Sulf-1 substrate that can be used to test the enzyme activity. In comparison with compound 2, the 1-O-methyl-modified 6′-O-sulfonamide trisaccharide 5 exhibits similar inhibitory property (IC50 = 0.27 μM) that is an appropriate molecule to complex with Sulf-1 for further 3D structural studies by X-ray single crystal diffraction technique. Compound 5 is also a potent candidate for new drug discovery against diseases related to Sulf-1 overexpression. The 6′-N-sulfate trisaccharide 6, which is not a Sulf-1 substrate, has been characterized as an inhibitor with IC50 = 4.6 μM for Sulf-1. The effectiveness of 5 to inhibit Sulf-1’s activity is 17 times better than 6.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
K-YT and Z-HT performed the experimental work and analyzed the spectral data. T-JC obtained Sulf-1 and conducted the IC50 measurements. S-CH provided the ideas. S-CH, P-HL, and K-YT wrote the manuscript.
Funding
This work was supported by the Ministry of Science and Technology of Taiwan (MOST 109-2639-M-001-004-ASP to S-CH). K-YT had a scholarship supported by the Program of Research Performance Enhancement via Students Entering Ph.D. Programs Straight from an Undergraduate/Master’s Program from National Taiwan University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.947475/full#supplementary-material
References
Chanalaris, A., Clarke, H., Guimond, S. E., Vincent, T. L., Turnbull, J. E., and Troeberg, L. (2019). Heparan Sulfate Proteoglycan Synthesis Is Dysregulated in Human Osteoarthritic Cartilage. Am. J. Pathology 189, 632–647. doi:10.1016/j.ajpath.2018.11.011
Chang, C.-H., Lico, L. S., Huang, T.-Y., Lin, S.-Y., Chang, C.-L., Arco, S. D., et al. (2014). Synthesis of the Heparin-Based Anticoagulant Drug Fondaparinux. Angew. Chem. Int. Ed. 53, 9876–9879. doi:10.1002/anie.201404154
Chiu, L.-T., Sabbavarapu, N. M., Lin, W.-C., Fan, C.-Y., Wu, C.-C., Cheng, T.-J. R., et al. (2020). Trisaccharide Sulfate and its Sulfonamide as an Effective Substrate and Inhibitor of Human Endo-O-Sulfatase-1. J. Am. Chem. Soc. 142, 5282–5292. doi:10.1021/jacs.0c00005
Chivu-Economescu, M., Necula, L. G., Matei, L., Dragu, D., Bleotu, C., Sorop, A., et al. (2022). Collagen Family and Other Matrix Remodeling Proteins Identified by Bioinformatics Analysis as Hub Genes Involved in Gastric Cancer Progression and Prognosis. Ijms 23, 3214. doi:10.3390/ijms23063214
El Masri, R., Crétinon, Y., Gout, E., and Vivès, R. R. (2020). HS and Inflammation: A Potential Playground for the Sulfs? Front. Immunol. 11. doi:10.3389/fimmu.2020.00570
El Masri, R., Seffouh, A., Lortat-Jacob, H., and Vivès, R. R. (2017). The "in and Out" of Glucosamine 6-O-Sulfation: the 6th Sense of Heparan Sulfate. Glycoconj. J. 34, 285–298. doi:10.1007/s10719-016-9736-5
Hanson, S. R., Best, M. D., and Wong, C.-H. (2004). Sulfatases: Structure, Mechanism, Biological Activity, Inhibition, and Synthetic Utility. Angew. Chem. Int. Ed. 43, 5736–5763. doi:10.1002/anie.200300632
Hu, Y.-P., Lin, S.-Y., Huang, C.-Y., Zulueta, M. M. L., Liu, J.-Y., Chang, W., et al. (2011). Synthesis of 3-O-Sulfonated Heparan Sulfate Octasaccharides that Inhibit the Herpes Simplex Virus Type 1 Host-Cell Interaction. Nat. Chem. 3, 557–563. doi:10.1038/nchem.1073
Hur, K., Han, T.-S., Jung, E.-J., Yu, J., Lee, H.-J., Kim, W. H., et al. (2012). Up-regulated Expression of Sulfatases (SULF1 and SULF2) as Prognostic and Metastasis Predictive Markers in Human Gastric Cancer. J. Pathol. 228, 88–98. doi:10.1002/path.4055
Justo, T., Smart, N., and Dhoot, G. K. (2022). Context Dependent Sulf1/Sulf2 Functional Divergence in Endothelial Cell Activity. Ijms 23, 3769. doi:10.3390/ijms23073769
Kreuger, J., and Kjellén, L. (2012). Heparan Sulfate Biosynthesis. J. Histochem Cytochem. 60, 898–907. doi:10.1369/0022155412464972
Lai, J., Chien, J., Staub, J., Avula, R., Greene, E. L., Matthews, T. A., et al. (2003). Loss of HSulf-1 Up-Regulates Heparin-Binding Growth Factor Signaling in Cancer. J. Biol. Chem. 278, 23107–23117. doi:10.1074/jbc.M302203200
Li, J.-P., and Kusche-Gullberg, M. (2016). Heparan Sulfate: Biosynthesis, Structure, and Function. Int. Rev. Cell Mol. Biol. 325, 215–273. doi:10.1016/bs.ircmb.2016.02.009
Lin, T.-S., Hsieh, C.-H., Kuo, C., Juang, Y.-P., Hsieh, Y. S. Y., Chiang, H., et al. (2020). Sulfation Pattern of Chondroitin Sulfate in Human Osteoarthritis Cartilages Reveals a Lower Level of Chondroitin-4-Sulfate. Carbohydr. Polym. 229, 115496. doi:10.1016/j.carbpol.2019.115496
MacKinnon, N., While, P. T., and Korvink, J. G. (2016). Novel Selective TOCSY Method Enables NMR Spectral Elucidation of Metabolomic Mixtures. J. Magnetic Reson. 272, 147–157. doi:10.1016/j.jmr.2016.09.011
Otsuki, S., Hanson, S. R., Miyaki, S., Grogan, S. P., Kinoshita, M., Asahara, H., et al. (2010). Extracellular Sulfatases Support Cartilage Homeostasis by Regulating BMP and FGF Signaling Pathways. Proc. Natl. Acad. Sci. U.S.A. 107, 10202–10207. doi:10.1073/pnas.0913897107
Otsuki, S., Taniguchi, N., Grogan, S. P., D'Lima, D., Kinoshita, M., and Lotz, M. (2008). Expression of Novel Extracellular Sulfatases Sulf-1 and Sulf-2 in Normal and Osteoarthritic Articular Cartilage. Arthritis Res. Ther. 10, R61. doi:10.1186/ar2432
Rosen, S. D., and Lemjabbar-Alaoui, H. (2010). Sulf-2: an Extracellular Modulator of Cell Signaling and a Cancer Target Candidate. Expert Opin. Ther. Targets 14, 935–949. doi:10.1517/14728222.2010.504718
Severmann, A.-C., Jochmann, K., Feller, K., Bachvarova, V., Piombo, V., Stange, R., et al. (2020). An Altered Heparan Sulfate Structure in the Articular Cartilage Protects against Osteoarthritis. Osteoarthr. Cartil. 28, 977–987. doi:10.1016/j.joca.2020.04.002
Shamdani, S., Chantepie, S., Flageollet, C., Henni-Chebra, N., Jouan, Y., Eymard, F., et al. (2020). Heparan Sulfate Functions Are Altered in the Osteoarthritic Cartilage. Arthritis Res. Ther. 22, 283. doi:10.1186/s13075-020-02352-3
Vivès, R. R., Seffouh, A., and Lortat-Jacob, H. (2014). Post-synthetic Regulation of HS Structure: The Yin and Yang of the Sulfs in Cancer. Front. Oncol. 3. doi:10.3389/fonc.2013.00331
Viviano, B. L., Paine-Saunders, S., Gasiunas, N., Gallagher, J., and Saunders, S. (2004). Domain-specific Modification of Heparan Sulfate by Qsulf1 Modulates the Binding of the Bone Morphogenetic Protein Antagonist Noggin. J. Biol. Chem. 279, 5604–5611. doi:10.1074/jbc.M310691200
Zulueta, M. M. L., Lin, S.-Y., Lin, Y.-T., Huang, C.-J., Wang, C.-C., Ku, C.-C., et al. (2012). α-Glycosylation by D-Glucosamine-Derived Donors: Synthesis of Heparosan and Heparin Analogues that Interact with Mycobacterial Heparin-Binding Hemagglutinin. J. Am. Chem. Soc. 134, 8988–8995. doi:10.1021/ja302640p
Keywords: heparan sulfate, glycosaminoglycans, endo-6-O-sulfatases, carbohydrate chemistry, inhibitors
Citation: Tseng K-Y, Tzeng Z-H, Cheng T-JR, Liang P-H and Hung S-C (2022) Design and Synthesis of 1-O- and 6′-C-Modified Heparan Sulfate Trisaccharides as Human Endo-6-O-Sulfatase 1 Inhibitors. Front. Chem. 10:947475. doi: 10.3389/fchem.2022.947475
Received: 18 May 2022; Accepted: 14 June 2022;
Published: 13 July 2022.
Edited by:
M. Carmen Galan, University of Bristol, United KingdomReviewed by:
Raghavendra Kikkeri Venkatanarayan, Indian Institute of Science Education and Research, Pune, IndiaGavin Miller, Keele University, United Kingdom
Copyright © 2022 Tseng, Tzeng, Cheng, Liang and Hung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pi-Hui Liang, cGhsaWFuZ0BudHUuZWR1LnR3; Shang-Cheng Hung, c2NodW5nQGdhdGUuc2luaWNhLmVkdS50dw==
 Kuei-Yao Tseng
Kuei-Yao Tseng Zheng-Hao Tzeng
Zheng-Hao Tzeng Ting-Jen Rachel Cheng
Ting-Jen Rachel Cheng Pi-Hui Liang
Pi-Hui Liang Shang-Cheng Hung
Shang-Cheng Hung