- 1Department of Biotechnology and Life Sciences, University of Insubria, Varese, Italy
- 2Department of Genetics and Biotechnology, Ivan Franko National University of Lviv, Lviv, Ukraine
Glycopeptide antibiotics are valuable natural metabolites endowed with different pharmacological properties, among them are dalbaheptides used to treat different infections caused by multidrug-resistant Gram-positive pathogens. Dalbaheptides are produced by soil-dwelling high G-C Gram-positive actinobacteria. Their biosynthetic pathways are encoded within large biosynthetic gene clusters. A non-ribosomally synthesized heptapeptide aglycone is the common scaffold for all dalbaheptides. Different enzymatic tailoring steps, including glycosylation, are further involved in decorating it. Glycosylation of dalbaheptides is a crucial step, conferring them specific biological activities. It is achieved by a plethora of glycosyltransferases, encoded within the corresponding biosynthetic gene clusters, able to install different sugar residues. These sugars might originate from the primary metabolism, or, alternatively, their biosynthesis might be encoded within the biosynthetic gene clusters. Already installed monosaccharides might be further enzymatically modified or work as substrates for additional glycosylation. In the current minireview, we cover recent updates concerning the genetics and enzymology behind the glycosylation of dalbaheptides, building a detailed and consecutive picture of this process and of its biological evolution. A thorough understanding of how glycosyltransferases function in dalbaheptide biosynthesis might open new ways to use them in chemo-enzymatic synthesis and/or in combinatorial biosynthesis for building novel glycosylated antibiotics.
Introduction
Among different bacterial phyla, the mycelia-forming members of actinobacteria‐broadly known as actinomycetes‐remain the best antibiotic providers (Bérdy, 2005; Hutchings et al., 2019). The biosynthesis of antibiotics involves many enzymes, which are encoded by co-localized genes‐biosynthetic gene clusters (BGCs) (Medema et al., 2015). BGCs undergo modular evolution, often exchanging operons and single genes coding for biosynthetic and modification enzymes (Medema et al., 2014). Genes for glycosyltransferases (GTs) are one such example, being found in different BGCs, with corresponding proteins having relaxed substrate specificity and consequently being able to modify different natural scaffolds (Salas and Méndez, 2007).
The astonishing variability of glycosylation patterns in one group of antibiotics led to their eponymous description as glycopeptide antibiotics (GPAs, Nicolaou et al., 1999). Natural GPAs amalgamate five types of related compounds, differing in chemical structures, where, paradoxically, only types I-IV‐also known as dalbaheptides (Parenti and Cavalleri, 1989)‐are glycosylated (Nicolaou et al., 1999). All dalbaheptides possess a non-ribosomal heptapeptide aglycone differing in amino acid (aa) composition, cross-linking, and decoration (Nicolaou et al., 1999); they inhibit the growth of Gram-positive bacteria by blocking cell wall maturation (Binda et al., 2014; Yushchuk et al., 2020a). Glycosylation and acylation (a step depending on glycosylation) of dalbaheptides contribute to their antimicrobial activities, favoring dimerization and membrane localization at the site of action (Gerhard et al., 1993; Mackay et al., 1994; Beauregard et al., 1995; Snyder et al., 1998). On the other hand, excessive glycosylation does not always bring pharmacological benefits: it seemed to induce platelet aggregation in patients treated with ristocetin (Howard and Firkin, 1971; Coller and Gralnick, 1977), which was consequently withdrawn from the clinical use (Gangarosa et al., 1958).
Dalbaheptides are clinically used as drugs of last resort against multidrug-resistant Gram-positive pathogens (Marcone et al., 2018). First-generation GPAs‐vancomycin and teicoplanin (produced by different Amycolatopsis spp. and Actinoplanes teichomyceticus ATCC 31121, respectively)‐have a long and reliable history of clinical application (Jovetic et al., 2010; Binda et al., 2014; Marcone et al., 2018). In turn, natural GPAs served as precursors for three second-generation semisynthetic and clinically used GPAs (Binda et al., 2014; Butler et al., 2014): dalbavancin derived from A40926 (produced by Nonomuraea gerenzanensis ATCC 39727) (Crotty et al., 2016), telavancin from chloroeremomycin (from Kibdelosporangium aridum A82846) (Klinker and Borgert, 2015), and oritavancin from vancomycin (Crotty et al., 2016).
Although the chemical variety of glycosyl groups decorating dalbaheptide aglycones is quite remarkable (Nicolaou et al., 1999), many aspects of the genetics behind their biosynthesis and incorporation remain obscure. In this minireview, we focus on those dalbaheptides whose BGC sequences are nowadays available, and for which some experimental evidence about their glycosylation steps is reported in the literature. The model BGCs are cep, bal, tei, vcm, and dbv, responsible for the production of chloroeremomycin (in K. aridum A82846) (van Wageningen et al., 1998), balhimycin (in Amycolatopsis balhimycina DSM 5908) (Shawky et al., 2007), teicoplanin (Li et al., 2004), vancomycin (in Amycolatopsis orientalis HCCB10007) (Xu et al., 2014), and A40926 (Sosio et al., 2003), respectively. BGC from Amycolatopsis sp. MJM2582 (Truman et al., 2014) represents the ristocetin biosynthetic pathway and was found also in other Amycolatopsis spp. (Spohn et al., 2014; Liu et al., 2021). Glycosylation-related genes from more recently described BGCs for UK-68,597 (auk from Actinoplanes sp. ATCC 53533) (Yim et al., 2014a), pekiskomycin (pek from Streptomyces sp. WAC1420) (Thaker et al., 2013), keratinimicin (ker from Amycolatopsis keratiniphila NRRL B-24117) (Xu et al., 2019), and A50926 (from Nonomuraea coxensis DSM 45129) (Yushchuk et al., 2021) are also reviewed. Overall, multiple recent findings on dalbaheptide glycosylation updated the overall picture and merit a proper review, outlining what is known and why it is still worthy of further investigations.
Delineating Steps in Dalbaheptide Glycosylation
The biosynthesis of dalbaheptides is generally divided into three distinct stages (Yim et al., 2014b, 2016; Yushchuk et al., 2020b), that is, 1) generation of non-proteinogenic aa pool, further utilized in 2) non-ribosomal biosynthesis of the oligopeptide aglycones (coupled with the oxidative cross-linking); fully cross-linked aglycones are further 3) modified in a variety of tailoring steps. All dalbaheptide BGCs encode GTs, tailoring enzymes significantly contributing to the structural variety of these antibiotics (Nicolaou et al., 1999). Non-glycosylated dalbaheptide A47934 (from Streptomyces toyocaensis NRRL 15009) is the only exception here; consistently, the corresponding BGC lacks GT genes (Pootoolal et al., 2002).
More in detail, different steps might be defined in the glycosylation process of dalbaheptides, layer by layer “wrapping” the aglycone. The first step includes the biosynthesis of non-conventional sugar donors for aglycone decoration. Indeed, while some dalbaheptides are decorated with sugars deriving from primary metabolism (e.g., α-d-mannose and N-acetylglucosamine (GlcNAc) in teicoplanin or A40926), aglycones of vancomycin, balhimycin, and chloroeremomycin are decorated with the non-conventional monosaccharides l-vancosamine, l-4-oxovancosamine, and l-epivancosamine, respectively. In a similar manner to the biosynthesis of non-proteinogenic aa, enzymes required for the biosynthesis of such non-conventional monosaccharides are encoded within dalbaheptide BGCs. The second step (often the last one) consists of O-glycosylation of the aromatic aa forming the aglycone. In the third one, the installed sugars might be further modified in minor or major ways (e.g., α-d-mannose O-acetylation and GlcNAc deacetylation in A40926 biosynthesis).
Biosynthesis of Non-Conventional Monosaccharides was Required for the Glycosylation of Dalbaheptides
Conventional sugars in GPAs‐from primary metabolism‐are d-mannose, d-glucose, d-arabinose, GlcNAc, and l-rhamnose. Non-conventional sugar residues include l-vancosamine, l-epivancosamine, l-4-oxovancosamine, l-ristosamine, and l-actinosamine. In addition to the aforementioned examples, l-vancosamine is present in Substitute with UK-68,597, while l-ristosamine is characteristic for ristocetin, and l-actinosamine for keratinimicin (Xu et al., 2019).
Biosynthesis of l-epivancosamine was initially studied in chloroeremomycin producer (van Wageningen et al., 1998). In vitro experiments (Chen et al., 2000; Kirkpatrick et al., 2000) demonstrated how five enzymes encoded within cep‐namely, EvaA-E‐transformed dTDP-4-oxo-6-deoxy-d-glucose into l-epivancosamine (Figure 1). Biosynthetic routes for aminosugars decorating other dalbaheptides were deduced from this model pathway. Initially activated substrate dTDP-4-oxo-6-deoxy-d-glucose is commonly derived from d-glucose-1-phosphate by the action of non-BGC-encoded enzymes (Figure 1). One notable exception is the UK-68,597 biosynthesis, since auk contains a gene for glucose 1-phosphate thymidylylransferase‐auk7 (Yim et al., 2014a)‐required for the D-glucose-1-phosphate activation. The presence of Auk7 likely positively contributes to the pool of l-vancosamine precursors in UK-68,597 biosynthesis.
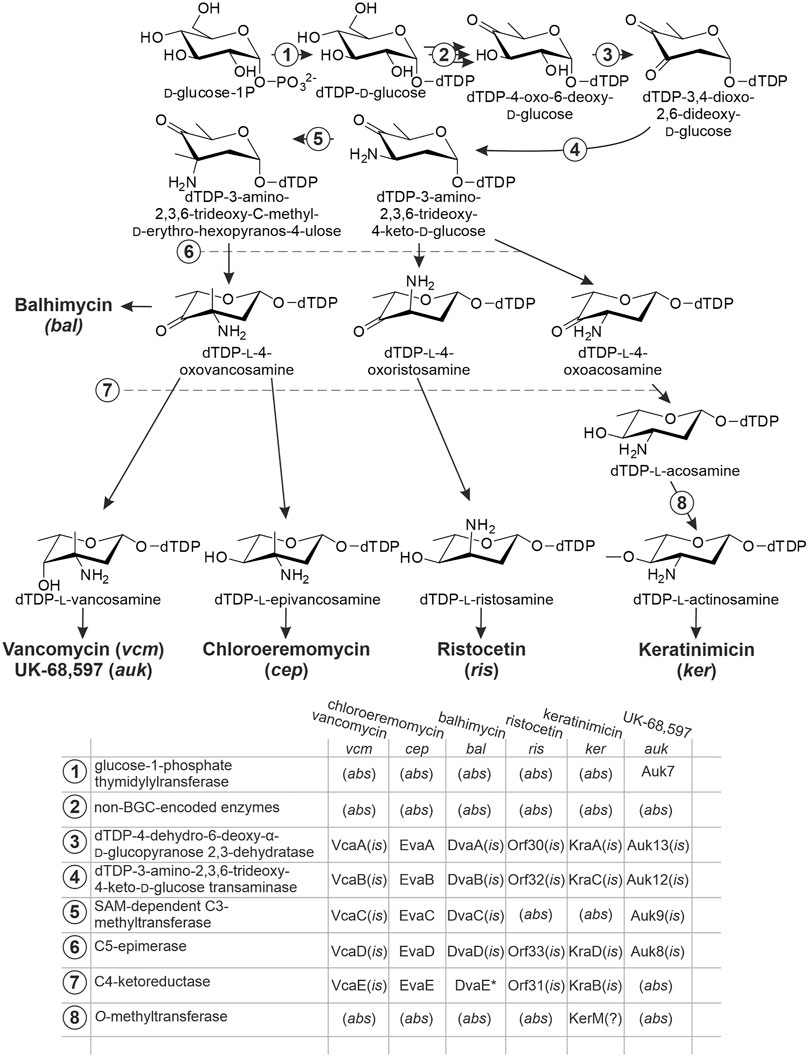
FIGURE 1. Enzymes involved in the biosynthesis of non-conventional aminosugars, decorating aglycones of some dalbaheptides. Biosynthetic pathway of l-epivancosamine serves as a model, since EvaA-B-C-D-E, coded within cep, are the only enzymes that were studied experimentally. Functions of all other enzymes were assigned by in silico comparison (is); (abs) indicates that the corresponding gene is absent from BGCs; (?) indicates that the assigned function is speculative, having no experimentally investigated prototype. Asterisk at DvaE indicates that this protein is mutated. Refer to the main text for more details.
vcm for vancomycin contains the orthologs of evaA-E genes, namely, vcaA-E (Xu et al., 2014) (Figure 1). The two sets of proteins are quite similar, sharing at least 75% of aa sequence identity (aa s.i.) in each pair (Xu et al., 2014). Nevertheless, minor differences in EvaE and VcaE seem to impact the function, making the first to convert dTDP-l-4-oxovancosamine into dTDP-l-epivancosamine, while the second yields dTDP-l-vancosamine (Figure 1). Instead, auk contains the orthologs of only evaA, evaB, evaC, and evaD (auk13, auk12, auk9, and auk8, respectively) (Yim et al., 2014a), lacking the ortholog of evaE: probably a functional homolog (or analog) of evaE resides outside the auk borders, finally contributing to l-vancosamine production. More understandable is the case of bal (Shawky et al., 2007), where evaA-E orthologs are named dvaA-E. Here, evaE ortholog dvaE is truncated, coding only the 99 aa C-terminal part of C4-ketoreductase (Donadio et al., 2005). Such truncated protein is non-functional, and consequently, the aminosugar biosynthesis terminates at the stage of dTDP-l-4-oxovancosamine (Figure 1). Biosynthetic pathways for l-ristosamine and l-actinosamine, coded within ris and ker, are more diverged. They both missed the EvaC ortholog (Spohn et al., 2014; Truman et al., 2014; Xu et al., 2019; Liu et al., 2021), resulting in the lack of a methyl group at the C3 position (Figure 1). Thus, biosynthesis of l-ristosamine might be attributed to the orthologs of EvaA, EvaB, EvaD, and EvaE‐Orf30, Orf32, Orf33, and Orf31, respectively (Truman et al., 2014). The same protein set is encoded within ker‐KraA-D (Xu et al., 2019). The major difference between l-actinosamine and l-ristosamine is O-methylation of the C4-position. It is still unknown how this methylation is achieved; however, the annotation of ker reveals the presence of a gene coding for an O-methyltransferase‐kerM (Xu et al., 2019). Since the aglycone of keratinimicin lacks any O-methylations, it seems plausible that KerM catalyzes the ultimate step of l-actinosamine biosynthesis (Figure 1).
In all cases described above the orthologs of evaA-E-B-D are most likely co-expressed, forming one operon, while the orthologs of evaC belong to a separate transcriptional unit (Shawky et al., 2007; Liu et al., 2021). auk is an exception, with auk8-9 (evaD-C) and auk12-13 (evaA-B) probably belonging to different operons (Yim et al., 2014a).
GTs Involved in Dalbaheptide Glycosylation
All GTs decorating aglycones of dalbaheptides belong to two families, according to the Carbohydrate-Active enZYmes Database (CAZy, http://www.cazy.org, Drula et al., 2022). GTs responsible for the installation of non-conventional aminosugars and conventional d-glucose, d-arabinose, GlcNAc, and l-rhamnose, belong to the GT1 family. These GTs require sugar substrates to be either dTDP- or UDP-activated, and share a unique two-domain structure (the so-called GT-B fold, Lairson et al., 2008), having C- and N-terminal Rossmann-like domains connected by a flexible linker region (Zhang et al., 2020). Recognition sites for the donor NDP-activated sugars are located at C-terminal domains (Chang et al., 2011), while N-terminal domains contain the acceptor binding site for dalbaheptide aglycone (Chang et al., 2011; Zhang et al., 2020). GTs of the second‐GT39‐family are responsible for the installation of d-mannose and require d-mannosyl-1-phosphoundecaprenol as a donor substrate. Large hydrophobic GT39-GTs are predicted as membrane-associated, having a GT-C fold with multiple transmembrane helices and intracellular active sites (Lairson et al., 2008).
In the aforementioned dalbaheptides, GT1-GTs attach sugar residues preferentially at AA-4 (4-hydroxyphenylglycine, Hpg) and AA-6 (β-hydroxytyrosine, Bht) of the aglycone or add additional monosaccharides to already existing mono/di/trisaccharides at AA-4. Fully cross-linked aglycones serve as acceptor substrates for GT1-GTs under physiological conditions, albeit some GT1-GTs were able to recognize partially cross-linked aglycones under certain experimental conditions, for instance, in mutasynthesis approaches (Weist et al., 2004; Butz et al., 2008). GT39-GTs attach mannose at AA-7 (3,5-dihydroxyphenylglycine, Dpg). The presence of multiple GTs within one pathway might result in the production of mixtures of related congeners, differing in glycosylation patterns. Dalbaheptides glycosylated at other AA positions were also described (Nicolaou et al., 1999), indicating that glycosylation might also occur at 1) AA-2 (Bht) and AA-1 (Hpg) in type II aglycone, and 2) AA-1 (Hpg) and AA-3 (Bht) in type III aglycone. Unfortunately, we currently lack any genomic information on the producers of these molecules, which would merit further investigations.
Type I dalbaheptides chloroeremomycin, balhimycin, and vancomycin served as the first models for experimental investigation of GT functions. Corresponding BGCs encode slightly different sets of GT1-GTs: GtfA, GtfB, and GtfC in cem; BgtfA, BgtfB, and BgtfC in bal; and GtfD and GtfE in vcm. Orthologous GTs GtfA and BgtfA install l-epivancosamine and l-4-oxovancosamine at AA-6 of chloroeremomycin and balhimycin, respectively. vcm does not encode GtfA ortholog, explaining why vancomycin is not glycosylated at AA-6. Peculiarly, this particular difference between vancomycin and chloroeremomycin seems to augment the antimicrobial activity of the latter, implying that l-epivancosamine at AA-6 facilitates cell wall binding (Nagarajan, 1993; Allen et al., 2002). GtfB, BgtfB, and GtfE are orthologs, glucosylating AA-4 in the biosynthesis of all three antibiotics (Figure 2A, Pelzer et al., 1999; Losey et al., 2001; Mulichak et al., 2001). Then, GtfC and GtfD attach l-epivancosamine or l-vancosamine to d-glucose at AA-4 in the biosynthesis of chloroeremomycin and vancomycin, respectively (Figure 2A, Losey et al., 2001, 2002; Mulichak et al., 2004). GtfC, GtfD, and BgtfC are orthologous proteins, but balhimycin lacks a disaccharide at AA-4, which could be found only in balhimycin V (a congener produced in residual amounts) (Pelzer et al., 1999; Stegmann et al., 2010). This might be due to the low affinity of BgtfC for the donor substrate‐l-4-oxovancosamine, produced as a consequence of DvaE mutation. More recently described type I dalbaheptide pekiskomycin is only glucosylated at AA-4, coherently with the only 1 GT encoded in pek: Pek28, which is a GtfB ortholog (Figure 2A) (Thaker et al., 2013).
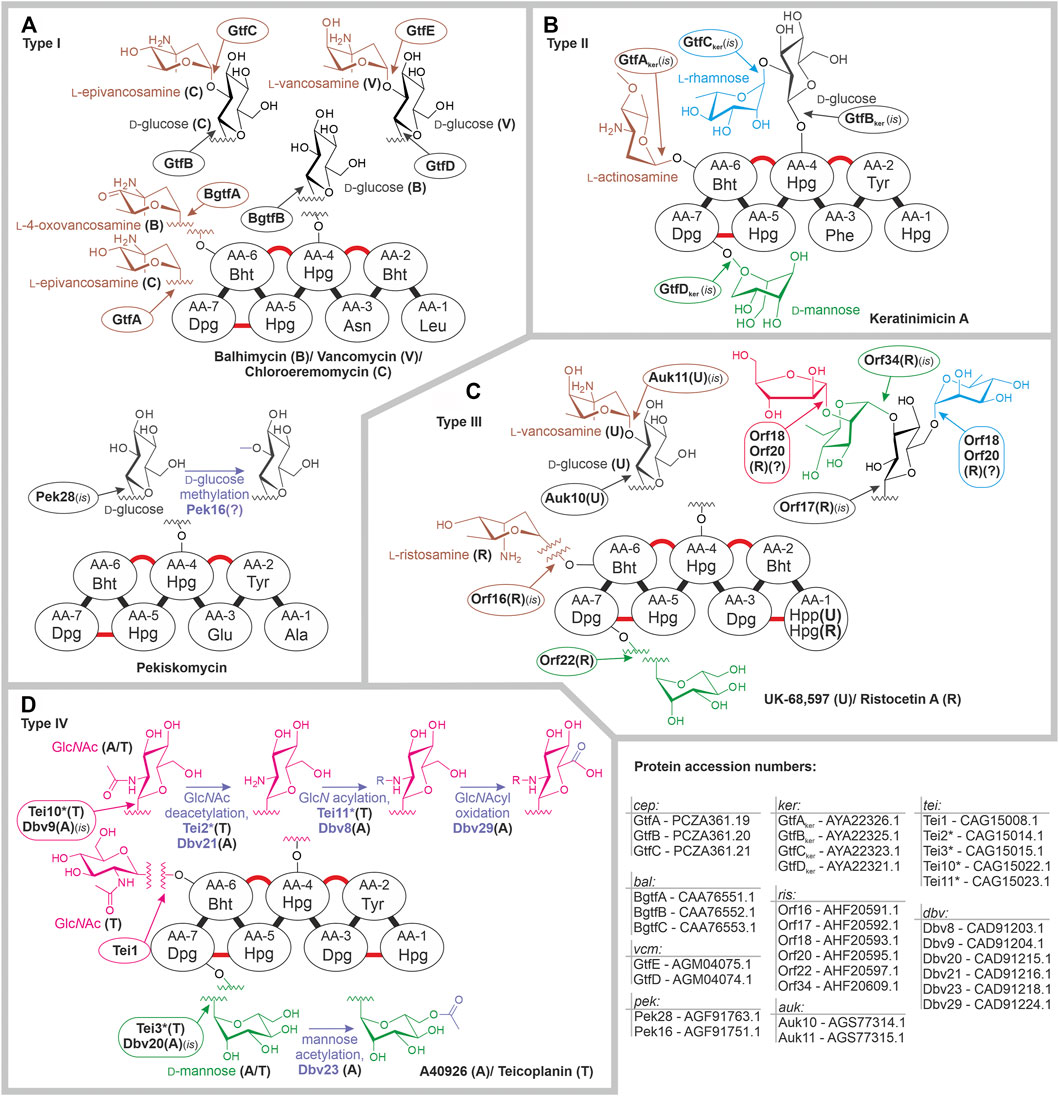
FIGURE 2. Glycosylation patterns of type I (A), type II (B), type III (C), and type IV (D) dalbaheptides and enzymes involved therein. Refer to the main text for more details. Dalbaheptide aglycones are depicted schematically with cross-links shown in red, and chlorination and sulfation sites are not shown. Aglycone amino acid abbreviations mean following: Leu‐leucine; Asn‐asparagine; Ala‐alanine; Glu‐glutamine; Phe‐phenylalanine; Tyr‐tyrosine; Bht‐ß-hydroxytyrosine; Dpg‐3,5-dihydroxyphenylglycine; Hpg‐4-hydroxyphenylglycine; Hpp‐4-hydroxyphenypyruvate; Tyr‐. GlcN stated for N-glucosamine, GlcNAcyl for N-acylglucosamine. All enzymes, whose functions were assigned by in silico comparison are marked with (is); (?) indicates that function was assigned in silico without experimentally investigated prototype or exact function remains unknown. For fast access to protein sequences mentioned in this figure, use following links: cep: GtfA‐PCZA361.19, GtfB‐PCZA361.20, GtfC‐PCZA361.21; bal: BgtfA‐CAA76551.1, BgtfB‐CAA76552.1, BgtfC‐CAA76553.1; vcm: GtfE‐AGM04075.1, GtfD‐AGM04074.1; pek: Pek28‐AGF91763.1, Pek16‐AGF91751.1; ker: GtfAker‐AYA22326.1, GtfBker‐AYA22325.1, GtfCker‐AYA22323.1, GtfDker‐AYA22321.1; ris: Orf16‐AHF20591.1, Orf17‐AHF20592.1, Orf18‐AHF20593.1, Orf20‐AHF20595.1; Orf22‐AHF20597.1, Orf34‐AHF20609.1; auk: Auk10‐AGS77314.1, Auk11‐AGS77315.1; tei: Tei1‐CAG15008.1, Tei2*‐CAG15014.1, Tei3*‐CAG15015.1, Tei10*‐CAG15022.1, Tei11*‐CAG15023.1; dbv: Dbv8‐CAD91203.1, Dbv9‐CAD91204.1, Dbv20‐CAD91215.1, Dbv21‐CAD91216.1, Dbv23‐CAD91218.1, Dbv29‐CAD91224.1.
So far, little is known about GTs decorating aglycones of type II dalbaheptides. ker (single type II BGC sequenced) carries three genes for GT1-GTs and one gene for GT39-GT (Xu et al., 2019). Among ker-encoded GT1-GTs, gtfAker, gtfBker, and gtfCker (ker was added to distinguish them from cep genes) are orthologs of gtfA, gtfB, and gtfC, respectively (Xu et al., 2019). Thus, GtfBker most likely installs d-glucose at AA-4, and l-rhamnose is then appended to d-glucose by GtfCker; this leaves GtfAker responsible for the attachment of l-actinosamine at AA-6 (Figure 2B). Single ker-encoded GT39-GT GtfDker most probably attaches d-mannose at AA-7 (Figure 2B).
More information is available on the GTs involved in the biosynthesis of type III dalbaheptides. Ristocetin BGC encodes 6 GTs, four of GT1 family and two belonging to the GT39 family (Truman et al., 2014). Phylogenetic reconstruction allowed to assign functions to 4 GTs, assuming that Orf16 attaches l-ristosamine at AA-6, Orf17‐d-glucose at AA-4, then appended with d-mannose by Orf34; finally, a second mannosyltransferase‐Orf22‐was expected to act at AA-7 (Figure 2C, Truman et al., 2014), as later confirmed by its heterologous expression in Streptomyces coelicolor carrying sta (Yim et al., 2016). Functions of Orf18 and Orf20 were not assigned, since these proteins were distantly related to known GTs (Truman et al., 2014). Another type III dalbaheptide‐UK-68,597‐is decorated with 2-L-vancosaminyl-d-glucose disaccharide at AA-4 (Yim et al., 2014a), while auk carries three genes for GT1-GTs. In vitro assay showed that Auk10 (GtfB ortholog) is responsible for glucosylation; in silico analysis then suggested l-vancosamine to be installed by Auk11 (GtfC ortholog, Figure 2C) (Yim et al., 2014a). Although the third GT, Auk14, is a GtfA ortholog, UK-68,597 lacks sugar residues at AA-6. In vitro assay suggested that Auk14 might be inactive or possess a very low affinity to substrates available in UK-68,597 biosynthesis, as observed with BgtfA (Yim et al., 2014a).
Glycosylation events that take place in type IV dalbaheptide biosynthesis are well defined (Figure 2D). tei and dbv BGCs encode three and two GTs, respectively. Accordingly, both antibiotics carry a GlcNAc moiety at AA-4 and d-mannose at AA-7, while teicoplanin aglycone is also decorated with another GlcNAc at AA-6. GtfB ortholog‐Tei10*‐installs the GlcNAc moiety at AA-4, as demonstrated from multiple in vivo and in vitro experiments (Li et al., 2004; Howard-Jones et al., 2007; Truman et al., 2009; Yushchuk et al., 2016, 2020b). Consistently, Dbv9‐a Tei10* ortholog‐is supposed to play the same role in A40926 glycosylation. Tei1 was shown to attach GlcNAc to teicoplanin aglycone at AA-6 (Li et al., 2004), whereas dbv lacks any Tei1 ortholog, explaining why A40926 has no sugars at AA-6. Finally, Tei3* was shown to be responsible for the decoration of teicoplanin aglycone with d-mannose at AA-7 (Yushchuk et al., 2016), implying that Dbv20 (Tei3* ortholog) has the same function in A40926 biosynthesis (Figure 2D). Interestingly, ramoplanin BGC (ramo), recently shown to be genetically related to tei (Waglechner et al., 2019), encodes a homolog of Tei3*‐Ramo29 (45% aa s.i., Chen et al., 2013). Ramoplanin is a clinically relevant peptide antibiotic produced by Actinoplanes ramoplaninifer ATCC 33076 (Marcone et al., 2017). Unlike dalbaheptides, it carries a 4-d-mannosyl-d-mannose disaccharide, instead of a single d-mannose residue. Ramo29 was shown to install the first d-mannose residue (Chen et al., 2013), but the GT responsible for the second mannosylation remains unknown. This merits further investigation, since such GT looks like a promising tool to further modify mannosylated dalbaheptides.
Concluding this section, it is interesting to report that genes for GTs tend to form one operon in type I-III BGCs (Shawky et al., 2007; Liu et al., 2021), being more scattered in type IV BGCs (Alduina et al., 2007; Yushchuk et al., 2019). It is also notable that genes for mannosyltransferases are present in different types of BGCs coming from distant actinobacterial lineages. This might indicate d-mannose residues at AA-7 to be an ancestral feature for all dalbaheptides.
Further Modification Occurring on Attached Sugar Residues
Some further modifications of attached sugars occur during the biosynthesis of dalbaheptides, although they are quite rare. The first example comes from pekiskomycin, having d-glucose methylated. pek encodes two methyltransferases. One of them‐Pek30‐was experimentally shown to methylate the N-terminus of A47934 aglycone (Yim et al., 2016), leaving the other‐Pek16‐possibly responsible for d-glucose methylation (Figure 2A). Another notable modification is the acylation of AA-4 GlcNAc in type IV dalbaheptides, such as teicoplanin and A40926. To achieve this modification, GlcNAc at AA-4 is first deacetylated with orthologous deacetylases Tei2*/Dbv21 (Ho et al., 2006; Truman et al., 2006), and N-glucosamine is then acylated with orthologous acyltransferases Tei11*/Dbv8 (Figure 2D) (Li et al., 2004; Kruger et al., 2005; Howard-Jones et al., 2007; Yushchuk et al., 2016). Peculiarly, orthologs of Tei2*/Dbv21 are present in many (if not in all) BGCs for non-acylated type I-III dalbaheptides. The one from cep‐CepI‐was studied in vitro and shown to be inactive due to a single aa substitution (Truman et al., 2008). The omnipresence of tei2* orthologs in type I-III BGCs induces to speculate that the N-acylglucosamine moiety at AA-4 is an ancestral feature, lost or modified in many evolutionary lineages of dalbaheptides.
Modifications of A40926 sugars do not end with acylation. N-acylglucosamine moiety is further oxidized to N-acylaminoglucuronic acid group by Dbv29 (Figure 2D) (Li et al., 2007). The biological role of such modification is unclear, although it seems to reduce the A40926 antimicrobial activity (Malabarba et al., 1995). Peculiarly, noc BGC in N. coxensis DSM 45129 lacks an ortholog for dbv29, coding the biosynthesis of non-oxidized A40926 analog‐dalbaheptide A50926 (Yushchuk et al., 2021). Finally, the d-mannose residue at AA-7 of A40926 is O-acetylated with Dbv23 (Figure 2D) (Sosio et al., 2010). This modification is unstable and fades away in the alkaline extraction of A40926 (Alt et al., 2019); once again, its biological role is unclear, although it might be important for the regulation of antibiotic export and self-resistance (Alduina et al., 2020).
Conclusion and Outlook
Further research of dalbaheptide glycosylation is important for several reasons. Understanding of GTs donor- (activated sugar) and acceptor- (aglycone) substrate specificities will allow further chemical derivatization of these scaffolds using in vitro chemo-enzymatic synthesis (Nakayama et al., 2014) or in vivo combinatorial biosynthesis (Yim et al., 2016). While the first approach has been widely investigated in the past for generating novel hybrid GPAs by combining natural and synthetic aglycones and sugars (as reviewed in Marcone et al., 2018), in vivo combinatorial biosynthesis is promising, but still rather limited in its applications. Alternatively, the two-domain architecture of GT1-GTs might be exploited to create “chimaeras” with an expanded functional repertoire (Truman et al., 2009). Finally, a better comprehension of glycosylation mechanisms will contribute to tracing out a more complete picture of dalbaheptide evolution. Unlike other aspects of glycopeptide biosynthesis (Donadio et al., 2005; Waglechner et al., 2019; Andreo-Vidal et al., 2021), phylogenomics of GTs and sugar modification enzymes has not been studied yet. We believe that such reconstruction might open new scenarios on the evolution of antibiotic biosynthetic pathways.
Author Contributions
KZ, FB, and OY collected data and articles and co-wrote the review; OY prepared the figures; and VF and FM supervised the work and co-wrote the review.
Funding
This work was supported by grant “Fondo di Ateneo per la Ricerca” to FM and FB and by the BG-09F grant of the Ministry of Education and Science of Ukraine to VF.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Consorzio Interuniversitario per le Biotecnologie for supporting FB congresses attendances.
References
Alduina, R., Piccolo, L. L., D'Alia, D., Ferraro, C., Gunnarsson, N., Donadio, S., et al. (2007). Phosphate-controlled Regulator for the Biosynthesis of the Dalbavancin Precursor A40926. J. Bacteriol. 189, 8120–8129. doi:10.1128/JB.01247-07
Alduina, R., Tocchetti, A., Costa, S., Ferraro, C., Cancemi, P., Sosio, M., et al. (2020). A Two-Component Regulatory System with Opposite Effects on Glycopeptide Antibiotic Biosynthesis and Resistance. Sci. Rep. 10, 1–12. doi:10.1038/s41598-020-63257-4
Allen, N. E., LeTourneau, D. L., Hobbs, J. N., and Thompson, R. C. (2002). Hexapeptide Derivatives of Glycopeptide Antibiotics: Tools for Mechanism of Action Studies. Antimicrob. Agents Chemother. 46, 2344–2348. doi:10.1128/AAC.46.8.2344-2348.2002
Alt, S., Bernasconi, A., Sosio, M., Brunati, C., Donadio, S., and Maffioli, S. I. (2019). Toward Single-Peak Dalbavancin Analogs through Biology and Chemistry. ACS Chem. Biol. 14, 356–360. doi:10.1021/acschembio.9b00050
Andreo-Vidal, A., Binda, E., Fedorenko, V., Marinelli, F., and Yushchuk, O. (2021). Genomic Insights into the Distribution and Phylogeny of Glycopeptide Resistance Determinants within the Actinobacteria Phylum. Antibiotics 10, 1533–1630. doi:10.3390/antibiotics10121533
Beauregard, D. A., Williams, D. H., Gwynn, M. N., and Knowles, D. J. (1995). Dimerization and Membrane Anchors in Extracellular Targeting of Vancomycin Group Antibiotics. Antimicrob. Agents Chemother. 39, 781–785. doi:10.1128/AAC.39.3.781
Binda, E., Marinelli, F., and Marcone, G. (2014). Old and New Glycopeptide Antibiotics: Action and Resistance. Antibiotics 3, 572–594. doi:10.3390/antibiotics3040572
Butler, M. S., Hansford, K. A., Blaskovich, M. A. T., Halai, R., and Cooper, M. A. (2014). Glycopeptide Antibiotics: Back to the Future. J. Antibiot. 67, 631–644. doi:10.1038/ja.2014.111
Butz, D., Schmiederer, T., Hadatsch, B., Wohlleben, W., Weber, T., and Süssmuth, R. D. (2008). Module Extension of a Non-ribosomal Peptide Synthetase of the Glycopeptide Antibiotic Balhimycin Produced by Amycolatopsis balhimycina. ChemBioChem 9, 1195–1200. doi:10.1002/cbic.200800068
Chang, A., Singh, S., Phillips, G. N., and Thorson, J. S. (2011). Glycosyltransferase Structural Biology and its Role in the Design of Catalysts for Glycosylation. Curr. Opin. Biotechnol. 22, 800–808. doi:10.1016/j.copbio.2011.04.013
Chen, H., Thomas, M. G., Hubbard, B. K., Losey, H. C., Walsh, C. T., and Burkart, M. D. (2000). Deoxysugars in Glycopeptide Antibiotics: Enzymatic Synthesis of TDP-L-Epivancosamine in Chloroeremomycin Biosynthesis. Proc. Natl. Acad. Sci. 97, 11942–11947. doi:10.1073/pnas.210395097
Chen, J.-S., Wang, Y.-X., Shao, L., Pan, H.-X., Li, J.-A., Lin, H.-M., et al. (2013). Functional Identification of the Gene Encoding the Enzyme Involved in Mannosylation in Ramoplanin Biosynthesis in Actinoplanes Sp. Biotechnol. Lett. 35, 1501–1508. doi:10.1007/s10529-013-1233-3
Coller, B. S., and Gralnick, H. R. (1977). Studies on the Mechanism of Ristocetin-Induced Platelet Agglutination. J. Clin. Invest. 60, 302–312. doi:10.1172/JCI108778
Crotty, M. P., Krekel, T., Burnham, C.-A. D., and Ritchie, D. J. (2016). New Gram-Positive Agents: the Next Generation of Oxazolidinones and Lipoglycopeptides. J. Clin. Microbiol. 54, 2225–2232. doi:10.1128/JCM.03395-15
Donadio, S., Sosio, M., Stegmann, E., Weber, T., and Wohlleben, W. (2005). Comparative Analysis and Insights into the Evolution of Gene Clusters for Glycopeptide Antibiotic Biosynthesis. Mol. Genet. Genomics 274, 40–50. doi:10.1007/s00438-005-1156-3
Drula, E., Garron, M.-L., Dogan, S., Lombard, V., Henrissat, B., and Terrapon, N. (2022). The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 50, D571–D577. doi:10.1093/nar/gkab1045
Gangarosa, E. J., Landerman, N. S., Rosch, P. J., and Herndon, E. G. (1958). Hematologic Complications Arising during Ristocetin Therapy. N. Engl. J. Med. 259, 156–161. doi:10.1056/nejm195807242590402
Gerhard, U., Mackay, J. P., Maplestone, R. A., and Williams, D. H. (1993). The Role of the Sugar and Chlorine Substituents in the Dimerization of Vancomycin Antibiotics. J. Am. Chem. Soc. 115, 232–237. doi:10.1021/ja00054a033
Ho, J.-Y., Huang, Y.-T., Wu, C.-J., Li, Y.-S., Tsai, M.-D., and Li, T.-L. (2006). Glycopeptide Biosynthesis: Dbv21/Orf2* from dbv/tcp Gene Clusters Are N-Ac-Glm Teicoplanin Pseudoaglycone Deacetylases and Orf15 from cep Gene Cluster Is a Glc-1-P Thymidyltransferase. J. Am. Chem. Soc. 128, 13694–13695. doi:10.1021/ja0644834
Howard, M. A., and Firkin, B. G. (1971). Ristocetin - A New Tool in the Investigation of Platelet Aggregation. Thromb. Haemost. 26, 362–369. doi:10.1055/s-0038-1653684
Howard-Jones, A. R., Kruger, R. G., Lu, W., Tao, J., Leimkuhler, C., Kahne, D., et al. (2007). Kinetic Analysis of Teicoplanin Glycosyltransferases and Acyltransferase Reveal Ordered Tailoring of Aglycone Scaffold to Reconstitute Mature Teicoplanin. J. Am. Chem. Soc. 129, 10082–10083. doi:10.1021/ja0735857
Hutchings, M. I., Truman, A. W., and Wilkinson, B. (2019). Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 51, 72–80. doi:10.1016/j.mib.2019.10.008
Jovetic, S., Zhu, Y., Marcone, G. L., Marinelli, F., and Tramper, J. (2010). β-Lactam and Glycopeptide Antibiotics: First and Last Line of Defense? Trends Biotechnol. 28, 596–604. doi:10.1016/j.tibtech.2010.09.004
Kirkpatrick, P. N., Scaife, W., Spencer, J. B., Williams, D. H., Hallis, T. M., and Liu, H.-w. (2000). Characterisation of a Sugar Epimerase Enzyme Involved in the Biosynthesis of a Vancomycin-Group Antibiotic. Chem. Commun. 17, 1565–1566. doi:10.1039/b004463f
Klinker, K. P., and Borgert, S. J. (2015). Beyond Vancomycin: the Tail of the Lipoglycopeptides. Clin. Ther. 37, 2619–2636. doi:10.1016/j.clinthera.2015.11.007
Kruger, R. G., Lu, W., Oberthür, M., Tao, J., Kahne, D., and Walsh, C. T. (2005). Tailoring of Glycopeptide Scaffolds by the Acyltransferases from the Teicoplanin and A-40,926 Biosynthetic Operons. Chem. Biol. 12, 131–140. doi:10.1016/j.chembiol.2004.12.005
Lairson, L. L., Henrissat, B., Davies, G. J., and Withers, S. G. (2008). Glycosyltransferases: Structures, Functions, and Mechanisms. Annu. Rev. Biochem. 77, 521–555. doi:10.1146/annurev.biochem.76.061005.092322
Li, T., Huang, F., Haydock, S., Mironenko, T., Leadlay, P., and Spencer, J. (2004). Biosynthetic Gene Cluster of the Glycopeptide Antibiotic Teicoplanin Characterization of Two Glycosyltransferases and the Key Acyltransferase. Chem. Biol. 11, 107–119. doi:10.1016/S1074-5521(04)00002-X
Li, Y.-S., Ho, J.-Y., Huang, C.-C., Lyu, S.-Y., Lee, C.-Y., Huang, Y.-T., et al. (2007). A Unique Flavin Mononucleotide-Linked Primary Alcohol Oxidase for Glycopeptide A40926 Maturation. J. Am. Chem. Soc. 129, 13384–13385. doi:10.1021/ja075748x
Liu, K., Hu, X.-R., Zhao, L.-X., Wang, Y., Deng, Z., and Tao, M. (2021). Enhancing Ristomycin A Production by Overexpression of ParB-like StrR Family Regulators Controlling the Biosynthesis Genes. Appl. Environ. Microbiol. 87, 1–19. doi:10.1128/AEM.01066-21
Losey, H. C., Jiang, J., Biggins, J. B., Oberthür, M., Ye, X.-Y., Dong, S. D., et al. (2002). Incorporation of Glucose Analogs by GtfE and GtfD from the Vancomycin Biosynthetic Pathway to Generate Variant Glycopeptides. Chem. Biol. 9, 1305–1314. doi:10.1016/S1074-5521(02)00270-3
Losey, H. C., Peczuh, M. W., Chen, Z., Eggert, U. S., Dong, S. D., Pelczer, I., et al. (2001). Tandem Action of Glycosyltransferases in the Maturation of Vancomycin and Teicoplanin Aglycones: Novel Glycopeptides,. Biochemistry 40, 4745–4755. doi:10.1021/bi010050w
Mackay, J. P., Gerhard, U., Beauregard, D. A., Maplestone, R. A., and Williams, D. H. (1994). Dissection of the Contributions toward Dimerization of Glycopeptide Antibiotics. J. Am. Chem. Soc. 116, 4573–4580. doi:10.1021/ja00090a005
Malabarba, A., Ciabatti, R., Scotti, R., Goldstein, B. P., Ferrari, P., Kurz, M., et al. (1995). New Semisynthetic Glycopeptides MDL 63,246 and MDL 63,042, and Other Amide Derivatives of Antibiotic A-40,926 Active against Highly Glycopeptide-Resistant VanA Enterococci. J. Antibiot. 48, 869–883. doi:10.7164/antibiotics.48.869
Marcone, G. L., Binda, E., Berini, F., and Marinelli, F. (2018). Old and New Glycopeptide Antibiotics: from Product to Gene and Back in the post-genomic Era. Biotechnol. Adv. 36, 534–554. doi:10.1016/j.biotechadv.2018.02.009
Marcone, G. L., Binda, E., Reguzzoni, M., Gastaldo, L., Dalmastri, C., and Marinelli, F. (2017). Classification of Actinoplanes Sp. ATCC 33076, an Actinomycete that Produces the Glycolipodepsipeptide Antibiotic Ramoplanin, as Actinoplanes ramoplaninifer Sp. Nov. Int. J. Syst. Evol. Microbiol. 67, 4181–4188. doi:10.1099/ijsem.0.002281
Medema, M. H., Kottmann, R., Yilmaz, P., Cummings, M., Biggins, J. B., Blin, K., et al. (2015). Minimum Information about a Biosynthetic Gene Cluster. Nat. Chem. Biol. 11, 625–631. doi:10.1038/nchembio.1890
Medema, M. H., Cimermancic, P., Sali, A., Takano, E., and Fischbach, M. A. (2014). A Systematic Computational Analysis of Biosynthetic Gene Cluster Evolution: Lessons for Engineering Biosynthesis. Plos Comput. Biol. 10, e1004016–12. doi:10.1371/journal.pcbi.1004016
Mulichak, A. M., Losey, H. C., Walsh, C. T., and Garavito, R. M. (2001). Structure of the UDP-Glucosyltransferase GtfB that Modifies the Heptapeptide Aglycone in the Biosynthesis of Vancomycin Group Antibiotics. Structure 9, 547–557. doi:10.1016/S0969-2126(01)00616-5
Mulichak, A. M., Lu, W., Losey, H. C., Walsh, C. T., and Garavito, R. M. (2004). Crystal Structure of Vancosaminyltransferase GtfD from the Vancomycin Biosynthetic Pathway: Interactions with Acceptor and Nucleotide Ligands,. Biochemistry 43, 5170–5180. doi:10.1021/bi036130c
Nagarajan, R. (1993). Structure-activity Relationships of Vancomycin-type Glycopeptide Antibiotics. J. Antibiot. 46, 1181–1195. doi:10.7164/antibiotics.46.1181
Nakayama, A., Okano, A., Feng, Y., Collins, J. C., Collins, K. C., Walsh, C. T., et al. (2014). Enzymatic Glycosylation of Vancomycin Aglycon: Completion of a Total Synthesis of Vancomycin and N- and C-Terminus Substituent Effects of the Aglycon Substrate. Org. Lett. 16, 3572–3575. doi:10.1021/OL501568T/SUPPL_FILE/OL501568T_SI_001.PDF
Nicolaou, K. C., Boddy, C. N. C., Bräse, S., and Winssinger, N. (1999). Chemistry, Biology, and Medicine of the Glycopeptide Antibiotics. Angew. Chem. Int. Ed. 38, 2096–2152. doi:10.1002/(sici)1521-3773(19990802)38:15<2096::aid-anie2096>3.0.co;2-f
Parenti, F., and Cavalleri, B. (1989). Proposal to Name the Vancomycin-Ristocetin like Glycopeptides as Dalbaheptides. J. Antibiot. 42, 1882–1883. doi:10.7164/antibiotics.42.1882
Pelzer, S., Süßmuth, R., Heckmann, D., Recktenwald, J., Huber, P., Jung, G., et al. (1999). Identification and Analysis of the Balhimycin Biosynthetic Gene Cluster and its Use for Manipulating Glycopeptide Biosynthesis in Amycolatopsis mediterranei DSM5908. Antimicrob. Agents Chemother. 43, 1565–1573. doi:10.1128/aac.43.7.1565
Pootoolal, J., Thomas, M. G., Marshall, C. G., Neu, J. M., Hubbard, B. K., Walsh, C. T., et al. (2002). Assembling the Glycopeptide Antibiotic Scaffold: The Biosynthesis of from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. 99, 8962–8967. doi:10.1073/pnas.102285099
Salas, J. A., and Méndez, C. (2007). Engineering the Glycosylation of Natural Products in Actinomycetes. Trends Microbiol. 15, 219–232. doi:10.1016/j.tim.2007.03.004
Shawky, R. M., Puk, O., Wietzorrek, A., Pelzer, S., Takano, E., Wohlleben, W., et al. (2007). The Border Sequence of the Balhimycin Biosynthesis Gene Cluster from Amycolatopsis balhimycina Contains bbr, Encoding a StrR-like Pathway-specific Regulator. J. Mol. Microbiol. Biotechnol. 13, 76–88. doi:10.1159/000103599
Snyder, N. J., Cooper, R. D. G., Briggs, B. S., Zmijewski, M., Mullen, D. L., Kaiser, R. E., et al. (1998). Enzymatic Deacylation of Teicoplanin Followed by Reductive Alkylation: Synthesis and Antibacterial Activity of New Glycopeptides. J. Antibiot. 51, 945–951. doi:10.7164/antibiotics.51.945
Sosio, M., Canavesi, A., Stinchi, S., and Donadio, S. (2010). Improved Production of A40926 by Nonomuraea Sp. Through Deletion of a Pathway-specific Acetyltransferase. Appl. Microbiol. Biotechnol. 87, 1633–1638. doi:10.1007/s00253-010-2579-2
Sosio, M., Stinchi, S., Beltrametti, F., Lazzarini, A., and Donadio, S. (2003). The Gene Cluster for the Biosynthesis of the Glycopeptide Antibiotic A40926 by Nonomuraea Species. Chem. Biol. 10, 541–549. doi:10.1016/S1074-5521(03)00120-0
Spohn, M., Kirchner, N., Kulik, A., Jochim, A., Wolf, F., Muenzer, P., et al. (2014). Overproduction of Ristomycin a by Activation of a Silent Gene Cluster in Amycolatopsis japonicum MG417-CF17. Antimicrob. Agents Chemother. 58, 6185–6196. doi:10.1128/AAC.03512-14
Stegmann, E., Frasch, H.-J., and Wohlleben, W. (2010). Glycopeptide Biosynthesis in the Context of Basic Cellular Functions. Curr. Opin. Microbiol. 13, 595–602. doi:10.1016/j.mib.2010.08.011
Thaker, M. N., Wang, W., Spanogiannopoulos, P., Waglechner, N., King, A. M., Medina, R., et al. (2013). Identifying Producers of Antibacterial Compounds by Screening for Antibiotic Resistance. Nat. Biotechnol. 31, 922–927. doi:10.1038/nbt.2685
Truman, A. W., Dias, M. V. B., Wu, S., Blundell, T. L., Huang, F., and Spencer, J. B. (2009). Chimeric Glycosyltransferases for the Generation of Hybrid Glycopeptides. Chem. Biol. 16, 676–685. doi:10.1016/j.chembiol.2009.04.013
Truman, A. W., Fan, Q., Röttgen, M., Stegmann, E., Leadlay, P. F., and Spencer, J. B. (2008). The Role of Cep15 in the Biosynthesis of Chloroeremomycin: Reactivation of an Ancestral Catalytic Function. Chem. Biol. 15, 476–484. doi:10.1016/j.chembiol.2008.03.019
Truman, A. W., Kwun, M. J., Cheng, J., Yang, S. H., Suh, J.-W., and Hong, H.-J. (2014). Antibiotic Resistance Mechanisms Inform Discovery: Identification and Characterization of a Novel Amycolatopsis Strain Producing Ristocetin. Antimicrob. Agents Chemother. 58, 5687–5695. doi:10.1128/AAC.03349-14
Truman, A. W., Robinson, L., and Spencer, J. B. (2006). Identification of a Deacetylase Involved in the Maturation of Teicoplanin. ChemBioChem 7, 1670–1675. doi:10.1002/cbic.200600308
Van Wageningen, A. A., Kirkpatrick, P. N., Williams, D. H., Harris, B. R., Kershaw, J. K., Lennard, N. J., et al. (1998). Sequencing and Analysis of Genes Involved in the Biosynthesis of a Vancomycin Group Antibiotic. Chem. Biol. 5, 155–162. doi:10.1016/S1074-5521(98)90060-6
Waglechner, N., McArthur, A. G., and Wright, G. D. (2019). Phylogenetic Reconciliation Reveals the Natural History of Glycopeptide Antibiotic Biosynthesis and Resistance. Nat. Microbiol. 4, 1862–1871. doi:10.1038/s41564-019-0531-5
Weist, S., Kittel, C., Bischoff, D., Bister, B., Pfeifer, V., Nicholson, G. J., et al. (2004). Mutasynthesis of Glycopeptide Antibiotics: Variations of Vancomycin's AB-Ring Amino Acid 3,5-Dihydroxyphenylglycine. J. Am. Chem. Soc. 126, 5942–5943. doi:10.1021/ja0499389
Xu, F., Wu, Y., Zhang, C., Davis, K. M., Moon, K., Bushin, L. B., et al. (2019). A Genetics-free Method for High-Throughput Discovery of Cryptic Microbial Metabolites. Nat. Chem. Biol. 15, 161–168. doi:10.1038/s41589-018-0193-2
Xu, L., Huang, H., Wei, W., Zhong, Y., Tang, B., Yuan, H., et al. (2014). Complete Genome Sequence and Comparative Genomic Analyses of the Vancomycin-Producing Amycolatopsis orientalis. BMC Genomics 15, 1. doi:10.1186/1471-2164-15-363
Yim, G., Kalan, L., Koteva, K., Thaker, M. N., Waglechner, N., Tang, I., et al. (2014a). Harnessing the Synthetic Capabilities of Glycopeptide Antibiotic Tailoring Enzymes: Characterization of the UK-68,597 Biosynthetic Cluster. ChemBioChem 15, 2613–2623. doi:10.1002/cbic.201402179
Yim, G., Thaker, M. N., Koteva, K., and Wright, G. (2014b). Glycopeptide Antibiotic Biosynthesis. J. Antibiot. 67, 31–41. doi:10.1038/ja.2013.117
Yim, G., Wang, W., Thaker, M. N., Tan, S., and Wright, G. D. (2016). How to Make a Glycopeptide: a Synthetic Biology Approach to Expand Antibiotic Chemical Diversity. ACS Infect. Dis. 2, 642–650. doi:10.1021/acsinfecdis.6b00105
Yushchuk, O., Binda, E., and Marinelli, F. (2020a). Glycopeptide Antibiotic Resistance Genes: Distribution and Function in the Producer Actinomycetes. Front. Microbiol. 11, 1–9. doi:10.3389/fmicb.2020.01173
Yushchuk, O., Horbal, L., Ostash, B., Marinelli, F., Wohlleben, W., Stegmann, E., et al. (2019). Regulation of Teicoplanin Biosynthesis: Refining the Roles of Tei Cluster-Situated Regulatory Genes. Appl. Microbiol. Biotechnol. 103, 4089–4102. doi:10.1007/s00253-019-09789-w
Yushchuk, O., Ostash, B., Pham, T. H., Luzhetskyy, A., Fedorenko, V., Truman, A. W., et al. (2016). Characterization of the post-assembly Line Tailoring Processes in Teicoplanin Biosynthesis. ACS Chem. Biol. 11, 2254–2264. doi:10.1021/acschembio.6b00018
Yushchuk, O., Ostash, B., Truman, A. W., Marinelli, F., and Fedorenko, V. (2020b). Teicoplanin Biosynthesis: Unraveling the Interplay of Structural, Regulatory, and Resistance Genes. Appl. Microbiol. Biotechnol. 104, 3279–3291. doi:10.1007/s00253-020-10436-y
Yushchuk, O., Vior, N. M., Andreo-Vidal, A., Berini, F., Rückert, C., Busche, T., et al. (2021). Genomic-led Discovery of a Novel Glycopeptide Antibiotic by Nonomuraea coxensis DSM 45129. ACS Chem. Biol. 16, 915–928. doi:10.1021/acschembio.1c00170
Keywords: glycopeptide antibiotics, dalbaheptides, ramoplanin, teicoplanin, A40926, glycosyltransferase, biosynthetic gene cluster
Citation: Yushchuk O, Zhukrovska K, Berini F, Fedorenko V and Marinelli F (2022) Genetics Behind the Glycosylation Patterns in the Biosynthesis of Dalbaheptides. Front. Chem. 10:858708. doi: 10.3389/fchem.2022.858708
Received: 20 January 2022; Accepted: 21 February 2022;
Published: 24 March 2022.
Edited by:
Zhongping Tan, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaReviewed by:
Max Julian Cryle, Monash University, AustraliaCopyright © 2022 Yushchuk, Zhukrovska, Berini, Fedorenko and Marinelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Flavia Marinelli, ZmxhdmlhLm1hcmluZWxsaUB1bmluc3VicmlhLml0
 Oleksandr Yushchuk
Oleksandr Yushchuk Kseniia Zhukrovska2
Kseniia Zhukrovska2 Francesca Berini
Francesca Berini Flavia Marinelli
Flavia Marinelli