- 1State Key Laboratory of Phytochemistry and Plant Resources in West China, CAS Center for Excellence in Molecular Plant Sciences, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2Hunan Provincial Key Laboratory for Synthetic Biology of Traditional Chinese Medicine, Hunan University of Medicine, Huaihua, China
- 3Savaid Medical School, University of Chinese Academy of Sciences, Beijing, China
- 4State Key Laboratory of Southwestern Chinese Medicine Resources, Innovative Institute of Chinese Medicine and Pharmacy, Chengdu University of Traditional Chinese Medicine, Chengdu, China
During a screening for antifungal secondary metabolites, six new mono-/bis-alkenoic acid derivatives (2–7) and one known alkenoic acid derivative (1) were isolated from an endophytic fungi Scopulariopsis candelabrum. Their chemical structures were identified by 1H-NMR, 13C-NMR, 2D NMR, and high-resolution mass spectrometry, as well as comparisons with previously reported literatures. Among them, fusariumesters C‒F (2–5) are bis-alkenoic acid derivatives dimerized by an ester bond, while acetylfusaridioic acid A (6) and fusaridioic acid D (7) are alkenoic acid monomers. All the isolates were submitted to an antifungal assay against Candida albicans and the corn pathogen Exserohilum turcicum using the filter paper agar diffusion method. As a result, only compound 1 decorating with β-lactone ring turned out to be active against these two tested fungi. The broth microdilution assay against Candida albicans showed the minimum inhibitory concentration (MIC) value of 1 to be 20 μg/ml, while the minimum inhibitory concentration value of the positive control (naystatin) was 10 μg/ml. And the half maximal inhibitory concentration (IC50) value (21.23 μg/ml) of 1 against Exserohilum turcicum was determined by analyzing its inhibition effect on the mycelial growth, using cycloheximide (IC50 = 46.70 μg/ml) as the positive control.
Introduction
Candida albicans, as an opportunistic pathogenic fungus, normally maintain symbiosis with the human body in the skin, oral cavity, and gastrointestinal tract (Mishra and Koh, 2021). When the body’s homeostasis is destroyed, C. albicans transforms into pathogenic fungi, causing various fungal diseases from superficial skin infections to life-threatening systemic infections (Pappas et al., 2009; Hall and Noverr, 2017). According to statistics, four hundred thousand people are infected with C, albicans every year, and 75% of women suffer from vulvovaginal candidiasis at least once in their lives (Fidel et al., 2004; Yang et al., 2014; Rajendran et al., 2016). Even with drug treatment, the fatality rate of invasive C. albicans infection is still close to 40% (Lohse et al., 2018; Whitesell et al., 2019). Among immunocompromised people such as chemotherapy and organ transplantation, the mortality rate of fungal diseases caused by C. albicans is 33–50% (Krcmery et al., 2000; Winston, 1999; Levesque et al., 2015). Therefore, infections caused by C. albicans are still nonnegligible threats to human health.
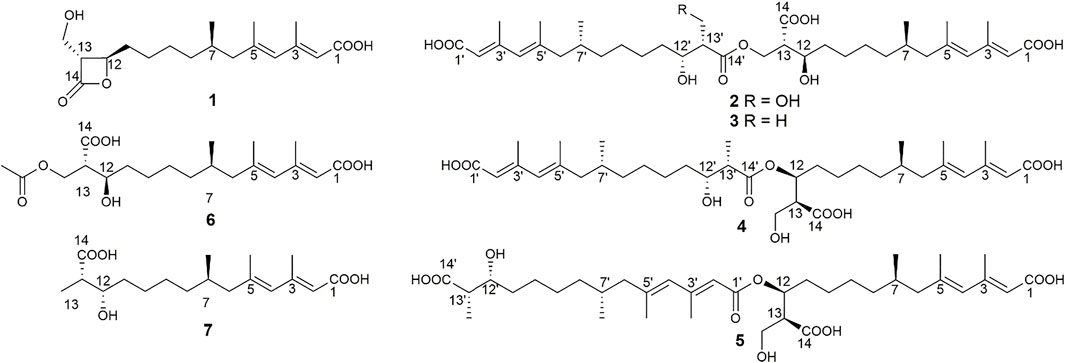
As an antifungal agent, fluconazole is widely used in the treatment of fungal diseases caused by C. albicans because of its low price, low toxicity, and high efficiency (Lu et al., 2017). However, the drug resistance of C. albicans caused by the widespread use of fluconazole is becoming an increasingly serious problem, and the discovery for new antifungal drugs has become more and more urgent (Whaley et al., 2016; Lu et al., 2017; Lu et al., 2021). Fungal secondary metabolites, as an important source of antifungal drugs (Baker et al., 2007; Di Santo, 2010; Cantrell et al., 2012; Schueffler and Anke, 2014), have attracted much more attention from the researchers. In the past 10 years, 25% of antifungal active compounds are derived from fungi (Aldholmi et al., 2019). Among all the fungal microbial resources, plant endophytic fungi were thought as the valuable resources for the discovery of antifungal agents (Uzma et al., 2018; Newman and Cragg, 2020). Recently, a program to discover antifungal constituents from endophytic fungi associated with characteristic food resources of Yunnan Province, China, was conducted in our lab. Accordingly, an antifungal screening of the strain fermentation extracts against C. albicans targeted an endophytic fungus from stems of tea trees, Scopulariopsis candelabrum KIB-int20. Secondary metabolites reported from the genus Scopulariopsis were mainly cyclodepsipeptides (such as scopularides A and B) and some dihydroquinolin-2-one-containing alkaloids (Yu et al., 2008; Shao et al., 2015; Elbanna et al., 2019). In this study, seven mono-/bis-alkenoic acid derivatives (1–7, Figure 1) were isolated from S. candelabrum KIB-int20 during a screening for antifungal secondary metabolites. We herein report the isolation, structure elucidation and antifungal activity of these polyketides.
Materials and Methods
General Experimental Procedures
Optical rotations were measured on an Autopol VI manufactured by Rudolph Research Analytical, Hackettstown, NJ, United States IR spectra were measured on a Nicolet iS10 FT-IR spectrometer (Thermo Fisher Scientific, United States) with KBr disks. NMR spectra were recorded in CDCl3 (δH 7.26 ppm, δC 77.16 ppm) or DMSO-d6 (δH 2.50 ppm, δC 39.52 ppm) using Bruker Avance III 600 or 800 MHz spectrometers (Bruker Corp, Switzerland). HR-ESI-MS analysis were carried out on a Shimadzu UPLC-IT-TOF mass spectrometer (Shimadzu Corp, Japan). Silica gel (100–200 mesh and 200–300 mesh, Qingdao Marine Chemical Inc, China) and Sephadex LH-20 (18–111 μm, Pharmacia Biotech Ltd, Sweden) were used for the chromatography column (CC). Precoated silica gel GF254 plates (0.20–0.25 mm in thickness, Qingdao Marine Chemical Inc, China) were used for thin-layer chromatography (TLC) analyses. Semipreparative HPLC was conducted on a Hitachi Chromaster system (Hitachi Ltd, Japan), equipped with a DAD detector and a YMC-Triart C18 column (250 × 10.0 mm i. d, 5 μm), using a flow rate of 3.0 ml/min at a column temperature at 28°C, and 0.1% (v/v) acetic acid was added to each HPLC mobile phase.
Strain Isolation and Cultivation
Strain S. candelabrum KIB-int20 was isolated from the stems of tea trees (Camellia sinensis (L.) O. Ktze) from Dali, Yunnan Province, China. It was identified as S. candelabrum by a combination of ITS sequence and fungal morphological identification. The internal transcribed spaces (ITS) region was amplified and sequenced using the general primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). The ITS region of the fungus was a 605 bp DNA sequence (GenBank No. OK445701), which showed 99% identity to the ITS sequence of strain S. candelabrum (GenBank No. LM652483.1).
S. candelabrum KIB-int20 was first inoculated on a PDA (filtrate of boiled fresh potatoes 200 g/L, dextrose 20 g/L, agar 20 g/L) plate for 5 days, and then transferred to several PDA plates for another 7 days culture. About one-sixth of agar blocks with fungi mycelium was inoculated into a tissue culture vessel (370 ml) containing the fermentation medium. For each tissue culture vessel, 20 g cargo rice, 10 g peptone and 12 ml water were added and sterilized at 121°C for 30 min immediately. The inoculated medium was statically cultivated for 1 month in a dark environment at room temperature. Strain S. candelabrum KIB-int20 was finally fermented with 5 kg of cargo rice in total.
Extraction and Isolation
The fermentation solid of S. candelabrum KIB-int20 was extracted with acetone (10 L×2, d×2) at room temperature. The extracts were concentrated to remove organic solvent. The aqueous residue was then partitioned with EtOAc (2.5 L×4) to obtain an oily crude extract (50 g). The extract was then subjected to silica gel CC eluting with petroleum ether−EtOAc (1:0, 10:1, 5:1, 2:1, 1:1, 1:2, 1:5, 1:10 and 0:1, v/v) to give nine fractions (A−I). An antifungal screening of each fraction against C. albicans was conducted, and fraction D turned out to be active. The main metabolites in each fraction were further analyzed by DAD-HPLC. Main metabolites in fractions D and E shared the same UV absorptions. In this way, fractions D and E were selected for further study. Selected fraction E (petroleum ether−EtOAc 1:1) was first separated by Sephadex LH-20 CC (CH2Cl2−CH3OH, 1:1), and divide it into six subfractions according to the detection results of thin layer chromatography (10% ethanol sulfate in EtOH was served as chromogenic agent). Further purification of these subfractions by semipreparative DAD-HPLC gave compounds 2 (78% methanol in H2O, tR = 19.8 min, 3.5 mg), 3 (78% methanol in H2O, tR = 33.0 min, 3.5 mg), 4 (78% methanol in H2O, tR = 38.0 min, 6.4 mg), and 5 (78% methanol in H2O, tR = 50.0 min, 5.4 mg). Another selected fraction D (petroleum ether−EtOAc, 2:1) was sequentially subjected to Sephadex LH-20 CC (CH2Cl2−CH3OH, 1:1) and semipreparative DAD-HPLC to afford 1 (65% methanol in H2O, tR = 32.5 min, 8.2 mg), 6 (65% methanol in H2O, tR = 34 min, 6.9 mg), and 7 (65% methanol in H2O, tR = 42.0 min, 4.4 mg).
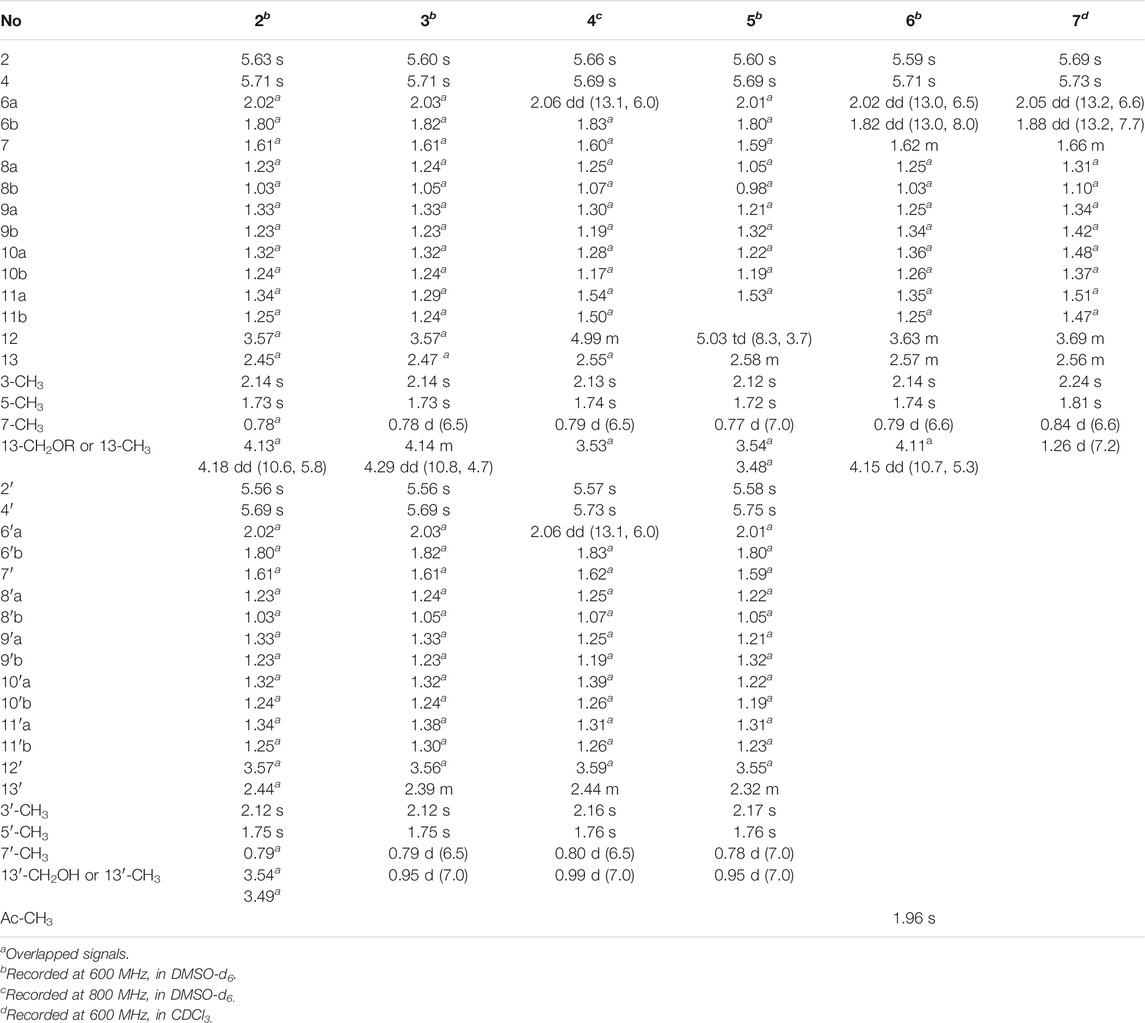
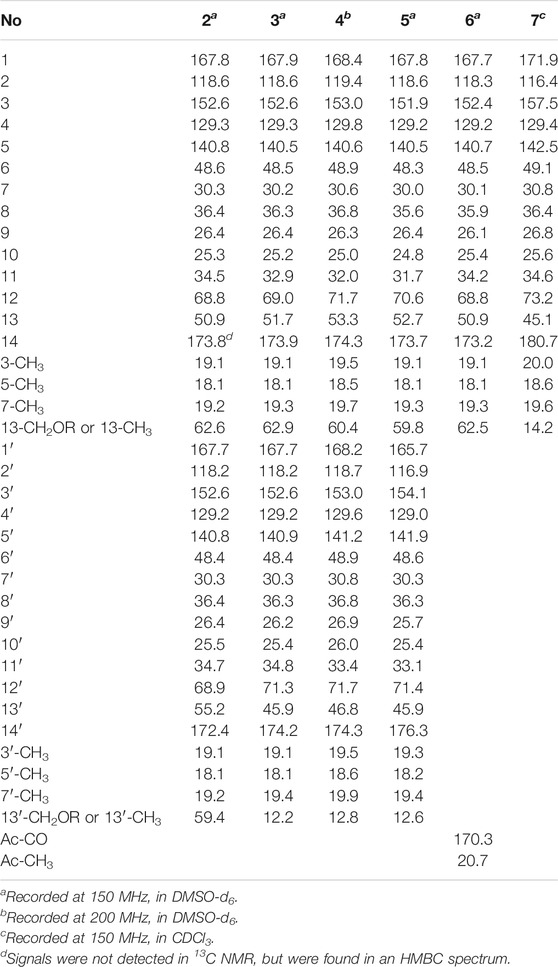
Fusariumester C (2): colorless oil [α]19.9D 4.5 (c 0.2, MeOH) UV (MeOH); λmax (log ε) 196 (3.99), 232 (3.59), 269 (3.93) nm; IR (KBr) νmax 3,419, 2,927, 2,856, 1712, 1,620, 1,382, 1,250, 1,176 cm₋1. HR-ESI-MS: m/z 665.3909 [M ‒ H]‒ (calcd for C36H57O11, 665.3906). 1H NMR (600 MHz, DMSO-d6) data see Table 1, and 13C NMR (150 MHz, DMSO-d6) data see Table 2.
Fusariumester D (3): colorless oil [α]19.9D 3.2 (c 0.2, MeOH) UV (MeOH); λmax (log ε) 196 (4.07), 232 (3.69), 270 (4.07) nm; IR (KBr) νmax 3,659, 3,433, 2,927, 2,857, 2,011, 1,711, 1,621, 1,530, 1,378, 1,343, 1,324, 1,251, 1,176 cm₋1. HR-ESI-MS: m/z 649.3952 [M‒H]‒ (calcd for C36H57O10, 649.3957). 1H NMR (600 MHz, DMSO-d6) data see Table 1, and 13C NMR (150 MHz, DMSO-d6) data see Table 2.
Fusariumester E (4): colorless oil [α]19.9D 1.0 (c 0.2, MeOH) UV (MeOH); λmax (log ε) 196 (4.22), 229 (3.86), 269 (4.32) nm; IR (KBr) νmax 2,925, 2,854, 2,644, 2,566, 1,687, 1,604, 1,381, 1,325, 1,253, 1,178 cm₋1. HR-ESI-MS: m/z 649.3959 [M‒H]‒ (calcd for C36H57O10, 649.3957). 1H NMR (800 MHz, DMSO-d6) data see Table 1, and 13C NMR (200 MHz, DMSO-d6) data see Table 2.
Fusariumester F (5): colorless oil [α]19.9D ‒ 6.0 (c 0.2, MeOH) UV (MeOH); λmax (log ε) 196 (4.35), 232 (3.96), 271 (4.38) nm; IR (KBr) νmax 3,420, 2,927, 2,857, 2,644, 1,712, 1,619, 1,381, 1,234, 1,150 cm₋1. HR-ESI-MS: m/z 649.3956 [M‒H]‒ (cald for C36H57O10, 649.3957). 1H NMR (600 MHz, DMSO-d6) data see Table 1, and 13C NMR (150 MHz, DMSO-d6) data see Table 2.
Acetylfusaridioic acid A (6): colorless oil [α]19.9D 5.7 (c 0.15, MeOH) UV (MeOH); λmax (log ε) 196 (3.85), 230 (3.48), 270 (3.95) nm; IR (KBr) νmax 3,412, 2,928, 2,859, 2,645, 1,740, 1,716, 1,618, 1,382, 1,250, 1,184 cm₋1. HR-ESI-MS: m/z 383.2074 [M‒H]‒ (calcd for C20H31O7, 383.2075). 1H NMR (600 MHz, DMSO-d6) data see Table 1, and 13C NMR (150 MHz, DMSO-d6) data see Table 2.
Fusaridioic acid D (7): colorless oil [α]19.9D 7.2 (c 0.2, MeOH) UV (MeOH); λmax (log ε) 196 (3.66), 230 (3.66), 269 (4.09) nm; IR (KBr) νmax 3,400, 2,929, 2,858, 2,640, 2,229, 2,195, 2,179, 2,164, 2,153, 2,113, 2,056, 2,023, 2.011, 1,970, 1,959, 1,692, 1,622, 1,377, 1,324, 1,251, 1,175 cm₋1. HR-ESI-MS: m/z 325.2023 [M‒H]‒ (calcd for C18H29O5, 325.2020). 1H NMR (600 MHz, CDCl3) data see Table 1, and 13C NMR (150 MHz, CDCl3) data see Table 2.
Antifungal Activity Assay
Rough Antifungal Activity Test: The rough antifungal activity of compounds 1−7 was measured by the filter paper agar diffusion method (Xu et al., 2015). 1 ml suspension (1 × 105 CFU cell or spore concentration) of C. albicans, or Exserohilum turcicum, Curvularia lunata, or Fusarium oxysporum in 20% glycerin was inoculated in a Petri dish containing PDA medium; autoclaved paper disks (6 mm diameter) were placed around the fungal inoculant on the same Petri dish, and each of the paper disks impregnated with 10 μg testing samples, nystatin (positive control) or an equivalent volume of methanol (blank control). Fungal inoculants were cultivated in dark at 30°C for 2 days, and then the size of the inhibition zones was analyzed. Each compound was retested three times.
Measurement of minimum inhibitory concentration (MIC) Values (Yu et al., 2016): A single colony of C. albicans on the SDA plate (1% peptone, 4% dextrose and 2% agar) was picked and inoculated into 5 ml YPD (1% yeast extract, 2% peptone and 2% dextrose) liquid medium and cultivated at 37°C, 200 r/min for 16 h to reach the logarithmic growth phase. According to the measured growth curve of C. albicans, the fungal inoculum was diluted with YPD liquid medium, ensuring the abundance of the strains was 3 × 107 CFU/ml. Compound 1, nystatin (positive control) and equivalent methanol (blank control) were dispensed in single wells and mixed with diluted fungal inoculum to make the final concentrations of tested compounds were 10, 20, 40, 80, 160 and 320 μg/ml in a single well, respectively. After 48 h of shaking culture at 37°C and 200 r/min, the results were determined visually. The MIC was defined as the lowest concentration where there was no visible growth of C. albicans. All the experiments were carried out in triplicate.
Measurement of half maximal inhibitory concentration (IC50) Values (Weidenborner et al., 1990): The IC50 of compound 1 against E. turcicum was evaluated using 48-well culture plates. The conidia used in these experiments were collected from the 7-day-old culture of fungi grown on PDA. The conidia were collected and the suspension was diluted with sterile water and mix 1:1 with PDB (filtrate of boiled fresh potatoes 200 g/L, dextrose 20 g/L) solution for activity test. 1 ml 1/2 PDB spore suspension was added to single wells, and 1, 2, 4, 8, 16 or 32 μL compound 1 or cycloheximide (10 mg/ml) was added to make the final concentration is 9.99–310.07 μg/ml in a single well. After the 48-well plate was cultured at 200 r/min and 30°C for 7 days, the mycelium at each concentration were collected, dried and weighed. The inhibition rates were treated by nonlinear regression analysis of logistic dose–respond curves (Graph Pad Prism eight statistic software) to get the IC50 value.
Results and Discussion
Structural Elucidation
A series of chromatographic methods were used for the isolation of monomeric compounds from the strain fermentation extracts, and diverse spectroscopic analyses were used for their structure elucidation. As a result, seven polyketides were isolated and identified, including one known compound, hymeglusin (1) (Kumagai et al., 1992; Tomoda et al., 1988), four new bis-alkenoic acid derivatives named fusariumesters C−F (2–5), and two new alkenoic acid monomers named acetylfusaridioic acid A (6) and fusaridioic acid D (7). The absolute configuration of hymeglusin (1) was previously determined by chemical degradation method and Mosher method (Chiang et al., 1988). The optical rotation of hymeglusin (1) was {[α]19.4D 24.56 (c 0.2, CHCl3)} in our project, which reported in the literature was [α]22D 10.6 (c 0.1, CHCl3) (Kanaida et al., 2021).
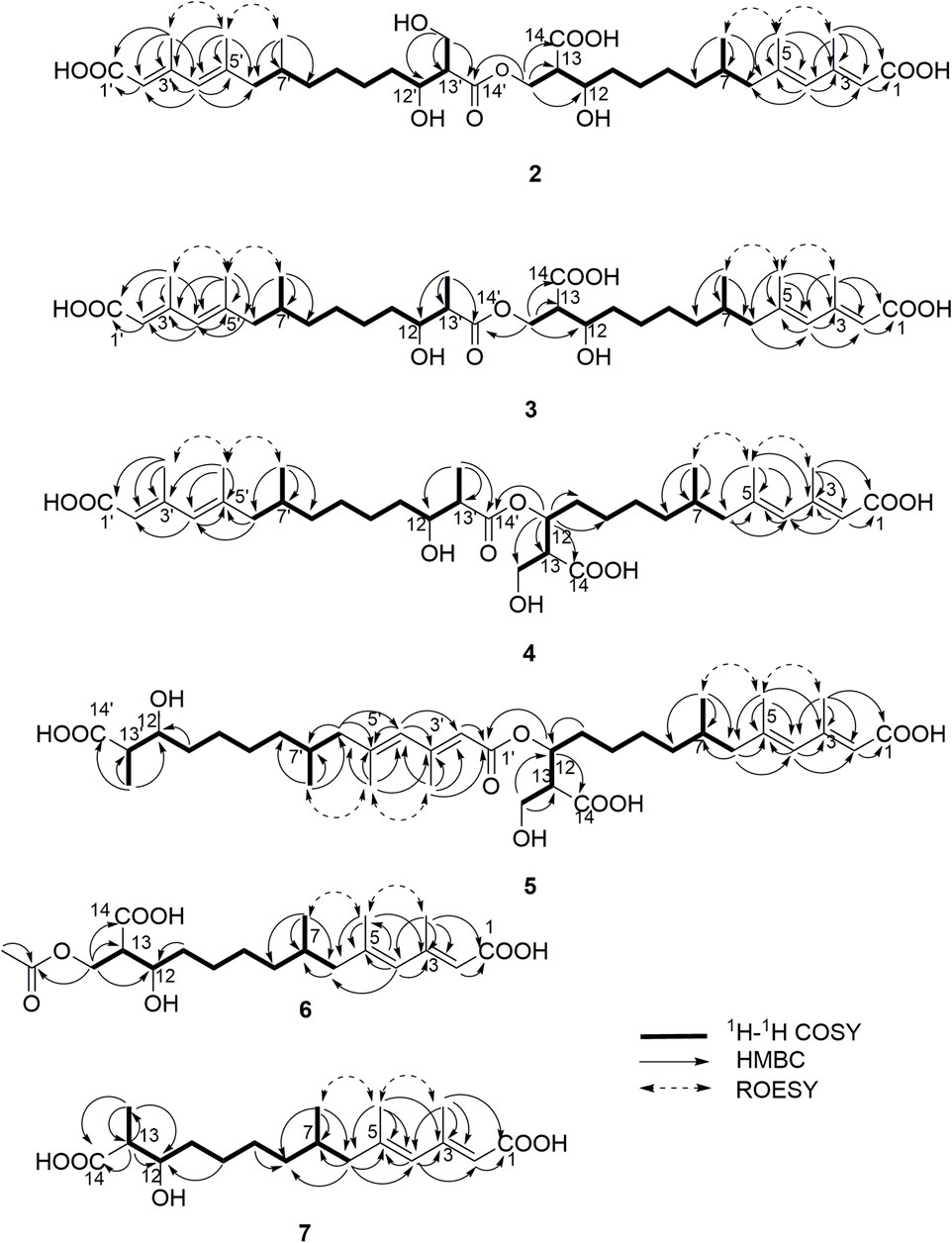
Compound 2 (fusariumester C) was isolated as colorless oil. Its molecular formula was determined to be C36H58O11 by HR-ESI-MS analysis (m/z 665.3909 [M ‒ H]‒, calcd for C36H57O11, 665.3906, Supplementary Figure S9), suggesting eight degrees of unsaturation. The IR spectrum of 2 displayed characteristic adsorptions for carbonyl groups and carbon-carbon double bonds at 1,712 and 1,620 cm−1, respectively. The 13C NMR data (Table 2) of 2 showed 36 carbon resonances, and all signals appeared in pairs. Moreover, each pair of the signals closely resembled those of fusaridioic acid A (Liu et al., 2018). Additionally, comparing the molecular weight (666 Da) of 2 with twice that of fusaridioic acid A (342 Da) yielded a difference of 18 Da. Accordingly, 2 was speculated to be an esterified dimer of fusaridioic acid A (Liu et al., 2018). Further detailed analysis of its NMR data (Supplementary Figures S3–8) supported this hypothesis. Based on the literature report (Liu et al., 2018), two 13C NMR signals at δC 167.7 and δC 167.8 were obviously assigned as carboxylic acid carbonyls connecting to quaternary olefinic carbons, while signals at δC 173.8 and δC 172.4 were assigned as aliphatic carboxylic acid carbonyl groups or ester carbonyl groups. The methines at δC 68.8 and δC 68.9 were attributed to hydroxy-substituted ones. The two methylenes at δC 62.6 and δC 59.4 were assigned to be oxygen-bearing ones, and the small difference between their chemical shifts may due to the formation of an ester bond for one of these two carbons. The 1H NMR data (Table 1 and Supplementary Figure S3) of 2 revealed four olefinic proton signals at δH 5.56, 5.63, 5.71, 5.69. Detailed HMBC correlations associated with these four above-mentioned protons established the presence of two pairs of diene moieties conjugating with terminal carboxyls (the fragments from C-1 to C-6, and from C-1′ to C-6′, Figure 2). The 1H NMR data of 2 also showed two overlapped doublets (δH 0.78 and 0.79) of methyls in the high field region (Supplementary Figure S3). Starting with these two above-mentioned methyl signals, two similar aliphatic carbon chains [7-Me(C-6)/C-7/C-8/C-9/C-10/C-11/C-12/C-13/13-CH2O and 7′-Me(C-6′)/C-7′/C-8′/C-9′/C-10′/C-11′/C-12′/C-13′/13′-CH2O, Figure 2] were deduced based on a combined analyses of its HSQC and 1H–1H COSY spectra. Lastly, the key HMBC correlations from the proton at δH 4.13 (one proton of 13-CH2OR) to the carbons of C-12, C-13, C-14 and C-14′ demonstrated that two molecules of fusaridioic acid A were dimerized via the ester bond built by 13-CH2OH and the carboxylic acid group at C-14′ (Figure 2). Thus, the planar structure of 2 was elucidated.
The configurations of the four double bonds in 2 were revealed by the analysis of its ROESY spectrum (Figure 2 and Supplementary Figure S8). However, the 1H NMR data of 2 were helpless for the determination of its stereochemistry because of signal overlapping. For the stereochemistry of reported mono-/bis-alkenoic acid derivatives, the configurations of C-7 (7′) and C-13 (13′) were conserved to be 7R (7′R) and 13S (13′S) (Liu et al., 2018; Niu et al., 2019; Tang et al., 2019), while the configuration of C-12 (12′) turned out to be 12 (12′) R or 12 (12′) S (Liu et al., 2018). Recently, it was reported that the configurations of fungal polyketides were conserved in general with few exceptions (Takino et al., 2021). Therefore, in view of the shared biosynthetic pathway of mono-/bis-alkenoic acid derivatives, as well as previous literature reports (Liu et al., 2018; Niu et al., 2019; Tang et al., 2019), the configurations of C-7 (7′) and C-13 (13′) in 2 were supposed to be 7R (7′R) and 13S (13′S), respectively. Compared with the 13C NMR and 1H NMR chemical shifts of C-12 (12′) in the dimers reported in the literature (Liu et al., 2018, Supplementary Table S1), the absolute configurations of C-12 (12′) in compound 2 were determined to be 12R (12′R). In this way, the chemical structure of 2 was identified as shown in Figure 1. Since three similar alkenoic acid dimers were given the trivial names of fusariumesters A1, A2, and B (Liu et al., 2018) previously, compound 2 was named as fusariumester C.
Compound 3 (fusariumester D) was isolated as colorless oil. Its molecular formula was confirmed to be C36H58O10 by HR-ESI-MS data (m/z 649.3952 [M ‒ H]‒, calcd for C36H57O10, 649.3957, Supplementary Figure S16), indicating eight degrees of unsaturation. The molecular weight of compound 3 (650 Da) was lower than that of compound 2 (666 Da) with a difference of 16 Da. The 1H and 13C NMR spectra of 3 closely resembled those of 2 (Table 1 and Table 2), and the comparison of their 1H and 13C NMR data (Supplementary Figures S10,11) indicated a methyl (13′-Me) in 3 instead of a hydroxymethyl (13′-CH2OH) in 2. This hypothesis was confirmed by HMBC (Supplementary Figure S13) correlations from protons of 13′-CH2OH to C-12′, C-13′, and C-14’ (Figure 2). Analogously, a group of HMBC correlations from the proton at δH 4.14 (one proton of 13-CH2OR) to C-12, C-13, C-14, and C-14′ proved that 3 shared the same esterified dimerization way as 2.
Compound 4 (fusariumester E) is a colorless viscous oil. Its molecular formula was determined to be same with that of 3 based on the HR-ESI-MS analysis (m/z 649.3959 [M ‒ H]‒, calcd for C36H57O10, 649.3957, Supplementary Figure S23). The 1H and 13C NMR data of 4 and 3 were highly similar (Table 1 and Table 2). The most striking differences between their 13C NMR data were that chemical shift of 13-CH2OR was upfield shifted from 62.9 ppm in 3 to 60.4 ppm in 4, and the chemical shift of C-12 was downfield shifted from 69.0 ppm in 3 to 71.7 ppm in 4. In view of the same molecular formula shared by 3 and 4, this phenomenon implied that 4 might possess a different dimerization site. Key HMBC correlations from H-12 to C-10, C-11, C-13, C-14 and C-14′ certified that an ester bond was built in 4 with the carboxylic acid group in one monomer at C-14′ and the hydroxyl at C-12 in another monomer (Figure 2).
Compound 5 (fusariumester F) was also isolated as colorless oil. Its molecular formula was speculated to be C36H58O10 by HR-ESI-MS analysis (m/z 649.3956 [M ‒ H]‒, cald for C36H57O10, 649.3957, Supplementary Figure S30). Comparing the 1H and 13C NMR data of 5 with those of 2–4 (Table 1 and Table 2) predicted 5 also to be a bis-alkenoic acid derivative but decorating with another new esterified dimerization way. A vital HMBC (Supplementary Figure S27) cross peak of H-12/C-1′ defined that the hydroxyl at C-12 in one monomer and the terminal carboxylic acid at C-1′ contributed to the dimerized ester bond in 5.
Compound 6 (acetylfusaridioic acid A) was isolated as colorless oil. Its elemental composition was determined to be C20H32O7 by HR-ESI-MS analysis (m/z 383.2074 [M ‒ H]‒, calcd for 383.2075 C20H31O7, Supplementary Figure S37), indicating five degrees of unsaturation. Its 1H NMR and HSQC data distinctly showed two olefinic protons (δH 5.59 and 5.71), two oxygenated gem-protons (δH 4.11 and 4.15), and four methyl groups (three singlets at δH 2.14, 1.96, 1.74, and one doublet at δH 0.79). Three down-field shifted carbonyls (δC 173.2, 170.3, 167.7), four olefinic carbons (δC 152.4, 140.7, 129.2, 118.3), one oxygen-bearing methylene (δC 62.5), and four methyl groups (δC 20.7, 19.3, 18.1, 19.1) were observed in the 13C NMR spectrum of 6. Examination of its detailed 13C NMR data with those of fusaridioic acid A (Liu et al., 2018) suggested that 6 was the product of acetylation at 13-CH2OH in fusaridioic acid A. Key HMBC (Supplementary Figure S34) correlations from protons (δH 4.11 and 4.15) of 13-CH2O moiety to the carbonyl (δC 170.3) of the acetyl group supported this deduction (Figure 2). Like compound 2, the configurations of C-7 and C-13 in 6 were supposed to be 7R and 13S based on a thought of conserved biosynthetic logic. And the C-12 (12′) absolute configurations in compounds 3, 4, 5, and 6 were determined to be 12R (12′R), 12S (12′R), 12S (12′R), and 12R, respectively, by comparisons of their proton NMR data of H-12 (12′) with literature reports (Supplementary Table S1).
Fusaridioic acid D (7) was found to possess the molecular formula C18H30O5 from the HR-ESI-MS data (m/z 325.2023 [M‒H]‒, calcd for 325.2020 C18H29O5, Supplementary Figure S44), corresponding to an unsaturation index of four. Detailed analyses of its 1H and 13C NMR data (Table 1 and Table 2) revealed that 7 is one of the monomers involved in compound 3. That’s to say the chemical structure of 7 is a dehydroxylation product of fusaridioic acid A (Liu et al., 2018). This hypothesis was further proved by key HMBC (Supplementary Figure S41) correlations from a methyl (δH 1.26) to C-14, C-13, and C-12 in 7 (Figure 2). By referring to the 13C NMR and 1H NMR chemical shifts of known compounds with similar structural units, the absolute configuration of C-12 in compound 7 was determined to be 12S (Supplementary Tables S1) (Ying and Hong, 2007; Bisek et al., 2008).
Evaluation of Antifungal Activity
The rough antifungal activity of compounds 1–7 were measured by the filter paper agar diffusion method (Xu et al., 2015). Compounds 2–7 showed no significant inhibitory activity against C. albicans, and only compound 1 could inhibit C. albicans (Supplementary Figure S49). The MIC value (20 μg/ml) of 1 against C. albicans was then determined by broth microdilution techniques (Yu et al., 2016), using nystatin (MIC = 10 μg/ml) as the positive control (Supplementary Figure S46). As the previous references reported the antifungal activity of alkenoic acids against plant pathogens (Liu et al., 2018; Niu et al., 2019; Tang et al., 2019), all the isolates were also submitted to an antifungal assay against three agricultural pathogenic fungi, Exserohilum turcicum, Curvularia lunata, and Fusarium oxysporum. As a result, only compound 1 showed a good inhibitory activity against E. turcicum, with an IC50 value (21.23 μg/ml) significantly lower than the positive control, cycloheximide (IC50 = 46.70 μg/ml) (Supplementary Figures S47,48). Except for hymeglusin (1), none of the isolated alkenoic acid derivatives could suppress the growth of tested pathogenic fungi. Therefore, it could be concluded that the β-lactone ring was a key moiety for the antifungal activity. Just as the previous reports, the antifungal activity of alkenoic acid derivatives is always accompanied by the appearance of the β-lactone ring (Liu et al., 2018; Niu et al., 2019; Tang et al., 2019). It was reported that the β-lactone ring played a key role in inhibiting fungal HMG-CoA (3-hydroxy-3-methylglutaryl-CoA) synthase activity (Greenspan et al., 1993; Tomoda et al., 2004; Skaff et al., 2012).
Conclusions
In recent years, 23 alkenoic acid derivatives were reported in total (Supplementary Figure S51) (Liu et al., 2018; Niu et al., 2019; Tang et al., 2019). Among them, most of the alkenoic acid derivatives are acyclic, and seven of them are decorating with terminal lactone rings, including β-lactone (such as fusarilactone A, fusarilactone B, hymeglusin, fusarisolin A, and fusariumester B), γ-lactone (such as fusarisolin B), and δ-lactone (such as fusarilactone C). For these reported alkenoic acids, oxidative modifications (carboxylic acids or hydroxyls) often occur at C-1, C-12, Me-13, and C-14. The stereochemistry of acyclic alkenoic acids is usually conserved with 7R and 13S, while the stereochemistry of C-12 with a hydroxy modification is hybrid with R (eg, fusarisolin D, fusaridioic acid A, and fusariumester A2) or S (eg, fusariumester A1 and L-660282) configuration. Preliminary biosynthetic study of 1233A (equal to F-244, L-659, 699, or hymeglusin) revealed that alkenoic acids are built via a type I polyketide synthase (PKS) logic (Kumagai et al., 1992; Kato et al., 2020). A plausible biosynthetic pathway of compounds 1–7 was also proposed in our project (Supplementary Figure S50).
To date, only three examples of dimerized alkenoic acid compounds (fusariumesters A1, A2, and B) have been reported (Liu et al., 2018), of which fusariumesters A1 and A2 are dimerized by an ester bond formed by the hydroxyl group at C-12 and the carboxyl group at C-14′, and fusariumester B is dimerized by an ester bond involving the hydroxyl group at C-12 and the carboxyl group at C-1′. In this paper, we reported six new mono-/bis-alkenoic acid derivatives (2–7) and one known alkenoic acid derivative (1) from an endophytic fungi S. candelabrum. Consistent with previous reports (Greenspan et al., 1987; Omura et al., 1987; Tomoda et al., 1988), the antifungal screening found that hymeglusin (1) with a β-lactone ring exhibited obvious activity against C. albicans (Tomoda et al., 1988) and E. turcicum. In addition, our discovery of these four new dimerized alkenoic acids (fusariumesters C−F, 2–5) expanded the structure diversity of this family of natural products. These alkenoic acid dimers may be formed via the esterification of the same or different monomers with the aid of one or more esterases (Xu et al., 2020). It is also possible that the dimerization is initiated by thioesterase catalysis (Du and Lou, 2010).
Data Availability Statement
All datasets for this study are included in the article/Supplementary Material.
Author Contributions
JT and XH contributed equally to this work. JT and XH isolated the compounds. JT, XH, and LW identified the chemical structures and prepared the original manuscript. M-HC isolated the strain and performed the strain fermentation. JT and ZW conducted the antifungal assay. YY identified the strain. JT, LW, ZY, J-PH, and S-XH revised the manuscript. S-XH, YY, J-PH, and LW provided financial supports for this project. S-XH designed and supervised the research. All authors approved of the final version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (Nos. 21977100 and 32000239), Yunnan Provincial Science and Technology Department (Nos. 2019FJ007, 2019FA034, and 202101AU070066), and Biological Resources Program of Chinese Academy of Sciences (No.KFJ-BRP-009).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.812564/full#supplementary-material
References
Aldholmi, M., Marchand, P., Ourliac-Garnier, I., Le Pape, P., and Ganesan, A. (2019). A Decade of Antifungal Leads from Natural Products: 2010-2019. Pharmaceuticals 12, 182. doi:10.3390/ph12040182
Baker, D. D., Chu, M., Oza, U., and Rajgarhia, V. (2007). The Value of Natural Products to Future Pharmaceutical Discovery. Nat. Prod. Rep. 24, 1225–1244. doi:10.1039/b602241n
Bisek, N., Wetzel, S., Arndt, H.-D., and Waldmann, H. (2008). Synthesis and Conformational Analysis of Stevastelin C3 Analogues and Their Activity against the Dual-specific Vaccina H1-Related Phosphatase. Chem. Eur. J. 14, 8847–8860. doi:10.1002/chem.200800692
Cantrell, C. L., Dayan, F. E., and Duke, S. O. (2012). Natural Products as Sources for New Pesticides. J. Nat. Prod. 75, 1231–1242. doi:10.1021/np300024u
Chiang, Y. C. P., Chang, M. N., Yang, S. S., Chabala, J. C., and Heck, J. V. (1988). Absolute Configuration of L659,699, a Novel Inhibitor of Cholesterol Biosynthesis. J. Org. Chem. 53, 4599–4603. doi:10.1021/Jo00254a042
Di Santo, R. (2010). Natural Products as Antifungal Agents against Clinically Relevant Pathogens. Nat. Prod. Rep. 27, 1084–1098. doi:10.1039/b914961a
Du, L., and Lou, L. (2010). PKS and NRPS Release Mechanisms. Nat. Prod. Rep. 27, 255–278. doi:10.1039/b912037h
Elbanna, A. H., Khalil, Z. G., Bernhardt, P. V., and Capon, R. J. (2019). Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian marine Fish Gastrointestinal Tract-Derived Fungi. Mar. Drugs 17, 475. doi:10.3390/md17080475
Fidel, P. L., Barousse, M., Espinosa, T., Ficarra, M., Sturtevant, J., Martin, D. H., et al. (2004). An Intravaginal Live Candida challenge in Humans Leads to New Hypotheses for the Immunopathogenesis of Vulvovaginal Candidiasis. Infect. Immun. 72, 2939–2946. doi:10.1128/IAI.72.5.2939-2946.2004
Greenspan, M. D., Bull, H. G., Yudkovitz, J. B., Hanf, D. P., and Alberts, A. W. (1993). Inhibition of 3-Hydroxy-3-Methylglutaryl-CoA Synthase and Cholesterol Biosynthesis by β-lactone Inhibitors and Binding of These Inhibitors to the Enzyme. Biochem. J. 289, 889–895. doi:10.1042/Bj2890889
Greenspan, M. D., Yudkovitz, J. B., Lo, C. Y., Chen, J. S., Alberts, A. W., Hunt, V. M., et al. (1987). Inhibition of Hydroxymethylglutaryl-Coenzyme A Synthase by L-659,699. Proc. Natl. Acad. Sci. 84, 7488–7492. doi:10.1073/pnas.84.21.7488
Hall, R. A., and Noverr, M. C. (2017). Fungal Interactions with the Human Host: Exploring the Spectrum of Symbiosis. Curr. Opin. Microbiol. 40, 58–64. doi:10.1016/j.mib.2017.10.020
Kanaida, M., Kimishima, A., Eguchi, S., Iwatsuki, M., Watanabe, Y., Honsho, M., et al. (2021). Total Syntheses and Chemical Biology Studies of Hymeglusin and Fusarilactone A, Novel Circumventors of β‐Lactam Drug Resistance in Methicillin‐Resistant Staphylococcus aureus. Chemmedchem 16, 2106–2111. doi:10.1002/cmdc.202100219
Kato, S., Motoyama, T., Uramoto, M., Nogawa, T., Kamakura, T., and Osada, H. (2020). Induction of Secondary Metabolite Production by Hygromycin B and Identification of the 1233A Biosynthetic Gene Cluster with a Self-Resistance Gene. J. Antibiot. 73, 475–479. doi:10.1038/s41429-020-0295-4
Krčméry, V., Frič, M., Pisarčiková, M., Huttová, M., Filka, J., Kralinský, K., et al. (2000). Fungemia in Neonates: Report of 80 Cases from Seven University Hospitals. Pediatrics 105, 913–915. doi:10.1542/peds.105.4.913
Kumagai, H., Tomoda, H., and Omura, S. (1992). Biosynthesis of Antibiotic 1233A (F-244) and Preparation of (14C) 1233A. J. Antibiot. 45, 563–567. doi:10.7164/antibiotics.45.563
Levesque, E., Paugam-Burtz, C., Saliba, F., Khoy-Ear, L., Merle, J.-C., Jung, B., et al. (2015). Fungal Complications after Candida preservation Fluid Contamination in Liver Transplant Recipients. Transpl. Int. 28, 1308–1316. doi:10.1111/tri.12633
Liu, S.-Z., Yan, X., Tang, X.-X., Lin, J.-G., and Qiu, Y.-K. (2018). New Bis-Alkenoic Acid Derivatives from a marine-derived Fungus Fusarium Solani H915. Mar. Drugs 16, 483. doi:10.3390/md16120483
Lohse, M. B., Gulati, M., Johnson, A. D., and Nobile, C. J. (2018). Development and Regulation of Single- and Multi-Species Candida Albicans Biofilms. Nat. Rev. Microbiol. 16, 19–31. doi:10.1038/nrmicro.2017.107
Lu, H., Shrivastava, M., Whiteway, M., and Jiang, Y. (2021). Candida Albicans Targets that Potentially Synergize with Fluconazole. Crit. Rev. Microbiol. 47, 323–337. doi:10.1080/1040841X.2021.1884641
Lu, M., Li, T., Wan, J., Li, X., Yuan, L., and Sun, S. (2017). Antifungal Effects of Phytocompounds on Candida Species Alone and in Combination with Fluconazole. Int. J. Antimicrob. Agents 49, 125–136. doi:10.1016/j.ijantimicag.2016.10.021
Mishra, A. A., and Koh, A. Y. (2021). The Microbial and Host Factors that Govern Candida Gastrointestinal Colonization and Dissemination. Curr. Opin. Microbiol. 63, 29–35. doi:10.1016/j.mib.2021.05.012
Newman, D. J., and Cragg, G. M. (2020). Plant Endophytes and Epiphytes: Burgeoning Sources of Known and "unknown" Cytotoxic and Antibiotic Agents? Planta Med. 86, 891–905. doi:10.1055/a-1095-1111
Niu, S., Tang, X.-X., Fan, Z., Xia, J.-M., Xie, C.-L., and Yang, X.-W. (2019). Fusarisolins A-E, Polyketides from the marine-derived Fungus Fusarium Solani H918. Mar. Drugs 17, 125. doi:10.3390/md17020125
Omura, S., Tomoda, H., Kumagai, H., Greenspan, M. D., Yodkovitz, J. B., Chen, J. S., et al. (1987). Potent Inhibitory Effect of Antibiotic 1233A on Cholesterol Biosynthesis Which Specifically Blocs 3-Hydroxy-3-Methylglutaryl Coenzyme A Synthase. J. Antibiot. 40, 1356–1357. doi:10.7164/antibiotics.40.1356
Pappas, P. G., Kauffman, C. A., Andes, D., Benjamin, D. K., Calandra, T. F., Edwards, J. E., et al. (2009). Clinical Practice Guidelines for the Management Candidiasis: 2009 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 503–535. doi:10.1086/596757
Rajendran, R., Sherry, L., Nile, C. J., Sherriff, A., Johnson, E. M., Hanson, M. F., et al. (2016). Biofilm Formation Is a Risk Factor for Mortality in Patients with Candida albicans Bloodstream infection-Scotland, 2012-2013. Clin. Microbiol. Infect. 22, 87–93. doi:10.1016/j.cmi.2015.09.018
Schueffler, A., and Anke, T. (2014). Fungal Natural Products in Research and Development. Nat. Prod. Rep. 31, 1425–1448. doi:10.1039/c4np00060a
Shao, C.-L., Xu, R.-F., Wang, C.-Y., Qian, P.-Y., Wang, K.-L., and Wei, M.-Y. (2015). Potent Antifouling marine Dihydroquinolin-2(1H)-One-Containing Alkaloids from the Gorgonian Coral-Derived Fungus Scopulariopsis sp. Mar. Biotechnol. 17, 408–415. doi:10.1007/s10126-015-9628-x
Skaff, D. A., Ramyar, K. X., Mcwhorter, W. J., Barta, M. L., Geisbrecht, B. V., and Miziorko, H. M. (2012). Biochemical and Structural Basis for Inhibition of enterococcus Faecalis Hydroxymethylglutaryl-CoA Synthase, Mvas, by Hymeglusin. Biochemistry 51, 4713–4722. doi:10.1021/bi300037k
Takino, J., Kotani, A., Ozaki, T., Peng, W., Yu, J., Guo, Y., et al. (2021). Biochemistry‐Guided Prediction of the Absolute Configuration of Fungal Reduced Polyketides. Angew. Chem. Int. Ed. 60, 23403–23411. doi:10.1002/anie.202110658
Tang, X.-X., Yan, X., Fu, W.-H., Yi, L.-Q., Tang, B.-W., Yu, L.-B., et al. (2019). New β-Lactone with Tea Pathogenic Fungus Inhibitory Effect from Marine-Derived Fungus MCCC3A00957. J. Agric. Food Chem. 67, 2877–2885. doi:10.1021/acs.jafc.9b00228
Tomoda, H., Kumagai, H., Takahashi, Y., Tanaka, Y., Iwai, Y., and Omura, S. (1988). F-244(1233A), a Specific Inhibitor of 3-Hydroxy-3-Methylglutaryl Coenzyme A Synthase. Taxonomy of Producing Strain, Fermentation, Isolation and Biological Properties. J. Antibiot. 41, 247–249. doi:10.7164/antibiotics.41.247
Tomoda, H., Ohbayashi, N., Morikawa, Y., Kumagai, H., and Ōmura, S. (2004). Binding Site for Fungal β-lactone Hymeglusin on Cytosolic 3-Hydroxy-3-Methylglutaryl Coenzyme A Synthase. Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids 1636, 22–28. doi:10.1016/j.bbalip.2003.11.005
Uzma, F., Mohan, C. D., Hashem, A., Konappa, N. M., Rangappa, S., Kamath, P. V., et al. (2018). Endophytic Fungi-Alternative Sources of Cytotoxic Compounds: a Review. Front. Pharmacol. 9, 309. doi:10.3389/fphar.2018.00309
Weidenbörner, M., Hindorf, H., Jha, H. C., and Tsotsonos, P. (1990). Antifungal Activity of Flavonoids against Storage Fungi of the Genus Aspergillus. Phytochemistry 29, 1103–1105. doi:10.1016/0031-9422(90)85412-9
Whaley, S. G., Berkow, E. L., Rybak, J. M., Nishimoto, A. T., Barker, K. S., and Rogers, P. D. (2016). Azole Antifungal Resistance in Candida albicans and Emerging Non-albicans Candida Species. Front. Microbiol. 7, 2173. doi:10.3389/fmicb.2016.02173
Whitesell, L., Robbins, N., Huang, D. S., Mclellan, C. A., Shekhar-Guturja, T., Leblanc, E. V., et al. (2019). Structural Basis for Species-Selective Targeting of Hsp90 in a Pathogenic Fungus. Nat. Commun. 10, 402. doi:10.1038/s41467-018-08248-w
Winston, D. J., Pakrasi, A., and Busuttil, R. W. (1999). Prophylactic fluconazole in liver transplant recipients. A randomized, double-blind, placebo-controlled trial. Ann. Intern. Med. 131, 729–737. doi:10.7326/0003-4819-131-10-199911160-00003
Xu, L., Wu, P., Wright, S. J., Du, L., and Wei, X. (2015). Bioactive Polycyclic Tetramate Macrolactams from Lysobacter Enzymogenes and Their Absolute Configurations by Theoretical ECD Calculations. J. Nat. Prod. 78, 1841–1847. doi:10.1021/acs.jnatprod.5b00099
Xu, Y., Minhazul, K. A. H. M., and Li, X. (2020). The Occurrence, Enzymatic Production, and Application of Ethyl Butanoate, an Important Flavor Constituent. Flavour Fragr J. 35, 601–615. doi:10.1002/ffj.3613
Yang, Z.-T., Wu, L., Liu, X.-Y., Zhou, M., Li, J., Wu, J.-Y., et al. (2014). Epidemiology, Species Distribution and Outcome of Nosocomial Candida spp.Bloodstream Infection in Shanghai. BMC Infect. Dis. 14, 241. doi:10.1186/1471-2334-14-241
Ying, Y., and Hong, J. (2007). Synthesis of Brasilibactin A and Confirmation of Absolute Configuration of β-hydroxy Acid Fragment. Tetrahedron Lett. 48, 8104–8107. doi:10.1016/j.tetlet.2007.09.112
Yu, Q., Ravu, R. R., Jacob, M. R., Khan, S. I., Agarwal, A. K., Yu, B.-Y., et al. (2016). Synthesis of Natural Acylphloroglucinol-Based Antifungal Compounds against Cryptococcus Species. J. Nat. Prod. 79, 2195–2201. doi:10.1021/acs.jnatprod.6b00224
Keywords: Alkenoic acid derivatives, polyketides, Microascaceae, Scopulariopsis candelabrum, antifungal activity, Candida albicans
Citation: Tang J, Huang X, Cao M-H, Wang Z, Yu Z, Yan Y, Huang J-P, Wang L and Huang S-X (2022) Mono-/Bis-Alkenoic Acid Derivatives From an Endophytic Fungus Scopulariopsis candelabrum and Their Antifungal Activity. Front. Chem. 9:812564. doi: 10.3389/fchem.2021.812564
Received: 10 November 2021; Accepted: 10 December 2021;
Published: 11 January 2022.
Edited by:
Naohiko Yoshikai, Tohoku University, JapanReviewed by:
Guangbo Ge, Shanghai University of Traditional Chinese Medicine, ChinaSabrin R. M. Ibrahim, Batterjee Medical College, Saudi Arabia
Copyright © 2022 Tang, Huang, Cao, Wang, Yu, Yan, Huang, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Wang, bGl3YW5nY3l5QGNkdXRjbS5lZHUuY24=; Sheng-Xiong Huang, c3hodWFuZ0BtYWlsLmtpYi5hYy5jbg==
 Jun Tang
Jun Tang Xueshuang Huang
Xueshuang Huang Ming-Hang Cao
Ming-Hang Cao Zhiyan Wang
Zhiyan Wang Zhiyin Yu
Zhiyin Yu Yijun Yan1
Yijun Yan1 Jian-Ping Huang
Jian-Ping Huang Li Wang
Li Wang Sheng-Xiong Huang
Sheng-Xiong Huang