
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 20 September 2021
Sec. Medicinal and Pharmaceutical Chemistry
Volume 9 - 2021 | https://doi.org/10.3389/fchem.2021.733350
 Canjie Li1†
Canjie Li1† Xin Sun1,2†
Xin Sun1,2† Wenjing Yin3
Wenjing Yin3 Zhaochun Zhan1
Zhaochun Zhan1 Qing Tang1
Qing Tang1 Wenzhi Wang1
Wenzhi Wang1 Xuefang Zhuo1
Xuefang Zhuo1 Zhongnan Wu2
Zhongnan Wu2 Haipeng Zhang3
Haipeng Zhang3 Yaolan Li1*
Yaolan Li1* Yubo Zhang1,3*
Yubo Zhang1,3* Guocai Wang1*
Guocai Wang1*Seven new clerodane diterpenoids, crassifolins Q−W (1–7), along with five known analogues (8–12), were isolated from the roots of Croton crassifolius. Their structures were identified by comprehensive spectroscopic analysis (UV, IR, NMR, and HR-ESI-MS), and their absolute configurations were determined by ECD spectra and X-ray crystallography. The activities of compounds 1–5 against inflammatory cytokines IL-6 and TNF-α levels on LPS-induced RAW 264.7 macrophages were assessed, and compound 5 showed the most significant activity with the secretion levels of IL-6 and TNF-α at 32.78 and 12.53%, respectively. Moreover, compounds 1–5 were screened for their anti-angiogenesis using a human umbilical vein endothelial cells in vitro mode; the results showed all of them exhibited obvious anti-angiogenesis activities, in particular, compound 5 showed the strongest anti-angiogenesis effect in the range of 6.25–50 μM.
The genus Croton (Euphorbiaceae) is a rich source of diterpenoids with interesting structural diversity such as clerodane-type (Campos et al., 2010; Khanitha and Damrong, 2011; Sun et al., 2014), kauritan-type (Dao et al., 2011; Suwancharoen et al., 2012), heliurane-type (Martha et al., 2011; Huang et al., 2014a), phorbol ester-type (Zhang et al., 2013; Corlay et al., 2014), and trachylobane-type (Martinsen et al., 2010). These diterpenoids exhibited remarkable biological characteristics including anti-inflammatory (Wang et al., 2015), anti-tumor (Fernandes et al., 2013; Young et al., 2013; Maslovskaya et al., 2015), anti-viral (Nina et al., 2014), anti-bacterial (Liu et al., 2014), and anti-oxidation activities (Mariana et al., 2013), which make them particularly valuable for phytochemical and pharmacological research studies. Croton crassifolius Geisel (Euphorbiaceae) is well known as “Jiguxiang” and mainly distributed in the south and southwest area of China, such as Guangxi, Guangdong, and Hainan provinces. The roots of the C. crassifolius are used as a traditional medicine for treatment of joint pain, rheumatic arthritis, stomach ache, pharyngitis, and jaundice (Boonyarathanakornkit et al., 1988). Previous phytochemical studies on C. crassifolius showed that the derivatives of diterpenoids were the main active ingredients (Huang et al., 2014b; Li et al., 2014).
In our early research studies, the investigations on C. crassifolius had led to the isolation of some new diterpenoids (Wang et al., 2012; Wang et al., 2016). However, the diterpenoids from their work showed relatively moderate biological activities including anti-HSV and anti-angiogenesis. In order to search more bioactive natural products from Croton plants, the phytochemical investigation on the roots of C. crassifolius was performed, leading to the isolation of seven new clerodane diterpenoids, named crassifolins Q−W (1–7), along with five known compounds, EBC-205 (8) (Maslovskaya et al., 2019), mallotucin B (9) (Takashi et al., 1976), crassin D (10) (Yuan et al., 2017), spiro [furan-3(2H),6'-(6H)naphtho (1,8-bc)furan]-2'a (2'H)-carboxylic acids (11) (Nakatsu et al., 1981; Li et al., 2019), and crassifolin H (12) (Qiu et al., 2016).
Considering the potential anti-inflammatory activity of clerodane diterpenoids (Zhang et al., 2019), we evaluated the inhibitory effects of diterpenoids 1–5 on inflammation by testing the secretion level of inflammatory cytokines IL-6 and TNF-α on LPS-stimulated RAW 264.7 cells. In earlier research papers, inflammation has anti-cancer effects, which can affect the host immune response, help the body recognize and kill cancer cells, and can be used in anti-cancer immunotherapy (Bryant, 1868). However, in recent years, more and more studies have shown that inflammation can promote the occurrence and development of cancer under most conditions and reduce the therapeutic effect of tumors (Francescone et al., 2014; Moses et al., 2018). Furthermore, the formation of new vessels and angiogenesis are important processes of tumor growth and metastasis. On this basis, we want to isolate compounds that can exhibit anti-inflammatory and anti-angiogenesis activities simultaneously, so as to act as lead compounds to interfere with inflammation and cancer. Moreover, a previous study (Wang et al., 2016) reported that some diterpenoids of this plant exhibited potential activity on anti-angiogenesis, which inspired us to conduct an anti-angiogenesis activity on compounds 1–5 using a human umbilical vein endothelial cell (HUVEC) in vitro mode. Herein, this article reported the isolation, structural elucidation, and evaluation of anti-inflammatory and anti-angiogenic activities of the new compounds.
Compound 1 was isolated as a colorless crystal. Its molecular formula was deduced as C20H26O5 on the basis of its HR-ESI-MS data [m/z 369.1660 (M + Na)+, calcd for C20H26O5Na, 369.1672]. The IR spectrum suggested the presence of hydroxyl (3,463 cm−1), carbonyl (1710 cm−1), and furan ring (1,461, 867 cm−1) in the structure of 1. The 1H NMR spectrum (Table 1) revealed the presence of three olefinic protons [δH 7.35 (1H, s), 7.22 (1H, s), and 6.29 (1H, s)] and two methyl groups [δH 1.30 (3H, s) and 1.12 (3H, s)]. The 13C NMR and DEPT spectra (Table 2) showed 20 carbons including two carboxyl carbons (δC 183.7 and 181.1), five quaternary carbons, seven methylenes, four methines, and two methyls, suggesting that 1 is a clerodane diterpenoid with a comparable structure to the known compound crassifolin B (Wang et al., 2012). The major differences between 1 and crassifolin B were the absence of a methyl (δC 16.3, C-17) and a γ-lactone ring, but the presence of a carboxylic acid group (δC 181.1) and a β-substituted furan ring, and the chemical shifts of C-13 to C-17 shifted from δC 171.2, 115.2, 174.2, 73.3, 16.3 to δC 125.3, 111.2, 142.9, 138.7, 181.1, respectively. These differences suggested that the γ-lactone in crassifolin B was replaced by a β-substituted furan ring in 1, which was verified by the 1H–1H COSY correlation between H-14 (δH 6.29) and H-15 (δH 7.35) and the HMBC cross peaks from H-12 (δH 2.04) to C-13 (δC 125.3)/C-14 (δC 111.2)/C-16 (δC 138.7) and from H-16 (δH 7.22) to C-13/C-14/C-15 (Figure 1). In addition, the 1H–1H COSY correlations of H-6 (δH 1.47)/H-7 (δH 1.92) and H-7/H-8 (δH 2.70), along with the HMBC cross peaks from H-8 to C-9 (δC 40.9)/C-17 (δC 181.1)/C-20 (δC 23.0) were observed, which could confirm that the carboxylic acid group was attached to C-8 (Figure 1). Furthermore, to establish the stereochemistry of 1, the single crystals were obtained in a chloroform system and subjected to an X-ray diffraction experiment with Cu Kα radiation. The crystal data [Flack parameter -0.1 (3), hooft parameter -0.3 (3)] determined the absolute configuration of 1 as 4R, 8R, 9S (Figure 2). Consequently, the structure was determined and named as crassifolin Q. Compound 2 was obtained as a colorless crystal and the HR-ESI-MS showed an [M + Na]+ ion peak at m/z 453.1506 (calcd for C23H26O8Na, 453.1520), consistent with the molecular formula of C23H26O8. The 1H NMR spectrum (Table 1) showed apparent signals for an aldehyde proton [δH 9.89 (1H, s)], three olefinic protons [δH 7.62 (1H, s), 7.54 (1H, s), and 6.50 (1H, s)], two oxygenated methines [δH 5.48 (1H, t, J = 9.0 Hz) and 5.43 (1H, t, J = 5.1 Hz)], one oxygenated methyl [δH 3.68 (3H, s)], and two methyls [δH 1.92 (3H, s) and 1.05 (3H, d, J = 6.4 Hz)]. The 13C NMR and DEPT spectra (Table 2) exhibited the presence of 23 carbons, including one carbonyl carbon (δC 196.7), eight quaternary carbons, six methines, five methylenes, and three methyls, indicating that 2 was also a clerodane diterpenoid. The analysis of 1D NMR data showed that 2 had the analogous planar structure as that crassifolin M (Wang et al., 2016), except for the absence of a carbonyl group (δC 195.7, C-6), but the presence of an oxygenated methine (δC 71.7) and an acetoxyl group (δC 170.1 and 21.1) in 2 and the chemical shifts of C-5, C-6, and C-7 shifted from δC 133.2, 195.7, 40.9 to δC 129.8, 71.7, 32.7, correspondingly. In addition, the acetoxyl group was attached to C-6, which was confirmed by 1H–1H COSY correlation between H-6 (δH 5.46) and H-7 (δH 2.10), together with HMBC cross peaks from H-22 (δH 1.92) to C-6 (δC 71.7)/C-21 (δC 170.1) (Figure 1). Moreover, the absolute configuration of 2 was determined as 4S, 6S, 8R, 9R, and 12S by a single X-ray diffraction experiment (Figure 2). Therefore, the structure of two was confirmed as showed in Figure 3 and named as crassifolin R. Compound 3, a yellow oil, which had the molecular formula C25H34O6 assigned by HR-ESI-MS (m/z 453.2234 [M + Na]+, calcd for C25H34O6Na, 453.2248). The IR spectrum indicated the presence of hydroxyl (3,419 cm−1) and ester carbonyl groups (1718, 1,259 cm−1). According to its 1H NMR spectrum (Table 1), compound 3 exhibited the presence of characteristic signals for two olefinic protons [δH 6.83 (1H, s, J = 7.0 Hz) and 5.82 (1H, s)], one methine [δH 1.78 (1H, m)], two oxygenated methylenes [δH 4.75 (2H, s), 4.38 (1H, dd, J = 11.0, 4.3 Hz), and 3.83 (1H, t, J = 9.2 Hz)], and four methyls [δH 1.78 (3H, s), 1.77 (3H, d, J = 7.0 Hz), 1.27 (3H, s), and 1.00 (3H, s)]. Comparison of 1D NMR data with those of crassifolin B (Wang et al., 2012) showed that they were similar to a typical clerodane diterpenoid skeleton. The differences between 3 and crassifolin B were the absence of a methyl (δC 16.2, C-17), but the presence of an oxygenated methylenes (δC 65.7) and a cis-2-methylbut-2-enoyloxy moiety (δC 168.4, 137.6, 128.8, 14.5, and 12.2). As a result, the chemical shifts of C-8 and C-17 shifted from δC 33.8 and 16.2 in crassifolin B to δC 39.0 and 65.7 in 3, respectively. The presence of cis-2-methylbut-2-enoyloxy moiety was confirmed by the 1H–1H COSY correlation between H3-24 (δH 1.77) and H-23 (δH 6.83), together with the HMBC cross peaks from H3-24 to C-22 (δC 128.8)/C-23 (δC 137.6), from H3-25 (δH 1.78) to C-21 (δC 168.4)/C-22 (Figure 1). In addition, the HMBC cross peaks from H-17 (δH 3.83) to C-21 indicated that the cis-2-methylbut-2-enoyloxy moiety was connected to C-17 (δC 65.7). Due to the influence of substituents on the chemical shift of olefinic hydrogen, the chemical shift of olefinic proton in cis-2-methylbut-2-enoyloxy moiety was greater than that of trans-2-methylbut-2-enoyloxy moiety (Pedro et al., 1989; Russel et al., 2014), and our 1H NMR spectrum of 3 was consistent with cis-2-methylbut-2-enoyloxy moiety. Herein, the 2-methylbut-2-enoyloxy moiety in compound 3 was cis-configuration. Moreover, the absolute configuration of 3 was established by quantum-chemical ECD calculations. The experimental ECD spectrum of 3 showed a positive cotton effect at 224 nm (Δε +50.9), which was compared with the predicted ECD spectrum of (4R, 8R, 9S)-3 and its enantiomer (Figure 4), indicating the absolute configuration of 3 was 4R, 8R, and 9S. Hence, the structure of 3 was established as Figure 3 and named as crassifolin S. Compound 4 was a white powder and showed an [M + Na]+ ion peak at m/z 355.1869 (calcd for C20H28O4Na, 355.1880) in the HR-ESI-MS spectrum, consistent with the molecular formula of C20H28O4. The 1H NMR spectrum of 4 (Table 1) exhibited one olefinic proton [δH 7.10 (1H, s)], one methine [δH 1.72 (1H, m)], and three methyls [δH 1.29 (3H, d, J = 6.8 Hz), 0.88 (3H, s), and 0.87 (3H, s)]. The NMR spectroscopic data of 4 resembled those of crassifolin B (Wang et al., 2012). A detailed comparison of the NMR spectra of 4 with crassifolin B indicated the connection position of C-12 and γ-lactone ring has changed, and the chemical shifts of C-12 to C-16 shifted from δC 23.7, 171.4, 115.1, 174.3, 73.3 to 20.5, 135.2, 174.5, 70.2, 143.9, respectively. The γ-lactone ring was confirmed by the 1H–1H COSY correlation between H-15 (δH 4.75) and H-16 (δH 7.10) along with the HMBC correlations from H-15 to C-13 (δC 135.2)/C-14 (δC 174.5)/C-16 (δC 143.9), from H-16 to C-14 (δC 174.5)/C-15 (δC 70.2). Moreover, the HMBC correlation from H-12 (δH 2.22) to C-13 was observed, which could confirm that the γ-lactone ring was attached to C-12 (Figure 1). The experimental ECD spectrum of 4 showed a positive cotton effect at 202 nm (Δε +35.0) and a negative cotton effect at 222 nm (Δε -42.1), which was similar to those of the calculated ECD spectrum for (4R, 8R, 9S)-4 (Figure 5), suggested the absolute configuration of 4 is 4R, 8R, and 9S. Thus, compound 4 was named crassifolin T. Compound 5 was isolated as a yellow oil and the molecular formula was determined to be C20H26O4 by HR-ESI-MS (m/z 353.1710 [M + Na]+, calcd for C20H24O6Na, 353.1723). The IR spectrum showed characteristic ester carbonyl group (1743, 1,236 cm−1). The 1H NMR spectrum of 5 (Table 1) showed one olefinic proton [δH 5.82 (1H, s)], one oxygenated methine [δH 4.99 (1H, t, J = 8.1 Hz)], and three methyls [δH 1.32 (3H, s), 1.02 (3H, s), and 0.98 (3H, d, J = 7.2 Hz)]. The NMR spectra of 5 exhibited several similarities to crassifolin B (Wang et al., 2012). The main differences were the absence of a carboxylic acid group (δC 183.7, C-18) and a methylene (δC 27.8, C-6), but the presence of an ester group (δC 180.7) and a tertiary carbon bearing oxygen (δC 74.7), and the chemical shifts of C-4, C-6, and C-18 shifted from 47.8, 27.8, 183.7 to 41.7, 74.7, 180.7, respectively. This indicated that a lactone ring was formed between C-4 and C-6, which was confirmed by the 1H–1H COSY between H-6 (δH 4.99) and H-7 (δH 1.63) and the HMBC cross peak from H-19 (δH 1.32) to C-18. In addition, the key NOESY correlations between H-6 and H3-17 (δH 0.98)/H3-20 (δH 1.02), indicated that H-6 and H-17/H-20 were α-orientated (Figure 6). The experimental ECD spectrum of 5 exhibited a positive cotton effect at 226 nm (Δε +10.1) and a negative cotton effect at 206 nm (Δε -2.0), which was similar to those of the calculated ECD spectrum for (4S, 6R, 8R, 9S)-5 (Figure 7). Thus, the structure of 5 was elucidated as 4S, 6R, 8R, 9S and compound 5 was named crassifolin U. Compound 6, a white powder, which had an [M + H]+ ion peak at m/z 373.1283, consistent with the molecular formula C20H20O7. The 1H NMR spectrum of 6 (Table 1) showed three olefinic protons [δH 7.46 (1H, s), 7.44 (1H, s), and 6.40 (1H, s)], one oxygenated methine [δH 5.90 (t, J = 7.5 Hz)], and two methyls [δH 3.68 (s) and 1.14 (d, J = 7.0 Hz)]. The 1D NMR data of 6 were similar to those of spiro [furan-3(2H),1' (2'H)-naphtha-lene]-5'-carboxylic acid (Rodríguez and Jimeno, 2004), and the main differences were the absence of two methines [δC 50.2 (C-5) and 50.0 (C-10)] and a methylene (δC 28.1, C-1), but the presence of two olefinic carbons (δC 144.9 and 145.9) and a carbonyl group (δC 199.9). Thus, it indicated that two methines were replaced by two olefinic carbons and the methylene was replaced by a carbonyl group. These were confirmed by HMBC cross peaks from H-2 (δH 2.56) to C-1 (δC 199.9), from H-7 (δH 2.44) to C-5 (δC 144.9), and from H-11 (δH 2.71) to C-10 (δC 145.9) (Figure 1). The single crystal were obtained in a methanol system and subjected to an X-ray diffraction experiment with Cu Kα radiation and the crystal data [Flack parameter -0.06 (3), hooft parameter -0.03 (3)] determined the absolute configuration of 6 is 4, 8, 9R, 12S (Figure 1). Compound 6 was named crassifolin V. Compound 7 was obtained as yellow oil and had the molecular formula C20H24O6. The 1H NMR spectrum of 7 (Table 1) exhibited three olefinic protons [δH 8.05 (1H, s), 7.41 (1H, s), and 6.75 (1H, s)], one methine [δH 1.97 (m)], and three methyls [δH 1.20 (s), 1.08 (s) and 0.99 (d, J = 6.5 Hz)]. According to the 1D NMR spectroscopic data, the planar structure of compound 7 showed several similarities to methyl 5a,10a-epoxy-2,12-dioxo-13 (16),14-enthalimandien18-oate (Marcos et al., 2003), and the main differences were the absence of a carbethoxy group, but the presence of a carboxylic acid group (δC 176.6, C-18). Thus, it indicated the carbethoxy group was replaced by a carboxylic acid group in 7, which was supported by the HMBC cross peaks from H-3 (δH 2.70) to C-4 (δC 51.0) and from H3-19 (δH 1.08) to C-4/C-5/C-18 (Figure 1). Moreover, the NOESY correlation between H-8 and H3-20 was not observed, which suggested that H3-20 and H3-17 were α-orientated; besides, the correlations of H3-20/H-1α, of H-1α/H-3α, and of H-3α/H3-19 indicated that H3-20 and H3-19 were α-orientated (Figure 6). The experimental ECD spectrum of 7 showed a positive cotton effect at 281 nm (Δε +2.1) and a negative cotton effect at 226 nm (Δε -7.0), which was similar to those of the calculated ECD spectrum for (4S, 5R, 8R, 9S, 10R)-7 (Figure 8), suggested the absolute configuration of 7 is 4S, 5R, 8R, 9S, 10R. Compound 7 was named crassifolin W.
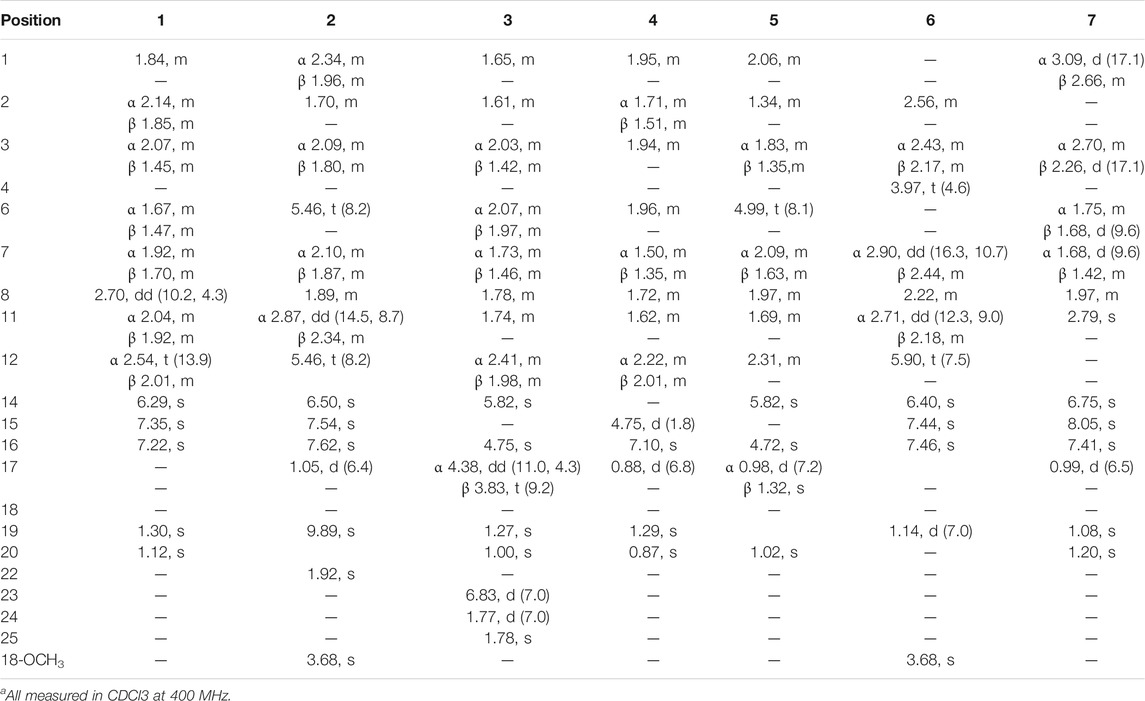
TABLE 1. 1H NMR data of 1–7 (δ in ppm, J in Hz)a.
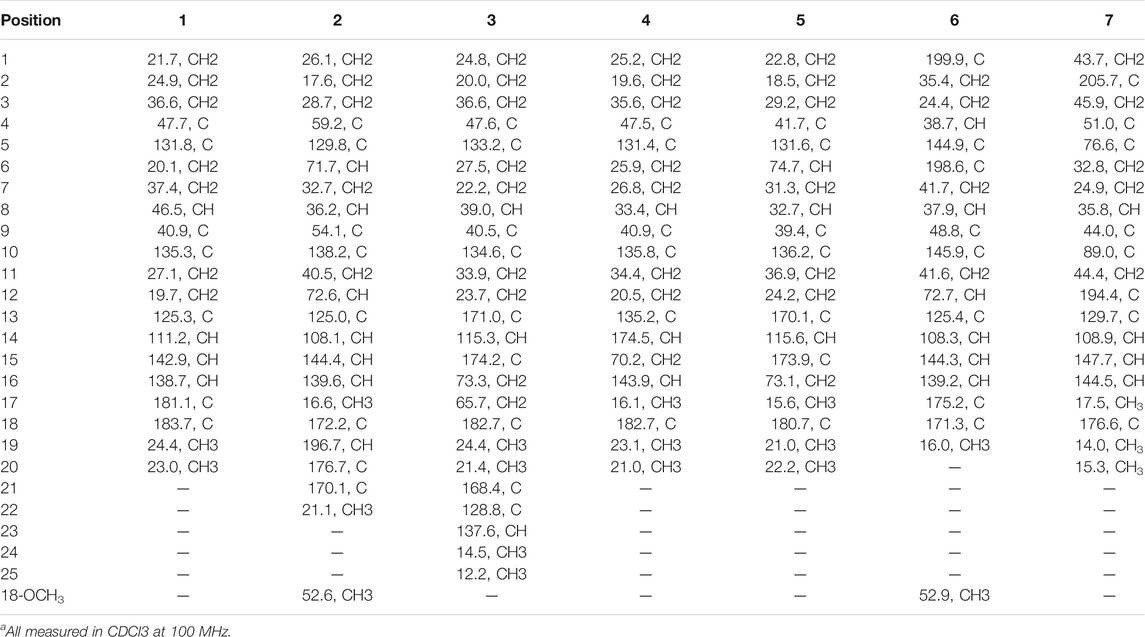
TABLE 2. 13C NMR data of 1–7 (δ in ppm)a.
In this paper, the new compounds we isolated from C. crassifolius all have the clerodane skeleton, which inspired us deriving their possible biosynthetic pathways. Biogenetically, geranylgeranyl diphosphate is the precursor of diterpenoids, and it converts to the halimane system first (Figure 9). The carbocations participate in methyl migration, thereby generates some unsaturated halimanes represented by ∆5,10-halimane (Maslovskaya et al., 2019). In this process, ∆5,10-halimane underwent oxidation to produce crassifolin B, or underwent oxidation and esterification to gain crotohalimaneic acid. On the one side, crassifolin B underwent substitution, transfer, and esterification to afford crassifolin S (3), crassifolin T (4), and crassifolin U (5), respectively. On the other side, crotohalimaneic acid underwent oxidation to obtain crassifolin Q (1) and crotohalimoneic acid. Crotohalimoneic acid underwent esterification and oxidation to get crassifolin R (2) and crassifolin V (6). However, the stereochemistry of C-18 and C-19 of compound 7 and 8 were different for other isolated compounds, which implied their skeleton were different from others.
According to previous reports (Wang et al., 2015; Maslovskaya et al., 2019), clerodane diterpenoids of Croton crassifolius had showed significant anti-inflammatory effect. Therefore, compounds (1–5) were tested for their activities against inflammatory cytokines IL-6 and TNF-α on RAW 264.7 cells. The cytotoxicities of compounds 1–5 were assessed by MTT assay on RAW 264.7 cells. As a result, compounds 1–5 at the concentration of 50 μM had no obvious cytotoxic activities on RAW 264.7 cells after 24 h treatment. In this study, ELISA was applied to evaluate the levels of two inflammatory cytokines IL-6 and TNF-α. As shown in Table 3, compounds 1–5 showed the secretion levels of IL-6 ranging from 32.78 to 77.88%, and the TNF-α ranging from 12.53 to 89.38%. Among them, compound 5 showed the most significant secretion levels of IL-6 and TNF-α at 32.78 and 12.53%, respectively.
Considering the potential anti-angiogenesis activity of diterpenoids from C. crassifolius (Huang et al., 2016; Wang et al., 2016), compounds 1–5 were evaluated for their anti-angiogenesis activities by using tube formation assay in HUVECs. In this assay, we evaluated anti-angiogenesis activities of compounds according to junction densities, vessel percentage areas, and average vessel lengths. Experimental results (Figure 10 and Supplementary Figure S71−Figure S74) showed that compounds 1–5 significantly inhibited angiogenesis by reducing the number of cavities formation. Among these compounds, compound 5 showed the strongest anti-angiogenesis effect, resulting in half maximal inhibitory concentration (IC50) values of junction densities, vessel percentage areas, and average vessel lengths of 7.20 ± 2.23, 48.27 ± 1.98, and 8.62 ± 2.89 μM, respectively. These results revealed that compounds 1–5 significantly inhibited the tube formation of HUVECs and showed their anti-angiogenesis activities. According to the results of experiments with biological activities, crassifolin U (5) showed better inhibitory activities in both inflammation and angiogenesis. Compared with other compounds, there were more lactone rings in the structure of compound 5, which implied that the lactone ring may be an important functional group for anti-inflammatory and anti-angiogenesis activities.
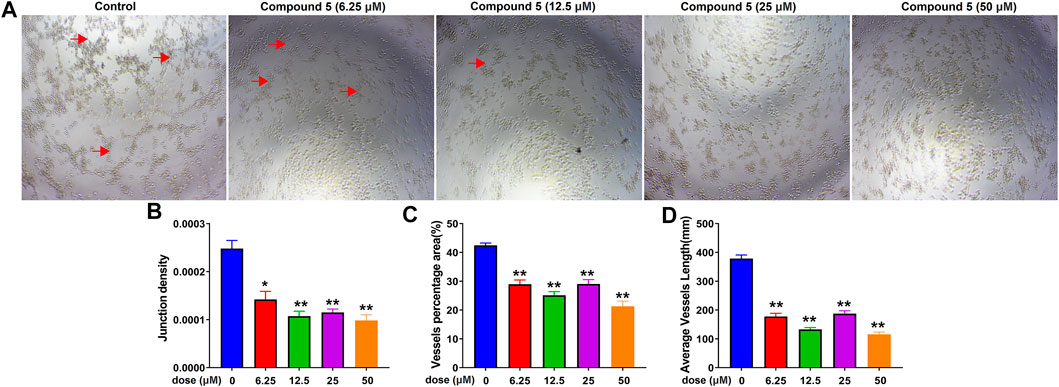
FIGURE 10. The inhibitory effect of compound 5 on angiogenesis. (A) Bright field images of HUVECs seeded on Matrigel for 24 h after treatment at different doses of compound 5; red arrows indicated the cavities formation during angiogenesis. (B), (C), (D) Quantitative analysis on junction densities, vessel percentage areas, and average vessel lengths, respectively. Data are expressed as mean ± SD of three independent experiments. *p < 0.05, **p < 0.005, ***p < 0.001 compared with the control group.
In conclusion, seven new clerodane diterpenoids were isolated from the roots of C. crassifolius. Their structures were identified by comprehensive analysis of spectroscopic data and X-ray crystallography. The anti-inflammatory activities of compounds 1–5 was determined according to two inflammatory cytokines IL-6 and TNF-α, and compound 5 showed the strongest anti-inflammatory ability with the secretion of IL-6 and TNF-α at 32.78 and 12.78%, respectively. Moreover, the results of experiments on anti-angiogenesis activities showed compounds 1–5 inhibited angiogenesis obviously and compound 5 showed the strongest anti-angiogenesis effect with doses in the range of 6.25–50 μM.
UV spectra were measured by a JASCO V-550 UV/VIS spectrophotometer (JASCO, Tokyo, Japan). IR spectra were measured by a JASCO FT/IR-480 plus FT-IR spectrometer with KBr pellets (JASCO, Tokyo, Japan). A JASCO P-1020 polarimeter was used for measuring the optical rotations (JASCO, Tokyo, Japan). HRESIMS data were accomplished using an Agilent 6210 ESI/TOF mass spectrometer (Agilent, Massachusetts, United States). ECD spectra were measured by an Applied Photophysics Chirascan plus CD (Applied Photophysics, United Kingdom). NMR spectra were obtained on a Bruker AV-400 spectrometer with TMS as the internal standard (Bruker, Karlsruhe, Germany), and the chemical shifts (δ) are expressed in ppm and coupling constants (J) in Hz. For column chromatography (CC), silica gel (200–300 mesh, Qingdao Marine Chemical Plant, Qingdao, P. R. China), ODS (50 μm, YMC, Kyoto, Japan), and Sephadex LH-20 (Pharmacia Biotech, Uppsala, Sweden) were used. Silica gel GF254 plates (Yantai Chemical Industry Research Institute, Yantai, China) were used for thin-layer chromatography (TLC). HPLC separations were performed using a COSMOSIL C18 preparative column (5 μm, 20 × 250 mm). All chemical reagents were purchased from Shandong Yuwang Chemical Company (Shandong, P. R. China).
The roots of C. crassifolius were collected from Yulin City, Guangxi province of China. The species was identified by Professor Guangxiong Zhou of Jinan University. A voucher specimen (No. 20200713) is deposited in the Institute of Traditional Chinese Medicine and Natural Products of Jinan University.
The air-dried roots of C. crassifolius (20.0 kg) were extracted three times with 95% ethanol at room temperature. The solution was evaporated under reduced pressure to get a residue (1,500.0 g), which was suspended in water and then partitioned with petroleum ether (PE, 55.6 g), dichloromethane (CH2Cl2, 1,005.3 g), and ethyl acetate (EtOAc, 180.9 g). The dichloromethane extract was chromatographed on a silica gel column chromatograph using the petroleum ether/ethyl acetate (100:0→0:100, v/v) solvent system. The fractions were examined by TLC and combined to give eight fractions (Fr. 1−Fr. 8). Fr. 6 (72.4 g) was subjected to a reversed silica gel column chromatography eluting with MeOH/H2O (30:70→100:0, v/v) to afford six subfractions (Fr. 6A−Fr. 6F). Fr. 6C (6.1 g) was separated by Sephadex LH-20 (CHCl3/MeOH, 1:1, v/v) and further purified by the preparative HPLC (MeOH/H2O, 70:30, v/v) to yield compounds 1 (20.2 mg), 2 (26.8 mg), 3 (23.6 mg), and 8 (19.8 mg). F. 6D (10.2 g) was purified by Sephadex LH-20 (CHCl3/MeOH, 1:1, v/v) to obtain compounds 4 (232.6 mg), 5 (22.4 mg), 6 (230.8 mg), 9 (332.5 mg), and 10 (33.8 mg). Fr. 6E (5.5 g) was separated by the preparative HPLC (MeOH/H2O, 60:40, v/v) to afford compound 7 (30.7 mg), 10 (35.5 mg), 11 (19.7 mg), and 12 (22.9 mg).
Crassifolin Q (1): colorless crystal (MeOH); mp 139–140°C; [α]25D +36.8 (c 1.0, MeOH); HR-ESI-MS m/z 369.1660 [M + Na]+ (calcd for C19H24O7Na, 369.1672); UV (MeOH) λmax 200 nm; IR (KBr) νmax 3,463, 2,956, 1710, 1,461, 1,355, 1,203, 1,037 cm−1; 1H and 13C NMR data see Tables 1, 2. Crassifolin R (2): colorless crystal (MeOH); mp 146–147°C; [α]25D +57.9 (c 1.0, MeOH); HR-ESI-MS m/z 453.1506 [M + Na]+ (calcd for C19H24O7Na, 453.1520); UV (MeOH) λmax 201 nm; IR (KBr) νmax 3,133, 2,965, 1737, 1,450, 1,249, 1,164, 1,018, 808, 748, 603 cm−1; 1H and 13C NMR data see Tables 1, 2. Crassifolin S (3): yellow oil; [α]25D +33.9 (c 1.0, MeOH); HR-ESI-MS m/z 453.2234 [M + Na]+ (calcd for C19H24O7Na, 453.2248); UV (MeOH) λmax 203 nm; IR (KBr) νmax 3,419, 2,942, 1718, 1,446, 1,357, 1,259, 1,155, 1,027, 867 cm−1; ECD (MeOH) λmax (Δε) 224 (+50.9) nm; 1H and 13C NMR data see Tables 1, 2. Crassifolin T (4): white powder; mp 135–136°C; [α]25D -48.6 (c 1.0, MeOH); HR-ESI-MS m/z 355.1869 [M + Na]+ (calcd for C19H24O7Na, 355.1880); UV (MeOH) λmax 201 nm; IR (KBr) νmax 2,946, 2,657, 1735, 1,448, 1,280, 1,195, 1,056, 923, 842 cm−1; ECD (MeOH) λmax (Δε) 201 (+35.0), 224 (−42.1) nm; 1H and 13C NMR data see Tables 1, 2. Crassifolin U (5): yellow oil; (α)25D +42.5 (c 1.0, MeOH); HR-ESI-MS m/z 353.1710 (M + Na)+ (calcd for C19H24O7Na, 353.1723); UV (MeOH) λmax 202 nm; IR (KBr) νmax 2,935, 1743, 1,639, 1,448, 1,236, 1,164, 1,014, 869 cm−1; ECD (MeOH) λmax (Δε) 226 (+10.1), 206 (−2.0) nm; 1H and 13C NMR data see Tables 1, 2. Crassifolin V (6): white powder; mp 144–147°C; [α]25D +22.7 (c 1.0, MeOH); HR-ESI-MS m/z 373.1283 [M + H]+ (calcd for C20H21O7, 373.1282); UV (MeOH) λmax 203 nm; IR (KBr) νmax 2,948, 2,898, 1745, 1,679, 1,434, 1,201, 1,160, 1,025 cm−1; ECD (MeOH) λmax (Δε) 263 (+42.5), 225 (−28.1) nm; 1H and 13C NMR data see Tables 1, 2. Crassifolin W (7): yellow oil; (α)25D -32.5 (c 1.0, MeOH); HR-ESI- MS m/z 361.1644 [M + H]+ (calcd for C20H25O6, 361.1646); UV (MeOH) λmax 200 nm; IR (KBr) νmax 3,436, 2,942, 2,879, 1783, 1720, 1,666, 1,562, 1,511, 1,390, 1,253, 1,155, 871 cm−1; ECD (MeOH) λmax (Δε) 280 (+2.1), 226 (−7.0) nm; 1H and 13C NMR data see Tables 1, 2.
Crystal data for 1: C20H26O5, M = 346.41, orthorhombic, space group P212121, a = 9.129 (5) Å, b = 24.185 (5) Å, c = 17.080 (5) Å, V = 3,771.66 (2) Å3, Z = 8, dx = 1.220 g/cm3, F (000) = 1,488.0, μ(Cu Kα) = 0.708 mm−1. Data collection was performed on a Gemini S Ultra using graphite monochromatic radiation (λ = 1.54184 Å), 2,188 unique reflections were collected to θmax = 74.98°, where 3,898 reflections were observed [F2 > 2σ(F2)]. The structure was solved by direct methods (SHELXS 97) and refined by full matrix least-squares on F2. Final R = 0.0525, Rw = 0.1726, and S = 1.046. Crystallographic data for this structure have been deposited with the Cambridge Crystallographic Data Center as CCDC 2078339 for crassifolin Q (1). Crystal data for 2: C23H26O8, M = 430.44, orthorhombic, space group P212121, a = 8.2304 (1) Å, b = 10.0512 (2) Å, c = 25.6929 (4) Å, V = 2,125.46 (6) Å3, Z = 4, dx = 1.345 g/cm3, F (000) = 912.0, μ(Cu Kα) = 0.850 mm−1. Data collection was performed on a Gemini S Ultra using graphite monochromatic radiation (λ = 1.54184 Å), 2,479 unique reflections were collected to θmax = 73.93°, where 4,308 reflections were observed [F2 > 2σ(F2)]. The structure was solved by direct methods (SHELXS 97) and refined by full matrix least-squares on F2. Final R = 0.0357, Rw = 0.1174, and S = 1.026. Crystallographic data for this structure have been deposited with the Cambridge Crystallographic Data Center as CCDC 2078342 for crassifolin R (2). Crystal data for 6: C20H21O7, M = 372.12, orthorhombic, space group P212121, a = 6.3217 (4) Å, b = 7.3898 (8) Å, c = 10.1209 (7) Å, V = 440.11 (7) Å3, Z = 1, dx = 1.405 g/cm3, F (000) = 196.0, μ(Cu Kα) = 0.895 mm−1. Data collection was performed on a Gemini S Ultra using graphite monochromatic radiation (λ = 1.54184 Å), 2,998 unique reflections were collected to θmax = 74.455°, where 3,600 reflections were observed [F2 > 2σ(F2)]. The structure was solved by direct methods (SHELXS 97) and refined by full matrix least-squares on F2. Final R = 0.0488, Rw = 0.1459, and S = 1.108. Crystallographic data for this structure have been deposited with the Cambridge Crystallographic Data Center as CCDC 2093010 for crassifolin V (6).
RAW 264.7 cells were purchased from American Type Culture Collection. Cells were seeded in a 96-well plate at the density of 5 × 104 cells/ml for 24 h and then the cells were treated with compounds 1–5 for 24 h. The mixture was then removed and each well of the plates was incubated with 30 μl of MTT (5 mg/ml) at 37°C for 4 h. After complete removal of the supernatant, DMSO (100 μl/well) was added into the plates to dissolve the formazan produced in the cells. The absorbance was recorded by a microplate reader at 570 nm.
To quantify the level of proinflammatory cytokines, RAW 264.7 cells were placed in a 24-well plate at a density of 2 × 105/ml for 24 h. Then, the cells were pretreated with 50 μM of isolated compounds 1–5 for 12 h before stimulation with LPS for another 12 h. The level of IL-6 and TNF-α in cell supernatants were quantified with ELISA kits according to the manufacturer's protocols. In this assay, data were expressed as mean ± SD. Statistical significance was considered when p value <0.05.
The effects of compounds on angiogenesis of endothelial cell in vitro were tested using the tube formation assay on Matrigel (BD Biosciences, MA, United States). In this assay, Matrigel (50 μl) was added to the wells of a pre-chilled 96-well plate and put at 37°C for 30 min to congeal. The prepared HUVECs with a density of 2 × 105 cells/well was added and treated with isolated compounds 1–5 at different concentrations. The image of tube formation was collected after 8 h. The junction densities, vessel percentage areas, and average vessel lengths were measured and recorded. In this assay, data were expressed as mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared with the control group.
The original contributions presented in the study are included in the article/Supplementary files, further inquiries can be directed to the corresponding authors.
CL, XS, and WY performed the experiments. CL, ZZ, and QT identified the structures. QT, WW, XZ, and ZW performed the ECD calculation. YL, YZ, and GW conceived and designed the experiments. CL wrote the paper. HZ and CL deduced the biosynthesis pathway. YZ and ZZ revised the paper. All authors have approved the final version of the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81803376, 81973190), the Guangdong Basic and Applied Basic Research Foundation (Nos. 2020B1515020033, 2020A1515110415), the Natural Science Foundation of Guangdong Province (No. 2018B030311020), the Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2017BT01Y036), the Pearl River S&T Nova Program of Guangzhou (201906010069), and the high performance public computing service platform of Jinan University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.733350/full#supplementary-material
Boonyarathanakornkit, L., Che, C.-t., Fong, H., and Farnsworth, N. (1988). Constituents of Croton Crassifolius Roots. Planta Med. 54, 61–63. doi:10.1055/s-2006-962339
Bryant, T. (1868). Remarks on Some Cases of Inflammation of the Breast Simulating Cancer. BMJ 2, 608–609. doi:10.1136/bmj.2.415.608
Campos, M. C. O., Salomão, K., Castro-Pinto, D. B., Leon, L. L., Barbosa, H. S., Maciel, M. A. M., et al. (2010). Croton Cajucara Crude Extract and Isolated Terpenes: Activity on Trypanosoma Cruzi. Parasitol. Res. 107, 1193–1204. doi:10.1007/s00436-010-1988-6
Corlay, N., Delang, L., Girard-Valenciennes, E., Neyts, J., Clerc, P., Smadja, J., et al. (2014). Tigliane Diterpenes from Croton Mauritianus as Inhibitors of Chikungunya Virus Replication. Fitoterapia 97, 87–91. doi:10.1016/j.fitote.2014.05.015
Dao, T.-T., Lee, K.-Y., Jeong, H.-M., Nguyen, P.-H., Tran, T. L., Thuong, P.-T., et al. (2011). Ent-Kaurane Diterpenoids fromCroton tonkinensisStimulate Osteoblast Differentiation. J. Nat. Prod. 74, 2526–2531. doi:10.1021/np200667f
Fernandes, V. C., Pereira, S. I. V., Coppede, J., Martins, J. S., Rizo, W. F., Beleboni, R. O., et al. (2013). The Epimer of Kaurenoic Acid from Croton Antisyphiliticus Is Cytotoxic toward B-16 and HeLa Tumor Cells through Apoptosis Induction. Genet. Mol. Res. 12, 1005–1011. doi:10.4238/2013.April.2.16
Francescone, R., Hou, V., and Grivennikov, S. I. (2014). Microbiome, Inflammation, and Cancer. Cancer J. 20, 181–189. doi:10.1097/PPO.0000000000000048
Huang, H.-L., Qi, F.-M., Yuan, J.-C., Zhao, C.-G., Yang, J.-W., Fang, F.-H., et al. (2014a). Labdane-type Diterpenoids from Croton Laevigatus. RSC Adv. 4, 39530–39540. doi:10.1039/c4ra04863f
Huang, W. H., Li, G. Q., Li, J. G., Ge, W., Chung, H. Y., Wang, G. C., et al. (2014b). Two New Clerodane Diterpenoids from Croton Crassifolius. Heterocycles 89, 1585–1593. doi:10.3987/COM-13-12882
Khanitha, P., and Damrong, S. (2011). Clerodane Diterpenoids and a Trisubstituted Furan from Croton Oblongifolius. Phytochem. Lett. 4, 147–150. doi:10.1016/j.phytol.2011.02.004
Li, F., Zhang, D.-B., Li, J.-T., He, F.-J., Zhu, H.-L., Li, N., et al. (2019). Bioactive Terpenoids from Croton Laui. Nat. Product. Res. 9, 1–9. doi:10.1080/14786419.2019.1675062
Li, H.-H., Qi, F.-M., Dong, L.-L., Fan, G.-X., Che, J.-M., Guo, D.-D., et al. (2014). Cytotoxic and Antibacterial Pyran-2-One Derivatives from Croton Crassifolius. Phytochem. Lett. 10, 304–308. doi:10.1016/j.phytol.2014.10.022
Liu, C.-P., Xu, J.-B., Zhao, J.-X., Xu, C.-H., Dong, L., Ding, J., et al. (2014). Diterpenoids from Croton Laui and Their Cytotoxic and Antimicrobial Activities. J. Nat. Prod. 77, 1013–1020. doi:10.1021/np500042c
Marcos, I. S., Hernández, F. A., Sexmero, M. J., DÍez, D., Basabe, P., Pedrero, A. B., et al. (2003). Synthesis and Absolute Configuration of (−)-chettaphanin I and (−)-chettaphanin II. Tetrahedron 59, 685–694. doi:10.1016/s0040-0(02)01564-8
Mariana, M. B. A., Francisco, C. M. C., Catia, A. A., Humberto, R. B., Rafael, S. D., Galba, M. C., et al. (2013). Antioxidant and Antimicrobial Activities of 7-Hydroxycalamenene-Rich Essential Oils from Croton Cajucara Benth. Molecules 18, 1128–1137. doi:10.3390/molecules18011128
Martha, S. F., Claudia, P. C., Sandra, J. M., Fabio, A., and Coralia, O. (2011). Cytotoxic Labdane Diterpenoids Isolated from the Hexane Fraction of the Croton Stipuliformis Stem Bark. Vitae 18, 173–182.
Martinsen, A., Baccelli, C., Navarro, I., Abad, A., Quetin-Leclercq, J., and Morel, N. (2010). Vascular Activity of a Natural Diterpene Isolated from Croton Zambesicus and of a Structurally Similar Synthetic Trachylobane. Vasc. Pharmacol. 52, 63–69. doi:10.1016/j.vph.2009.11.002
Maslovskaya, L. A., Savchenko, A. I., Gordon, V. A., Reddell, P. W., Pierce, C. J., Boyle, G. M., et al. (2019). New Halimanes from the Australian Rainforest Plant Croton Insularis. Eur. J. Org. Chem. 2019, 1058–1060. doi:10.1002/ejoc.201801548
Maslovskaya, L. A., Savchenko, A. I., Gordon, V. A., Reddell, P. W., Pierce, C. J., Parsons, P. G., et al. (2015). EBC-316, 325-327, and 345: New Pimarane Diterpenes from Croton insularis Found in the Australian Rainforest. Aust. J. Chem. 68, 652–659. doi:10.1071/CH14550
Moses, C., Garcia-Bloj, B., Harvey, A. R., and Blancafort, P. (2018). Hallmarks of Cancer: The CRISPR Generation. Eur. J. Cancer 93, 10–18. doi:10.1016/j.ejca.2018.01.002
Nakatsu, T., Ito, S., and Kawashima, T. (1981). Mallotucin C and D, Two Diterpenic Lactones from Mallotus Repandus. Heterocycles 15, 241–244. doi:10.3987/S-1981-01-0241
Nina, C., Leen, D., Emmanuelle, G. V., Johan, N., Patricia, C., Jacqueline, S., et al. (2014). Tigliane Diterpenes from Croton Mauritianus as Inhibitors of Chikungunya Virus Replication. Fitoterapia 97, 87–91. doi:10.1016/j.fitote.2014.05.015
Qiu, M., Cao, D., Gao, Y., Li, S., Zhu, J., Yang, B., et al. (2016). New Clerodane Diterpenoids from Croton Crassifolius. Fitoterapia 108, 81–86. doi:10.1016/j.fitote.2015.11.016
Rodríguez, B., and Jimeno, M. L. (2004). 1H and13C NMR Spectral Assignments and Conformational Analysis of 14 19-Nor-Neoclerodane Diterpenoids. Magn. Reson. Chem. 42, 605–616. doi:10.1002/mrc.1384
Sun, Y., Wang, M., Ren, Q., Li, S., Xu, J., Ohizumi, Y., et al. (2014). Two Novel Clerodane Diterpenenes with NGF-Potentiating Activities from the Twigs of Croton Yanhuii. Fitoterapia 95, 229–233. doi:10.1016/j.fitote.2014.03.012
Suwancharoen, S., Chonvanich, O., Roengsumran, S., and Pornpakakul, S. (2012). Seco-kaurane Skeleton Diterpenoids from Croton Oblongifolius. Chem. Nat. Compd. 48, 583–586. doi:10.1007/s10600-012-0317-y
Takashi, K., Tokyo, T. N., and Yoshimasa, F. (1976). Diterpenic Lactones of Mallotus Repandus. Heterocycles 5, 227–232.
Wang, G.-C., Li, J.-G., Li, G.-Q., Xu, J.-J., Wu, X., Ye, W.-C., et al. (2012). Clerodane Diterpenoids from Croton Crassifolius. J. Nat. Prod. 75, 2188–2192. doi:10.1021/np300636k
Wang, J.-F., Yang, S.-H., Liu, Y.-Q., Li, D.-X., He, W.-J., Zhang, X.-X., et al. (2015). Five New Phorbol Esters with Cytotoxic and Selective Anti-inflammatory Activities from Croton Tiglium. Bioorg. Med. Chem. Lett. 25, 1986–1989. doi:10.1016/j.bmcl.2015.03.017
Wang, J.-J., Chung, H. Y., Zhang, Y.-B., Li, G.-Q., Li, Y.-L., Huang, W.-H., et al. (2016). Diterpenoids from the Roots of Croton Crassifolius and Their Anti-angiogenic Activity. Phytochemistry 122, 270–275. doi:10.1016/j.phytochem.2015.12.011
Young, H. S., Myung, S. L., Eun, Y. C., Phuong, T. T., Nguyen, M. K., and Song, I. S. (2013). An Ent-Kaurane Diterpenoid from Croton Tonkinensis Induces Apoptosis by Regulating AMP-Activated Protein Kinase in SK-HEP1 Human Hepatocellular Carcinoma Cells. Biol. Pharm. Bull. 36, 158–164. doi:10.1248/bpb.b12-00873
Yuan, Q.-Q., Tang, S., Song, W.-B., Wang, W.-Q., Huang, M., and Xuan, L.-J. (2017). Crassins A-H, Diterpenoids from the Roots of Croton Crassifolius. J. Nat. Prod. 80, 254–260. doi:10.1021/acs.jnatprod.6b00425
Zhang, D.-B., Tang, Z.-S., Xie, P., Liang, Y.-N., Yu, J.-G., Zhang, Z., et al. (2019). A Pair of New Neo-Clerodane Diterpenoid Epimers from the Roots of Croton Crassifolius and Their Anti-inflammatory. Nat. Product. Res. 34, 2945–2951. doi:10.1080/14786419.2019.1601193
Keywords: Euphorbiaceae, Croton crassifolius, clerodane diterpenoids, anti-inflammatory activity, anti-angiogenic activity
Citation: Li C, Sun X, Yin W, Zhan Z, Tang Q, Wang W, Zhuo X, Wu Z, Zhang H, Li Y, Zhang Y and Wang G (2021) Crassifolins Q−W: Clerodane Diterpenoids From Croton crassifolius With Anti-Inflammatory and Anti-Angiogenesis Activities. Front. Chem. 9:733350. doi: 10.3389/fchem.2021.733350
Received: 30 June 2021; Accepted: 02 August 2021;
Published: 20 September 2021.
Edited by:
Simone Brogi, University of Pisa, ItalyReviewed by:
Fateh V Singh, VIT University, IndiaCopyright © 2021 Li, Sun, Yin, Zhan, Tang, Wang, Zhuo, Wu, Zhang, Li, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaolan Li, dGxpeWxAam51LmVkdS5jbg==; Yubo Zhang, eWJ6aGFuZzk5QDEyNi5jb20=; Guocai Wang, dHdhbmdndW9jYWlAam51LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.