
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 27 July 2021
Sec. Analytical Chemistry
Volume 9 - 2021 | https://doi.org/10.3389/fchem.2021.715890
This article is part of the Research Topic Methodological Advances of MALDI Mass Spectrometry-Based Techniques in Organic and Biomedical Analysis View all 10 articles
Rapid diagnostics of bacterial infection is the key to successful recovery and eradication of the disease. Currently, identification of bacteria is based on the detection of highly abundant proteins, mainly ribosomal proteins, by routine MALDI-TOF mass spectrometry. However, relying solely on proteins is limited in subspecies typing for some pathogens. This is the case for, for example, the mycobacteria belonging to the Mycobacterium abscessus (MABS) complex, which is classified into three subspecies, namely, M. abscessus subsp. abscessus, M. abscessus subsp. bolletii, and M. abscessus subsp. massiliense. Being able to detect bacteria accurately and rapidly at the subspecies level could not only reliably identify the pathogen causing the disease but also enable better antibiotic stewardship. For instance, M. abscessus subsp. abscessus and M. abscessus subsp. bolletii possess a functional erm41 (erythromycin ribosomal methylation gene 41) gene, whilst M. abscessus subsp. massiliense does not, resulting in differences in macrolide antibiotic (e.g., clarithromycin and azithromycin) susceptibilities. This presents a challenge for physicians when designing an appropriate treatment regimen. To address this challenge, in addition to proteins, species-specific lipids have now been considered as a game changer in clinical microbiology diagnostics. However, their extraction can be time-consuming, and analysis requires the use of apolar toxic organic solvents (e.g., chloroform). Here, we present a new method to accurately detect species and subspecies, allowing the discrimination of the mycobacteria within the MABS complex and relying on the use of ethanol. We found that a combination of the matrix named super-DHB with 25% ethanol with a bacterial suspension at McFarland 20 gave robust and reproducible data, allowing the discrimination of the bacteria within the MABS complex strains tested in this study (n = 9). Further investigations have to be conducted to validate the method on a larger panel of strains for its use in diagnostic laboratories.
Matrix-assisted laser desorption ionization/time-of-flight (MALDI-TOF) mass spectrometry (MS), a cost-effective, rapid, and accurate method for microbial identification based on protein signatures, has revolutionized microbiological diagnostics (Croxatto et al., 2012; Clark et al., 2013; Singhal et al., 2015). However, protein profiling by MALDI-TOF MS possesses some limitations when it comes to an efficient typing for some closely related species and subspecies (Kostrzewa et al., 2019; Welker et al., 2019).
That is the case for the bacterium named Mycobacterium abscessus, which is part of a complex of very similar organisms. The M. abscessus complex (MABS complex) is classified into three subspecies, namely, M. abscessus subsp. abscessus, M. abscessus subsp. bolletii, and M. abscessus subsp. massiliense. M. abscessus is found to be present within 80% of the respiratory isolates of rapidly growing mycobacteria and demonstrates resistance to many antibiotic classes like aminoglycosides, β-lactams, and macrolides (Johansen et al., 2020). For example, M. abscessus subsp. abscessus and M. abscessus subsp. bolletii possess a functional erm41 (erythromycin ribosomal methylation gene 41) gene, whilst M. abscessus subsp. massiliense does not (Kim et al., 2010; Ryu et al., 2016; Ryan and Byrd, 2018). This results in different macrolide antibiotic (i.e., clarithromycin and azithromycin) susceptibilities. Therefore, identification of the different subspecies is important in the clinical environment. As mentioned earlier, clarithromycin is more effective against M. abscessus subsp. massiliense lung infections, while resistance is commonly observed in M. abscessus subsp. abscessus and M. abscessus subsp. bolletii isolates (Adekambi et al., 2006; Esther et al., 2010; Kim et al., 2010; Koh et al., 2011; Harada et al., 2012). This presents a challenge for physicians when designing an appropriate treatment regimen (Koh et al., 2011; Brown-Elliott et al., 2012; Harada et al., 2012; Kothavade et al., 2013).
Current available tools, such as PCR, sequencing of multiple genes, and whole genome sequencing, are time-consuming, relatively expensive, and require experts for the sample preparation and data analysis, which is not ideal for routine clinical use despite offering excellent subspecies identification (Nash et al., 2009; Kim et al., 2010; Koh et al., 2011). That is why having a diagnostic tool that allows early MABS complex identification would be ideal for subspecies identification and therefore guiding drug prescription (Ryu et al., 2016). In order to introduce this tool across clinical laboratories worldwide, it needs to be simple, rapid, inexpensive, and highly specific. Therefore, the continuous development of laboratory techniques for subspecies-level discrimination between bacterial species alongside the identification of their antibiotic-resistant counterparts is of utmost importance to best assist physicians in the administration of an appropriate drug regimen.
MALDI-TOF MS offers an alternative to the methods cited earlier. Indeed, several studies report on the use of MALDI-TOF MS to detect bacteria within the MABS complex (Tseng et al., 2013; Fangous et al., 2014; Panagea et al., 2015; Kehrmann et al., 2016; Luo et al., 2016). Those approaches rely on a multistep sample preparation consisting of the inactivation of the bacteria, followed by mechanical lysis in order to have access to intracellular proteins that serve as markers. However, the multistep strategy combined with extensive data analysis used to detect the MABS complex can be improved in order to ease routine microbiology laboratory practice.
In terms of addressing this unmet need, species-specific lipid fingerprints could represent an attractive approach when it comes to microbial identification by MALDI-ToF MS. Lipids are highly abundant in bacteria and structurally diverse. Several studies have reported successfully on the use of microbial lipidomics by MALDI-ToF MS and their uses in academic laboratories and diagnostic laboratories for accurate microbial identification, making lipid fingerprinting a complementary approach to protein-based fingerprinting in order to tackle the challenge of microbial differentiation by MALDI-ToF MS (Shu. et al., 2012; Voorhees et al., 2013; Cox et al., 2015; Larrouy-Maumus and Puzo, 2015; Leung et al., 2017; Gonzalo et al., 2020; Lellman and Cramer, 2020).
Nevertheless, those methods, based on lipid fingerprints (cited earlier), rely on the use of highly toxic organic solvents (e.g., CHCl3, CH3OH, or a mixture of CHCl3 and CH3OH), hampering an easy implementation of the method in routine clinical laboratories due to safety concerns. Also, those solvents have a relative polarity ranging from 0.259 to 0.762, which could have an impact on the selectivity of the species-specific lipids to be extracted and detected by MALDI-TOF MS, influencing the outcome of bacterial identification. To address this challenge, we investigated the use of ethanol (CH3CH2OH, relative polarity 0.654) as a commonly used organic solvent widely used in routine clinical microbiology laboratories worldwide as an alternative to chloroform and methanol to extract species-specific lipids for the rapid identification of bacteria belonging to the MABS complex.
M. abscessus subsp. abscessus ATCC 19977, M. abscessus subsp. abscessus M02004436, M. abscessus subsp. abscessus M07001466, M. abscessus subsp. bolletii M09013950, M. abscessus subsp. bolletii M04009973, M. abscessus subsp. bolletii H123680049, M. abscessus subsp. massiliense M10010169, M. abscessus subsp. massiliense M07012434, and M. abscessus subsp. massiliense H120280022 obtained from the National Mycobacterium Reference-NMRS-South (United Kingdom) were cultured in Middlebrook 7H9 broth liquid medium supplemented with 0.5 g/L Fraction V (bovine serum albumin), 0.05% tyloxapol, 0.2% dextrose, 0.2% glycerol, and 10 mM NaCl. The inoculated cultures were then incubated at 37°C for 2 days. Bacterial pellets were heat-killed at 95°C for 30 min before leaving the BSL3 containment area.
100 µl of bacterial suspension (from liquid- or agar-based media) was placed into 1.5 ml microtubes that contain 200 µl of double-distilled water. The bacteria were then washed twice with 200 μl of double-distilled water and suspended at various McFarland standards ranging from 5 to 40 using a McFarland tube densitometer (Grant-Bio®). Then, 0.5 µl of the resuspended pellet was pipetted onto the MALDI target plate and mixed with 1.2 µl of the MALDI matrix (Figure 1). The matrix used consisted of a 9:1 mixture of 2,5-dihydroxybenzoic acid and 2-hydroxy-5-methoxybenzoic acid (super-DHB, Sigma-Aldrich) at a concentration of 10 mg/ml in ethanol at various percentages ranging from 0 to 100%. Additionally, for external calibration, 0.5 µl of calibration peptide mixture was loaded along with 0.5 µl of the given calibration matrix (peptide calibration standard II, Bruker Daltonics, Germany). The samples were loaded onto a disposable MBT 96 Biotarget plate (Bruker Part-No. 1840375).
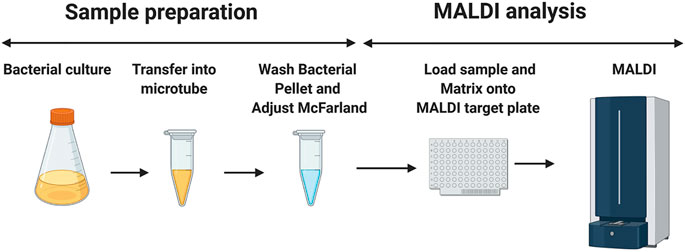
FIGURE 1. Schematic diagram of the sample preparation before the MALDI mass spectrometry measurement. 1.5 ml of mycobacterial culture is aliquoted in a microtube. The mycobacterial suspension is then washed twice with double-distilled water and adjusted to the desired McFarland suspension. 0.5 μL of this preparation is loaded into the MALDI target plate, followed by the addition of 1.2 μl of the matrix (super-DHB solubilized at 10 mg/ml in ethanol) and mixing on the MALDI MBT 96 Biotarget plate. Once dried, the mass spectra are acquired in the linear positive ion mode. The image has been created using BioRender.
MS analyses were performed on an MALDI Biotyper Sirius system (Bruker Daltonics, Germany). The mass spectra were scanned in the range of m/z 1,100–1,600. The mass profiles were acquired using FlexControl 3.4 software (Bruker Daltonics, Germany). The spectra were recorded in the linear positive ion mode (laser intensity 95%, ion source 1 = 10.00 kV, ion source 2 = 8.98 kV, lens = 3.00 kV, detector voltage = 2652 V, pulsed ion extraction = 150 ns). Each spectrum corresponded to an ion accumulation of 5,000 laser shots randomly distributed on the spot. The spectra obtained were processed with default parameters using FlexAnalysis v.3.4 software (Bruker Daltonics, Germany).
In this study, we optimized the number of bacteria loaded onto the MALDI target plate and the solvent to solubilize the matrix prior to MS analyses in the positive ion mode to address the major challenge in subspecies typing by MALDI-TOF MS inside the MABS complex.
By using the reference strain M. abscessus subsp. abscessus ATCC 19977, we found that a suspension of bacteria at McFarland 20 combined with the matrix super-DHB solubilized in 25% ethanol at a final concentration of 10 mg/ml was most appropriate for our experiments. We chose to use super-DHB as a matrix due to its versatility for the analysis of lipids and glycolipids (Schiller et al., 2007). This was decided following observation of the raw mass spectra obtained across 5 McFarland dilutions (5, 10, 20, 30, and 50) and 5 percentages of ethanol used to solubilize the matrix (10, 25, 50, 70, and 100%). Regardless of the McFarland dilution and percentage of ethanol used in the study, the positive ion mass spectra of M. abscessus subsp. abscessus ATCC 19977 showed two sets of peaks starting at m/z 1201.8 up to m/z 1299.9 and starting at m/z 1375.9 up to m/z 1445.9, distant of 14 mass units with the most intense peaks at m/z 1259.9 and m/z 1405.9 assigned to potassium cationized–diglycosylated glycopeptidolipids ([M + K]+m/z 1259.9) and triglycosylated glycopeptidolipids ([M + K]+m/z 1405.9) (Figure 2A), which are known to make up more than 70% of the surface-exposed mycobacterial lipids. Glycopeptidolipids have been demonstrated to play a key role in mycobacterial physiology and pathogenicity (Byrd and Lyons, 1999; Howard et al., 2006; Catherinot et al., 2007; Ripoll et al., 2007; Whang et al., 2017; Gutierrez et al., 2018; Tran et al., 2019; Kurz and Rivas-Santiago, 2020; Jackson et al., 2021). Such lipid profiles are similar in terms of lipid species detected to the ones that have been obtained in the literature using conventional methods (Ripoll et al., 2007; Gonzalo et al., 2020); this indicates that as a solvent (to solubilize the matrix), ethanol produces efficient extraction and co-crystallization, and thereby desorption and ionization of surface lipids on the MALDI target plate. A possible mechanism is that ethanol could act as an eraser enabling the selective on-target extraction of the surface-exposed di- and triglycosylated glycopeptidolipids and their co-crystallization with the matrix, allowing their desorption and ionization.
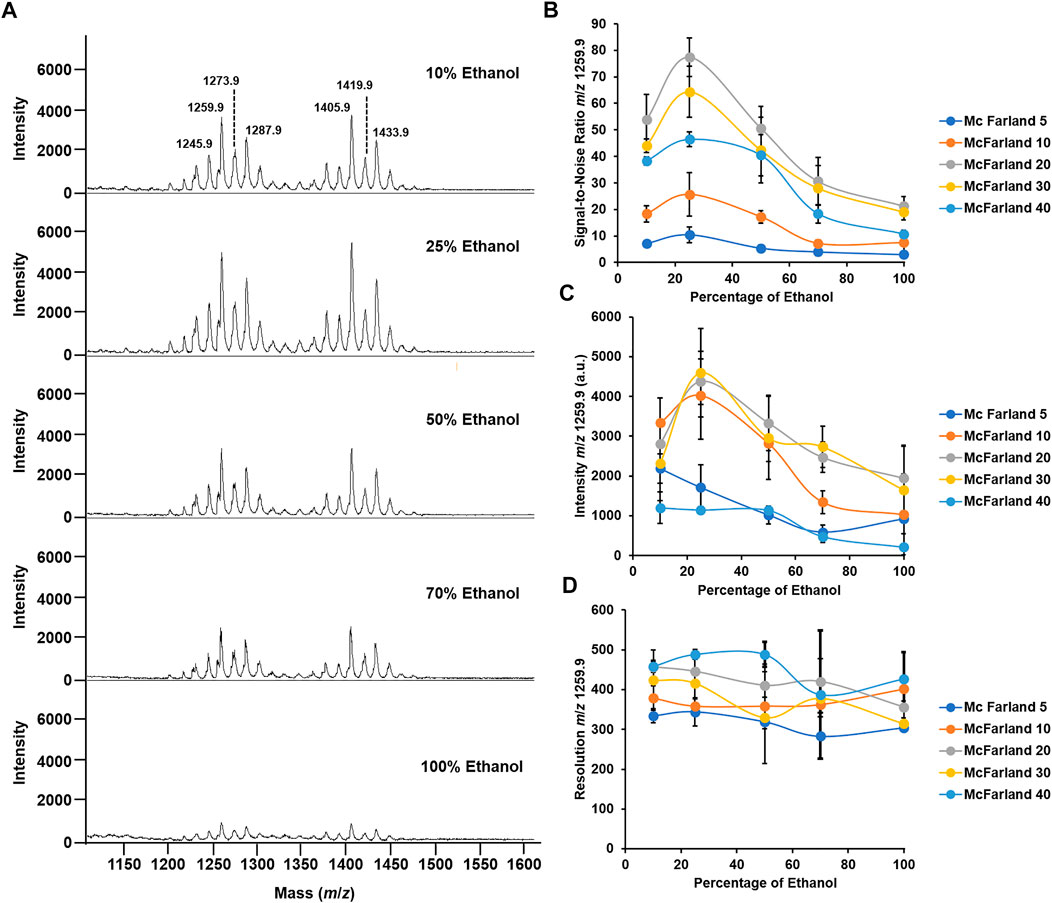
FIGURE 2. Optimization of McFarland and percentage of ethanol for the optimal mass spectrum signal. (A) Mass spectra acquired in the positive ion mode of M. abscessus subsp. abscessus ATCC 19977 set at McFarland 20 and for a percentage of ethanol to solubilize the matrix ranging from 10 to 100%. (B) Graph depicting the changes in the S/N ratio in the function of the percentage of ethanol across 5 McFarland dilutions. (C) Graph depicting the changes in the intensity of the peak at m/z 1259.9 in the function of the percentage of ethanol across 5 McFarland dilutions. (D) Graph depicting the changes in the resolution of the peak at m/z 1259.9 in the function of the percentage of ethanol across 5 McFarland dilutions. Experiments have been performed in biological and technical triplicates. Error bars indicate the standard deviation.
In addition, here, we aimed to have the highest quality signal in order to enhance reproducible and robust data. To decide which bacterial suspension and percentage of ethanol were appropriate for performing the analysis, we recorded the signal-to-noise (S/N), resolution, and intensity of the dominant peak at m/z 1259.9. First, regarding the S/N, as seen in Figure 2B, at McFarland 5, the S/N is ∼10 across all percentages of ethanol tested. Interestingly, when using a bacterial suspension of McFarland 10, the S/N increases when passing from 10 to 25% ethanol, going from S/N 18 to 25, and then decreases and plateaus at S/N 7.5 for a percentage of ethanol of 100%. A similar pattern is observed when using higher McFarland values. Indeed, for a bacterial suspension set at McFarland 20, at 10% ethanol, the S/N is ∼50, which is almost 3 times higher than it is when using McFarland 10, and it even increases up to S/N ∼80 for a value of 25% ethanol before decreasing gradually to S/N ∼20 for 100% ethanol. However, when using higher bacterial suspension, for example, McFarland 30 or 40, the S/N tends to decrease across all percentages of ethanol tested compared to the S/N observed for McFarland 20, going for S/N to at 25% ethanol to S/N ∼60 and S/N ∼40 for the same percentage of ethanol. Such observations could be explained by the ratio of the matrix to the sample required for the optimal co-crystallization of the molecules of interest with the matrix on the MALDI target plate and allowing their optimal desorption and ionization (Smolira and Wessely-Szponder, 2015; Wiangnon and Cramer, 2015; Wang et al., 2016; Boesl, 2017). In other words, the fact that higher McFarland or higher percentage of ethanol did not generate good quality spectra could be explained by the poor co-crystallization with the matrix. That could lead to a nonhomogeneous spot on the MALDI target plate, precluding any transfer of the energy provided by the laser to the matrix in allowing the desorption of the molecules of interest (Dreisewerd, 2003; Wang et al., 2016).
A similar conclusion can be drawn for the intensity of the signal. As seen in Figure 2C, for both McFarland 5 and McFarland 40, the signal intensity remains low across all the percentages of ethanol tested, with an intensity of ∼1,000 a.u. However, for intermediate McFarland values, when going from 10% ethanol to 25% ethanol, the intensity of the signal increases from ∼2,000 a.u. to 5,000 a.u. and then decreases gradually when reaching 100% ethanol. That agrees with the S/N data where McFarland 20 and 25% ethanol are the parameters required for the optimal quality signal.
Regarding the resolution, neither McFarland nor the percentage of ethanol seems to influence that parameter which has its range from 300 to 500 with no significative differences (Figure 2D).
Taken together, a bacterial suspension set at McFarland 20 and the use of the super-DHB solubilized at 10 mg/ml in 25% ethanol gave the most appropriate quality data for bacterial species identification by routine MALDI-TOF MS.
To test if that optimized methodological approach has the potential to address the challenge of subspecies typing in the MABS complex, we applied the developed method to M. abscessus subsp. abscessus ATCC 19977, M. abscessus subsp. abscessus M02004436, M. abscessus subsp. abscessus M07001466, M. abscessus subsp. bolletii M09013950, M. abscessus subsp. bolletii M04009973, M. abscessus subsp. bolletii H123680049, M. abscessus subsp. massiliense M10010169, M. abscessus subsp. massiliense M07012434, and M. abscessus subsp. massiliense H120280022. . As seen in Figure 3 for the mass spectra for the M. abscessus subsp. abscessus strains, the mass spectra for the M. abscessus subsp. bolletii strains and M. abscessus subsp. massiliense strains are composed of two sets of peaks starting at m/z 1201.8 up to m/z 1299.9 and starting at m/z 1375.9 up to m/z 1445.9, distant of 14 mass units with the most intense peaks assigned to potassium cationized–diglycosylated glycopeptidolipids and triglycosylated glycopeptidolipids. However, as opposed to the mass spectrum of M. abscessus subsp. abscessus strains, where the ratio of diglycosylated glycopeptidolipids/triglycosylated glycopeptidolipids is ∼1:1, in the mass spectra of M. abscessus subsp. bolletii strains, the ratio of diglycosylated glycopeptidolipids/triglycosylated glycopeptidolipids is ∼1:0.2 and for M. abscessus subsp. massiliense strains, the ratio of diglycosylated glycopeptidolipids/triglycosylated glycopeptidolipids is ∼1:0.5 (Figure 3). Taken together, those data suggest that the lipid fingerprint has the potential to discriminate between mycobacteria within the MABS complex.
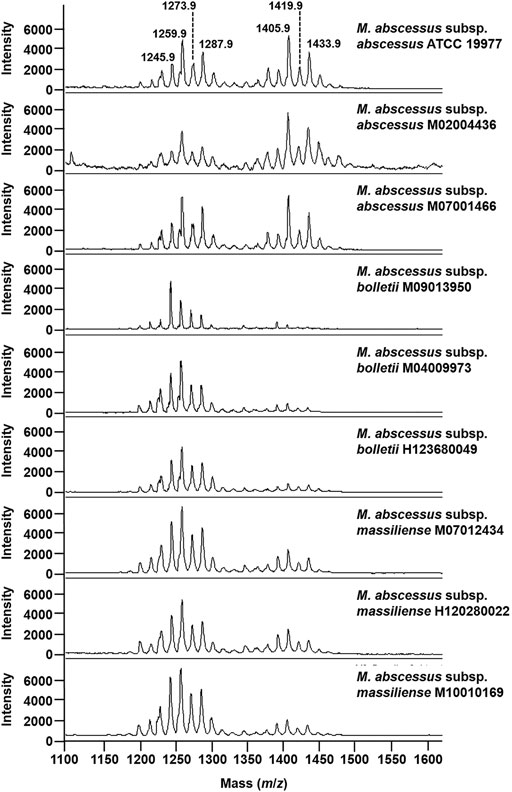
FIGURE 3. Mass spectra of mycobacteria belonging to the M. abscessus complex. Mass spectra were acquired in the positive ion mode with a bacterial suspension set at McFarland 20 and using the super-DHB as a matrix solubilized at 10 mg/ml in 25% ethanol.
Here, we provide a new and simple method for the detection of subspecies-specific lipids, applied to the MABS complex. The method is easy to put in place, requires a minimal number of steps, and is performed on a routine MALDI mass spectrometer in the positive ion mode. One of the limitations of the study resides in the use of a very limited amount of strains (n = 9). Further studies are now required to test and validate that new approach in the rapid detection of the mycobacteria belonging to the MABS complex, using characterized clinical isolates and other bacterial species and subspecies.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
GL-M and MJK conceived the study, participated in its design, and performed the experiments. AB and FD provided bacterial strains. All authors discussed, reviewed, and approved the final manuscript.
This study was supported by the MRC Confidence in Concept Fund and the ISSF Wellcome Trust Grant 105603/Z/14/Z (GL-M).
GL-M and FD are coinventors of the method “Screening With Mass Spectrometry For Mycobacteria Prior To Cardiothoracic Surgery,” for which a patent has been filed by Imperial Technology Transfer (International Publication Number WO2021064426A1). Author MK is employed by Bruker Daltonics GmbH & Co. KG (Bremen, Germany).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adekambi, T., Berger, P., Raoult, D., and Drancourt, M. (2006). rpoB Gene Sequence-Based Characterization of Emerging Non-tuberculous Mycobacteria with Descriptions of Mycobacterium Bolletii Sp. Nov., Mycobacterium Phocaicum Sp. Nov. And Mycobacterium Aubagnense Sp. Nov. Int. J. Syst. Evol. Microbiol. 56 (Pt 1), 133–143. doi:10.1099/ijs.0.63969-0
Boesl, U. (2017). Time-of-flight Mass Spectrometry: Introduction to the Basics. Mass. Spec. Rev. 36 (1), 86–109. doi:10.1002/mas.21520
Brown-Elliott, B. A., Nash, K. A., and Wallace, R. J. (2012). Antimicrobial Susceptibility Testing, Drug Resistance Mechanisms, and Therapy of Infections with Nontuberculous Mycobacteria. Clin. Microbiol. Rev. 25 (3), 545–582. doi:10.1128/CMR.05030-11
Byrd, T. F., and Lyons, C. R. (1999). Preliminary Characterization of a Mycobacterium Abscessus Mutant in Human and Murine Models of Infection. Infect. Immun. 67 (9), 4700–4707. doi:10.1128/IAI.67.9.4700-4707.1999
Catherinot, E., Clarissou, J., Etienne, G., Ripoll, F., Emile, J.-F., Daffé, M., et al. (2007). Hypervirulence of a Rough Variant of the Mycobacterium Abscessus Type Strain. Infect. Immun. 75 (2), 1055–1058. doi:10.1128/IAI.00835-06
Clark, A. E., Kaleta, E. J., Arora, A., and Wolk, D. M. (2013). Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry: a Fundamental Shift in the Routine Practice of Clinical Microbiology. Clin. Microbiol. Rev. 26 (3), 547–603. doi:10.1128/CMR.00072-12
Cox, C. R., Jensen, K. R., Saichek, N. R., and Voorhees, K. J. (2015). Strain-Level Bacterial Identification by CeO2-Catalyzed MALDI-TOF MS Fatty Acid Analysis and Comparison to Commercial Protein-Based Methods. Sci. Rep. 5, 10470. doi:10.1038/srep10470
Croxatto, A., Prod'hom, G., and Greub, G. (2012). Applications of MALDI-TOF Mass Spectrometry in Clinical Diagnostic Microbiology. FEMS Microbiol. Rev. 36 (2), 380–407. doi:10.1111/j.1574-6976.2011.00298.x
Dreisewerd, K. (2003). The Desorption Process in MALDI. Chem. Rev. 103 (2), 395–426. doi:10.1021/cr010375i
Esther, C. R., Esserman, D. A., Gilligan, P., Kerr, A., and Noone, P. G. (2010). Chronic Mycobacterium Abscessus Infection and Lung Function Decline in Cystic Fibrosis. J. Cystic Fibrosis 9 (2), 117–123. doi:10.1016/j.jcf.2009.12.001
Fangous, M.-S., Mougari, F., Gouriou, S., Calvez, E., Raskine, L., Cambau, E., et al. (2014). Classification Algorithm for Subspecies Identification within the Mycobacterium Abscessus Species, Based on Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 52 (9), 3362–3369. doi:10.1128/JCM.00788-14
Gonzalo, X., Broda, A., Drobniewski, F., and Larrouy-Maumus, G. (2021). Performance of Lipid Fingerprint-Based MALDI-ToF for the Diagnosis of Mycobacterial Infections. Clin. Microbiol. Infect. 27, 912.e1–912.e2. doi:10.1016/j.cmi.2020.08.027
Gutiérrez, A. V., Viljoen, A., Ghigo, E., Herrmann, J.-L., and Kremer, L. (2018). Glycopeptidolipids, a Double-Edged Sword of the Mycobacterium Abscessus Complex. Front. Microbiol. 9, 1145. doi:10.3389/fmicb.2018.01145
Harada, T., Akiyama, Y., Kurashima, A., Nagai, H., Tsuyuguchi, K., Fujii, T., et al. (2012). Clinical and Microbiological Differences between Mycobacterium Abscessus and Mycobacterium Massiliense Lung Diseases. J. Clin. Microbiol. 50 (11), 3556–3561. doi:10.1128/JCM.01175-12
Howard, S. T., Rhoades, E., Recht, J., Pang, X., Alsup, A., Kolter, R., et al. (2006). Spontaneous Reversion of Mycobacterium Abscessus from a Smooth to a Rough Morphotype Is Associated with Reduced Expression of Glycopeptidolipid and Reacquisition of an Invasive Phenotype. Microbiology (Reading) 152 (Pt 6), 1581–1590. doi:10.1099/mic.0.28625-0
Jackson, M., Stevens, C. M., Zhang, L., Zgurskaya, H. I., and Niederweis, M. (2021). Transporters Involved in the Biogenesis and Functionalization of the Mycobacterial Cell Envelope. Chem. Rev. 121 (9), 5124–5157. doi:10.1021/acs.chemrev.0c00869
Johansen, M. D., Herrmann, J.-L., and Kremer, L. (2020). Non-tuberculous Mycobacteria and the Rise of Mycobacterium Abscessus. Nat. Rev. Microbiol. 18 (7), 392–407. doi:10.1038/s41579-020-0331-1
Kehrmann, J., Wessel, S., Murali, R., Hampel, A., Bange, F.-C., Buer, J., et al. (2016). Principal Component Analysis of MALDI TOF MS Mass Spectra Separates M. Abscessus (Sensu Stricto) from M. Massiliense Isolates. BMC Microbiol. 16, 24. doi:10.1186/s12866-016-0636-4
Kim, H.-Y., Kim, B. J., Kook, Y., Yun, Y.-J., Shin, J. H., Kim, B.-J., et al. (2010). Mycobacterium Massiliense Is Differentiated from Mycobacterium Abscessus and Mycobacterium Bolletii by Erythromycin Ribosome Methyltransferase Gene (Erm) and Clarithromycin Susceptibility Patterns. Microbiol. Immunol. 54 (6), 347–353. doi:10.1111/j.1348-0421.2010.00221.x
Koh, W.-J., Jeon, K., Lee, N. Y., Kim, B.-J., Kook, Y.-H., Lee, S.-H., et al. (2011). Clinical Significance of Differentiation ofMycobacterium massiliensefromMycobacterium Abscessus. Am. J. Respir. Crit. Care Med. 183 (3), 405–410. doi:10.1164/rccm.201003-0395OC
Kostrzewa, M., Nagy, E., Schröttner, P., and Pranada, A. B. (2019). How MALDI-TOF Mass Spectrometry Can Aid the Diagnosis of Hard-To-Identify Pathogenic Bacteria - the Rare and the Unknown. Expert Rev. Mol. Diagn. 19 (8), 667–682. doi:10.1080/14737159.2019.1643238
Kothavade, R. J., Dhurat, R. S., Mishra, S. N., and Kothavade, U. R. (2013). Clinical and Laboratory Aspects of the Diagnosis and Management of Cutaneous and Subcutaneous Infections Caused by Rapidly Growing Mycobacteria. Eur. J. Clin. Microbiol. Infect. Dis. 32 (2), 161–188. doi:10.1007/s10096-012-1766-8
Kurz, S. G., and Rivas-Santiago, B. (2020). Time to Expand the Picture of Mycobacterial Lipids: Spotlight on Nontuberculous Mycobacteria. Am. J. Respir. Cel Mol. Biol. 62 (3), 275–276. doi:10.1165/rcmb.2019-0324ED
Larrouy-Maumus, G., and Puzo, G. (2015). Mycobacterial Envelope Lipids Fingerprint from Direct MALDI-TOF MS Analysis of Intact Bacilli. Tuberculosis 95 (1), 75–85. doi:10.1016/j.tube.2014.11.001
Lellman, S. E., and Cramer, R. (2020). Bacterial Identification by Lipid Profiling Using Liquid Atmospheric Pressure Matrix-Assisted Laser Desorption/ionization Mass Spectrometry. Clin. Chem. Lab. Med. 58 (6), 930–938. doi:10.1515/cclm-2019-0908
Leung, L. M., Fondrie, W. E., Doi, Y., Johnson, J. K., Strickland, D. K., Ernst, R. K., et al. (2017). Identification of the ESKAPE Pathogens by Mass Spectrometric Analysis of Microbial Membrane Glycolipids. Sci. Rep. 7 (1), 6403. doi:10.1038/s41598-017-04793-4
Luo, L., Liu, W., Li, B., Li, M., Huang, D., Jing, L., et al. (2016). Evaluation of Matrix-Assisted Laser Desorption Ionization−Time of Flight Mass Spectrometry for Identification of Mycobacterium Abscessus Subspecies According to Whole-Genome Sequencing. J. Clin. Microbiol. 54 (12), 2982–2989. doi:10.1128/JCM.01151-16
Nash, K. A., Brown-Elliott, B. A., and Wallace, R. J. (2009). A Novel Gene, Erm (41), Confers Inducible Macrolide Resistance to Clinical Isolates of Mycobacterium Abscessus but Is Absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 53 (4), 1367–1376. doi:10.1128/AAC.01275-08
Panagea, T., Pincus, D. H., Grogono, D., Jones, M., Bryant, J., Parkhill, J., et al. (2015). Mycobacterium Abscessus Complex Identification with Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 53 (7), 2355–2358. doi:10.1128/JCM.00494-15
Ripoll, F., Deshayes, C., Pasek, S., Laval, F., Beretti, J.-L., Biet, F., et al. (2007). Genomics of Glycopeptidolipid Biosynthesis in Mycobacterium Abscessus and M. chelonae. BMC Genomics 8, 114. doi:10.1186/1471-2164-8-114
Ryan, K., and Byrd, T. F. (2018). Mycobacterium Abscessus: Shapeshifter of the Mycobacterial World. Front. Microbiol. 9, 2642. doi:10.3389/fmicb.2018.02642
Ryu, Y. J., Koh, W.-J., and Daley, C. L. (2016). Diagnosis and Treatment of Nontuberculous Mycobacterial Lung Disease: Clinicians' Perspectives. Tuberc. Respir. Dis. 79 (2), 74–84. doi:10.4046/trd.2016.79.2.74
Schiller, J., Süß, R., Fuchs, B., Müller, M., Petković, M., Zschörnig, O., et al. (2007). The Suitability of Different DHB Isomers as Matrices for the MALDI-TOF MS Analysis of Phospholipids: Which Isomer for what Purpose? Eur. Biophys. J. 36 (4-5), 517–527. doi:10.1007/s00249-006-0090-6
Shu, X., Li, Y., Liang, M., Yang, B., Liu, C., Wang, Y., et al. (2012). Rapid Lipid Profiling of Bacteria by Online MALDI-TOF Mass Spectrometry. Int. J. Mass Spectrom. 321-322, 71–76. doi:10.1016/j.ijms.2012.05.016
Singhal, N., Kumar, M., Kanaujia, P. K., and Virdi, J. S. (2015). MALDI-TOF Mass Spectrometry: an Emerging Technology for Microbial Identification and Diagnosis. Front. Microbiol. 6, 791. doi:10.3389/fmicb.2015.00791
Smolira, A., and Wessely-Szponder, J. (2015). Importance of the Matrix and the Matrix/sample Ratio in MALDI-TOF-MS Analysis of Cathelicidins Obtained from Porcine Neutrophils. Appl. Biochem. Biotechnol. 175 (4), 2050–2065. doi:10.1007/s12010-014-1405-1
Tran, T., Bonham, A. J., Chan, E. D., and Honda, J. R. (2019). A Paucity of Knowledge Regarding Nontuberculous Mycobacterial Lipids Compared to the Tubercle bacillus. Tuberculosis 115, 96–107. doi:10.1016/j.tube.2019.02.008
Tseng, S.-P., Teng, S.-H., Lee, P.-S., Wang, C.-F., Yu, J.-S., and Lu, P.-L. (2013). Rapid Identification ofM. abscessusandM. Massilienseby MALDI-TOF Mass Spectrometry with a Comparison to Sequencing Methods and Antimicrobial Susceptibility Patterns. Future Microbiol. 8 (11), 1381–1389. doi:10.2217/fmb.13.115
Voorhees, K. J., Jensen, K. R., McAlpin, C. R., Rees, J. C., Cody, R., Ubukata, M., et al. (2013). Modified MALDI MS Fatty Acid Profiling for Bacterial Identification. J. Mass. Spectrom. 48 (7), 850–855. doi:10.1002/jms.3215
Wang, C.-C., Lai, Y.-H., Ou, Y.-M., Chang, H.-T., and Wang, Y.-S. (2016). Critical Factors Determining the Quantification Capability of Matrix-Assisted Laser Desorption/ionization- Time-Of-Flight Mass Spectrometry. Phil. Trans. R. Soc. A. 374 (2079), 20150371. doi:10.1098/rsta.2015.0371
Welker, M., Van Belkum, A., Girard, V., Charrier, J.-P., and Pincus, D. (2019). An Update on the Routine Application of MALDI-TOF MS in Clinical Microbiology. Expert Rev. Proteomics 16 (8), 695–710. doi:10.1080/14789450.2019.1645603
Whang, J., Back, Y. W., Lee, K.-I., Fujiwara, N., Paik, S., Choi, C. H., et al. (2017). Mycobacterium Abscessus Glycopeptidolipids Inhibit Macrophage Apoptosis and Bacterial Spreading by Targeting Mitochondrial Cyclophilin D. Cell Death Dis. 8 (8), e3012. doi:10.1038/cddis.2017.420
Keywords: MALDI, lipids, diagnostics, mycobacteria, solvent
Citation: Jia Khor M, Broda A, Kostrzewa M, Drobniewski F and Larrouy-Maumus G (2021) An Improved Method for Rapid Detection of Mycobacterium abscessus Complex Based on Species-Specific Lipid Fingerprint by Routine MALDI-TOF. Front. Chem. 9:715890. doi: 10.3389/fchem.2021.715890
Received: 27 May 2021; Accepted: 28 June 2021;
Published: 27 July 2021.
Edited by:
Gerard Bolbach, Sorbonne Universités, FranceReviewed by:
Tao Yu, North Carolina State University, United StatesCopyright © 2021 Jia Khor, Broda, Kostrzewa, Drobniewski and Larrouy-Maumus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerald Larrouy-Maumus, Zy5sYXJyb3V5LW1hdW11c0BpbXBlcmlhbC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.