
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Chem., 01 June 2021
Sec. Analytical Chemistry
Volume 9 - 2021 | https://doi.org/10.3389/fchem.2021.705458
This article is part of the Research TopicAdvances in Nucleic Acid-Based Biosensors and ImagingView all 7 articles
It is well known that cancer incidence and death rates have been growing, but the development of cancer theranostics and therapeutics has been a challenging work. Recently, nucleic acid probe–based fluorescent sensing and imaging have achieved remarkable improvements in a variety of cancer management techniques, credited to their high sensitivity, good tolerance to interference, fast detection, and high versatility. Herein, nucleic acid probe–based fluorescent sensing and imaging are labeled with advanced fluorophores, which are essential for fast and sensitive detection of aberrant nucleic acids and other cancer-relevant molecules, consequently performing cancer early diagnosis and targeted treatment. In this review, we introduce the characteristics of nucleic acid probes, summarize the development of nucleic acid probe–based fluorescent sensing and imaging, and prominently elaborate their applications in cancer diagnosis and treatment. In discussion, some challenges and perspectives are elaborated in the field of nucleic acid probe–based fluorescent sensing and imaging.
Nucleic acids (DNA and RNA) are one of the most essential components for organisms. Nucleic acid mutations, such as DNA translocations (Javadekar and Raghavan, 2015), small insertions and deletions (indels), and single-nucleotide polymorphisms (SNPs) (Mohlendick et al., 2019), are frequent events during cancer progression. Rapid progress in fluorescence-based nucleic acid probes is beneficial for studying the structural and conformational polymorphisms of nucleic acids and further investigating their variability, internal dynamics, and interactions with proteins, metabolites, and targeting drugs at the sub-molecular level (Sinkeldam et al., 2010; Michel et al., 2020).
Specific nucleic acid probes hold with a particular sequence, and they can recognize a broad range of targets, such as metal ions, small organic molecules, proteins, and even viruses or cells (Xiang and Lu, 2011; Huang et al., 2019; Bai et al., 2020). Nucleic acid probes mainly include DNA and RNA probes. DNA probes are useful tools for elaborating the biological processes of nucleic acid amplification, ligation, duplication, and transcription (Wu et al., 2014; Jia et al., 2015). SNPs are the most common DNA variations, and the multi-color SNP probes can discriminate four SNP variants with unique fluorescence colors, but the ideal multi-color SNP probes remain to be explored (Obliosca et al., 2013). RNA probes are responsible for severe DNA interference due to the similar structures between DNA and RNA, which restrains the exploration of RNA probes (Wang et al., 2016; Yao et al., 2018).
Generally, the specific structures of nucleic acid probes are beneficial to fabricate molecular computing devices, nanobiotechnology, and biomedical technology (Pu et al, 2014), like nucleic acid probe–based fluorescent sensing and imaging platforms. Fluorophores are crucial for the improvement of nucleic acid probe–based fluorescent sensing and imaging (Ebrahimi et al., 2014). Organic or conventional fluorophores have low quantum yield and poor photostability (Xu et al., 2018). Importantly, an increasing number of novel fluorescent nanomaterials have been developed, such as quantum dots (QDs), silver nanoclusters (AgNCs), gold nanoparticles (AuNPs), upconversion nanomaterials, and cationic conjugated polymers (CCPs) (Xu et al., 2016; Borghei et al., 2019). These fluorophores are characterized with better brightness, photostability, and size-tunable fluorescence spectrum and can directly or indirectly recognize specific targets with different patterns, such as hydrogen bonds, single-stranded DNA (ssDNA)/RNA hybridization, aptamer–target binding, enzyme inhibition, and enzyme-mimicking activity (Adinolfi et al., 2017).
To date, cancer mortality has been increasing around the word, chemotherapy and radiotherapy are widely used in cancer clinical treatment, but long-term treatment with chemotherapy drugs and radiotherapy can lead to multi-drug resistance, bone marrow suppression, and other adverse reactions (Kovacs et al., 2018; Zhou et al., 2018). Therefore, the development of effective treatment and early diagnosis becomes the key to decrease the death rate (He et al., 2019; Zhao et al., 2019). Since nucleic acid probe–based fluorescent sensing and imaging are attractive ways to identify the status of disease development, they have been widely investigated for usage in the early diagnosis and targeted treatment of various cancers by transforming biorecognition events into an amplified fluorescence signal (Mo et al., 2017; Lou et al., 2019; Li et al., 2021). Such tools allow for direct molecular recognition between nucleic acid probes and tested targets in living cells and tissues, which can quantitatively discriminate the amount and position of mutated DNA by producing an easily recordable and interpretable fluorescence signal (Hamd-Ghadareh et al., 2017).
Taken together, nucleic acid probe–based fluorescent sensing and imaging platforms are of great benefit to detect the positions and concentrations of cancer-relevant targets (Gao et al., 2018). Thereby, we would comprehensively elaborate the application of nucleic acid probe–based fluorescent sensing and imaging platforms in cancer diagnosis and therapy. In order to facilitate the development of innovative nucleic acid probe–based fluorescent sensing and imaging systems, some challenges and perspectives would be discussed.
There are numerous outstanding fluorophores utilized for nucleic acid probe–based sensing, comprising QDs, carbon dots (CDs), AgNCs, AuNPs, CCPs, and upconversion nanomaterials. Presently, the fluorescence intensity-based measurement is extensively used, in which the fluorescence intensity varies based on the levels of targets, leading to the accurate and quantitative measurement of cancer-relevant molecules. Prospectively, conjugation of nucleic acid probe–based fluorescent sensing with high-throughput microdevices, such as lateral flow devices, microfluidics, and microarrays, has shown distinguished advantages in cancer point-of-care diagnosis and oncogene-guided individual therapy (Figure 1).
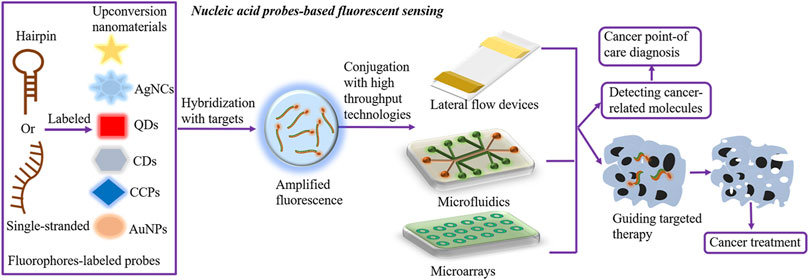
FIGURE 1. Nucleic acid probe–based fluorescent sensing. Hairpin or single-stranded nucleic acid probes are labeled with fluorophores to construct nucleic acid probe–based fluorescent sensing platforms. Effective fluorophores comprise QDs, CDs, AgNCs, AuNPs, CCPs, and upconversion nanomaterials. In the presence of targets, fluorophore-labeled probes hybridize targets and transmit the amplified fluorescence signal, further detecting the levels of cancer-relevant molecules and facilitating oncogene-guided individual therapy. In addition, the conjugation of fluorescent sensing with high-throughput microdevices, such as lateral flow devices, microfluidics, and microarrays, has shown distinguished advantages in cancer point-of-care diagnosis. AgNCs, silver nanoclusters; AuNPs, gold nanoparticles; CDs, carbon dots; QDs, quantum dots; CCPs, cationic conjugated polymers.
Fluorescence biosensors are valuable tools for early diagnosing of cancer with precise and in situ monitoring of the spatiotemporal changes of miRNAs or proteins and identifying DNA mutations, such as single labeled molecular beacons (MBs) (FAM-MBs, with carboxyfluorescein and without quencher), a label-free beacon (AIE-MBs, without fluorogen and quencher), enzyme/nanomaterial-free and dual amplification, peptide nucleic acid (PNA), flow cytometry (FCM), nucleic acid aptamers–CDs, and enzymatic reaction–modified fluorescence sensing (Table 1).
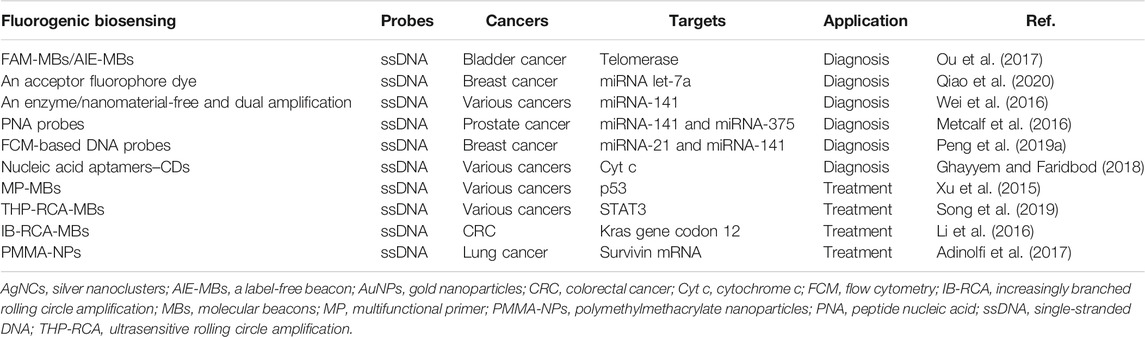
TABLE 1. Application of nucleic acid probe–based fluorescent sensing in cancer diagnosis and treatment.
Graphene oxide (GO) is a typical nanomaterial that holds exceptional optical, electrical, mechanical, and chemical properties (Lin et al., 2014). It has attracted enormous attention in the study of DNA-based sensors by interacting with ssDNA through π–π stacking interactions (Tang et al., 2015). Human telomerase has been considered a promising cancer marker. The single labeled FAM-MBs are designed to detect telomerase activity with the aid of GO. To further simplify this structure, the more sensitive label-free AIE-MBs are constructed to monitor telomerase activity based on the enhanced fluorescence production (Ou et al., 2017). But the label-free AIE-MBs could carry a high signal-to-background ratio. Presently, their applications in bladder cancer diagnosis have been reported (Ou et al., 2017).
MicroRNAs (miRNAs) serve as ponderable serum cancer biomarkers due to their functions in modulating oncogenic pathways (Ndzi et al., 2019). Numerous nucleic acid probe–based fluorescent sensors have a great value in evaluating the serum concentrations of miRNAs (Yamamura et al., 2012). For instance, there are two programmable DNA probes labeled with either a donor or an acceptor fluorophore dye. In the presence of targets, the fluorescent sensing platform contributes to fluorescence resonance energy transfer (FRET) and signal amplification with a cascade hybridization reaction. The assay can sensitively detect the concentration of miRNA let-7a at single-cell resolution and discriminate let-7a from other highly homologous miRNAs in different molecular subtypes of breast cancer (Qiao et al., 2020). Nevertheless, the target-triggered and self-assembly character offers a high signal-to-noise ratio and strong read-out ratio, subsequently allowing for an effective detection at low abundance.
miRNA-141 is important for accelerating epithelial to mesenchymal transition (EMT) (Bhardwaj et al., 2017). An enzyme/nanomaterial-free and dual amplified strategy is developed for highly sensitive detection of miRNA-141 by combining hybridization chain reaction (HCR) and catalytic hairpin assembly (CHA) amplification (Wei et al., 2016). The HCR and CHA synergistically generate a remarkably amplified fluorescence signal, and the fluorescence signal intensity represents the concentration of the miRNA-141 target. Meanwhile, the platform can differentiate miRNA-141 from its family members and be expanded for designing DNA hairpin probes (Wei et al., 2016).
In addition, a novel PNA probe–based fluorogenic biosensor is designed to selectively target the miRNA-141 biomarker in serum without amplification step. In this system, PNAs are engineered with uncharged oligonucleotide analogs. And PNAs are capable of hybridizing to complementary targets with high affinity and specificity, further analyzing the concentrations of circulating miRNA-141 and miRNA-375, which have been applied for sensitively diagnosing prostate cancer (Pca) (Metcalf et al., 2016). In addition, fluorophore-labeled PNA probes are able to quantitatively and specifically detect multiplexed miRNAs in living cancer cells when conjugated with the nano metal–organic framework (NMOF) vehicle, and the release of PNAs from the NMOF would lead to the recovery of fluorescence (Wu et al., 2015). Innovatively, the interaction between immobilized PNA probes and DNA targets leads to enzyme-catalyzed pigmentation, allowing for simple visual read-out with up to 100% accuracy (Jirakittiwut et al., 2020).
Furthermore, a simpler DNA probe sensor has been creatively presented through integrating with FCM, which is based on the double key “unlocked mechanism” and the fluorescence enrichment signal amplification (Peng et al., 2019a; Oldham et al., 2020). In the sensor, fluorescent particle (FS)–labeled hairpin DNA probes (HDs) serve as the lock of “unlocked mechanism” and specifically hybridize with the probes on polystyrene (PS) microparticles. In the presence of miRNA targets, both miRNA targets and duplex-specific nuclease (DSN) act as the double key to specifically unlock HDs and increase the enrichment of HDs on PS microparticles. Then, the unlocked fluorescent probes lead to the enrichment of the fluorescent signal (Peng et al., 2019b). The FCM-based DNA probe sensor is allowed to measure miRNA-21 and miRNA-141 in breast cancer blood samples with higher sensitivity (Peng et al., 2019a). The whole procedure does not need a complex purification process, indicating a simplified FCM-based nucleic acid probe fluorescent sensing platform.
Protein also exerts enormous functions in diverse pathological activities, which provides effective targets for cancer diagnosis. For example, cytochrome c (Cyt c), a heme protein, is a significant biomarker for apoptosis (Chen et al., 2021). Cyt c–specific nucleic acid aptamers have the strong binding affinity to Cyt c. The detection of Cyt c relies on the interaction of nucleic acid aptamers with fluorescent CDs. In the presence of Cyt c, the interaction between nucleic acid aptamers and Cyt c would result in the release of CDs and fluorescence production, and the intensity of fluorescence is proportional to the concentration of Cyt c (Ghayyem and Faridbod, 2018). Therefore, the nucleic acid aptamer–CD sensing platform could be used for detecting various cancer-related proteins through designing target-specific nucleic acid aptamers, but CDs can only adsorb ssDNA probes via π–π interaction.
Nucleic acids serve as substrates of nucleic acid enzymes, and enzymatic reaction–mediated fluorescence sensing is a versatile avenue to improve the sensitivity of cancer diagnosis when combining with target-dependent cycling amplification (Allinson, 2010). In detail, when probe strands recognize the target DNA strands, the probe–target complexes are instantly digested by a specific enzyme to emit a fluorescence signal. Then, the released target DNA strands immediately react with another probe and give out a stronger signal (Zuo et al., 2010). Exonuclease III (Exo III) is one of the DNA-repair enzymes Chen et al. (2019a), and it is inclined to be recognized by MBs. MB-labeled fluorescence probes can effectively cleave Exo III and stimulate DNA-dependent signal recycling amplification, further testifying DNA mutation and diagnosing cancer (Zuo et al., 2010; Chen et al., 2019b).
In this work, a novel and low-background fluorescent sensor platform is developed to detect nucleic acids based on the combination of δ-FeOOH nanosheets with Exo III–assisted target-recycling signal amplification. δ-FeOOH nanosheets, as the quenchers, are conjugated with the dye-labeled ssDNA probes. The dye-labeled ssDNA probes integrate with the DNA targets to form a double-strand DNA complex (dsDNA). Then, the dye-labeled ssDNA probes in the dsDNA complex will be gradually hydrolyzed into short fragments by Exo III, and the fluorescence signal is recovered due to the weaker bind affinity between short fragments and δ-FeOOH nanosheets (Wu et al., 2020). Markedly, the most suitable environment should be provided for boosting Exo III activity, and this sensing platform would become a universal approach for optimizing the early detection of DNA mutation.
Abnormal changes in tumor suppressor genes, oncogenes, and other molecules are found in various cancers. Thus, precisely targeting these aberrant molecules via nucleic acid probe–based fluorescent sensing is prospective for guiding and optimizing cancer gene–based individual treatment. There are several noble fluorescence sensing strategies, including multifunctional primer–integrated MBs (MP-MBs), ultrasensitive rolling circle amplification (THP-RCA), increasingly branched rolling circle amplification (IB-RCA), and polymethylmethacrylate nanoparticle (PMMA-NP)–modified MBs (Table 1).
p53 is an essential tumor suppressor, and targeting p53 mutation should also be concerned (D'Orazi et al., 2021). Presently, the MP-MB probe has been developed to detect p53 gene. Compared with the traditional MBs, MP-MBs can not only selectively identify the targets and sensitively transmit a hybridization signal but also act as the primer during enzymatic polymerization. Specifically, hybridization of MP-MBs with p53 gene can restore the fluorescence intensity and provoke the pre-locked primer by changing the molecular configuration of MP-MBs, further targeting p53 mutation and instructing p53 gene–guided individual therapy (Xu et al., 2015). MP-MBs do not require any chemical modification, and with less species requirement, they have wider sequence diversity and preserved intrinsic bioactivity.
STAT3 is a potent proto-oncogene, and screening STAT3 gene is useful for cancer therapy (Kryczek et al., 2014). A novel THP-RCA strategy is designed to ultrasensitively detect human proto-oncogenes via conjugating with target-catalyzed hairpin structure–mediated padlock cyclization. For the system, hairpin probe (HP) 1 is formed as the cyclization template and RCA reaction primer and HP2 is the padlock probe. The two probes fold into a hairpin structure via self-hybridization. In the presence of STAT3 DNA, HP2 hybridizes with HP1 in an end-to-end manner. Then, HP2 is cyclized by ligase on the HP1 template; the cyclized HP2 enables the RCA and generates a long tandem ssDNA product that is capable of hybridizing with considerable quantity of MBs. Subsequently, the amplified fluorescence value represents the ultrasensitive detection of STAT3 gene (Song et al., 2019). Moreover, the sensing system is suitable for target detection in human serum.
Similarly, IB-RCA is constructed for highly sensitively detecting and targeting the colorectal cancer (CRC) gene, Kras gene codon 12, which comprises a padlock probe (PP) and an MB (Li et al., 2016). The PP is circularized after hybridization with the DNA target, while the stem of the MB is opened by the DNA target. The newly opened MB hybridizes with the circularized PP to generate a long tandem ssDNA product, consequently triggering the next RCA reactions and producing a dramatically amplified fluorescence signal (Li et al., 2016). It is worth noting that IB-RCA efficiently transduces the fluorescence signal in a simpler way compared with conventional amplification methods.
In addition, targeting mRNAs is the other cardinal avenue to cancer treatment. Survivin is an overexpressed anti-apoptotic protein and considered a pharmacological target for effective anticancer therapy (Meiners et al., 2021). Survivin MBs can selectively detect survivin mRNA through embedding into the cells with the assistance of Lipofectamine, but MBs might be degraded by enzymes in vivo (Bishop et al., 2015). In order to overcome this problem, biocompatible core–shell PMMA-NPs serve as the carrier of MBs to specifically target survivin mRNA in A549 human lung adenocarcinoma epithelial cells, which suppresses cancer cell proliferation (Adinolfi et al., 2017). PMMA-NPs consist of a fluorescein-modified hydrophobic PMMA core and an external hydrophilic shell functionalized with primary amine groups and quaternary ammonium salts. Interestingly, the PMMA-NP carrier has higher biocompatibility, lower cytotoxicity to healthy cells, higher biological inertness, lower synthesis costs, and higher selectivity, as well as prolonging the drug half-life in the human body compared with classical transfection reagents such as Lipofectamine, which extend the application of PMMA-NPs (Brandts et al., 2021).
At present, even if nucleic acid probe–based fluorescent sensing has made significant progress in cancer diagnosis and therapy, it cannot detect the cancer-relevant targets in situ. It is worth noting that nucleic acid probe–based fluorescent imaging can visualize the cancer target expression, composed of visualizing the changes in molecule conformation, locating surface molecules, and targeting cancer cells in living samples with a high spatiotemporal resolution, resulting in an elevated efficiency of cancer diagnosis and treatment (Figure 2). Due to the excellent functions of RNA during cancer development, the investigation of multi-fluorophore color RNA probes is required for understanding the correlation of gene expression and interaction between nucleic acids (Okamoto, 2011).
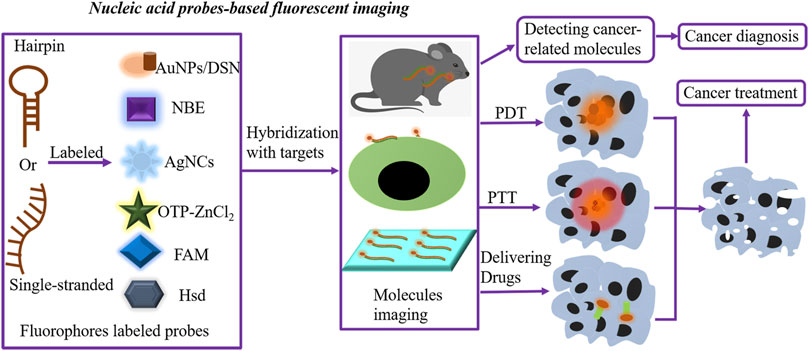
FIGURE 2. Nucleic acid probe–based fluorescent imaging. Hairpin or single-stranded nucleic acid probes are labeled with fluorophores to form nucleic acid probe–based fluorescent imaging platforms. Novel fluorophores include AuNPs/DSN, AgNC-MBs, FAM, OTP-ZnCl2, Hsd, and NBE. In the presence of targets, fluorophore-labeled probes hybridize targets, further performing molecular imaging and locating molecules expressed on the surface of cells or tissues and targeting cancer cells in living samples. The fluorescent imaging platforms can detect cancer-related molecules, resulting in an elevated efficiency of cancer diagnosis. Meanwhile, these imaging methods are utilized for delivering anticancer drugs and guiding PDT and PTT, further killing cancer cells by in situ imaging of low-abundance biomarkers. AgNCs, silver nanoclusters; AuNPs, gold nanoparticles; MBs, molecular beacons; DSN, double-specific nuclease; PDT, photodynamic therapy; PTT, photothermal therapy.
miRNAs have become ideal and noninvasive cancer biomarkers. To accomplish better and faster miRNA imaging, Au nanoparticles (AuNPs)/double-specific nuclease (DSN), AgNC-generating MBs (AgNC-MBs), reduced graphene oxide (rGO), FAM, OTP-ZnCl2, Hsd, and NBE-modified fluorescent probes are applied for fabricating imaging platforms and measuring mutant-type targets in diagnosis of various cancers (Figure 2; Table 2).
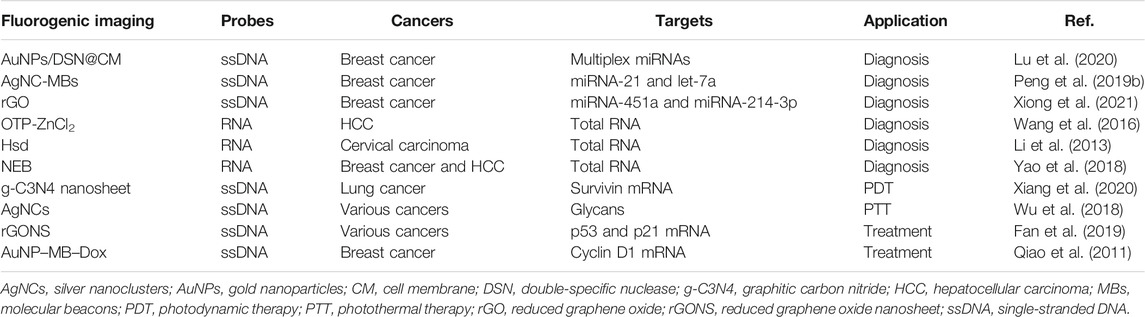
TABLE 2. Application of nucleic acid probe–based fluorescent imaging in cancer diagnosis and treatment.
Highly efficient cellular transfection and intracellular signal amplification are basis for low-abundance miRNA imaging (Chen et al., 2019a; Lu et al., 2020). A study uses AuNPs/DSN to encapsulate the functional cancer cell membrane (CM) vesicle, and AuNPs are modified with three types of fluorescent probes. The AuNPs/DSN@CM can specifically target the cancer cell, and the internalized AuNPs/DSN@CM further recognizes the miRNA targets and induces DSN-based recycle signal amplification, leading to simultaneous detection of multiple miRNAs. This approach has successfully analyzed and monitored the dynamic changes in oncogenic miRNAs in breast cancer cells with high sensitivity (Lu et al., 2020). Compared with traditional AuNPs, AuNPs/DSN@CM exhibits the higher transfection efficiency, biocompatibility, and specificity.
Favorably, the fluorescent AgNC-MBs are economical alternatives for detecting multiple nucleic acids (Del Bonis-O'Donnell et al., 2016; Huang et al., 2018). However, most of AgNC-MBs have limited versatility; the reason is that fluorescence properties of DNA-AgNCs will be severely damaged when the AgNC-stabilizing sequence is embedded into the MB sequence. Based on toehold-mediated DNA strand displacement, a new type of AgNC-MB is constructed by combining with total internal reflection fluorescence (TIRF)–based single-molecule fluorescence imaging (Peng et al., 2019b). The AgNC-MB platform can simultaneously measure two breast cancer–related miRNAs (miRNA-21 and let-7a) and distinguish the mutant-type targets at low abundance (Peng et al., 2019a). In addition, miRNA-451a and miRNA-214-3p are meaningful biomarkers for breast cancer; the novel rGO-modified DNA nanoprobe is prepared for simultaneous dual-color imaging of miRNA-451a and miRNA-214-3p (Xiong et al., 2021). Above all, the AgNC-MBs and rGO-modified imaging platforms provide versatile methods for sensitively and simultaneously imaging multiple miRNA biomarkers. Notably, AgNCs have been employed in single-molecule microscopy, molecular logic devices, and metal ion sensing (Adinolfi et al., 2017).
Currently, fluorescent RNA probes are developing. The OTP-ZnCl2 complex has a better interaction with nucleolus RNA than DNA, and it can stably insert into the inside of RNA based on the hydrogen bonds between OTP-ZnCl2 and RNA, between the oxime group and the base pair of RNA (Wang et al., 2016). Because of the outstanding cell permeability, low cytotoxicity, and counterstain compatibility, OTP-ZnCl2 has become a favorable dye for designing selective RNA fluorescent probes and two-photon fluorescence imaging. In this study, OTP-ZnCl2–based fluorescent RNA probes are allowed for accurate RNA imaging within hepatocellular carcinoma (HCC) cells (Wang et al., 2016). In addition, the near-infrared and cell-impermeant fluorescent dye Hsd is utilized to modify RNA probes owing to its selective response to RNA, and it can enter into the living cells for selective RNA staining and imaging with low cytotoxicity and fluorescence quantum yield. But the process needs the assistance of cucurbit[7]uril (CB7) to strengthen the potential of Hsd in cancer diagnosis (Li et al., 2013). Besides, NBE, an NIR fluorescent probe, has no response to DNA. NBE-modified RNA probes are utilized for fulfilling excellent RNA imaging in live breast cancer and HCC cells with good photostability, high selectivity, and fast response to RNA, which facilitates the diagnosis of various cancers according to RNA contents (Yao et al., 2018).
Likewise, nucleic acid probe–based fluorescent imaging is an available approach for guiding cancer treatment and improving the therapeutic efficacy with in situ imaging of low-abundance nucleic acid targets. Currently, some imaging methods are utilized for delivering anticancer drugs and guiding photodynamic therapy (PDT) and photothermal therapy (PTT) (Figure 2; Table 2).
Recently, the water-dispersible graphitic carbon nitride (g-C3N4) nanosheet has been considered an excellent nanocarrier functioned with CHA amplification, and it is applied for self-tracking transfection of DNA hairpin probes (Xiang et al., 2020; Lin et al., 2021a). The cancer-related mRNAs will efficiently initiate the DNA hairpin probes, ultimately leading to an amplified fluorescence signal via hybridization and mRNA displacement. Then, the enhanced fluorescence imaging will sensitively analyze the low-abundance cancer-relevant mRNAs, directly track the location, and guide precise PDT of cancers upon light irradiation (Xiang et al., 2020). Presently, the g-C3N4 nanosheet–based nanoassembly has been used for low-abundance survivin mRNA imaging and anticancer PDT, which do not show obvious side effects (Xiang et al., 2020).
As we all know, aberrant alterations of glycans are involved in many types of cancers. Herein, DNA-stabilized AgNC probes have been presented for label-free fluorescence imaging of cell surface glycans and fluorescence-guided PTT. In this pattern, surface glycans are specifically labeled by DNA-AgNC fluorescent probes via the dibenzocyclooctyne (DBCO)-functioned and DNA-initiated hybridization chain reaction (HCR). Then, DNA-AgNC probes produce the amplified signal, subsequently killing cancer cells and inhibiting cancer growth due to the remarkable photothermal properties of the HCR. Furthermore, DNA-AgNCs can dramatically reduce the cost and the instability of fluorescent dyes, and the HCR prevents the introduction of excessive azido-sugars and ensures apparent fluorescence. These results present the high value of the fluorescence imaging nanoplatform in visualizing specific glycans and guiding anticancer PTT (Wu et al., 2018).
The p53 and p21 genes play vital roles in blocking cancer development; it is important to monitor mRNA levels of the two markers (Lei et al., 2020). Herein, a reduced graphene oxide nanosheet (rGONS)–modified nanosystem is constructed for in situ and real-time p53 and p21 mRNA imaging by adsorbing the FAM-labeled p21 probe (P21) and Cy5-labeled p53 probe (P53). Once the two fluorescence probes hybridize with corresponding targets, the formation of DNA/RNA duplexes directly facilitates the release of probes from the rGONS surface and then restores the fluorescence signal (Fan et al., 2019). Therefore, the nanosystem in situ reveals a p53 and p21 mRNA–related regulatory process, which is practicable for drug screening and therapy evaluation in clinics.
Doxorubicin (Dox) is a common anticancer drug, and fluorophore-modified Dox is essential for cancer targeted therapy by intercalation within DNA/RNA. Herein, Dox carriers directly impact the therapeutic efficiency, and MB-functionalized AuNPs are identified as superior carriers to deliver fluorescent Dox. When MBs selectively interact with mRNA targets, fluorescent Dox is released from the AuNP–MB–Dox complex. The released Dox is positively correlated with the quantities of mRNA targets (Qiao et al., 2011). This strategy selectively detects the concentration of cyclin D1 mRNA in breast cancer and induces cyclin D1 + breast cancer cell apoptosis. Obviously, AuNP-MBs are the ideal carriers for transporting anticancer drugs, as they can specifically interact with cancer-relevant mRNA targets and kill cancer cells with lower side effects (Qiao et al., 2011).
All in all, the applications of fluorescent biosensors and imaging technologies are increasingly widespread. However, there are some defects that need to be improved: i) The nucleic acid probes might lose the ability to hybridize with target strands when the target sequences form secondary structures such as hairpins or quadruplexes, subsequently disturbing the surrounding sequences (Ming et al., 2019). Thus, an open strand–based model is required for eliminating the influence of complicated secondary structures. The model would be conducted for observing low-abundance DNA mutations in cancer samples, further improving cancer gene–based individual therapy. ii) We should also focus on the development of other fluorescence signaling techniques, such as lifetime, correlation spectroscopy, polarization, and localization; they are excellent carriers for delivering information and exploring molecular interactions and DNA structures, which will make significant advancements in cancer diagnostics and theranostics (Su et al., 2012; Adinolfi et al., 2017).
Some nucleic acid probe–based fluorescent sensing platforms are simultaneously labeled with fluorogen and quencher. The synthesis of both fluorogen and quencher is complex, and the relative distance between fluorogen and quencher is difficult to control, which may lead to false-positive and false-negative results (Ou et al., 2017). In order to realize more specific molecular recognition and more accurate quantification of target molecules, studies are supposed to focus on designing new nucleic acid probes with more chemical functionalities and less nonspecific interactions. Even though fluorescent probe–labeled nucleic acid aptamers exert brilliant functions with low cytotoxicity and high specificity, the performance of aptamer-based sensors remains to be improved due to fast nuclease degradation, rapid renal excretion, and weaker binding affinity (Tan et al., 2019).
Nucleic acid fluorescent probe–based imaging technology has attracted widespread attention, which can display quantitative maps according to the concentrations of target molecules in living samples. Although numerous nucleic acid probes have sufficient sensitivity and selectivity for in vitro imaging of various targets, there are several scientific and technical challenges to in situ and in vivo fluorescence imaging: i) the complexity of tumor microenvironment (TME) might cause damage to normal cells and non-target molecules (Lin et al., 2021b) and ii) the high background of enzymatic catalysis dramatically decreases the feasibility of in vivo fluorescence imaging (Ferrero et al., 2021). Thus, novel fluorophores with high quantum yield need to be explored for eliminating the background signal and reducing the perturbation to normal biological processes, which is vital for monitoring the enzymatic processes with greater temporal and spatial resolution. Currently, a number of aptamer-based methods for in vivo fluorescence imaging have been reported, such as fluorescent dyes, QDs, or upconversion nanoparticle–labeled aptamers (Bagalkot et al., 2007; Kim et al., 2012).
Although nucleic acid probe–based fluorescent sensing and imaging systems have made some progress, several drawbacks need to be ameliorated, including low sample throughput, defective reproducibility, insufficient quantitation accuracy, high operation costs, complicated procedures, and long assay period (Fang et al., 2019). To solve these deficiencies, miniaturized and automated nanodevices would be rapidly developed via integrating fluorescence-labeled nucleic acid probes with high-throughput technologies, such as lateral flow devices, microfluidic chips, or microarray chips (Fang et al., 2019). These creative nanodevices would achieve tremendous advancements in cancer diagnostics and theranostics through real-time monitoring of biological processes, rapidly identifying targets and characterizing enzymes in a complex system (Fang et al., 2019). Nevertheless, how to assist them to exert more sophisticated functions in complicated biological environments remains to be explored.
GH, LW, and YF conceived and wrote the article. JY and CS revised and reviewed the article. All authors contributed to the article and approved the submitted version.
This work was supported by “Hunan Cancer Hospital Climb Plan.”
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AgNCs, silver nanoclusters; AuNPs, gold nanoparticles; CDs, carbon dots; CM, cell membrane; CRC, colorectal cancer; Cyt c, cytochrome c; DSN, double-specific nuclease; FCM, flow cytometry; g-C3N4, graphitic carbon nitride; HCC, hepatocellular carcinoma; IB-RCA, increasingly branched rolling circle amplification; MBs, molecular beacons; MP, multifunctional primer; PDT, photodynamic therapy; PMMA-NPs, polymethylmethacrylate nanoparticles; PNA, peptide nucleic acid; PTT, photothermal therapy; rGO, reduced graphene oxide; rGONS, reduced graphene oxide nanosheet; ssDNA, single-stranded DNA; THP-RCA, ultrasensitive rolling circle amplification; TME, tumor microenvironment.
Adinolfi, B., Pellegrino, M., Giannetti, A., Tombelli, S., Trono, C., Sotgiu, G., et al. (2017). Molecular beacon-decorated Polymethylmethacrylate Core-Shell Fluorescent Nanoparticles for the Detection of Survivin mRNA in Human Cancer Cells. Biosens. Bioelectron. 88, 15–24. doi:10.1016/j.bios.2016.05.102
Allinson, S. L. (2010). DNA End-Processing Enzyme Polynucleotide Kinase as a Potential Target in the Treatment of Cancer. Future Oncol. 6 (6), 1031–1042. doi:10.2217/fon.10.40
Bagalkot, V., Zhang, L., Levy-Nissenbaum, E., Jon, S., Kantoff, P. W., Langer, R., et al. (2007). Quantum Dot−Aptamer Conjugates for Synchronous Cancer Imaging, Therapy, and Sensing of Drug Delivery Based on Bi-fluorescence Resonance Energy Transfer. Nano Lett. 7 (10), 3065–3070. doi:10.1021/nl071546n
Bai, Y., Shu, T., Su, L., and Zhang, X. (2020). Functional Nucleic Acid-Based Fluorescence Polarization/anisotropy Biosensors for Detection of Biomarkers. Anal. Bioanal. Chem. 412 (25), 6655–6665. doi:10.1007/s00216-020-02754-x
Bhardwaj, M., Sen, S., Chosdol, K., Sharma, A., Pushker, N., Kashyap, S., et al. (2017). miRNA-200c and miRNA-141 as Potential Prognostic Biomarkers and Regulators of Epithelial-Mesenchymal Transition in Eyelid Sebaceous Gland Carcinoma. Br. J. Ophthalmol. 101 (4), 536–542. doi:10.1136/bjophthalmol-2016-309460
Bishop, C. J., Kozielski, K. L., and Green, J. J. (2015). Exploring the Role of Polymer Structure on Intracellular Nucleic Acid Delivery via Polymeric Nanoparticles. J. Controlled Release 219, 488–499. doi:10.1016/j.jconrel.2015.09.046
Borghei, Y.-S., Hosseini, M., Ganjali, M. R., and Ju, H. (2019). A Unique FRET Approach toward Detection of Single-Base Mismatch DNA in BRCA1 Gene. Mater. Sci. Eng. C 97, 406–411. doi:10.1016/j.msec.2018.12.049
Brandts, I., Barría, C., Martins, M. A., Franco-Martínez, L., Barreto, A., Tvarijonaviciute, A., et al. (2021). Waterborne Exposure of Gilthead Seabream (Sparus aurata) to Polymethylmethacrylate Nanoplastics Causes Effects at Cellular and Molecular Levels. J. Hazard. Mater. 403, 123590. doi:10.1016/j.jhazmat.2020.123590
Chen, F., Yin, S., Luo, B., Wu, X., Yan, H., Yan, D., et al. (2021). VDAC1 Conversely Correlates with Cytc Expression and Predicts Poor Prognosis in Human Breast Cancer Patients. Oxidative Med. Cell Longevity 2021, 7647139. doi:10.1155/2021/7647139
Chen, J., Yang, H.-H., Yin, W., Zhang, Y., Ma, Y., Chen, D., et al. (2019a). Metastable Dumbbell Probe-Based Hybridization Chain Reaction for Sensitive and Accurate Imaging of Intracellular-specific MicroRNAs In Situ in Living Cells. Anal. Chem. 91 (7), 4625–4631. doi:10.1021/acs.analchem.8b05920
Chen, P., Huang, K., Zhang, P., Sawyer, E., Wu, Z., Wei, X., et al. (2019b). Exonuclease III-Assisted Strand Displacement Reaction-Driven Cyclic Generation of G-Quadruplex Strategy for Homogeneous Fluorescent Detection of Melamine. Talanta 203, 255–260. doi:10.1016/j.talanta.2019.05.020
D'Orazi, G., Cordani, M., and Cirone, M. (2021). Oncogenic Pathways Activated by Pro-inflammatory Cytokines Promote Mutant P53 Stability: Clue for Novel Anticancer Therapies. Cell Mol Life Sci. 78 (5), 1853–1860. doi:10.1007/s00018-020-03677-7
Del Bonis-O'Donnell, J. T., Vong, D., Pennathur, S., and Fygenson, D. K. (2016). A Universal Design for a DNA Probe Providing Ratiometric Fluorescence Detection by Generation of Silver Nanoclusters. Nanoscale 8 (30), 14489–14496. doi:10.1039/c6nr03827a
Ebrahimi, S., Akhlaghi, Y., Kompany-Zareh, M., and Rinnan, Å. (2014). Nucleic Acid Based Fluorescent Nanothermometers. ACS Nano 8 (10), 10372–10382. doi:10.1021/nn5036944
Fan, J., Tong, C., Dang, W., Qin, Y., Liu, X., Liu, B., et al. (2019). An rGONS-Based Biosensor for Simultaneous Imaging of P53 and P21 mRNA in Living Cells. Talanta 204, 20–28. doi:10.1016/j.talanta.2019.05.087
Fang, X., Zheng, Y., Duan, Y., Liu, Y., and Zhong, W. (2019). Recent Advances in Design of Fluorescence-Based Assays for High-Throughput Screening. Anal. Chem. 91 (1), 482–504. doi:10.1021/acs.analchem.8b05303
Ferrero, G. O., Faba, E. M. S., and Eimer, G. A. (2021). Biodiesel Production from Alternative Raw Materials Using a Heterogeneous Low Ordered Biosilicified Enzyme as Biocatalyst. Biotechnol. Biofuels 14 (1), 67. doi:10.1186/s13068-021-01917-x
Gao, P., Mei, C., He, L., Xiao, Z., Chan, L., Zhang, D., et al. (2018). Designing Multifunctional Cancer-Targeted Nanosystem for Magnetic Resonance Molecular Imaging-Guided Theranostics of Lung Cancer. Drug Deliv. 25 (1), 1811–1825. doi:10.1080/10717544.2018.1494224
Ghayyem, S., and Faridbod, F. (2018). A Fluorescent Aptamer/carbon Dots Based Assay for Cytochrome C Protein Detection as a Biomarker of Cell Apoptosis. Methods Appl. Fluoresc. 7 (1), 015005. doi:10.1088/2050-6120/aaf0ca
Hamd-Ghadareh, S., Salimi, A., Fathi, F., and Bahrami, S. (2017). An Amplified Comparative Fluorescence Resonance Energy Transfer Immunosensing of CA125 Tumor Marker and Ovarian Cancer Cells Using green and Economic Carbon Dots for Bio-Applications in Labeling, Imaging and Sensing. Biosens. Bioelectron. 96, 308–316. doi:10.1016/j.bios.2017.05.003
He, J., Li, C., Ding, L., Huang, Y., Yin, X., Zhang, J., et al. (2019). Tumor Targeting Strategies of Smart Fluorescent Nanoparticles and Their Applications in Cancer Diagnosis and Treatment. Adv. Mater. 31 (40), e1902409. doi:10.1002/adma.201902409
Huang, N.-H., Li, R.-T., Fan, C., Wu, K.-Y., Zhang, Z., and Chen, J.-X. (2019). Rapid Sequential Detection of Hg2+ and Biothiols by a Probe DNA-MOF Hybrid Sensory System. J. Inorg. Biochem. 197, 110690. doi:10.1016/j.jinorgbio.2019.04.004
Huang, S., Yao, H., Wang, W., Zhang, J.-R., and Zhu, J.-J. (2018). Correction: Highly Sensitive Fluorescence Quantification of Intracellular Telomerase Activity by Repeat G-Rich DNA Enhanced Silver Nanoclusters. J. Mater. Chem. B 6 (32), 5313. doi:10.1039/c8tb90112k
Javadekar, S. M., and Raghavan, S. C. (2015). Snaps and Mends: DNA Breaks and Chromosomal Translocations. FEBS J. 282 (14), 2627–2645. doi:10.1111/febs.13311
Jia, Y., Zuo, X., Lou, X., Miao, M., Cheng, Y., Min, X., et al. (2015). Rational Designed Bipolar, Conjugated Polymer-DNA Composite beacon for the Sensitive Detection of Proteins and Ions. Anal. Chem. 87 (7), 3890–3894. doi:10.1021/ac504690y
Jirakittiwut, N., Munkongdee, T., Wongravee, K., Sripichai, O., Fucharoen, S., Praneenararat, T., et al. (2020). Visual Genotyping of Thalassemia by Using Pyrrolidinyl Peptide Nucleic Acid Probes Immobilized on Carboxymethylcellulose-Modified Paper and Enzyme-Induced Pigmentation. Microchim Acta 187 (4), 238. doi:10.1007/s00604-020-4197-8
Kim, J. K., Choi, K.-J., Lee, M., Jo, M.-h., and Kim, S. (2012). Molecular Imaging of a Cancer-Targeting Theragnostics Probe Using a Nucleolin Aptamer- and microRNA-221 Molecular beacon-conjugated Nanoparticle. Biomaterials 33 (1), 207–217. doi:10.1016/j.biomaterials.2011.09.023
Kovács, N., Szigeti, K., Hegedűs, N., Horváth, I., Veres, D. S., Bachmann, M., et al. (2018). Multimodal PET/MRI Imaging Results Enable Monitoring the Side Effects of Radiation Therapy. Contrast Media Mol. Imaging 2018, 5906471. doi:10.1155/2018/5906471
Kryczek, I., Lin, Y., Nagarsheth, N., Peng, D., Zhao, L., Zhao, E., et al. (2014). IL-22+CD4+ T Cells Promote Colorectal Cancer Stemness via STAT3 Transcription Factor Activation and Induction of the Methyltransferase DOT1L. Immunity 40 (5), 772–784. doi:10.1016/j.immuni.2014.03.010
Lei, K., Li, W., Huang, C., Li, Y., Alfason, L., Zhao, H., et al. (2020). Neurogenic Differentiation Factor 1 Promotes Colorectal Cancer Cell Proliferation and Tumorigenesis by Suppressing the P53/p21 axis. Cancer Sci. 111 (1), 175–185. doi:10.1111/cas.14233
Li, H., Xu, J., Wang, Z., Wu, Z.-S., and Jia, L. (2016). Increasingly Branched Rolling circle Amplification for the Cancer Gene Detection. Biosens. Bioelectron. 86, 1067–1073. doi:10.1016/j.bios.2016.07.095
Li, L., Xu, S., Yan, H., Li, X., Yazd, H. S., Li, X., et al. (2021). Nucleic Acid Aptamers for Molecular Diagnostics and Therapeutics: Advances and Perspectives. Angew. Chem. Int. Ed. 60 (5), 2221–2231. doi:10.1002/anie.202003563
Li, Z., Sun, S., Yang, Z., Zhang, S., Zhang, H., Hu, M., et al. (2013). The Use of a Near-Infrared RNA Fluorescent Probe with a Large Stokes Shift for Imaging Living Cells Assisted by the Macrocyclic Molecule CB7. Biomaterials 34 (27), 6473–6481. doi:10.1016/j.biomaterials.2013.05.020
Lin, F., Shao, Y., Wu, Y., and Zhang, Y. (2021a). NIR Light-Propelled Janus-Based Nanoplatform for Cytosolic-Fueled microRNA Imaging. ACS Appl. Mater. Inter. 13 (3), 3713–3721. doi:10.1021/acsami.0c21071
Lin, K. Y., Hin Lam, C., Lin, X. H., Hsu, J. I., Fan, S. Y., Gupta, N. K., et al. (2021b). Improved Stabilities of Labeling Probes for the Selective Modification of Endogenous Proteins in Living Cells and In Vivo. Chem. Asian J. 16, 937–948. doi:10.1002/asia.202100060
Lin, Y., Ren, J., and Qu, X. (2014). Catalytically Active Nanomaterials: a Promising Candidate for Artificial Enzymes. Acc. Chem. Res. 47 (4), 1097–1105. doi:10.1021/ar400250z
Lou, Y.-F., Peng, Y.-B., Luo, X., Yang, Z., Wang, R., Sun, D., et al. (2019). A Universal Aptasensing Platform Based on Cryonase-Assisted Signal Amplification and Graphene Oxide Induced Quenching of the Fluorescence of Labeled Nucleic Acid Probes: Application to the Detection of Theophylline and ATP. Microchim Acta 186 (8), 494. doi:10.1007/s00604-019-3596-1
Lu, H., Guo, K., Cao, Y., Yang, F., Wang, D., Dou, L., et al. (2020). Cancer Cell Membrane Vesicle for Multiplex MicroRNA Imaging in Living Cells. Anal. Chem. 92 (2), 1850–1855. doi:10.1021/acs.analchem.9b03764
Meiners, A., Bäcker, S., Hadrović, I., Heid, C., Beuck, C., Ruiz-Blanco, Y. B., et al. (2021). Specific Inhibition of the Survivin-CRM1 Interaction by Peptide-Modified Molecular Tweezers. Nat. Commun. 12 (1), 1505. doi:10.1038/s41467-021-21753-9
Metcalf, G. A. D., Shibakawa, A., Patel, H., Sita-Lumsden, A., Zivi, A., Rama, N., et al. (2016). Amplification-Free Detection of Circulating microRNA Biomarkers from Body Fluids Based on Fluorogenic Oligonucleotide-Templated Reaction between Engineered Peptide Nucleic Acid Probes: Application to Prostate Cancer Diagnosis. Anal. Chem. 88 (16), 8091–8098. doi:10.1021/acs.analchem.6b01594
Michel, B. Y., Dziuba, D., Benhida, R., Demchenko, A. P., and Burger, A. (2020). Probing of Nucleic Acid Structures, Dynamics, and Interactions with Environment-Sensitive Fluorescent Labels. Front. Chem. 8, 112. doi:10.3389/fchem.2020.00112
Ming, Z., Chen, Q., Chen, N., Lin, M., Liu, N., Hu, J., et al. (2019). Eliminating the Secondary Structure of Targeting Strands for Enhancement of DNA Probe Based Low-Abundance point Mutation Detection. Analytica Chim. Acta 1075, 137–143. doi:10.1016/j.aca.2019.05.015
Mo, L., Li, J., Liu, Q., Qiu, L., and Tan, W. (2017). Nucleic Acid-Functionalized Transition Metal Nanosheets for Biosensing Applications. Biosens. Bioelectron. 89 (Pt 1), 201–211. doi:10.1016/j.bios.2016.03.044
Möhlendick, B., Schmid, K. W., and Siffert, W. (2019). The GNAS SNP c.393C>T (Rs7121) as a Marker for Disease Progression and Survival in Cancer. Pharmacogenomics 20 (7), 553–562. doi:10.2217/pgs-2018-0199
Ndzi, E. N., Indu Viswanath, A. N., Adzemye, N. G., Tamgue, O., Nsongka, M. V., Nair, A. S., et al. (2019). Upregulated Bovine Tuberculosis microRNAs Trigger Oncogenic Pathways: An In Silico Perception. Int. J. Mycobacteriol 8 (1), 70–74. doi:10.4103/ijmy.ijmy_9_19
Obliosca, J. M., Liu, C., and Yeh, H.-C. (2013). Fluorescent Silver Nanoclusters as DNA Probes. Nanoscale 5 (18), 8443–8461. doi:10.1039/c3nr01601c
Okamoto, A. (2011). ECHO Probes: a Concept of Fluorescence Control for Practical Nucleic Acid Sensing. Chem. Soc. Rev. 40 (12), 5815–5828. doi:10.1039/c1cs15025a
Oldham, R. A. A., Faber, M. L., Keppel, T. R., Buchberger, A. R., Waas, M., Hari, P., et al. (2020). Discovery and Validation of surfaceN-Glycoproteins in MM Cell Lines and Patient Samples Uncovers Immunotherapy Targets. J. Immunother. Cancer 8 (2), e000915. doi:10.1136/jitc-2020-000915
Ou, X., Hong, F., Zhang, Z., Cheng, Y., Zhao, Z., Gao, P., et al. (2017). A Highly Sensitive and Facile Graphene Oxide-Based Nucleic Acid Probe: Label-free Detection of Telomerase Activity in Cancer Patient's Urine Using AIEgens. Biosens. Bioelectron. 89 (Pt 1), 417–421. doi:10.1016/j.bios.2016.05.035
Peng, M., Fang, Z., Na, N., and Ouyang, J. (2019a). A Versatile Single-Molecule Counting-Based Platform by Generation of Fluorescent Silver Nanoclusters for Sensitive Detection of Multiple Nucleic Acids. Nanoscale 11 (35), 16606–16613. doi:10.1039/c9nr04608a
Peng, W., Zhao, Q., Chen, M., Piao, J., Gao, W., Gong, X., et al. (2019b). An Innovative "unlocked Mechanism" by a Double Key Avenue for One-Pot Detection of microRNA-21 and microRNA-141. Theranostics 9 (1), 279–289. doi:10.7150/thno.28474
Pu, F., Ren, J., and Qu, X. (2014). Nucleic Acids and Smart Materials: Advanced Building Blocks for Logic Systems. Adv. Mater. 26 (33), 5742–5757. doi:10.1002/adma.201401617
Qiao, G., Zhuo, L., Gao, Y., Yu, L., Li, N., and Tang, B. (2011). A Tumor mRNA-dependent Gold Nanoparticle-Molecular beacon Carrier for Controlled Drug Release and Intracellular Imaging. Chem. Commun. 47 (26), 7458–7460. doi:10.1039/c1cc11490e
Qiao, L., Wu, C., Cai, Z., Wu, X., Wu, P., and Cai, C. (2020). Cascade Signal Amplification Sensing Strategy for Highly Specific and Sensitive Detection of Homologous microRNAs in Different Molecular Subtypes of Breast Cancer. Analytica Chim. Acta 1093, 86–92. doi:10.1016/j.aca.2019.09.038
Sinkeldam, R. W., Greco, N. J., and Tor, Y. (2010). Fluorescent Analogs of Biomolecular Building Blocks: Design, Properties, and Applications. Chem. Rev. 110 (5), 2579–2619. doi:10.1021/cr900301e
Song, H., Yang, Z., Jiang, M., Zhang, G., Gao, Y., Shen, Z., et al. (2019). Target-catalyzed Hairpin Structure-Mediated Padlock Cyclization for Ultrasensitive Rolling circle Amplification. Talanta 204, 29–35. doi:10.1016/j.talanta.2019.05.057
Su, X., Xiao, X., Zhang, C., and Zhao, M. (2012). Nucleic Acid Fluorescent Probes for Biological Sensing. Appl. Spectrosc. 66 (11), 1249–1261. doi:10.1366/12-06803
Tan, Y., Li, Y., and Tang, F. (2019). Nucleic Acid Aptamer: A Novel Potential Diagnostic and Therapeutic Tool for Leukemia. Ott 12, 10597–10613. doi:10.2147/OTT.S223946
Tang, L., Wang, Y., and Li, J. (2015). The Graphene/nucleic Acid Nanobiointerface. Chem. Soc. Rev. 44 (19), 6954–6980. doi:10.1039/c4cs00519h
Wang, H., Tian, X., Du, W., Zhang, Q., Guan, L., Wang, A., et al. (2016). A Two-Photon Fluorescent RNA Probe Screened from a Series of Oxime-Functionalized 2,2′:6′,2′′-terpyridine ZnX2(X = Cl, Br, I) Complexes. J. Mater. Chem. B 4 (28), 4818–4825. doi:10.1039/c6tb01202g
Wei, Y., Zhou, W., Li, X., Chai, Y., Yuan, R., and Xiang, Y. (2016). Coupling Hybridization Chain Reaction with Catalytic Hairpin Assembly Enables Non-enzymatic and Sensitive Fluorescent Detection of microRNA Cancer Biomarkers. Biosens. Bioelectron. 77, 416–420. doi:10.1016/j.bios.2015.09.053
Wu, J., Li, N., Yao, Y., Tang, D., Yang, D., Ong’achwa Machuki, J., et al. (2018). DNA-stabilized Silver Nanoclusters for Label-free Fluorescence Imaging of Cell Surface Glycans and Fluorescence Guided Photothermal Therapy. Anal. Chem. 90 (24), 14368–14375. doi:10.1021/acs.analchem.8b03837
Wu, L., Ren, J., and Qu, X. (2014). Target-responsive DNA-Capped Nanocontainer Used for Fabricating Universal Detector and Performing Logic Operations. Nucleic Acids Res. 42 (21), e160. doi:10.1093/nar/gku858
Wu, T., Li, X., Fu, Y., Ding, X., Li, Z., Zhu, G., et al. (2020). A Highly Sensitive and Selective Fluorescence Biosensor for Hepatitis C Virus DNA Detection Based on δ-FeOOH and Exonuclease III-Assisted Signal Amplification. Talanta 209, 120550. doi:10.1016/j.talanta.2019.120550
Wu, Y., Han, J., Xue, P., Xu, R., and Kang, Y. (2015). Nano Metal-Organic Framework (NMOF)-based Strategies for Multiplexed microRNA Detection in Solution and Living Cancer Cells. Nanoscale 7 (5), 1753–1759. doi:10.1039/c4nr05447d
Xiang, M.-H., Li, N., Liu, J.-W., Yu, R.-Q., and Jiang, J.-H. (2020). A Tumour mRNA-Triggered Nanoassembly for Enhanced Fluorescence Imaging-Guided Photodynamic Therapy. Nanoscale 12 (16), 8727–8731. doi:10.1039/d0nr00941e
Xiang, Y., and Lu, Y. (2011). Using Personal Glucose Meters and Functional DNA Sensors to Quantify a Variety of Analytical Targets. Nat. Chem. 3 (9), 697–703. doi:10.1038/nchem.1092
Xiong, X., Dang, W., Luo, R., Long, Y., Tong, C., Yuan, L., et al. (2021). A Graphene-Based Fluorescent Nanoprobe for Simultaneous Imaging of Dual miRNAs in Living Cells. Talanta 225, 121947. doi:10.1016/j.talanta.2020.121947
Xu, H., Li, D., Zhao, Y., Wang, X., Li, D., and Wang, Y. (2018). Sodium 4-mercaptophenolate Capped CdSe/ZnS Quantum Dots as a Fluorescent Probe for pH Detection in Acidic Aqueous media. Luminescence 33 (2), 410–416. doi:10.1002/bio.3428
Xu, J., Dong, H., Shen, W., He, S., Li, H., Lu, Y., et al. (2015). New Molecular beacon for P53 Gene point Mutation and Significant Potential in Serving as the Polymerization Primer. Biosens. Bioelectron. 66, 504–511. doi:10.1016/j.bios.2014.12.008
Xu, Q., Lou, X., Wang, L., Ding, X., Yu, H., and Xiao, Y. (2016). Rapid, Surfactant-free, and Quantitative Functionalization of Gold Nanoparticles with Thiolated DNA under Physiological pH and its Application in Molecular Beacon-Based Biosensor. ACS Appl. Mater. Inter. 8 (40), 27298–27304. doi:10.1021/acsami.6b08350
Yamamura, S., Yatsushiro, S., Yamaguchi, Y., Abe, K., Shinohara, Y., and Kataoka, M. (2012). Detection of miRNA in Cell Cultures by Using Microchip Electrophoresis with a Fluorescence-Labeled Riboprobe. Sensors 12 (6), 7576–7586. doi:10.3390/s120607576
Yao, Q., Li, H., Xian, L., Xu, F., Xia, J., Fan, J., et al. (2018). Differentiating RNA from DNA by a Molecular Fluorescent Probe Based on the "Door-Bolt" Mechanism Biomaterials. Biomaterials 177, 78–87. doi:10.1016/j.biomaterials.2018.05.050
Zhao, X., Ning, Q., Mo, Z., and Tang, S. (2019). A Promising Cancer Diagnosis and Treatment Strategy: Targeted Cancer Therapy and Imaging Based on Antibody Fragment. Artif. Cell Nanomedicine, Biotechnol. 47 (1), 3621–3630. doi:10.1080/21691401.2019.1657875
Zhou, Y., Zhen, M., Guan, M., Yu, T., Ma, L., Li, W., et al. (2018). Amino Acid Modified [70] Fullerene Derivatives with High Radical Scavenging Activity as Promising Bodyguards for Chemotherapy protection. Sci. Rep. 8 (1), 16573. doi:10.1038/s41598-018-34967-7
Keywords: nucleic acid probes, fluorescent sensing, fluorescent imaging, cancer diagnosis, cancer therapy
Citation: Huang G, Su C, Wang L, Fei Y and Yang J (2021) The Application of Nucleic Acid Probe–Based Fluorescent Sensing and Imaging in Cancer Diagnosis and Therapy. Front. Chem. 9:705458. doi: 10.3389/fchem.2021.705458
Received: 05 May 2021; Accepted: 17 May 2021;
Published: 01 June 2021.
Edited by:
Zhihe Qing, Changsha University of Science and Technology, ChinaCopyright © 2021 Huang, Su, Wang, Fei and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinfeng Yang, eWFuZ2ppbmZlbmdAaG5jYS5vcmcuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.