- 1Institute of Bioengineering, College of Chemical and Biological Engineering, Zhejiang University, Hangzhou, China
- 2Hangzhou Global Scientific and Technological Innovation Center, Zhejiang University, Hangzhou, China
- 3Hwa Chong Institution, Singapore, Singapore
Pseudomonas putida KT2440 has become an attractive chassis for heterologous expression with the development of effective genetic manipulation tools. Improving the level of transcriptional regulation is particularly important for extending the potential of P. putida KT2440 in heterologous expression. Although many strategies have been applied to enhance the heterologous expression level in P. putida KT2440, it was still at a relatively low level. Herein we constructed a T7-like expression system in P. putida KT2440, mimicking the pET expression system in Escherichia coli, which consisted of T7-like RNA polymerase (MmP1) integrated strain and the corresponding expression vector for the heterologous expression enhancement. With the optimization of the insertion site and the copy number of RNA polymerase (RNAP), the relative fluorescence intensity (RFI) of the super-folder green fluorescent protein (sfGFP) was improved by 1.4-fold in MmP1 RNAP integrated strain. The induction point and IPTG concentration were also optimized. This strategy was extended to the gene-reduced strain EM42 and the expression of sfGFP was improved by 2.1-fold. The optimal RNAP integration site was also used for introducing T7 RNAP in P. putida KT2440 and the expression level was enhanced, indicating the generality of the integration site for the T7 expression system. Compared to other inducible expression systems in KT2440, the heterologous expression level of the Mmp1 system and T7 system were more than 2.5 times higher. Furthermore, the 3.6-fold enhanced expression level of a difficult-to-express nicotinate dehydrogenase from Comamonas testosteroni JA1 verified the efficiency of the T7-like expression system in P. putida KT2440. Taken together, we constructed and optimized the T7-like and T7 expression system in P. putida, thus providing a set of applicable chassis and corresponding plasmids to improve recombinant expression level, expecting to be used for difficult-to-express proteins.
Introduction
Pseudomonas putida KT2440, a non-pathogenic soil bacteria, is an attractive chassis for heterologous expression (Timmis, 2002; Dammeyer et al., 2011; Poblete-Castro et al., 2012; Lieder et al., 2015) due to its rapid growth (Martins Dos Santos et al., 2004), metabolic diversity (Poblete-Castro et al., 2012; Nikel et al., 2014, Nikel et al., 2016), rapid generation ability of nicotinamide adenine dinucleotide (Ebert et al., 2011), the operability of genetic manipulation (Martínez-García and de Lorenzo, 2019) and robustness to extreme environments (Nikel and de Lorenzo, 2018).
Many efforts have been made to enhance the heterologous expression level in P. putida KT2440 by regulating the transcriptional level or translation level. To improve the integration expression, the constitutive promoter libraries have been constructed (Zobel et al., 2015) and the T7 RNA polymerase has been integrated into the genome of P. putida KT2440 (Herrero et al., 1993; Christina et al., 2012). Additionally, to enhance the plasmid expression, suites of broad-host-expression plasmids (Calero et al., 2016; Cook et al., 2018), a library of synthetic promoters (Elmore et al., 2017), and the dual-inducible duet-expression system have been generated (Gauttam et al., 2020). The modification of the ribosome-binding site (Calero et al., 2016) and codon optimization (Dammeyer et al., 2011) were also carried out to enhance the expression at the translational level. However, the expression level of P. putida KT2440 is still low, limiting the application of P. putida as a chassis for heterologous expression in industry.
T7 RNA polymerase expression system is an important and high-efficient expression system in model strains such as Escherichia coli (Wang et al., 2018; Shilling et al., 2020). However, the T7 RNA polymerase-mediated expression system is not always efficient for some non-model microbial strains (Zhao et al., 2017). For example, the T7 expression system constructed in P. putida KT2440 was low efficiency and was rarely used (Christina et al., 2012). Recently, broad-host T7-like systems including the MmP1 expression system have been developed in Halomonas sp. TD01 and P. entomophila LAC31 (Zhao et al., 2017). Furthermore, the MmP1 expression system constructed in the plasmid has successfully contributed to the enhancement of heterologous expression of nicotinate dehydrogenase (NDHase) in C. testosteroni CNB-2 (Lu et al., 2020). Given the high efficiency and versatility, it is supposed that the T7-like system might be suitable for the heterologous expression in P. putida KT2440.
In this study, we attempted to construct the T7-like expression system and evaluate the heterologous expression level in P. putida KT2440. MmP1 RNAP cassette was successfully integrated into the genome of P. putida KT2440, which significantly improved the expression level of super-folder green fluorescent protein (sfGFP). The insertion site and the copy number of MmP1 RNAP cassette were then optimized to improve the efficiency of the MmP1 expression system. We successfully obtained the T7-like expression system in P. putida KT2440, which consisted of the MmP1 RNAP integrated strain and corresponding plasmid. We also utilized the strategy to obtain a T7 expression system in P. putida KT2440 with T7 RNAP inserted in the optimal site. These expression systems were much more efficient than those previously reported in P. putida KT2440. The Mmp1 expression system was also constructed in the P. putida EM42. The expression level of difficult-to-express NDHase was successfully enhanced by using the MmP1 expression system in both P. putida KT2440 and P. putida EM42, demonstrating the high efficiency of the expression system.
Materials and Methods
Strains, Culture Conditions, and Chemicals
E. coli strain DH5α was used for molecular cloning and plasmid propagation throughout this study. P. putida KT2440 and EM42 were used for the characterization of T7 and MmP1 RNAP-promoter pairs. Luria-Bertani (LB) medium was used for culturing E. coli and P. putida. Antibiotics were supplemented at the following concentrations when needed: for E. coli, 50 μg/ml kanamycin, 15 μg/ml tetracycline; for P. putida, 100 μg/ml kanamycin, 25 μg/ml tetracycline, 100 μg/ml spectinomycin, 50 μg/ml gentamicin. The inducer for Ptrc promoter was isopropyl β-D-1-thiogalactopyranoside (IPTG) and 3-methylbenzoate was for Xyls/Pm, arabinose was for AraC/ParaB, and rhamnose was for RhaS/Prha. E. coli was grown at 37°C, and P. putida KT2440 and EM42 were incubated at 30°C. The DNA polymerase PrimeSTAR® Max was purchased from Takara Bio Inc (Dalian, China). The Green Taq Mix and the One Step Cloning Kit were purchased from Vazyme Biotechnology (Nanjing, China). The Fast Digest DpnI and the PageRuler™ Prestained Protein Ladder 10–180 kDa were purchased from Thermo Scientific (United States).
Construction of Plasmids
All the strains and plasmids used in this study are listed in Supplementary Table 1. All the integration strains are listed in Supplementary Table 1. All the primers used to construct vectors are listed in Supplementary Table 2. Plasmid pSEVA-gRNAF was used as the backbone for plasmid construction. Plasmid pSEVA64-vdh was derived from pSEVA-gRNAF by replacing the original N20 sequence with “GTGGAACGCCCCGGTGATAC” via inverse PCR using primers vdh-gF/vdh-gR. The 800 bp length upstream and downstream homologous arms of vdh acquired from the Pseudomonas Genome Database were amplified using primers vdh-1F/vdh-ZR, vdh-ZF/vdh-2R, and connected via overlap-extension (SOE) PCR. The N20 sequence was designed by CasOT (Xiao et al., 2014). A similar method was applied to construct the plasmids pSEVA64-5003 and pSEVA64-5007. T7-like RNAP cassette and promoter were cloned from the plasmid pMmP1-p321, which were donated by the Chen team (Zhao et al., 2017). The RNAP module of T7 and MmP1 was inserted between the upstream homologous arm and the downstream homologous arm to generate the gene inserting vector pSEVA64-T7-vdh, pSEVA64-MmP1-vdh, pSEVA64-MmP1-5003, pSEVA64-T7-5003, and pSEVA64-Mmp1-5007. The verification plasmids containing RNAP modules and their promoter modules (the reporter protein is the sfGFP) were assembled on the plasmid backbone pSEVA64 (Martínez-Garćía et al., 2015) using One Step Cloning Kit to create the plasmids pSEVA64-DE-pRO, pSEVA64-MmP1-pRO. Removed the RNAP module of pSEVA64-DE-pRO, pSEVA64-MmP1-pRO to create the plasmids pSEVA64-DE-sfGFP and pSEVA64-MmP1-sfGFP. Similarly, the other five inducible expression systems (LacIq-Ptrc, LacIq-Ptrc, Xyls/Pm, AraC/ParaB, RhaS/Prha) were assembled on the upstream of sfGFP with the plasmid backbone pSEVA64 to create the plasmid pSEVA644-sfGFP, pSEVA644R-sfGFP, pSEVA648-sfGFP, pSEVA6410-sfGFP, and pSEVA64-Rha-sfGFP.
Construction of Platform Strains
The platform strains including KTVT and KTVM were constructed by integrating the RNAP module of T7 and MmP1 into the vdh site of the genome of P. putida KT2440, respectively. The integration was carried out by CRISPR/Cas9 system (Sun et al., 2018). The integrated strain KTCM, KTFM, EMCM, EMFM were constructed by integrating the RNAP module of MmP1 into the PP_5003 and the PP_5007 site of P. putida KT2440 and EM42, respectively. The other four P. putida KT2440 mutants, KTCVM, KTVFM, KTCFM, KTCFVM were constructed by the different combinations of the integration sites. The integrated strain KTCT was generated by introducing the T7 RNAP cassette into the PP_5003 site of P. putida KT2440.
Coarse Characterization
Plasmids pSEVA64-MmP1-pRO and pSEVA64-MmP1-sfGFP were transformed into P. putida KT2440, KTVM, respectively, using the electroporation method (Figure 1). Plasmids pSEVA64-DE-pRO and pSEVA64-DE-sfGFP were transformed into P. putida KT2440, KTVT, respectively. The candidate strains were screened in selection plates and identified by colony PCR. These strains were incubated in LB medium at 30°C, and 1 mM IPTG was added for the induction expression when the optical density reached OD600 = 0.4 (about 3 h). After 24 h of incubation, cells were harvested by centrifugation at 4°C and 12,000 × g for 10 min. The cell pellets were washed twice with phosphate buffer (pH 7.4) and then diluted two times. 200 μ of the diluted liquid was added into the 96-well microtiter plates and measured the absolute fluorescence intensity (AFI) and cell density (OD600) by a microplate reader using the excitation wavelength at 488 nm and fluorescence emission at 510 nm. Finally, the relative fluorescence intensity (RFI) was calculated by the AFI/OD600.
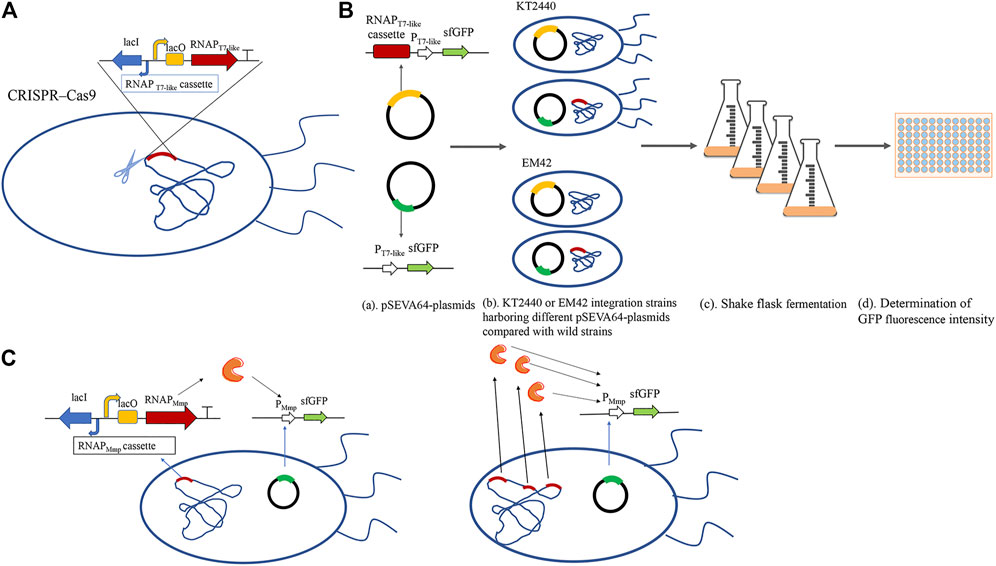
FIGURE 1. The schematic diagram of the MmP1 expression system constructed in P. putida. (A) Construction of integrated strain by integrating the RNAP cassette into nonessential gene of the genome in P. putida by CRISPR/Cas9; (B) Flowchart about the flask shake fermentation of sfGFP with different RNA polymerase cassette; (a) Construction of verified plasmids: the upper contains both RNAP cassette and promoter modules; the lower contains only promoter modules; (b) The verified plasmids were into wild-type strains and RNAP integrated strains, respectively; (c) Shake flask experiments were carried out by inoculating with 2% of the seed cultures at 30°C; (d) Cells were harvested and diluted two times. 200 μl of the diluted liquid was added into the 96-well microtiter plates and measured the absolute fluorescence intensity (AFI) and cell density (OD600) by microplate reader using the excitation wavelength at 488 nm and fluorescence emission at 510 nm. Finally, the relative fluorescence intensity (RFI) was calculated by the AFI/OD600.; (C)Construction of different copy numbers of MmP1 RNAP integrated in the genome of P. putida.
Optimization of Induction Condition of Different Inducible Expression Systems in P. putida KT2440
For the Mmp1 expression system, the RNAP-integrated strains harboring the corresponding verification plasmids were grown in 5 ml LB with 50 μg/ml gentamicin overnight at 30°C. Then, shake flask experiments were carried out by inoculating 2% of the seed cultures at 30°C. 1 mM IPTG was added in the shake flasks at 0, 2, 4, 6, and 8 h after the inoculation at 30°C, respectively. The AFI and OD600 of the cultures were measured after 24 h of inoculation to obtain the optimal induction point. Next, the optimal IPTG concentration was obtained using a similar method. Different IPTG concentrations (1, 10, 25, 75, 100, 250, 500, 750, 1,000, 1,500, 2,000, 2,500, and 3,000 μM) were added at 4 h (the optimal induction point) after inoculating in shake flasks at 30°C. Then the optimal IPTG concentration was obtained by comparing the RFI of sfGFP.
For other inducible expression systems, P. putida KT2440 strains containing sfGFP expression plasmids (pSEVA644-sfGFP, pSEVA644R-sfGFP, pSEVA648-sfGFP, pSEVA6410-sfGFP, pSEVA64-Rha-sfGFP) were grown overnight in 5 ml LB with 50 μg/ml gentamicin. 2% of the seed cultures were inoculated into shake flask. Varying amounts of inducer were added 4 h after inoculating: IPTG (0, 250, 500, 750, 1,000, 1,500 μM) for pSEVA644-sfGFP, pSEVA644R-sfGFP, 3-methylbenzoate (0, 0.25, 0.5, 0.75, 1 mM) for pSEVA648-sfGFP, arabinose (0, 10, 50, 80, 100, 120 mM) for pSEVA6410-sfGFP, rhamnose (0, 3, 5, 10, 15 mM) for pSEVA64-Rha-sfGFP. Optimal inducer concentrations were obtained by comparing the RFI of sfGFP.
The NDHase Activity Assay
The engineered strains (KTCM and EMCM), as well as the wild-type strains (KT2440 and EM42) harboring pSEVA64-JA-sfGFP plasmid, were cultured in 5 ml LB overnight at 30°C. 1 ml of the seed cultures were used to inoculate in 250 ml shake flasks. Then 0.5 mM IPTG was added at 4 h after inoculation for the induction expression at 18°C for 20 h. After the shake flask fermentation, 4 ml of the samples were harvested by centrifugation at 4°C and 4,000 rpm for 10 min. The cell pellets were resuspended with 1 ml of 0.1 M 3-cyanpyridine. To measure NDHase activity, 0.05 mM PMS was added into 1 ml resuspension with magnetic stirring for 2 h at 30°C. Cells were heated at 99°C for 5 min to terminate the reaction. High-performance liquid chromatography (HPLC) with a Pntulips QS-C18 column (4.6 mm × 250 mm, 5 μm) was used to detect the production of 6-hydroxy-3-cyanopyridine (6-HCP). Detection was performed using a wavelength of 260 nm with 1 ml/min of the liquid phase containing acetonitrile and water (10:90 v/v). One unit (U) of NDHase activity was defined as the amount of enzyme required to produce 1 μmol of 6-HCP in 1 min under the test conditions.
Result
Construction of MmP1 Expression System in P. putida KT2440
To study the effect of T7-like expression system containing T7-like RNAP cassette and T7-like promoter module in P. putida KT2440, the Mmp1 RNAP cassette was inserted either in the genome or plasmid. The Mmp1 RNAP cassette was first integrated into the vdh site (locus tag is PP_3357) in the genome by CRISPR/Cas9, resulting in the integrated strain KTVM (Figure 1A and Supplementary Figure 1). The corresponding plasmid containing PMmP1 promoter and sfgfp was transformed into the integrated strain KTVM to examine the effect of constructed system. The RNAP cassette coupled with sfgfp was also inserted in the plasmid pSEVA64, which was then transformed into P. putida KT2440.
The absolute fluorescence intensity (AFI) and cell density (OD600) were determined after shake-flask fermentation (Figure 1B). When the MmP1 RNAP cassette was expressed in the plasmid, the RFI value was slightly higher than that inserted into the genome (Figure 2). However, the large-size expression plasmid might decrease the transformation frequency (Walker and Klaenhammer, 2002), affect the quantity of the plasmid DNA isolated from the host strain (Yang and Yang, 2012), and was difficult to construct than smaller size plasmids. Although RNAP cassette inserted in the plasmid provided relatively higher sfGFP expression, considering the RNAP integrated into the genome was genetically stable and convenient to manipulate as the development of the genome editing technology. Hence, the MmP1 RNAP cassette integrated into the genome of P. putida KT2440 was chosen for further enhancing the heterologous expression.
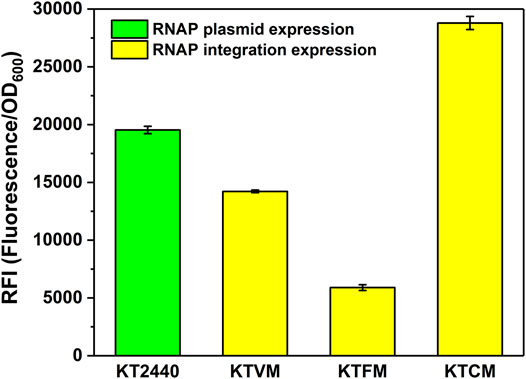
FIGURE 2. Optimization of the MmP1 RNAP casette integrated into P. putida KT2440 with different integration sites. KT2440: KT2440 harboring the plasmid containing the MmP1 RNAP module and sfGFP with MmP1 promoter; KTVM: KTVM with MmP1 RNAP cassette integrated into vdh site of KT2440 harboring the plasmid containing sfGFP with MmP1 promoter; KTFM: KTFM with MmP1 RNAP cassette integrated into phaC1 site of KT2440 harboring the plasmid containing sfGFP with MmP1 promoter; KTCM: KTCM with MmP1 RNAP cassette integrated into phaF site of KT2440 harboring the plasmid containing sfGFP with MmP1 promoter.
Optimization of the Mmp1 Expression System in P. putida KT2440
Optimization of the Insertion Site of MmP1 RNAP Cassette
The insertion sites were optimized to further improve the MmP1 expression system in P. putida KT2440 (Supplementary Table 3). Three nonessential genes including phaC1, phaF, vdh were selected as the insertion sites, which have been used to integrate other heterologous genes (Gong et al., 2016). The modified strains were named KTCM, KTFM, and KTVM with RNAP cassette inserted in phaC1, phaF, and vdh, respectively. Interestingly, the expression of sfGFP was improved 2-fold in KTCM relative to that in KTVM. Compared to the plasmid expression style, the expression of the sfGFP in KTCM was increased by 1.4-fold (Figure 2), whereas that in KTFM was decreased by 3.3-fold.
Optimization of the Copy Number of the MmP1 RNAP Cassette
We also investigated the effect of the copy number of MmP1 RNAP cassette integrated into P. putida KT2440 on expression. Two copies of RNAP integrated strains were constructed including KTCFM with insertion sites of phaC1 and phaF, KTCVM with insertion sites of phaC1 and vdh, KTFVM with insertion sites of phaF and vdh, and three copies of RNAP integration strain, KTCFVM was constructed with the insertion sites of phaC1, phaF, and vdh (Figure 1C). Among them, the two-copy strain KTCVM displayed the highest expression level of sfGFP, 1.3-fold higher than the RNAP plasmid expression system (Figure 3). However, it was still slightly lower than the KTCM (Figure 2). Thus, the strain KTCM was selected for further investigation, and the MmP1 expression system in P. putida KT2440 consisting of the host strain KTCM and the plasmid pSEVA64-MmP1-sfGFP carrying PMmP1 promoter corresponding to MmP1 RNAP.
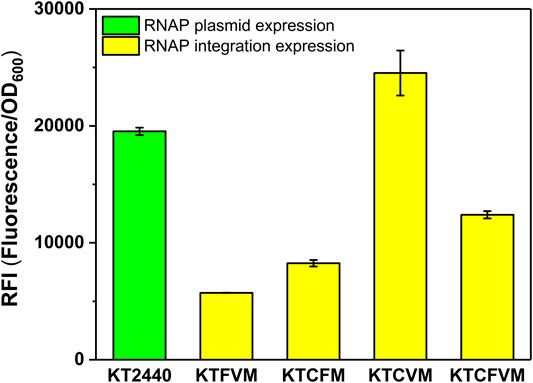
FIGURE 3. The effect of different copy numbers of Mmp1 RNAP on expression. KT2440: KT2440 harboring the plasmid containing the MmP1 RNAP module and sfGFP with MmP1 promoter; KTFVM: KTFVM with insertion sites of phaF and vdh harboring the plasmid containing sfGFP with MmP1 promoter; KTCFM: KTCFM with insertion sites of phaC1 and phaF harboring the plasmid containing sfGFP with MmP1 promoter; KTCVM: KTCVM with insertion sites of phaC1 and vdh harboring the plasmid containing sfGFP with MmP1 promoter; KTCFVM: KTCFVM with insertion sites of phaC1, phaF and vdh harboring the plasmid containing sfGFP with MmP1 promoter.
Construction of the MmP1 Expression System in P. putida EM42
To verify the versatility of the MmP1 expression system, we extended the MmP1 expression system to the gene-reduced strain EM42. The optimal RNAP integration site (phaC1) was utilized in EM42 resulting in the integrated strain EMCM. As a result, the RFI in EMCM was about 2.1-fold higher than that of the MmP1 RNAP plasmid expression in EM42 (Figure 4).
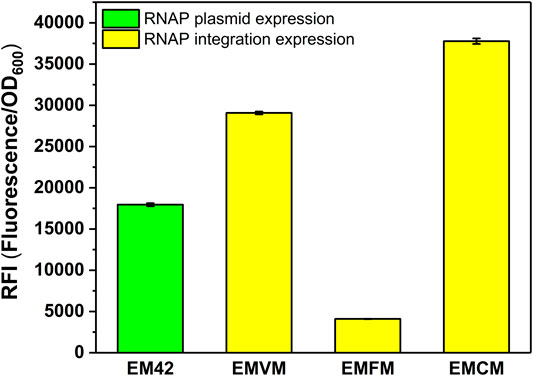
FIGURE 4. RFI of the MmP1 RNAP cassette integrated into P. putida EM42 with different integration sites. EM42: EM42 harboring the plasmid containing the MmP1 RNAP module and sfGFP with MmP1 promoter; EMVM: EMVM with insertion sites of vdh harboring the plasmid containing sfGFP with MmP1 promoter; EMFM: EMFM with insertion sites of phaF harboring the plasmid containing sfGFP with MmP1 promoter; EMCM: EMCM with insertion sites of phaC1 harboring the plasmid containing sfGFP with MmP1 promoter.
Verification of the Commonality of the Strategy to Improve Expression Level With T7 RNAP
Similar to the construction of KTVM, KTCM, we introduced the T7 RNAP cassette into the genome of KT2440 to obtain KTVT and KTCT. To test whether the strategy of increasing heterologous expression level applied to T7 RNAP, the corresponding plasmid containing sfGFP was transformed into the integrated strains KTVT and KTCT. It turns out that the optimal insertion site was also effective for T7 RNAP. As shown in Figure 5, the RFI in KTCT was 1.4-fold higher than that in KTVT.
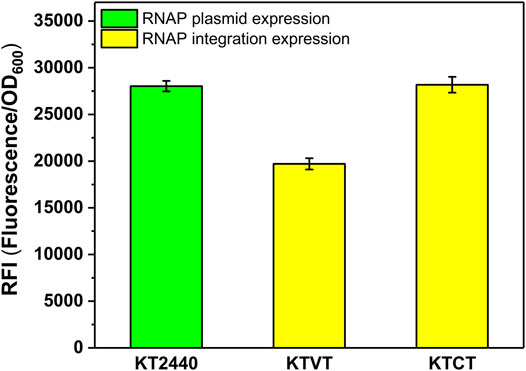
FIGURE 5. RFI of the T7 RNAP cassette integrated into P. putida KT2440 with different integration sites. KT2440: KT2440 harboring the plasmid containing the T7 RNAP module and sfGFP with T7 promoter; KTVT: KTVT with T7 RNAP cassette integrated into vdh site of KT2440 harboring the plasmid containing sfGFP with T7 promoter; KTCT: KTCT with T7 RNAP cassette integrated into phaC1 site of KT2440 harboring the plasmid containing sfGFP with T7 promoter.
Comparing the Optimized MmP1 Expression System and the T7 Expression System With Other Expression Systems in P. putida KT2440
The optimized MmP1 and T7 expression systems were compared with other inducible expression systems in P. putida KT2440 to understand its effect. The induction conditions were first optimized. The optimization of the induction condition of KT2440 was performed under different induction points (0–8 h) by using the MmP1 expression system. As shown in Supplementary Figure 4A, when IPTG was added at the induction point 4 h after inoculation, the final RFI value was highest, which was determined to be the optimal induced point (Supplementary Figure 4A).
Different inducer concentrations were tested for the effect on expression level and the optimal inducer concentration for six inducible expression systems in P. putida was 0.5 mM IPTG for MmP1 expression system, 1 mM IPTG for LacIq/ptac expression system, 2 mM IPTG for LacIq/ptrc expression system, 80 mM arabinose for AraC/ParaB expression system, 5 mM rhamnose for RhaS/Prha expression system, 0.25 mM 3-methylbenzoate for Xyls/Pm expression system (Supplementary Figure 4). The optimal condition of the T7 expression system was considered the same as that of the MmP1 expression system.
The optimized induction conditions were then utilized to compare the expression level of different inducible systems in P. putida KT2440. As shown in Figure 6, a high expression level was achieved by the MmP1 expression system showing 2.5-fold, 2.6-fold, 6.6-fold, 9.3-fold, and 10.1-fold improvement compared to RhaS/Prha, LacIq-Ptrc AraC/ParaB Xyls/Pm, and LacIq-Ptac, respectively. The expression level of the optimized T7 expression system was slightly higher than MmP1 by 1.2-fold. The expression level of RhaS/Prha was followed by the LacIq-Ptrc expression system and the AraC/ParaB expression system was lower than the LacIq-Ptrc expression system, almost half of the RhaS/Prha expression system. LacIq-Ptac and Xyls/Pm showed a weak expression level. All expression systems showed more than 10-fold improved sfGFP expression in induced conditions compared to uninduced conditions, except for Xyls/Pm, specifically, the ratio of induced expression to uninduced expression was 73, 82, 43, 35, 123, and 15 for T7, Mmp1, RhaS/Prha, LacIq-Ptrc, AraC/ParaB, and LacIq-Ptac expression system, respectively.
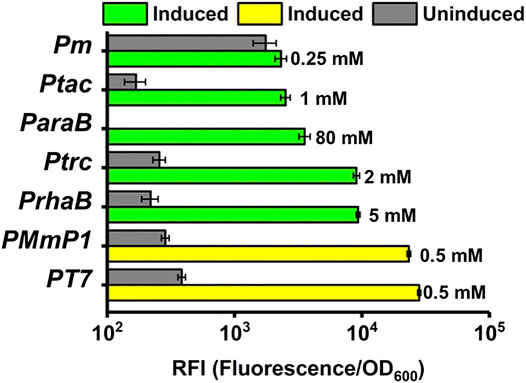
FIGURE 6. The RFI of the T7 and MmP1 expression system compared with the other inducible expression systems. The number to the right of each column represents the optimized inducer concentration.
Verification of the Effectiveness of the MmP1 Expression System by the Expression of NDHase
The difficult-to-express NDHase from C. testosteroni JA1 was expressed using the MmP1 expression system both in KTCM and EMCM compared with KT2440 (Supplementary Figure 5). The activity of the NDHase using 3-cyanopyridine as substrate in the KTCM was 82.1 U/L, 3.6-fold higher than that in KT2440. The activity of the NDHase in the EMCM was 50.4 U/L, which is 2.2-fold higher than that in EM42 (Figure 7), exhibiting the excellent performance of the MmP1 expression system.
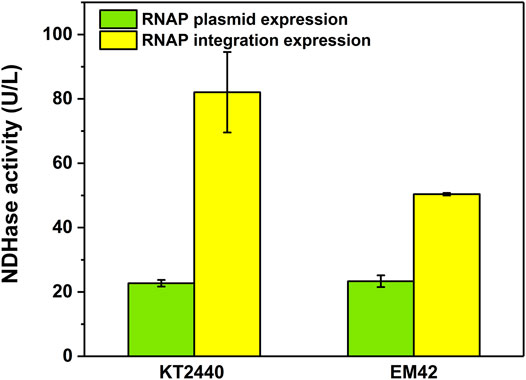
FIGURE 7. The activity of NDHase in the MmP1 expression system with the integration site of PP_5003 compared with the plasmid expression both in P. putida KT2440 and P. putida EM42.
Discussion
In previous research, the T7-like system MmP1 dramatically increased the GFP expression level in P. entomophila LAC31 (Zhao et al., 2017). Here, we would like to explore whether the T7-like system integrated into the genome of P. putida could also improve the expression level. We introduced the Mmp1 RNAP expression system both in the plasmid with the most effective origin of replication pRO1600/ColE1 (Damalas et al., 2020) and in the genome of P. putida. It was found that MmP1 RNAP was highly specific to the MmP1 promoter in P. putida KT2440 and the expression system was strictly regulated by IPTG (Supplementary Figure 2) with 82-fold induction. Previously, Zhao et al. found that one more Lac operator (lacO) introduced into the plasmid was necessary for the MmP1 expression system in Halomonas sp. TD01 (Zhao et al., 2017). However, lacO was not necessary for the MmP1 expression system in P. putida KT2440. Therefore, the MmP1 expression system was appropriate and convenient for heterologous expression in P. putida KT2440.
To optimize the MmP1 expression system in P. putida, we investigated the effect of insertion site and copy number on the heterologous protein expression. Three integration sites including vdh, phaC1, and phaF (locus tag is PP_3357 PP_5003 and PP_5007) were selected, which were reported to be nonessential genes and available insertion sites in P. putida (Gong et al., 2016). The RNAP cassette inserted in phaC1 could significantly increase the expression of the sfGFP by 1.4-fold in KTCM, compared with the RNAP plasmid expression. Additionally, the RFI of sfGFP in KTCFM, KTFVM, and KTCFVM was lower than that in KT2440, indicating that the increased copy number of RNAP was not useful for the expression. Therefore, integrating one copy of the RNAP cassette in the site of phaC1 in P. putida KT2440 was determined to be best for the improvement of expression level. And the MmP1 expression system in P. putida KT2440 was established, containing the host strain KTCM and plasmid pSEVA64-MmP1-sfGFP carrying its associated promoter PMmP1.
The induction conditions were then optimized by changing the induced point and the IPTG concentrations. It was finally determined that IPTG concentration of 0.5 mM and the induced point of 4 h after the inoculation provided the highly efficient heterologous expression in P. putida KT2440.
We then evaluated the versatility of the optimized MmP1 expression system in the genome-reduced strain P. putida EM42 showing much higher levels of recombinant protein production compared to P. putida KT2440 (Martínez-García et al., 2014). The MmP1 RNAP was inserted in the optimal integration site (phaC1) of the gene-reduced strain EM42 and successfully increased the expression of sfGFP by 2.1-fold in EMCM (Figure 4).
The optimal RNAP integration site (phaC1) was also used for introducing T7 RNAP in P. putida KT2440 and the expression level was enhanced as well, indicating the generality of the integration site for the T7 expression system. The MmP1 and T7 expression system were then compared with the existing expression systems in P. putida KT2440 (Calero et al., 2016; Martínez-García and de Lorenzo, 2017; Cook et al., 2018; Damalas et al., 2020). T7 and MmP1 expression systems both showed excellent performance than other cases and were nearly three times the LacIq-Ptrc expression system, although the LacIq-Ptrc expression system in P. putida KT2440 was improved almost 2-fold by switching PlacIq with weaker constitutive promoters (Cook et al., 2018). This demonstrated that the MmP1 expression system constructed in this study was extremely effective.
The expression level of RhaS/Prha was followed by the AraC/ParaB expression system, consistent with previous reports (Calero et al., 2016), but opposite to Damalas et al. (2020). The Xyls/Pm expression system seemed to be the weakest, which was similar to the previous reports (Damalas et al., 2020), but opposite to the findings of Calero et al. (2016). This could be due to the different origin of replication and the different ribosome-binding sites we used. In the previous study, T7 RNAP was integrated into the downstream of the glmS gene showing a lower expression level than RhaS/Prha and AraC/ParaB expression system (Calero et al., 2016). In this study, we chose different insertion sites for T7 RNAP and ended up with more than 3-fold higher expression levels than other existing inducible expression systems (Figures 4, 5), suggesting that the insertion site was critical to effect of the RNAP.
To further confirm the utility of the established MmP1 expression system in P. putida, we performed the heterologous expression assay by using the difficult-to-express protein NDHase from C. testosteroni JA1. To our delight, the expression level of NDHase in both the engineered KTCM and EMCM were highly improved compared to the original strains. By using 3-cyanopyridine as substrate, the activities of whole-cell KTCM and EMCM were 82.1 and 50.4 U/L, which was almost 7- and 4-fold higher than that in the P. entomophila L48 (Yang et al., 2010).
Hence, we established the MmP1 and T7 expression systems in P. putida and provided a set of useful chassis (KTCM, KTCT, and EMCM) and corresponding plasmids (pSEVA64-MmP1-sfGFP and pSEVA64-DE-sfGFP) to express difficult-to-express proteins in synthetic biology. These expression systems also widened the application of T7-like RNA polymerase-based transcriptional regulation on the genome of P. putida. In addition, the strategy of optimizing the insertion sites discussed in the present study could potentially be used for enhancing the heterologous expression level for other non-model microbial strains.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
TL, JS, SJ, LY, and JW conceived and designed the experiments. TL carried out the experiments and drafted the manuscript. All authors analyzed the data. SJ, SS, and JW discussed and revised the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the National Key R&D Program of China (No.2019YFA09005000) and the National Natural Science Foundation of China (No. 21676240).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.664967/full#supplementary-material
References
Calero, P., Jensen, S. I., and Nielsen, A. T. (2016). Broad-Host-Range ProUSER Vectors Enable Fast Characterization of Inducible Promoters and Optimization Ofp-Coumaric Acid Production in Pseudomonas putida KT2440. ACS Synth. Biol. 5, 741–753. doi:10.1021/acssynbio.6b00081
Christina, S., Thies, S., Link, O., Isabell, C., Knops, K., Wilhelm, S., et al. (2012). Novel Broad Host Range Shuttle Vectors for Expression in Escherichia coli , Bacillus Subtilis and Pseudomonas putida. J. Biotechnol. 161, 71–79. doi:10.1016/j.jbiotec.2012.02.020
Cook, T. B., Rand, J. M., Nurani, W., Courtney, D. K., Liu, S. A., and Pfleger, B. F. (2018). Genetic Tools for Reliable Gene Expression and Recombineering in Pseudomonas putida. J. Ind. Microbiol. Biotechnol. 45, 517–527. doi:10.1007/s10295-017-2001-5
Damalas, S. G., Batianis, C., Martin‐Pascual, M., Lorenzo, V., and Martins dos Santos, V. A. P. (2020). SEVA 3.1: Enabling Interoperability of DNA Assembly Among the SEVA, BioBricks and Type IIS Restriction Enzyme Standards. Microb. Biotechnol. 13, 1793–1806. doi:10.1111/1751-7915.13609
Dammeyer, T., Steinwand, M., Krüger, S., Dübel, S., Hust, M., and Timmis, K. N. (2011). Efficient Production of Soluble Recombinant Single Chain Fv Fragments by a Pseudomonas putida Strain KT2440 Cell Factory. Microb. Cell Fact 10, 11. doi:10.1186/1475-2859-10-11
dos Santos, V. A. P. M., Heim, S., Moore, E. R. B., Strätz, M., and Timmis, K. N. (2004). Insights into the Genomic Basis of Niche Specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6, 1264–1286. doi:10.1111/j.1462-2920.2004.00734.x
Ebert, B. E., Kurth, F., Grund, M., Blank, L. M., and Schmid, A. (2011). Response of Pseudomonas putida KT2440 to Increased NADH and ATP Demand. Appl. Environ. Microbiol. 77, 6597–6605. doi:10.1128/AEM.05588-11
Elmore, J. R., Furches, A., Wolff, G. N., Gorday, K., and Guss, A. M. (2017). Development of a High Efficiency Integration System and Promoter Library for Rapid Modification of Pseudomonas putida KT2440. Metab. Eng. Commun. 5, 1–8. doi:10.1016/J.METENO.2017.04.001
Gauttam, R., Mukhopadhyay, A., and Singer, S. W. (2020). Construction of a Novel Dual-Inducible Duet-Expression System for Gene (Over)expression in Pseudomonas putida. Plasmid 110, 102514. doi:10.1016/j.plasmid.2020.102514
Gong, T., Liu, R., Zuo, Z., Che, Y., Yu, H., Song, C., et al. (2016). Metabolic Engineering of Pseudomonas putida KT2440 for Complete Mineralization of Methyl Parathion and γ-Hexachlorocyclohexane. ACS Synth. Biol. 5, 434–442. doi:10.1021/acssynbio.6b00025
Herrero, M., de Lorenzo, V., Ensley, B., and Timmis, K. N. (1993). A T7 RNA Polymerase-Based System for the Construction of Pseudomonas Strains with Phenotypes Dependent on TOL-Meta Pathway Effectors. Gene 134, 103–106. doi:10.1016/0378-1119(93)90181-2
Lieder, S., Nikel, P. I., de Lorenzo, V., and Takors, R. (2015). Genome Reduction Boosts Heterologous Gene Expression in Pseudomonas putida. Microb. Cell Fact. 14, 23–14. doi:10.1186/s12934-015-0207-7
Lu, Z.-H., Yang, L.-R., and Wu, J.-P. (2020). Efficient Heterologous Expression of Nicotinate Dehydrogenase in Comamonas Testosteroni CNB-2 with Transcriptional, Folding Enhancement Strategy. Enzyme Microb. Tech. 134, 109478. doi:10.1016/j.enzmictec.2019.109478
Martínez-García, E., Aparicio, T., Goñi-Moreno, A., Fraile, S., and de Lorenzo, V. (2015). SEVA 2.0: An Update of the Standard European Vector Architecture for De-/re-construction of Bacterial Functionalities. Nucleic Acids Res. 43, D1183–D1189. doi:10.1093/nar/gku1114
Martínez-García, E., and de Lorenzo, V. (2017). Molecular Tools and Emerging Strategies for Deep Genetic/genomic Refactoring of Pseudomonas. Curr. Opin. Biotechnol. 47, 120–132. doi:10.1016/j.copbio.2017.06.013
Martínez-García, E., and de Lorenzo, V. (2019). Pseudomonas putida in the Quest of Programmable Chemistry. Curr. Opin. Biotechnol. 59, 111–121. doi:10.1016/j.copbio.2019.03.012
Martínez-García, E., Nikel, P. I., Aparicio, T., and de Lorenzo, V. (2014). Pseudomonas 2.0: Genetic Upgrading of P. Putida KT2440 as an Enhanced Host for Heterologous Gene Expression. Microb. Cell Fact. 13, 159. doi:10.1186/s12934-014-0159-3
Nikel, P. I., Chavarría, M., Danchin, A., and de Lorenzo, V. (2016). From Dirt to Industrial Applications: Pseudomonas putida as a Synthetic Biology Chassis for Hosting Harsh Biochemical Reactions. Curr. Opin. Chem. Biol. 34, 20–29. doi:10.1016/j.cbpa.2016.05.011
Nikel, P. I., and de Lorenzo, V. (2018). Pseudomonas putida as a Functional Chassis for Industrial Biocatalysis: From Native Biochemistry to Trans-metabolism. Metab. Eng. 50, 142–155. doi:10.1016/j.ymben.2018.05.005
Nikel, P. I., Martínez-García, E., and De Lorenzo, V. (2014). Biotechnological Domestication of Pseudomonads Using Synthetic Biology. Nat. Rev. Microbiol. 12, 368–379. doi:10.1038/nrmicro3253
Poblete-Castro, I., Becker, J., Dohnt, K., dos Santos, V. M., and Wittmann, C. (2012). Industrial Biotechnology of Pseudomonas putida and Related Species. Appl. Microbiol. Biotechnol. 93, 2279–2290. doi:10.1007/s00253-012-3928-0
Shilling, P. J., Mirzadeh, K., Cumming, A. J., Widesheim, M., Köck, Z., and Daley, D. O. (2020). Improved Designs for pET Expression Plasmids Increase Protein Production Yield in Escherichia coli. Commun. Biol. 3. doi:10.1038/s42003-020-0939-8
Sun, J., Wang, Q., Jiang, Y., Wen, Z., Yang, L., Wu, J., et al. (2018). Genome Editing and Transcriptional Repression in Pseudomonas putida KT2440 via the Type II CRISPR System. Microb. Cell Fact. 17, 41. doi:10.1186/s12934-018-0887-x
Timmis, K. N. (2002). Pseudomonas putida: a Cosmopolitan Opportunist Par Excellence. Environ. Microbiol. 4, 779–781. doi:10.1046/j.1462-2920.2002.00365.x
Walker, S. a., and Klaenhammer, T. R. (2002). The Effect of Increasing Plasmid Size on Transformation Efficiency in Escherichia coli. J. Exp. Microbiol. Immunol. 2, 207–223.
Wang, W., Li, Y., Wang, Y., Shi, C., Li, C., Li, Q., et al. (2018). Bacteriophage T7 Transcription System: an Enabling Tool in Synthetic Biology. Biotechnol. Adv. 36, 2129–2137. doi:10.1016/j.biotechadv.2018.10.001
Xiao, A., Cheng, Z., Kong, L., Zhu, Z., Lin, S., Gao, G., et al. (2014). CasOT: A Genome-Wide Cas9/gRNA Off-Target Searching Tool. Bioinformatics 30, 1180–1182. doi:10.1093/bioinformatics/btt764
Yang, J., and Yang, Y. (2012). Plasmid Size Can Affect the Ability of Escherichia coli to Produce High-Quality Plasmids. Biotechnol. Lett. 34, 2017–2022. doi:10.1007/s10529-012-0994-4
Yang, Y., Chen, T., Ma, P., Shang, G., Dai, Y., and Yuan, S. (2010). Cloning, Expression and Functional Analysis of Nicotinate Dehydrogenase Gene Cluster From Comamonas Testosteroni JA1 That Can Hydroxylate 3-Cyanopyridine. Biodegradation 21, 593–602. doi:10.1007/s10532-010-9327-2
Zhao, H., Zhang, H. M., Chen, X., Li, T., Wu, Q., Ouyang, Q., et al. (2017). Novel T7-like Expression Systems Used for Halomonas. Metab. Eng. 39, 128–140. doi:10.1016/j.ymben.2016.11.007
Keywords: Pseudomonas putida KT2440, chassis, heterologous expression, synthetic biology, RNA polymerase, difficult-to-express protein
Citation: Liang T, Sun J, Ju S, Su S, Yang L and Wu J (2021) Construction of T7-Like Expression System in Pseudomonas putida KT2440 to Enhance the Heterologous Expression Level. Front. Chem. 9:664967. doi: 10.3389/fchem.2021.664967
Received: 08 February 2021; Accepted: 05 July 2021;
Published: 16 July 2021.
Edited by:
Jian Dong Cui, Tianjin University of Science and Technology, ChinaReviewed by:
Francisco Solano, University of Murcia, SpainJay D. Keasling, University of California, Berkeley, United States
Copyright © 2021 Liang, Sun, Ju, Su, Yang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuyun Ju, anVzaHV5dW5Aemp1LmVkdS5jbg==; Jianping Wu, d2pwQHpqdS5lZHUuY24=
 Tianxin Liang1
Tianxin Liang1 Jun Sun
Jun Sun Lirong Yang
Lirong Yang Jianping Wu
Jianping Wu