- Division of Agricultural Chemicals, ICAR-Indian Agricultural Research Institute, New Delhi, India
Despite the emergence of novel biotechnological and biological solutions, agrochemicals continue to play an important role in crop protection. Fungicide resistance is becoming a major problem; numerous cases of fungicide resistance have occurred worldwide in the last decade, resulting in the loss of several fungicides. The discovery of new molecules has therefore assumed critical importance in crop protection. In our quest for biologically active molecules, we herein report the synthesis of a series of twenty-one 3-Iodochromone derivatives (4a–4u), in a two-step process by condensation of 2-hydroxyacetophenone derivatives (2a–2u) with N,N-dimethylformamidedimethylacetal yielding enaminones (3a–3u), followed by cyclization with iodine to corresponding 3-iodochromones. Characterization of these compounds was done by IR, 1H NMR, 13C NMR, and LC-HRMS techniques. All synthesized compounds were screened for their fungicidal activity against Sclerotium rolfsii. Among these 6,8-Dichloro-3-iodochromone 4r was found to be most active (ED50 = 8.43 mg L−1). 2D-Quantitative Structural Activity Relationship (2D-QSAR) analysis was also performed by generating three different models viz., Multiple Linear Regression (MLR, Model 1), Principal Component Regression (PCR, Model 2), and Partial Least Squares (PLS, Model 3). Predictive power and statistical significance of these models were assessed with external and internal validation and leave one-out cross-validation was used for verification. In QSAR study, MLR (Model 1) was found to be best having correlation coefficient (r2) 0.943, cross-validated correlation coefficient (q2) 0.911 and r2pred 0.837. It was observed that DeltaEpsilonC, T_2_Cl_6, T_2_F_6, T_T_F_3, and ZCompDipole are the major descriptors which influence the fungicidal activity of 3-Iodochromone derivatives. The physicochemical parameters were estimated by the VLifeMDS 4.6 software. The QSAR study results will be helpful for structure optimization to improve the activity.
Introduction
The growth of human civilization has been closely related to crop production, and plant diseases have been a concern for human being perhaps since plants were cultivated more than 10,000 years ago. As a consequence of plant diseases, world agriculture faces an estimated loss of 18% annually amounting to approximately 1,300 billion INR (Oerke, 2006). Sclerotium rolfsii Sacc. is a devastating soilborne fungus that infects more than five hundred agricultural and horticultural plant species around the world, causing root rot, stem rot, collar rot, willow, and foot rot diseases (Aycock, 1966; Punja, 1985). Crucifers, cucurbits, and legumes are its most common hosts. The fungus is of considerable economic significance because it causes 10–100 percent crop loss in different crops. Due to the formation of excessive sclerotia it may persist in soil for several years (Punja, 1985). Chemical crop protection measures continue to play an important role in agribusiness in spite of the emergence of novel biotechnological and biological solutions. Resistance to fungicides is becoming a major problem generating disease control problems in many crops. In the last decade, numerous cases of fungicide resistance have occurred worldwide, leading to loss of several fungicides (Hollomon, 2015). Therefore, the discovery of new molecules has assumed critical importance to combat the fungal infections.
Chromone is a group of naturally occurring compounds, reported, mainly in plants. The chromone moiety is a pharmacophore in a large number of natural and synthetic bioactive molecules. The chromone scaffold is present in plant’s secondary metabolites: flavones and isoflavones. Chromones are reported to have anti-tumor, anti-inflammatory and anti-fungal activities, and inhibitory activities, toward, phosphatases, kinases, cyclooxygenases, aromatases, acetylcholinesterases, and monoamine oxidases (Gasparova et al., 1997; Gaspar et al., 2011, Gaspar et al., 2014).
Our group has been actively involved in developing new crop protection products (Kaushik et al., 2019; Yadav et al., 2019). In our quest for biologically active molecules, we herein reported the synthesis of a series of iodochromones and their evaluation against S. rolfsii. A QSAR study was carried out with the objective to find the molecular properties which affect the fungicidal activity.
Experimental
Chemicals and Instruments
Chemicals were purchased from industrial manufacturers and, unless otherwise specified, were used without any further purification. Precoated Merck-silica gel 60F254 plates were used for thin layer chromatography (TLC); UV cabinet was used to detect developed plates. Column chromatography was performed with 100–200 mesh silica gels. Melting points were recorded by Buchi M-560 instrument and were uncorrected. The IR spectroscopy was done with PerkinElmer 2000 FT-IR spectrometer; KBr disc were used for samples preparation. The 1H NMR and 13C spectra were recorded on a Jeol alpha-400 and at 100.6 MHz, respectively, using TMS as an internal standard. The chemical shift values were on δ scale and the coupling constants (J) were in Hz. Signals from OH groups in 1HNMR spectra were verified by removing them by shaking in D2O.
High Resolution Mass Spectrometry (HRMS) was performed by AB SCIEX Triple TOFTM 5600+ equipped with Turboion Spray (TIS), SCIEX ExionLC, and PDA detector. Compounds were separated through C-18 column (2.7 µm, 4.6 × 100 mm) by eluting with methanol and water (98:2, v/v) at 0.3 ml/min at 40°C.
ED50 values were estimated with the SPSS statistical package. The whole computational work was carried out by using VLifeMDS QSAR plus 4.6 software using the Lenovo PC having window 8.1 operating system and Intel (R) Celeron (R) processor.
Synthesis
Synthesis of Substituted 2-Hydroxyacetophenones (2a–2n)
2-hydroxyacetophenone and bromoalkanes or iodoalkanes of different chain lengths were taken in a molar ratio of 1:1.2 and stirred continuously for 6 h at 60°C in the presence of K2CO3 and acetone. Reaction was supervised by thin layer chromatography (TLC) with ethyl acetate: hexane (3:7).
General Method for Synthesis of Substituted 3-Dimethylamino-1-(2-hydroxyphenyl)propenones (3a–3u)
A mixture of substituted 2-hydroxyacetophenones (2a–2u) (1.2 mmol) and N,N-dimethylformamidedimethylacetal (2.4 mmol, 2 molequiv) was heated at 90°C overnight and allowed to cool. The reaction was worked up by removing solvent using vacuum evaporation on Heidolph rotary evaporator Hei-VAP, and the pure product were obtained by column chromatography with hexane: ethyl acetate (85:15). Compounds 3f, 3g, 3h, 3i, 3j, 3k, 3l, 3m, 3n, and 3o were reported for the first time in the literature.
Spectral Analysis of Synthesized 3-Dimethylamino-1-(2-hydroxyphenyl)propenones (3a–3u)
3-Dimethylamino-1-(2-hydroxyphenyl)propenone (3a)
It was obtained as a brown solid in 82% yield; m.p.:127–128°C, Rf: 0.42 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,710 (O-H stretch), 2,949 (aliphatic C-H), 1,635 (C=O), 1,565 (C=C stretch), 1,455 (C-H bending of CH2), and 1,264 (C-O). 1H NMR (400 MHz, CDCl3): δ 2.87 (3H, s, N-CH3), 3.12 (3H, s, N-CH3), 5.73 (1H, d, J = 12, H-3), 6.62–6.73 (4H, m, ArH), and 7.88 (1H, d, J = 12.4, H-2).13C NMR (100.6 MHz, CDCl3): 190.31 (C-1), 162.10 (C-2′), 154.05 (C-3), 133.97 (C-4″), 130.08 (C-6″), 122.31 (C-1″), 121.42.84 (C-5″), 116.21 (C-3″), and 89.75 (C-2). HR-MS for C11H13NO2 [M + H]+m/z: Calcd 192.1091; Observed 192.1087. The major mass fragments observed were C7H5O3+ (137), C5H8O+ (98), and C4H10N+ (72).
3-Dimethylamino-1-(2-hydroxy-4-methoxyphenyl)propenone (3b)
It was obtained as bright light yellow solid in 87% yield; m.p.: 137–140°C, Rf: 0.42 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,711 (O-H stretch), 2,953 (aliphatic C-H), 1,630 (C=O), 1,561 (C=C stretch), 1,458 (C-H bending of CH2), and 1,266 (C-O). 1HNMR (400 MHz, CDCl3): δ 2.95 (3H, s, N-CH3), 3.13 (3H, s, N-CH3), 3.98 (2H, t, J = 6.8, H-1′), 5.67 (1H, d, J = 12, H-2), 6.33–6.37 (2H, m, H-5′ and H-3′), 7.58 (1H, d, J = 8.8 H-6″), and 7.85 (1H, d, J = 12.4, H-3).13C NMR (100.6 MHz, CDCl3): 190.81 (C-1), 165.53 (4C″), 164.06 (2C″), 154.05 (C-3), 129.79 (C-6″), 113.72 (C-1″), 106.84 (C-5′), 101.53 (C-3″), and 89.75 (C-2). HR-MS for C12H15NO3 [M + H]+m/z: Calcd 222.1124; Observed 222.1118. The major mass fragments observed were C8H7O3+(151), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(2-hydroxy-4-ethoxyphenyl)propenone (3c)
It was obtained as bright light yellow solid in 85% yield; m.p.: 140–142°C, Rf: 0.43 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,708 (O-H stretch), 2,951 (aliphatic C-H), 1,642 (C=O), 1,563 (C=C stretch), 1,455 (C-H bending of CH2), and 1,258 (C-O). 1HNMR (400 MHz, CDCl3): δ 1.41 (3H, t, J = 6.8, H-2′), 4.07 (2H, q, H-1′), 2.95 (3H, s, N-CH3), 3.12 (3H, s, N-CH3), 3.98 (2H, t, J = 6.8, H-1′), 5.69 (1H, d, J = 9.6, H-2), 6.35–6.39 (2H, m, H-5″ and H-3″), 7.60 (1H, d, J = 8.8 H-6″), and 7.85 (1H, d, J = 9.6, H-3). 13C NMR (100.6 MHz, CDCl3): 190.83 (C-1), 165.56 (C-4″), 164.09 (C-2″), 154.07 (C-3), 129.80 (C-6″), 113.77 (C-1″), 106.86 (C-5″), 101.57 (C-3″), 89.78 (2C), 68.20 (C-1′), and 15.07 (C-2′). HR-MS for C13H17NO3 [M + H]+m/z: Calcd 236.1281; Observed 236.1268. The major mass fragments observed were C9H9O3+(165), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(2-hydroxy- 4-propoxyphenyl)propenone (3d)
It was obtained as orange yellow solid in 81% yield; m.p.: 121–124°C, Rf: 0.45 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,709 (O-H stretch), 2,949 (aliphatic C-H), 1,641 (C=O), 1,569 (C=C stretch), 1,461 (C-H bending of CH2), and 1,268 (C-O). 1HNMR (400 MHz, CDCl3): δ 1.06 (3H, t, J = 7.2, H-3′), 1.73–1.88 (2H, m, H-2′), 2.95 (3H, s, N-CH3), 3.12 (3H, s, N-CH3), 3.99 (2H, t, J = 6.8, H-1′), 5.69 (1H, d, J = 9.6, H-2), 6.35–6.39 (2H, m, H-5″ and H-3″), 7.62 (1H, d, J = 8.8 H-6″), and 7.84 (1H, d, J = 9.6, H-3).13C NMR (100.6 MHz, CDCl3): 190.81 (C-1), 165.53 (C-4″), 164.06 (C-2″), 154.05 (C-3), 129.81 (C-6″), 113.62 (C-1″), 106.65 (C-5″), 101.53 (C-3″), 89.75 (C-2), 68.18 (C-1′), 31.07 (C-2′), and 15.60 (C-3′). HR-MS for C14H19NO3 [M + H]+m/z: Calcd 250.1437; Observed 250.1424. The major mass fragments observed were C10H11O3+(179), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(2-hydroxy-4-isopropoxyphenyl)propenone (3e)
It was obtained as orange yellow solid in 77% yield; m.p.: 143–145°C, Rf: 0.48 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,740 (O-H stretch), 2,954 (aliphatic C-H), 1,647 (C=O), 1,563 (C=C stretch), 1,458 (C-H bending of CH2), and 1,263 (C-O). 1HNMR (400 MHz, CDCl3): δ 1.30 (1H, d, J = 4.8 Hz, CH3), 4.63–4.65 (1H, m, H-1′), 2.95 (3H, s, N-CH3), 3.12 (3H, s, N-CH3), 5.69 (1H, d, J = 12.4, H-2), 6.31–6.37 (2H, m, H-5″ and H-3″), 7.61 (1H, d, J = 8.8 H-6″), and 7.82 (1H, d, J = 12.4, H-3).13C NMR (100.6 MHz, CDCl3): 190.90 (C-1), 164.93 (C-4″), 164.08 (C-2″), 154.34 (C-3), 130.09 (C-6″), 114.31 (C-1″), 106.91 (C-5″), 101.75 (C-3″), 89.87 (C-2), 69.22 (C-1′), and 22.65 (iCH3). HR-MS for C14H19NO3 [M + H]+m/z: Calcd 250.1437; Observed 250.1424. The major mass fragments observed were C10H11O3+(179), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(4-Butoxy-2-hydroxyphenyl)propenone (3f)
It was obtained as lemon yellow solid in 88% yield; m.p.: 103–105°C Rf: 0.51 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,716 (O-H stretch), 2,955 (aliphatic C-H), 1,638 (C=O), 1,566 (C=C stretch), 1,465 (C-H bending of CH2), and 1,256 (C-O). 1HNMR (400 MHz, CDCl3): δ 0.96 (3H, t, J = 7.2, H-4′), 1.43–1.78 (2H, m, H-2′ and H-3′), 2.95 (3H, s, N-CH3), 3.16 (3H, s, N-CH3), 3.98 (2H, t, J = 6.8, H-1′), 5.63 (1H, d, J = 12, H-2), 6.32–6.38 (2H, m, H-5″ and H-3″), 7.55 (1H, d, J = 8.8 H-6″), and 7.83 (1H, d, J = 12.4, H-3).13C NMR (100.6 MHz, CDCl3): 190.79 (C-1), 165.49 (C-4″), 164.05 (C-2″), 154.01 (C-3), 129.76 (C-6″), 113.77 (C-1″), 106.80 (C-5″), 101.54 (C-3″), 89.85 (C-2), 68.13 (C-1′), 31.52 (C-2′), 25.70 (C-3′), and 14.29 (C-4′). HR-MS for C15H21NO3 [M + H]+m/z: Calcd 264.1594; Observed 264.1585. The major mass fragments observed were C11H13O3+(193), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(4-Pentyloxy-2-hydroxyphenyl)propenone (3g)
It was obtained as lemon yellow solid in 78% yield; m.p.: 98–100°C, Rf: 0.53 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,707 (O-H stretch), 2,943 (aliphatic C-H), 1,642 (C=O), 1,576 (C=C stretch), 1,468 (C-H bending of CH2), and 1,267 (C-O). 1HNMR (400 MHz, CDCl3): δ 0.91 (3H, t, J = 6.8, H-5′), 1.31–1.79 (6H, m, H-2′,H-3′and H-4′), 2.95 (3H, s, N-CH3), 3.15 (3H, s, N-CH3), 3.96 (2H, t, J = 6.8, H-1′), 5.66 (1H, d, J = 12.4, H-2), 6.34–6.40 (2H, m, H-5″ and H-3″), 7.59 (1H, d, J = 8.8 H-6″), and 7.84 (1H, d, J = 12.4, H-3). 13C NMR (100.6 MHz, CDCl3): 190.83 (C-1), 165.54 (C-4″), 164.01 (C-2″), 154.03 (C-3), 129.71 (C-6″), 113.70 (C-1″), 106.82 (C-5″), 101.51 (C-3″), 89.83 (C-2), 68.17 (C-1′), 31.50 (C-2′), 28.93 (C-3′), 25.66 (C-4′), and 14.20 (C-5′). HR-MS for C16H23NO3 [M + H]+m/z: Calcd 278.1750; Observed 278.1765. The major mass fragments observed were C12H15O3+(207), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(4-Hexyloxy-2-hydroxyphenyl)propenone (3h)
It was obtained as pale yellow solid in 75% yield; m.p.: 87–92°C, Rf: 0.60 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,718 (O-H stretch), 2,937 (aliphatic C-H), 1,639 (C=O), 1,569 (C=C stretch), 1,470 (C-H bending of CH2), and 1,264 (C-O). 1HNMR (400 MHz, CDCl3): δ 0.90 (3H, t, J = 6.8, H-6′), 1.30–1.79 (8H, m, H-2′, H-3′, H-4′ and H-5′), 2.95 (3H, s, N-CH3), 3.16 (3H, s, N-CH3), 3.98 (2H, t, J = 6.8, H-1′), 5.69 (1H, d, J = 12, H-2), 6.35–6.39 (2H, m, H-5″ and H-3″), 7.60 (1H, d, J = 8.8 H-6″), and 7.84 (1H, d, J = 12.4, H-3).13C NMR (100.6 MHz, CDCl3): 190.80 (C-1), 165.58 (C-4″), 164.03 (C-2″), 154.05 (C-3), 129.74 (C-6″), 113.73 (C-1″), 106.86 (C-5″), 101.53 (C-3″), 89.87 (C-2), 68.19 (C-1′), 31.57 (C-2′), 28.95 (C-3′), 25.67 (C-4′), 22.65 (C-5′), and 14.19 (C-6′). HR-MS for C17H25NO3 [M + H]+m/z: Calcd 292.1907; Observed 292.1912. The major mass fragments observed were C13H17O3+(221), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(4-Heptyloxy-2-hydroxyphenyl)propenone (3i)
It was obtained as pale yellow solid in 80% yield; m.p.: 90–92°C, Rf: 0.50 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,709 (O-H stretch), 2,941 (aliphatic C-H), 1,631 (C=O), 1,572 (C=C stretch), 1,472 (C-H bending of CH2), and 1,266 (C-O). 1HNMR (400 MHz, CDCl3): δ 0.88 (3H, t, J = 6.8, H-7′), 1.23–1.78 (10H, m, H-2′, H-3′, H-4′, H-5′and H-6′), 2.92 (3H, s, N-CH3), 3.14 (3H, s, N-CH3), 3.95 (2H, t, J = 6.8, H-1′), 5.66 (1H, d, J = 12, H-2), 6.35–6.39 (2H, m, H-5″ and H-3″), 7.55 (1H, d, J = 8.8 H-6″), and 7.82 (1H, d, J = 12.4, H-3). 13C NMR (100.6 MHz, CDCl3): 190.81 (C-1), 165.59 (C-4″), 164.05 (C-2″), 154.07 (C-3), 129.72 (C-6″), 113.74 (C-1″), 106.89 (C-5″), 101.54 (C-3″), 89.89 (C-2), 68.16 (C-1′), 31.57–22.66 (C-2′, C-3′, C-4′, C-5′, C-6′), and 14.20 (C-7′). HR-MS for C18H27NO3 [M + H]+m/z: Calcd 306.2063; Observed 306.2059. The major mass fragments observed were C14H19O3+(235), C7H5O3+(137), C5H8O+(98), and C4H10N+(72).
3-Dimethylamino-1-(2-hydroxy-4-octyloxyphenyl)propenone (3j)
It was obtained as yellow solid in 76% yield; m.p.: 70–72°C, Rf: 0.45 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,718 (O-H stretch), 2,936 (aliphatic C-H), 1,641 (C=O), 1,579 (C=C stretch), 1,475 (C-H bending of CH2), and 1,262 (C-O). 1HNMR (400 MHz, CDCl3): δ 0.90 (3H, t, J = 6.8, H-8′), 1.24–1.78 (12H, m, H-2′, H-3′, H-4′, H-5′, H-6′, and H-7′), 2.92 (3H, s, N-CH3), 3.13 (3H, s, N-CH3), 3.94 (2H, t, J = 6.8, H-1′), 5.67 (1H, d, J = 12, H-3), 6.35–6.39 (2H, m, H-5″ and H-3″), 7.58 (1H, d, J = 8.8 H-6″), and 7.82 (1H, d, J = 12.4, H-2).13C NMR (100.6 MHz, CDCl3): 190.80 (C-1), 165.57 (C-4″), 164.07 (C-2″), 154.04 (C-3), 129.74 (C-6″), 113.72 (C-1″), 106.90 (C-5″), 101.56 (C-3″), 89.90 (C-2), 68.15 (C-1′), 31.86–22.65 (C-2′, C-3′, C-4′, C-5′, C-6′, C-7′), and 14.46 (C-8′). HR-MS for C19H29NO3 [M + H]+m/z: Calcd 320.2220; Observed 320.2214. The major mass fragments observed were C15H21O3+(249), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(2-hydroxy-4-nonyloxyphenyl)propenone (3k)
It was obtained as yellow solid in 86% yield; m.p.: 78–80°C, Rf: 0.49 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,711 (O-H stretch), 2,945 (aliphatic C-H), 1,642 (C=O), 1,593 (C=C stretch), 1,478 (C-H bending of CH2), and 1,265 (C-O).1HNMR (400 MHz, CDCl3): δ 0.86 (3H, t, J = 6.8, H-9′), 1.26–1.78 (12H, m, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′, and H-8′), 2.92 (3H, s, N-CH3), 3.13 (3H, s, N-CH3), 3.98 (2H, t, J = 6.8, H-1′), 5.64 (1H, d, J = 12, H-2), 6.35–6.39 (2H, m, H-5″ and H-3″), 7.59 (1H, d, J = 8.8 H-6″), and 7.87 (1H, d, J = 12.4, H-3). 13C NMR (100.6 MHz, CDCl3): 190.77 (C-1), 165.56 (C-4″), 164.03 (C-2″), 154.01 (C-3), 129.75 (C-6″), 113.69 (C-1″), 106.91 (C-5″), 101.55 (C-3″), 89.86 (C-2), 68.14 (C-1′), 31.88–22.63 (C-2′, C-3′, C-4′, C-5′, C-6′, C-7′, C-8′), and 14.45 (C-9′). HR-MS for C20H31NO3 [M + H]+m/z: Calcd 334.2376; Observed 334.2382. The major mass fragments observed were C16H23O3+(263), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino 1-(4-decyloxy-2-hydroxyphenyl)propenone (3l)
It was obtained as yellow solid in 83% yield; m.p.:76–79°C, Rf: 0.54 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,708 (O-H stretch), 2,955 (aliphatic C-H), 1,629 (C=O), 1,586 (C=C stretch), 1,498 (C-H bending of CH2), and 1,272 (C-O). 1HNMR (400 MHz, CDCl3): δ 0.86 (3H, t, J = 6.8, H-10′), 1.25–1.78 (16H, m, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′, H-8′, and H-9′), 2.92 (3H, s, N-CH3), 3.13 (3H, s, N-CH3), 3.94 (2H, t, J = 6.8 H-1′) 5.68 (1H, d, J = 12, H-2), 6.33–6.37 (2H, m, H-5″ and H-3″) 7.56 (1H, d, J = 8.8 H-6″), and 7.83 (1H, d, J = 12.4, H-3).13C NMR (100.6 MHz, CDCl3): 190.81 (C-1), 165.59 (C-4″), 164.05 (C-2″), 154.03 (C-3), 129.73 (C-6″), 113.74 (C-1″), 106.93 (C-5″), 101.58 (C-3″), 89.89 (C-2), 68.18 (C-1′), 31.97–22.76 (C-3′, C4′, C5′, C-6′, C-7′, C-8′, C-9′), and 14.19 (C-10′). HR-MS for C21H33NO3 [M + H]+m/z: Calcd 348.2533; Observed 348.2550. The major mass fragments observed were C17H25O3+(277), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino 1-(4-dodecyloxy-2-hydroxyphenyl)propenone (3m)
It was obtained as yellow solid in 81% yield; m.p.: 86–88°C, Rf: 0.51 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,757 (O-H stretch), 2,945 (aliphatic C-H), 1,634 (C=O), 1,539 (C=C stretch), 1,495 (C-H bending of CH2), and 1,278 (C-O). 1HNMR (400 MHz, CDCl3): δ 0.87 (3H, t, J = 6.8, H-12′), 1.26–1.77 (20H, m, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′, H-8′, H-9′, H-10′, and H-11′), 2.93 (3H, s, N-CH3), 3.15 (3H, s, N-CH3), 3.94 (2H, t, J = 6.8, H-1′), 5.69 (1H, d, J = 12, H-2), 6.33–6.37 (2H, m, H-5″ and H-3″) 7.58 (1H, d, J = 7.2, H-6″), and 7.84 (1H, d, J = 12.4, H-3). 13C NMR (100.6 MHz, CDCl3): 190.67 (C-1), 165.59 (C-4″), 164.04 (C-2″), 154.03 (C-3), 129.74 (C-6″), 113.73 (C-1″), 106.93 (C-5″), 101.57 (C-3″), 89.89 (C-2), 68.18 (C-1′), 31.94–22.73 (C-3′, C-4′, C-5′, C-6′, C-7′, C-8′, C-9′, C-10′, C-11′), and 14.20 (C-12′). HR-MS for C23H37NO3 [M + H]+m/z: Calcd 376.2846; Observed 376.2858. The major mass fragments observed were C19H29O3+(305), C7H5O3+(137), C5H8O+(98), and C4H10N+(72).
3-Dimethylamino 1-(4-tridecyloxy-2-hydroxyphenyl)propenone (3n)
It was obtained as yellow solid in 72% yield; m.p.: 87–90°C, Rf: 0.49 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,770 (O-H stretch), 2,941 (aliphatic C-H), 1,630 (C=O), 1,545 (C=C stretch), 1,497 (C-H bending of CH2), and 1,287 (C-O). 1HNMR (400 MHz, CDCl3): δ 0.86 (3H, t, J = 6.8, H-13′), 1.24–1.79 (22H, m, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′, H-8′, H-9′, H-10′, H-11′, and H-12′), 2.92 (3H, s, N-CH3), 3.14 (3H, s, N-CH3), 3.98 (2H, t, J = 6.8, H-1′), 5.66 (1H, d, J = 12, H-2), 6.32–6.35 (2H, m, H-5″ and H-3″), 7.56 (1H, d, J = 8.8, H-6″), and 7.80 (1H, d, J = 12.4, H-3). 13C NMR (100.6 MHz, CDCl3): 190.69 (C-1), 165.60 (C-4″), 164.05 (C-2″), 154.02 (C-3), 129.73 (C-6″), 113.74 (C-1″), 106.94 (C-5″), 101.58 (C-3″), 89.90 (C-2), 68.18 (C-1′), 32–22.77 (C-3′, C4′, C5′, C-6′, C-7′, C-8′, C-9′, C-10′, C-11′, C-12′), and 14.19 (C-13′). HR-MS for C24H39NO3 [M + H]+m/z: Calcd 390.3002; Observed 390.3010. The major mass fragments observed were C20H31O3+(319), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino 1-(4-tetradecyloxy-2-hydroxyphenyl)propenone (3o)
It was obtained as yellow solid in 78% yield; m.p.: 76–81°C, Rf: 0.50 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,763 (O-H stretch), 2,934 (aliphatic C-H), 1,645 (C=O), 1,536 (C=C stretch), 1,489 (C-H bending of CH2), and 1,278 (C-O). 1HNMR (400 MHz, CDCl3): δ 0.87 (3H, t, J = 6.8, H-14′), 1.26–1.80 (24H, m, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′, H-8′, H-9′, H-10′, H-11′ H-12′, and H-13′), 2.92 (3H, s, N-CH3), 3.19 (3H, s, N-CH3), 3.99 (2H, t, J = 6.8, H-1′), 5.69 (1H, d, J = 12, H-2), 6.31–6.36 (2H, m, H-5″ and H-3″), 7.55 (1H, d, J = 8.8, H-6″), and 7.83 (1H, d, J = 12.4, H-3). 13C NMR (100.6 MHz, CDCl3): 190.71 (C-1), 165.57 (C-4″), 164.03 (C-2″), 154.08 (C-3), 129.71 (C-6″), 113.70 (C-1″), 106.92 (C-5″), 101.55 (C-3″), 89.93 (C-2), 68.15 (C-1′), 31.97–22.71 (C-3′, C4′, C5′, C-6′, C-7′, C-8′, C-9′, C-10′, C-11′, C-12′, C-13′), and 14.16 (C-14′). HR-MS for C25H41NO3 [M + H]+m/z: Calcd 404.3159; Observed 404.3143. The major mass fragments observed were C21H33O3+(333), C7H5O3+(137), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino 1-(5-bromo-2-hydroxyphenyl)propenone (3p)
It was obtained as bright yellow solid in 87% yield; m.p.: 128–132°C, Rf: 0.50 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,735 (O-H stretch), 2,923 (aliphatic C-H), 1,638 (C=O), 1,548 (C=C stretch), 1,437 (C-H bending of CH2), and 1,258 (C-O). 1HNMR (400 MHz, CDCl3): δ 2.95 (3H, s, N-CH3), 3.18 (3H, s, N-CH3), 5.68 (1H, d, J = 9.6, H-2), 7.06 (1H, d, J = 7.2 Hz, H-3″) 7.43–7.47 (1H, m, H-4″), 7.78 (1H, d, J = 2, H-6″), and 7.90 (1H, d, J = 9.6, H-3). 13C NMR (100.6 MHz, CDCl3): 190.75 (C-1), 164.10 (C-2′), 154.06 (C-3), 134.44 (C-4′), 129.56 (C-6′), 124.63 (C-1′), 123.20 (C-3′), 118.73 (C-5′), and 89.93 (C-2). HR-MS for C11H11BrNO2 [M + H]+m/z: Calcd 269.0045; Observed 269.0029. The major mass fragments observed were C7H3BrO2+(197), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(5-chloro-2-hydroxyphenyl)propenone (3q)
It was obtained as bright yellow solid in 78% yield; m.p.: 125–128°C, Rf: 0.53 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,739 (O-H stretch), 2,925 (aliphatic C-H), 1,644 (C=O), 1,555 (C=C stretch), 1,429 (C-H bending of CH2), and 1,258 (C-O). 1HNMR (400 MHz, CDCl3): δ 2.93 (3H, s, N-CH3), 3.19 (3H, s, N-CH3), 5.69 (1H, d, J = 9.6, H-2), 6.81 (1H, d, J = 7.2 Hz, H-3″), 7.01–7.05 (1H, m, H-4″), 7.69 (1H, d, J = 2, H-6″), and 7.84 (1H, d, J = 9.6, H-3). 13C NMR (100.6 MHz, CDCl3): 190.63 (C-1), 158.67 (C-2), 154.19 (C-2″), 134.45 (C-4″), 131.26 (C-6″), 130.55 (C-5″), 121.10 (C-1″), 119.25 (C-3″), and 86.91 (C-3). HR-MS for C12H13ClNO2 [M + H]+m/z: Calcd 239.0707; Observed 239.0712. The major mass fragments observed were C7H3ClO2+(153), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino 1-(3,5-dichloro-2-hydroxyphenyl)propenone (3r)
It was obtained as bright yellow solid in 82% yield; m.p.: 137–141°C, Rf: 0.48 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,730 (O-H stretch), 2,918 (aliphatic C-H), 1,628 (C=O), 1,539 (C=C stretch), 1,422 (C-H bending of CH2), and 1,266 (C-O). 1HNMR (400 MHz, CDCl3): δ 2.99 (3H, s, N-CH3), 3.21 (3H, s, N-CH3), 5.64 (1H, d, J = 9.6, H-2), 7.41 (1H, d, J = 2, H-4″) 7.53 (1H, d, J = 2, H-6″), and 7.92 (1H, d, J = 9.6, H-3). 13C NMR (100.6 MHz, CDCl3): 190.66 (C-1), 164.10 (C-2″), 154.06 (C-3), 134.44 (C-4″), 130.60 (C-6″), 126.63 (C-5″), 122.20 (C-1″), 119.67 (C-3″), and 89.93 (C-2). HR-MS for C11H11Cl2NO2 [M + H]+m/z: Calcd 260.0239; Observed 260.0224. The major mass fragments observed were C7H3Cl2O2+(188), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(2-hydroxy-5-methylphenyl)propenone (3s)
It was obtained as bright yellow solid in 84% yield; m.p.: 105–109°C, Rf: 0.52 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,742 (O-H stretch), 2,922 (aliphatic C-H), 1,639 (C=O), 1,550 (C=C stretch), 1,431 (C-H bending of CH2), and 1,253 (C-O). 1HNMR (400 MHz, CDCl3): δ 2.97 (3H, s, N-CH3), 3.18 (3H, s, N-CH3), 5.61 (1H, d, J = 9.6, H-2), 6.27(1H, d, J = 7.2 Hz, H-3″) 7.01–7.05 (1H, m, H-4″), 7.81 (1H, d, J = 2, H-6″), and 7.89 (1H, d, J = 9.6, H-3). 13C NMR (100.6 MHz, CDCl3): 190.75 (C-1), 164.10 (C-2″), 154.06 (C-3), 135.20 (C-4″), 130.46 (C-6″), 129.54 (C-5″), 124.10 (C-1″), 119.12 (C-3″), and 89.93 (C-2). HR-MS for C12H15NO2 [M + H]+m/z: Calcd 206.1175; Observed 206.1169. The major mass fragments observed were C8H7O2+(133), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(5-fluoro-2-hydroxyphenyl)propenone (3t)
It was obtained as pale yellow solid in 88% yield; m.p.: 140–145°C, Rf: 0.45 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,740 (O-H stretch), 2,928 (aliphatic C-H), 1,641 (C=O), 1,557 (C=C stretch), 1,435 (C-H bending of CH2), and 1,258 (C-O). 1HNMR (400 MHz, CDCl3): δ 2.96 (3H, s, N-CH3), 3.15 (3H, s, N-CH3), 5.57 (1H, d, J = 9.6, H-2), 6.29 (1H, d, J = 7.2 Hz, H-3″) 7.03–7.07 (1H, m, H-4″), 7.82 (1H, d, J = 2, H-6″), and 7.87 (1H, d, J = 9.6, H-3). 13C NMR (100.6 MHz, CDCl3): 190.63 (C-1), 161.047 (C-5″), 156.13 (C-2″), 154.32 (C-2), 127.05 (C-1″), 121.10 (C-4″), 117.23 (C-6″), 115.04 (C-3′), and 89.83 (C-2). HR-MS for C11H12FNO2 [M + H]+m/z: Calcd 210.0924; Observed 210.0920. The major mass fragments observed were C21H33O3+(333), C7H4FO2+(139), C6H4FO+(111), C5H8NO+(98), and C4H10N+(72).
3-Dimethylamino-1-(5-chloro-2-hydroxy-4-methyl-phenyl)propenone (3u)
It was obtained as bright yellow solid in 80% yield; m.p.: 131–135°C, Rf: 0.49 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,742 (O-H stretch), 2,922 (aliphatic C-H), 1,639 (C=O), 1,550 (C=C stretch), 1,431 (C-H bending of CH2), and 1,253 (C-O).1HNMR (400 MHz, CDCl3): δ 2.41 (3H, CH3), 2.96 (3H, s, N-CH3), 3.10 (3H, s, N-CH3), 5.65 (1H, d, J = 9.6, H-2), 6.38 (1H, d, J = 7.2 Hz, H-3″),7.71 (1H, d, J = 2, H-6″), and 7.87 (1H, d, J = 9.6, H-3). 13C NMR (100.6 MHz, CDCl3): 190.71 (C-1), 158.67 (C-2), 155.15 (C-2″), 144.33 (C-4″), 131.53 (C-6″), 129.31 (C-5″), 122.10 (C-1′), 117.32 (C-3″), and 86.87 (C-3). HR-MS for C12H14ClNO2 [M + H]+m/z: Calcd 240.0785; Observed 240.0779. The major mass fragments observed were C8H7ClO2+(169), C5H8NO+(98), and C4H10N+(72).
General Method for the Synthesis of 3-Iodochromones (4a–4u)
The chromones were synthesized by taking substituted 3-(dimethylamino)-1-(2-hydroxyphenyl) propenone (0.54 mmol) in CHCl3 (15 ml) and iodine (1.09 mmol, 2 mol equiv), followed by stirring the mixture at 25°C for 8 h (Scheme 1) (Gammill, 1979). The formation of products was confirmed by thin-layer chromatography (TLC) in ethyl acetate:hexane (3:7) solvent system. The solution was washed with saturated Na2S2O3 (15 ml), and the aqueous layer was extracted with CHCl3 (20 ml). The solvent was removed using Heidolph rotary evaporator Hei-VAP. The synthesized chromones were purified by column chromatography to afford the pure chromones in 67–89% yield. Compounds 4f, 4g, 4h, 4i, 4j, 4k, 4l, 4m, 4n, and 4o are reported for the first time in the literature.
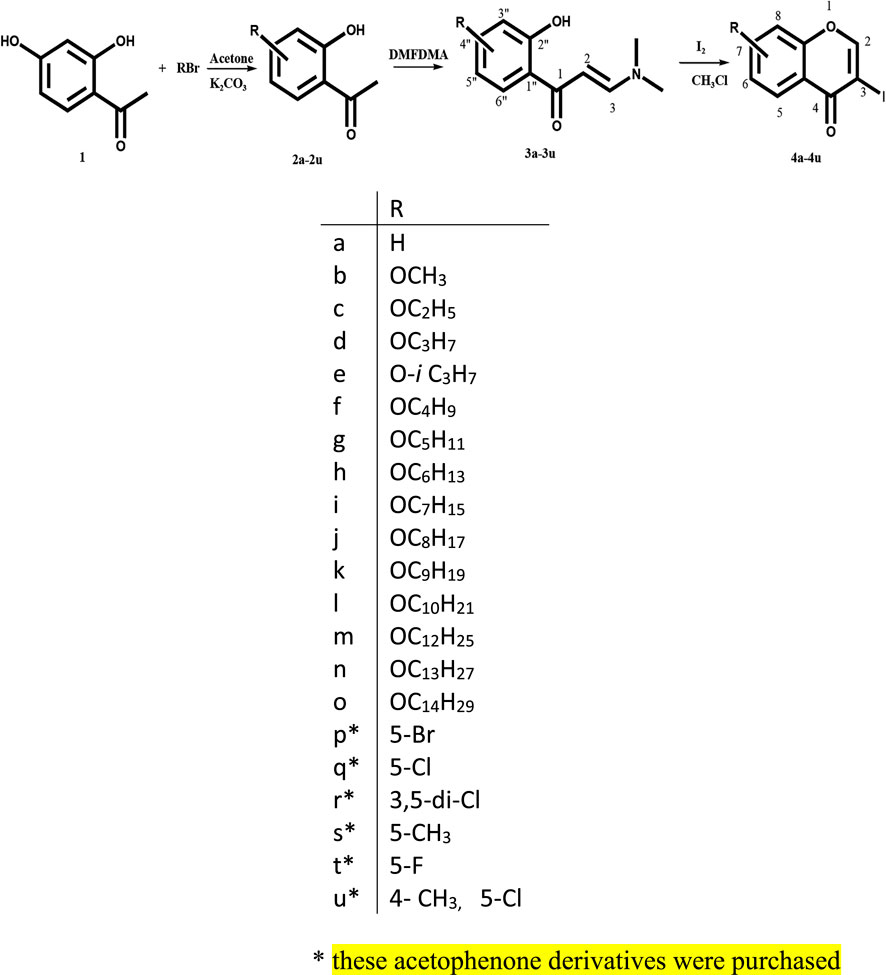
SCHEME 1. Synthesis of 3-iodochromones (Gammill, 1979).
3-Iodochromone (4a)
It was obtained as a white solid in 79% yield; m.p.: 102–103°C; Rf: 0.49 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,071 (aromatic C-H stretch), 2,932 (aliphatic C-H stretch), 1,646 (C=O stretch), 1,617(aromatic C=C stretch), 1,561 (pyrone ring C=C stretch), 1,426 (C-H bending of CH2), and 1,260 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 6.83 (1H, d, J = 2.4 Hz, H-8), 6.93–6.98 (1H, m, H-6, H-7), 8.08 (1H, d, J = 8.8 Hz, H-5), and 8.17 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 175.44 (C-4), 158.93 (C-2), 155.99 (C-9), 134.11 (C-7), 126.32 (C-5), 123.70 (C-10), 118.30 (C-6), 101.33 (C-8), and 94.78 (C-1). HR-MS for C9H5IO2 [M + H]+m/z: Calcd 272.9407; Observed 272.9406. The major mass fragment observed was C9H6IO3+ (288).
3-Iodo-7-methoxychromone (4b)
It was obtained as white solid in 89% yield; m.p.: 140–145°C, Rf: 0.51 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,068 (aromatic C-H stretch), 2,934 (aliphatic C-H stretch), 1,642 (C=O stretch), 1,612 (aromatic C=C stretch), 1,559 (pyrone ring C=C stretch), 1,434 (C-H bending of CH2), and 1,258 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 3.87 (3H, s, OCH3), 6.80 (1H, d, J = 2.4 Hz, H-8), 6.95–6.97 (1H, m, H-6), 8.10 (1H, d, J = 8.8 Hz, H-5), and 8.18 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 174.72 (C-4), 164.36 (C-7), 158.03 (C-2), 157.27 (C-9), 127.71 (C-5), 117.53 (C-6), 115.11 (C-10), 100.51 (C-8), and 55.99 (OCH3). HR-MS for C10H7IO3 [M + H]+m/z: Calcd 302.9512; Observed 302.9518. The major mass fragment observed was C9H6IO3+ (288).
Ethoxy-3-Iodochromone (4c)
It was obtained as white solid in 82% yield; m.p.: 100–105°C, Rf: 0.47 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,065 (aromatic C-H stretch), 2,946 (aliphatic C-H stretch), 1,640 (C=O stretch), 1,617 (aromatic C=C stretch), 1,565 (pyrone ring C=C stretch), 1,436 (C-H bending of CH2), and 1,266 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 1.42 (3H, t, J = 6.8 Hz, H-2′), 4.04 (2H, q, J = 6.8 Hz, H-1′), 6.29 (1H, d, J = 2.4 Hz, H-8), 6.34–6.36 (1H, m, H-6), 7.82 (1H, d, J = 8.8 Hz, H-5), and 7.93(1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 173.13 (C-4), 165.53 (C-7), 163.56 (C-2), 162.82 (C-9), 130.79 (C-5), 113.84 (C-10), 106.64 (C-6), 101.43 (C-8), 89.49 (C-3), 63.79 (C-1′), and 15.00 (C-2′). HR-MS for C11H9IO3 [M + H]+m/z: Calcd 316.9669; Observed 316.9652. The major mass fragment observed was C9H6IO3+(288).
3-Iodo-7-propoxychromone (4d)
It was obtained as yellowish white solid in 79% yield; m.p.: 88–91°C, Rf: 0.46 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,071 (aromatic C-H stretch), 2,944 (aliphatic C-H stretch), 1,642 (C=O stretch), 1,614 (aromatic C=C stretch), 1,559 (pyrone ring C=C stretch), 1,443 (C-H bending of CH2), and 1,256 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 1.06 (3H, t, J = 7.2, H-3′), 1.73–1.88 (2H, m, H-2′), 3.99 (2H, t, J = 6.4 Hz, H-1′), 6.79 (1H, d, J = 2.4 Hz, H-8), 6.95–6.98 (1H, m, H-6), 8.11 (1H, d, J = 9.2 Hz, H-5), and 8.18 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.56 (C-4), 164.41 (C-7), 163.56 (C-2), 162.78 (C-9), 131.48 (C-5), 113.55 (C-10), 106.37 (C-6), 101.25 (C-8), 89.51 (C-3), 63.71 (C-1′), 31.42 (C-2′), and 14.98 (C-3′). HR-MS for C12H11IO3 [M + H]+m/z: Calcd 330.9825; Observed 330.9820. The major mass fragment observed was C9H6IO3+(288).
3-Iodo-7-isopropoxychromone (4e)
It was obtained as yellowish white solid in 76% yield; m.p.: 67–70°C, Rf: 0.50 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,061 (aromatic C-H stretch), 2,956 (aliphatic C-H stretch), 1,639 (C=O stretch), 1,612 (aromatic C=C stretch), 1,566 (pyrone ring C=C stretch), 1,453 (C-H bending of CH2), 1,265 and (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 1.31 (1H, d, J = 4.8 Hz, CH3), 4.66–4.68 (1H, m, H-1′), 6.83 (1H, d, J = 2 Hz, H-8), 6.96–6.98 (1H, m, H-6), 8.38 (1H, s, H-2), 8.15 (1H, d, J = 7.2 Hz, H-5), and 8.22 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.61 (C-4), 163.82 (C-7), 157.99 (C-9), 157.32 (C-2), 127.97 (C-5), 116.32 (C-6), 115.27 (C-10), 101.43 (C-8), 87.10 (C-3), 71.02 (C-1′), and 21.84 (isopropyl CH3). HR-MS for C12H11IO3 [M + H]+m/z: Calcd 330.9825; Observed 330.9817. The major mass fragment observed was C9H6IO3+(288).
7-Butoxy-3-iodochromone (4f)
It was obtained as white solid in 81% yield; m.p.: 86–88°C, Rf: 0.57 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,059 (aromatic C-H stretch), 2,945 (aliphatic C-H stretch), 1,643 (C=O stretch), 1,607 (aromatic C=C stretch), 1,575 (pyrone ring C=C stretch), 1,493 (C-H bending of CH2), and 1,247 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 0.99 (3H, t, J = 6, H-4′), 1.47–1.81 (4H, m, H-2′ and 3′), 4.04 (2H, t, J = 5.2 Hz, H-1′), 6.80 (1H, d, J = 2 Hz, H-8), 6.95–6.98 (1H, m, H-6), 8.02 (1H, d, J = 7.2 Hz, H-5), and 8.19 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.42 (C-4), 163.82 (C-7), 158.99 (C-2), 157.97 (C-9), 127.48 (C-5), 116.07 (C-6), 115.30 (C-10), 101.33 (C-8), 87.53 (C-3), 68.78 (C-1′), 30.95 (C-2′), 19.18 (C-3′), and 14.16 (C-4′). HR-MS for C13H13IO3 [M + H]+m/z: Calcd 344.9982; Observed 344.9974. The major mass fragment observed was C9H6IO3+(288).
3-iodo-7-pentyloxychromone (4g)
It was obtained as yellowish white solid in 86% yield; m.p.: 77–80°C, Rf: 0.52 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,059 (aromatic C-H stretch), 2,922 (aliphatic C-H stretch), 1,637 (C=O stretch), 1,603 (aromatic C=C stretch), 1,570 (pyrone ring C=C stretch), 1,471 (C-H bending of CH2), and 1,253 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 0.93 (3H, t, J = 6.8, H-5′), 1.36–1.83 (6H, m, H-2′, H-3′, and H-4′), 4.02 (2H, t, J = 6.4 Hz, H-1′), 6.79 (1H, d, J = 2.4 Hz, H-8), 6.95–6.97 (1H, m, H-6), 8.11 (1H, d, J = 8.8 Hz, H-5), and 8.18 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.70 (C-4), 164.05 (C-7), 158.04 (C-9), 157.26 (C-2), 128.05 (C-5), 115.71 (C-6), 115.59 (C-10), 100.54 (C-8), 87.20 (C-3), 68.93 (C-1′), 31.55 (C-2′), 28.93 (C-3′), 25.67 (C-4′), and 14.19 (C-5′). HR-MS for C14H15IO3 [M + H]+m/z: Calcd 359.0138; Observed 359.0152. The major mass fragment observed was C9H6IO3+(288).
7-Hexyloxy-3-iodochromone (4h)
It was obtained as white solid in 81% yield; m.p.: 88–90°C, Rf: 0.48 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,063 (aromatic C-H stretch), 2,919 (aliphatic C-H stretch), 1,635 (C=O stretch), 1,623 (aromatic C=C stretch), 1,571 (pyrone ring C=C stretch), 1,478 (C-H bending of CH2), and 1,257 (C-O Stretch). 1H NMR (400 MHz, CDCl3): 1.42 (3H, t, J = 6.8 Hz), 4.06 (2H, q, J = 6.8 Hz), 6.74 (1H, s), 6.90–6.93 (1H, m), 8.05 (1H, d, J = 9.2 Hz), and 8.17 (1H, s). 13C NMR (100.6 MHz, CDCl3): 172.70 (C-4), 163.98 (C-7), 158.03 (C-9), 157.27 (C-2), 128.04 (C-5), 115.73 (C-6), 115.56 (C-10), 100.53 (C-8), 87.20 (C-3), 68.92 (C-1′), 31.57 (C-2′), 28.96 (C-3′), 25.69 (C-4′), 22.65 (C-5′), and 14.20 (C-6′). HR-MS for C15H17IO3 [M + H]+m/z: Calcd 373.0295; Observed 373.0241. The major mass fragment observed was C9H6IO3+(288).
7-Heptyloxy-3-iodochromone (4i)
It was obtained as white solid in 79% yield; m.p.: 73–76°C, Rf: 0.56 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,083 (aromatic C-H stretch), 2,921 (aliphatic C-H stretch), 1,645 (C=O stretch), 1,612 (aromatic C=C stretch), 1,577 (pyrone ring C=C stretch), 1,463 (C-H bending of CH2), and 1,259 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 0.89 (3H, t, J = 6.8, H-7′), 1.27–1.82 (10H, m, H-2′, H-3′, H-4′, H-5′, H-6′), 4.02 (2H, t, J = 6.8 Hz, H-1′), 6.79 (1H, d, J = 2.4 Hz, H-8), 6.95–6.98 (1H, m, H-6), 8.12 (1H, d, J = 8.8 Hz, H-5),and 8.18 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.70 (C-4), 163.99 (C-7), 158.05 (C-9), 157.27 (C-2), 128.05 (C-5), 115.72 (C-6), 115.59 (C-10), 100.54 (C-8), 87.20 (C-3), 68.93 (C-1′), 31.57–22.67 (C-2′, C-3′, C-4′, C-5′,C-6′), and 14.19 (C-7′). HR-MS for C16H19IO3 [M + H]+m/z: Calcd 387.0451; Observed 387.0446. The major mass fragment observed was C9H6IO3+(288).
3-Iodo-7-Octyloxychromone (4j)
It was obtained as yellowish white solid in 76% yield; m.p.: 73–74°C, Rf: 0.49 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,045 (aromatic C-H stretch), 2,923 (aliphatic C-H stretch), 1,642 (C=O stretch), 1,615 (aromatic C=C stretch), 1,593 (pyrone ring C=C stretch), 1,468 (C-H bending of CH2), and 1,266 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 0.87 (3H, t, J = 6.8, H-8′), 1.24–1.84 (12H, m, H-2′, H-3′, H-4′, H-5′, H-6′, and H-7′), 4.03 (2H, t, J = 6.8 Hz, H-1′), 6.79 (1H, d, J = 2 Hz, H-8), 6.95–6.98 (1H, m, H-6), 8.12 (1H, d, J = 8.8 Hz, H-5), and 8.18 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.46 (C-4), 163.87 (C-7), 159.07 (C-2), 158.05 (C-9), 127.54 (C-5), 116.12 (C-6), 115.36 (C-10), 101.40 (C-8), 87.55 (C-3), 69.06 (C-1′), 31.86–22.66 (C-2′, C-3′, C-4′, C-5′, C-6′, C-7′), and 14.46 (C-8′). HR-MS for C17H21IO3 [M + H]+m/z: Calcd 401.0608; Observed 401.0601. The major mass fragment observed was C9H6IO3+(288).
3-Iodo-7-nonyloxychromone (4k)
It was obtained as white solid in 84% yield; m.p.: 78–80°C, Rf: 0.56 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,080 (aromatic C-H stretch), 2,918 (aliphatic C-H stretch), 1,636 (C=O stretch), 1,617 (aromatic C=C stretch), 1,587 (pyrone ring C=C stretch), 1,460 (C-H bending of CH2), and 1,264 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 0.87 (3H, t, J = 6.8, H-9′), 1.26–1.82 (14H, m, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′, and H-8′), 4.03 (2H, t, J = 6.4 Hz, H-1′), 6.79 (1H, d, J = 2 Hz, H-8), 6.95–6.97 (1H, m, H-6), 8.11 (1H, d, J = 8.8 Hz, H-5), and 8.18 (1H, s, H-2).13C NMR (100.6 MHz, CDCl3): 172.70 (C-4), 164.06 (C-7), 158.03 (C-9), 157.29 (C-2), 128.06 (C-5), 115.73 (C-6), 115.58 (C-10), 100.55 (C-8), 87.20 (C-3), 68.92 (C-1′), 31.94–22.73 (C-3′, C4′, C5′, C-6′, C-7′, C-8′), and 14.19 (C-9′). HR-MS for C18H23IO3 [M + H]+m/z: Calcd 415.0764; Observed 415.0764. The major mass fragment observed was C9H6IO3+(288).
7-Decyloxy-3-iodochromone (4l)
It was obtained as cream solid in 80% yield; m.p.: 68–71°C, Rf: 0.46 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,085 (aromatic C-H stretch), 2,925 (aliphatic C-H stretch), 1,641 (C=O stretch), 1,615 (aromatic C=C stretch), 1,591 (pyrone ring C=C stretch), 1,465 (C-H bending of CH2), and 1,278 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 0.90 (3H, t, J = 6.8, H-10′), 1.30–1.82 (16H, m, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′, H-8′ and H-9′), 4.02 (2H, t, J = 6.8 Hz, H-1′), 6.78 (1H, d, J = 2 Hz, H-8), 6.94–6.97 (1H, m, H-6), 8.10 (1H, d, J = 8.8 Hz, H-5), and 8.17 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.70 (C-4), 164.08 (C-7), 158.05 (C-9), 157.26 (C-2), 128.07 (C-5), 115.72 (C-6), 115.59 (C-10), 100.55 (C-8), 87.20 (C-3), 68.93 (C-1′), 31.96–22.75 (C-3′, C4′, C5′, C-6′, C-7′, C-8′, C-9′), and 14.19 (C-10′). HR-MS for C19H25IO3 [M + H]+m/z: Calcd 429.0921; Observed 429.0946. The major mass fragment observed was C9H6IO3+(288).
7-Dodecyloxy-3-iodochromone (4m)
It was obtained as white solid in 73% yield; m.p.: 58–60°C, Rf: 0.46 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,082 (aromatic C-H stretch), 2,922 (aliphatic C-H stretch), 1,639 (C=O stretch), 1,618 (aromatic C=C stretch), 1,589 (pyrone ring C=C stretch), 1,462 (C-H bending of CH2), and 1,268 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 0.87 (3H, t, J = 6.8, H-12′), 1.26–1.83 (20H, m, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′, H-8′, H-9′, H-10′ and H-11′), 4.02 (2H, t, J = 6.4 Hz, H-1′), 6.79 (1H, d, J = 2.4 Hz, H-8), 6.95–6.97 (1H, m, H-6), 8.11 (1H, d, J = 9.2 Hz, H-5), and 8.18 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.75 (C-4), 163.99 (C-7), 158.03 (C-9), 157.28 (C-2), 128.05 (C-5), 115.73 (C-6), 115.56 (C-10), 100 (C-8), 87.24 (C-3), 68.90 (C-1′), 31.94–22.74 (C-3′, C4′, C5′, C-6′, C-7′, C-8′, C-9′, C-10′, C-11′), and 14.20 (C-12′). HR-MS for C21H29IO3 [M + H]+m/z: Calcd 457.1234; Observed 457.1205. The major mass fragment observed was C9H6IO3+(288).
3-Iodo-7-tridecyloxychromone (4n)
It was obtained as white solid in 83% yield; m.p.: 67–70°C, Rf: 0.46 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,087 (aromatic C-H stretch), 2,920 (aliphatic C-H stretch), 1,643 (C=O stretch), 1,619 (aromatic C=C stretch), 1,599 (pyrone ring C=C stretch), 1,472 (C-H bending of CH2), and 1,273 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 0.91 (3H, t, J = 5.6, H-13′), 1.28–1.87 (22H, m, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′, H-8′, H-9′, H-10′, H-11′, and H-12′), 4.07 (2H, t, J = 5.2 Hz, H-1′),6.84 (1H, d, J = 2 Hz, H-8), 7.00–7.02 (1H, m, H-6), 8.16 (1H, d, J = 7.2 Hz, H-5), and 8.23 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.62 (C-4), 163.97 (C-7), 158.00 (C-9), 157.23 (C-2), 128.00 (C-5), 115.72 (C-6), 115.53 (C-10), 100.51 (C-8), 87.21 (C-3), 68.92 (C-1′), 32.00–22.78 (C-3′, C4′, C5′, C-6′, C-7′, C-8′, C-9′, C-10′, C-11′, C-12′), and 14.22 (C-13′). HR-MS for C22H31IO3 [M + H]+m/z: Calcd 457.1234; Observed 457.1257. The major mass fragment observed was C9H6IO3+(288).
3-Iodo-7-tetradecyloxychromone (4o)
It was obtained as white solid in 80% yield; m.p.: 67–70°C, Rf: 0.43 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,081 (aromatic C-H stretch), 2,928 (aliphatic C-H stretch), 1,647 (C=O stretch), 1,625 (aromatic C=C stretch), 1,588 (pyrone ring C=C stretch), 1,469 (C-H bending of CH2), and 1,277 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 0.90 (3H, t, J = 5.6, H-14′), 1.25–1.84 (24H, m, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′, H-8′, H-9′, H-10′, H-11′, H-12′, and H-13′), 4.05 (2H, t, J = 5.2 Hz, H-1′), 6.85 (1H, d, J = 2 Hz, H-8), 6.95–6.99 (1H, m, H-6), 8.18 (1H, d, J = 7.2 Hz, H-5), and 8.21 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.60 (C-4), 164.93 (C-7), 158.04 (C-9), 157.20 (C-2), 128.04 (C-5), 115.67 (C-6), 115.50 (C-10), 100.53 (C-8), 87.20 (C-3), 68.91 (C-1′), 32.01–22.77 (C-3′, C4′, C5′, C-6′, C-7′, C-8′, C-9′, C-10′, C-11′, C-12′, C-13′), and 14.18 (C-14′). HR-MS for C23H33IO3 [M + H]+m/z: Calcd 485.1547; Observed 485.1533. The major mass fragment observed was C9H6IO3+(288).
7-Bromo-3-iodochromone (4p)
It was obtained as dark brown solid in 79% yield; m.p.: 105–110°C, Rf: 0.54 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,073 (aromatic C-H stretch), 2,919 (aliphatic C-H stretch), 1,636 (C=O stretch), 1,614 (aromatic C=C stretch), 1,542 (pyrone ring C=C stretch), 1,438 (C-H bending of CH2), and 1,283 (C-O Stretch). 1H NMR (400 MHz, CDCl3): 7.41 (1H, d, J = 7.2 Hz, H-8), 7.80–7.82 (1H, m, H-7), 8.32 (1H, s, H-2), and 8.39 (1H, d, J = 2 Hz, H-5). 13C NMR (100.6 MHz, CDCl3): 172.07 (C-4), 158.11 (C-2), 155.44 (C-9), 137.74 (C-8), 132.53 (C-5), 124.43 (C-10), 120.18 (C-8), 118.17 (C-8), and 86.88 (C-3). HR-MS for C9H4BrIO2 [M + H]+m/z: Calcd 350.8512; Observed 350.8529. The major mass fragment observed was C9H5O2+(145).
6-Chloro-3-iodochromone (4q)
It was obtained as bright yellow solid in 85% yield; m.p.: 132–136°C, Rf: 0.49 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,076 (aromatic C-H stretch), 2,922 (aliphatic C-H stretch), 1,639 (C=O stretch), 1,618 (aromatic C=C stretch), 1,531 (pyrone ring C=C stretch), 1,444 (C-H bending of CH2), and 1,255 (C-O Stretch). 1H NMR (400 MHz, CDCl3): 6.93 (1H, d, J = 7.2 Hz, H-8), 7.15–7.27 (1H, m, H-7), 8.32 (1H, s, H-2), and 8.39 (1H, d, J = 2 Hz, H-5). 13C NMR (100.6 MHz, CDCl3): 172.37 (C-4), 159.73 (C-2), 154.51 (C-9), 134.75 (C-7), 130.64 (C-6), 124.63 (C-5), 122.25 (C-10), 121.21 (C-8), and 86.69 (C-3). HR-MS for C9H4ClIO2 [M + H]+m/z: Calcd 306.9170; Observed 306.9163. The major mass fragment observed was C9H5O2+(145).
6, 8-Dichloro-3-iodochromone (4r)
It was obtained as cream solid in 81% yield; m.p.: 143–145°C, Rf: 0.61 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,051 (aromatic C-H stretch), 2,923 (aliphatic C-H stretch), 1,641 (C=O stretch), 1,615 (aromatic C=C stretch), 1,538 (pyrone ring C=C stretch), 1,421(C-H bending of CH2), and 1,274 (C-O Stretch). 1H NMR (400 MHz, CDCl3): δ 7.78 (1H, d, J = 2 Hz, H-5), 8.14 (1H, d, J = 2 Hz, H-7), and 8.38 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 171.83 (C-4), 157.70 (C-2), 150.63 (C-9), 134.41 (C-7), 131.64 (C-6), 124.69 (C-5), 124.43 (C-10), 123.26 (C-8), and 86.91 (C-3). HR-MS for C9H3Cl2IO2 [M + H]+m/z: Calcd 340.8627; Observed 340.8614. The major mass fragment observed was C9H5O2+(145).
3-Iodo-6-methylchromone (4s)
It was obtained as yellow solid in 67% yield; m.p.: 110–116°C, Rf: 0.55 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,075 (aromatic C-H stretch), 2,921 (aliphatic C-H stretch), 1,642 (C=O stretch), 1,618 (aromatic C=C stretch), 1,540 (pyrone ring C=C stretch), 1,453 (C-H bending of CH2), and 1,267 (C-O Stretch). 1H NMR (400 MHz, CDCl3): 2.41 (3H, CH3), 6.28 (1H, d, J = 6 Hz, H-8), 7.32 (1H, d, J = 8.8 Hz, H-7), 7.81 (1H, d, J = 2 Hz, H-5), and 8.22 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 173.44 (C-4), 157.72 (C-2), 154.46 (C-9), 136.13 (C-6), 135.45 (C-7), 125.82 (C-5), 121.49 (C-10), 117.81 (C-8), 86.72 (C-3), and 21.09 (PhCH3). HR-MS for C10H7IO2 [M + H]+m/z: Calcd 286.9563; Observed 286.9549. The major mass fragment observed was C10H7O2+(159).
6-Fluoro-3-iodochromone (4t)
It was obtained as light yellow solid in 81% yield; m.p.: 123–126°C, Rf: 0.57 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,063 (aromatic C-H stretch), 2,942 (aliphatic C-H stretch), 1,647 (C=O stretch), 1,616 (aromatic C=C stretch), 1,553 (pyrone ring C=C stretch), 1,436 (C-H bending of CH2), and 1,257 (C-O Stretch). 1H NMR (400 MHz, CDCl3): 7.41 (1H, d, J = 7.2 Hz, H-8), 7.80–7.82 (1H, m, H-7), 8.32 (1H, s, H-2), and 8.39 (1H, d, J = 2 Hz, H-5). 13C NMR (100.6 MHz, CDCl3): 172.77 (C-4), 161.04 (C-6), 158.01 (C-2), 152.45 (C-9), 122.84 (C-7), 120.46 (C-8), 111.52 (C-5), and 86.69 (C-3). HR-MS for C9H4ClIO2 [M + H]+m/z: Calcd 290.9262; Observed 290.9258. The major mass fragment observed was C9H5O2+(145).
6-Chloro-3-iodo-7-methylchromone (4u)
It was obtained as light yellow solid in 83% yield; m.p.: 120–125°C, Rf: 0.50 (ethyl acetate: hexane, 3:7). IR (cm−1): 3,067 (aromatic C-H stretch), 2,918 (aliphatic C-H stretch), 1,638 (C=O stretch), 1,612 (aromatic C=C stretch), 1,538 (pyrone ring C=C stretch), and 1,259 (C-O Stretch). 1H NMR (400 MHz, CDCl3): 2.47 (3H, CH3), 7.33 (1H, s, H-8), 8.15 (1H, s, H-5), and 8.23 (1H, s, H-2). 13C NMR (100.6 MHz, CDCl3): 172.37 (C-4), 159.73 (C-2), 154.51 (C-9), 145.53 (C-7), 131.33 (C-5), 129.12 (C-6), 123.01 (C-10), 120.13 (C-8), and 86.59 (C-3). HR-MS for C10H6ClIO2 [M + H]+m/z: Calcd 320.9173; Observed 320.9158. The major mass fragment observed was C10H7O2+(159).
Bioefficacy
Test Fungus
The test fungus S. rolfsii ITCC 6474 was procured from Indian Type Culture Collection (ITCC) center, Division of Plant Pathology, ICAR-Indian Agricultural Research Institute, New Delhi-110012, India and kept at 27°C for at least 4–7 days on Potato Dextrose Agar (PDA) slant. The fungus was subcultured in Petri plates for further bioassay studies.
In vitro Fungicidal Activity
A stock solution (1, 000 mgL−1) of each synthesized compound was prepared in DMSO. Preliminary screening was carried out at different concentrations. A final bioassay was conducted at five different concentrations namely 1,000, 500, 250, 125, and 62.50 mg L−1 of 4i–4n, 4p, 4s, and 4u chromones, and all other chromones were tested at 100, 50, 25, 12.5, and 6.25 mg L−1, respectively. All concentrations were tested in triplicates. Commercially available fungicide Mancozeb (technical) was taken as positive control.
An in vitro antifungal bioassay was carried out on PDA medium by poisoned food technique (Nene and Thapliyal, 1979). Fungal growth (colony diameter) was measured and percentage inhibition was calculated by Abbott’s formula (Abbott, 1925).
where C = colony diameter (mm) of the control and T = colony diameter (mm) of the test plate.
Corrected percentage inhibition (IC) was calculated by given formula.
where I = Percentage inhibition, CF = (90-C)/C × 100, 90 is the diameter (mm) of the Petri plate, and C is the growth of the fungus (mm) in control.
ED50 (mg L−1) values (Effective Dose for 50% inhibition) were calculated using SPSS statistical Package (v16.0).
Quantitative Structure Activity Relationship
QSAR analysis was done by taking negative logarithm of observed ED50 (mgL−1) [pED50 = −log (ED50)] as dependent variable and 2D descriptors (Table 1) as independent variables. 2D Structures of compounds were drawn in Chemdraw Ultra 7.0 software and converted to 3D structures. A total of 239 2D descriptors were determined encoding different molecular structural characteristics consisting of electronic, spatial, thermodynamic, and structural descriptors, for example, element count, atomic valence connectivity index (chiV), path cluster, estate number, retention index (chi), chain path count, logP, semi-empirical path count, molecular cluster, molecular weight, topological index, and refractivity. Descriptors were calculated by geometry optimization and energy minimization carried out by the batch energy minimization method in the Merck molecular force field (MMFF) at RMS gradient (criteria for convergence) 0.01, distance dependent dielectric 1, and the number of cycles (max) 1, 000. Different Baumann alignment-independent (AI) descriptors were also calculated. All computational work was carried out with the help of VLifeMDS QSAR plus 4.6 software using the Lenovo PC with Windows 8.1 operating system and the Intel (R) Celeron (R) processor.
Training and Test Set
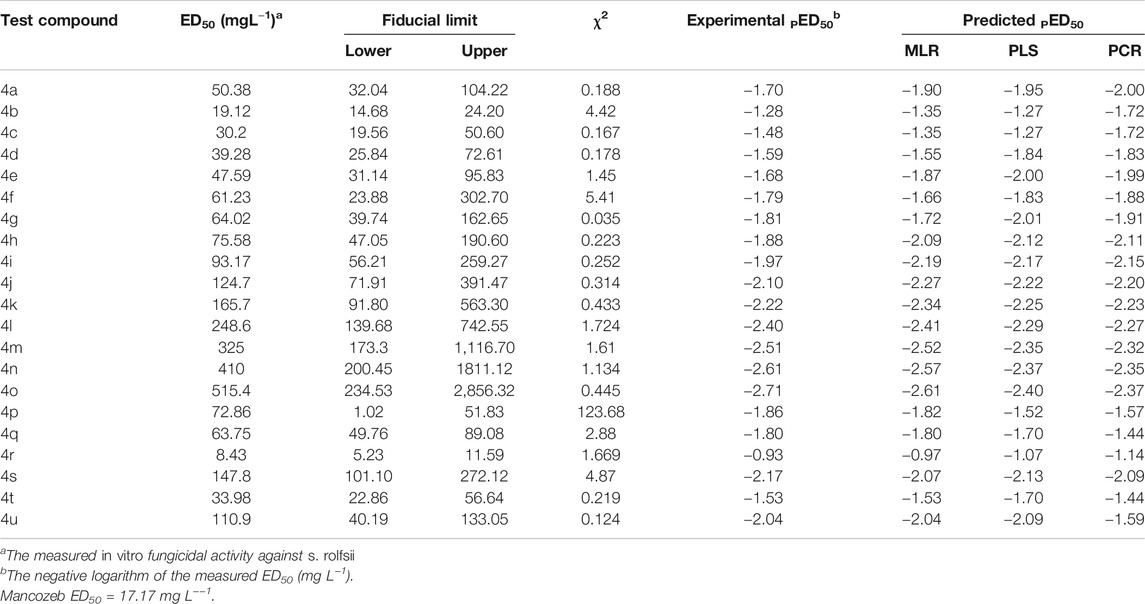
The entire data of 21 compounds were divided into a training set (14 compounds) and a test set (4 compounds), and 3 compounds were taken for validation with the help of the sphere exclusion method (Hudson et al., 1996). Unicolumn statistics were used to check the accuracy of selection of training and test sets, as the maximum value of the training set was greater than that of the test set and the minimum value of the training set was less than that of the test set (Table 2).
Regression Analysis
Regression analysis was done with three model building methods, MLR, PCR, and PLS. Various 2D descriptors were taken as independent variables and pED50 values as dependent variables by taking cross-correlation limit as 0.5; five variables in the final equation and r2 as the term selection criteria, F-test “in” at 4 and “out” at 3.99, r2 and F-test value. Values were fixed at 0 for variance cutoff, 10 for random iterations, and auto scaling for scaling. Developed QSAR models were assessed with the help of statistical parameters such as n = total number of compounds used in regression, k = total number of descriptors in a model, r2 = regression coefficient, q2 = cross-validated correlation coefficient, F = F-test (Fisher test value) for statistical significance, pred_r2 = predictive squared correlation coefficients, pred_r2se = coefficient of correlation of predicted data set, and r2 se and q2 se = standard error (SE) of estimation.
Multiple Linear Regression Analysis
MLR defines linear relationship between a single response variable and a number of predictor variables. In the present work, pED50 fungicidal activity was response variable and 2D descriptors were predictor variables. In this method, regression coefficients values (r2) were calculated by the least squares curve fitting method. In regression analysis, conditional mean of the dependent variable (pED50) Y depends on (descriptors) X (Eq. 1).
where Y = dependent variable, “x”s = independent variables, “b”s = regression coefficients for “x”s, and “c” = regression constant or intercept (Devillers, 1996; Croux and Joossens, 2005).
Principal Component Regression Method
In this method, the whole data were divided into principal components (PCs), smaller sets having major details of the large set. The main aim of PCR is to find out the values of a dependent variable with the help of selected PCs of independent variables. These PCs were not correlated, but had a basic linear relationship of original variables. The data were rotated into a new set of axes in such a way that first few axes showed greatest variability within the data. First PC (PC1) had maximum possible variation in the data, and each successive PC was taken perpendicular to preceding PCs and represent highest of the outstanding variance. The PC value is calculated by rotation of each point to a particular axis. A new group of axes for the data was chosen on the basis of a descending value data variance. Principal component analysis (PCA) also describes the fashion of similarity of the observations and the variables by exhibiting them as points in maps. PCR gives a mechanism for obtaining structure in datasets.
Partial Least Squares Regression Method
The partial least squares (PLS) test correlation between a set of dependent variables (Y) and a set of predictor variables (X). The main aim of PLS regression is helpful in describing the common structure by estimating the biological activity (dependent variables Y) from descriptors (X) (Huberty, 1994). PLS developed orthogonal components based on the relationship between predictors and respective outputs, while retaining highest variance of independent variables.
Validation of The QSAR Model
The QSAR model was validated with Leave-one-out (LOO) cross validation, by dividing training dataset into equal size subsets after eliminating one biological activity data (number of subsets = number of data points). These subsets were used to develop the model for calculating predicted activity (value of response variable of excluded data). Since in LOO subset all the data points were serially considered as predicted, the mean value of predicted activity was similar for LOO q2 and r2. After elimination of the next data point, the same procedure was repeated until all data points were removed. Thus, three statistically significant models were developed by LOO cross-validation. (Kubyani, 1994). Eq. 2 was used for calculating q2.
where Ypred = predicted, Yact = actual, Ymean = mean values of the pED50, and Σ (Ypred − Yact)2 = predictive residual error sum of squares (PRESS). External validation has also been performed to verify model validity, which tests how well the equation generalizes. A training set was used to develop an adjustment model for predicting activities of test set members. The predictive performance of equations was determined by q2, and coefficients of predictive squared correlation (pred _r2). pred_r2 was calculated by Eq. 3.
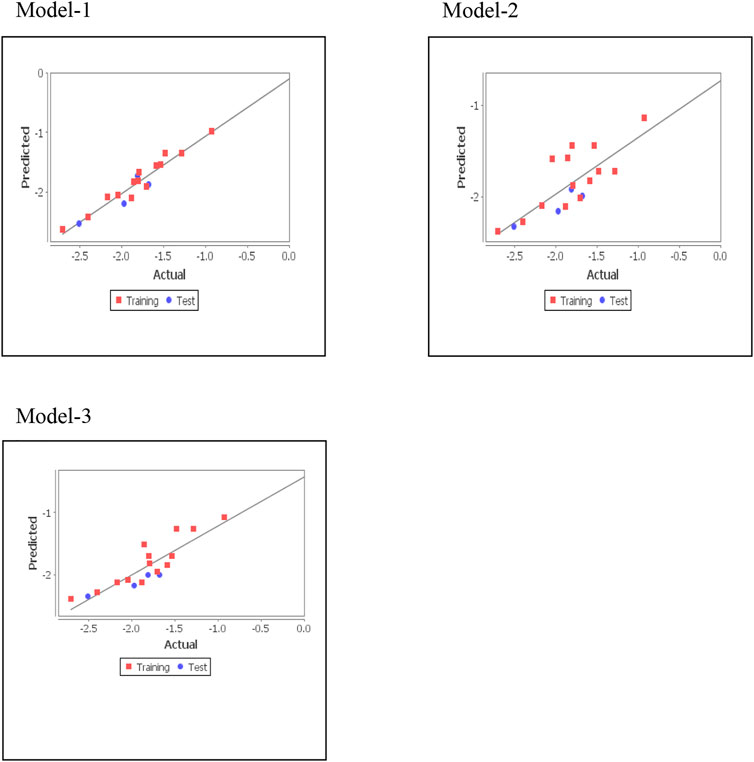
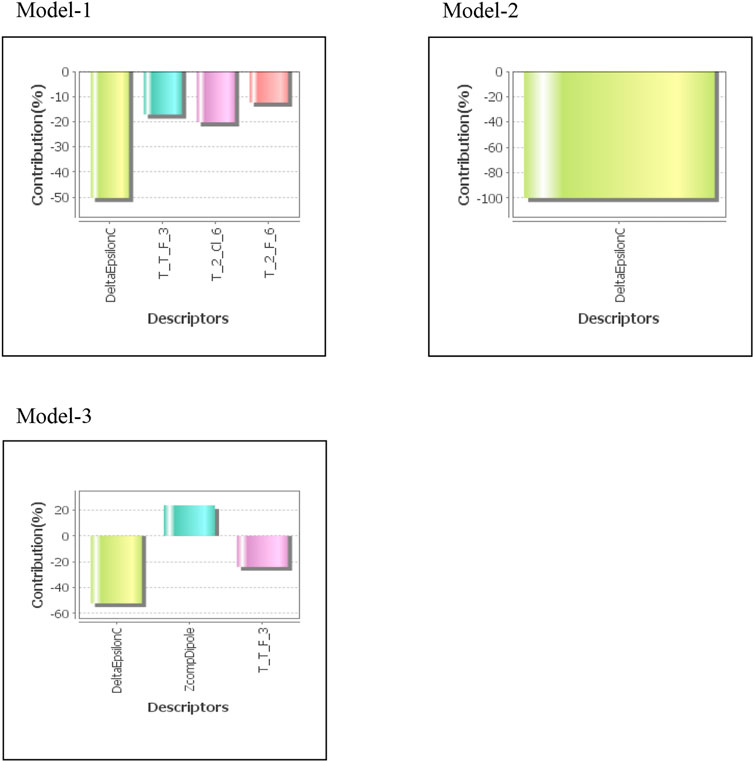
where Ypred(Test) = predicted activity and YTest = observed activity of test set compounds and YTraining = mean activity value of the training set. Statistical significance of model was validated by the fitness plot (Figure 1) and it was also supported by closeness of observed and predicted activity (Table 3). The magnitude of different descriptors employed for developing QSAR models were present in contribution charts (Figure 2).
Result and Discussion
Synthesis and Characterization
In this study, total 21 compounds (4a–4u) were synthesized out of which 10 compounds (4f, 4g, 4h, 4i, 4j, 4k, 4l, 4m, 4n, and 4o) were novel. The compounds synthesized by the above method were obtained in the yield ranging from 67 to 89%. Characterization of compounds was done by IR, 1H NMR 13C NMR, and LC-HRMS. In enaminones (3a–3u), peaks at δ 5.57–5.73 (1H, d, J = 8.8 Hz, H-2) and δ 7.55–7.92 (1H, d, J = 8.8 Hz, H-3) as two doublets for two protons with J = 8.8 Hz each were representative peaks of olefinic bond in 1H-NMR spectrum of all the compounds, and confirms the formation of enaminone. In 13C-NMR, the peaks at δ 89.75–89.93 (C-2) and 154.01–158.67 (C-3) for HC = CH and at δ 190.31–190.90 for C=O were conspicuous for all the compounds. The higher chemical shifts values of H-3 and C-3 than H-2 and C-2 were due to carbonyl moiety, which polarizes the C=C double bond. In IR spectra, stretching of (C=O) at 1,628–1,647 and (C=C) at 1,539–1,593 cm−1 supported the NMR data.
In case of chromones (4a–4u), peaks at δ 7.93–8.38 (1H, s, H-2) in 1H NMR and 157.20–163.56 (C-2) and 171.83–175.44 (C-4, C=O) in 13C NMR spectra of all compounds confirms synthesis of chromone derivatives. Stretching of (C=O) at 1,635–1,647 and (pyrone ring, C=C) at 1,531–1,599 cm−1 in IR spectra justified the NMR data.
In Vitro Fungicidal Activity
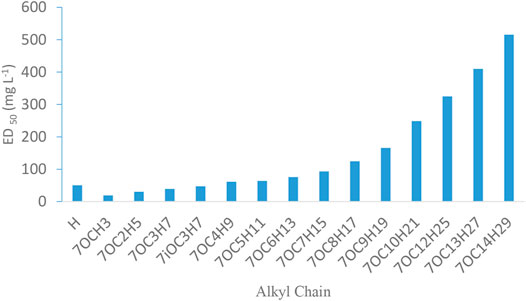
In vitro evaluation showed that all the tested compounds (4a–4u) exhibited promising fungicidal activity against S. rolfsii (Table 3), and compound 4r was found to be most active (ED50 = 8.43 mgL−1) which was better than Mancozeb (ED50 = 17.17 mgL−1), a commercial fungicide. It was observed that with increasing length of alkyl chain, fungicidal activity of alkoxy chromones (4a–4n) significantly decreased (Figure 3). Among halo chromone derivatives (4p, 4q, 4r, 4t, and 4u) 6, 8 dichlro derivative (4r) exhibited the highest and 4 fluoro derivative (4u) the least activity.
2D-QSAR Study
Three statistically significant QSAR models viz. Model-1 (MLR) Eq. 4, Model-2 (PCR) Eq. 5, and Model-3 (PLS) Eq. 6 were developed in 2D-QSAR study of fungicidal activity.
Model-1 (MLR)
where n = 14, DF = 9, r2 = 0.9434, q2 = 0.9117, F_test = 37.5361, r2_se = 0.1290, q2_se = 0.1612, pred_r2 = 0.8374, and pred_r2se = 0.1730.
Model-2 (PCR)
where n = 14, DF = 12, r2 = 0.5979, q2 = 0.4509, F_test = 17.8453, r2_se = 0.2979, q2_se = 0.3482, pred_r2 = 0.6877, and pred_r2se = 0.2397.
Model-3 (PLS)
where n = 14, DF = 11, r2 = 0.8006, q2 = 0.6167, F_test = 22.0866, r2_se = 0.2191, q2_se = 0.3038, pred_r2 = 0.6186, and pred_r2se = 0.2649.
In above QSAR models, correlation coefficient (r2) was used to calculate biological activity variance by multiplying with 100. The predictive ability (q2) of generated QSAR models was assessed by LOO (Left-out-one) method. F is the ratio of variance of models and that of error in regression. Models with a higher F value and lower SE of estimation (r2se and q2se) were considered statistically significant. External validation with pred_r2 > 0.3, established the predictive power of the QSAR model. Among these three models, the MLR model was found best as revealed by q2, r2, higher values of F-test, and pred_r2. The high q2 value is the best indicator of 2D QSAR’s reliability since only a high r2 could be due to data overfitting. Quite often, a q2 value of more than 0.5 is considered appropriate. (Golbraikh and Tropsha, 2002; Doweyko, 2004; Ponce et al., 2004).
The developed models showed that fungicidal activity was inversely related to descriptors, DeltaEpsilonC and AI descriptor, T_2_Cl_6, T_2_F_6, and T_T_F_3 and directly related to ZcompDipole. Two descriptors viz. DeltaEpsilonC and T_2_Cl_6 significantly (∼70%) impact the fungicidal activity of test compounds. Alignment Independent (AI) descriptors were estimated, as explained in Baumann’s paper (Balaban, 1982), on the basis of molecular topology, type of bond, and atom. Every atom was given a minimum of one and a maximum of three attributes. Molecular topology (T) was designated as the first attribute, followed by atom symbol and atoms linked with multiple (double or triple) bonds as second and third attribute, respectively. Then, selective distance count statistics, which counts all the fragments between the first atom and the last atom isolated by a graph distance, for all combinations of various attributes were calculated. Graph distance is the least number of atoms across the path joining two atoms in molecular structure. For example, selective distance count statistic “AB2” (e.g., TOPO2N3) counts all the fragments between a start atom with attribute “A” (e.g., “2” a double bonded atom) and an end atom with attribute “B” (e.g., “N”) separated by a graph distance 3. Topological indices are numerical values associated with chemical constitutions which establish correlation between biological activity and chemical structure. AI descriptors in this study were calculated with the help of attributes namely, 2 (atom with double bond), 3 (atom with double bond), C (Carbon), N (Nitrogen), O (Oxygen), S (Sulfur), H (Hydrogen), F (Fluorine), Cl (Chlorine), and Br (Bromine) with distance ranging from 0 to 7. DeltaEpsilonC is a measure of contribution of electronegativity. The result revealed that it is negatively correlated with fungicidal activity of the test compounds.
Conclusion
The study revealed that all test compounds showed fungicidal activity against S. rolfsii., but compound 4r showed the highest activity. The QSAR study determined quantitative correlation between fungicidal activity and structural/physicochemical properties of test compounds. The variables in developed model equations established that structural, molecular shape analysis, electronic, and thermodynamic descriptors played a major role in fungicidal activity of the compounds. In the case of MLR and PLS, the overall prediction was found to be around 94 and 80%, respectively. The 2D-QSAR study revealed that results of MLR analysis exhibited significant predictive power and reliability than the other two methods (PCR and PLS). Information and understanding of descriptors influencing fungicidal activity of these chromones could be used for structure optimization to improve activity.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
PK and NS conceptualized the idea; VR did HRMS interpretation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author (PK) is thankful to ICAR-IARI, New Delhi, India for providing all the facilities for research work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.636882/full#supplementary-material.
References
Abbott, W. S. (1925). A method of computing the effectiveness of an insecticide. J. Econ. Entomol. 18, 265–267. doi:10.1093/jee/18.2.265a
Aycock, R. (1966). Stem rot and other diseases caused by Sclerotium rolfsii. Raleigh, NC: North Carolina State University.
Balaban, A. T. (1982). Highly discriminating distance-based topological index. Chem. Phys. Lett. 89, 399–404. doi:10.1016/0009-2614(82)80009-2
Croux, C., and Joossens, K. (2005). Influence of observations on the misclassification probability in quadratic discriminant analysis. J. Multivar. Anal. 96, 384–403. doi:10.1016/j.jmva.2004.11.001
Deising, H. B., Reimann, S., Peil, A., and Weber, W. E. (2002). “Disease management of rusts and powdery mildews,” in The mycota XI. Application in agriculture. Editor F. Kempken (Berlin, Germany: Springer), 243–269.
Devillers, J. (1996). Neuronal network in QSAR and drug design. London, United Kingdom: Academic Press.
Doweyko, A. M. (2004). 3D-QSAR illusions. J. Comput. Aided Mol. Des. 18, 587–596. doi:10.1007/s10822-004-4068-0
Gammill, R. B. (1979). A new and efficient synthesis of 3-halogenated 4H-1-Benzopyran-4-ones. Synthesis 1979, 901–903. doi:10.1055/s-1979-28869
Gaspar, A., Matos, M. J., Garrido, J., Uriarte, E., and Borges, F. (2014). Chromone: a valid scaffold in medicinal chemistry. Chem. Rev. 114, 4960–4992. doi:10.1021/cr400265z
Gaspar, A., Silva, T., Yáñez, M., Vina, D., Orallo, F., Ortuso, F., et al. (2011). Chromone, a privileged scaffold for the development of monoamine oxidase inhibitors. J. Med. Chem. 54, 5165–5173. doi:10.1021/jm2004267
Gasparová, R., Lácová, M., El-Shaaer, H. M., and Odlerová, Z. (1997). Synthesis and antimycobacterial activity of some new 3-heterocyclic substituted chromones. Farmaco 52, 251–253.
Golbraikh, A., and Tropsha, A. (2002). Beware of q2!. J. Mol. Graph. Model. 20, 269–276. doi:10.1016/s1093-3263(01)00123-1
Hollomon, D. W. (2016). Fungicide resistance: facing the challenge—a review. Plant Protect. Sci. 51, 170–176. doi:10.17221/42/2015-PPS
Hudson, B. D., Hyde, R. M., Rahr, E., Wood, J., and Osman, J. (1996). Parameter based methods for compound selection from chemical databases. Quant. Struct. Act. Relat. 15, 285–289. doi:10.1002/qsar.19960150402
Kaushik, P., Sarkar, D. J., Chander, S., Rana, V. S., and Shakil, N. A. (2019). Insecticidal activity of phenolic acid amides against brown planthopper (BPH), Nilaparvata lugens (Stål) and their QSAR analysis. J. Environ. Sci. Health B 54, 489–497. doi:10.1080/03601234.2019.1574174
Kubyani, H. (1994). Variable selection in QSAR studies: an evolutionary algorithm. Quant. Struct. Act. Relat. 13, 285–294. doi:10.1002/qsar.19940130306
Nene, Y. L., and Thapliyal, P. N. (1979). in “Fungicides in plant disease control,” in Evaluation of fungicides. Editors Y. L. Nene, and P. N. Thapliyal (New Delhi, India: Oxford and IBH Publishing Co.), 406–428.
Ponce, Y. M., Garit, J. A. C., Torrens, F., Zaldivar, V. R., and Castro, E. A. (2004). Atom, atom-type, and total linear indices of the “molecular pseudograph’s atom adjacency matrix”: application to QSPR/QSAR studies of organic compounds. Molecules 9, 1100–1123. doi:10.3390/91201100
Punja, Z. K. (1985). The biology, ecology, and control of Sclerotium rolfsh. Annu. Rev. Phytopathol. 23, 97–127. doi:10.1146/annurev.py.23.090185.000525
Keywords: 3-iodochromone, fungicidal activity, sclerotium rolfsii, QSAR, multiple linear regression, principle component analysis, partial least square
Citation: Kaushik P, Shakil NA and Rana VS (2021) Synthesis, Biological Evaluation, and QSAR Studies of 3-Iodochromone Derivatives as Potential Fungicides. Front. Chem. 9:636882. doi: 10.3389/fchem.2021.636882
Received: 11 December 2020; Accepted: 17 February 2021;
Published: 30 April 2021.
Edited by:
Simone Brogi, University of Pisa, ItalyReviewed by:
Rajeev K. Singla, Sichuan University, ChinaAndrei I. Khlebnikov, Tomsk Polytechnic University, Russia
Copyright © 2021 Kaushik, Shakil and Rana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Parshant Kaushik, cGFyc2hhbnRhZ3JpY29AZ21haWwuY29t
 Parshant Kaushik
Parshant Kaushik Najam A. Shakil
Najam A. Shakil Virendra S. Rana
Virendra S. Rana