- 1Department of Pure and Industrial Chemistry, University of Nigeria, Nsukka, Nigeria
- 2Organic Synthesis and Process Chemistry, Council for Scientific and Industrial Research-India Institute of Chemical Technology, Hyderabad, India
- 3Department of Pharmaceutical and Medicinal Chemistry, University of Nigeria, Nsukka, Nigeria
- 4Department of Industrial Chemistry, Evangel University Akaeze, Enugu, Nigeria
The increase of antimicrobial resistance (AMR) and antimalarial resistance are complex and severe health issues today, as many microbial strains have become resistant to market drugs. The choice for the synthesis of new dipeptide-carboxamide derivatives is as a result of their wide biological properties such as antimicrobial, anti-inflammatory, and antioxidant activities. The condensation reaction of substituted benzenesulphonamoyl pentanamides with the carboxamide derivatives using peptide coupling reagents gave targeted products (8a-j). The in silico antimalarial and antibacterial studies showed good interactions of the compounds with target protein residues and a higher dock score in comparison with standard drugs. In the in vivo study, compound 8j was the most potent antimalarial agent with 61.90% inhibition comparable with 67% inhibition for Artemisinin. In the in vitro antimicrobial activity, compounds 8a and 8b (MIC 1.2 × 10−3 M and 1.1 × 10−3 M) were most potent against S. aureus; compound 8a, 8b, and 8j with MIC 6.0 × 10−3 M, 5.7 × 10−4 M, and 6.5 × 10−4 M, respectively, were the most active against B. subtilis; compound 8b (MIC 9.5 × 10−4 M) was most active against E.coli while 8a, 8b and 8d were the most active against S. typhi. Compounds 8c and 8h (MIC 1.3 × 10−3 M) each were the most active against C. albicans, while compound 8b (MIC 1.3 × 10−4 M) was most active against A. niger.
Introduction
Infectious diseases and their resistance to many available commercial drugs have remained the most challenging task for human existence. Sulfonamides consist of major drug components called sulfa drugs. The functional group –SO2NH2 in sulfonamides enables it to possess many pharmacological properties such as antibacterial (Qadir et al., 2015), antifungal (Jyothi and Madhavi, 2018), antimalarial (Maloy Kumar et al., 2018), anticancer (Abdelaziz et al., 2015), inhibitors of human carbonic anhydrase I (hCA I) and human carbonic anhydrase II (hCA II, Kilicaslan et al., 2016), anti-HIV (Jiao et al., 2010), and many others. Carboxamides are also ubiquitous functionality in medicate particles as pharmacophores (Montalbetti and Falque, 2005). Carboxamides have been accounted for as antimicrobial and antioxidant (Eze et al., 2019), carbonic anhydrase enzyme inhibitor and antioxidant (Deniz et al., 2019), anticancer (Kumar et al., 2009), anthelmintic (Ugwu et al., 2018a), antitubercular (Ugwu et al., 2014), antitrypanosomal (Ugwu et al., 2018b), and anti-inflammatory and analgesic (Ugwu et al., 2018c) agents.
Day and Greenfield (2004) reported that peptides are resourceful pharmacophores as they play important roles within the physical body and other organisms. As a result of good properties of peptides such as solubility, permeability, and bioavailability, many short peptide derivatives possess the ability to bind to membrane receptors (Qi et al., 2010; Thompson et al., 2012). Khavinson and Anisimov (2000) reported Lys-Glu based dipeptide as an antitumor agent. In 2004, Nitta et al. (2004) reported the neuroprotective effect of Leu-Ile. In the work of Kayser and Meisel (1996), Tyr-Gly was reported to reinforce the proliferation of peripheral blood lymphocytes. Peptides have also been reported as an antimicrobial (Jatinder et al., 2015), carbonic anhydrase I, II, IV, and XII inhibitor (Zehra Küçükbay et al., 2016; Küçükbay et al., 2019), antiplasmodial (Amit et al., 2015; Jatinder et al., 2015), and antihypertensive agent (Kitts and Weiler, 2003). In a continuation of our work, we synthesized novel Leu-Val dipeptide carboxamide scaffolds bearing sulfonamide moieties with potent antimalarial and antimicrobial properties. The quest for leu-val combination skeleton was as a result of antimalarial properties of ala-gly dipeptides (Ugwuja et al., 2019), antimalarial and antioxidant property of val-val dipeptides by Ezugwu et al. (2020), and also an antimalarial property of quinine derivatives containing some amino acid, dipeptide, or tripeptide (Panda et al., 2013).
Experimental General
The chemicals and solvents used were purchased from Aldrich (Sigma-Aldrich) and AVRA Chemicals Pvt. Ltd. (Hyderabad, India) and used without purification. 1H-NMR and 13C-NMR spectra were recorded on Advance 300, 400, and 500 MHz spectrometers in DMSO-d6 using TMS as internal standard. FT-IR spectra were recorded on Thermo Nicollet Nexus 670 spectrometer. Mass spectra were obtained on Agilent LCMS instrument. HRMS were measured on Agilent Technologies 6510, Q-TOFLC/MS ESI-Technique. Melting points were determined in open glass capillary tubes on a Stuart melting point apparatus and are uncorrected. All experiments were carried out at Dr. B. China Raju's Laboratory, Organic Synthesis and Processing Chemistry Division, CSIR-Indian Institute of Chemical Technology, Hyderabad, India. All reactions were monitored by thin layer chromatography (TLC) on precoated silica gel 60 F254 (mesh); sports were visualized under UV light and in oven with Ninhydrin. Merck neutral aluminum oxide activated (60–325 mesh) was used for chromatography.
Chemistry
General Procedure for the Synthesis of Substituted Benzenesulfonamoyl Pentanamides
Appropriate substituted benzenesulfonyl chloride (1a-c, 1.82 mmol) was added in portions for 1 h to an aqueous solution of L-leucine (1.5 mmol) containing sodium carbonate (Na2CO3, 1.82 mmol) at −5°C. The slurry formed was stirred at room temperature for 4 h (TLC (MeOH/DCM, 1:9 monitored). The mixture was acidified to pH 2 (Ugwu et al., 2014; Ezugwu et al., 2020).
General Procedure for the Synthesis of Carbmate Derivatives (6a-f)
To a solution of Boc-valine (3.0 g, 13.82 mmol) in dichloromethane (20 mL) was added triethylamine (20.7 mmol), EDC.HCl (16.0 mmol), HOBt (13.82 mmol) at 0°C, and substituted aniline (13.82 mmol) was added after stirring for 15 min. The resulting mixture was warmed to room temperature and stirred for 19–24 h (TLC monitored). Upon completion, the crude products were obtained after an aqueous work-up and purified by column chromatography (ethyl acetate/hexane = 5:95, Ezugwu et al., 2020).
Synthesis of Carboxamide Derivatives (7a-e)
Dichloromethane/trifluoroacetic acid (1:1%) was added to compounds 6a-c and stirred at room temperature for 1 h (TLC monitored). The products were obtained after evaporating the solvent under reduced pressure. The solid TFA salts were precipitated on the addition of diethylether and dried (Ezugwu et al., 2020).
General Procedures for Synthesis of Leu-Val Dipeptides
To a solution of substituted benzenesulfonamoyl pentanamides (1.0 mmol) in dichloromethane (20 mL) was added triethylamine (20.7 mmol), EDC.HCl (16.0 mmol), HOBt (13.82 mmol) at 0°C, and carboxamide derivatives (7a-e, 1.0 mmol) were added after stirring for 15 min. The resulting mixture was warmed to room temperature and stirred for 19–24 h (TLC monitored). Upon completion, the crude products were obtained after an aqueous work-up and purified by column chromatography (ethyl acetate/hexane = 5:95, Ezugwu et al., 2020).
(S)-N-({(S)-1-[(4-Methylphenyl)amino]-3-methyl-1-oxobutan-2-yl})-4-methyl-2-(4-nitrophenylsulfonamido)pentanamide (8a)
Yield (0.315 g, 65.9%), light yellow solid, M.p = 172–17°C. FTIR (KBr, cm−1): 3,312, 3,261, and 3,194 (3NH), 2,965, 2,921 (C-H Aliphatic), 1,642, 1,640 (2C=O, amide), 1,533, 1,450 (C=C-Aromatic), 1,354 (SO2), 1,170, 1,090 (C-N). 1H-NMR (400 MHz, DMSO) δ9.91 (s, 1H, NH of amide), 8.43 (dd, J = 8.5, 6.8 Hz, 1H, SO2-NH), 8.33 (d, J = 8.8 Hz, 2H, Ar-H), 8.12 (d, J = 8.6 Hz, 1H, NH of amide), 8.03 (dd, J = 8.9, 2.0 Hz, 2H, Ar-H), 7.44 (d, J = 8.4 Hz, 2H, Ar-H), 7.09 (d, J = 8.3 Hz, 2H, Ar-H), 4.04–3.95 (m, 2H, CH-C=O), 2.24 (s, 3H, CH3-Ar), 1.91–1.80 [m, 1H, CH-(CH3)2], 1.69–1.58 [m, 1H, CH-(CH3)2], 1.41–1.30 (m, 2H, CH2), 0.85 (d, J = 6.6 Hz, 3H, CH3), 0.79 (d, J = 6.5 Hz, 3H, CH3), 0.73 [dd, J = 6.6, 4.0 Hz, 6H (CH3)2]. 13C-NMR (101 MHz, DMSO) δ171.21, 169.85 (2C=O), 149.73, 147.17, 136.66, 132.73, 129.52, 128.72, 124.62, 119.62 (eight aromatic carbons), 58.78, 55.12, 42.29, 31.20, 24.34, 23.40, 21.59, 20.92, 19.45, 18.69 (10 aliphatic carbons). ESI-MS: m/z, 505 [M+H]+. HRMS-ESI: calcd. For C24H32N4O6S [M+H]+ 505.2121; Found 505.2125.
(S)-N-({(S)-1-[(4-Chlorophenyl)amino]-3-methyl-1-oxobutan-2-yl})-4-methyl-2-(4-nitrophenylsulfonamido)pentanamide (8b)
Yield (0.356 g, 71.6%), light yellow solid, M.p = 183–184°C. FTIR (KBr, cm−1): 3,300, 3,270, and 3,107 (3NH), 2,962, 2,871 (C-H Aliphatic), 1,643, 1,602 (2C=O, amide), 1,534, 1,497, 1,461 (C=C-Aromatic), 1,397, 1,348, 1,245, 1,207 (SO2), 1,164, 1,089, 1,013 (C-N). 1H-NMR (400 MHz, DMSO) δ10.18 (s, 1H, NH of amide), 8.48–8.27 (m, 3H, SO2-NH + Ar-H), 8.10 (dd, J = 55.7, 1.2 Hz, 3H, NH of amide + Ar-H), 7.60 (d, J = 1.4 Hz, 2H, Ar-H), 7.35 (d, J = 0.8 Hz, 2H, Ar-H), 4.07 (t, J = 46.6 Hz, 2H, 2[CH-C=O]), 2.06–1.80 (m, 1H, CH-[CH3]2), 1.63 (d, J = 6.6 Hz, 1H, CH[CH3]2), 1.40–1.28 (m, 2H, CH2), 0.78 (dd, J = 34.3, 10.6 Hz, 12H [CH3]4).13C-NMR (101 MHz, DMSO) δ171.27, 170.29 (2C=O), 149.72, 147.16, 138.10, 129.10, 128.71, 127.39, 124.62, 121.11 (eight aromatic carbons), 58.88, 55.12, 42.26, 31.10, 24.34, 23.46, 21.60, 19.44, 18.67 (nine aliphatic carbons). ESI-MS: m/z, 525 [M+H]+. HRMS-ESI: calcd. For C23H29N4O6S [M+H]+ 525.1575; Found 525.1584.
(S)-N-({(S)-1-[(4-Isopropylphenyl)amino]-3-methyl-1-oxobutan-2-yl})-4-methyl-2-(4-nitrophenylsulfonamido)pentanamide (8c)
Yield (0.36 g, 72%), cream solid, M.p = 126–127°C. FTIR (KBr, cm−1): 3,330, 3,306, 3,101 (3NH), 2,962, 2,871 (C-H Aliphatic), 1,643, 1,609 (2C=O, amide), 1,533, 1,462 (C=C-Aromatic), 1,348, 1,246 (SO2), 1,163, 1,088, 1,017 (C-N). 1H-NMR (400 MHz, DMSO) δ9.90 (s, 1H, NH of amide), 8.38 (d, J = 8.9 Hz, 1H, SO2-NH), 8.33 (d, J = 8.9 Hz, 2H, Ar-H), 8.09 (d, J = 8.8 Hz, 1H, NH of amide), 8.03 (d, J = 8.9 Hz, 2H, Ar-H), 7.45 (d, J = 8.6 Hz, 2H, Ar-H), 7.15 (d, J = 8.5 Hz, 2H, Ar-H), 4.05–3.94 (m, 2H, 2[CH-C=O]), 2.82 (dt, J = 13.8, 6.9 Hz, 1H, CH-[CH3]2), 1.84 (dd, J = 13.7, 6.8 Hz, 1H, CH-[CH3]2), 1.69–1.57 (m, 1H, CH-[CH3]2), 1.41–1.26 (m, 2H, CH2), 1.16 (d, J = 6.9 Hz, 6H [CH3]2), 0.86–0.71 (m, 12H [CH3]4).13C-NMR (101 MHz, DMSO) δ171.21, 169.87 (2C=O), 149.74, 147.21, 143.91, 136.92, 128.72, 126.87, 124.63, 119.72 (eight aromatic carbons), 58.76, 55.08, 42.30, 33.36, 31.20, 24.43, 24.34, 23.48, 21.78, 21.61, 19.44, 18.71 (12 aliphatic carbons). ESI-MS: m/z, 533[M+H]+. HRMS-ESI: calcd. For C26H36N4O6S [M+H]+ 533.2434; Found 533.2445.
(S)-N-({(S)-1-[(3-Fluorophenyl)amino]-3-methyl-1-oxobutan-2-yl})-4-methyl-2-(4-nitrophenylsulfonamido)pentanamide (8d)
Yield (0.43 g, 89.6%), light yellow solid, M.p = 135–136°C. FTIR (KBr, cm−1): 3,302, 3,102 (NH), 2,963, 2,871 (C-H Aliphatic), 1,644, 1,610 (2C=O, amide), 1,536, 1,492, 1,442 (C=C-Aromatic), 1,348, 1,211 (SO2), 1,162, 1,088, 1,013 (C-N). 1H-NMR (400 MHz, DMSO) δ10.22 (s, 1H, NH of amide), 8.38 (dd, J = 34.0, 8.6 Hz, 3H, SO2-NH + Ar-H), 8.13 (d, J = 8.2 Hz, 1H, NH of amide), 8.03 (d, J = 8.4 Hz, 2H, Ar-H), 7.55 (d, J = 11.5 Hz, 1H, Ar-H), 7.40–7.21 (m, 2H, Ar-H), 6.88 (d, J = 7.6 Hz, 1H, Ar-H), 4.12–3.87 (m, 2H, CH-C=O), 1.93–1.80 [m, 1H, CH-(CH3)2], 1.64 [d, J = 5.1 Hz, 1H, CH-(CH3)2], 1.35 (ddd, J = 17.3, 11.6, 6.7 Hz, 2H, CH2), 0.87–0.72 [m, 12H (CH3)4].13C-NMR (101 MHz, DMSO) δ171.32, 170.53 (2C=O), 163.77, 161.37, 149.72, 147.18, 140.86, 140.75, 130.91, 130.82, 128.72, 124.61, 115.32, 110.41, 110.20, 106.46, 106.19 (aromatic carbons), 58.84, 55.04, 42.25, 31.09, 24.33, 23.46, 21.57, 19.43, 18.63 (nine aliphatic carbons). ESI-MS: m/z, 509 [M+H]+. HRMS-ESI: calcd. For C23H29N3FO4S [M+H]+ 509.1870; Found 509.1870.
(S)-N-({(S)-1-[(4-Methylphenyl)amino]-3-methyl-1-oxobutan-2-yl})-4-methyl-2-(4-methylphenylsulfonamido)pentanamide (8e)
Yield (0.48 g, 96%), white solid, M.p = 191–192°C. FTIR (KBr, cm−1): 3,319, 3,267, 3,110 (3NH), 2,960, 2,910 (C-H Aliphatic), 1,643, 1,606 (2C=O, amide), 1,528, 1,456 (C=C-Aromatic), 1,377, 1,333, 1,244, 1,202 (SO2), 1,158, 1,088 (C-N). 1H-NMR (300 MHz, DMSO) δ9.93 (s, 1H, NH of amide), 7.99 (d, J = 8.5 Hz, 1H, SO2-NH), 7.89 (d, J = 9.0 Hz, 1H, NH of amide), 7.64 (d, J = 8.2 Hz, 2H, Ar-H), 7.48 (d, J = 8.4 Hz, 2H, Ar-H), 7.26 (d, J = 8.1 Hz, 2H, Ar-H), 7.11 (d, J = 8.3 Hz, 2H, Ar-H), 4.07 (t, J = 7.9 Hz, 1H, CH-C=O), 3.82 (td, J = 9.3, 5.3 Hz, 1H, CH-C=O), 2.26 (d, J=7.6 Hz, 6H, 2[CH3-Ar]), 1.86 (dt, J = 13.4, 6.7 Hz, 1H, CH-[CH3]2), 1.64–1.52 (m, 1H, CH[CH3]2), 1.30 (dd, J = 17.0, 8.4 Hz, 2H, CH2), 0.83–0.74 (m, 9H [CH3]3), 0.72 (d, J = 6.5 Hz, 3H, CH3). 13C-NMR (101 MHz, DMSO) δ171.64, 169.89 (2C=O), 142.78, 138.58, 136.73, 132.76, 129.70, 129.57, 127.09, 119.65 (eight aromatic carbons), 58.64, 55.18, 42.47, 31.44, 24.27, 23.47, 21.67, 21.37, 20.93, 19.50, 18.74 (11 aliphatic carbons). ESI-MS: m/z, 474 [M+H]+. HRMS-ESI: calcd. For C25H36N3O4S [M+H]+ 474.2427; Found 474.2424.
(S)-N-({(S)-1-[(4-Isopropylphenyl)amino]-3-methyl-1-oxobutan-2-yl})-4-methyl-2-(4-methylphenylsulfonamido)pentanamide (8f)
Yield (0.40 g, 75.7%), solid, M.p = 102–103°C. FTIR (KBr, cm−1): 3,313, 3,261, 3,105 (3NH), 3,050 (C-H-aromatic), 2,962, 2,871 (C-H Aliphatic), 1,645, 1,604 (2C=O, amide), 1,533, 1,458, 1,416 (C=C Aromatic), 1,378, 1,331, 1,204 (SO2), 1,158, 1,089 (C-N). 1H-NMR (300 MHz, DMSO) δ9.94 (s, 1H, NH of amide), 7.98 (d, J = 8.5 Hz, 1H, SO2-NH), 7.88 (d, J = 8.9 Hz, 1H, NH of amide), 7.65 (d, J = 8.0 Hz, 2H, Ar-H), 7.50 (d, J = 8.2 Hz, 2H, Ar-H), 7.26 (d, J = 7.9 Hz, 2H, Ar-H), 7.17 (d, J = 8.3 Hz, 2H, Ar-H), 4.08 (t, J = 7.8 Hz, 1H, CH-C=O), 3.82 (dd, J = 14.0, 8.8 Hz, 1H, CH-C=O), 2.83 (dt, J = 13.6, 6.8 Hz, 1H, CH-[CH3]2), 2.27 (s, 3H, CH3-Ar-H), 1.87 (dd, J = 13.4, 6.6 Hz, 1H, CH-[CH3]2), 1.68–1.49 (m, 1H, CH-[CH3]2), 1.42–1.26 (m, 2H, CH2), 1.17 (d, J = 6.8 Hz, 6H [CH3]2), 0.79 (dd, J = 9.8, 6.2 Hz, 9H [CH3]3), 0.72 (d, J = 6.4 Hz, 3H, CH3).13C-NMR (101 MHz, DMSO) δ171.64, 169.91 (2C=O), 143.94, 142.76, 138.62, 137.00, 129.70, 127.09, 126.90, 119.74 (eight aromatic carbons) 58.65, 55.18, 42.49, 33.37, 31.44, 24.44, 24.27, 23.47, 21.69, 21.34, 19.49, 18.75 (12 aliphatic carbons). ESI-MS: m/z, 502 [M+H]+. HRMS-ESI: calcd. For C27H39N3O4S [M+H]+ 502.2740; Found 502.2766.
(S)-N-({(S)-1-[(3-Fluorophenyl)amino]-3-methyl-1-oxobutan-2-yl})-4-methyl-2-(4-methylphenylsulfonamido)pentanamide (8g)
Yield (0.32 g, 64%), solid, M.p = 166–167°C. FTIR (KBr, cm−1): 3,327, 3,275, 3,185 (3NH), 2,962 (C-H Aliphatic), 1,680, 1,645 (2C=O, amide), 1,549, 1,491, 1,446 (C=C-Aromatic), 1,318, 1,212 (SO2), 1,157, 1,090 (C-N). 1H-NMR (300 MHz, DMSO) δ10.27 (s, 1H, NH of amide), 8.05 (d, J = 8.4 Hz, 1H, SO2-NH), 7.89 (d, J = 9.0 Hz, 1H, NH of amide), 7.62 (dd, J = 16.4, 9.9 Hz, 3H,ArH), 7.38–7.23 (m, 4H, Ar-H), 6.89 (t, J = 8.1 Hz, 1H, Ar-H), 4.06 (t, J = 7.8 Hz, 1H, CH-C=O), 3.84 (dd, J = 14.3, 9.3 Hz, 1H, CH-C=O), 2.28 (s, 3H, CH3-Ar), 1.88 (dd, J = 13.5, 6.7 Hz, 1H, CH-[CH3]2), 1.58 (d, J = 6.2 Hz, 1H, CH-[CH3]2), 1.36–1.24 (m, 2H, CH2), 0.79 (dd, J = 12.0, 4.8 Hz, 9H [CH3]3), 0.73 (d, J = 6.5 Hz, 3H, CH3). 13C-NMR (101 MHz, DMSO) δ171.77, 170.61 (2C=O), 163.79, 161.39, 142.76, 140.94, 140.83, 138.62, 130.97, 130.88, 129.69, 127.09, 115.38, 110.43, 110.22, 106.50, 106.23 (aromatic carbons), 58.81, 55.09, 42.43, 31.27, 24.27, 23.47, 21.67, 21.35, 19.48, 18.71 (10 aliphatic carbons). ESI-MS: m/z, 478 [M+H]+. HRMS-ESI: calcd. For C24H32N3FO4S [M+H]+ 478.2176; Found 478.2181.
(S)-N-({(S)-1-[(4-Bromophenyl)amino]-3-methyl-1-oxobutan-2-yl})-4-methyl-2-((4-methylphenyl)sulfonamido)pentanamide (8h)
Yield (0.31 g, 81%), off-white solid, M.p = 140–142°C. FTIR (KBr, cm−1): 3,335, 3,269, 3,106 (3NH), 2,962, 2,873 (C-H Aliphatic), 1,687, 1,637 (2C=O, amide), 1,537, 1,489, 1,458 (C=C-Aromatic), 1,396, 1,315, 1,242 (SO2), 1,155, 1,088, 1,009 (C-N). 1H-NMR (400 MHz, DMSO) δ10.20 (s, 1H, NH of amide), 8.05 (d, J = 8.4 Hz, 1H, SO2-NH), 7.88 (d, J = 8.8 Hz, 1H, NH of amide), 7.65 (d, J = 8.2 Hz, 2H, Ar-H), 7.58 (d, J = 8.9 Hz, 2H, Ar-H), 7.49 (d, J = 8.9 Hz, 2H, Ar-H), 7.26 (d, J = 8.2 Hz, 2H, Ar-H), 4.07 (t, J = 7.9 Hz, 1H, CH-C=O), 3.83 (dd, J = 13.8, 8.9 Hz, 1H, CH-C=O), 2.27 (s, 3H, CH3-Ar), 1.88 (dp, J = 13.2, 6.6 Hz, 1H, CH-[CH3]2), 1.59 (tt, J = 13.2, 6.5 Hz, 1H, CH-[CH3]2), 1.38–1.24 (m, 2H, CH2), 0.89–0.76 (m, 9H [CH3]3), 0.73 (d, J = 6.5 Hz, 3H, CH3). 13C-NMR (101 MHz, DMSO) δ171.72, 170.39 (2C=O), 142.75, 138.61, 132.04, 129.68, 127.08, 126.10, 121.55, 115.41 (eight aromatic carbons), 58.67, 55.15, 42.44, 31.28, 24.27, 23.46, 21.68, 21.37, 19.49, 18.73 (10 aliphatic carbons). ESI-MS: m/z, 537 [M+H]+. ESI-HRMS: calcd. For C24H32N3BrO4S [M+Na]+ 560.1195; Found 560.1202.
(S)-N-({(S)-1-[(4-Chlorophenyl)amino]-3-methyl-1-oxobutan-2-yl})-4-methyl-2-((4-methylphenyl)sulfonamido)pentanamide (8i)
Yield (0.145 g, 61%), off-white solid, M.p = 177–179°C. FTIR (KBr, cm−1): 3,333, 3,272, 3,154 (3NH), 2,962 (C-H Aliphatic), 1,682, 1,643 (2C=O, amide), 1,540, 1,495, 1,459 (C=C-Aromatic), 1,397, 1,316, 1,247 (SO2), 1,156, 1,090, 1,012 (C-N). 1H-NMR (400 MHz, DMSO) δ10.16 (s, 1H, NH of amide), 8.01 (d, J = 8.5 Hz, 1H, SO2-NH), 7.86 (d, J = 9.1 Hz, 1H, NH of amide), 7.71–7.59 (m, 4H, Ar-H), 7.39–7.34 (m, 2H, Ar-H), 7.26 (d, J = 8.0 Hz, 2H, Ar-H), 4.07 (t, J = 7.9 Hz, 1H, CH-C=O), 3.83 (td, J = 9.4, 5.2 Hz, 1H, CH-C=O), 2.27 (s, 3H, CH3-Ar), 1.88 (tt, J = 13.7, 6.8 Hz, 1H, CH-[CH3]2), 1.59 (dt, J = 18.3, 6.6 Hz, 1H, CH-[CH3]2), 1.39–1.23 (m, 2H, CH2), 0.87–0.76 (m, 9H [CH3]3), 0.72 (d, J = 6.5 Hz, 3H, CH3). 13C-NMR (126 MHz, DMSO) δ171.73, 170.36 (2C=O), 142.75, 138.63, 138.16, 129.69, 129.16, 127.09, 126.10, 121.16 (eight aromatic carbons), 58.76, 55.10, 42.44, 31.30, 24.27, 23.48, 21.68, 21.37, 19.49, 18.74 (10 aliphatic carbons). ESI-MS: m/z, 494 [M+H]+, HRMS-ESI: calcd. For C24H32N3ClO4S [M+H]+ 494.1880; Found 494. 1879.
(S)-N-({(S)-1-[(Phenyl) amino]-3-methyl-1-oxobutan-2-yl})-4-methyl-2-(4-methylphenylsulfonamido)pentanamide (8j)
Yield (0.44 g, 91%), solid, M.p = 160–161°C. FTIR (KBr, cm−1): 3,307, 3,259, 3,194 (3NH), 3,058 (C-H-aromatic) 2,962, 2,921 (C-H Aliphatic), 1,643, 1,603 (2C=O, amide), 1,541, 1,446 (C=C Aromatic), 1,380, 1,324, 1,245, 1,205 (SO2), 1,159, 1,090 (C-N). 1H-NMR (400 MHz, DMSO) δ10.01 (s, 1H, NH of amide), 7.99 (d, J = 8.5 Hz, 1H, SO2-NH), 7.87 (d, J = 9.0 Hz, 1H, NH of amide), 7.65 (d, J = 8.1 Hz, 2H, Ar-H), 7.60 (d, J = 7.8 Hz, 2H, Ar-H), 7.31 (t, J = 7.8 Hz, 2H, Ar-H), 7.26 (d, J = 8.0 Hz, 2H, Ar-H), 7.05 (t, J = 7.3 Hz, 1H, Ar-H), 4.10 (t, J = 7.8 Hz, 1H, CH-C=O), 3.83 (td, J = 9.2, 5.2 Hz, 1H, CH-C=O), 2.27 (s, 3H, CH3-Ar), 1.88 (dt, J = 13.4, 6.7 Hz, 1H, CH[CH3]2), 1.59 (dd, J = 12.5, 6.2 Hz, 1H, CH-[CH3]2), 1.39–1.23 (m, 2H, CH2), 0.80 (dd, J = 11.1, 6.3 Hz, 9H [CH3]3), 0.73 (d, J = 6.5 Hz, 3H, CH3). 13C-NMR (101 MHz, DMSO) δ171.68, 170.17 (2C=O), 142.77, 139.24, 138.61, 129.69, 129.23, 127.10, 123.84, 119.62 (eight aromatic carbons), 58.68, 55.17, 42.48, 31.41, 24.28, 23.47, 21.68, 21.36, 19.51, 18.73 (10 aliphatic carbons). ESI-MS: m/z, 460 [M+H]+, HRMS-ESI: calcd. For C24H33N3O4S [M+H]+ 460.2270; Found 460.2269.
In silico Methodology
Physicochemical Properties
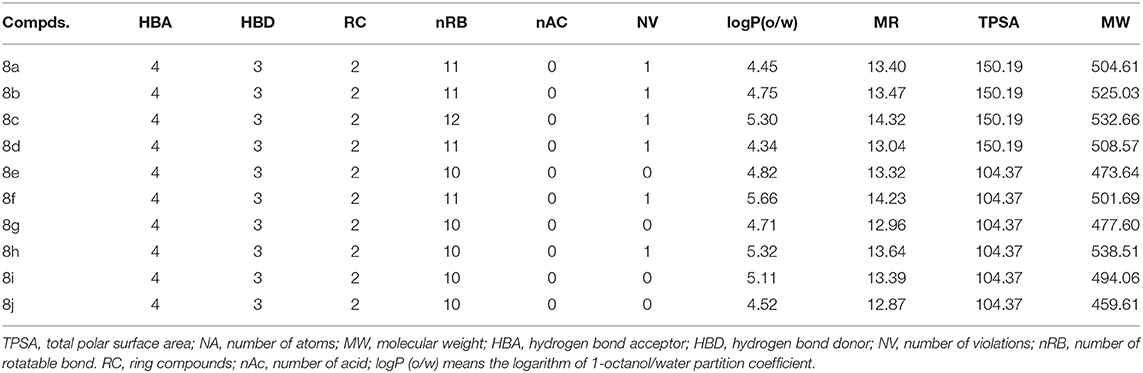
The drug-likeness of the synthesized compounds are shown in Table 1. The molecular parameters calculated include molecular weight (MW), partition coefficient (log P), hydrogen bond acceptor (HBA), hydrogen bond donor (HBD), topological polar surface area (TPSA), number of a rotatable bond (nRB), and molar refractivity (MR). The drug-likeness was determined using Lipinski's rule of five.
Molecular Docking
In this study, proteins essential for malaria and bacterial infections were evaluated. The protein targets were plasmepsin II (PDB ID: 1SME) from P. falciparum, and (PDB ID: 5MMN) for antimalarial and antimicrobial studies, respectively, were obtained from the protein data bank.
Plasmepsin II from P. falciparum has been used as a novel target for antimalarial drug development because of its role as hemoglobin-degrading enzyme (Silva et al., 1996). Targeting the GyrB/ParE ATP-binding sites is an emerging approach in discovery of resistant bacteria. These ATP-binding sites located on bacterial DNA gyrase (5MMN) and topoisomerase IV, making them potent drug targets (Panchaud et al., 2017).
The 3D crystal structures of protein targets and co-crystallized ligands were from (https://www.rcsb.org/). The co-crystallized ligands were used to validate the docking protocols by redocking them into the active binding sites of the receptors. The structures of the molecules were drawn using ChemSketch. Further purification of the protein and ligands were furnished utilizing the discovery studio to erase various chains, the water of crystallization from the protein, and reduced the energy of the structures. Discovery Studio Visualizer, v16.1.0.15350 was used to envisage the interactions of the prepared ligands into the binding cavity of the protein receptors after docking. SwissADME was used to predict the physicochemical properties of the compounds.
Antimicrobial Evaluation
The microorganisms below were isolated clinically from University of Nigeria, Nsukka at Department of Pharmaceutical Microbiology and Biotechnology laboratory. Staphylococcus aureus, Escherichia coli, Bacillus subtilis, Salmonella typhi, Candida albicans, and Aspergillus niger.
Standardization of the test organism suspension
The standardization of the microorganisms was completed using 0.5 MacFaland turbid equivalent.
Control test (standard)
The standard antibiotics used were ciprofloxacin and fluconazole.
Experimental
Into a sterile petri dish containing a suspension of the stock (4 mL, 50 mg/mL) was added double strength sterile molten agar (16.0 mL) and mixed thoroughly to obtain 1 mg/mL solution. Lower concentrations (0.1–0.9 mg/mL) were calculated from the equation C1V1= C2V2. The plates were allowed to gel and divided into seven parts. The test microbes were patterned on the plates, labeled, and kept in an incubator at 37°C for 24 h and 35°C for 48 h, respectively, for antibacterial and antifungal activities. Further incubation of the plates for 24 h at 37°C and 48 h at 25°C, respectively, used to test for bactericidal and fungicidal activities.
In vivo Anti-malaria Test
Experimental Design and Treatment of Mice
The antiplasmodial activity were assessed by adopting the methods of Okokon and Nwafor (2009) and Ezugwu et al. (2020).
Results and Discussion
Chemistry
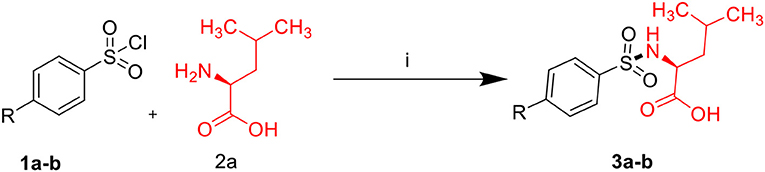
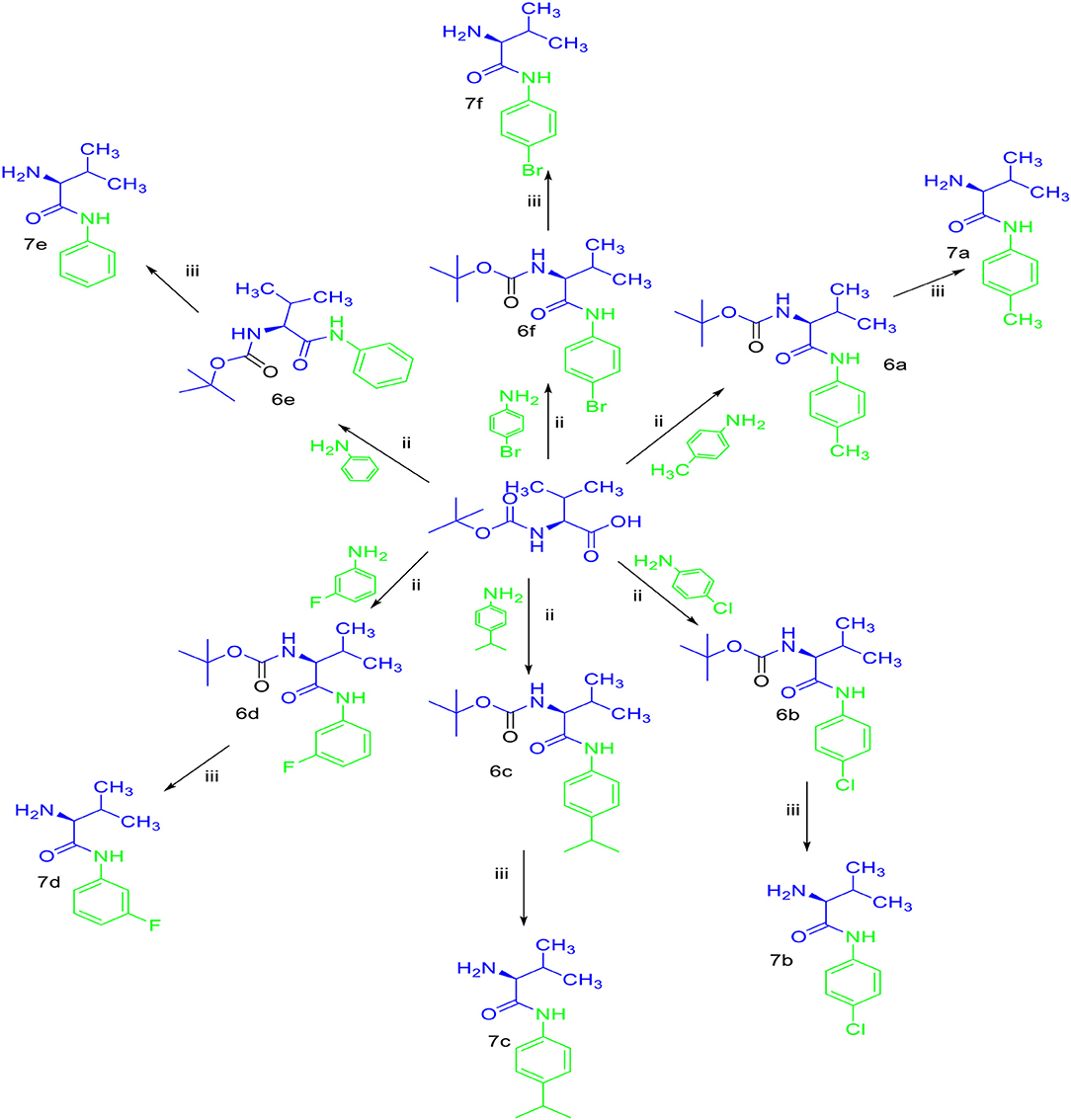
Sulfonamides and peptides are two important pharmacophores as found in the literature and as such are sorted after functionalities in the drive to combat drug resistance by organisms. The reported compounds showed good binding interaction in the active site of the target proteins as shown in Figures 1, 2. To synthesize the compounds (8a-j) we adopted the use of classical peptide coupling reagent, 1-hydroxybenzotriazole (HOBt) and 1-ethyl-3-(3′-dimethylaminopropyl) carbodiimide hydrochloride (EDC.HCl) in the amidation of compounds (7a-e) with substituted benzenesulfonamides derived from L-leucine. Also, the activation of the carboxylic acid group of leucine was enhanced with HOBt as the EDC.HCI alone could not activate the carboxylic acid functionality. The use of HOBt and EDC.HCl was also recommended to reduce the risk of racemization. In our work, we synthesized and characterized molecules containing sulfonamide, carboxamide, and Dipeptides moieties. The reaction of substituted benzenesulfonyl chloride (1a-b) with L-leucine afforded substituted benzenesulphonamoyl alkanamides (3a-b) in Scheme 1. The reaction of commercially available Boc-protected valine with substituted amines using EDC.HCl, HOBt, and triethylamine (TEA) in DCM afforded the carbamate derivatives of valine (6a-e) in Scheme 2. Compounds (7a-e) were synthesized through the reaction of compound (6a-e) with DCM/ TFA (1:1%) for 1 h, respectively. The amidation of compound (3a-b) with the TFA salt of unprotected amides (7a-e) using peptide coupling reagents EDC.HCl, HOBt, TEA, to afford the desired products (8a-j) in Scheme 3. In the Infrared (IR) spectrum of 8a, bands between ~3,312 and ~3,194 cm−1 are for N-H while ~1,642 and ~1,640 cm−1 are for the two carbonyls of amide, respectively. In the 1H-Nuclear magnetic resonance (NMR) spectrum of 8a, the methylene group of leucine displayed a multiplet at δ1.30–1.41 and –methane group of leucine and valine showed multiplet at δ3.95–4.04 due to the interactions with the near amide group protons. The typical NH resonance of the sulfonamide part of the dipeptide conjugates was detected at the δ8.43 ppm region as a doublet peak. The other two NH resonances of the diamide were observed at δ9.91 ppm and 8.12 ppm as singlet and doublet, respectively. The aromatic protons were observed at δ8.33, 8.03 pp, and 7.44 ppm, 7.09 ppm region as doublet and doublet peaks, respectively, for the eight aromatic protons. In the 13C-Nuclear magnetic resonance (NMR) spectrum, two peaks at 169.85 and 171.21 ppm for the carbonyl carbons of the amide groups, eight peaks ranging from 119.62 to 149.73 ppm for aromatic carbons, and 10 peaks ranging from 18.69 to 58.78 ppm for aliphatic carbons confirmed the formation of 8a, which was also supported by its high-resolution mass spectrometer (HRMS) peak spectrum with a peak at m/z 505.2125 for [M+H]+. All other compounds were in agreement with their structures.
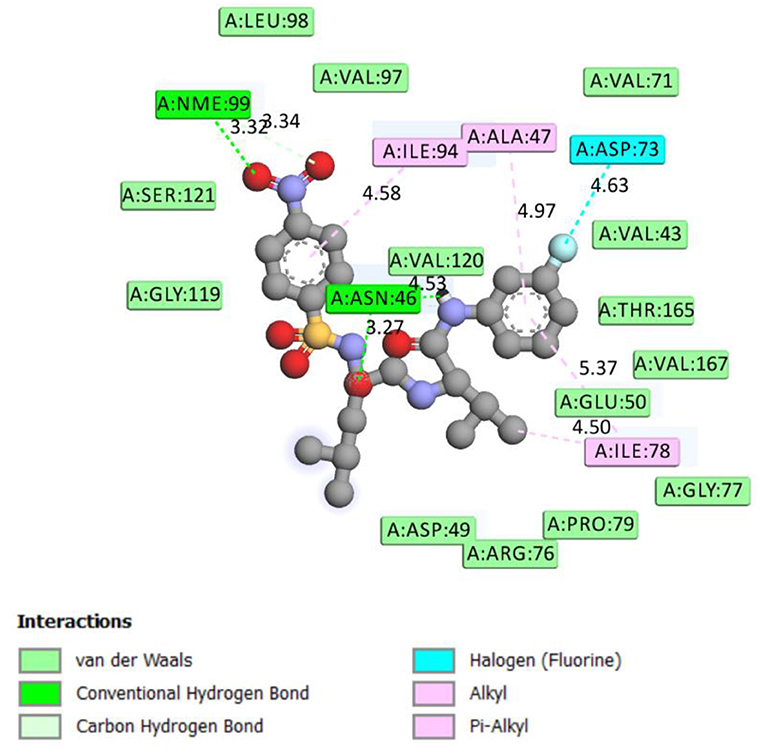
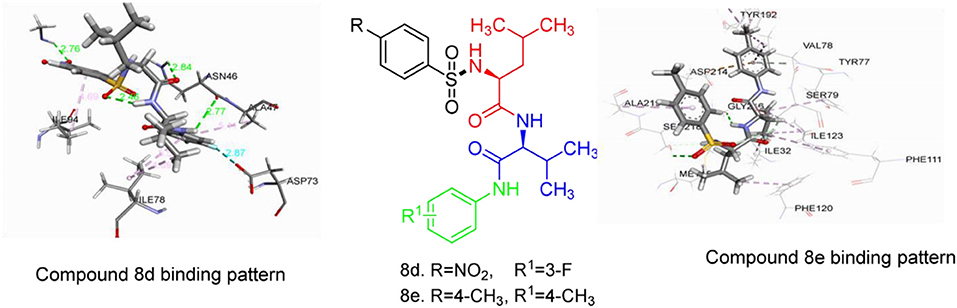
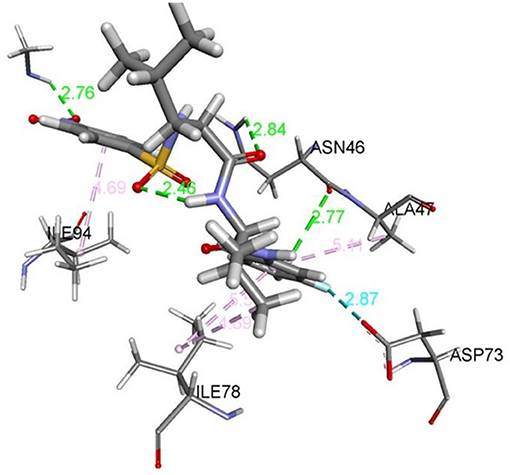
Figure 1. 2D representation of binding interaction of compound 8d and the amino acid residues of 5MMN.
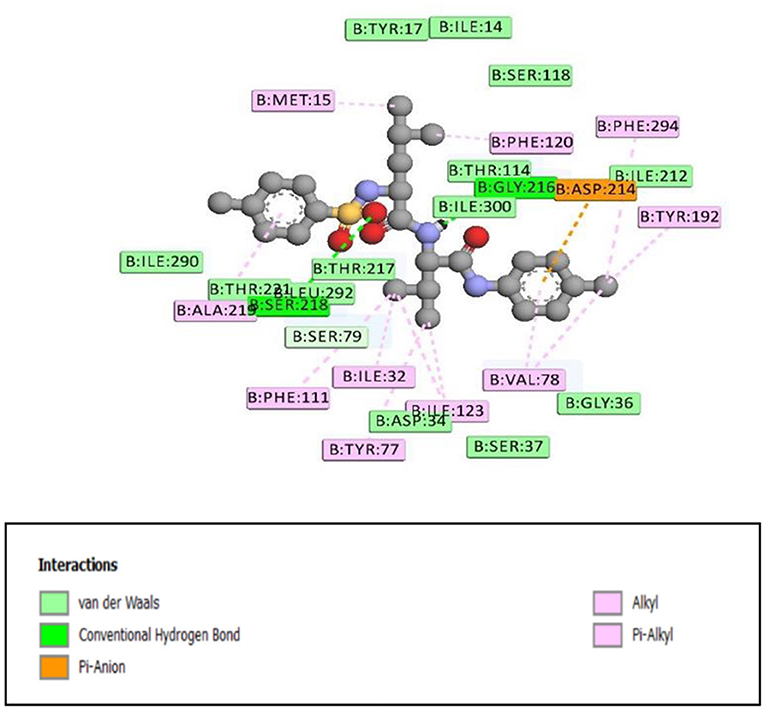
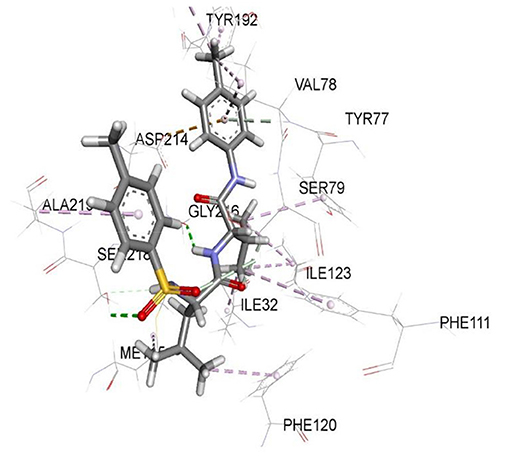
Figure 2. 2D representation of binding interaction of compound 8e and the amino acid residues of 1SME.
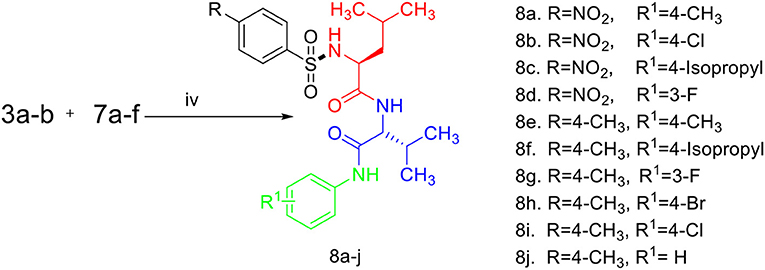
Scheme 3. Synthesis of compounds (8a-j). (i) Na2CO3, H2O, HCl, −5–0°C, r.t, 4 h. (ii) EDC.HCl, HOBt, TEA, DCM, r.t, 19–24 h. (iii) TFA/DCM (1:1%). (iv) EDC.HCI, HOBt, TEA, r.t, 19–24 h.
Physicochemical Properties Results
The pharmacokinetic properties were evaluated in the form of hydrogen bond donor ≤5, hydrogen bond acceptor ≤10, molecular weight value of ≤500, and partition coefficient (Log P) value ≤5. The results (Table 1) showed that the compounds would not pose oral bioavailability, transport, and permeability problems.
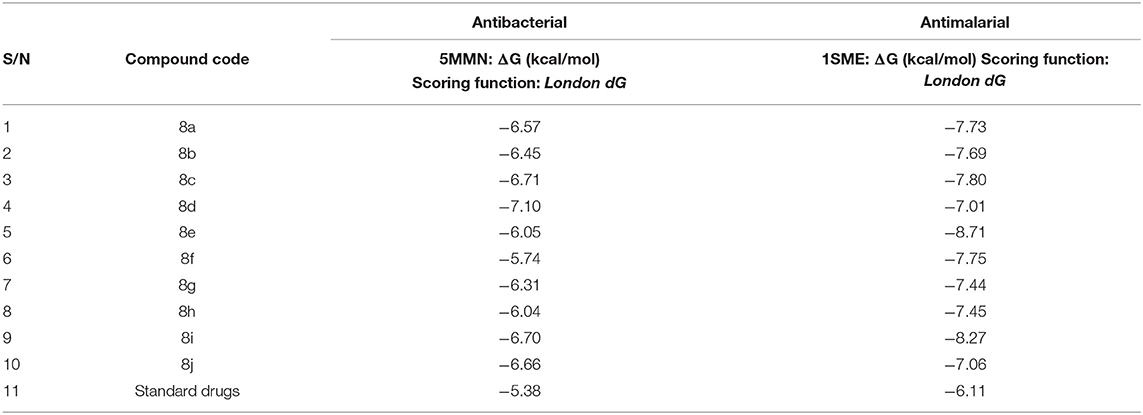
Molecular Docking Results
The data in Table 2 reveals the binding energy of the synthesized dipeptides docked into the binding sites of the 5MMN and 1SME. The binding affinities of the leu-val dipeptides with the receptors were significant when compared to the ciprofloxacin and Chloroquine, respectively. Compounds 8d and 8e exhibited similar in silico antibacterial (−7.10 kcal/mol) and in silico antimalarial activity (−8.71 kcal/mol) as the standard (ciprofloxacin) (−5.38 kcal/mol) and Chloroquine (−6.11 kcal/mol), respectively. We moved extra to gain perception into the character of the binding interactions between the compounds and the receptors. Figures 3, 4 shows the stereo view of compounds 8d and 8e in the binding cavity of 5MMN and 1SME, while Figures 1, 2 illustrate how the atoms of compounds 8d and 8e interacted with the amino acid residues of 5MMN and 1SME, respectively. The binding interaction of 8d with 5MMN in the active site is shown in Figures 1, 3. The O-atom of 8d interacts with HD22 ASN 46 at an intermolecular distance of 2.84 Å. Another O-atoms interacted with H of NME 99 (2.76 Å), HA of ASN 46 at 2.70 Å, and with HH31 of NME 99 at an intermolecular distance of 2.84 Å. There is hydrogen bond interaction with O-atom of 8d at 2.46 Å, while another H-atom interacted with O of ASN 46 (2.77 Å). There was an F-atom of compound 8d interaction with OD2 of ASP 73 (2.87 Å).
There were five hydrogen bonds formed in this interaction. In Figure 2, different atoms of compound 8e interacted with the ISME receptor. O-atom of compound 8e interacted with HG of SER 218, GLY 216, HB2 of SER 79, and HB3 of SER 79 at an intermolecular distance of 2.79, 2.46, 3.07, and 2.55 Å, respectively, through hydrogen bond interaction. Also, compound 8e through pi donor hydrogen bond interaction had contact with H VAL 78 at a distance of 3.21 Å. Other hydrophobic interactions with 8d, 8e, and conserved amino acid residues were also shown in Figures 1, 2, respectively.
Table 3 further validated the docking protocols and provided insights on the nature of the binding interactions of both the native ligands and the hit molecules with the amino acid residues in the binding sites of the receptors. Here the major amino acid residues involved in the interactions were considered. In 1SME receptor, both the native ligand (pepstatin) and compound 8e interacted with almost the same amino acide residues in its binding site. For example, pepstatin and 8e interacted with GLY 216 through H-bonding at distances of 3.75 and 2.46 Å, respectively. This further shows that the compound 8e occupied the same binding site as the native ligand, thereby validating the docking. We also noted that 8e was closer to interact with GLY 216 than the native ligand. This probably could have resulted in the higher binding affinity of 8e. This trend was also observed with most of the amino acid residues interacting with the molecules.
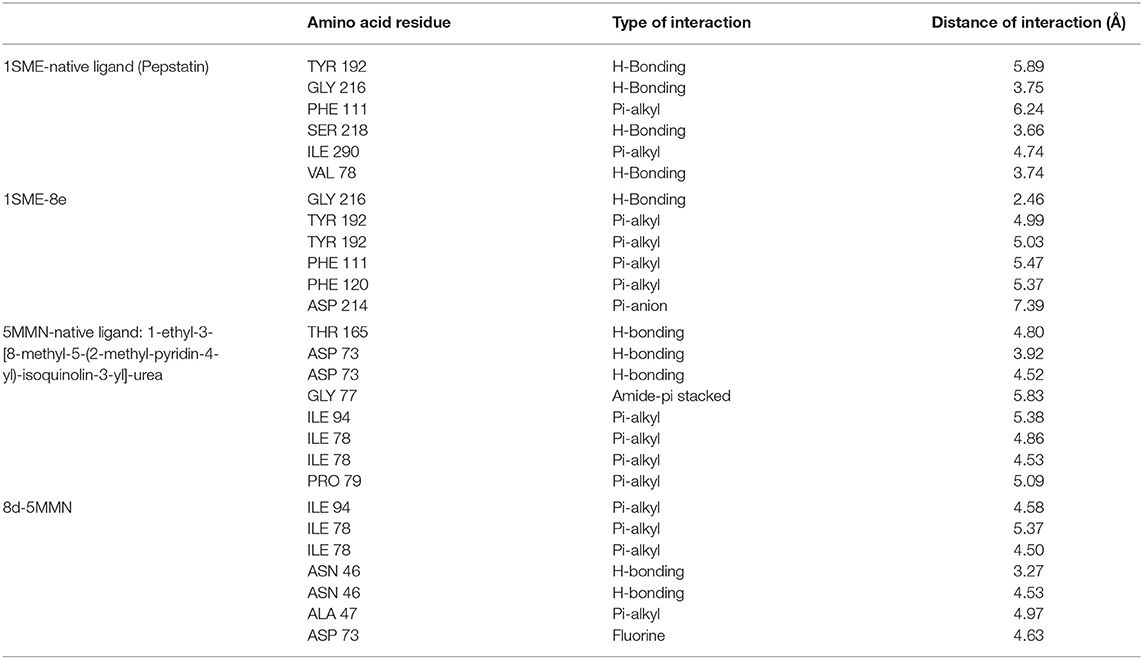
Table 3. Comparison of binding interactions of the native ligands and hit molecule (8e) to the binding sites of 1SME and 5MMN.
In vitro Antimicrobial Activities
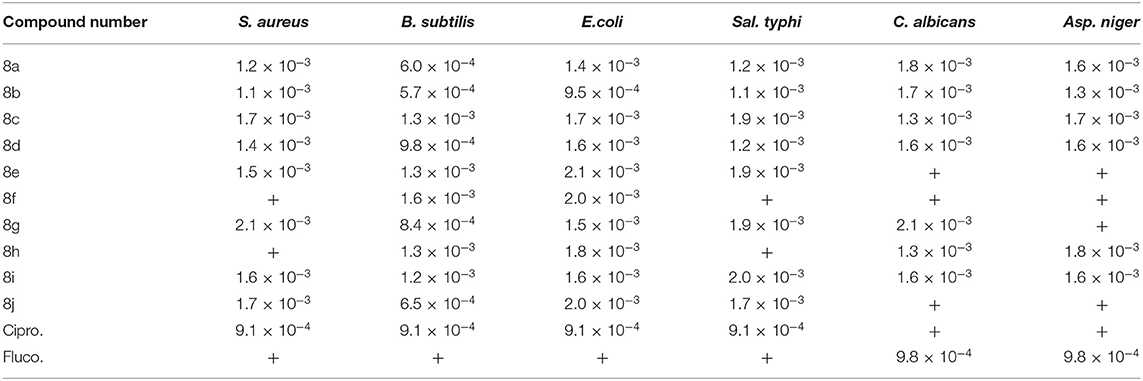
Table 4 comprises of minimum inhibitory concentration (MIC) of the synthesized compounds. The in vitro antibacterial properties of the compounds and Ciprofloxacin were evaluated against (staphylococcus aureus and Bacillus subtilis) and (Salmonella typhi and Escherichia coli) by the agar dilution method as Gram-positive and Gram-negative organisms, respectively. The in vitro antifungal properties of leu-val dipeptides synthesized were deduced by the agar dilution method against two fungal strains (Candida albicans and Aspergillus niger) and fluconazole as reference drug. The figures in Table 4 revealed that compound 8a with MIC value of 1.2 × 10−3 M displayed comparable anti-bacterial activity against S. aureus to standard (9.1 × 10−4 M). Compounds 8a, 8b, 8g, and 8j with MIC values ranging from (5.7 × 10−4M−8.4 × 10−4 M) have the same or comparable activity with the standard drug (9.1 × 10−4 M) against B. subtilis. All the compounds showed inhibition against these two Gram-positive organisms except compounds 8f and 8h, which are resistant to S. aureus. For the Gram-negative bacteria, it was revealed compounds 8b with MIC value of 9.5 × 10−4 M showed activity against E. coli. All other compounds inhibits the growth of E. coli though less than the standard.
The data in Table 4 revealed that the synthesized compounds showed good activity against S. typhi though less than the standard except for compounds 8f and 8h that are resistant to S. typhi. Further evaluation of the data revealed that compounds 8a and 8b are more potent as antibacterial agents when compared with other synthesized leu-val dipeptides derivatives. More so, it was revealed that the synthesized compounds showed better activity against C. albicans and A. niger though less active when compared with fluconazole except for compounds 8e, 8f, and 8j that do not have any inhibition against C. albicans and compounds 8e, 8f, 8g, and 8j that are resistant against A. niger.
In vivo Antimalarial
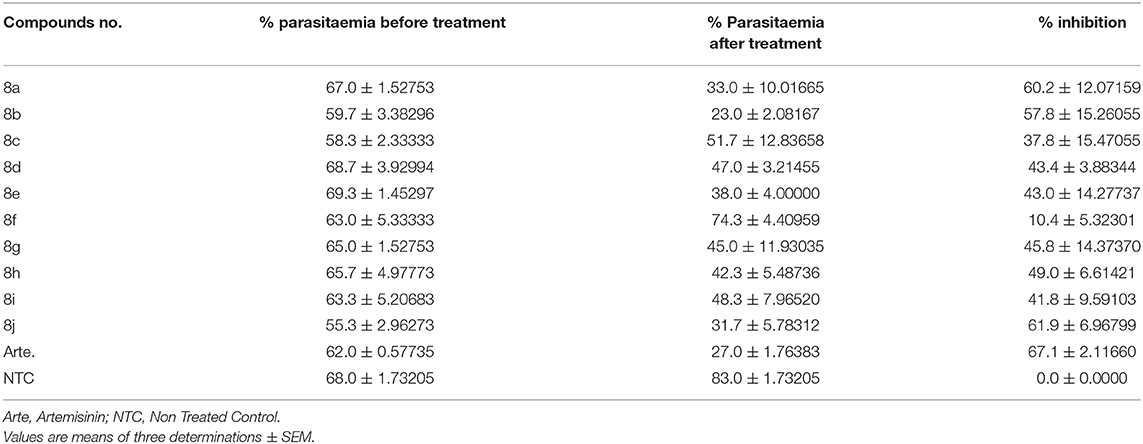
The leu-val dipeptides synthesized were evaluated for in vivo antimalarial activity against P. berghei NK (65 Strain) poison mice. The animal ethics committee, Veterinary Medicine Department, University of Nigeria, Nsukka gave permission and approval for the use of animals in this experiment (PG/PhD/16/80697). The percentage inhibition of parasite was calculated from the equation [(A-B)/A] × 100 (Ugwuja et al., 2019; Ezugwu et al., 2020); where A = parasitemia of the untreated group and B = parasitemia of the tested group. In this study, the compounds with the percentage inhibition below 30 are inactive, 30–40 are partially active, and 40 and above are regarded to be active. From Table 5 it was revealed that compounds 8a, 8b, 8d, 8e, 8g, 8h, 8i, and 8j which have 43.30–61.90% inhibition were active in comparable with the standard drug (with 67% inhibition). The analysis evaluation of the compounds synthesized revealed that compound 8j (61.90%) was the most potent antimalarial when compared with others, looking at the structure-activity relationship on the 4-nitrophenylsulfonamide hybrids (8a-d). The effect of 4-methyl, 4-chloro, 4-Isopropyl, and 3-fluoro substituent on the N-phenylacetamide was studied, and it was revealed that 4-methyl-N-phenylacetamide derivative (8a, 60.2%) was the most potent inhibitor, for P. berghei followed by 8b (57.8%) and 8d (43.4%), and 8c were considered moderately active. The effects of 4-methyl, 4-bromo, 4-chloro, 4-Isopropyl, and 3-fluoro substituent on the N-phenylacetamide among p-methylbenzenesulfonamide hybrids (8d-j) revealed that compounds 8j and 8h with 61.90 and 49.80% inhibition, respectively, were more active.
Conclusion
In this paper, we have described an approach to obtain leu-val based dipeptide derivatives that were tested for their antimalarial and antimicrobial properties. The results obtained showed that among the leu-val dipeptides synthesized, compound 8j was more active against P. berghei. Compound 8b was the most to inhibit growth of E. coli, compound 8a and 8b were most active against S. aureus, compounds 8a, 8b, and 8d were most active against S. typhi, B. subtilis is inhibited most with compounds 8a, 8b, and 8j. Compounds 8c, 8h, and 8b were the most active synthesized compounds against C. albicans and A. niger, respectively.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee, Veterinary Medicine Department, University of Nigeria, Nsukka.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by CSIR-TWAS, Postgraduate award, Grant/Award Number: 22/FF/CSIR-TWAS/2017.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors express their sincere gratitude to The World Academy of Science (TWAS) and Council for Science and Industrial Research (CSIR), New Delhi for the joint sponsorship of the experimental work at CSIR-India Institute of Chemical Technology, Hyderabad, Telangana, India under the 2017 CSIR-TWAS Postgraduate Fellowship Award, Department of Veterinary Anatomy, Faculty of Veterinary Medicine, University of Nigeria, Nsukka for the in vivo antimalarial analysis, Department of Pharmaceutical and Medicinal Chemistry, University of Nigeria, Nsukka for in vitro antimicrobial studies.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2020.583926/full#supplementary-material
References
Abdelaziz, A. M., Yu, M., Li, P., Zhong, L., Singab, A. N. B., Hanna, A. G., et al. (2015). Synthesis and evaluation of 5-chloro-2-methoxy-N-(4-sulphamoylphenyl) benzamide derivatives as anti-cancer agents. Med. Chem. 5, 253–260. doi: 10.4172/2161-0444.1000272
Amit, M., Rahul, P. G., Sunil, B., Nathan, E. G., Ben, M. D., Abhay, T. S., et al. (2015). Antiplasmodial activity of short peptide-based compounds. RSC Adv. 5:22674. doi: 10.1039/C5RA00779H
Day, T., and Greenfield, S. A. (2004). Bioactivity of a peptide derived from acetylcholinesterase in hippocampal organotypic cultures. Exp. Brain Res. 155:500. doi: 10.1007/s00221-003-1757-1
Deniz, Ü. P., Zehra Küçükbay, F., Küçükbay, H., Angeli, A., and Supuran, C. T. (2019). Synthesis carbonic anhydrase enzyme inhibition and antioxidant activity of novel benzothiazole derivatives incorporating glycine, methionine, alanine, and phenylalanine moieties. J. Enzyme Inhib. Med. Chem. 34:343349. doi: 10.1080/14756366.2018.1553040
Eze, F. U., Okoro, U. C., Ugwu, D. I, and Okafor, S. N. (2019). Biological activity evaluation of some new benzenesulphonamide derivatives. Front. Chem. 7:634. doi: 10.3389/fchem.2019.00634
Ezugwu, J. A., Okoro, U. C, Ezeokonkwo, M. A, Bhimapaka, C., Okafor, S. N., Ugwu, D. I., et al. (2020). Synthesis and biological evaluation of Val–Val dipeptide–sulfonamide conjugates. Arch. Pharm. 353:e2000074. doi: 10.1002/ardp.202000074
Jatinder, P. K. G., Singh, S., and Sethi, N. (2015). Synthesis, characterization and antimicrobial activity ofprotected dipeptides and their deprotected analogs. Orient. J. Chem. 31, 417–421. doi: 10.13005/ojc/310149
Jiao, Z. G., He, H. Q., Zeng, C. C., Tan, J. J., Hu, L. M., and Wang, C. X. (2010). Design, synthesis and anti-HIV integrase evaluation of N-(5-chloro-8-hydroxy-2Styrylquinolin-7-yl) benzenesulfonamide derivatives. Molecules 15, 1903–1917. doi: 10.3390/molecules15031903
Jyothi, B., and Madhavi, N. (2018). Synthesis and biological screening of pyrimidine linked benzene sulfonamide derivatives. Int. J. Pharm. Sci. Res. 9, 5534–5543. doi: 10.13040/IJPSR.0975-8232
Kayser, H., and Meisel, H. (1996). Stimulation of human peripheral blood lymphocytes by bioactive peptides derived from bovine milk proteins. FEBS Lett. 383, 18–20. doi: 10.1016/0014-5793(96)00207-4
Khavinson, V. K., and Anisimov, V. N. (2000). Synthetic dipeptide vilon (LLys-L-Glu) increases life span and inhibits a development of spontaneous tumors in mice. Doklady Akad. Nauk. 372, 421–423.
Kilicaslan, S., Arslan, M., Ruya, Z., Bilen, C., Adem, E., Nahit, G., et al. (2016). Synthesis and evaluation of sulfonamide-bearing thiazole as carbonic anhydrase isoforms hCA I and hCA II. J. Enzyme Inhib. Med. Chem. 31, 1300–1305. doi: 10.3109/14756366.2015.1128426
Kitts, D. D., and Weiler, K. (2003). Bioactive proteins and peptides from food sources. Applications of bioprocesses used in isolation and recovery. Curr. Pharm. Design 9, 1309–1323. doi: 10.2174/1381612033454883
Küçükbay, H., Bugday, N., Küçükbay, F. Z., Berrino, E., Bartolucci, G., Del Prete, S., et al. (2019). Synthesis and carbonic anhydrase inhibitory properties of novel 4-(2-aminoethyl)benzenesulphonamide-dipeptide conjugates. Bioorg. Chem. 83, 414–423. doi: 10.1016/j.bioorg.2018.11.003
Kumar, V., Mudgal, M. M., Rani, N., Jha, A., Jaggi, M., Singh, A. T., et al. (2009). Synthesis of functionalized amino acid derivatives as new pharmacophores for designing anticancer agents. J. Enzyme Inhib. Med. Chem. 24, 763–770. doi: 10.1080/14756360802362975
Maloy Kumar, P., Panda, G., Srivastava, K., and Puri, S. K. (2018). Design, synthesis and antimalarial activity of benzene and isoquinoline sulfonamide derivatives. Bioorg. Med. Chem. Lett. 18, 776–781. doi: 10.1016/j.bmcl.2007.11.038
Montalbetti, C. A. G. N., and Falque, V. (2005). Amide bond formation and peptide coupling. Tetrahedron 61:10852. doi: 10.1016/j.tet.2005.08.031
Nitta, A., Nishioka, H., Fukumitsu, H., Furukawa, Y., Sugiura, H., Shen, L., et al. (2004). Hydrophobic dipeptide Leu-Ile protects against neuronal death by inducing brainderived neurotrophic factor and glial cell line-derived neurotrophic factor synthesis. J. Neurosci. Res. 78, 250–258. doi: 10.1002/jnr.20258
Panchaud, P., Bruyère, T., Blumstein, A. C., Bur, D., Chambovey, A., Ertel, E. A., et al. (2017). Discovery and optimization of isoquinoline ethyl ureas as antibacterial agents. J. Med. Chem. 60, 3755–3775. doi: 10.1021/acs.jmedchem.6b01834
Panda, S. S., Ibrahim, M. A., Küçükbay, H., Meyers, M. J., Sverdrup, F. M., El-Feky, S. A., et al. (2013). Synthesis and antimalarial bioassay of quinine – peptide conjugates. Chem. Biol. Drug Des. 82:361. doi: 10.1111/cbdd.12134
Qadir, M. A., Ahmed, M., Aslam, H., Waseem, S., and Shafiq, M. I. (2015). Amidine sulfonamides and benzene sulfonamides: synthesis and their biological evaluation. J. Chem. 15:8. doi: 10.1155/2015/524056
Qi, Z., Verma, R., Gehring, C., Yamaguchi, Y., Zhao, Y., Ryan, C. A., et al. (2010). Ca2þ signaling by plant Arabidopsis thaliana pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMPactivated Ca2þ channels. Proc. Natl. Acad. Sci. U.S.A. 107:21193 doi: 10.1073/pnas.1000191107
Silva, A. M., Lee, A. Y., Gulnik, S. V., Maier, P., Collins, J., Bhat, T. N., et al. (1996). Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 93:10034. doi: 10.1073/pnas.93.19.10034
Thompson, A. A., Liu, W., Chun, E., Katritch, V., Wu, H., Vardy, E., et al. (2012). Structure of the nociceptin/ orphanin FQ receptor in complex with a peptide mimetic. Nature 485:495. doi: 10.1038/nature11085
Ugwu, D. I., Ezema, B. E., Eze, F. U., and Ugwuja, D. I. (2014). Synthesis and structural activity relationship study of antitubercularcarboxamides. Int. J. Med. Chem. 2014, 1–18. doi: 10.1155/2014/614808
Ugwu, D. I., Okoro, U. C., and Mishra, N. K. (2018a). Synthesis, characterization and anthelmintic activity evaluation of pyrimidine derivatives bearing carboxamide and sulphonamide moieties. J. Serbian Chem. Soc. 83, 401–409. doi: 10.2298/JSC170127109U
Ugwu, D. I., Okoro, U. C., and Mishra, N. K. (2018b). Synthesis, characterization and in vivo antitrypanosomal activities of new carboxamides bearing quinoline moiety. PLoS ONE 13:e0191234. doi: 10.1371/journal.pone.0191234
Ugwu, D. I., Okoro, U. C., Ukoha, P. O., Gupta, A., and Okafor, S. N. (2018c). Novel anti-inflammatory and analgesic agents: synthesis, molecular docking and in vivo studies. J. Enzyme Inhib. Med. Chem. 33, 405–415. doi: 10.1080/14756366.2018.1426573
Ugwuja, D. I., Okoro, U. C., Soman, S. S., Soni, R., Okafor, S. N., Ugw, D. I., et al. (2019). New peptide derived antimalaria and antimicrobial agents bearing sulphonamide moiety. J. Enzyme Inhib. Med. Chem. 34, 1388–1399. doi: 10.1080/14756366.2019.1651313
Keywords: antimalarial, antimicrobial, benzenesulfonamide, Leu-Val dipeptide, in silico studies
Citation: Ezugwu JA, Okoro UC, Ezeokonkwo MA, Bhimapaka CR, Okafor SN, Ugwu DI, Ekoh OC and Attah SI (2020) Novel Leu-Val Based Dipeptide as Antimicrobial and Antimalarial Agents: Synthesis and Molecular Docking. Front. Chem. 8:583926. doi: 10.3389/fchem.2020.583926
Received: 15 July 2020; Accepted: 12 October 2020;
Published: 24 November 2020.
Edited by:
Simona Rapposelli, University of Pisa, ItalyReviewed by:
Leonardo L. G. Ferreira, University of São Paulo, BrazilMarcus Scotti, Federal University of Paraíba, Brazil
Copyright © 2020 Ezugwu, Okoro, Ezeokonkwo, Bhimapaka, Okafor, Ugwu, Ekoh and Attah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James A. Ezugwu, amFtZXMuZXp1Z3d1QHVubi5lZHUubmc=
 James A. Ezugwu
James A. Ezugwu Uchechukwu C. Okoro1
Uchechukwu C. Okoro1 Mercy A. Ezeokonkwo
Mercy A. Ezeokonkwo Sunday N. Okafor
Sunday N. Okafor David I. Ugwu
David I. Ugwu