- 1Laboratory of Applied Chemistry and Environment (LCAE), Faculty of Sciences, University Mohammed Premier, Oujda, Morocco
- 2Laboratory of Biochemistry (LB), Department of Biology, Faculty of Sciences, University Mohamed Premier, Oujda, Morocco
- 3Centre Régional des Métiers de l'Education et de Formation Oujda, Oriental, Morocco
- 4Department of Chemistry, College of Science, King Saud University, Riyadh, Saudi Arabia
- 5Department of Chemistry, College of Science, Ibb University, Ibb, Yemen
- 6Department of Chemistry, Science College, An-Najah National University, Nablus, Palestine
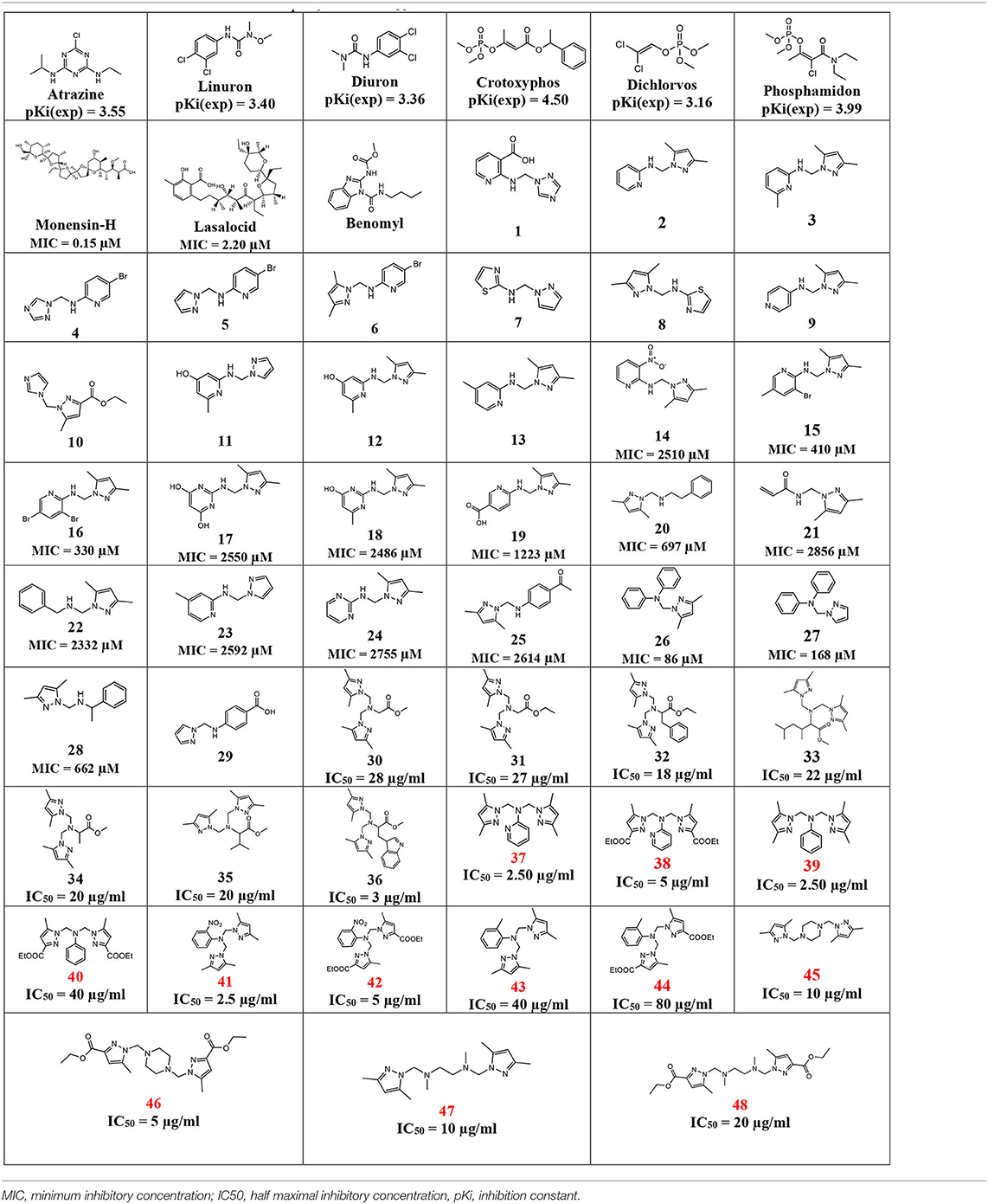
Twelve recent compounds, incorporating several heterocyclic moieties such as pyrazole, thiazole, triazole, and benzotriazole, made in excellent yield up to 37–99.6%. They were tested against Fusarium oxysporum f. sp. albedinis fungi (Bayoud disease), where the best results are for compounds 2, 4, and 5 with IC50 = 18.8–54.4 μg/mL. Density functional theory (DFT) study presented their molecular reactivity, while the docking simulations to describe the synergies between the trained compounds of dataset containing all the tested compounds (57 molecules) and F. oxysporum phytase domain (Fophy) enzyme as biological target. By comparing the results of the docking studies for the Fophy protein, it is found that compound 5 has the best affinity followed by compounds 2 and 4, so there is good agreement with the experimental results where their IC50 values are in the following order: 74.28 (5) < 150 (2) < 214.10 (4), using Blind docking/virtual screening of the homology modeled protein and two different tools as Autodock Vina and Dockthor web tool that gave us predicted sites for further antifungal drug design.
Introduction
Fusarium oxysporum f. sp. albedinis (F.o.a.) displays the leading dangerous agent among all pathogens of date palm plant, notably in North Africa (Freeman and Maymon, 2000). Thus, infections appear in the vascular wilt of Phoenix dactylifera. It is also called Bayoud disease, which is frequently fatal and kills plants in 6 months to 2 years. In the horticultural field, F. oxysporum is one of the important fungus organisms raised in cultivated lands. It makes up 40–70% of the entire fusarial flora. It is represented by several diversified forms in terms of morphology and physiology. These forms are saprophytes or parasites of many plants and represent various levels of virulence. Bayoud disease has destroyed more than 15 million Moroccan and Algerian P. dactylifera trees (Diana et al., 1995; Hakkou et al., 2004). Unfortunately, to this time, no remedial treatment exists against this fungus, except some limited methods such as land disinfection (Thangavelu and Gopi, 2015), resistant strains propagation (Joshi, 2018), and practice, which have a significance because they reduce this disease's impact. One of the important five-membered hetero atomic rings, where nitrogen and sulfur are separated by one carbon, is 1,3-thiazole, prepared by original strategies. It is a pharmacophore, a privileged scaffold, in many compounds with several biological activities (Reis et al., 2011; Fadda et al., 2012; Alegaon et al., 2014; Bekhit et al., 2015; Varghese et al., 2016; El-Naggar and Abdel-Mottaleb, 2017; El-Sayed and Ismail, 2019; Nayak and Gaonkar, 2019; Pricopie et al., 2019). Pyrazole is a five-membered heterocyclic with two adjacent nitrogen atoms, common in a variety of commercial compounds applied in many industrial fields (Pongor et al., 2004; Elayyachy et al., 2005; Bouabdallah et al., 2006; Dawood and Abdel-Wahab, 2012; Abrigach et al., 2014; Singh et al., 2014, 2016; Akhtar et al., 2017). Another five-membered heterocyclic is 1,2,4-triazole; has three nitrogen atoms at positions 1, 2, and 4 of the ring; and used as a pharmacophore core linked to other compounds, offering different pharmacological activities (Touzani et al., 2003; Sahu et al., 2013; Barbuceanu et al., 2014; Cetin and Gecibesler, 2015; Elbelghiti et al., 2016; Shaikh et al., 2016; Singh et al., 2016; Dalloul et al., 2017). The pyridine may be a six-membered heterocyclic containing merely one nitrogen atom and again documented for several applications (Fadda et al., 2012; Hu et al., 2014; Sun et al., 2014; El-Naggar and Abdel-Mottaleb, 2017; Wei et al., 2019). The pyrimidine is a six-membered heterocyclic having two nitrogen atoms at positions 1 and 3 of the ring, and it has been of significant interest in many applications (Gatta et al., 1990; Iaroshenko et al., 2011; El-Adasy, 2017; Thangarasu et al., 2019). However, because of insufficient information on the pathogenesis of the F.o.a. fungus, numerous computational approaches have usually used to understand further the mechanism of action for this disease antifungal. In this background, docking simulation (Prabhudeva et al., 2019) remains one of the most powerful tools to give an atomistic insight into molecular recognition by predicting the strength of molecule protein–binding modes. The chosen target is the F. oxysporum phytase domain (Fophy) enzyme, a protein that plays versatile roles in agricultural and feeding fields. It catalyzes the degradation of phytate (an essential constituent of grains, cereals, and oilseeds) into inorganic phosphorus and myoinositol phosphate derivatives. Inhibition of the Fophy enzyme can affect the expansion of the fungus indirectly by preventing the phytate degradation, well-established as a robust chelating agent readily binding to covalent metal ions and renders them insoluble and therefore unavailable for absorption. These characteristics made both these proteins prospective potential targets to develop new anti-F. oxysporum inhibitors (Gontia-Mishra et al., 2014). And keeping in mind the biological significance of heterocyclic ligands, we described the synthesis of new heterocyclic compounds, used as potent antimicrobial agents, in our study. A molecular docking approach was used for the best antifungal derivatives against Fophy. The structure of this approach was constructed using the homology model that has been previously reported in the literature (Soundararajan et al., 2011; Abrigach et al., 2014; Tighadouini et al., 2019; Toubi et al., 2019), to achieve better insight into the ligand–receptor binding interactions and direct future synthesis. In case that there is no cofactor, blind docking, and virtual screening are used in this study for site prediction and protocol validation using Autodock Vina (Seeliger and de Groot, 2010) and Dockthor (Santos et al., 2020).
Materials and Methods
Chemicals and Instruments
All the chemicals used in this study were of analytical grades (Aldrich, purity >99%). Melting points were measured with Koffler bank and the FTIR analysis with the FTIR-8400S spectrometer using KBr pellets. We recorded the 1H and 13C nuclear magnetic resonance (NMR) spectra on AVANCE 300, 400, and 500 MHz from BRUKER. The in vitro anti-Fusarium activity was tested by the agar diffusion technique.
Synthesis of the Pyrazole and Triazole
General Procedure for Preparing Compounds 1–12
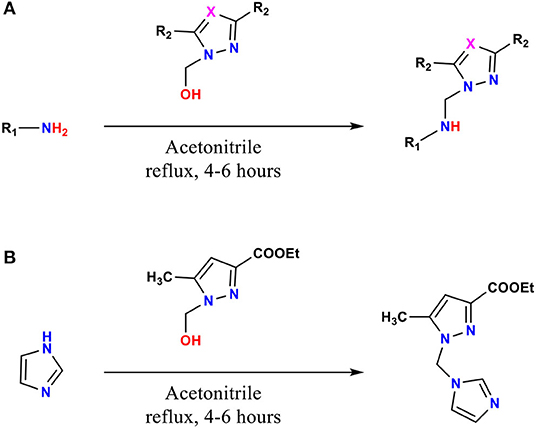
The ligands 1–9, 11, and 12 were prepared by condensation of different monoamines with pyrazole or 1,2,4-triazole methanol derivatives (Pathway A, Figure 1), whereas 10 were prepared by condensation of imidazole with ethyl 1-(hydroxymethyl)-5-methyl-1H-pyrazole-3-carboxylate (Pathway B, Figure 1), according to the method described in the literature (Kaddouri et al., 2017, 2019, 2020).
2-(((1H-1,2,4-triazol-1-yl)methyl)amino) nicotinic acid, 1: Viscous product; 1H NMR [500 MHz, dimethyl sulfoxide (DMSO)-d6]: δ 8.33 (m, 1H), 7.09 (s, 4H), 4.94 (s, 2H); 13C NMR (125 MHz, DMSO-d6): δ 168.54, 159.67, 151.10, 144.17, 140.26, 113.46, 111.81, 54.
N-((3,5-dimethyl-1H-pyrazol-1-yl) methyl)pyridin-2-amine, 2: Yield, 89.85%; mp 122–124°C; 1H NMR (500 MHz, CDCl3): δ 8.03 (d, J = 5.1 Hz, 1H), 7.33–7.29 (m, 1H), 6.56 (d, J = 7.2 Hz, 1H), 6.46 (d, J = 8.3 Hz, 1H), 5.51 (s, 1H), 5.48 (s, 1H), 2.34 (s, 3H), 2.13 (s, 1H), 2.10 (s, 3H); 13C NMR (125 MHz, CDCl3): δ 156.53, 148.18, 147.39, 139.70, 137.59, 114.31, 109.06, 105.29, 54.42, 13.49, 11.12; CG-MS: m/z (%) = 281.2 [M+DMSO] + [calcd. for C11H14N4 [M+DMSO] + 280.26].
N-((3,5-dimethyl-1H-pyrazol-1-yl) methyl)-6-methylpyridin-2-amine, 3: Yield, 30%; mp: 136°C−138°C; 1H NMR (500 MHz, CD2Cl2): δ 7.42 (t, J = 10 Hz, 1H), 7.21 (dd, J = 10 Hz, 1H), 6.40 (d, J = 5 Hz, 1H), 6.25 (d, J = 10 Hz, 1H), 5.64 (s, 1H), 5.43 (s, 2H), 2.36 (s, 3H), 2.27 (s, 3H), 2.06 (s, 3H), 13C NMR (125 MHz, CD2Cl2) δ: 157.01, 155.95, 146.57, 139.51, 137.98, 112.49, 105.95, 104.97, 53.79, 24.39, 13.78, 11.17; CG-MS: m/z (%) = 281.2 [M+ACN+Na]+ [calcd. for C12H16N4 [M+ACN+Na]+ 280.29].
N-((1H-1,2,4-triazol-1-yl)methyl)-5-bromopyridin-2-amine, 4: Yield, 62.14%; mp: 64–66°C; 1H NMR (500 MHz, DMSO-d6): δ 8.56 (s, 1H), 8.15 (s, 1H), 7.95 (s, 1H), 6.64 (d, J = 8.9 Hz, 1H), 5.67 (d, J = 6.9, 1H); 13C NMR (125 MHz, DMSO-d6) δ: 155.68, 151.11, 147.63, 144.07, 139.78, 111.06, 107.64, 54.55; CG-MS: m/z (%) = 370.9 [M+2ACN+CH3OH+2H]+ [calcd. for C8H8BrN5 [M+2ACN+CH3OH+2H]+ 370.09].
N-((1H-pyrazol-1-yl)methyl)-5-bromopyridin-2-amine, 5 (Abrigach et al., 2014): Yield, 62.36%; mp: 114–116°C; 1H NMR (500 MHz, DMSO-d6): δ 8.14 (s, 1H), 7.98 (t, J = 6.9 Hz, 1H), 7.78 (d, J = 2.3 Hz, 1H), 7.63 (d, J = 8.9 Hz, 1H), 7.44 (d, J = 1.8 Hz, 1H), 6.62 (d, J = 8.9 Hz, 1H), 6.22 (dd, J = 2.3 Hz, 1H), 5.60 (d, J = 6.9 Hz, 2H); 13C NMR (125 MHz, DMSO-d6): δ 156.11, 147.63, 139.62, 138.55, 129.47, 110.78, 107.16, 105.16, 56.44; CG-MS: m/z (%) = 372.8 [M+DMSO+K+2H]+ [calcd. for C9H9BrN4 [M+DMSO+K+2H]+ 372.1].
5-Bromo-N-((3,5-dimethyl-1H-pyrazol-1-yl)methyl) pyridin-2-amine, 6 (Abrigach et al., 2014): Yield, 93.6%; mp: 150–152°C; 1H NMR (500 MHz, DMSO-d6): δ 8.11 (s, 1H), 7.84 (t, J = 6.6 Hz, 1H), 7.60 (d, J = 8.9 Hz, 1H), 6.64 (d, J = 8.9 Hz, 1H), 5.76 (s, 1H), 5.42 (d, J = 6.6 Hz, 2H), 2.56 (s, 3H), 2.34 (s, 3H); 13C NMR (125 MHz, DMSO-d6): δ 156.06, 147.44, 139.47, 110.66, 101.75, 53.42, 13.29, 10.67; CG-MS: m/z (%) = 281.0 [M]+ [calcd. for C11H13BrN4 [M]+ 281.16].
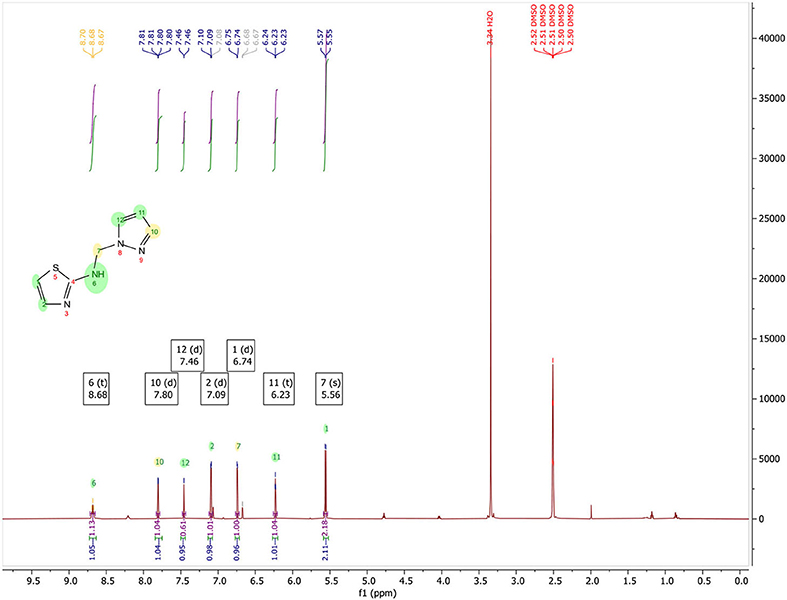
N-((1H-pyrazol-1-yl)methyl)thiazol-2-amine, (Figure 3): Yield, 53.08%; mp: 108–110°C; 1H NMR (500 MHz, DMSO-d6) in Figure 4: δ 8.68 (t, J = 6.7 Hz, 1H), 7.80 (d, J = 2.3 Hz, 1H), 7.46 (d, J = 1.1 Hz, 1H), 7.09 (d, J = 3.6 Hz, 1H), 6.74 (d, J = 3.6 Hz, 1H), 6.23 (t, J = 2.0 Hz, 1H), 5.56 (s, 2H); 13C NMR (125 MHz, DMSO-d6) in Figure 5: δ 168.11, 139.33, 139.02, 130.47, 108.52, 105.76, 59.40, CG-MS: m/z (%) = 213 [M+CH3OH]+ [calcd. for C7H8N4S [M+CH3OH]+ 212.23].
N-((3,5-dimethyl-1H-pyrazol-1-yl)methyl)thiazol-2-amine, 8: Yield, 37.12%; mp: 152–154°C; 1H NMR (500 MHz, CDCl3): δ 7.05 (d, J = 3.6 Hz, 1H), 6.78 (s, 2H), 6.44 (d, J = 3.6 Hz, 1H), 5.70 (s, 1H), 5.45 (s, 2H), 2.35 (s, 3H); 2.14 (s, 3H); 13C NMR (125 MHz, CDCl3): δ 167.52, 148.68, 140.07, 138.67, 108.07, 105.57, 56.74, 13.46, 11.11; CG-MS: m/z (%) = 293.1 [M+2ACN+2H]2 + [calcd. for C9H12N4S [M+2ACN+2H]+ 292.28].
N-((3,5-dimethyl-1H-pyrazol-1-yl)methyl)pyridin-4-amine, 9 (Abrigach et al., 2014): Yield, 29.79%; mp: 154–156°C; 1H NMR (300 MHz, MeOD): δ 8.06 (d, J = 1.5 Hz, 2H), 6.81 (d, J = 1.5 Hz, 2H), 6.79 (t, J = 1.5 Hz, 1H), 6.55 (s, 1H), 5.39 (s, 2H), 2.32 (s, 3H), 2.17 (s, 3H); 13C NMR (75 MHz, MeOD): δ 155.50, 148.49, 147.45, 139.91, 108.89, 105.87, 65.58, 11.91, 9.64; CG-MS: m/z (%) = 281.4 [M+DMSO+H]+ [calcd. for C11H14N4 [M+DMSO+H]+ 281.26].
Ethyl 1-((1H-imidazol-1-yl)methyl)-5-methyl-1H-pyrazole-3-carboxylate, 10: Yield, 99.6%; mp: 74–76°C; 1H NMR (500 MHz, DMSO-d6): δ 7.68 (d, J = 1.0 Hz, 1H), 7.04 (d, J = 1.0 Hz, 3H), 6.50 (d, J = 0.9 Hz, 2H), 4.26 (q, J = 7.1 Hz, 2H), 2.26 (s, 3H), 1.28 (s, 3H); 13C NMR (125 MHz, DMSO-d6): δ 161.63, 135.11, 121.64, 106.56, 59.89, 14.14, 10.68; CG-MS: m/z (%) = 310 [M+ACN+CH3OH+H]+ [calcd. for C11H14N4O2 [M+ACN+CH3OH+H]+ 309.26].
2-(((1H-pyrazol-1-yl)methyl)amino)-6-methylpyridin-4-ol, 11 (Abrigach et al., 2014): Yield, 93.82%; mp: 238–240°C, 1H NMR (500 MHz, DMSO-d6): δ 11.3 (s, 1H), 6.71 (s, 1H), 6.11 (s, 2H), 5.18 (s, 2H), 1.89 (s, 3H), 1.80 (m, 1H), 1.76 (s, 3H), 1.63 (s, 3H); 13C NMR (125 MHz, DMSO-d6): δ 164.80, 155.47, 153.96, 138.96, 129.30, 105.64, 100.34, 73.24, 23.60; CG-MS: m/z (%) = 234.1 [M+H]+ [calcd. for C11H15N5O [M+H]+ 233.28].
2-(((3,5-dimethyl-1H-pyrazol-1-yl)methyl)amino)-6-methylpyridin-4-ol, 12: Yield, 90.65%; mp: 100–102°C, 1H NMR (500 MHz, DMSO-d6): δ 11.00 (s, 1H), 7.76 (t, J = 2.3 Hz, 1H), 7.46 (t, J = 2.1 Hz, 1H), 5.56 (d, H = 12 Hz, 1H), 5.39 (s, 1H), 5.36 (d, J = 3.5 Hz, 1H), 1.98 (s, 3H); 13C NMR (125 MHz, DMSO-d6): δ 175.56, 172.05, 171.78, 163.62, 161.41, 99.54, 25.16, 23.20, 22.41; CG-MS: m/z (%) = 206.1 [M+H]+ [calcd. for C9H11N5O [M+H]+ 205.22].
Biological Evaluation
Anti-Fusarium Assay
First, the F.o.a. was isolated from Bouffagous Gharas date palm from Figuig in Morocco that is infected by the vascular fusariosis according to the protocol described by Benabbes et al. (2015), who followed the protocol of Locke and Colhoun (1974). Then, DMSO solution of each ligand was made at a concentration (4 mg/mL) and was thus employed for the preparation of various concentrations from potato dextrose agar (PDA) solutions with different volumes (50, 160, and 500 μL). The Petri plates were prepared with 8.7-cm diameter using 10 mL (Neri et al., 2006). After that, cultivated F.o.a. was transplanted onto the solid PDA with a pellet form in each plate center and again was incubated at 28°C for 5 days. The results were expressed in percentage (%) of inhibition, calculated from the measured width of F.o.a., compared to the positive control having only DMSO, which has no inhibition on F.o.a. (Hmouni et al., 1996). The experiments are repeated three times (three independent experiments, n = 3 with SEM ±).
where D0 = diameter in centimeters of F.o.a. in the control and Dx = diameter in centimeters of F.o.a. in the test.
Positive control: PDA+500 μL of DMSO; negative control: PDA+F.o.a.
After that, several tests were done to find the volume of the ligand to 50% of inhibition experimentally not doing the linear regression, with the objective to calculate the concentration of ligand needed to inhibit 50% of the F.o.a. (Radi et al., 2015; Tighadouni et al., 2016; Koudad et al., 2019; Tighadouini et al., 2020).
Theoretical Investigations
DFT Calculations
The DFT study was performed using Gaussian 09W software (Frisch et al., 2009) by the DFT (Eschrig, 2003; Capelle, 2006; Van Mourik et al., 2014; Domingo et al., 2016; Contreras-García and Yang, 2018) method with three functional parameters of Becke associated to the functional correlation gradient corrected by Lee Yang Parr (B3LYP) (Becke and Becke, 1993; Becke, 2014) and the exchange correlation in combination with 6–31 G (d, p) orbital basis sets for all atoms, with no symmetrical constrains. The molecular electrostatic potential (MEP) surfaces are generated in default parameters as total density and electrostatic potential (ESP) with the self-consistent field matrix for cubes and surface map generation.
Ligand Preparation
The compounds of interest for further docking purposes are geometrically optimized applying the DFT method, and the frequency was calculated for ground state verification after saving them as mole files later included in open label to convert them into pdbqt files that will be incorporated in Autodock Vina for virtual screening.
Protein Preparation and Active Site Selection
The modeled proteins structure of Fophy considered as target reported in the literature (Abrigach et al., 2018; Kaddouri et al., 2019; Tighadouini et al., 2019; Toubi et al., 2019) was prepared in Autodock 4 default parameters, and the whole protein was used as grid for blind docking–virtual screening (Table 1) with Perl as launcher of virtual screening for all the ligands in Autodock Vina (Seeliger and de Groot, 2010).

Table 1. Blind docking simulation parameters and virtual screening configuration using Autodock Vina.
For the docking validation, Dockthor (Santos et al., 2020), a web tool for ligand–protein docking, was used for blind docking/virtual screening of the active compounds in the whole Fophy protein chain, and the parameters for this is described in Table 2.
Dataset Preparation
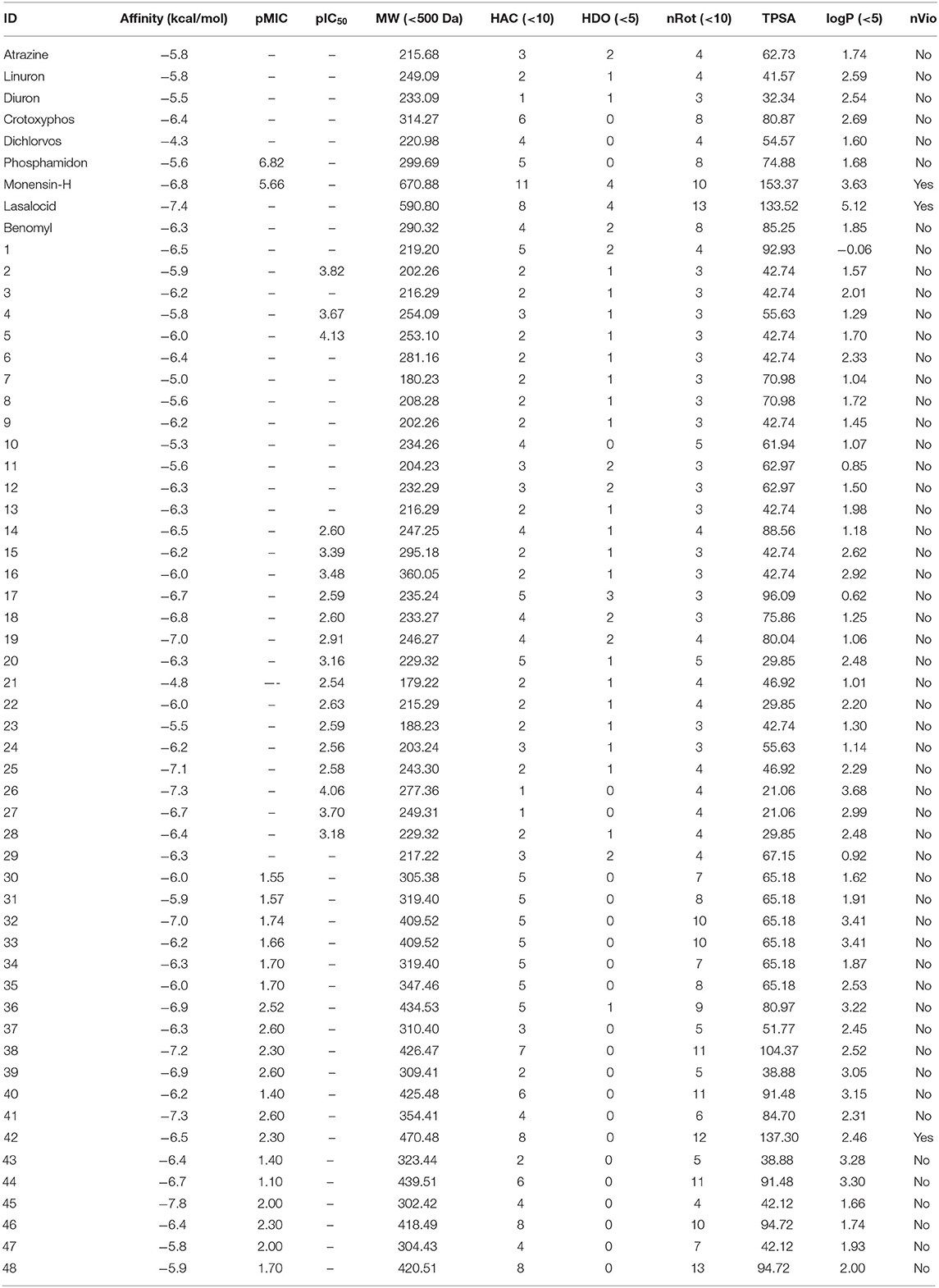
The selected compounds are all the testes against F.o.a., collected from the references (Waring et al., 2002; Radi et al., 2012, 2015; Smaail et al., 2012; Boussalah et al., 2013; Loth et al., 2015; Abrigach et al., 2017, 2018; Tighadouini et al., 2018, 2019; Koudad et al., 2019; Toubi et al., 2019).
In Table 3, this datatset contained 57 compounds, where 6 of them are tested against phytase inhibitors with Ki values that are converted to pKi between 3.16 for Dichlorvos and 4.50 for crytoxyphos. The dataset compounds structure is collected in with their pKi, IC50, or MIC values.
ADME Predictions
The ADME (adsorption, distribution, metabolism, and exertion) properties are depicted from SwissADME web tool (Hou et al., 2004; Arnott and Planey, 2012; Daina et al., 2017).
Results and Discussion
Chemistry
The mono-alkylated ligands based on pyrazole and 1,2,4-triazole are prepared (Figure 2), whereas compounds 5, 6, 9, and 11 were described in the literature (Touzani et al., 2003; Kaddouri et al., 2019, 2020). Several physicochemical analysis methods that are described in section Synthesis of the Pyrazole and Triazole Ligands characterized the prepared ligands.
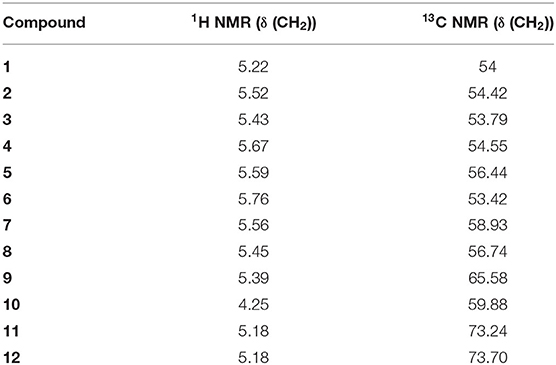
All compounds were characterized by 1H and 13C NMR. Table 4 illustrates the chemical shifts of CH2 in the examined ligands, where there is no significant difference between the studied ligands, and their structures are unique.
The chemical shifts of the mono-alkylated ligands 1–12 are located in the regions 5.18–5.76 ppm for 1H NMR, indicating a doublet peak that takes place because of the coupling of CH2 with the nearest free proton NH, and 53.42–73.7 ppm for 13C NMR, except for ligand 10, which carries an original chemical structure. For ligand 10, the 1H NMR chemical shift is 4.25 ppm with a single peak, and the 13C NMR chemical shift is 59.88 ppm.
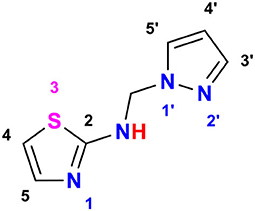
As an example, for the characterization of the studied compounds, N-((1H-pyrazol-1-yl)methyl)thiazol-2-amine, 7 in Figure 3 is displayed as follows:
To formulate this compound, 1.5 g of 2-aminothiazole (14.9 mmol) and 1.47 g of (1H-pyrazol-1-yl) methanol (14.9 mmol) were mixed in acetonitrile under reflux for 4 h, and the solvent was evaporated and then recrystallized in diethyl ether after filtration to have the final product (1.42 g, 53.08%): mp 108–110°C.
In Figure 4, 1H NMR (DMSO-d6, 500 MHz) spectrum of N-((1H-pyrazol-1-yl)methyl)thiazol-2-amine encloses the following peaks:
• Triplet integration at 8.68 ppm with JH−H = 6.7 Hz for the proton of the NH.
• Doublet integration at 7.80 ppm with JH−H = 2.3 Hz for the proton in position 3′.
• Doublet integration at 7.46 ppm with JH−H = 1.1 Hz for the proton in position 5′.
• Doublet integration at 7.09 ppm with JH−H = 3.6 Hz for the proton in position 5.
• Doublet di-doublet integration at 6.74 ppm with JH−H = 3.6 Hz for the proton in position 4.
• Triplet integration at 6.23 ppm with JH−H = 2.0 Hz for the proton in position 4′.
• Singlet integration at 5.56 ppm for the protons of the CH2.
In Figure 5, NMR 13C (DMSO-d6, 125 MHz) of N-((1H-pyrazol-1-yl)methyl)thiazol-2-amine contain the following peaks at:
• 168.11 for the carbone C2;
• 139.33 for the carbone C3′;
• 139.02 for the carbone C5;
• 130.47 for the carbone C5′;
• 108.52 for the carbone C4;
• 105.76 for the carbone C4′;
• 59.40 for the carbone CH2.
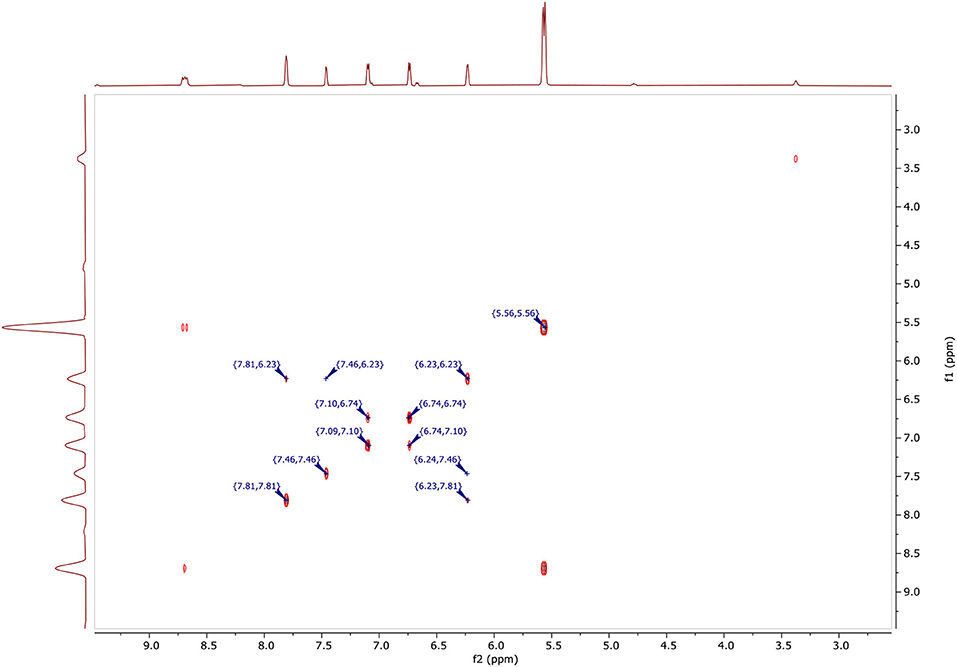
In Figure 6, two-dimensional NMR COSY (DMSO-d6, 400 MHz) δ ppm contains the following spots at:
• (5.56, 5.56) for CH2.
• (6.23, 6.23), (7.46, 6.23), (7.81, 6.23), (6.24, 7.46), and (6.23, 7.81) for CH (4′).
• (6.71, 6.74), (7.10, 6.74), and (6.71, 7.10) for CH (4).
• (7.09, 7.10), (7.10, 6.74), and (6.71, 7.10) for CH (5).
• (7.46, 7.46), (7.46, 6.23), and (6.24, 7.46) for CH (5′).
• (7.81, 7.81), (7.81, 6.23), and (6.23, 7.81) for CH (3′).
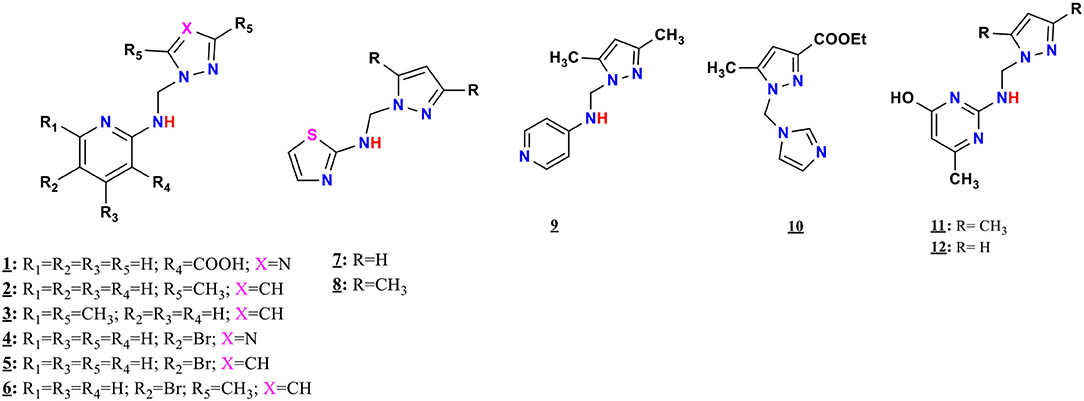
Figure 6. Synthesis route and chemical structures of the mono-alkylated pyrazole and triazole ligands.
Biological Assay and Lipophilicity Study
The preliminary results established that ligands 2, 4, and 5 displayed a significant inhibitory effect on F.o.a., in volumes over 50 μL, i.e., at concentrations higher than 16 μg/mL. To affirm that effect, we retested those ligands in three independent experiments (n = 3 experiments with ±SEM) to get the minimum inhibitory concentrations (MICs) experimentally.
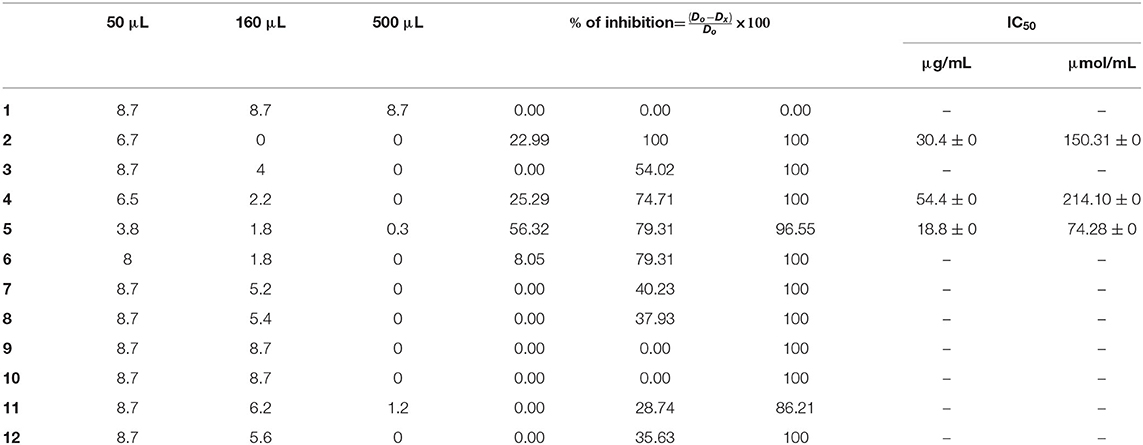
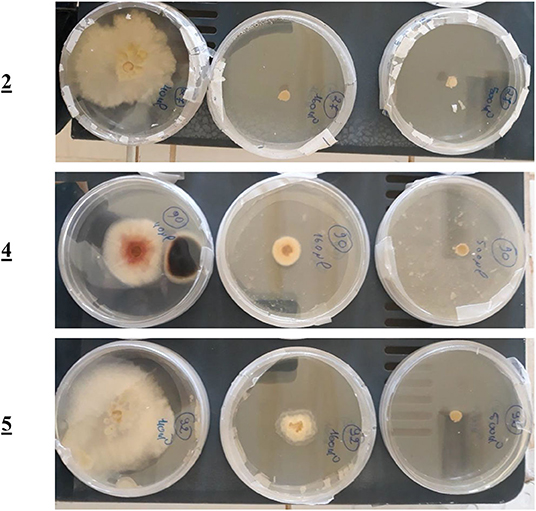
For the mono-alkylated ligands, Table 5 demonstrates that ligand 2 carries a 6.7-cm diameter of the strain in 50 μL (Figure 7) that resembled 22.99% inhibition. It kills F.o.a., appearing from 160 μL, at 100% inhibition. Ligand 3 has the same structure as ligand 2 but methyl group at the α position of the pyridine ring. This causes its inactivity, exhibiting no inhibition in 50 μL. Ligand 1 shows no inhibition at all. Ligands 4 (Figure 7), 5, and 6 show a respectable percentage of inhibition in 50 μL, corresponding to 25.29, 56.32, and 8.05%, respectively. The highest inhibition was seen in ligand 5 with IC50 = 18.8 μg/mL (Figure 7); it has no substituents on the pyrazole moiety, contrary to the substituted one; ligand 6 has a less percentage of inhibition. The results also show no meaningful difference, in terms of inhibition efficiency, between ligands 7 and 8, even if there are two methyl groups at positions 3 and 5 on the pyrazole moiety in compound 8. Ligands 9 and 10 kill F.o.a. in 500 μL with 100% of inhibition, showing no inhibition in other volumes. Ligands 11 and 12 show acceptable percentages of inhibition, starting from 160 μL, at 28.74%, and 35.63%, respectively, and the finest result obtained is by the non-substituted pyrazole moiety in compound 12. These compounds have identical or better results than compounds described in the literature by Tighadouini et al. (2019), where their compound 9 has 76% of inhibition in 200 μL, whereas compound 5 has 79.31% in 160 μL.
Table 6 shows the three best anti-Fusarium candidates IC50 values, where the highest value is for ligand 5 having IC50 = 18.8 μg/mL and IC50 = 74.28 μmol/L, substituted pyridine ring with Brome.
The lipophilicity (Veber et al., 2002; Leeson and Springthorpe, 2007; Podunavac-Kuzmanovic et al., 2008; Mannhold et al., 2009; Arnott and Planey, 2012; Hadda et al., 2014; Sima et al., 2017) characteristic of a ligand is the most important key in drug design and discovery, contributing to the ADMET-Tox (Gleeson et al., 2011; Glaab, 2016; Kauthale et al., 2017; Dhandapani and Balachandar, 2019) (administration, distribution, metabolism, excretion, and toxicity) of drugs and providing us insights about their solubility, cell membrane permeability, and so on (Arnott and Planey, 2012). It is expressed as the LogP of a compound with two different solvents: 1-octanol showing the lipid membrane and water as the model for cytoplasm (Holladay, 2015).
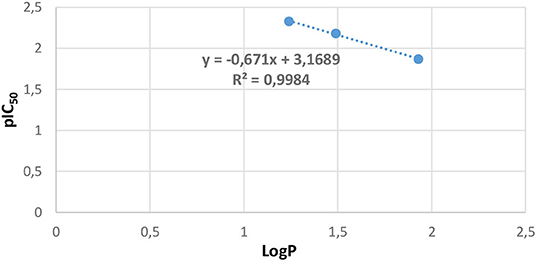
In our study, the LogP values of compounds 2, 4, and 5 were calculated handling the “Marvin sketch 19.13” software (https://chemaxon.com/products/marvin). The results are summed up in Table 6, which demonstrates that ligand 5 is the most effective with the biggest value of lipophilicity (LogP = 1.93); the order of inhibition efficiency IC50 correlated well with the following lipophilicity values order: 5 > 2 > 4. The LogP values were in the optimum region (LogP = 3), and hence, all three ligands could pass through the lipid membrane by the intense decrease in their IC50 values; for instance, for ligand 4, IC50 = 214.10 μmol/L and LogP = 1.24. Based on these results, lipophilicity and antifungal activity correlation plot are represented in Figure 8, and the analysis of these results established a notable linear correlation between pIC50 and LogP according to the following equation: y = −0.671x + 3.1689, where x: LogP, y: pIC50, and correlation coefficient of R2 = 0.9984.
To sum up, the results proved a correlation between the lipophilicity and antifungal activity of the ligands, but further precise experiments recommended to figure out the unfamiliar mechanism of action for the investigated ligands.
Theoretical Investigations
DFT Calculations
MEP may be a truly capable tool utilized to calculate or envision the reactive zones of nucleophilic and electrophilic attacks on a molecular system. It is often generated by mapping the ESP onto the isoelectron density surface of the molecule, providing us the chance to know the distribution of the electronic charge all over the structure. MEP mapping remains helpful in figuring out the synergy of a molecule with its environment and monitoring the hydrogen binding interactions, for its biological recognition processes (Kryachko, 2013; Alpaslan et al., 2019).
In our research, the optimized structures are in their global minima because of its positive frequencies values, MEP maps of compounds 2, 4, and 5 generated based on their density functional theory (DFT) optimized geometries; displayed in Figure 9. The negative ESP regions of compound 2 mainly concentrated over the nitrogen atoms of the pyrazole ring with a value of −1.174 eV, whereas the negative charges of compound 4 were situated on the nitrogen atoms of the triazole ring with a value of −1.612 eV. Compound 5 has negative charges located on the nitrogen atoms of the pyrazole ring with a value of −0.492 eV greater than the negative charge of compound 9 reported by Tighadouini et al. (2019), which has −1.850 eV. The positive charges of three ligands 2, 4, and 5 located were in the NH region of the N-C-N junction with values of 0.989, 1.612, and 1.324 eV, respectively. These results permitted us valuable information on the potential sites engaged in interactions between hydrogen bonds and the amino acid residues of protein receptors.
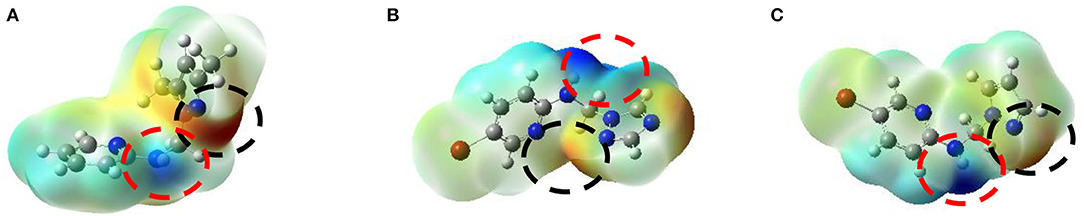
Figure 9. MEP surfaces for the three best anti-Fusarium candidates 2 (A), 4 (B), and 5 (C), where the negative regions (black circle) are related to electrophilic reactivity, whereas the positive regions (red circle) are for nucleophilic reactivity.
Blind Docking/Virtual Screening
In this study, the rotatable bonds of the ligand flexibility were allowed, while the protein was adopted as a rigid structure. As a first step of this study, the three-dimensional structures of the homology modeled Fophy protein and its template, which is Aspergillus niger phytase (PDB:3K4P) (Gontia-Mishra et al., 2014; Toubi et al., 2019), were aligned using Pymol software (Seeliger and de Groot, 2010), and it is presented in Figure 10.
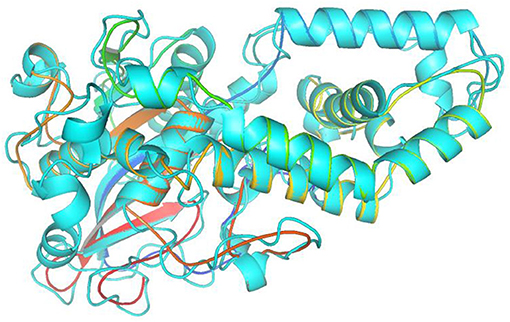
Figure 10. Structure alignment of homology modeled Fophy protein (colored) and its template 3K4P (blue).
As presented, there is good alignment between the two structures with smooth change; forward blind docking/virtual screening in both structures is provided using the active compounds against F.o.a., where the dataset compounds are the ones in Table 7.
From Table 7, only 31 compounds have pMIC values between 2.54 and 7.10, with 42 having pIC50 values between 1.09 and 4.69; thus, there were 73 compounds in dataset of 48 compounds other than the nine first compounds as bactericides and insecticides. The pKi values were from 2.74 and 5.83, with 97% of the compounds not violating the Lipinski's rule of 5, which makes it a good database for future in vivo tests.
For the Blind docking with virtual screening, the results obtained from Autodock Vina are presented in Figure 11 and Table 8.
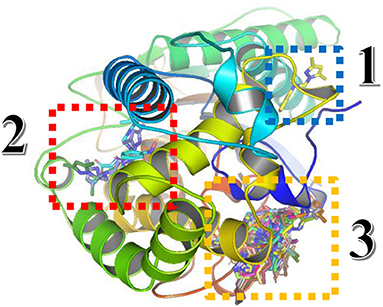
Figure 11. Three-dimensional presentation of the three binding sites found by Blind docking/virtual screening.
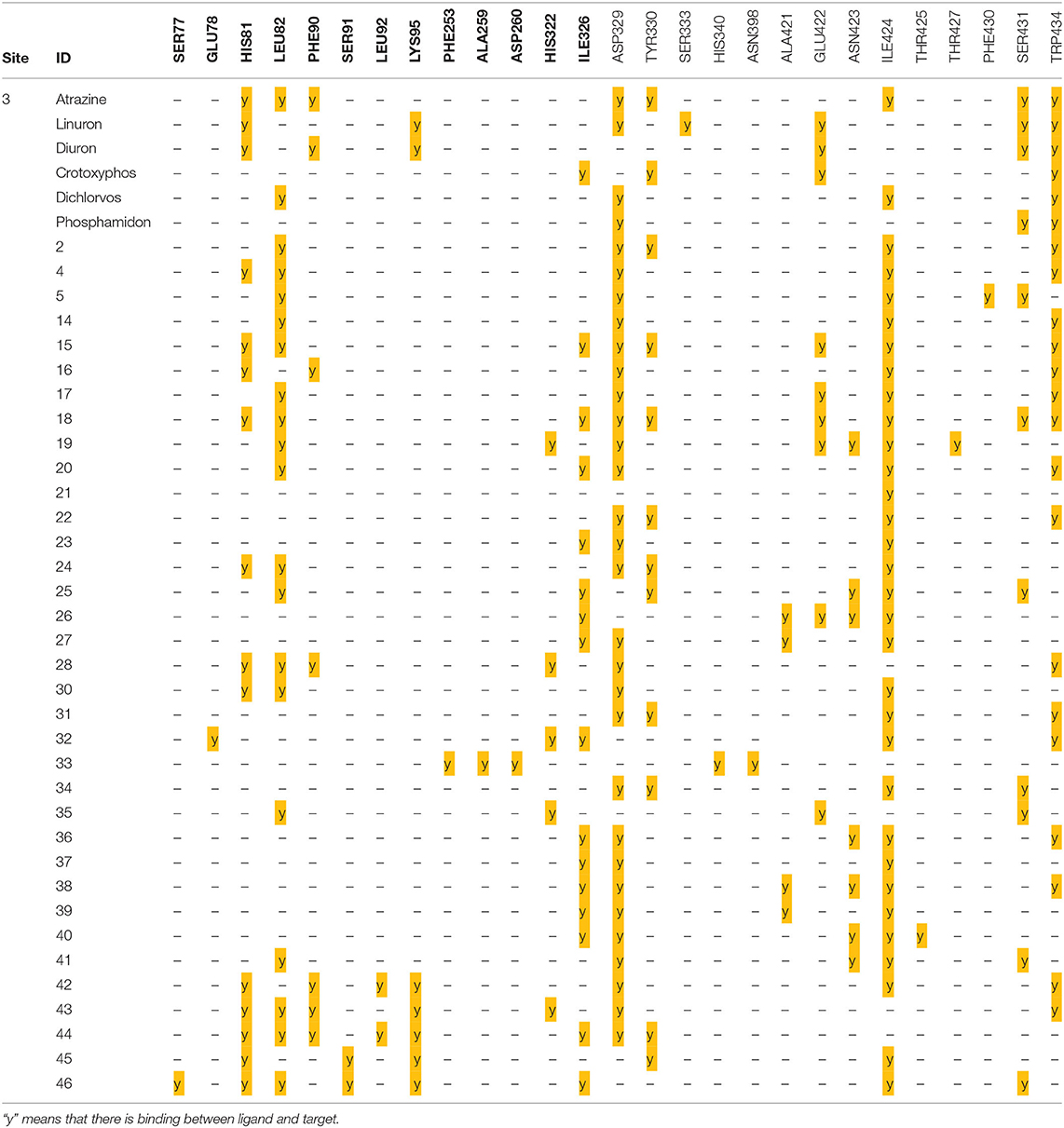
From the results above, the majority of the ligands screened are binding into the third active site, whereas only compound 3 is in the first site and the compounds monensin-H; 33, 54, and 89 are in the second one, but with which residues the compounds are interacting in the third site; thus, the following data were collected as shown in Table 8.
From data above, the site contains the following residues: SER77, GLU78, HIS81, LEU82, PHE90, SER91, LEU92, LYS95, PHE253, ALA259, ASP260, HIS322, ILE326, ASP329, TYR330, SER333, HIS340, ASN398, ALA421, GLU422, ASN423, ILE424, THR425, THR427, PHE430, SER431, and TRP434, where 15.09% of the bonds are with ILE424, 14.62% with ASP329, 10.37% with TRP434, 9.43% with LEU82, and 7.5% with ILE326.
For more specific study, the modes of binding interactions for ligands 2, 4, and 5, which are in the third site of the Fophy protein, are presented in Figure 12. Based on the docking results, ligand 5 reached the strongest affinity of −6.0 kcal/mol, whereas ligand 2 showed −5.9 kcal/mol, and ligand 4 showed −5.8 kcal/mol.
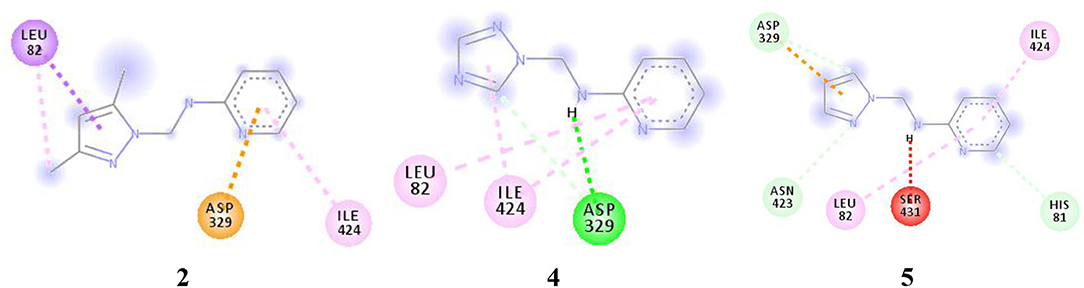
Figure 12. The two-dimensional interactions of ligand 2, 4, and 5 with the chosen active site in the Fophy protein.
For more specific study, the modes of binding interactions for ligands 2, 4, and 5, which are in the third site of the Fophy protein, are presented in Figure 12. Based on the docking results, ligand 5 reached the strongest affinity of −6.0 kcal/mol, whereas ligand 2 showed −5.9 kcal/mol, and ligand 4 showed −5.8 kcal/mol.
As presented in Figure 12 and Table 9, the studied compounds have multiple bonds, but only compounds 5 and 4 have carbon hydrogen bonds, whereas the strongest stay for the compounds with distance ranges from 1.59 to 5.40 Å for all bonds.
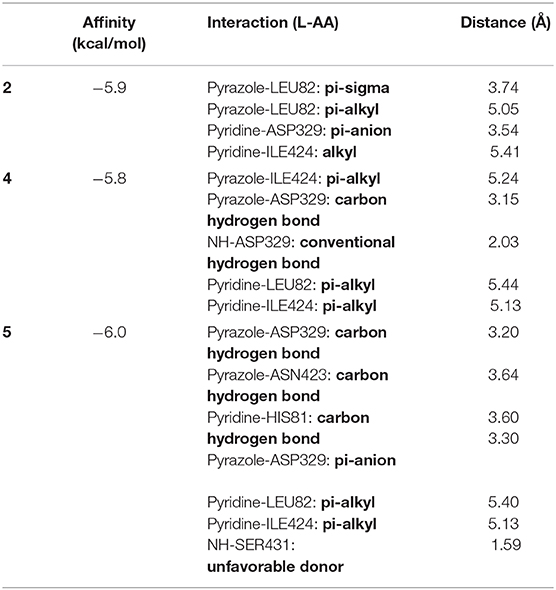
Table 9. The binding interactions between the best-studied ligands and the Fophy protein selected active site.
By comparing the results of the docking studies for the Fophy protein, it is found that compound 5 has the best affinity followed by compounds 2 and 4, so there is good agreement with the experimental results where their IC50 values are in the following order: 74.28 (5) < 150 (2) < 214.10 (4), but needs more investigations with much more compounds to build a model for phytase inhibitor prediction.
For protocol validation, blind docking/virtual screening was done for the same ligands and parameters with A. niger phytase (PDB:3K4P), which is the homology modeling template of the studied Fophy protein, implemented in Autodock Vina, and the results are presented in Figure 13.
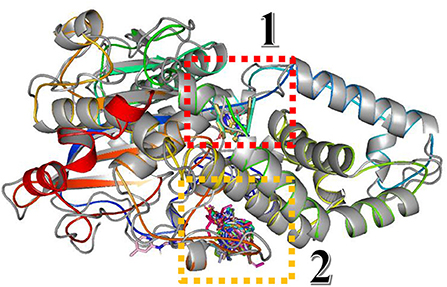
Figure 13. Structure alignment of Blind docking/virtual screening results on Aspergillus niger phytase protein and Fophy.
As presented in Figure 13, there are two different sites instead of three found in Fophy protein; compound 3 in the third one is eliminated because it is not active against F.o.a., and commonly most of the ligands screened are in the same site (the third one for Fophy protein and the second for A. niger phytase protein), which is approved also from all ligand visualizations using Autodock Vina and Dockthor as presented in Figure 14.
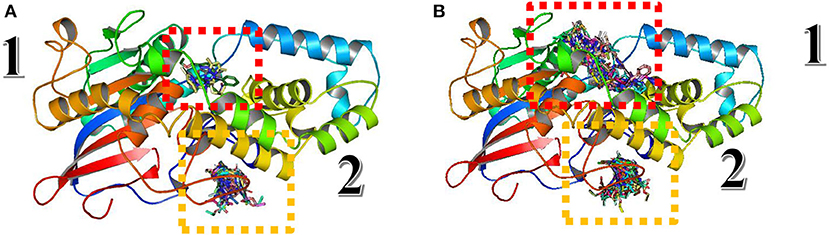
Figure 14. Blind docking/virtual screening of Fophy protein using Autodock Vina (A) and Dockthor (B).
Conclusion
Series of 12 pyrazole- and triazole-based ligands were prepared in good yield up to 99.6% and characterized using 1H and 13C NMR spectroscopy. The preliminary antifungal test screening against F.o.a. proved that ligands 2, 4, and 5 showed close to total inhibition of the fungus with an appreciable increase in their efficiency, starting from low concentrations. This encouraged us to study their reactivity, using DFT, and binding affinity, using the Fophy protein to show ligand–protein interactions, as described in the literature. From the results obtained using computational methods such as DFT studies of MEP surfaces, we found that ligand 2 had negative potential electrostatic regions mainly concentrated over the nitrogen atoms of the pyrazole ring with a value of −1.174 eV, whereas compound 4 had negative charges on the nitrogen atoms of the triazole ring with a value of −1.612 eV. Ligand 5 had negative charges on the nitrogen atoms of the pyrazole with a value of −0.492 eV. The positive charges of three ligands 2, 4, and 5 were in the NH region of the N-C-N junction with values of 0.989, 1.612, and 1.324 eV, respectively. These results gave us valuable information about the potential sites involved in interactions between hydrogen bonds and the amino acid residues of the protein receptors, correlating well with the docking results. Using Blind docking/virtual screening, the predicted site contains the following residues: SER77, GLU78, HIS81, LEU82, PHE90, SER91, LEU92, LYS95, PHE253, ALA259, ASP260, HIS322, ILE326, ASP329, TYR330, SER333, HIS340, ASN398, ALA421, GLU422, ASN423, ILE424, THR425, THR427, PHE430, SER431, TRP434, where 15.09% of the bonds are with ILE424, 14.62% with ASP329, 10.37% with TRP434, 9.43% with LEU82, and 7.5% with ILE326. In the binding mode of interactions for ligands 2, 4, and 5, ligand 5 reached the strongest affinity of −6.0 kcal/mol, with the strongest carbon hydrogen bonds with distance range of 1.59 to 5.40 Å for all bonds, whereas ligand 2 showed −5.9 kcal/mol, and ligand 4 showed −5.8 kcal/mol. For docking validation, the same protocol using Autodock Vina and another protocol using the Dockthor web tool give us the same predicted sites on Fophy protein.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Author Contributions
YK conceived and designed the chemistry experiments, perform the theoretical investigations experiments as DFT and molecular docking, interpreted all the data, and wrote the paper. SO and RB conceived, designed, and performed the biological experiments. FA performed the homology modeling of the studied protein and revised the paper. ME, AA, NA-Z, and IW revised the manuscript. RT supervised the work and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Project number (RSP-2020/78).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledged Researchers Supporting Project number (RSP-2020/78), King Saud University, Riyadh, Saudi Arabia.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2020.559262/full#supplementary-material
Abbreviations
DFT, Density Functional Theory; Fophy, Fusarium oxysporum phytase domain; F.o.a., Fusarium oxysporum f. sp. albedinis; NMR, Nuclear Magnetic Resonance; ppm, Part per million; % inh, Percentage of inhibition; D0, Diameter in cm of F.o.a. in the control; Dx, Diameter in cm of F.o.a. in the test; MIC, Minimum Inhibition Concentration; IC50, The half maximal inhibitory concentration; LogP, Lipophilicity; ADMET-Tox, Administration, Distribution, Metabolism, Excretion, and Toxicity; MEP, Molecular Electrostatic Potential; Val, Valine; Arg, Arginine; Leu, Leucine; Glu, Glutamic acid; Lys, Lysine; His, Histidine; KBr, Potassium bromide; δ, Chemical shift; DMSO-d6, Deuterated Dimethyl sulfoxide; CDCl3, Deuterated chloroform; CD2Cl2, Deuterated dichloromethane; MeOH, Deuterated Methanol; DCM, Dichloromethane; B3LYP, 3 parameters of Becke with functional correlation gradient corrected by Lee Yang Parr.
References
Abrigach, F., Karzazi, Y., Benabbed, R., Elyoubi, M., Khoutoul, M., Taibi, N., et al. (2017). Synthesis, biological screening, POM and 3D-QSAR analyses of some novel pyrazolic compounds. Med. Chem. Res. 26, 1784–1795. doi: 10.1007/s00044-017-1888-8
Abrigach, F., Khoutoul, M., Merghache, S., Oussaid, A., Lamsayah, M., Zarrouk, A., et al. (2014). Antioxidant activities of N-((3,5-dimethyl-1H-pyrazol-1-yl)methyl)pyridin-4-amine derivatives. Der. Pharma. Chem. 6, 280–285.
Abrigach, F., Rokni, Y., Takfaoui, A., Khoutoul, M., and Doucet, H. (2018). In vitro screening, homology modeling and molecular docking studies of some pyrazole and imidazole derivatives biomedicine & pharmacotherapy in vitro screening, homology modeling and molecular docking studies of some pyrazole and imidazole derivatives. Biomed. Pharmacother. 103, 653–661. doi: 10.1016/j.biopha.2018.04.061
Akhtar, J., Khan, A. A., Ali, Z., Haider, R., and Shahar Yar, M. (2017). Structure-activity relationship (SAR) study and design strategies of nitrogen-containing heterocyclic moieties for their anticancer activities. Eur. J. Med. Chem. 125, 143–189. doi: 10.1016/j.ejmech.2016.09.023
Alegaon, S. G. K. R. A., Garg, M. K., Dushyant, K., and Vinod, D. (2014). 1,3,4-Trisubstituted pyrazole analogues as promising anti-inflammatory agents. Bioorg. Chem. 54, 51–59. doi: 10.1016/j.bioorg.2014.04.001
Alpaslan, G. B. B., Demir, N., Tümer, Y., Yapar, G., Yildirim, N., Yildiz, M., et al. (2019). Synthesis, characterization, biological activity and theoretical studies of a 2-amino-6-methoxybenzothiazole-based fluorescent schiff base. J. Mol. Struct. 1180, 170–178. doi: 10.1016/j.molstruc.2018.11.065
Arnott, J. A., and Planey, S. L. (2012). The influence of lipophilicity in drug discovery and design. Expert Opin. Drug Discov. 7, 863–875. doi: 10.1517/17460441.2012.714363
Barbuceanu, S. F., Ilies, D. C., Saramet, G., Uivarosi, V., Draghici, C., and Radulescu, V. (2014). Synthesis and antioxidant activity evaluation of new compounds from hydrazinecarbothioamide and 1,2,4-triazole class containing diarylsulfone and 2,4-difluorophenyl moieties. Int. J. Mol. Sci. 15, 10908–10925. doi: 10.3390/ijms150610908
Becke, A. D. (2014). A new mixing of hartree–fock and local density-functional theories. J. Chem. Phys. 98:1372. doi: 10.1063/1.464304
Becke, A. D., and Becke, A. D. (1993). Density functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98:5648. doi: 10.1063/1.464913
Bekhit, A. A., Hassan, A. M., Abd El Razik, H. A., El-Miligy, M. M., El-Agroudy, E. J., and Bekhit Ael, D. (2015). New heterocyclic hybrids of pyrazole and its bioisosteres: design, synthesis and biological evaluation as dual acting antimalarial-antileishmanial agents. Eur. J. Med. Chem. 94, 30–44. doi: 10.1016/j.ejmech.2015.02.038
Benabbes, R., Lahmass, I., Souna, F., El Youbi, M., Saalaoui, E., Hakkou, A., et al. (2015). In vitro inhibitory effect of the extract powder of rosemary (Rosmarinus officinalis), oleander (Nerium oleander), grenadier (Punica granatum) on the growth of Fusarium oxysporum fs albidinis and in vivo test Antagonist fungi on the incidence and the control of vascular wilt disease of date palm in palm grove in figuig south of morocco. Adv. Environ. Biol. 9, 126–132.
Bouabdallah, I., Touzani, R., Zidane, I., and Ramdani, A. (2006). Synthesis of new tripodal ligand: N.N-bis[(1,5-dimethylpyrazol-3-yl)methyl]benzylamine. Catal. Commun. 8, 707–712. doi: 10.1016/j.catcom.2006.08.034
Boussalah, N., Touzani, R., Souna, F., Himri, I., Bouakka, M., Hakkou, A., et al. (2013). Antifungal activities of amino acid ester functional pyrazolyl compounds against Fusarium oxysporum f. sp. albedinis and Saccharomyces cerevisiae yeast. J. Saudi Chem. Soc. 17, 17–21. doi: 10.1016/j.jscs.2011.02.016
Capelle, K. (2006). A bird's-eye view of density-functional theory. arXiv. doi: 10.1590/S0103-97332006000700035
Cetin, A., and Gecibesler, I. (2015). Evaluation as antioxidant agents of 1,2,4-triazole derivatives: effects of essential functional groups. J. Appl. Pharm. Sci. 5, 120–126. doi: 10.7324/JAPS.2015.50620
Contreras-García, J., and Yang, W. (2018). Perspective: chemical information encoded in electron density. Wu Li Hua Xue Xue Bao 34, 567–580. doi: 10.3866/PKU.WHXB201801261
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7:42717. doi: 10.1038/srep42717
Dalloul, H. M., EI-nwairy, K., Shorafa, A. Z., and Samaha, A. A. (2017). Synthesis and biological activities of some new spiro 1, 2, 4-triazole derivatives having sulfonamide moiety. Org. Commun. 4, 280–287. doi: 10.25135/acg.oc.27.17.08.046
Dawood, K. M., and Abdel-Wahab, B. F. (2012). ChemInform abstract: synthesis and applications of bipyrazole systems. ChemInform 43, 491–545. doi: 10.3998/ark.5550190.0013.112
Dhandapani, M., and Balachandar, S. (2019). Biological action of molecular adduct pyrazole:trichloroacetic acid on candida albicans and ctDNA - A combined experimental, Fukui functions calculation and molecular docking analysis. J. Mol. Struct. 1184, 129–138. doi: 10.1016/j.molstruc.2019.02.006
Diana, F., Maurice, L., Ouinten, M., and Jean-Paul, G. (1995). Le Bayoud du Palmier Dattier: Une Maladie Qui Menace la Phoeniciculture. Phytoma: la Défense des Végétaux, 36–40.
Domingo, L. R., Ríos-Gutiérrez, M., and Pérez, P. (2016). Applications of the conceptual density functional theory indices to organic chemistry reactivity. Molecules 21:748. doi: 10.3390/molecules21060748
El-Adasy, A. M. (2017). Synthesis characterization, antioxidant and quantum chemical calculations of some new thiophene, diazepine and pyrimidine derivatives containing sulfamoyl moiety. Int. J. Chem. Stud. 5, 872–886.
Elayyachy, M., Elkodadi, M., Aouniti, A., Ramdani, A., Hammouti, B., Malek, F., and Elidrissi, A. (2005). New bipyrazole derivatives as corrosion inhibitors for steel in hydrochloric acid solutions. Mater. Chem. Phys. 93, 281–285. doi: 10.1016/j.matchemphys.2005.03.059
Elbelghiti, M., Karzazi, Y., Dafali, A., Hammouti, B., Bentiss, F., Obot, I. B., Bahadur, I., et al. (2016). Experimental, quantum chemical and monte carlo simulation studies of 3,5-disubstituted-4-amino-1,2,4-triazoles as corrosion inhibitors on mild steel in acidic medium. J. Mol. Liq. 218, 281–293. doi: 10.1016/j.molliq.2016.01.076
El-Naggar, A. M., and Abdel-Mottaleb, M. S. A. (2017). Novel thiazole derivatives of medicinal potential : synthesis and modeling. J. Chem. 2017:4102796. doi: 10.1155/2017/4102796
El-Sayed, A. A., and Ismail, M. F. (2019). Synthesis and in-vitro antioxidant and antitumor evaluation of novel pyrazole-based heterocycles. J. Iran. Chem. Soc. 16, 921–937. doi: 10.1007/s13738-018-1566-x
Eschrig, H. (2003). The Fundamentals of Density Functional Theory. Dresden: Institute for Solid State and Materials Research Dresden and University of Technology Dresden. 204–204.
Fadda, A. A., Berghot, M. A., Amer, F. A., Badawy, D. S., and Bayoumy, N. M. (2012). Synthesis and antioxidant and antitumor activity of novel pyridine, chromene, thiophene and thiazole derivatives. Arch. Pharm. 345, 378–385. doi: 10.1002/ardp.201100335
Freeman, S., and Maymon, M. (2000). Reliable detection of the fungal pathogenfusarium oxysporum f . sp. albedinis, causal agent of bayoud disease of date palm, using molecular techniques. Phytoparasitica 28, 341–348. doi: 10.1007/BF02981829
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Cheeseman, J. R., Scalmani, G., et al. (2009). Gaussian 09, Revision A.02. Gaussian 09, Inc, Wallingford, CT.
Gatta, F., Perotti, F., Gradoni, L., Gramiccia, M., Orsini, S., Palazzo, G., et al. (1990). Synthesis of some 1-(dihydroxypropyl)pyrazolo[3,4-d]-pyrimidines and in vivo evaluation of their antileishmanial and antitrypanosomal activity. Eur. J. Med. Chem. 25, 419–424. doi: 10.1016/0223-5234(90)90005-N
Glaab, E. (2016). Building a virtual ligand screening pipeline using free software : a survey building a virtual ligand screening pipeline using free software : a survey. Brief Bioinform. 17, 352–366. doi: 10.1093/bib/bbv037
Gleeson, M. P., Hersey, A., Montanari, D., and Overington, J. (2011). Probing the links between in vitro potency, ADMET and physicochemical parameters. Nat. Rev. Drug Discov. 10, 197–208. doi: 10.1038/nrd3367
Gontia-Mishra, I., Kumar Singh, V., Tripathi, N., Sasidharan, S., and Tiwari, S. (2014). Computational identification, homology modelling and docking analysis of phytase protein from Fusarium oxysporum. Biologia 69, 1283–1294. doi: 10.2478/s11756-014-0447-8
Hadda, T. B., Khardli, F. Z., Mimouni, M., Daoudi, M., Kerbal, A., Salgado-Zamora, H., et al. (2014). Impact of geometric parameters, charge, and lipophilicity on bioactivity of armed quinoxaline, benzothiaole, and benzothiazine: pom analyses of antibacterial and antifungal activity. Phosphorus Sulfur Silicon Related Elements 189, 753–761. doi: 10.1080/10426507.2013.855763
Hakkou, A., Chakroune, K., Souna, F., and Bouakka, M. (2004). “La Fusariose Vasculaire du Palmier Dattier (Bayoud): Méthodes de lutte,” in Communication Presented at the Multidisciplinary Conference on the Ecological and Cultural Bases of the Oasis of Figuig (Oujda). Available online at: http://hdl.handle.net/10045/26115 (accessed February 24, 2012).
Hmouni, A., Hajlaoui, M. R., and Mlaiki, A. (1996). Résistance de botrytis cinerea aux benzimidazoles et aux dicarboximides dans les cultures abritées de tomate en Tunisie. EPPO Bull. 26, 697–705. doi: 10.1111/j.1365-2338.1996.tb01513.x
Holladay, M. W. (2015). The organic chemistry of drug design and drug action: third edition. Drug Dev. Res. 75, 471–472.
Hou, T. J., Xia, K., Zhang, W., and Xu, X. J. (2004). ADME evaluation in drug discovery. Part 4. prediction of aqueous solubility based on atom contribution approach. J. Chem. Int. Comput. Sci. 44, 266–275. doi: 10.1021/ci034184n
Hu, Y., Zhang, J., Yu, C., Li, Q., Dong, F., Wang, G., et al. (2014). Synthesis, characterization, and antioxidant properties of novel inulin derivatives with amino-pyridine group. Int. J. Biol. Macromol. 70, 44–49. doi: 10.1016/j.ijbiomac.2014.06.024
Iaroshenko, V. O., Specowius, V., Vlach, K., Vilches-Herrera, M., Ostrovskyi, D., Mkrtchyan, S., et al. (2011). A general strategy for the synthesis of difluoromethyl-containing pyrazoles, pyridines and pyrimidines. Tetrahedron 67, 5663–5677. doi: 10.1016/j.tet.2011.05.085
Joshi, R. (2018). A review of Fusarium oxysporum on its plant interaction and industrial use. J. Med. Plants Stud. 6, 112–115. doi: 10.22271/plants.2018.v6.i3b.07
Kaddouri, Y., Abrigach, F., Mechbal, N., Karzazi, Y., El Kodadi, M., Aouniti, A., et al. (2019). Pyrazole compounds: synthesis, molecular structure, chemical reactivity, experimental and theoretical DFT FTIR spectra. Mater. Today Proc. 13, 956–963. doi: 10.1016/j.matpr.2019.04.060
Kaddouri, Y., Abrigach, F., Yousfi, E. B., El Kodadi, M., and Touzani, R. (2020). New thiazole, pyridine and pyrazole derivatives as antioxidant candidates: synthesis, DFT calculations and molecular docking study. Heliyon 6:e03185. doi: 10.1016/j.heliyon.2020.e03185
Kaddouri, Y. A. T., Abrigach, F., El Azzouzi, M., Zarrouk, A., and Hammouti, B. (2017). Tridentate pyrazole ligands: Synthesis, characterization and corrosion inhibition properties with theoretical investigations. JMES. 8, 845–856.
Kauthale, S., Tekale, S., Damale, M., Sangshetti, J., and Pawar, R. (2017). Synthesis, antioxidant, antifungal, molecular docking and ADMET studies of some thiazolyl hydrazones. Bioorg. Med. Chem. Lett. 27, 3891–3896. doi: 10.1016/j.bmcl.2017.06.043
Koudad, M., El Hamouti, C., Elaatiaoui, A., Dadou, S., Oussaid, A., Abrigach, F., et al. (2019). Synthesis, crystal structure, antimicrobial activity and docking studies of new imidazothiazole derivatives. J. Iran. Chem. Soc. 17, 297–306. doi: 10.1007/s13738-019-01766-4
Kryachko, E. S. (2013). Applications of Density Functional Theory to Biological and Bioinorganic Chemistry. Berlin; Heidelberg: Springer-Verlag (2013). doi: 10.1007/978-3-642-32750-6
Leeson, P. D., and Springthorpe, B. (2007). The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 6, 881–890. doi: 10.1038/nrd2445
Locke, T., and Colhoun, J. (1974). Contributions to a method of testing oil palm seedlings for resistance to Fusarium oxysporum Schl. f. sp. elaeidis toovey. Phytopathol. J. 79, 77–92. doi: 10.1111/j.1439-0434.1974.tb02691.x
Loth, B. M., Tir Touil, A., Len, C., and Verstraete, W. (2015). Antifungal activity of synthesized dithiocarbamate derivatives on Fusarium oxysporum f sp albedinis in algeria. J. Chem. Pharm. Res. 7, 49–54.
Mannhold, R., Poda, G. I., Ostermann, C., and Tetko, I. V. (2009). Calculation of molecular lipophilicity: state-of-the-art and comparison of log P methods on more than 96,000 compounds. J. Pharm. Sci. 98, 861–893. doi: 10.1002/jps.21494
Nayak, S., and Gaonkar, S. L. (2019). A review on recent synthetic strategies and pharmacological importance of 1,3-thiazole derivatives. Mini Rev. Med. Chem. 19, 215–238. doi: 10.2174/1389557518666180816112151
Neri, F., Mari, M., and Brigati, S. (2006). Control of penicillium expansum by plant volatile compounds. Plant Pathol. 55, 100–105. doi: 10.1111/j.1365-3059.2005.01312.x
Podunavac-Kuzmanovic, S. O., Cvetkovic, D. D., and Barna, D. J. (2008). The effect of lipophilicity on the antibacterial activity of some 1-benzylbenzimidazole derivatives. J. Serbian Chem. Soc. 73, 967–978. doi: 10.2298/JSC0810967P
Pongor, S. D. L., CemaŽar, M., Zahariev, S., and Guarnaccia, C. (2004). Solvent-free synthesis of azole carboximidamides. Tetrahedron. Lett. 45, 9423–9426. doi: 10.1016/j.tetlet.2004.10.114
Prabhudeva, M. G., Vivek, H. K., and Ajay kumar, K. (2019). Synthesis of novel pyrazole carboxamides using reusable catalyst as antimicrobial agents and molecular docking studies. Chem. Data Collect. 20, 100–193. doi: 10.1016/j.cdc.2019.100193
Pricopie, A. I., Focşan, M., Ionuţ, I., Marc, G., Vlase, L., Găină, L. I., et al. (2019). Design and synthesis of novel 1,3-thiazole and 2-hydrazinyl-1,3-thiazole derivatives as anti-candida agents: in vitro antifungal screening, molecular docking study, and spectroscopic investigation of their binding interaction with bovine serum albumin. Molecules 24:1079. doi: 10.3390/molecules24193435
Radi, S., Tighadouini, S., Feron, O., Riant, O., Bouakka, M., Benabbes, R., et al. (2015). Synthesis of novel beta-keto-enol derivatives tethered pyrazole, pyridine and furan as new potential antifungal and anti-breast cancer agents. Molecules 20, 20186–20194. doi: 10.3390/molecules201119684
Radi, S., Toubi, Y., Hakkou, A., Souna, F., Himri, I., and Bouakka, M. (2012). Synthesis, antibacterial and antifungal activities of Novel N.N'-bipyrazole piperazine derivatives. Lett. Drug Des. Discov. 9, 853–857. doi: 10.2174/157018012803307941
Reis, R. E. C. A., Cecília, M., De Souza, B. V., Francisco, V., Montenegro, R. C., Jérsia, A., et al. (2011). Synthesis and anticancer activities of some novel 2- (benzo [d] thiazol-2-yl) - 8-substituted-2 H -pyrazolo [4, 3- c] quinolin-3 (5H)-ones. Eur. J. Med. Chem. 46, 1448–1452. doi: 10.1016/j.ejmech.2011.01.066
Sahu, N., Sahu, J. K., and Kaushik, A. (2013). A review on ‘triazoles ’: their chemistry and pharmacological potentials. Curr. Res. Pharm. Sci. 03, 108–113.
Santos, K. B., Guedes, I. A., Karl, A. L. M., and Dardenne, L. E. (2020). Highly flexible ligand docking: benchmarking of the dockthor program on the LEADS-PEP protein-peptide data set. J. Chem. Inf. Model. 60, 667–683. doi: 10.1021/acs.jcim.9b00905
Seeliger, D., and de Groot, B. L. (2010). Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 24, 417–422. doi: 10.1007/s10822-010-9352-6
Shaikh, M. H., Subhedar, D. D., Khan, F. A. K., Sangshetti, J. N., and Shingate, B. B. (2016). 1,2,3-Triazole incorporated coumarin derivatives as potential antifungal and antioxidant agents. Chin. Chem. Lett. 27, 295–301. doi: 10.1016/j.cclet.2015.11.003
Sima, I. A., Kot-Wasik, A., Wasik, A., Namieśnik, J., and Sârbu, C. (2017). Assessment of lipophilicity indices derived from retention behavior of antioxidant compounds in RP-HPLC. Molecules 22:550. doi: 10.3390/molecules22040550
Singh, N., Shah, P., Dwivedi, H., Mishra, S., Tripathi, R., Sahasrabuddhe, A. A., et al. (2016). Integrated machine learning, molecular docking and 3D-QSAR based approach for identification of potential inhibitors of trypanosomal N-myristoyltransferase. Mol. Biosyst. 12, 3711–3723. doi: 10.1039/C6MB00574H
Singh, R. N., Rawat, P., and Sahu, S. (2014). Synthesis, characterization and computational study on ethyl 4-(3-Furan-2yl-acryloyl)-3,5-dimethyl-1H-pyrrole-2-carboxylate. J. Mol. Struct. 1076, 437–445. doi: 10.1016/j.molstruc.2014.07.074
Smaail, R., Yahya, T., Imad, H., Abdelkader, H., Faiza, S., Imane, H., et al. (2012). Synthesis, antibacterial and antifungal activities of some new bipyrazolic tripodal derivatives. Res. J. Chem. Sci. 2, 40–44.
Soundararajan, P., Sakkiah, S., Sivanesan, I., Lee, K. W., and Jeong, B. R. (2011). Macromolecular docking simulation to identify binding site of FGB1 for antifungal compounds. Bull. Korean Chem. Soc. 32, 3675–3681. doi: 10.5012/bkcs.2011.32.10.3675
Sun, G. X., Yang, M. Y., Shi, Y. X., Sun, Z. H., Liu, X. H., Wu, H. K., et al. (2014). Microwave assistant synthesis, antifungal activity and DFT theoretical study of some novel 1,2,4-triazole derivatives containing pyridine moiety. Int. J. Mol. Sci. 15, 8075–8090. doi: 10.3390/ijms15058075
Thangarasu, P., Manikandan, A., and Thamaraiselvi, S. (2019). Discovery, synthesis and molecular corroborations of medicinally important novel pyrazoles; drug efficacy determinations through in silico, in vitro and cytotoxicity validations. Bioorg. Chem. 86, 410–419. doi: 10.1016/j.bioorg.2019.02.003
Thangavelu, R., and Gopi, M. (2015). Field suppression of fusarium wilt disease in banana by the combined application of native endophytic and rhizospheric bacterial isolates possessing multiple functions. Phytopathol. Mediterr. 54, 241–252.
Tighadouini, S., Benabbes, R., Tillard, M., Eddike, D., Haboubi, K., Karrouchi, K., et al. (2018). Synthesis, crystal structure, DFT studies and biological activity of (Z)-3-(3-bromophenyl)-1-(1,5-dimethyl-1H-pyrazol-3-yl)-3-hydroxyprop-2-en-1-one. Chem. Cent. J. 12:122. doi: 10.1186/s13065-018-0492-4
Tighadouini, S., Radi, S., Abrigach, F., Benabbes, R., Eddike, D., and Tillard, M. (2019). Novel β-keto-enol pyrazolic compounds as potent antifungal agents. Design, synthesis, crystal structure, DFT. Homology modeling, and docking studies. J. Chem. Inf. Model. 59, 1398–1409. doi: 10.1021/acs.jcim.8b00828
Tighadouini, S., Radi, S., Benabbes, R., Youssoufi, M. H., Shityakov, S., El Massaoudi, M., et al. (2020). Synthesis, biochemical characterization, and theoretical studies of novel beta-keto-enol pyridine and furan derivatives as potent antifungal agents. ACS Omega 5, 17743–17752. doi: 10.1021/acsomega.0c02365
Tighadouni, S., Radi, S., Sirajuddin, M., Akkurt, M., Özdemir, N., Ahmad, M., et al. (2016). In vitro antifungal, anticancer activities and POM analyses of a novel bioactive schiff base 4-{[(E)-furan-2-ylmethylidene]amino}p-henol: synthesis, characterization and crystal structure. J. Chem. Soc. Pakistan 38, 157–165.
Toubi, Y. F. A., Radi, S., Souna, F., Hakkou, A., Alsayari, A., Bin Muhsinah, A., et al. (2019). Synthesis, antimicrobial screening, homology modeling, and molecular docking studies of a new series of schiff base derivatives as prospective fungal inhibitor candidates. Molecules 24:3250. doi: 10.3390/molecules24183250
Touzani, R., Garbacia, S., Lavastre, O., Yadav, V. K., and Carboni, B. (2003). Efficient solution phase combinatorial access to a library of pyrazole- and triazole-containing compounds. J. Comb. Chem. 5, 375–378. doi: 10.1021/cc030100b
Van Mourik, T., Buhl, M., and Gaigeot, M. P. (2014). Density functional theory across chemistry, physics and biology. Philos. Trans. A Math. Phys. Eng. Sci. 372:20120488. doi: 10.1098/rsta.2012.0488
Varghese, N., Jacob, J., Mythri, M., Nija, B., and Sheeba Jasmine, T. S. (2016). Synthesis of thiazole derivatives- a review. World J. Pharm. Pharm. Sci. 5, 624–636.
Veber, D. F., Johnson, S. R., Cheng, H.-Y., Smith, B. R., Ward, K. W., and Kopple, K. D. (2002). Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 45, 2615–2623. doi: 10.1021/jm020017n
Waring, M. J., Ben-Hadda, T., Kotchevar, A. T., Ramdani, A., Touzani, R., Elkadiri, S., et al. (2002). 2,3-Bifunctionalized quinoxalines: synthesis, DNA interactions and evaluation of anticancer, anti-tuberculosis and antifungal activity. Molecules 7, 641–656. doi: 10.3390/70800641
Keywords: pyrazole, triazole, Fusarium oxysporum, antifungal, DFT, docking
Citation: Kaddouri Y, Abrigach F, Ouahhoud S, Benabbes R, El Kodadi M, Alsalme A, Al-Zaqri N, Warad I and Touzani R (2020) Mono-Alkylated Ligands Based on Pyrazole and Triazole Derivatives Tested Against Fusarium oxysporum f. sp. albedinis: Synthesis, Characterization, DFT, and Phytase Binding Site Identification Using Blind Docking/Virtual Screening for Potent Fophy Inhibitors. Front. Chem. 8:559262. doi: 10.3389/fchem.2020.559262
Received: 05 May 2020; Accepted: 16 October 2020;
Published: 11 December 2020.
Edited by:
Simone Brogi, University of Pisa, ItalyReviewed by:
Stevan Armakovic, University of Novi Sad, SerbiaMehdi Khoshneviszadeh, Shiraz University of Medical Sciences, Iran
Anna Biernasiuk, Medical University of Lublin, Poland
Copyright © 2020 Kaddouri, Abrigach, Ouahhoud, Benabbes, El Kodadi, Alsalme, Al-Zaqri, Warad and Touzani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yassine Kaddouri, eS5rYWRkb3VyaUB1bXAuYWMubWE=; Rachid Touzani, ci50b3V6YW5pQHVtcC5hYy5tYQ==
 Yassine Kaddouri
Yassine Kaddouri Farid Abrigach1
Farid Abrigach1 Ali Alsalme
Ali Alsalme Nabil Al-Zaqri
Nabil Al-Zaqri Ismail Warad
Ismail Warad