
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 20 October 2020
Sec. Supramolecular Chemistry
Volume 8 - 2020 | https://doi.org/10.3389/fchem.2020.00814
This article is part of the Research Topic Materials for Electroanalysis Based on Advanced Frameworks View all 11 articles
 Marzieh Alizadeh1
Marzieh Alizadeh1 Parviz Aberoomand Azar1
Parviz Aberoomand Azar1 Sayed Ahmad Mozaffari2*
Sayed Ahmad Mozaffari2* Hassan Karimi-Maleh3
Hassan Karimi-Maleh3 Ali-Mohammad Tamaddon4
Ali-Mohammad Tamaddon4An ultrasensitive DNA electrochemical biosensor based on the carbon paste electrode (CPE) amplified with ZIF-8 and 1-butyl-3-methylimidazolium methanesulfonate (BMIMS) was fabricated in this research. The DNA/BMIMS/ZIF-8/CPE was used for the selective determination of a mitoxantrone anticancer drug in aqueous solution, resulting in a good catalytic effect and a powerful ability for determining mitoxantrone. Also, the interaction of the mitoxantrone anticancer drug with guanine bases of ds-DNA was used as a powerful strategy in the suggested biosensor, which was confirmed with docking investigation. Docking study of mitoxantrone into the ds-DNA sequence showed the intercalative binding mode of mitoxantrone into the nitrogenous-based pairs of ds-DNA. The effective factors such as ds-DNA concentration, temperature, buffer types, and incubation time were also optimized for the fabricated mitoxantrone biosensor. The results showed that, under optimum conditions (T = 25°C; incubation time=12 min; pH= 4.8 acetate buffer solution and [DNA] = 50 mg/L), the DNA/BMIMS/ZIF-8/CPE could be used in mitoxantrone assay in a concentration ranging from 8.0 nM to 110 μM with a detection limit of 3.0 nM. In addition, recovery data between 99.18 and 102.08% were obtained for the determination of mitoxantrone in the injection samples using DNA/ZIF-8/BMIMF/CPE as powerful biosensors.
Mitoxantrone (developed in the 1980's) is one of the famous anthracycline anti-cancer agents with a wide range of applications in the treatment of breast cancer, acute myelogenous leukemia, and Non-Hodgkin's lymphoma (Lenk et al., 1987; Vollmer et al., 2010). This drug stays in the body for a long time (elimination t1/2 = 75 h) and has various side effects such as low blood counts, nausea, vomiting, weakness, low blood pressure, and hair loss (Scott and Figgitt, 2004). Moreover, the highest concentrations of mitoxantrone were detected in the heart, liver, and thyroid (Fox, 2004). Assay of anticancer drugs in biological samples such as blood is one of the most important strategies available to monitor the harmful effects of such drugs on the body (Bolanowska et al., 1983; Baghayeri et al., 2014, 2017, 2018a; Beitollahi et al., 2014; Veisi et al., 2015; Fouladgar, 2018). From the methods reported for measuring the pharmaceutical and biological compounds, electrochemical methods with more advantages such as simplicity of analysis method, low cost, and fast analysis are considered to be more important compared to the other methods (Yuan et al., 2013, 2017; Mozaffari et al., 2014; Eren et al., 2015; Movaghgharnezhad and Mirabi, 2019). Intercalative binding between DNA and mitoxantrone has been proven by the Li group (Li et al., 2005). Accordingly, this makes it possible to design the DNA-based electrochemical biosensors for selective analysis of the drug (Tiwari and Sharma, 2020).
Moreover, the DNA-based biosensors have been reported as powerful tools with a high selectivity for the analytical determination of many compounds, especially for anticancer drugs (Brett et al., 1998; Gooding, 2002; Ozsoz et al., 2003; Ensafi et al., 2011). In this regard, the specific interaction of anticancer drugs with adenine and guanine bases in the complex structure of DNA has been used as an appropriate analytical factor to design new biosensors in the analysis of anticancer drugs (Karimi-Maleh et al., 2018; Khodadadi et al., 2019; Yin et al., 2019). In addition, the sensitivity of the DNA-based biosensors is very low at surface of bare electrodes due to the presence of low-conductivity ds-DNA on the sensor surface (Li et al., 2017, 2020). Correspondingly, this point was introduced as one of the most important problems caused by the conventional DNA-based biosensors (Karimi-Maleh et al., 2020c). To overcome this problem, the DNA-based biosensors are typically amplified using the high-conductivity modifiers such as conductor polymers, organic and inorganic compounds, ionic liquids, and nanomaterials (Cheraghi et al., 2017; Sanati and Faridbod, 2017; Baghayeri et al., 2019; Faridbod and Sanati, 2019).
Nanomaterials such as nanoparticles, nanotubes, and nano porous compounds showed many advantages in different fields and also created a new approach to science (Rahmanian et al., 2015; Xu et al., 2018; Yuan et al., 2019, 2020; Karimi-Maleh et al., 2020a). Accordingly, Nano porous materials like zeolitic imidazolate frameworks (ZIF) are a new type of nanomaterials with a high surface area and metal ion in center and imidazolate linkers (Quang Khieu et al., 2018). Recently, many researchers focused on the usage of ZIF and especially ZIF-8 for the electrochemical applications (Wang et al., 2015). ZIF-8 is very stable in water and other aqueous solutions and could be used as mediator for the fabrication of electrochemical sensors for analysis of electroactive compound in water solution (Banerjee et al., 2008). Although the high surface area of ZIF compounds makes creating a more active surface area for electrochemical sensors possible, its low electrical conductivity is one of the most important problems of its high use in electrochemical sensors (Lu et al., 2020). Therefore, to eliminate this problem, the simultaneous usage of these materials with compounds that have a high electrical conductivity such as conductive polymers and ionic liquids, is recommended (Baghayeri et al., 2018b; Jin et al., 2018; Chen et al., 2019).
Ionic liquids are highly conductive and are a green type of organic compound with a wide range application in different scientific fields (Marr and Marr, 2016; Osada et al., 2016; Atta et al., 2019; Tahernejad-Javazmi et al., 2019; Arabali et al., 2020). Moreover, due to the high conductivity and wide electrochemical range windows, ionic liquids were used as amplifiers with a high quality in the fabrication of electrochemical sensors (Bijad et al., 2013; Beytur et al., 2018; Li et al., 2019; Hojjati-Najafabadi et al., 2020). There are many published scientific papers for the application of ionic liquid coupled with other nanomaterials to create a high quality electrochemical sensor in the environmental and biological compounds analyses (Karimi-Maleh et al., 2020b).
Based on the scientific information reported in previous studies, this study developed a high-sensitivity electrochemical biosensor in terms of the use of DNA as a recognition element for analyzing the mitoxantrone anti-cancer drug. To improve the sensitivity of the DNA-based biosensor, the electrode surface was amplified with ZIF-8 and BMIMF as the conductive modifiers with a high surface. The results showed a good selectivity for the analysis of mitoxantrone anti-cancer drug in drugs samples. The docking investigation confirmed the intercalation interaction between the guanine base and mitoxantrone anti-cancer drugs.
Electrochemical investigation was performed by electrochemical workstation model Ivium-Vertex connected to an electrochemical Cell (Azar electrode Company, Iran). Moreover, the I-V signals were displayed based on the Ag/AgCl/KClsat reference electrode's potential. Mitoxantrone hydrochloride, ZIF-8, BMIMF, and DNA (Calf Thymus) were purchased from Sigma-Aldrich. Also, carbon powder and paraffin oil were obtained from Merck Company. In addition, phosphoric acid, boric acid, acetic acid, and Tris hydrochloride were purchased from Across Company. Notably, the stock solution of mitoxantrone hydrochloride (0.001 M) was prepared by dissolving 0.517 g mitoxantrone hydrochloride in 100 mL distillated water under the stirring conditions.
The BMIMS/ZIF-8/CPE was prepared by mixing ZIF-8 with carbon powder as the powder components in the ratio 5:95 (w/w), and paraffin oil and BMIMS as the liquid binders in the ratio 8:2 (v/v). Accordingly, these ratios of powder and binder components were optimized by recording voltammograms of solution containing 1.0 mM [Fe(CN)6]3−, 4− at the surface of electrodes with different ratios of the components. Also, the stability of ZIF-8 as aqueous solution helps for a repeatable electrochemical response in electroanalytical systems.
DNA/BMIMS/ZIF-8/CPE was perpetrated by the addition of 10 μL of ds-DNA solution (50 mg/L) prepared into acetate buffer (0.5 M, pH 4.8) using a dropwise strategy. Notably, this value was optimized by recording ds-DNA at the surface of DNA/BMIMS/ZIF-8/CPE in the concentration ranged between 10 and 60 mg/L.
To study the intercalation of mitoxantrone hydrochloride with ds-DNA at the surface of DNA/BMIMS/ZIF-8/CPE, the electrode was immersed into a solution containing Tris-HCL buffer solution (pH = 7.4) with mitoxantrone hydrochloride and then remained for 12 min under the stringing condition. Afterward, the electrode was washed with the acetate buffer solution and the differential pulse voltammograms of electrode was then recorded in acetate solution (0.5 M, pH 4.8).
In this work, the molecular docking study is performed to evaluate the affinity of mitoxantrone drug in the active site of DNA hexamer d(CGATCG)2 containing an intercalation gap (PDB ID:1Z3F). For a comprehensive investigation of the binding orientation analysis, the best conformer with the lowest root mean square deviation (RMSD) value of 0 Å and the highest binding energy value is selected. The docking of mitoxantrone into the DNA sequence suggests the intercalation of the aromatic rings of mitoxantrone drug between cytosine and guanine base pairs of DNA with a binding energy of −6.7 kcal/mol as shown in Figure 1, curve a. The docked model reveals that the hydrogen and oxygen atoms of mitoxantrone drug have participated as the donor and acceptor to form four intermolecular hydrogen bonds (HBs) with base pairs of DNA (see Figure 1, curve b). It is found that the interaction of oxygen (O5) atoms of drug molecule with H22 atom of deoxyguanosine (DG6 of chain B) of DNA leads to O5…H22-N2 conventional HB with distance of 2.6 Å. Also, the H36 atom of drug as the proton donor interacts with O4′ atom of deoxyribose sugar moiety linked to guanine (DG6 of chain B) as a proton acceptor with a distance of 2.3 Å. Furthermore, the hydrogen atom of the hydroxyl terminal group of drug molecule is bonded to the second and forth oxygen atoms of deoxycytosine (DC5 of chain B) of hexamer of DNA, i.e., O2…H60-O6 and O4′…H60-O6 with the O…H distances of 2.4 and 2.2 Å, respectively. The hydrogen bond angles are 109.5°, 110.5°, 134.5°, and 163.6° for O5…H22-N2, O2…H60-O6, O4′…H60-O6 and O4′…H36-N8, respectively. In addition, the intermolecular interactions between carbonyl groups of mitoxantrone drug and oxygen atoms of DNA sequence, i.e., O2 atom of DC5 chain B, O4′ atom of DG6 chain B and O4′ atom of DG2 chain A with the respective O…O bond lengths of 3.3, 3.5 and 3.4 Å are observed. The docking study approves the interaction between mitoxantrone drug and guanine residues of DNA contributes in the formation of the stable mitoxantrone-DNA complex.
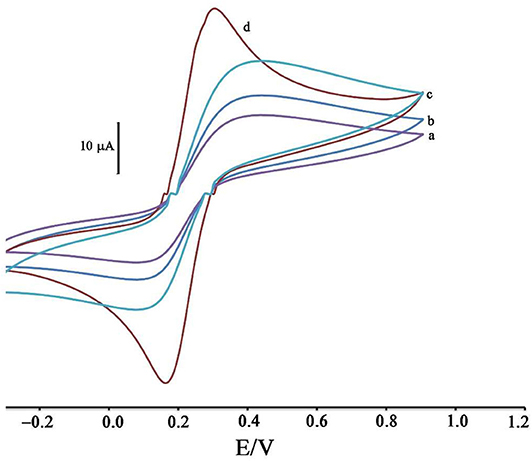
Figure 1. Cyclic voltammograms of solution 1.0 mM [Fe(CN)6]3−/4− in the presence of KCl 1.0 M at surface of (a) CPE, (b) ZIF-8/CPE, (c) BMIMS/CPE and (d) BMIMS/ZIF-8/CPE.
Mitoxantrone (12.5 mg/12.5 mL) was purchased from a local pharmacy and then used as a real sample with no pretreatment. The standard addition method was used for analyzing the mitoxantrone concentration in the injection sample using DNA/BMIMS/ZIF-8/CPE.
The modification of the CPE surface with BMIMS and ZIF-8 was investigated by recording the cyclic voltammograms of the solution containing 1.0 mM [Fe(CN)6]3−, 4−. By moving CPE (Figure 1, curve a) to ZIF-8/CPE (Figure 1, curve b), a little improvement was obtained in the oxidation signal of [Fe(CN) 6]3−, 4− redox solution, which can be related to the creation of a high surface area of ZIF-8 at surface of CPE. After the addition of BMIMS and at a surface of BMIMS/CPE (Figure 1, curve c), the oxidation current of CPE was increase from 14.65 to 24.4 μA that is relative to high conductivity of IL.
After modification of CPE with ZIF-8 and BMIMS, a sharp redox signal with an oxidation current 34.8 μA was observed relative to [Fe(CN)6]3−/4− that can be associated with the synergic effects of BMIMS and ZIF-8 at surface of CPE. This amplification can be created under a high sensitivity condition to determine mitoxantrone at surface of DNA/BMIMS/ZIF-8/CPE. In addition, active surface area of CPE, ZIF-8/CPE, BMIMS/CPE and BMIMS/ZIF-8/CPE were calculated about 0.121, 0.163, 0.184, and 0.22 cm2 by solution containing 1.0 mM [Fe(CN)6]3−/4− and results confirmed that mediators could be increased active surface area of CPE.
Figure 2 displays the ds-DNA signal of DNA/BMIMS/ZIF-8/CPE in the absence (curve a) and in the presence of 35.0 and 80.0 μM mitoxantrone (curves b & c), respectively. According to the data reported, the oxidation signal of ds-DNA decreased from 6.2 to 3.87 μA and 2.02 μA in the presence of 35.0 and 80.0 μM mitoxantrone, respectively. Furthermore, the peak potential related to the guanine base shifted from 816 to 825 mV and 845 mV along with the increase of mitoxantrone concentration confirming the intercalation interaction between mitoxantrone and guanine base in ds-DNA structure.
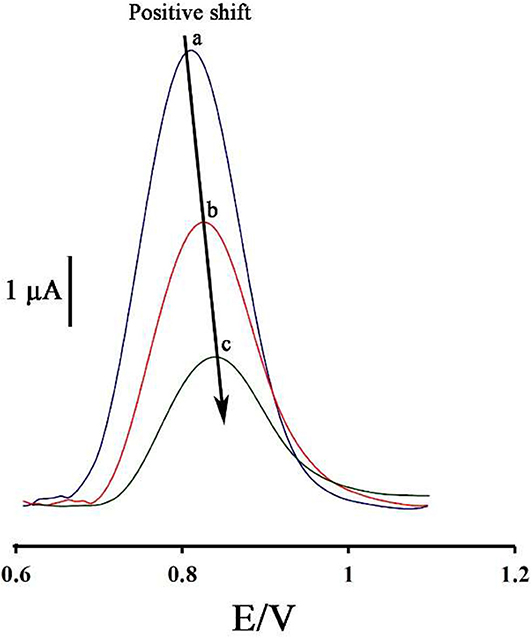
Figure 2. Differential pulse voltammograms of DNA/BMIMS/ZIF-8/CPE in the absence (a) and in the presence of 35.0 μM (b) and 80.0 μM (c) mitoxantrone, respectively.
As can be seen, along with increasing of the mitoxantrone concentration, the oxidation current of ds-DNA decreased. Accordingly, this point can be selected as an analytical factor for the determination of mitoxantrone concentration in the solution.
In order to create the best analytical conditions, it is important to optimize the significant factors in the analytical behavior of the biosensor. Therefore, the initial concentration of dsDNA, temperature, buffer types, and incubation time should be optimized.
Figure 3 displays the oxidation current of ds-DNA compared to the initial concentration of ds-DNA during the modification process.
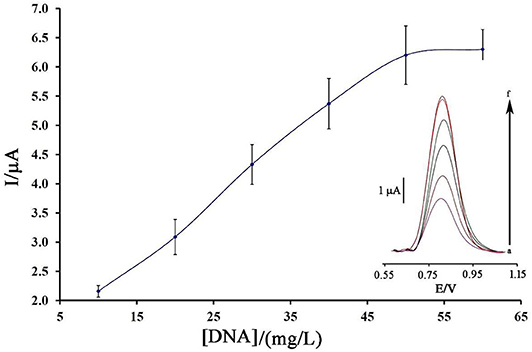
Figure 3. Plot of the oxidation current of ds-DNA compared to its concentration at surface of BMIMS/ZIF-8/CPE (n = 4). Inset) Relative differential pulse voltammograms of ds-DNA at surface of BMIMS/ZIF-8/CPE.
As can be seen, along with the increase of the initial concentration of ds-DNA, the oxidation signal of DNA/BMIMS/ZIF-8/CPE increased to a concentration of 50 mg/L, which then remained constant. Correspondingly, this point confirms that, in the solution containing 50 mg/L, the electrode surface of BMIMS/ZIF-8/CPE was saturated by ds-DNA, and also the maximum signal can be observed.
Temperature is known as one of the important factors in the fabrication of a DNA biosensor. In this regard, the changes in the ambient temperature of the test can affect the stability of the DNA at the electrode surface. Therefore, in this research, this factor was optimized. As can be seen in Figure 4A, by increasing the ambient temperature from 15 to 25°C, the ds-DNA signal has increased, and then, along with increasing the temperature up to 35°C, this signal has decreased. This point confirms that, in high temperatures, the electrode surface cannot keep ds-DNA at surface of BMIMS/ZIF-8/CPE.
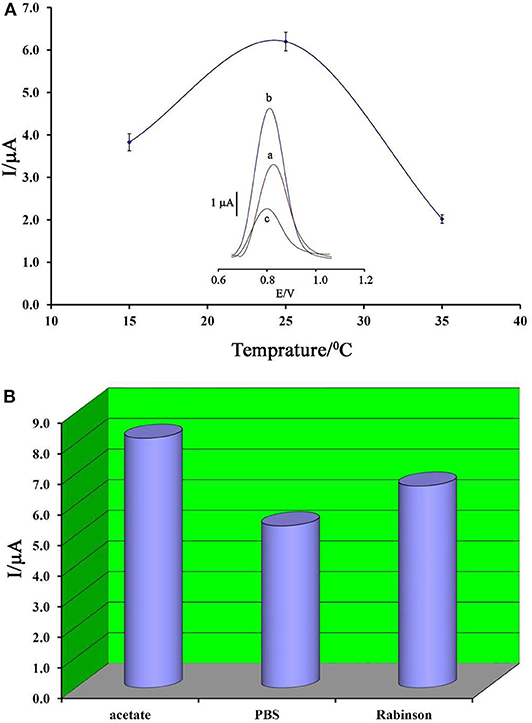
Figure 4. (A) Plot of oxidation current of ds-DNA compared to changing in temperature (n = 4). Inset) Differential pulse voltammograms of ds-DNA/BMIMS/ZIF-8/CPE recorded at (a) 15°C, (b) 25°C, and (c) 35°C. (B) Diagram of oxidation current of ds-DNA compared to the type of buffer recorded under the optimum conditions.
Accordingly, the decreased viscosity of binders in the carbon paste matrix could be considered as one of the main reasons for this point. In addition, in low temperatures, the activity of ds-DNA was low and DNA/BMIMS/ZIF-8/CPE showed a low oxidation signal.
In addition, the type of buffer is one of the main factors in the deposition of ds-DNA at surface of BMIMS/ZIF-8/CPE. Therefore, the effect of acetate buffer, Britton–Robinson buffer, and phosphate buffer solutions on the deposition step of ds-DNA were investigated. As can be seen in Figure 4B, the best oxidation signal relative to ds-DNA can be detected in the solution containing acetate buffer solution and this buffer was selected as the best condition in the next step of the experiment. Moreover, the interference between the phosphate groups of phosphate buffers or Britton–Robinson buffer can be considered as the most important factor in the creation of a weak ds-DNA signal in these buffers.
Notably, incubation time is an important factor in the final step of ds-DNA biosensor application in determining anticancer drugs. Also, the low time of the intercalation step does not allow the biosensor to have a proper interaction between the guanine base and the anticancer drugs. Also, in the long term, it is possible to release the ds-DNA-drug into solution and saturation guanine sites in ds-DNA by anticancer drugs. In this regard, the recorded data showed that 12 min is a suitable incubation time for this study (Figure 5).
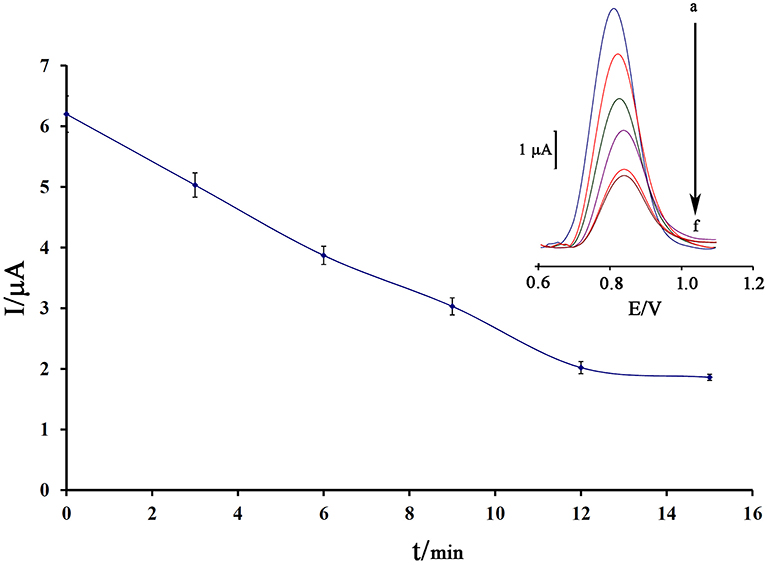
Figure 5. Effect of the incubation time of 80.0 μM mitoxantrone with DNA/BMIMS/ZIF-8/CPE on the guanine oxidation signal (n = 4). Inset Relative DPVs in incubation time investigation.
To investigate the repeatability of the DNA biosensor, four different DNA/BMIMS/ZIF-8/CPE which were fabricated by the same procedure and oxidation signal of guanine were also recorded at the surface of fabricated electrodes (Figure 6). The obtained results showed the relative standard deviation of about 2.7 and 3.1% in the current and potential of guanine signal for four electrodes that are acceptable values for a novel DNA-biosensor.
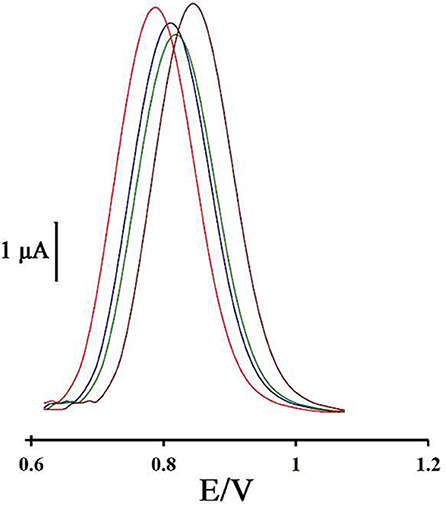
Figure 6. Differential pulse voltammogram of four DNA/ZIF-8/BMIMF/CPE prepared under the same condition.
In this research, the molecular docking study was performed to evaluate the affinity of Mitoxantrone (MTX) drug in the active site of DNA hexamer d (CGATCG) 2 containing an intercalation gap (PDB ID: 1Z3F). For conducting a comprehensive investigation on the binding orientation analysis, the best conformer with the lowest root mean square deviation (RMSD) value of 0 Å and also with the highest binding energy value, was selected. Docking of MTX into DNA sequence suggests the intercalation of the aromatic rings of Mitoxantrone drug between the cytosine and guanine base pairs of DNA with a binding energy of −6.7 kcal/mol, as shown in Figure 7A. The docked model revealed that, the hydrogen and oxygen atoms of MTX drug are participating as the donor and acceptor, to form four intermolecular hydrogen bonds (HBs) with base pairs of DNA, respectively (see Figure 7B). It was found that the interaction of oxygen (O5) atoms of the drug molecule with H22 atoms of deoxyguanosine (DG6 of chain B) of DNA leads to O5…H22-N2 conventional HB with a distance of 2.6 Å. Also, the H36 atom of the drug, as the proton donor, interacts with O4′ atom of deoxyribose sugar moiety linked to guanine (DG6 of chain B), as a proton acceptor with a distance of 2.3 Å. Furthermore, the hydrogen atom of the hydroxyl terminal group of drug molecule is bonded to the second and forth oxygen atoms of deoxycytosine (DC5 of chain B) of hexamer of DNA, i.e., O2…H60-O6 and O4′…H60-O6 with the O…H distances of 2.4 and 2.2 Å, respectively. Moreover, the hydrogen bond angles are 109.5°, 110.5°, 134.5°, and 163.6° for O5…H22-N2, O2…H60-O6, O4′…H60-O6, and O4′…H36-N8, respectively. In addition, the intermolecular interactions between carbonyl groups of MTX drug and oxygen atoms of DNA sequence, such as O2 atom of DC5 chain B, O4′ atom of DG6 chain B, and O4′ atom of DG2 chain A with the respective O…O bond lengths of 3.3, 3.5, and 3.4 Å were observed. The docking study approves the interaction between mitoxantrone drug and guanine residues of DNA that contributes in the formation of the stable MTX-DNA complex.
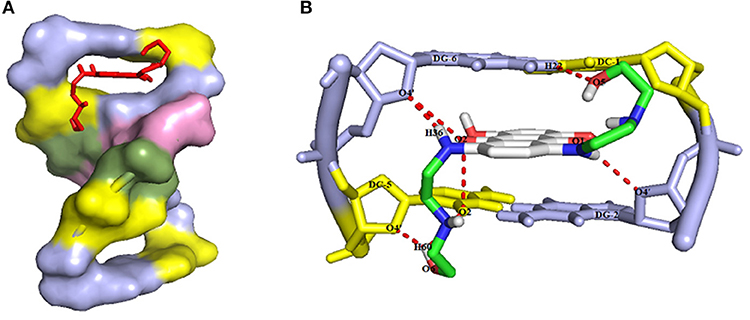
Figure 7. (A) The intercalation of MTX drug (red color) into the guanine and cytosine base pairs of DNA receptor (deoxcytosine DC and deoxyguanosine DG in yellow and blue light colors, respectively), and (B) The intermolecular hydrogen bond interactions between MTX drug and DNA hexamer d(CGATCG)2 with the numbering atoms discussed earlier in the text.
Using a decreasing trend in DNA signals and its relationship with the concentration of mitoxantrone, a linear dynamic range from 8.0 nM to 110 μM with the equation of ΔIpa = 0.052 Cmitoxantrone + 0.589 (R2 = 0.991) and a detection limit of 3.0 nM was calculated to determine mitoxantrone at surface of DNA/BMIMS/ZIF-8/CPE (Figure 8).
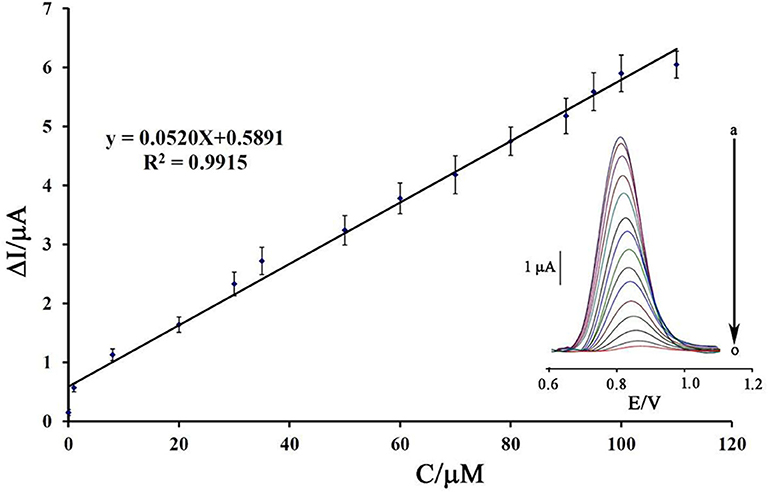
Figure 8. Plot of net current vs. mitoxantrone concentration (n = 4). Inset) DP voltammograms of DNA/BMIMS/ZIF-8/CPE in the presence of (a) 0.0; (b) 0.008; (c) 1.0; (d) 8.0; (e) 20.0; (f) 30.0; (g) 35.0; (h) 50.0; (i) 60.0; (j) 70.0; (k) 80.0; (l) 90.0; (m) 95; (n) 100, and (o) 110 μM mitoxantrone.
Binding energy between guanine and mitoxantrone can be determined by equation 1 as follows:
Where m is the binding number and Ka is the association equilibrium constant. Using the slope of recording plot in Figure 9 and equation 1, the values of m and Ka were determined to be 0.334 and 1.737 × 103 M−1, respectively.
The selectivity of DNA/BMIMS/ZIF-8/CPE, as a new biosensor, was investigated in the presence some usual organic and inorganic interference with an acceptable error of 5% in current and potential. The obtained results showed 1,000-fold of ions such as Na+, Li+, Br−, and , 500-fold of methionine, alanine, and phenylalanine, and also 400-fold of vitamin C and vitamin B2 had no interference in determination of 20 μM mitoxantrone using the suggested biosensor.
In addition, the ability of DNA/BMIMS/ZIF-8/CPE was checked as a new biosensor for determination of mitoxantrone in the injection samples using the standard addition method. The results are presented in Table 1 and recovery data between 99.18 and 102.08% confirmed the high performance ability of DNA/BMIMS/ZIF-8/CPE for determination of mitoxantrone in real samples.
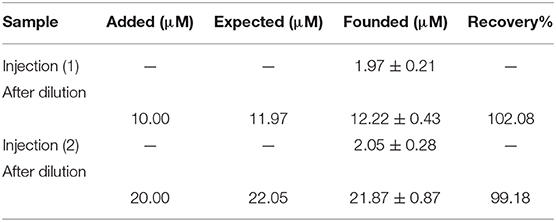
Table 1. The results related to real sample analysis of mitoxantrone by DNA/BMIMS/ZIF-8/CPE (n = 4).
In this study, a high performance DNA biosensor amplified with ZIF-8 and 1-butyl-3-methylimidazolium methanesulfonate was made-up as a new analytical tool to determine mitoxantrone anticancer drug. The presence of ZIF-8 helps in high loading of ds-DNA and also in improving the quality of the sensor in optimum conditions (T = 25°C; incubation time = 12 min; pH = 4.8 acetate buffer solution and [DNA] = 50 mg/L). In addition, the BMIMS helped as a conductive binder for improving the sensitivity of sensor for trace level analysis of mitoxantrone anticancer drug. Moreover, the DNA/BMIMS/ZIF-8/CPE was successfully used for nano-molar determination of mitoxantrone (LOD = 3.0 nM). In addition, recovery data 99.18–102.08% confirmed the high performance ability of DNA/BMIMS/ZIF-8/CPE as a new biosensor to determine mitoxantrone in the injection samples.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
MA: experimental part. PA and SM: characterization of electrochemical results. HK-M: written of paper and electrode modification design. A-MT: Checking the English level of paper and helping in intercalation investigation. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Arabali, V., Malekmohammadi, S., and Karimi, F. (2020). Surface amplification of pencil graphite electrode using CuO nanoparticle/polypyrrole nanocomposite: a powerful electrochemical strategy for determination of tramadol. Microchem. J. 158:105179. doi: 10.1016/j.microc.2020.105179
Atta, N. F., Galal, A., and Hassan, S. H. (2019). Ultrasensitive determination of nalbuphine and tramadol narcotic analgesic drugs for postoperative pain relief using nano-cobalt oxide/ionic liquid crystal/carbon nanotubes-based electrochemical sensor. J. Electroanal. Chem. 839, 48–58. doi: 10.1016/j.jelechem.2019.03.002
Baghayeri, M., Alinezhad, H., Fayazi, M., Tarahomi, M., Ghanei-Motlagh, R., and Maleki, B. (2019). A novel electrochemical sensor based on a glassy carbon electrode modified with dendrimer functionalized magnetic graphene oxide for simultaneous determination of trace Pb (II) and Cd (II). Electrochim. Acta 312, 80–88. doi: 10.1016/j.electacta.2019.04.180
Baghayeri, M., Amiri, A., Alizadeh, Z., Veisi, H., and Hasheminejad, E. (2018a). Non-enzymatic voltammetric glucose sensor made of ternary NiO/Fe3O4-SH/para-amino hippuric acid nanocomposite. J. Electroanal. Chem. 810, 69–77. doi: 10.1016/j.jelechem.2018.01.007
Baghayeri, M., Mahdavi, B., Hosseinpor-Mohsen Abadi, Z., and Farhadi, S. (2018b). Green synthesis of silver nanoparticles using water extract of Salvia leriifolia: antibacterial studies and applications as catalysts in the electrochemical detection of nitrite. Appl. Organomet. Chem. 32:e4057. doi: 10.1002/aoc.4057
Baghayeri, M., Maleki, B., and Zarghani, R. (2014). Voltammetric behavior of tiopronin on carbon paste electrode modified with nanocrystalline Fe50Ni50 alloys. Mater. Sci. Eng. C 44, 175–182. doi: 10.1016/j.msec.2014.08.023
Baghayeri, M., Rouhi, M., Lakouraj, M. M., and Amiri-Aref, M. (2017). Bioelectrocatalysis of hydrogen peroxide based on immobilized hemoglobin onto glassy carbon electrode modified with magnetic poly (indole-co-thiophene) nanocomposite. J. Electroanal. Chem. 784, 69–76. doi: 10.1016/j.jelechem.2016.12.006
Banerjee, R., Phan, A., Wang, B., Knobler, C., Furukawa, H., O'Keeffe, M., et al. (2008). High-throughput synthesis of zeolitic imidazolate frameworks and application to CO2 capture. Science 319, 939–943. doi: 10.1126/science.1152516
Beitollahi, H., Tajik, S., Mohammadi, S. Z., and Baghayeri, M. (2014). Voltammetric determination of hydroxylamine in water samples using a 1-benzyl-4-ferrocenyl-1H-[1, 2, 3]-triazole/carbon nanotube-modified glassy carbon electrode. Ionics 20, 571–579. doi: 10.1007/s11581-013-1004-0
Beytur, M., Kardaş, F, Akyildirim, O., Özkan, A., Bankoglu, B, Yüksek, H., Yola, M. L., et al. (2018). A highly selective and sensitive voltammetric sensor with molecularly imprinted polymer based silver@ gold nanoparticles/ionic liquid modified glassy carbon electrode for determination of ceftizoxime. J. Mol. Liquids 251, 212–217. doi: 10.1016/j.molliq.2017.12.060
Bijad, M., Karimi-Maleh, H., and Khalilzadeh, M. A. (2013). Application of ZnO/CNTs nanocomposite ionic liquid paste electrode as a sensitive voltammetric sensor for determination of ascorbic acid in food samples. Food Analy. Methods 6, 1639–1647. doi: 10.1007/s12161-013-9585-9
Bolanowska, W., Gessner, T., and Preisler, H. (1983). A simplified method for determination of daunorubicin, adriamycin, and their chief fluorescent metabolites in human plasma by high-pressure liquid chromatography. Cancer Chemother. Pharmacol. 10, 187–191. doi: 10.1007/BF00255759
Brett, A. O., Macedo, T., Raimundo, D., Marques, M., and Serrano, S. (1998). Voltammetric behaviour of mitoxantrone at a DNA-biosensor. Biosens. Bioelectron. 13, 861–867. doi: 10.1016/S0956-5663(98)00053-0
Chen, Y., Huang, W., Chen, K., Zhang, T., Wang, Y., and Wang, J. (2019). Facile fabrication of electrochemical sensor based on novel core-shell PPy@ ZIF-8 structures: enhanced charge collection for quercetin in human plasma samples. Sens Actuators B Chem. 290, 434–442. doi: 10.1016/j.snb.2019.04.006
Cheraghi, S., Taher, M. A., Karimi-Maleh, H., and Faghih-Mirzaei, E. (2017). A nanostructure label-free DNA biosensor for ciprofloxacin analysis as a chemotherapeutic agent: an experimental and theoretical investigation. N. J. Chem. 41, 4985–4989. doi: 10.1039/C7NJ00609H
Ensafi, A. A., Taei, M., Rahmani, H., and Khayamian, T. (2011). Sensitive DNA impedance biosensor for detection of cancer, chronic lymphocytic leukemia, based on gold nanoparticles/gold modified electrode. Electrochim. Acta 56, 8176–8183. doi: 10.1016/j.electacta.2011.05.124
Eren, T., Atar, N., Yola, M. L., and Karimi-Maleh, H. (2015). A sensitive molecularly imprinted polymer based quartz crystal microbalance nanosensor for selective determination of lovastatin in red yeast rice. Food Chem. 185, 430–436. doi: 10.1016/j.foodchem.2015.03.153
Faridbod, F., and Sanati, A. L. (2019). Graphene quantum dots in electrochemical sensors/biosensors. Curr. Anal. Chem. 15, 103–123. doi: 10.2174/1573411014666180319145506
Fouladgar, M. (2018). CuO-CNT nanocomposite/ionic liquid modified sensor as new breast anticancer approach for determination of doxorubicin and 5-fluorouracil drugs. J. Electrochem. Soc. 165, B559–B564. doi: 10.1149/2.1001811jes
Fox, E. J. (2004). Mechanism of action of mitoxantrone. Neurology 63, S15–S18. doi: 10.1212/WNL.63.12_suppl_6.S15
Gooding, J. J. (2002). Electrochemical DNA hybridization biosensors. Electroanalysis 14, 1149–1156. doi: 10.1002/1521-4109(200209)14:17<;1149::AID-ELAN1149>;3.0.CO;2-8
Hojjati-Najafabadi, A., Rahmanpour, M. S., Karimi, F., Zabihi-Feyzaba, H., Malekmohammad, S., Agarwal, S., et al. (2020). Determination of tert-Butylhydroquinone using a nanostructured sensor based on CdO/SWCNTs and ionic liquid. Int. J. Electrochem. Sci. 15, 6969–6980. doi: 10.20964/2020.07.85
Jin, Y. F, Ge, C. Y, Li, X. B, Zhang, M., Xu, G. R, and Li, D. H (2018). A sensitive electrochemical sensor based on ZIF-8–acetylene black–chitosan nanocomposites for rutin detection. RSC Adv. 8, 32740–32746. doi: 10.1039/C8RA06452K
Karimi-Maleh, H., Karimi, F., Alizadeh, M., and Sanati, A. L. (2020c). Electrochemical sensors, a bright future in the fabrication of portable kits in analytical systems. Chem. Rec. 20, 682–692. doi: 10.1002/tcr.201900092
Karimi-Maleh, H., Bananezhad, A., Ganjali, M. R., Norouzi, P., and Sadrnia, A. (2018). Surface amplification of pencil graphite electrode with polypyrrole and reduced graphene oxide for fabrication of a guanine/adenine DNA based electrochemical biosensors for determination of didanosine anticancer drug. Appl. Surf. Sci. 441, 55–60. doi: 10.1016/j.apsusc.2018.01.237
Karimi-Maleh, H., Karimi, F., Malekmohammadi, S., Zakariae, N., Esmaeili, R., Rostamnia, S., et al. (2020b). An amplified voltammetric sensor based on platinum nanoparticle/polyoxometalate/two-dimensional hexagonal boron nitride nanosheets composite and ionic liquid for determination of N-hydroxysuccinimide in water samples. J. Mol. Liquids 310:113185. doi: 10.1016/j.molliq.2020.113185
Karimi-Maleh, H., Shafieizadeh, M., Taher, M. A., Opoku, F., Kiarii, E. M., Govender, P. P., et al. (2020a). The role of magnetite/graphene oxide nano-composite as a high-efficiency adsorbent for removal of phenazopyridine residues from water samples, an experimental/theoretical investigation. J. Mol. Liquids 298:112040. doi: 10.1016/j.molliq.2019.112040
Khodadadi, A., Faghih-Mirzaei, E., Karimi-Maleh, H., Abbaspourrad, A., Agarwal, S., and Gupta, V. K. (2019). A new epirubicin biosensor based on amplifying DNA interactions with polypyrrole and nitrogen-doped reduced graphene: experimental and docking theoretical investigations. Sens Actuators B Chem. 284, 568–574. doi: 10.1016/j.snb.2018.12.164
Lenk, H., Müller, U., and Tanneberger, S. (1987). Mitoxantrone: mechanism of action, antitumor activity, pharmacokinetics, efficacy in the treatment of solid tumors and lymphomas, and toxicity. Anticancer Res. 7, 1257–1264.
Li, N., Ma, Y., Yang, C., Guo, L., and Yang, X. (2005). Interaction of anticancer drug mitoxantrone with DNA analyzed by electrochemical and spectroscopic methods. Biophys. Chem. 116, 199–205. doi: 10.1016/j.bpc.2005.04.009
Li, X., Song, J., Xue, Q., Zhao, H., Liu, M., Chen, B., et al. (2017). Sensitive and selective detection of the p53 gene based on a triple-helix magnetic probe coupled to a fluorescent liposome hybridization assembly via rolling circle amplification. Analyst 142, 3598–3604. doi: 10.1039/C7AN01255A
Li, X., Yin, F., Xu, X., Liu, L., Xue, Q., Tong, L., et al. (2020). A facile DNA/RNA nanoflower for sensitive imaging of telomerase RNA in living cells based on “zipper lock-and-key” strategy. Biosens. Bioelectron. 147:111788. doi: 10.1016/j.bios.2019.111788
Li, Y., Ji, Y., Ren, B., Jia, L., Ma, G., and Liu, X. (2019). Carboxyl-functionalized mesoporous molecular sieve/colloidal gold modified nano-carbon ionic liquid paste electrode for electrochemical determination of serotonin. Mater. Res. Bull. 109, 240–245. doi: 10.1016/j.materresbull.2018.10.002
Lu, S., Hummel, M., Chen, K., Zhou, Y., Kang, S., and Gu, Z. (2020). Synthesis of Au@ ZIF-8 nanocomposites for enhanced electrochemical detection of dopamine. Electrochem. Commun. 144:106715. doi: 10.1016/j.elecom.2020.106715
Marr, P. C., and Marr, A. C. (2016). Ionic liquid gel materials: applications in green and sustainable chemistry. Green Chem. 18, 105–128. doi: 10.1039/C5GC02277K
Movaghgharnezhad, S., and Mirabi, A. (2019). Advanced nanostructure amplified strategy for voltammetric determination of folic acid. Int. J. Electrochem. Sci. 14, 10956–10965. doi: 10.20964/2019.12.79
Mozaffari, S. A., Rahmanian, R., Abedi, M., and Amoli, H. S. (2014). Urea impedimetric biosensor based on reactive RF magnetron sputtered zinc oxide nanoporous transducer. Electrochim. Acta 146, 538–547. doi: 10.1016/j.electacta.2014.08.105
Osada de Vries, H., Scrosati, B., and Passerini, S. (2016). Ionic-liquid-based polymer electrolytes for battery applications. Angew. Chem. Int. Edn. 55, 500–513. doi: 10.1002/anie.201504971
Ozsoz, M., Erdem, A., Kara, P., Kerman, K., and Ozkan, D. (2003). Electrochemical biosensor for the detection of interaction between arsenic trioxide and DNA based on guanine signal. Electroanalysis 15, 613–619. doi: 10.1002/elan.200390077
Quang Khieu, D., Thi Thanh, M., Vinh Thien, T., Hai Phong, N., Hoang van, D., Dinh Du, P., et al. (2018). Synthesis and voltammetric determination of Pb (II) using a ZIF-8-based electrode. J. Chem. 2018, 1–12. doi: 10.1155/2018/5395106
Rahmanian, R., Mozaffari, S. A., and Abedi, M. (2015). Disposable urea biosensor based on nanoporous ZnO film fabricated from omissible polymeric substrate. Mater. Sci. Eng. C 57, 387–396. doi: 10.1016/j.msec.2015.08.004
Sanati, A. L., and Faridbod, F. (2017). Electrochemical determination of methyldopa by graphene quantum dot/1-butyl-3-methylimidazolium hexafluoro phosphate nanocomposite electrode. Int. J. Electrochem. Sci. 12, 7997–8005. doi: 10.20964/2017.09.71
Scott, L. J., and Figgitt, D. P. (2004). Mitoxantrone. CNS Drugs 18, 379–396. doi: 10.2165/00023210-200418060-00010
Tahernejad-Javazmi, F., Shabani-Nooshabadi, M., and Karimi-Maleh, H. (2019). 3D reduced graphene oxide/FeNi3-ionic liquid nanocomposite modified sensor; an electrical synergic effect for development of tert-butylhydroquinone and folic acid sensor. Comp. B Eng. 172, 666–670. doi: 10.1016/j.compositesb.2019.05.065
Tiwari, G., and Sharma, D. (2020). Electronic structure, spectra analysis and nano-range interactions of mitoxantrone with RNA base pairs: an anticancer drug. Mater. Today Proc. 29, 844–849. doi: 10.1016/j.matpr.2020.05.014
Veisi, H., Eshbala, F. H., Hemmati, S., and Baghayeri, M. (2015). Selective hydrogen peroxide oxidation of sulfides to sulfones with carboxylated multi-walled carbon nano tubes (MWCNTs-COOH) as heterogeneous and recyclable nanocatalysts under organic solvent-free conditions. RSC Adv. 5, 10152–10158. doi: 10.1039/C4RA14964E
Vollmer, T., Stewart, T., and Baxter, N. (2010). Mitoxantrone and cytotoxic drugs' mechanisms of action. Neurology 74, S41–S46. doi: 10.1212/WNL.0b013e3181c97f5a
Wang, Y., Zhang, Y., Hou, C., and Liu, M. (2015). Magnetic Fe3O4@ MOFs decorated graphene nanocomposites as novel electrochemical sensor for ultrasensitive detection of dopamine. RSC Adv. 5, 98260–98268. doi: 10.1039/C5RA20996J
Xu, C., Zhang, S., Zang, H., Yuan, B., Fan, R., Zhang, N., et al. (2018). A simple and facile electrochemical sensor for sensitive detection of histidine based on three-dimensional porous Ni foam. Int. J. Electrochem. Sci. 13, 9794–9802. doi: 10.20964/2018.10.60
Yin, F., Liu, L., Sun, X., Hou, L., Lu, Y., Xue, Q., et al. (2019). A facile deoxyuridine/biotin-modified molecular beacon for simultaneous detection of proteins and nucleic acids via a label-free and background-eliminated fluorescence assay. Analyst 144, 5504–5510. doi: 10.1039/C9AN01016E
Yuan, B., Sun, P., Zhao, L.-,j, Zhang, D., Zhang, Y., Qi, C., Niu, Y., et al. (2020). Pd nanoparticles supported on 1, 10-phenanthroline-5, 6-dione modified graphene oxide as superior bifunctional electrocatalyst for highly sensitive sensing. J. Electroanal. Chem. 861:113945. doi: 10.1016/j.jelechem.2020.113945
Yuan, B., Wang, H., Cai, J., Peng, Y., Niu, Y., Chen, H., et al. (2019). A novel oxidation-reduction method for highly selective detection of cysteine over reduced glutathione based on synergistic effect of fully fluorinated cobalt phthalocyanine and ordered mesoporous carbon, Sens. Actuators B Chem. 288, 180–187. doi: 10.1016/j.snb.2019.02.114
Yuan, B., Xu, C., Deng, D., Xing, Y., Liu, L., Pang, H., et al. (2013). Graphene oxide/nickel oxide modified glassy carbon electrode for supercapacitor and nonenzymatic glucose sensor. Electrochim. Acta 88, 708–712. doi: 10.1016/j.electacta.2012.10.102
Yuan, B., Xu, C., Zhang, R., Lv, D., Li, S., Zhang, D., et al. (2017). Glassy carbon electrode modified with 7, 7, 8, 8-tetracyanoquinodimethane and graphene oxide triggered a synergistic effect: Low-potential amperometric detection of reduced glutathione. Biosens. Bioelectron. 96, 1–7. doi: 10.1016/j.bios.2017.04.026
Keywords: mitoxantrone, ZIF-8, 1-butyl-3-methylimidazolium methanesulfonate, modified electrode, ds-DNA biosensor, drug analysis
Citation: Alizadeh M, Azar PA, Mozaffari SA, Karimi-Maleh H and Tamaddon A-M (2020) A DNA Based Biosensor Amplified With ZIF-8/Ionic Liquid Composite for Determination of Mitoxantrone Anticancer Drug: An Experimental/Docking Investigation. Front. Chem. 8:814. doi: 10.3389/fchem.2020.00814
Received: 28 June 2020; Accepted: 03 August 2020;
Published: 20 October 2020.
Edited by:
Baiqing Yuan, Ludong University, ChinaCopyright © 2020 Alizadeh, Azar, Mozaffari, Karimi-Maleh and Tamaddon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sayed Ahmad Mozaffari, bW96YWZmYXJpQGlyb3N0Lmly
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.