- 1Department of Pharmacy, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 2Haimen People's Hospital, Nantong, China
- 3School of Pharmacy, China Pharmaceutical University, Nanjing, China
Human NAD(P)H: quinone oxidoreductase (hNQO1) is an important biomarker for human malignant tumors. Detection of NQO1 accurately is of great significance to improve the early diagnosis of cancer and prognosis of cancer patients. In this study, based on the covalent assembly strategy, hNQO1-activated fluorescent probes 1 and 2 are constructed by introducing coumarin precursor 2-cyano-3-(4-(diethylamino)-2-hydroxyphenyl) acrylic acid and self-immolative linkers. Under reaction with hNQO1 and NADH, turn-on fluorescence appears due to in-situ formation of the organic fluorescent compound 7-diethylamino-3-cyanocoumarin, and fluorescent intensity changes significantly. Probe 1 and 2 for detection of hNQO1 are not interfered by other substances and have low toxicity in cells. In addition to quantitative detection of hNQO1 in vitro, they have also been successfully applied to fluorescent imaging in living cells.
Introduction
The presence of overexpressed reductase in solid tumor cells is the key to studying cancer progression. Rapid and selective reporting of cancer-related enzymes and other biologically active substances in cells will enable people to better understand tumor progression and invasiveness at the molecular level and methods of cancer diagnosis and treatment based on precision medicine. It also helps to study the targeted activation of chemotherapy drugs in tumors (Blum et al., 2007).
NAD(P)H: quinone oxidoreductase 1 (NQO1) is a cytoplasmic flavin protease that is widely distributed in the human body and can catalyze a two-electron reduction of various quinones to hydroquinone in a manner dependent on NAD(P)H (Ernster, 1967; Zhu et al., 2007; Bianchet et al., 2008; Siegel et al., 2012; Ross and Siegel, 2017; Beaver et al., 2019). Depending on the chemical properties of the hydroquinone formed, the reduction process may be a detoxification process or a biological activation process. In addition to participating in the detoxification metabolism of exogenous toxic substances in human body (Bianchet et al., 2008), NQO1 has also been found to participate in other biological processes, such as antioxidant function (Wefers et al., 1984), superoxide scavenging (Siegel et al., 2004), and regulation of the stability of specific proteins under oxidative stress (Nioi and Hayes, 2004; Dinkova-Kostova and Talalay, 2010). In many human solid tumors, the expression level is 5–200 times that of normal tissues (Zhang et al., 2018). For example, the expression of NQO1 in the liver of normal people is not abundant, but the expression level of NQO1 in the liver cancer is significantly higher than that of normal tissues by nearly 50 times (Aleksunes et al., 2006; Rougée et al., 2016). NQO1 is also highly expressed in other tumor cells, such as breast cancer (Marin et al., 1997; Yang et al., 2014), lung cancer (Siegel et al., 1998; Bey et al., 2007; Li et al., 2015), prostate cancer (Dong et al., 2010), gastric cancer (Lin et al., 2014), colon cancer (Ji et al., 2014), pancreatic cancer (Ough et al., 2005), head and neck cancer (Li et al., 2016), and cholangiocarcinoma (Wakai et al., 2011). In addition, high expression of NQO1 is closely related to the occurrence and development of cancer, tumor invasion, resistance to chemotherapy, and poor prognosis of patients.
To assess the activity of hNQO1 in living tumor model accurately and to distinguish metastasis in healthy tissue based on fluorescent methods immediately, we need to develop enzyme-activated fluorescent probes that respond quickly and have high sensitivity (Frangioni, 2008; Lee et al., 2010; Zhang J. J. et al., 2019). This type of probe can not only perform non-invasive detection on patients, but also keep normal cells intact. Real-time fluorescent imaging can achieve the purpose of constant monitoring and diagnosis. A new design strategy, the principle of covalent assembly, is used in the design and application of probes (Wu and Anslyn, 2005; Peng et al., 2012; Park and Kim, 2013; Quesneau et al., 2019). This strategy is mainly based on the reaction of non-fluorescent compounds triggered or catalyzed by the analyte, and then releases oxygen anions, which then attack the carbonyl carbon in the molecule to undergo a lactonization reaction, thereby forming a fluorescent group in situ. The advantage of this strategy is that it can greatly reduce the signal generated by the fluorescent probe itself and improve the target-to-background ratio.
Recently, a variety of NQO1-specific fluorescent probes have been reported (Punganuru et al., 2018; Kong et al., 2019; Park et al., 2019; Yuan et al., 2019; Zhang C. Y. et al., 2019; Zhu et al., 2019), but few of these probes were designed based on the principle of covalent assembly. Therefore, in order to improve the target-to-background ratio of the fluorescent probe, we adopted the principle of covalent assembly and designed NQO1-activited fluorescent probes 1 and 2 with trimethyl-locked quinone propionic acid (Q3PA) as the recognition group, coumarin precursor 2-cyano-3-(4-(diethylamino) 2-Hydroxyphenyl) acrylic acid as the fluorescent signal group and p-hydroxybenzyl alcohol or 2-mercaptoethanol as the spontaneously cleavable linker. These two probes not only have high selectivity and sensitivity against NQO1, but also have been successfully used in intracellular imaging, showing good potential in cancer diagnosis.
Materials and Methods
Instruments and Materials
1H NMR was recorded on Bruker AV-300 (300 MHz 1H, 126 MHz 13C) spectrometer. The solvents were CDCl3 (TMS as internal standard) and methanol-d4. High-resolution mass spectrometry (HRMS) was measured on Agilent 6230 mass spectrometer (ESI as ion source). The fluorescent spectrum was obtained using Shimadzu RF6000 fluorescence spectrophotometer. The absorbance was measured using Bio-Tek Synergy H1 multi-mode microplate reader. Fluorescent images were captured on Zeiss LSM700 laser confocal fluorescence microscope. hNQO1 was purchased from Sigma and NADH was purchased from Aladdin. RPMI 1640 medium, penicillin, streptomycin, and trypsin–EDTA were purchased from Gibco. All chemicals and solvents used in synthesis were purchased commercially and used without further purification unless noted otherwise.
The Synthesis of Probe 1 and 2
The synthesis of the raw material trimethyl-locked quinone propionic acid (Q3PA) refers to the synthesis methods in the existing literature. The synthesis of probe 1 and 2 can be available in Supporting Information.
Sensitivity of Probe 1 and 2 to NQO1
The probe is dissolved in DMSO to prepare a probe stock solution with a concentration of 10 mM for use, and stored in a refrigerator at 4°C. The NADH (100 μM) and the hNQO1 solutions (0.0625, 0.125, 0.25, 0.5, 1, 2, 4, 8, and 16 μg/mL) were prepared, respectively. The probe stock solution was diluted to 10 μM with PBS (10 mM, pH = 7.4) containing 0.025% DMSO. Then 100 μM NADH and different concentrations of NQO1 (0–16 μg/mL) are added to mix evenly and placed in a water bath at 37°C for 10 min. For all fluorescence spectroscopy studies, the fluorescent emission spectrum from 450 to 600 nm were measured and recorded with an excitation wavelength of 435 nm.
Selectivity of Probe 1 and 2 to NQO1
Another important parameter for evaluating the performance of newly designed fluorescent probes is high selectivity. Therefore, in order to evaluate the selectivity of these two probes, ions (Ca2+, K+, Mg2+, Na+, Cl−), amino acids (Arg, Ala, Cys, Gly, GSH, His, Tyr) and NADH were selected as detection targets. The probe stock solution was diluted to 10 μM in PBS (10 mM, pH = 7.4) containing 0.025% DMSO, and various interference reagents (100 μM) prepared in advance were added. The mixed solution was placed in a water bath at 37°C and reacted for 10 min. The fluorescent emission spectrum from 450 to 600 nm were measured and recorded with an excitation wavelength of 435 nm.
Cytotoxicity Assay
A549 cell culture: 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin were added to the 1640 medium and mixed as the culture medium for A549 cells. A549 cells with appropriate density and culture medium were added to T-25 flasks and cultured in a constant temperature incubator (37°C, 5% CO2). During passage, cells were digested with 0.25% trypsin digestion solution (containing 0.02% EDTA). The supernatant was discarded by centrifugation, and then medium was added to resuspend the cells. Finally, a small amount of cells was added to the T-25 flask for cultivation.
The CCK-8 experiment was used to evaluate the cytotoxicity of these two probes. The grown A549 cells were seeded into 96-well plates (10,000 cells/well) and incubated in a constant temperature incubator for 24 h. After discarding the original medium, 100 μL of 1640 medium containing different concentrations of probe (0, 2, 4, 6, 8, 12, 16, and 20 μM) was added and incubated for 24 h. The medium was then discarded and rinsed three times with PBS solution, and each well was filled with 1640 medium containing CCK-8 solution. After incubating the 96-well plate for about 20–60 min, the absorbance at 490 nm was measured. The cell viability was calculated using the following formula.
As: absorbance of cells incubated with probes of various concentrations; Ac: absorbance of cells incubated with 0 μM probe; Ab: absorbance of wells with medium containing CCK-8 solution.
Bioimaging Application
The grown A549 cells were seeded into a confocal dish at an appropriate density and divided into two groups: the probe group and the dicoumarin group. Both groups of cells were cultured in a constant temperature incubator for 24 h. The medium in the dicoumarin group was discarded, and then the medium containing 100 μM dicoumarin (DIC) was added and incubated for 12 h. After discarding the original medium of these two groups of cells, the medium containing 20 μM probe was added to continue incubation for 3 h. After rinsing with PBS solution for three times, fluorescent images were taken under a confocal laser scanning microscopy and fluorescent signals were recorded.
Results and Discussion
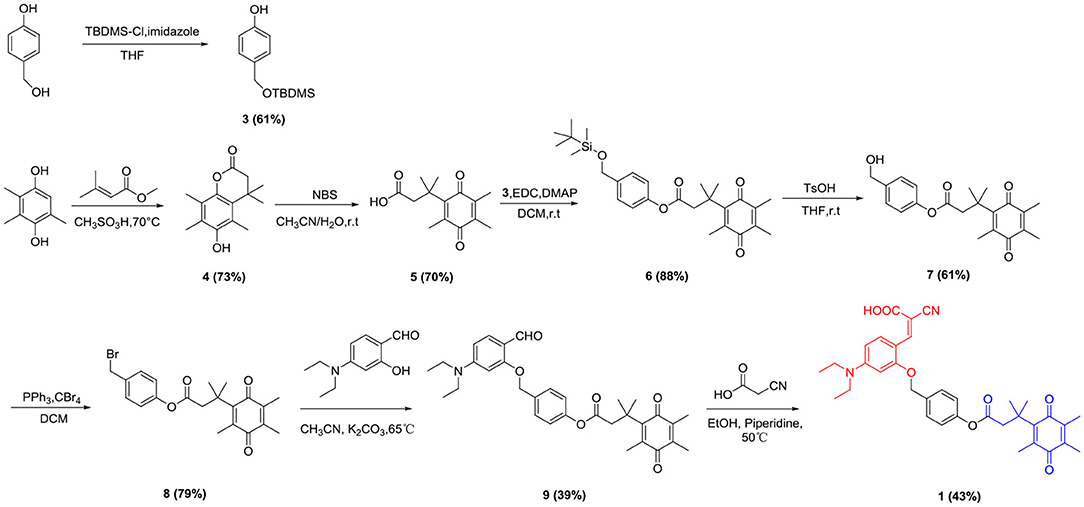
Synthesis Route of the Probe 1 and Probe 2
Trimethyl-locked quinone propionic acid (Q3PA) is an excellent enzyme substrate group, which can be quickly recognized and reduced by the NQO1 enzyme and undergo an intramolecular esterification reaction, which can quickly release the fluorescent group. Therefore, Q3PA was selected as the recognition group of the probe 1 and 2. Covalent-assembly based probes are in a non-fluorescent state before being triggered. After being activated by specific enzymes, fluorescent reporter groups are formed in situ. Such probes have high target background ratio and sensitivity and coumarin which was commonly used as organic fluorescent group has excellent spectral properties. Therefore, the coumarin precursor compound 2-cyano-3-(4-(diethylamino)-2-hydroxyphenyl) acrylic acid was selected as fluorescent signal group and was connected to Q3PA via p-hydroxybenzyl alcohol or 2-mercaptoethanol linker to design and synthesize novel fluorescent probe 1 and probe 2 for detecting hNQO1. The detailed synthesis of probe 1 was outlined in Scheme 1 and the detailed synthesis of probe 2 was outlined in Scheme 2. The structures of all intermediates and the final product were confirmed by nuclear magnetic resonance (NMR) and high resolution mass spectrometry (HRMS).
Spectrum Properties
It has been reported that NADH can react with dyes such as rhodamine, methylene blue, and resorcinol, thereby interfering with the fluorescent detection of NQO1 (Best et al., 2016). However, as shown in Figure S1, probe 1 and probe 2 showed only weak fluorescence with a maximum excitation wavelength at 435 nm, and the mixture with NADH also showed slightly enhanced fluorescence which indicated that the determination of hNQO1 by utilizing probe 1 and 2 will not be interfered by NADH. And absorbance spectra of probe 1 (A) and probe 2 (B) in the presence of hNQO1 and NADH were also investigated (Figure S2).
To further explore the ability of probe 1 and probe 2 for detecting hNQO1, their sensitivity to hNQO1 under the optimal enzyme reaction conditions were investigated. Probe 1 (10 μM) and probe 2 (10 μM) were mixed with NADH (100 μM) and different concentrations of hNQO1 (0–16 μg/mL), respectively. The fluorescent spectrum was measured. As shown in Figure 1A, the fluorescent intensity of probe 1 was extremely weak under excitation at 435 nm. After adding 0.5 μg/mL hNQO1, the fluorescence at 490 nm began to change significantly and increased with the addition of hNQO1. When the concentration of hNQO1 was 16 μg/mL, the fluorescent intensity increased about 10-folds. In addition, the fluorescence intensity of probe 1 at 490 nm has a good linear relationship (R2 = 0.9914) with the concentration of hNQO1 (0.125–4 μg/mL) (Figure 1B).
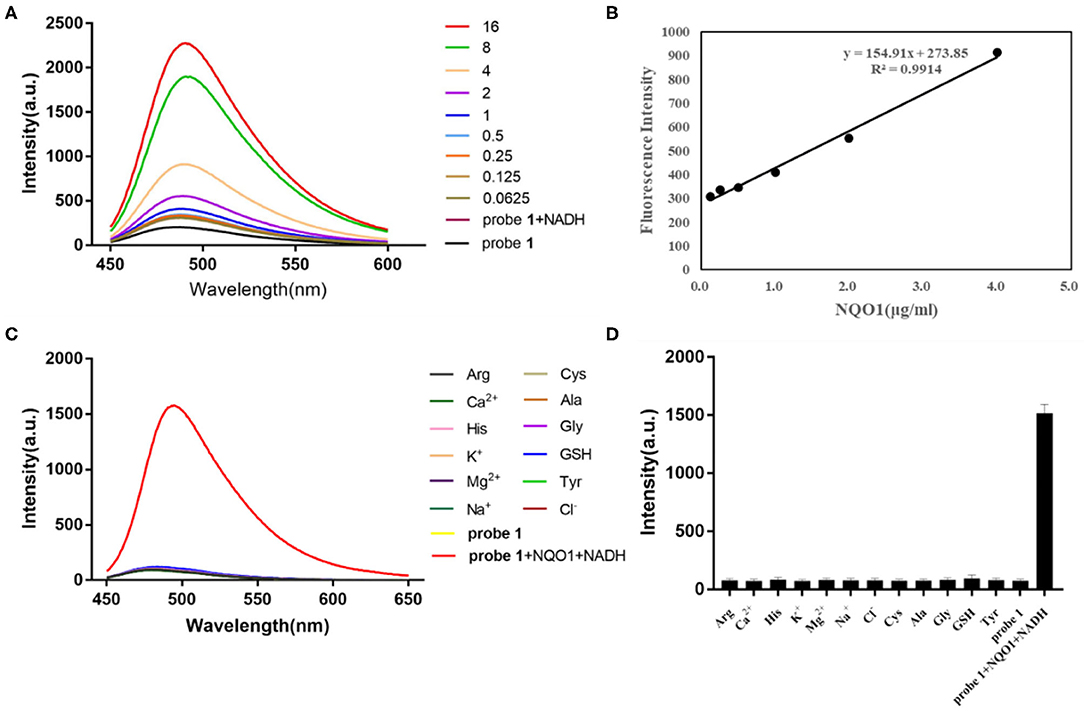
Figure 1. (A) Fluorescent spectrum of probe 1 (10 μM) with different concentrations of hNQO1 (0–16 μg/mL) and NADH (100 μM) in PBS buffer (10 mM, pH = 7.4). λex = 435 nm, excitation slit width = 5.0 nm, emission slit width = 3.0 nm. (B) Linear relationship (R2 = 0.9914) between the fluorescent intensity of probe 1 at 490 nm and the concentration of hNQO1 (0.125–4 μg/mL). (C) Fluorescence responses of probe 1 (10 μM) upon adding with NQO1 (16 μg/mL), NADH (100 μM) and other biologically-relevant species (100 μM) in PBS buffer (10 mM, pH = 7.4). (D) Bars represent the fluorescent intensity of probe 1 at 490 nm with hNQO1 and other biologically relevant substances.
Similarly, the fluorescent intensity of probe 2 at 490 nm increased sharply with the addition of hNQO1 (Figure 2). When the concentration of hNQO1 was 8 μg/mL, the fluorescent intensity was close to the peak and increased by nearly 7-folds compared with the fluorescence generated by probe itself. Moreover, a linear functional relationship (R2 = 0.9983) was obtained between the concentration of hNQO1 (0.25–4 μg/mL) and the fluorescence intensity of probe 2 at 490 nm. These results indicated that probe 1 and probe 2 have high sensitivity to NQO1, and can be used for further selective experiments and biological imaging applications.
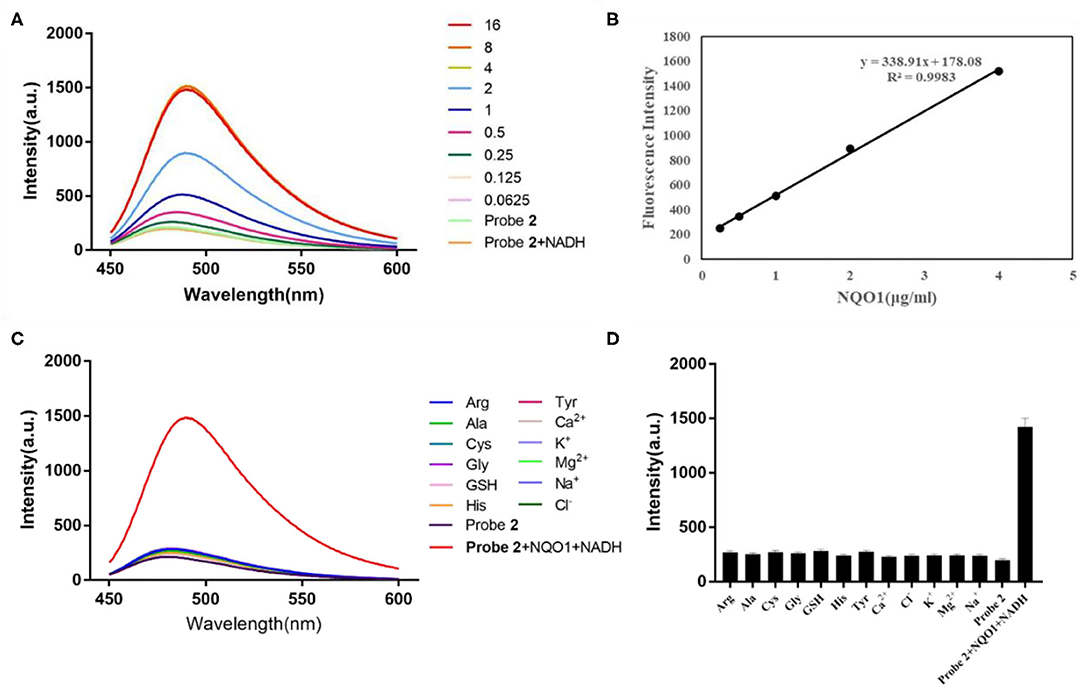
Figure 2. (A) Fluorescent spectrum of probe 2 (10 μM) after adding different concentrations of hNQO1 (0–16 μg/mL) and NADH (100 μM) in PBS buffer (10 mM, pH = 7.4). λex = 435 nm, excitation slit width = 3.0 nm, emission slit width = 5.0 nm. (B) Linear relationship (R2 = 0.9983) between the fluorescent intensity of probe 2 at 490 nm and the concentration of hNQO1 (0.125–4 μg/mL). (C) Fluorescence responses of probe 2 (10 μM) upon adding with NQO1 (16 μg/mL), NADH (100 μM) and other biologically-relevant species (100 μM) in PBS buffer (10 mM, pH = 7.4). (D) Bars represent the fluorescent intensity of probe 2 at 490 nm with hNQO1 and other biologically relevant substances.
Selectivity Evaluation
Another important parameter for evaluating the performance of newly designed fluorescent probes is high selectivity. It can be clearly observed in Figure 1C that the fluorescence spectrum of the solution did not show significant changes when probe 1 was incubated with interfering substances other than hNQO1. However, probe 1 incubated with hNQO1 showed a significant fluorescence emission peak at 490 nm, and the fluorescence intensity was significantly enhanced. In addition, Figure 1D showed that the change in fluorescence intensity of probe 1 caused by hNQO1 was nearly 20 times that of other interferences. Similarly, except for hNQO1, the fluorescence of probe 2 after adding other interfering substances was almost unchanged, and the fluorescent intensity of probe 2 caused by hNQO1 is nearly 6 times that of other interferents. These results indicated that regardless of the presence of other biologically relevant substances, probe 1 and probe 2 can be used as fluorescent turn-on probes to detect the activity of hNQO1 in cells with high selectivity.
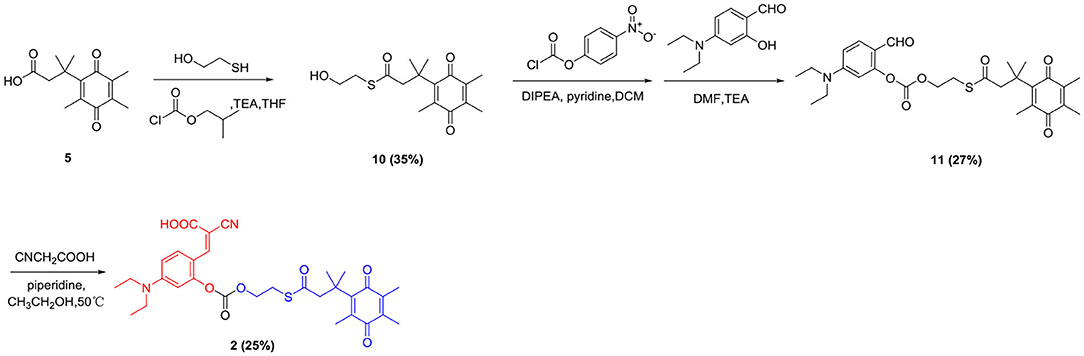
Reaction Mechanism
As shown in Scheme 3, probe 1 and probe 2 had almost no fluorescence. In the presence of NADH, hNQO1 played a role of two-electron reduction, reducing the quinone in probe 1 to hydroquinone, and then a cascade reaction occurred. The self- cleavage of the connecting linker led to the release of 4-methylenecyclohex-2,5-dien-1-one and the in-situ formation of the fluorescent compound 7-diethylamino-3-cyanocoumarin (compound 12). Compound 12 is composed of three parts, an electron donor part composed of a diethylamino group, a conjugated part composed of benzopyran, and an electron acceptor part composed of a carbonyl group and a cyano group. This D-π-A structure makes it have excellent spectral properties. The mechanism of probe 2 for selective identification and detection of hNQO1 is similar to that of probe 1. After responding to hNQO1, 1,3-oxathiolan-2-one and 7-diethylamino-3-cyanocoumarin were released and the fluorescence turned on. In addition, the results of HRMS confirmed the mechanism of probe 1 and probe 2 with the addition of hNQO1 and NADH. Fluorescent product compound 12 can be detected in the solution after the reaction of these two probes with hNQO1.
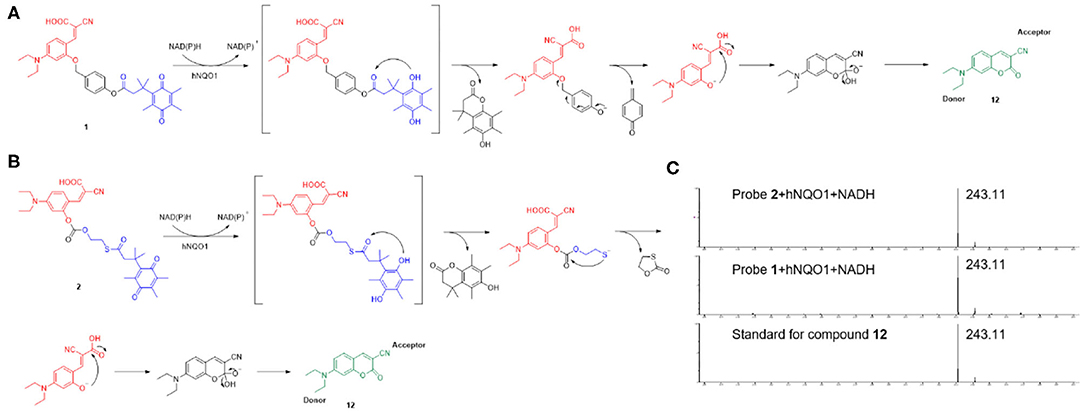
Scheme 3. The potential response mechanism of probe 1 (A) and probe 2 (B) for hNQO1 detection and the structure of compound 12. (C) HRMS spectra of standard for compound 12 and the reaction mixture of probe 1 and probe 2 with the addition of hNQO1 and NADH.
Cytotoxicity Evaluation
Considering the effectiveness and selectivity of probe 1 and probe 2 in detecting the activity of hNQO1 in vitro, we further studied the ability of these two probes to identify and distinguish tumor cells from normal cells with imaging in living cells. First, in order to evaluate the compatibility of the probes with biological systems, we used the CCK-8 method to investigate the toxic effects of these two probes on A549 cells. As shown in Figure 3, the viability of A549 cells incubated with probe 1 or probe 2 at different concentrations was above 80%. These results indicated that probe 1 and probe 2 have low toxicity and can be further used for biological imaging applications.
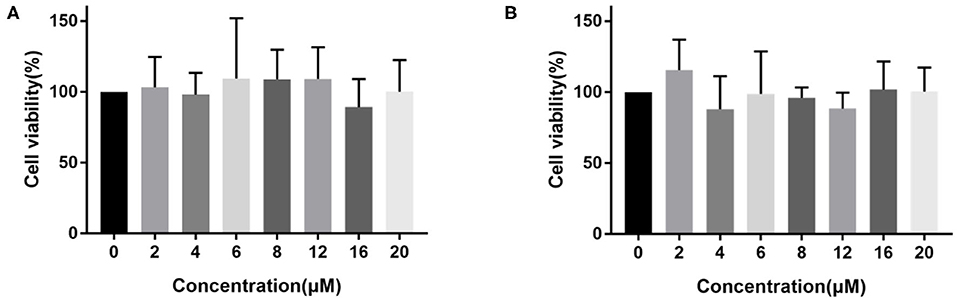
Figure 3. The viability of A549 cells incubated with different concentrations (0–20 μM) of probe 1 (A) and probe 2 (B).
Fluorescence Imaging in Cancer Cells
We next investigate the ability of these two probes to detect hNQO1 in living cells. A549 cells with high expression of hNQO1 were selected as detection objects and co-incubated with the probe for fluorescence confocal imaging. At the same time, A549 cells were pretreated with the hNQO1 inhibitor dicoumarol and co-incubated with the probe for fluorescence confocal imaging.
As can be seen from Figure 4, the A549 cells incubated with probe 1 showed obvious green fluorescence, and the fluorescence was mainly distributed in the cytoplasm. This indicates that probe 1 can penetrate the cell membrane into the cell and can also be triggered by hNQO1 to form a coumarin derivative in situ, releasing strong fluorescence. In contrast, after the cells were treated with dicoumarol, the hNQO1 activity in the cells was significantly inhibited. The lack of hNQO1 activation caused probe 1 to be unable to form a fluorophore in situ and eventually showed only weak fluorescence. This also confirmed that probe 1 can only be activated in the presence of hNQO1, resulting in an enhanced fluorescence signal. Similarly, as can be seen from Figure 5, probe 2 also showed significant green fluorescence in A549 cells, while the control group showed only weak fluorescence. In conclusion, probe 1 and probe 2 have good selectivity and sensitivity to hNQO1 in living cells, and are expected to be used for early diagnosis of cancer.
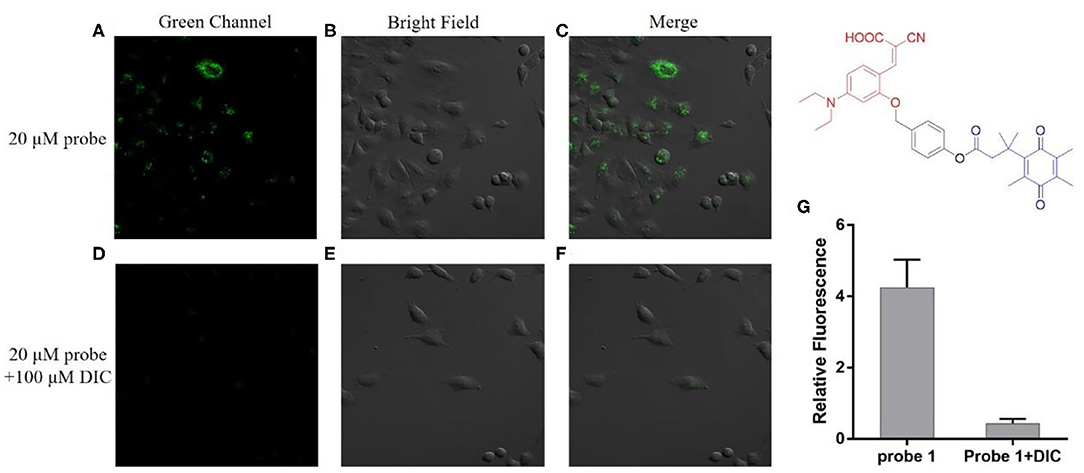
Figure 4. Fluorescence confocal imaging of A549 cells after incubation with probe 1 (A–C) and A549 cells pretreated with dicoumarol after incubation with probe 1 (D–F). λex = 488 nm. (G) Relative fluorescence intensity of the images from (A–F).
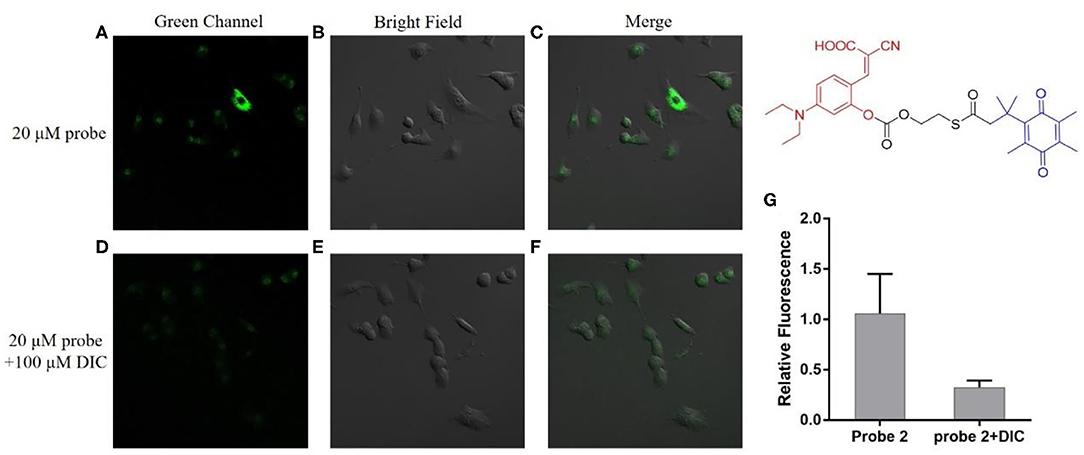
Figure 5. Fluorescence confocal imaging of A549 cells after incubation with probe 2 (A–C) and A549 cells pretreated with dicoumarol after incubation with probe 2 (D–F). λex = 488 nm. (G) Relative fluorescence intensity of the images from (A–F).
Conclusion
In summary, based on the principle of covalent assembly, we used NQO1-specific Q3PA as the recognition group, coumarin precursor (Z)-2-cyano-3-(4-(diethylamino)-2-hydroxyphenyl)acrylic acid as the fluorescent signal group, and p-hydroxybenzyl alcohol or 2-mercaptoethanol as the connecting linker to design and synthesize the enzyme-activated fluorescent probe 1 and probe 2. These two probes were excellent in selectivity and sensitivity. They could be quickly recognized and reduced by NQO1, and were not interfered by NADH and other biological related substances including ions and amino acids. Probe 1 and probe 2 showed weak fluorescence when excited at 435 nm. When they were incubated with NQO1 and NADH, an obvious emission peak at 490 nm was observed at the same excitation wavelength, and the fluorescent intensity increased by nearly 7–10 times. This change was due to the in-situ formation and release of the fluorescent compound 7-diethylamino-3-cyanocoumarin. Probe 1 has a good linear relationship with the concentration of hNQO1 in the range of 0.125–4 ug/ml, while probe 2 has a linear relationship with the concentration of hNQO1 in the range of 0.25–4 ug/ml. These two probes have good water solubility, good stability, and low toxicity to cells. In addition to quantitative detection of hNQO1 activity in vitro, they have also been used in fluorescence imaging in living cells successfully. Compared with dicoumarol-treated A549 cells, probes 1 and 2 showed significantly enhanced green fluorescent signals in A549 cells which have high expression of NQO1. This result shows that the human cancer cells with different hNQO1 activity levels can be easily detected and differentiated according to the fluorescent intensity displayed by probe 1 and probe 2. Therefore, NQO1-activated probe 1 and probe 2 have better application prospects in early clinical diagnosis and evaluation of drug efficacy in preclinical studies.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
XX and CG directed the experiments and wrote the paper. JH and LC did the experiments. YZ helped JH in some experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities 2632019ZD20 (XX) and National Talent Project for technical heritage of Traditional Chinese medicine 2014181 (CG).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2020.00756/full#supplementary-material
References
Aleksunes, L. M., Goedken, M., and Manautou, J. E. (2006). Up-regulation of NAD(P)H quinone oxidoreductase 1 during human liver injury. World J. Gastroenterol. 12, 1937–1940. doi: 10.3748/wjg.v12.i12.1937
Beaver, S. K., Mesa-Torres, N., Pey, A. L., and Timson, D. J. (2019). NQO1: a target for the treatment of cancer and neurological diseases, and a model to understand loss of function disease mechanisms. Biochim. Biophys. Acta Protein Proteonomics 1867, 663–676. doi: 10.1016/j.bbapap.2019.05.002
Best, Q. A., Johnson, A. E., Prasai, B., Rouillere, A., and McCarley, R. L. (2016). Environmentally robust rhodamine reporters for probe-based cellular detection of the cancer-linked oxidoreductase hNQO1. ACS Chem. Biol. 11, 231–240. doi: 10.1021/acschembio.5b00792
Bey, E. A., Bentle, M. S., Reinicke, K. E., Dong, Y., Yang, C. R., Girard, L., et al. (2007). An NQO1- and PARP-1-mediated cell death pathway induced in non-small-cell lung cancer cells by beta-lapachone. Proc. Natl. Acad. Sci. U. S. A. 104, 11832–11837. doi: 10.1073/pnas.0702176104
Bianchet, M. A., Erdemli, S. B., and Amzel, L. M. (2008). Structure, function, and mechanism of cytosolic quinone reductases. Vitam. Horm. 78, 63–84. doi: 10.1016/S0083-6729(07)00004-0
Blum, G., von Degenfeld, G., Merchant, M. J., Blau, H. M., and Bogyo, M. (2007). Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat. Chem. Biol. 3, 668–677. doi: 10.1038/nchembio.2007.26
Dinkova-Kostova, A. T., and Talalay, P. (2010). NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 501, 116–123. doi: 10.1016/j.abb.2010.03.019
Dong, Y., Bey, E. A., Li, L. S., Kabbani, W., Yan, J. S., Xie, X. J., et al. (2010). Prostate cancer radiosensitization through poly(ADP-Ribose) polymerase-1 hyperactivation. Cancer Res, 70, 8088–8096. doi: 10.1158/0008-5472.CAN-10-1418
Frangioni, J. V. (2008). New technologies for human cancer imaging. J. Clin. Oncol. 26, 4012–4021. doi: 10.1200/JCO.2007.14.3065
Ji, L. L., Wei, Y. Z., Jiang, T., and Wang, S. Y. (2014). Correlation of Nrf2, NQO1, MRP1, cmyc and p53 in colorectal cancer and their relationships to clinicopathologic features and survival. Int. J. Clin. Exp. Pathol. 7, 1124–1131 doi: 10.1016/j.prp.2014.02.001
Kong, F. P., Li, Y., Yang, C., Li, X., Wu, J. L., Liu, X. J., et al. (2019). A fluorescent probe for simultaneously sensing NTR and hNQO1 and distinguishing cancer cells. J. Mater. Chem. B. 7, 6822–6827. doi: 10.1039/C9TB01581G
Lee, S., Xie, J., and Chen, X. Y. (2010). Activatable molecular probes for cancer imaging. Curr. Top. Med. Chem. 10, 1135–1144. doi: 10.2174/156802610791384270
Li, L. S., Reddy, S., Lin, Z. H., Liu, S. P., Park, H., Chun, S. G., et al. (2016). NQO1-mediated tumor-selective lethality and radiosensitization for head and neck cancer. Mol. Cancer Ther. 15, 1757–1767. doi: 10.1158/1535-7163.MCT-15-0765
Li, Z. L., Zhang, Y., Jin, T. F., Men, J. G., Lin, Z. H., et al. (2015). NQO1 protein expression predicts poor prognosis of non-small cell lung cancers. BMC Cancer 15, :207. doi: 10.1186/s12885-015-1227-8
Lin, L. J., Qin, Y. Z., Jin, T. F., Liu, S. P., Zhang, S. N., et al. (2014). Significance of NQO1 overexpression for prognostic evaluation of gastric adenocarcinoma. Exp. Mol. Pathol. 96, 200–205. doi: 10.1016/j.yexmp.2013.12.008
Marin, A., Lopez de Cerain, A., Hamilton, E., Lewis, A., D., Martinez-Penuela, J. M., et al. (1997). DT-diaphorase and cytochrome B5 reductase in human lung and breast tumours. Br. J. Cancer. 76, 923–929. doi: 10.1038/bjc.1997.485
Nioi, P., and Hayes, J. D. (2004). Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat. Res. 555, 149–171. doi: 10.1016/j.mrfmmm.2004.05.023
Ough, M., Lewis, A., Bey, E. A., Gao, J., Ritchie, J. M., et al. (2005). Efficacy of beta-lapachone in pancreatic cancer treatment: exploiting the novel, therapeutic target NQO1. Cancer Biol. Ther. 4, 95–102. doi: 10.4161/cbt.4.1.1382
Park, J., and Kim, Y. (2013). An improved fluorogenic substrate for the detection of alkaline phosphatase activity. Bioorg. Med. Chem. Lett. 23, 2332–2335. doi: 10.1016/j.bmcl.2013.02.063
Park, S. Y., Won, M., Kang, C., Kim, J. S., and Lee, M. H. (2019). A coumarin-naphthalimide hybrid as a dual emissive fluorescent probe for hNQO1. Dyes Pigment. 164, 341–345. doi: 10.1016/j.dyepig.2019.01.050
Peng, Y., Dong, Y. M., Dong, M., and Wang, Y. W. (2012). A selective, sensitive, colorimetric, and fluorescence probe for relay recognition of fluoride and Cu(II) ions with “off-on-off” switching in ethanol-water solution. J. Org. Chem. 77, 9072–9080. doi: 10.1021/jo301548v
Punganuru, S. R., Madala, H. R., Arutla, V., and Srivenugopal, K. S. (2018). Cancer-specific biomarker hNQO1-activatable fluorescent probe for imaging cancer cells in vitro and in vivo. Cancers. 10, :470. doi: 10.3390/cancers10120470
Quesneau, V., Roubinet, B., Renard, P. Y., and Romieu, A. (2019). Reinvestigation of the synthesis of “covalent-assembly” type probes for fluoride ion detection. Identification of novel 7-(diethylamino)coumarins with aggregation-induced emission properties. Tetrahedron Lett. 60, :8. doi: 10.26434/chemrxiv.9742688
Ross, D., and Siegel, D. (2017). Functions of NQO1 in cellular protection and CoQ10 metabolism and its potential role as a redox sensitive molecular switch. Front. Physiol. 8, :595. doi: 10.3389/fphys.2017.00595
Rougée, L. R., Riches, Z., Berman, J. M., and Collier, A. C. (2016). The ontogeny and population variability of human hepatic NADPH dehydrogenase quinone oxido-reductase 1 (NQO1). Drug Metab Dispos. 44, 967–974. doi: 10.1124/dmd.115.068650
Siegel, D., Franklin, W. A., and Ross, D. (1998). Immunohistochemical detection of NAD(P)H:quinone oxidoreductase in human lung and lung tumors. Clin. Cancer Res. 4, 2065–2070
Siegel, D., Gustafson, D. L., Dehn, D. L., Han, J. Y., Boonchoong, P., Berliner, L. J., et al. (2004). NAD(P)H:quinone oxidoreductase 1: role as a superoxide scavenger. Molecular Mol. Pharmacology Pharmacol. 65, 1238–1247. doi: 10.1124/mol.65.5.1238
Siegel, D., Yan, C., and Ross, D. (2012). NAD(P)H:quinone oxidoreductase 1 (NQO1) in the sensitivity and resistance to antitumor quinones. Biochem. Pharmacol. 83, 1033–1040. doi: 10.1016/j.bcp.2011.12.017
Wakai, T., Shirai, Y., Sakata, J., Matsuda, Y., Korita, P. V., Takamura, M., et al. (2011). Prognostic significance of NQO1 expression in intrahepatic cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 4, 363–370 doi: 10.5754/hge11175
Wefers, H., Komai, T., Talalay, P., and Sies, H. (1984). Protection against reactive oxygen species by NAD(P)H: quinone reductase induced by the dietary antioxidant butylated hydroxyanisole (BHA). Decreased hepatic low-level chemiluminescence during quinone redox cycling. FEBS Lett. 169, 63–66. doi: 10.1016/0014-5793(84)80290-2
Wu, Q. Y., and Anslyn, E. V. (2005). Heavy metal analysis using a Heck-catalyzed cyclization to create coumarin. J. Mater. Chem. 15, 2815–2819. doi: 10.1039/b502199p
Yang, Y., Zhang, Y., Wu, Q. Y., Cui, X. L., Lin, Z. H., Liu, S. P., et al. (2014). Clinical implications of high NQO1 expression in breast cancers. J. Exp. Clin. Cancer Res. 33, :14. doi: 10.1186/1756-9966-33-14
Yuan, Z. W., Xu, M. J., Wu, T. Z., Zhang, X. Y., Shen, Y. Z., Ernest, U., et al. (2019). Design and synthesis of NQO1 responsive fluorescence probe and its application in bio-imaging for cancer diagnosis. Talanta. 198, 323–329. doi: 10.1016/j.talanta.2019.02.009
Zhang, C. Y., Zhang, Q. Z., Zhang, K., Li, L. Y., Pluth, M. D., et al. (2019). Dual-biomarker-triggered fluorescence probes for differentiating cancer cells and revealing synergistic antioxidant effects under oxidative stress. Chem. Sci. 10, 1945–1952. doi: 10.1039/C8SC03781G
Zhang, J. J., Chai, X. Z., He, X. P., Kim, H. J., Yoon, J., and Tian, H. (2019). Fluorogenic probes for disease-relevant enzymes. Chem. Soc. Rev. 48, 683–722. doi: 10.1039/C7CS00907K
Zhang, K. J., Chen, D., Ma, K., Wu, X. X., Hao, H. P., and Jiang, S. (2018). NAD(P)H:quinone oxidoreductase 1 (NQO1) as a therapeutic and diagnostic target in cancer. J. Med. Chem. 61, 6983–7003. doi: 10.1021/acs.jmedchem.8b00124
Zhu, H., Jia, Z., Mahaney, J. E., Ross, D., Misra, H. P., Trush, M. A., et al. (2007). The highly expressed and inducible endogenous NAD(P)H:quinone oxidoreductase 1 in cardiovascular cells acts as a potential superoxide scavenger. Cardiovasc. Toxicol. 7, 202–211. doi: 10.1007/s12012-007-9001-z
Keywords: NQO1, fluorescent probe, cell imaging, covalent assembly, living cell
Citation: Han J, Cheng L, Zhu Y, Xu X and Ge C (2020) Covalent-Assembly Based Fluorescent Probes for Detection of hNQO1 and Imaging in Living Cells. Front. Chem. 8:756. doi: 10.3389/fchem.2020.00756
Received: 23 June 2020; Accepted: 21 July 2020;
Published: 26 August 2020.
Edited by:
Guangle Niu, Shandong University, ChinaReviewed by:
Yuncong Chen, Nanjing University, ChinaRuoyao Zhang, Hong Kong University of Science and Technology, Hong Kong
Copyright © 2020 Han, Cheng, Zhu, Xu and Ge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowei Xu, eHdAY3B1LmVkdS5jbg==; Chaoliang Ge, Z2VjaGFvbGlhbmdAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Jialing Han2†
Jialing Han2† Longhao Cheng
Longhao Cheng Xiaowei Xu
Xiaowei Xu