
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 21 April 2020
Sec. Medicinal and Pharmaceutical Chemistry
Volume 8 - 2020 | https://doi.org/10.3389/fchem.2020.00270
This article is part of the Research Topic Peptide/Polyketide Molecules From Marine Macro and/or Microorganisms View all 7 articles
 Lu Wang1†
Lu Wang1† Xianyan Zhang1†
Xianyan Zhang1† Kaijin Zhang1
Kaijin Zhang1 Xiaomin Zhang1
Xiaomin Zhang1 Tianjiao Zhu1
Tianjiao Zhu1 Qian Che1
Qian Che1 Guojian Zhang1,2*
Guojian Zhang1,2* Dehai Li1,2*
Dehai Li1,2*Overexpression of the PbrlaeA gene of the fungus Penicillium brocae HDN-12-143 resulted in the isolation of four compounds including fumigatin chlorohydrin (1), whose configuration has not been reported before, and one new compound iso-fumigatin chlorohydrin (2). All structures including absolute configurations were elucidated on the basis of comprehensive spectroscopic data, 13C NMR calculations, and ECD calculations. Compounds 1 and 2 exhibited cytotoxic activity against HL-60 with IC50 of 18.63 and 24.83 μM.
LaeA is a broad-domain factor that can regulate the production of secondary metabolites (Keller et al., 2006; Kosalkova et al., 2009; Sarikaya et al., 2010). Overexpression of LaeA can enhance, or activate, the expression of gene clusters and produce new secondary metabolites (Jiang et al., 2016). During our recent work within the scope of exploring chemical diversity of fungal strains using tools manipulating the LaeA factor, the fungus Penicillium brocae HDN-12-143, isolated from sediment collected in the South China Sea, was selected as a candidate host to overexpress the LaeA gene due to its simple secondary metabolites background in comparison with its rich biosynthetic gene clusters, defined on the basis of bioinformatic analysis from the genome sequence, which shows a promising potential for secondary metabolite production (Figure S2).
For a start, we retrieved a native global regulator LaeA in Penicillium brocae 12-143 (PbrLaeA) by using LocalBLAST and InterPro analysis. Further overexpression of PbrLaeA in Penicillium brocae 12-143 led to the isolation of four compounds, including fumigatin chlorohydrin (1), whose configuration has not been reported before, as well as a new compound—iso-fumigatin chlorohydrin (2). Herein, we will report the biosynthetic pathway activation by overexpression of global regulator PbrLaeA, the isolation and characterization of resulting compounds, as well as the biological activity evaluation results.
The wild type and mutant strains of Penicillium brocae HDN12-143 were cultured on Difco™ Potato Dextrose Agar (Becton, Dickinson and Company, Sparks, USA). Full genome sequencing and assembly of HDN12-143 was manipulated by the Beijing Genomics Institute. PCR amplifications and verifications were performed on a T100™ Thermal Cycler (Bio-Rad Laboratories Inc., Singapore). Agarose electrophoresis was conducted on DYY-6C Type Electrophresis Apparatus (Liuyi Biotechnology, Beijing, China) and analyzed by SensiAnsys (Peiqing Science & Technology, Shanghai, China). UV spectra were recorded on Beckman DU 640 spectrophotometer (Beckman Coulter Inc., Brea, CA, USA). IR spectra were taken on Bruker tensor-27 spectrophotometer in KBr discs (Bruker Corporation, Billerica, MA, USA). Specific rotations were measured on JASCO P-1020 digital polarimeter (JASCO Corporation, Tokyo, Japan). ESIMS were obtained on Thermo Scientific LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) or Micromass Q-TOF ULTIMA GLOBAL GAA076 LC Mass spectrometer (Wasters Corporation, Milford, MA, USA). CD spectra were measured on JASCO J-715 spectropolarimeter (JASCO Corporation, Tokyo, Japan). NMR spectra were recorded on Agilent 500 MHz DD2 spectrometer using TMS as internal standard, and chemical shifts were recorded as δ-values (Agilent Technologies Inc., Santa Clara, CA, USA). Semi-preparative HPLC was performed on an ODS column [HPLC (YMC-Pack ODS-A, 10 × 250 mm, 5 μm, 3 mL/min)] (YMC Co., Ltd., Kyoto, Japan). Medium-pressure preparation liquid chromatography (MPLC) was performed on a Bona-Agela CHEETAHTM HP100 (Beijing Agela Technologies Co., Ltd., Beijing, China). Column chromatography (CC) was performed with silica gel (200–300 mesh, Qingdao Marine Chemical Inc., Qingdao, China), and Sephadex LH-20 (Amersham Biosciences, San Francisco, CA, USA), respectively.
The wild type fungus HDN-12-143 was isolated from a sediment sample collected in the South China Sea (China). The strain was incubated in potato dextrose agar medium at 28°C for 4 days, following which, it was incubated using the CTAB method (Tang et al., 2017) to obtain the genomic DNA library. By using classical microscopic analysis and ITS sequence alignment, HDN-12-143 was identified as Penicillium brocae (GenBank accession number MN410885). P. brocae HDN-12-143 was deposited at the Key Laboratory of Marine Drugs, the Ministry of Education of China, School of Medicine and Pharmacy, Ocean University of China, Qingdao, People's Republic of China.
The genomic DNA of Penicillium brocae HDN12-143 was analyzed on antiSMASH (https://fungismash.secondarymetabolites.org/). Following this, the sequence of PbrlaeA was identified by Localblast (Altschul et al., 1997) and analyzed with the reported LaeA obtained in NCBI (https://www.ncbi.nlm.nih.gov/). For the evolutionary relationship analysis, the amino acid sequences of PbrlaeA and other LaeA homologs from different penicillium species obtained from NCBI were aligned using the ClustalW (http://www.clustal.org/). The phylogenetic tree was constructed via the MEGA7 software (http://www.megasoftware.net/). The conserved domain of the PbrlaeA protein was scanned by using InterProScan tool (https://www.ebi.ac.uk/interpro/search/sequence-search).
The integrative vector pHyg, which mainly contains a constitutive promoter gpdA, ampicillin and hygromycin resistant genes, was digested with restriction endonucleases KpnI and XbaI (New England Biolabs, NEB). The PbrlaeA gene was amplified from the genomic DNA library of the P. brocae HDN-12-143 using specific primers containing KpnI and SpeI restriction sites (Table S1) via PCR catalyzed by TransStart® Fastpfu DNA Polymerase (Transgen Biotech, Beijing, China). The PCR products were confirmed as correct using PCR analysis catalyzed by 2×EasyTaq® PCR SuperMix (Transgen Biotech, Beijing, China). After being digested with the above-mentioned corresponding endonucleases, the PbrlaeA gene was placed downstream of gpdA promoter within the pHyg vector to generate pHyg-PbrlaeA (Figure S4). The recombinant vector was transformed into E. coli Trans1-T1 competent cell to amplify plasmids for transformation.
The strain P. brocae HDN-12-143 was inoculated on PDA plates and cultured at 28°C for 4 days to grow fresh spores. Collected mycelium into 50 mL Tween buffer (50 μL Tween20 into 50 mL ddH2O) and spores are isolated by using 40 μm cell strainer (Solarbio Science & Technology, Bejing, China). Fresh spores were added into 50 mL PDB + YE medium (20% potato, 2% dextrose, and 0.4% yeast extract) in 250 mL Erlenmeyer flasks and germinated at 28 °C and 180 rpm for about 8 h. The germinated spores were collected by centrifugation at 4,000 rpm for 15 min, washed by 20 mL osmotic buffer (1.2 M MgSO4, 10 mM sodium phosphate, pH 5.8) twice, following which the germinated spores were suspended into 10 mL of osmotic buffer containing 30 mg lysing enzymes from Trichodema harzianum (SIGMA-ALDRICH, USA) and 20 mg Yatalase (TaKaRa, Japan), transferred into an empty sterile bottle and cultured in a shaker of 28°C at 80 rpm for 6 h to form protoplast. After enzymolysis, the mixture was transferred into a centrifuge tube and covered with a isopyknic protoplast trapping buffer (0.6 M sorbitol, 0.1 M pH 7.0 Tris-HCl) softly. After centrifugation at 4,000 rpm for 15 min at 4°C, protoplasts were collected in the interface of the above two buffers. Following this, the protoplasts were collected and washed by 20 mL STC buffer (1.2 M sorbitol, 10 mM CaCl2, 10 mM pH 7.5 Tris-HCl), and then resuspended in 2 mL STC buffer for subsequent transformation.
The pHyg-PbrlaeA was dissolved in 50 μL STC buffer after amplification, extraction and freeze-drying. Fungal transformation was carried out by mixing 100 μL of protoplasts and 50 μL of pHyg-PbrlaeA solution; the mixture was gently mixed with a pipet tip and incubated for 60 min on ice. Next, 600 μL of PEG solution (60% PEG, 50 mM calcium chloride and 50 mM pH 7.5 Tris-HCl) was added to the mixture, following which, the mixture was incubated at room temperature for 25 min. The mixture was gently spread on the regeneration PSA (PDA medium with 1.2 M sorbitol and 200 μg/mL hygromycin) medium and incubated at 28°C for 3 days (Ohashi et al., 2017).
The hygromycin-resistant regenerated strains were transferred onto new PDA plates with 200 μg/mL hygromycin, respectively, for the second screening. The colonies that could grow were recognized as putative mutants.
The putative OE::PbrlaeA mutants and the wild-type strain were cultured on PDA media for 4 days at 28°C in an incubator for further genomic DNA extraction. PCR analysis to verify the gene insertion was carried out using two pairs of primers, as shown in Table S1 and Figure S5 (primers gpda-1 and YZ-LaeA-F to verify the upstream of the PbrlaeA gene, primers gpda-2 and YZ-LaeA-R to verify the downstream of the PbrlaeA gene). Following this screening, one transformant was recognized as the desired mutant.
The OE::PbrlaeA mutant strain was cultured under rotary shaker condition at 180 rpm at 28°C in 500 mL Erlenmeyer flasks containing 120 mL of liquid medium containing glucose (2%) and potato (20%) dissolved in naturally collected seawater (Huiquan Bay, Yellow Sea, Qiangdao, China). After 9 days shaker, the whole broth (30 L) was filtered through a cheesecloth to separate supernatant and mycelia. The former was extracted three times with EtOAc (3 × 30 L), while the latter was extracted three times with methanol (3 × 10 L) and concentrated under reduced pressure to afford an aqueous solution, which was then extracted three times with EtOAc (3 × 5 L). All EtOAc solutions were combined and concentrated under reduced pressure to obtain the extract (10 g).
The extract was applied to a silica gel (300–400 mesh) column and was separated into 12 fractions (fraction 1 to fraction 12) with a step gradient elution of CH2Cl2-MeOH. Following this, fraction 5 was purified by MPLC, giving nine subfractions (fraction 5–1 to fraction 5–9). Fractions 5–6 were applied on semipreparative HPLC (20:80 MeOH–H2O, 3 mL/min) to afford compound 1 (8 mg) and compound 2 (20 mg). Fractions 5–9 were further purified by semipreparative HPLC (25:75 MeOH–H2O, 3 mL/min) to yield compound 3 (8 mg). Then, fraction 9 was separated and purified by MPLC to obtain nine subfractions (fractions 9–1 to fractions 9–9). Fractions 9–6 were applied on semipreparative HPLC (20:80 MeOH–H2O to 100:0 MeOH–H2O) to get compound 4 (8 mg).
Conformational searches were performed, employing the systematic procedure implemented in Spartan'14 using the MMFF (Merck molecular force field). All MMFF minima were reoptimized with DFT calculations at the B3LYP/6-31+G(d) level using the Gaussian09 program (Frisch et al., 2010). The geometry was optimized starting from various initial conformations, with vibrational frequency calculations confirming the presence of minima. Time-dependent DFT calculations were performed on lowest-energy conformations (>5% population) for each configuration using 20 excited states and using a polarizable continuum model for MeOH. ECD spectra were generated using the program SpecDis (Bruhn et al., 2011) by applying a Gaussian band shape with a 0.30 eV width and 10 blue shifts to facilitate comparison to the experimental data. The 13C NMR chemical shifts of compounds 1 and 2 were calculated with the GIAO method at the B3LYP/6-311G (2d, p) levels in the Gaussian09 program (Frisch et al., 2010).
The cytotoxicity assays were evaluated by the MTT method against the K562 and HL-60 cancer cell lines, and the SRB method against the H975, MGC803, and HO-8910 cancer cell lines. All of the biological evaluations were carried out as previously reported in the References (Yu et al., 2016; Gao et al., 2018).
A LaeA analog named PbrlaeA (GeneBank No. MN410885) was identified via Localblast using AnlaeA (Q6TLK5.1) from Aspergillus nidulans as a query. The total size of the PbrlaeA gene is 1291 bp and the predicted open reading frame (ORF) is 1,122 bp, which may encode a polypeptide of 373 amino acids. Protein sequence alignment and phylogenetic analysis indicated that PbrlaeA was an S-adenosyl-L-methionine-dependent methyltransferase and closely related to PblaeA from Penicillium brasilianum (Figure S3).
The PbrlaeA gene fragments added by KpnI and SpeI (isocaudamer of XbaI) were amplified from genomic DNA of the strain P. brocae HDN-12-143 by using specific primers (Table S1) and ligated into the vector pHyg using restriction sites KpnI and XbaI. The recombinant plasmid was transformed into P. brocae HDN-12-143 and screened with 200 μg/mL hygromycin in PDA medium. The mutant strain was further verified by diagnostic PCR (Figure S5). After being fermented with PDB medium under shaking condition at 28°C for 9 days, the high performance liquid chromatography (HPLC) analysis of the extract of the OE::PbrlaeA mutant strain showed a series of new peaks compared with the extract of the wild type P. brocae HDN-12-143 strain (Figure 1), which indicated the production of new secondary metabolites.
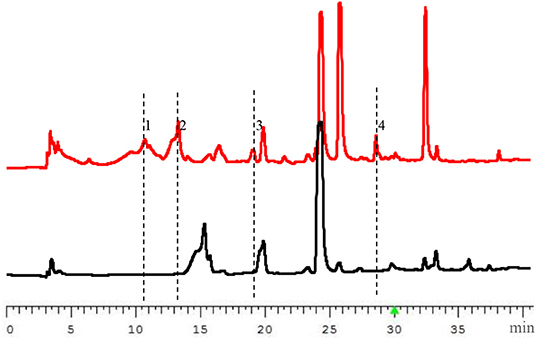
Figure 1. HPLC profiles of the extracts from wild type (WT, the lower diagram) and OE::PbrlaeA (MT, the upper diagram) strains of P. brocae HDN-12-143 (220 nm).
To confirm the structures of these newly produced secondary metabolites, the OE::PbrlaeA mutant strain was scaled up cultured (30 L) under rotary shaker condition at 180 rpm at 28°C. The extract (10 g) was fractionated by column chromatography, including silica gel, MPLC and semipreparative HPLC, leading to the isolation of compounds 1–4.
Compounds 1 and 2 were obtained as yellowish oil. The molecular formula of 1 and 2 were all established to be C8H9O5Cl deduced by the [M – H]− ion at m/z 219.0072 and 219.0071 (calcd for C8H8O5Cl: 219.0066) in the HRESIMS. The 1H NMR and 13C NMR spectroscopic data of compound 1 displayed one methyl, one oxymethyl, one methine, five quaternary carbons including two sp2 quaternary carbons and two ketone carbonyls. Based on the HMBC correlations from H1-2 to C-1, C-6, C-3, and C-4, from H3-7 to C-2, C-3, and C-4, and from H3-8 to C-6 (Figure 2), two possible planar structures a and b, where the locations of chlorine and hydroxyl groups were left undetermined (Figure 3A), were proposed for 1. Further 13C NMR calculation method was adopted to confirm the accurate structure (Pierens, 2014). Calculation of the 13C NMR chemical shifts of a and b at the B3LYP/6-311G (2d, p) levels in CD3OD were obtained, and the experimental chemical shifts agreed well with the calculated data of a (R2 = 0.9946) (Figure 3B), assigning the planar structure of compound 1 as a (fumigatin chlorohydrin). Further comparing of the optical rotation data of 1 −102.6 (c 0.10, H2O) with that of fumigatin chlorohydrin ([α]D −160 in H2O for fumigatin chlorohydrin] (Yamamoto et al., 1970) confirmed 1 as fumigatin chlorohydrin. The planar structure of fumigatin chlorohydrin was first reported in 1970 from the fungus Aspergillus Fumigatus (Yamamoto et al., 1970). However, the report only covered the 1H NMR data of fumigatin chlorohydrin, meanwhile the configuration and activity were not included. In the present work, we describe the absolute configuration for the first time.
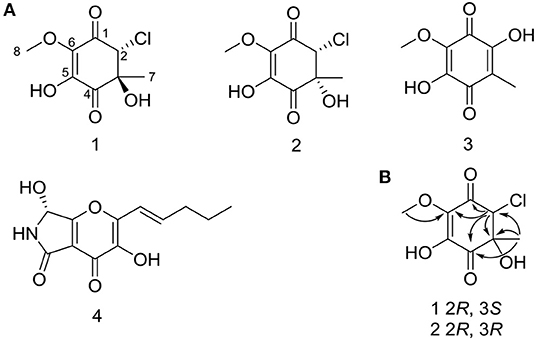
Figure 2. (A) Structures of 1–4 from mutant strain OE::PbrLaeA P. brocae HDN-12-143. (B) Key HMBC correlations of 1 and 2.
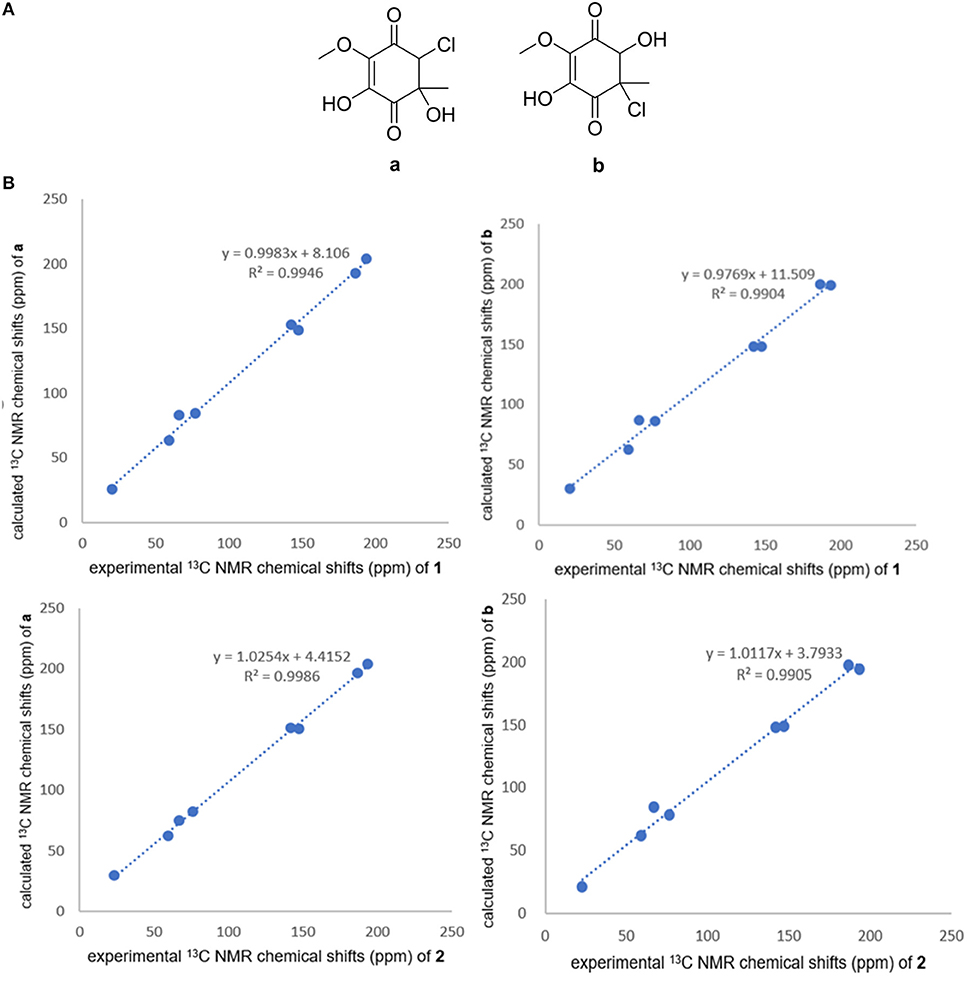
Figure 3. (A) Structures of a and b. (B) Correlation plots of experimental 13C NMR chemical shifts of 1 and 2 versus the corresponding calculated data for the proposed structures a and b.
Compound 2 had the same planar structure as 1 based on NMR and calculation of the 13C NMR chemical shifts, while the minor variations in chemical shifts of C-2, C-3, and C-7 (δC: 66.0, 76.6, and 20.2 in 1; 67.0, 76.2, and 22.8 in 2) revealed different configurations at the stereocenters. To establish the absolute configuration, the electronic circular dichroism (ECD) spectra of (2R, 3S), (2R, 3R), (2S, 3R), and (2S, 3S) were simulated using density functional theory (DFT) calculations performed at the B3LYP/6-31+G(d) level. The absolute configurations of (2R, 3S)-1 and (2R, 3R)-2 were unambiguously determined by the almost identical curves between the computational ECD curve (2R, 3S)-1 and (2R, 3R)-2 and the experimental one (Figure S1; Figure 4). Thus, compound 2 was confirmed to be a diastereoisomer of fumigatin chlorohydrin and named as iso-fumigatin chlorohydrin. From a biosynthetic view, fumigatin-like phenols are fundamentally derived from polyketide synthase biosynthetic (PKS) pathways, which adopted acetyl-CoA as precursor (Packter and Glover, 1965; Pacter, 1966). With regards to compounds 1–3, the specific enzymes responsible for the tailoring steps, like oxidation and halogenation on the aromatic skeleton, remain an intriguing topic for further investigation.
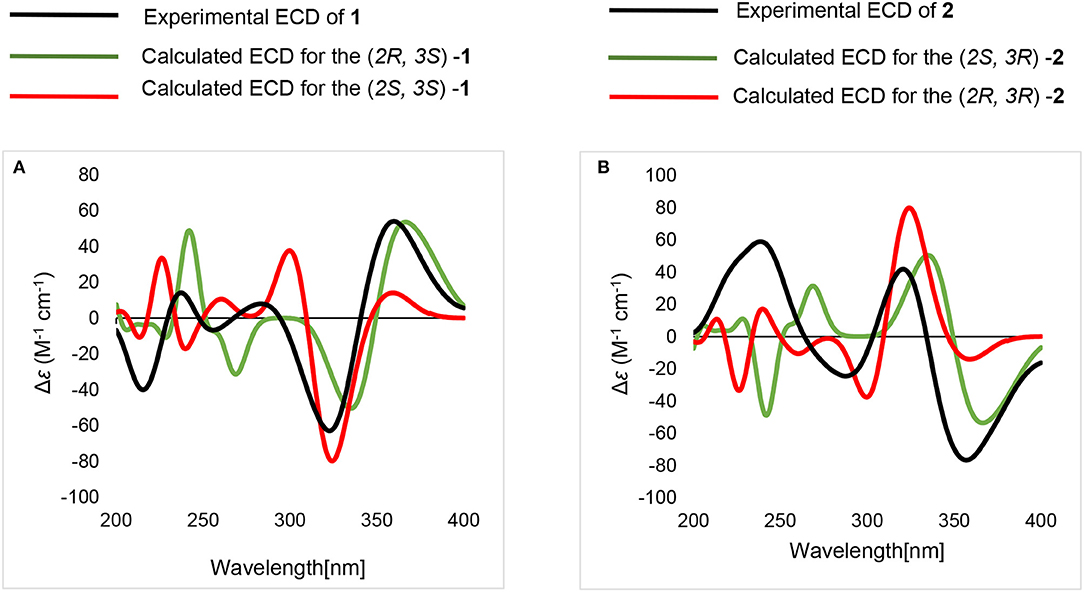
Figure 4. Calculated and experimental ECD spectra. (A) calculated and experimental ECD of (2R, 3S)-1, (2S, 3S)-1. (B) calculated and experimental ECD of (2S, 3R)-2, (2R, 3R)-2.
−6.9 (c 0.10, MeOH); UV (MeOH) λ max (log ε) 232 (1.8), 310 (2.8) nm; IR (KBr) νmax 3,417, 2,936, 1,716, 1,636, 1,457, 1,385, 1,328, 1,110, 1,029 cm−1; 1H NMR (CD3OD, 500 MHz) and 13C NMR (CD3OD, 125 MHz) data are shown in Table 1, HRESIMS m/z: 219.0072 [M – H]− (calcd. for C8H8O5Cl: 219.0066).
−15.3 (c 0.10, MeOH); UV (MeOH) λ max (log ε) 232 (1.8), 310 (2.8) nm; IR (KBr) νmax 3,445, 2,952, 1,676, 1,625, 1,447, 1,358, 1,242, 1,054, 985, 960 cm−1; 1H NMR (CD3OD, 500 MHz) and 13C NMR (CD3OD, 125 MHz) data are shown in Table 1, HRESIMS m/z: 219.0071 [M–H]− (calcd. for C8H8O5Cl: 219.0066).
The known compounds were identified as spinulosin (3) (Anslow and Raistrick, 1938) and pyranonigrin F (4) (Meng et al., 2015) through comparison of the NMR and MS data with the reported ones.
The MS, 1D and 2D NMR spectra for compounds 1–2 are available as Supplementary Material.
All the compounds were tested for their cytotoxicity against five cancer cell lines (HL-60, K562, H1975, MGC803 and HO-8910) (Yu et al., 2016). Compounds 1 and 2 exhibited weak activities against HL-60 with IC50 of 18.63 and 24.83 μM (Table S2). Besides, compounds 1 and 2 were also tested for the antimicrobial activity against Bacillus subtilis and antioxidant capacities by DPPH free radical scavenging assay, but no activity was observed (IC50 > 100 μM) (Yu et al., 2016; Gao et al., 2018).
Four compounds were isolated from the fungus Penicillium brocae HDN-12-143 by overexpression of the LaeA family gene of PbrlaeA. Among them, the planar structure of 1 and 2 was determined by comprehensive spectroscopic data with 13C NMR calculations. The absolute configuration was determined by calculating the ECD. Compounds 1 and 2 exhibited weak cytotoxicity against HL-60 with IC50 of 18.63 and 24.83 μM. The above study showed that overexpression of the global regulator LaeA analogs could be an efficient method to activate the silent biosynthetic pathway of marine-derived Penicillium strains, generating chemical diversity in their secondary metabolites profile.
The datasets generated for this study can be found in the GeneBank No. MN410885.
The contributions of the respective authors are as follows: LW drafted the work and performed the fermentation, extraction, as well as the isolation. XianZ constructed the plasmids and was involved in the acquisition of mutant strains, performed the biological evaluations, and bioinformatic analysis. KZ was involved in the acquisition of mutant strains and bioinformatic analysis. XiaoZ performed the biological evaluations. TZ and QC contributed to checking and confirming all the procedures of the isolation and identification. GZ and DL designed the study, supervised the laboratory work, and contributed to the critical reading of the manuscript.
This work was financially supported by the NSFC-Shandong Joint Fund (U1906212 and U1606403), the Pilot National Laboratory for Marine Science and Technology (2018SDKJ0401-2 and 2016ASKJ08-02), the National Natural Science Foundation of China Major Project for Discovery of New Leading Compounds (81991522), the National Science and Technology Major Project for Significant New Drugs Development (2018ZX09735004), the Fundamental Research Funds for the Central Universities (201941001), Project funded by China Postdoctoral Science Foundation (2017M622286), and the Taishan Scholar Youth Expert Program in Shandong Province (tsqn201812021).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2020.00270/full#supplementary-material
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anslow, W. K., and Raistrick, H. (1938). Studies in the biochemistry of micro-organisms Fumigatin (3-hydroxy-4-methoxy-2:5-toluquinone), and spinulosin (3:6-dihydroxy-4-methoxy-2:5-toluquinone), metabolic products respectively of Aspergillus fumigatus Fresenius and Penicillium spinulosum Thom. Biochem. J. 32, 687–696. doi: 10.1042/bj0320687
Bruhn, T., Schaumlöffel, A., Hemberger, Y., and Bringmann, G. S. (2011). SpecDis, version 1.53. Würzburg: University of Würzburg.
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., et al. (2010). Gaussian 09, Revision B.01, Gaussian, Inc., Wallingford, CT.
Gao, C. Z., Guo, Z. Y., Lu, X. Z., Chen, H. Y., Liu, L. W., Yu, Z. G., et al. (2018). Hexaricins, pradimicin-like polyketides from a marine sediment-derived Streptosporangium sp. and Their Antioxidant Effects. J. Nat. Prod. 81, 2069–2074. doi: 10.1021/acs.jnatprod.8b00397
Jiang, T., Wang, M., Li, L., Si, J., Song, B., Zhou, C., et al. (2016). Overexpression of the global regulator LaeA in Chaetomium globosum leads to the biosynthesis of chaetoglobosin Z. J. Nat. Prod. 79, 2487–2494. doi: 10.1021/acs.jnatprod.6b00333
Keller, N., Bok, J., Chung, D., Perrin, R. M., and Shwab, E. K. (2006). LaeA, a global regulator of Aspergillus toxins. Med. Mycol. 44, 83–85. doi: 10.1080/13693780600835773
Kosalkova, K., García-Estrada, C., Ullan, R. V., Godio, R. P., Feltrer, R., Teijeira, F., et al. (2009). The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 91, 214–225. doi: 10.1016/j.biochi.2008.09.004
Meng, L. H., Li, X. M., Liu, Y., and Wang, B. G. (2015). Polyoxygenated dihydropyrano [2, 3-c] pyrrole-4, 5-dione derivatives from the marine mangrove-derived endophytic fungus Penicillium brocae MA-231 and their antimicrobial activity. Chin. Chem. Lett. 26, 610–612. doi: 10.1016/j.cclet.2015.01.024
Ohashi, M., Liu, F., Hai, Y., Chen, M., Tang, M. C., Yang, Z., et al. (2017). SAM-dependent enzyme-catalysed pericyclic reactions in natural product biosynthesis. Nature 549, 502–506. doi: 10.1038/nature23882
Packter, N. M., and Glover, J. (1965). Biosynthesis of 14C fumigatin in Aspergillus fumigatus, fresenius. Biochim. Biophys. Acta. 100, 50–56. doi: 10.1016/0304-4165(65)90426-5
Pacter, N. (1966). Studies on the biosynthesis of phenols in fungi. Biochem. J. 98, 353–359. doi: 10.1042/bj0980353
Pierens, G. K. (2014). 1H and 13C NMR scaling factors for the calculation of chemical shifts in commonly used solvents using density functional theory. J. Comput. Chem. 35, 1388–1394. doi: 10.1002/jcc.23638
Sarikaya, B. O., Bayram, O., and Braus, G. H. (2010). LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 6:e1001226. doi: 10.1371/journal.pgen.1001226
Tang, M. C., Cui, X., He, X., Ding, Z., Zhu, T., Tang, Y., et al. (2017). Late-stage terpene cyclization by an integral membrane cyclase in the biosynthesis of isoprenoid epoxycyclohexenone natural products. Org. Lett. 19, 5376–5379. doi: 10.1021/acs.orglett.7b02653
Yamamoto, Y., Shinya, M., and Oohata, Y. (1970). Studies on the metabolic products of a strain of Aspergillus fumigatus (DH 413). IV. Biosynthesis of toluquinones and chemical structures of new metabolites. Chem. Pharm. Bull. 18, 561–569. doi: 10.1248/cpb.18.561
Keywords: genome mining, Penicillium brocae, overexpression, global regulator PbrlaeA, silent gene cluster, polyketide, fumigatin chlorohydrin
Citation: Wang L, Zhang X, Zhang K, Zhang X, Zhu T, Che Q, Zhang G and Li D (2020) Overexpression of Global Regulator PbrlaeA Leads to the Discovery of New Polyketide in Fungus Penicillium Brocae HDN-12-143. Front. Chem. 8:270. doi: 10.3389/fchem.2020.00270
Received: 22 December 2019; Accepted: 19 March 2020;
Published: 21 April 2020.
Edited by:
Valeria Costantino, University of Naples Federico II, ItalyReviewed by:
Bin Yu, Zhengzhou University, ChinaCopyright © 2020 Wang, Zhang, Zhang, Zhang, Zhu, Che, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guojian Zhang, emhhbmdndW9qaWFuQG91Yy5lZHUuY24=; Dehai Li, ZGVoYWlsaUBvdWMuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.