- The Key Laboratory of Life-Organic Analysis, Key Laboratory of Pharmaceutical Intermediates and Analysis of Natural Medicine, School of Chemistry and Chemical Engineering, Institute of Anticancer Agents Development and Theranostic Application, Qufu Normal University, Qufu, China
Herein, we report the synthesis, characterization and anticancer activity of a series of half-sandwich iridiumIII imidazole and benzimidazole N-heterocyclic carbene (NHC) anticancer complexes, and the general formula of which can be expressed as [(η5-Cpx)Ir(C∧N)Cl]Cl (Cpx: pentamethylcyclopentadienyl (Cp*) or biphenyl derivatives (Cpxbiph); C∧N: imidazole and benzimidazole NHC chelating ligands). Compared with cis-platin, these complexes showed interesting antitumor activity against A549 cells. Complexes could bind to bovine serum albumin (BSA) by means of static quenching mode, catalyze the oxidation of nicotinamide adenine dinucleotide (NADH) and increase the levels of reactive oxygen species (ROS). Meanwhile, these complexes could arrest the cell cycles of A549 cells and influence the mitochondrial membrane potential significantly. Due to the inherent luminescence property, laser confocal test show that complexes could enter cells followed an energy-dependent mechanism and effectively accumulate in lysosome (the value of Pearson's co-localization coefficient is 0.70 after 1 h), further destroy lysosome integrity and induce apoptosis.
Introduction
When cell growth is uncontrollable and abnormal proliferation occurs, malignant tumors are formed (Valastyan and Weinberg Robert, 2011; Grossi et al., 2016). Most forms of disseminated tumors are currently untreatable, although developments in chemotherapy over the past years have led to the treatment of various cancers (Thota et al., 2018). Cis-platin exerts good and broad-spectrum anticancer effects (Deubel and Lau, 2006; Romero-Canelón and Sadler, 2013; Muhammad and Guo, 2014), but, when used extensively, platinum-based drugs may promote drug resistance and instability; become insoluble in biological media; and become toxic to blood and gastrointestinal tract (Ma et al., 2014; Liu et al., 2015). Thus, various organometallic anticancer drugs, such as tin, iridium, ruthenium, and osmium anticancer complexes, have been developed (Sun and Che, 2009; Lu et al., 2015; Zeng et al., 2017; Ge et al., 2019; Li et al., 2019).
IrIII complexes are inert, half-sandwich IrIII complexes (a type of [(Cpx)Ir(L∧L')Z]X) show good anticancer activities (Richens, 2005), especially for tumors resistant to platinum-based drugs (Singh et al., 2010). These complexes are composed of cyclopentadienyl ligands (Cpx), chelating ligands (L∧L'), leaving groups (Z), and counter ions (X), each of which has an inherent effect on the anticancer activity of these complexes (Liu et al., 2011a). The types and positions of substituents on L∧L'-chelating ligands are the most studied and finely regulated for the effective modulation of targeted sites, the lipid solubility and even the anticancer activities of these complexes (Yellol et al., 2013; Zamora et al., 2015; Hao et al., 2019). The anticancer activity of a simple half-sandwich IrIII complex increases nearly 10 times when the bipyridine chelating ligand (N∧N) is replaced by a phenyl pyridine ligand (C∧N) (Liu and Sadler, 2014). The IC50 value (the concentration at which the growth of 50% of the cells is inhibited) of IrIII complex can be reduced by nearly 100 times by controlling the binding mode of bipyridine chelate ligand substituents and the number of triphenylamine (TPA) units (He et al., 2019).
As a chelating ligand, N-heterocyclic carbene (NHC) provides an s-donor, which not only ensures the high stability of a metal complex, but also prevents its hydrolyzation in various physiological media. Transition metal complexes containing NHC have been widely used in the field of catalysis (Powell et al., 2010; Schuh et al., 2012; Pellei et al., 2015). These complexes affect different catalytic modes and even change the progress of the cyclic metal complex catalytic process (Zou et al., 2018). Metal iridium complexes containing NHC targets lysosomes and mitochondria selectively, induces lysosome damage, alters mitochondrial membrane potential, blocks the cell growth cycle, and promotes apoptosis (Han et al., 2018). In the present study, a series of half-sandwich IrIII NHC complexes were synthesized and characterized in detail, and NHC pro-ligands were coordinated with metal iridium in the mode of C∧N (Figure 1). MTT assay was used to assess the anticancer activity of IrIII NHC complexes in A549 cells lines. The anticancer mechanism of these complexes were also studied by flow cytometer and laser scanning confocal microscope. The results indicated that half-sandwich IrIII NHC complexes may be promising candidate for anticancer drugs and broaden the research scope in this field.
Results and Discussion
Synthesis and Characterization
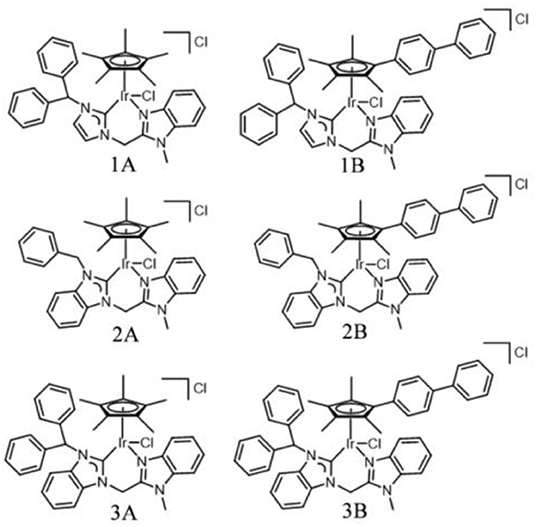
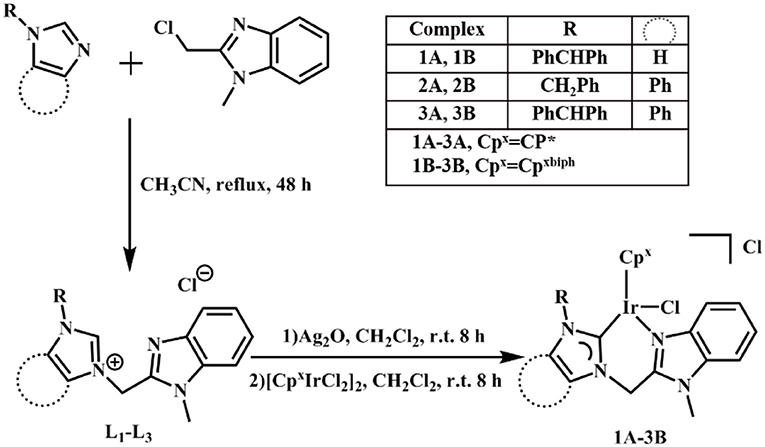
NHC pro-ligands and a series of half-sandwich IrIII NHC complexes of the type [(η5-Cpx)Ir(C∧N)Cl]Cl (1A-3B) containing Cp ring and its biphenyl derivative (Scheme 1), were synthesized and characterized for the first time. Then, 1-diphenylmethylimidazole, 1-benzylbenzimidazole and 1-benzhydrylbenzi- midazole were synthesized with imidazole and benzimidazole as raw materials and then reacted with 2-(chloromethyl)-1-methylbenzimidazole (Figure S1). The products were named L1, L2, and L3, respectively. The target complexes were obtained by catalyzing the reaction between the chelating ligands (L1–L3) and dimers (Cp* and Cpxbiph) in dichloromethane with the use of silver oxide (Ag2O) at ambient temperature and good yields of 59–69%. The target complexes were isolated as Cl salt and characterized by 1H NMR (Figure S2), 13C NMR (Figure S3), mass spectroscopy (Figure S4) and elemental analysis. The 1H NMR spectra and MS spectral data are provided in the support information, as shown in Tables S1, S2. At the same time, we characterized the carbon in the complex, in which the peak shift of C-Ir is between 148.66 and 167.58 ppm, and the peak shift of NCN is between 138.56 and 150.24 ppm. After simple purification, the complexes were isolated as powdered and non-hygroscopic solids. The complexes were highly soluble in dichloromethane, chloroform, and dimethyl sulfoxide; partially soluble in methanol; and insoluble in ether, petroleum ether, and n-hexane.
M-OH2 complexes usually have higher anticancer activities than M-Cl complexes (Liu et al., 2011b). Hydrolysis behavior of complex 3B in 50% CD3OD-d4/50% D2O (v/v) and 50% MeOH/50% H2O (v/v) was monitored by 1H NMR (Figure S5) and ultraviolet-visible spectrophotometry (UV-Vis; Figure S6) at 298 K, respectively. No obvious changes were observed in the 1H NMR and UV-Vis spectra, which indicating that IrIII NHC complexes did not hydrolyze and the structure was stable high water content. Thus, the IrIII NHC complexes are sufficiently stable for the bioassay.
Cytotoxicity Test
Lung cancer has a high mortality rate in developed and developing countries, therefore, A549 cells were selected as the model of biological research (Jemal et al., 2011). The antiproliferative activities of half-sandwich IrIII complexes [(Cpx)Ir(L∧L')Z]Cl are affected by changes in cyclopentadienyl or chelating ligand structures of the complexes (Liu and Sadler, 2014). The antiproliferative activities of half-sandwich IrIII complexes 1A–3B against A549 cancer cells after 24 h are shown in Table 1 and Figure 2. These complexes exhibit better activity than imidazolium salts (L1–L3) and [CpxIrCl2]2 (IC50: > 100), meanwhile, 1A–3B had higher IC50 values than cis-platin (5.9–18.2 μM), especially complex 3B (3.6 times of cis-platin).

Table 1. IC50 values of complexes 1A–3B and cis-platin against A549 cancer cells after 24 h exposure.
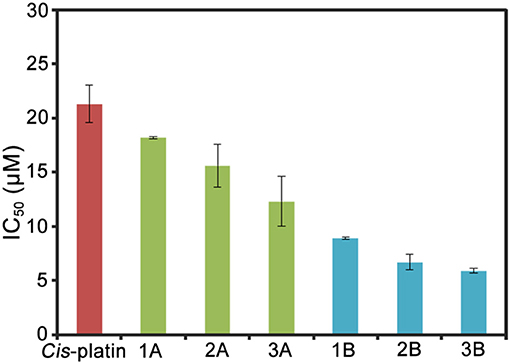
Figure 2. Histograms of IC50 values for complexes 1A–3B and cis-platin against A549 cells after 24 h.
This result is consistent with the previous study, which showed that a Cpxbiph group with strong electron donor capacity effectively improved the anticancer potential of IrIII complexes (Wang et al., 2017). 2A and 2B formed large conjugated systems with strong electron donor capacities after benzene rings were introduced to their imidazole groups and thus showed better anticancer activity than 1A and 1B, where benzene rings were on the terminal methyl groups 3A and 3B showed the best anticancer potential because large organic ligands, which effectively enhanced the lipophilicity of the complexes. 2A, 2B, and 3B had logP values (oil/water partition coefficient) were 0.92, 1.37, and 1.56, respectively, which further confirmed the sizes of the hydrophilic and lipophilic IrIII NHC complexes (Zhang et al., 2018).
Protein Bingding Assay
Serum albumin (SA) has significant binding properties and is an important drug delivery medium in vivo (Chen et al., 2016; Esteghamat-Panah et al., 2017). Bovine serum albumin (BSA) has the structural homology with human serum albumin (HSA), can be easily purified, and shows good stability and is thus an economical and effective protein binding model (Naz et al., 2017). The interactions of complexes with BSA were analyzed by using UV-Vis absorption and fluorescence spectrum (Figure 3).
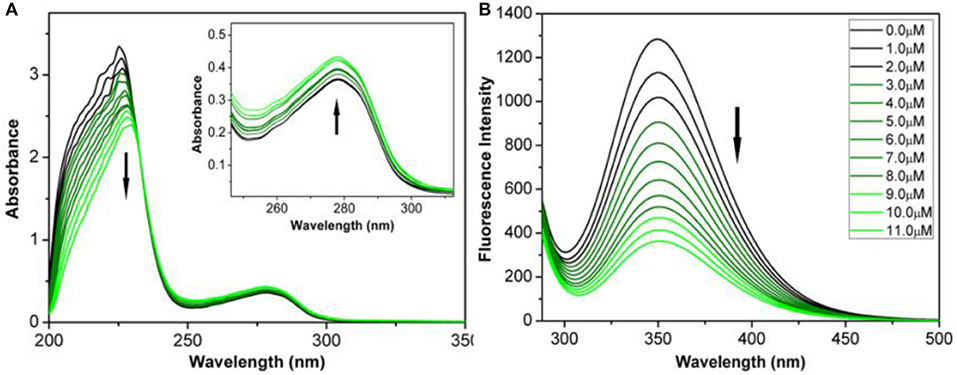
Figure 3. (A) UV-Vis spectrum of BSA reacted with complex 3B (0–11 μM) in Tris-HCl/NaCl buffer solution of pH=7.2. Inset: Wavelength range: 250–300 nm. (B) Fluorescence spectra (λex = 280 nm; λem = 343 nm) of complex 3B (0–11 μM) reacted with BSA. Arrows: The change tendency of UV-Vis spectrum and fluorescence intensity with the addition of complex 3B.
As shown in Figure 3A and Figure S7A, the maximum absorption at 228 nm (the absorption of BSA) decreased when the concentrations of complexes 1B–3B increased. The decrease indicated that the complexes combined with BSA through alpha-helical interference. The absorption peaks increased gradually at 278 nm. This result demonstrated that the complexes changed the microenvironments of the three aromatic acid residues (tryptophan, tyrosine and phenylalanine) in BSA (Zhang et al., 2015; Baral et al., 2017).
The molecular environment information near the fluorophore molecule was obtained by synchronous fluorescence spectroscopy at a low concentration (Jayabharathi et al., 2011). The use of Δλ = 15 nm and Δλ = 60 nm corresponds to the spectral characteristics of tyrosine residues and tryptophan residues, respectively. As shown in Figure S8, with the increase of the complexes, the synchronous fluorescence intensity decreased gradually. When Δλ = 15 nm, the emission wavelength red shifted to 1~3 nm (285 nm). Meanwhile, no change was observed when Δλ = 60 nm. Thus, tyrosine was more affected than tryptophan by the binding of IrIII NHC complexes to BSA.
The properties of the bonds between the complexes and BSA were further examined by fluorescence emission spectrum. The fluorescence spectra were calibrated for “internal filter” effect correction (Pacheco and Bruzzone, 2013). As shown in Figure 3B and Figure S7B, the fluorescence intensity of BSA (~343 nm) was quenched obviously when the concentrations of complexes 1B–3B increased. The possible quenching mechanism can be interpreted by using the Stern-Volmer equation and the Scatchard equation (Chatterjee and Mukherjee, 2014), Ksv(Stern-Volmer quenching constant), Kq(quenching rate constant), Kb (banding constant), and n (number of banding site) were then calculated. The Kq values of complexes 1B–3B were 1.82 × 1012, 2.34 × 1012 and 2.66 × 1012 M−1 s−1 (Figure S9 and Table S3), respectively, which were about two orders of magnitude higher than the value of a pure dynamic quenching mechanism (2.0 × 1010 M−1 s−1). The results indicated that the half-sandwich IrIII NHC complexes interacted with BSA in a static quenching mode. Additionally, 3B has the largest Kb (1.13 × 10−4 M−1) and n (1.31), which were consistent with the results of MTT assay that complex 3B has better best anticancer activity than 1B–3B. The results indicate that IrIII NHC complexes can be effectively combined with BSA, and thus, BSA can be considered as an excellent carrier for delivering these anticancer complexes in vivo.
Apoptosis Assay
The effect of IrIII complexes inducing apoptosis was determined by exposing the A549 cells to complex 3B with a concentration of 0.5 × IC50, 1.0 × IC50 and 2.0 × IC50 for 24 h. The cells were stained with AnnexinV/ Propidium Iodide, then detected by flow cytometry (Rubbiani et al., 2011). As shown in Figure 4 and Table S4, complex 3B induced apoptosis in a concentration-dependent manner, and this condition was mainly observed in early apoptosis and late apoptosis. At 2.0 × IC50, 97.6% (early apoptosis 56.2% + late apoptosis 41.4%) of the cells underwent apoptosis, and no considerable increase in increased necrotic population was detected. These results indicated that the IrIII NHC complexes induced apoptosis effectively and exhibited favorable anticancer activity.
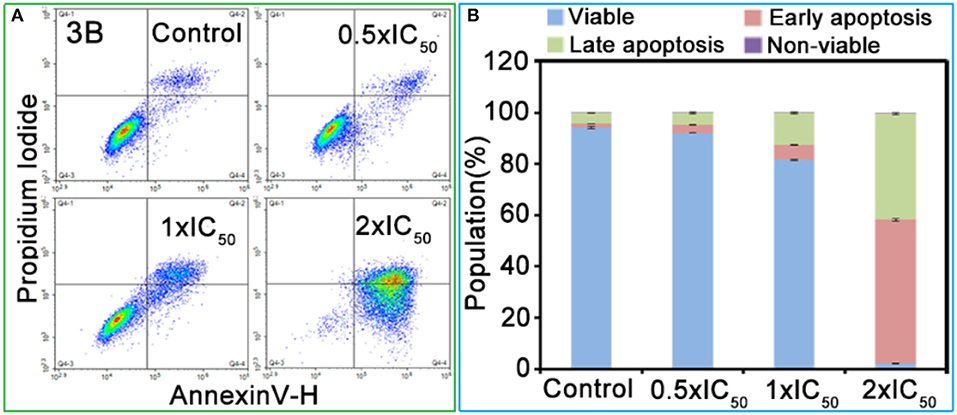
Figure 4. (A) Apoptosis analysis of A549 cancer cells after 24 h of exposure to complex 3B at 310 K determined by flow cytometry using Annexin V-FITC/PI staining. (B) Histograms were the populations of A549 cells in four stages treated by complex 3B. Data are quoted as mean ± SD of three replicates.
Reactive oxygen species (ROS) are mainly produced and stored by the mitochondria and play an important role in the regulation cell apoptosis and represent a pathway for oxidation anticancer mechanisms (Trachootham et al., 2009). After the 24 h treatment using complex 3B, the ROS levels in the A549 cells changed (Figure S10). When the concentration of complex 3B changed from 0.25 × IC50 to 0.5 × IC50, the ROS levels in the A549 cells were 1.3 and 1.6 times of the ROS level of the negative control (Table S5). This result is consistent with the result of the nicotinamide adenine dinucleotide (NADH) catalytic test. NADH (reduced coenzyme) can provide hydrides for IrIII complexes and lead to the production of ROS, which providing a possibility anticancer mechanism of oxidation. To investigate the catalytic ability of complex for NADH, the reactions of complex 3B (ca. 1 μM) with NADH (100 μM) in 60% MeOH/40% H2O (v/v) were monitored by UV-Vis spectrum at 298 K (Figure S11). The conversion of NADH to NAD+ was detected by measuring the changes at 339 and 259 nm (the absorption peak of NADH and NAD+, respectively). As shown, with the increase of the concentration of 3B, there is a noticeable reduce and increase at 339 and 259 nm, respectively, which confirmed the catalytic activity of these complexes.
The relationship between anticancer mechanism and cell cycle arrest was determined by analyzing the cell cycle arrest of complex 3B in A549 cells through flow cytometry. Compared with the untreated cells, the A549 cells were mainly arrested in the sub-G1 and G2/M, and the cell population increased from 52.0 to 66.8% and from 11.1 to 20.1%, respectively, when the concentration of complex 3B increased from 0.25 × IC50 to 2.0 × IC50 (Figure S12 and Table S6). The result suggested that complex 3B may blocked the synthesis of RNA, ribosomes, and several proteins (Senese et al., 2014). The 6.9% increase in the percentage of cells in the S phase indicated that complex 3B inhibited the synthesis of DNA, histone, and some DNA replication-related enzymes and that IrIII NHC complexes arrested tumor cell cycle at multiple stages and induced apoptosis (Sabharwal and Schumacker, 2014).
The mitochondria are the main energy-producing sites in cells and play an essential role in apoptosis. Once the mitochondrial membrane is damaged, releases signals, and the damage leads to mitochondrial dysfunction and then induces apoptosis (Nichi et al., 2016). The degree of mitochondrial dysfunction can be evaluated by measuring the changes in mitochondrial membrane potential (MMP; Gómez-Sintes et al., 2016). The change in the green/red fluorescence intensity ratio for JC-1 (an ideal fluorescent probe widely used for detecting MMP) reflects the depolarization of MMP. As shown in Figure 5 and Table S7, the population of mitochondrial membrane depolarized cells changed from 11.0 to 77.4% with the increase of complex 3B (from 0.25 × IC50 to 2.0 × IC50). Increase in the ratio of JC-1 green/red fluorescence intensity (Figure 5B) confirmed that the IrIII NHC complexes affect the integrity of the mitochondrial membrane and induce apoptosis.
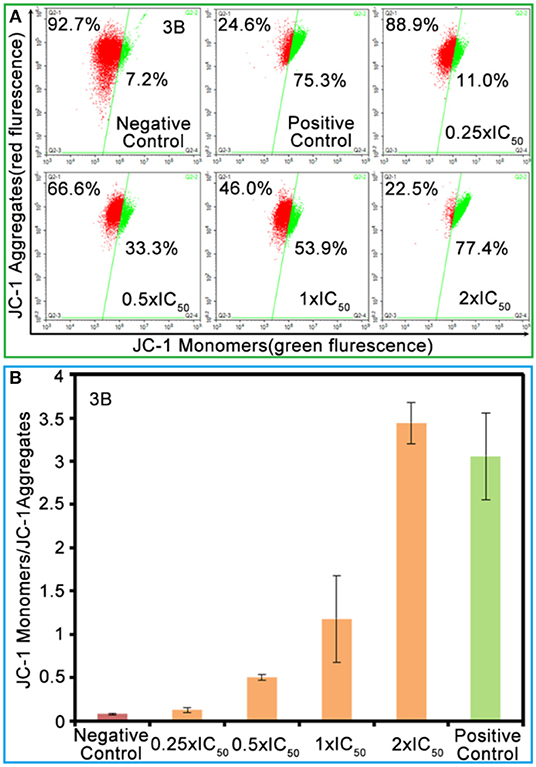
Figure 5. (A) The green/red fluorescence intensity ratio for JC-1 induced by complex 3B at the concentrations of 0.25 × IC50, 0.5 × IC50, 1.0 × IC50, and 2.0 × IC50. (B) Histograms of JC-1 monomer (green)/JC-1 aggregates (red) at different concentrations of complex 3B. Data were referenced as average ±SD. of three repeated tests.
Cellular Localization and Uptake Mechanism
The co-localization of IrIII NHC complexes was observed by laser confocal microscopy. Lyso Tracker Red DND-99 (LTRD) and Mito Tracker Deep Red (MTDR) were used. As shown in Figure 6, complex 3B was assimilated effectively by the A549 cells, and the complex mainly targeted lysosomes. The Pearson's co-localization coefficients (PCCs) after 1 h and 6 h were 0.70 and 0.81, respectively, however, which for mitochondria were 0.08 and 0.13. As the waste disposal center, lysosomes play an important role in many physiological processes and signal transduction pathways (Boya and Kroemer, 2008). These organelles are acidic (pH 4.5–5.5) and contain abundant nitrogen atoms (containing lone electron pairs). Thus, NHC pro-ligands provide convenient conditions for the lysosomes targeting. Interestingly, the IrIII NHC complex did not lead immediately induced A549 cells death. This feature is convenient for observing the effects the complexes on lysosome morphology.
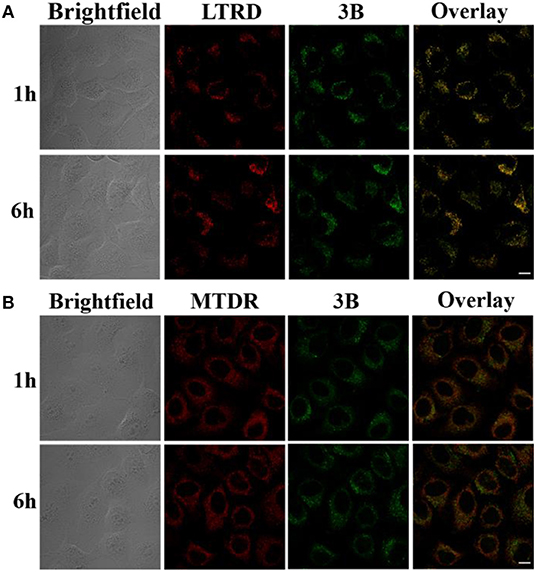
Figure 6. (A) Confocal images of A549 cells labeled by complex 3B (10 μM) and LTRD (75 nM, 1 h) after 1 h and 6 h (collected from 549 to 651 nm and excited at 594 nm). (B) Confocal images of A549 cells labeled by complex 3B (10 μM) and MTDR (500 nM, 30 min) after 1 h and 6 h (collected from 610 to 756 nm and excited at 644 nm). Complex 3B is excited at 488 nm. Scale bar: 20 μm.
The destruction of the lysosomal membrane destroys the integrity of the lysosome and induces cell death (Saftig and Klumperman, 2009; Zhitomirsky and Assaraf, 2017). Acridine orange (AO), an effective staining material for acidic organelles, was used to detect lysosomal damage induced by complex 3B. Then, whether the damage is the result of lysosome targeting specificity was determined. Green fluorescence represents the binding of AO to RNA in the nucleus or cytoplasm, whereas red fluorescence shows aggregation in the lysosome (Yamabe et al., 2016). As shown in Figure 7, A549 cells subjected to AO alone showed significant red fluorescence in their lysosomes, but the intensity of the red fluorescence decreased quickly after the cells were exposed to complex 3B with concentrations of 1.0 × IC50 and 2.0 × IC50 for 6 h, which was the result of lysosomal damage, which was mainly attributed to the introduction of nitrogen-containing ligands into the complexes. The ligands increased the total alkalinity of the molecules, which easily accumulated and damaged the integrity of the acidic lysosomes (Daum et al., 2017). Additionally, the intracellular iridium contents of 3B in the cytosol, nucleus, nuclear chromatin and cytoskeleton fractions isolated from A549 cells were determined by inductively coupled plasma mass spectrometry (ICP-MS) after 24 h exposure. 3B is mainly accumulated in the cytosol (Table S8), which further confirm the results of lysosomal damage. Meanwhile, all these explains why these complexes did not target the mitochondria, but the fact that mitochondrial dysfunction occurred, including the elevation of intracellular ROS levels and the depolarization of MMP. The results indicated that IrIII NHC complexes can induce apoptosis by lysosomal damage.
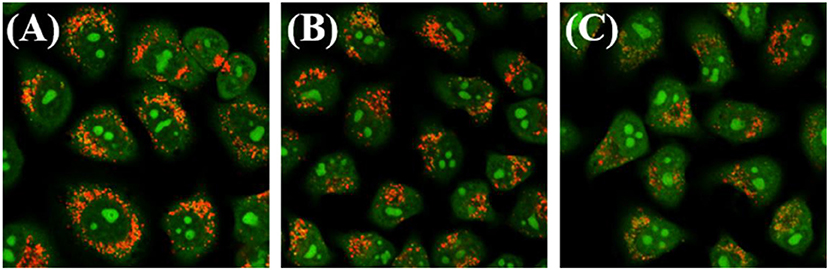
Figure 7. Observation of lysosomal disruption in A549 cells loaded with complex 3B for 6 h at 37°C, then stained with acridine orange (AO) (5 μM) at 37°C for 15 min. The cells were treated with (A) only AO; (B) AO and complex 3B (1.0 × IC50); (C) AO and complex 3B (2.0 × IC50). Data were collected at 510 ± 20 nm (green) and 625 ± 20 nm (red) after excited at 488 nm. Scale bar: 20 μm.
Small molecule drugs permeate cell membranes through different modes of action, mainly including energy-independent and energy-dependent mechanisms (Li et al., 2011). When the A549 cells were pretreated at 4°C or with carbonyl cyanide 3-chlorophenyl-hydrazone (CCCP, a metabolic inhibitor), their uptake efficiency decreased considerably after they were incubated with complex 3B (Figure 8), although no considerable change was observed in the level for complex 3B after pretreatment at 37°C or with chloroquine (an endocytosis inhibitor, which could inhibit endoderm acidification). Therefore, we conclude that the target IrIII complex enters cells through an energy-dependent mechanism, including hydrophobic factors, which are also important in entering cells (Ye et al., 2017).
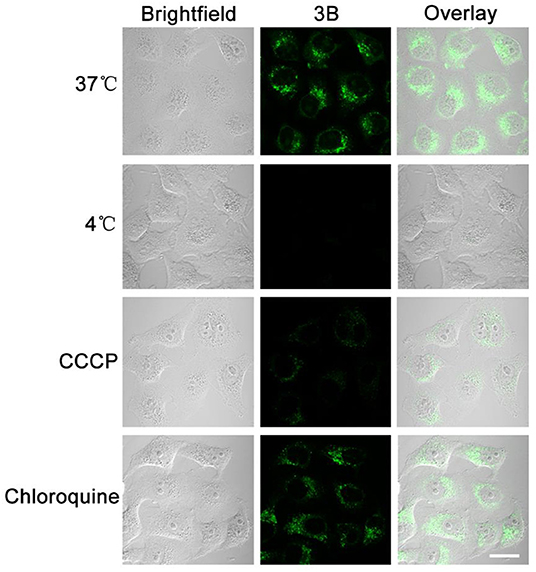
Figure 8. A549 cells were cultured at 37, 4°C, metabolic inhibitor (CCCP, 50 μM) and chloroquine (50 μM), and the uptake of complex 3B (10 μM, 30 min) was observed by confocal microscopy. The fluorescence was excited at 488 nm and collected from 549 nm to 651 nm. Scale bar: 20 μm.
Conclusions
A series of half-sandwich IrIII NHC complexes [(η5-Cpx)Ir(C∧N)Cl]Cl were synthetized and characterized. Each C∧N chelating ligand formed a six-member ring with a metal center (IrIII), and the existence of benzene ring connected imidazole ring at different locations can effectively enhanced the anticancer activity of the complexes. All the complexes showed potential anticancer activity toward A549 cells, and had better anticancer activity than cis-platin (the clinical anticancer drug). Moreover, the complexes interacted with BSA by static quenching mode, catalyzed the conversion of NADH to NAD+, induced ROS production, changed mitochondrial membrane potential, disrupted the cell cycle, and eventually induced apoptosis. The laser confocal test showed that complexes entered the cells followed through an energy-dependent mechanism, effectively targeted lysosomes, further destroyed the integrity of the lysosomes, and led to cell death. These results prompted us to further explore the medicinal value of the half-sandwich IrIII complex with additional NHC.
Experimental Section
General Procedure
The imidazole, benzimidazole, diphenylmethyl chloride, benzyl chloride, N-methyl-1, 2-phenylenediamine, diphenyl methane, ditertbutyl peroxide and the appropriate dimer [(η5-Cpx)IrCl2]2 includes [(η5-C5Me5)IrCl2]2 and [(η5-C5Me4-C6H4C6H5)IrCl2]2. 2-(chloromethyl)-1-methyl benzimidazol, 1-Diphenyl- methylimidazole, 1-Benzylbenzimidazole, 1-Benzhydryl-benzimidazole were prepared according to literature procedures (specific synthesis steps and experimental data in support information) (Nobre and Monteiro, 2004; Corberán et al., 2006). Nitrogen was used as the inert gas filling.
Synthesis of Chelating Ligands
1-diphenylmethylimidazole (1.17 g, 5.0 mmol) was added to a solution of 2-chloromethyl-1-mehylbenzenimdazole (0.48 g, 2.5 mmol) in acetonitrile. The reaction solution was distilled to 10 mL by vacuum rotary evaporation after reflex for 48 h, and 10 mL ether was added to precipitate the product. Then white powder product (L1) was filtered, washed with ether three times and dried in air. Yield: 76.6%. 1H NMR (500 MHz, DMSO-d6) δ 9.37 (s, 1H, NCHN), 7.97 (s, 1H, imidazole-H), 7.84 (s, 1H, imidazole-H), 7.61 (dd, J = 7.6, 5.4 Hz, 2H, Ar-H), 7.47 (dq, J = 14.4, 7.1 Hz, 6H, Ar-H), 7.30 (dd, J = 14.5, 7.5 Hz, 6H, Ar-H), 7.23 (t, J = 7.6 Hz, 1H, NCH), 5.89 (s, 2H, NCH2C), 3.85 (s, 3H, NCH3).
Chelating ligands L2 and L3 were synthesized using 1-benzylbenzimidazole and 1-benzhydryl-benzimidazole with 2-chloromethyl-1-mehylbenzenimdazole by the same method, respectively, the data were as follows:
L2: Yield: 72.1%. 1H NMR (500 MHz, DMSO-d6) δ 10.22 (s, 1H, NCHN), 8.14–8.10 (m, 1H, Ar-H), 8.05–8.00 (m, 1H, Ar-H), 7.69–7.66 (m, 2H, Ar-H), 7.64 (d, J = 8.1 Hz, 1H, Ar-H), 7.56 (d, J = 7.1 Hz, 2H, Ar-H), 7.53 (d, J = 8.0 Hz, 1H, Ar-H), 7.45 (t, J = 7.3 Hz, 2H, Ar-H), 7.40 (d, J = 7.2 Hz, 1H, Ar-H), 7.32–7.28 (m, 1H, Ar-H), 7.22–7.18 (m, 1H, Ar-H), 6.28 (s, 2H, NCH2), 5.91 (s, 2H, NCH2C), 3.97 (s, 3H, NCH3).
L3: Yield: 72.3%. 1H NMR (500 MHz, DMSO-d6) δ 9.69 (s, 1H, NCHN), 8.13 (d, J = 8.1 Hz, 1H, NCH), 7.78 (d, J = 8.1 Hz, 1H, Ar-H), 7.73 (s, 1H, Ar-H), 7.70–7.61 (m, 3H, Ar-H), 7.52 (t, J = 7.9 Hz, 5H, Ar-H), 7.49–7.42 (m, 6H, Ar-H), 7.28 (t, J = 7.7 Hz, 1H, Ar-H), 7.20 (t, J = 7.6 Hz, 1H, Ar-H), 6.23 (s, 2H, NCH2C), 3.91 (s, 3H, NCH3).
Synthesis of the Complexes (1A−3B)
General method: In a round bottom flask, silver oxide (2.4 eq) and (L1-L3) (2.0 eq) were added to a solvent of dichloromethane. After 8 h, the mixture was filtered through celite. The combined filtrates were added dropwise to a solution of dichloromethane containing Dimer ([(η5-Cpx)IrCl2]2, 1.0 eq) (Weaver et al., 2011). The solution was stirred for 8 h at room temperature, filtered with celite and washed with dichloromethane for three times. The solvent was removed by rotary evaporator and yellowish solids were obtained by crystallization in the solution of 10% dichloromethane/50% n-hexane. The data were listed as follows:
[(η5-Cp*)Ir(L1)Cl]Cl (1A): Yield: 65.4%. 1H NMR (500 MHz, DMSO-d6) δ 7.83–7.78 (m, 2H, imidazole-H), 7.73 (d, J = 7.9 Hz, 1H, NCH), 7.52–7.40 (m, 8H, Ar-H), 7.32 (d, J = 2.1 Hz, 1H, Ar-H), 7.24 (dd, J = 5.1, 1.7 Hz, 3H, Ar-H), 7.20 (d, J = 7.4 Hz, 2H, Ar-H), 6.00 (d, J = 16.6 Hz, 1H, NCH2C), 5.06 (d, J = 16.6 Hz, 1H, NCH2C), 4.08 (s, 3H, NCH3), 1.44 (s, 15H, Cp*-H). 13C NMR (126 MHz, DMSO) δ 154.03 (C-Ir), 149.42 (NCN), 140.79, 139.79, 138.37, 135.27, 130.09, 129.13, 128.72, 128.53, 127.75, 124.76, 124.37, 123.73, 122.47, 119.18, 112.71, 90.97, 64.96, 45.04, 31.87, 9.45 ppm. Elemental analysis: Found: C, 54.13; H, 4.79; N, 7.24%, calcd for C35H37Cl2IrN4: C, 54.12; H, 4.80; N, 7.21%. ESI-MS (m/z): calcd for C35H37ClIrN4: 741.23 [M-Cl]+; Found: 741.01.
[(η5-Cp*)Ir(L2)Cl]Cl (2A): Yield: 59.2%. 1H NMR (500 MHz, DMSO-d6) δ 8.23 (d, J = 8.3 Hz, 1H, Ar-H), 7.83 (d, J = 8.0 Hz, 1H, Ar-H), 7.66 (d, J = 7.9 Hz, 1H, Ar-H), 7.47 (dd, J = 11.2, 4.1 Hz, 1H, Ar-H), 7.44–7.38 (m, 2H, Ar-H), 7.26 (s, 5H, Ar-H), 7.20 (t, J = 7.4 Hz, 1H, Ar-H), 6.98 (d, J = 8.2 Hz, 1H, Ar-H), 6.28 (d, J = 17.0 Hz, 1H, NCH2), 6.00 (d, J = 15.3 Hz, 1H, NCH2), 5.78 (d, J = 15.4 Hz, 1H, NCH2C), 5.29 (d, J = 16.9 Hz, 1H, NCH2C), 4.20 (s, 3H, NCH3), 1.70 (s, 15H, Cp*-H). 13C NMR (126 MHz, DMSO) δ 148.66 (C-Ir), 138.56 (NCN), 136.40, 135.28, 135.09, 128.67, 128.03, 123.90, 112.92, 112.75, 92.56, 91.70, 52.30, 32.06, 9.39, 8.72 ppm. Elemental analysis: Found: C, 52.74; H, 4.71; N, 7.49%, calcd for C33H35Cl2IrN4: C, 52.79; H, 4.70; N, 7.46%. ESI-MS (m/z): calcd for C33H35ClIrN4: 715.22 [M-Cl]+; Found: 714.95.
[(η5-Cp*)Ir(L3)Cl]Cl (3A): Yield: 67.9%. 1H NMR (500 MHz, DMSO-d6) δ 8.30 (d, J = 8.4 Hz, 1H, Ar-H), 7.98 (s, 1H, NCH), 7.83 (d, J = 8.1 Hz, 1H, Ar-H), 7.77 (d, J = 8.0 Hz, 1H, Ar-H), 7.50 (dd, J = 22.0, 7.0 Hz, 4H, Ar-H), 7.45–7.41 (m, 1H, Ar-H), 7.37 (t, J = 7.7 Hz, 1H, Ar-H), 7.31 (d, J = 7.8 Hz, 2H, Ar-H), 7.24–7.14 (m, 5H, Ar-H), 7.07 (t, J = 8.0 Hz, 1H, Ar-H), 6.72 (d, J = 8.3 Hz, 1H, Ar-H), 6.29 (d, J = 16.8 Hz, 1H, NCH2C), 5.21 (d, J = 16.9 Hz, 1H, NCH2C), 4.21 (s, 3H, NCH3), 1.46 (s, 15H, Cp*-H). 13C NMR (126 MHz, DMSO) δ 156.63 (C-Ir), 150.24 (NCN), 146.24, 141.10, 134.17, 133.79, 133.12, 132.15, 127.18, 126.55, 123.28, 122.14, 118.90, 66.23, 48.65, 34.79, 13.46 ppm. Elemental analysis: Found: C, 56.61; H, 4.78; N, 6.79 %, calcd for C39H39Cl2IrN4: C, 56.65; H, 4.75; N, 6.78 %. ESI-MS (m/z): calcd for C39H39ClIrN4: 789.55 [M-Cl]+; Found: 789.01.
[(η5-Cpxbiph)Ir(L1)Cl]Cl (1B): Yield: 66.1%. 1H NMR (500 MHz, DMSO-d6) δ 7.85–7.71 (m, 9H, Ar-H), 7.52 (t, J = 7.7 Hz, 2H, imidazole-H), 7.49–7.45 (m, 1H, NCH), 7.41 (dd, J = 14.4, 7.1 Hz, 2H, Ar-H), 7.35 (d, J = 2.1 Hz, 1H, Ar-H), 7.32 (dd, J = 6.9, 2.8 Hz, 2H, Ar-H), 7.24–7.17 (m, 4H, Ar-H), 7.11 (s, 1H, Ar-H), 7.05 (t, J = 7.8 Hz, 2H, Ar-H), 6.48 (d, J = 7.7 Hz, 2H, Ar-H), 6.03 (d, J = 16.7 Hz, 1H, NCH2C), 5.14 (d, J = 16.6 Hz, 1H, NCH2C), 4.10 (s, 3H, NCH3), 1.90 (s, 3H, Cpxbiph-H), 1.64 (s, 3H, Cpxbiph-H), 1.43 (s, 3H, Cpxbiph-H), 1.34 (s, 3H, Cpxbiph-H). 13C NMR (126 MHz, DMSO) δ 152.57 (C-Ir), 149.12 (NCN), 140.73, 140.41, 139.89, 139.34, 138.05, 135.34, 131.39, 130.75, 129.83, 129.62, 128.59, 128.29, 127.81, 127.42, 127.06, 124.85, 124.37, 123.73, 122.53, 119.47, 112.79, 101.14, 97.67, 97.03, 83.35, 80.92, 65.25, 31.88, 11.37, 9.95, 9.69 ppm. Elemental analysis: Found: C, 60.35; H, 4.72; N, 6.10%, calcd for C46H43Cl2IrN4: C, 60.38; H, 4.74; N, 6.12%. ESI-MS (m/z): calcd for C46H43ClIrN4: 879.28 [M-Cl]+; Found: 879.43.
[(η5-Cpxbiph)Ir(L2)Cl]Cl (2B): Yield: 63.7%. 1H NMR (500 MHz, DMSO-d6) δ 8.25 (d, J = 8.4 Hz, 1H, Ar-H), 7.84 (d, J = 8.3 Hz, 1H, Ar-H), 7.76–7.64 (m, 7H, Ar-H), 7.59 (d, J = 8.2 Hz, 1H, Ar-H), 7.56–7.36 (m, 6H, Ar-H), 7.32 (t, J = 7.6 Hz, 1H, Ar-H), 7.22–7.10 (m, 3H, Ar-H), 6.97–6.90 (m, 2H, Ar-H), 6.32 (d, J = 17.0 Hz, 1H, NCH2), 5.80 (d, J = 15.7 Hz, 1H, NCH2), 5.42 (d, J = 15.8 Hz, 1H, NCH2C), 5.35 (d, J = 17.0 Hz, 1H, NCH2C), 4.22 (s, 3H, NCH3), 1.90 (s, 2H, Cpxbiph-H), 1.83 (d, J = 11.3 Hz, 3H, Cpxbiph-H), 1.74 (s, 3H, Cpxbiph-H), 1.70 (d, J = 9.8 Hz, 4H, Cpxbiph-H). 13C NMR (126 MHz, DMSO) δ 167.58 (C-Ir), 148.68 (NCN), 138.49, 136.40, 135.28, 135.09, 133.85, 128.67, 128.03, 124.81, 123.83, 119.19, 112.84, 112.59, 92.56, 91.70, 52.30, 42.04, 32.08, 31.43, 22.54, 14.45, 9.39 ppm. Elemental analysis: Found: C, 59.41; H, 4.66; N, 6.29%, calcd for C44H41Cl2IrN4: C, 59.45; H, 4.65; N, 6.30 %. ESI-MS (m/z): calcd for C44H41ClIrN4: 853.26 [M-Cl]+; Found: 853.32.
[(η5-Cpxbiph)Ir(L3)Cl]Cl (3B): Yield: 69.0%. 1H NMR (500 MHz, DMSO-d6) δ 8.31 (d, J = 8.5 Hz, 1H, Ar-H), 7.86 (d, J = 8.2 Hz, 1H, NCH), 7.81 (d, J = 7.2 Hz, 2H, Ar-H), 7.73 (dd, J = 13.9, 6.8 Hz, 6H, Ar-H), 7.54 (t, J = 7.7 Hz, 2H, Ar-H), 7.43 (ddt, J = 28.1, 12.8, 7.4 Hz, 5H, Ar-H), 7.27 (t, J = 7.4 Hz, 1H, Ar-H), 7.18 (dd, J = 8.8, 4.6 Hz, 1H, Ar-H), 7.14 (d, J = 4.3 Hz, 3H, Ar-H), 7.05 (dd, J = 15.6, 7.7 Hz, 3H, Ar-H), 6.58 (dd, J = 8.0, 4.2 Hz, 3H, Ar-H), 6.32 (d, J = 16.9 Hz, 1H, NCH2C), 5.28 (d, J = 16.8 Hz, 1H, NCH2C), 4.24 (s, 3H, NCH3), 1.89 (s, 3H, Cpxbiph-H), 1.69 (s, 3H, Cpxbiph-H), 1.39 (s, 3H, Cpxbiph-H), 1.33 (s, 3H, Cpxbiph-H). 13C NMR (126 MHz, DMSO) δ 165.72 (C-Ir), 149.11 (NCN), 140.46, 139.88, 138.92, 138.21, 136.57, 135.55, 135.16, 134.01, 131.11, 130.74, 129.66, 129.26, 128.90–128.18, 127.99, 127.65, 127.45, 127.15, 124.86, 123.87, 119.26, 114.68, 113.22, 112.79, 102.22, 98.12, 97.38, 85.56, 65.56, 63.28, 42.16, 32.28, 31.43, 22.54, 11.11, 9.81 ppm. Elemental analysis: Found: C, 62.24; H, 4.69; N, 5.84 %, calcd for C50H45Cl2IrN4: C, 62.23; H, 4.70; N, 5.81%. ESI-MS (m/z): calcd for C50H45ClIrN4: 929.24 [M-Cl]+; Found: 929.40.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
XL, YH, and XG conceived the research and conducted the experiments. ZL directed the project and co-wrote the paper.
Funding
We thank the University Research Development Program of Shandong Province (J18KA082), the National Natural Science Foundation of China (Grant No. 21671118), and the Taishan Scholars Program for support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2020.00182/full#supplementary-material
References
Baral, A., Satish, L., Das, D. P., Sahoo, H., and Ghosh, M. K. (2017). Construing the interactions between MnO2 nanoparticle and bovine serum albumin: insight into the structure and stability of a protein–nanoparticle complex. New J. Chem. 41, 8130–8139. doi: 10.1039/C7NJ01227F
Boya, P., and Kroemer, G. (2008). Lysosomal membrane permeabilization in cell death. Oncogene 27, 6434–6451. doi: 10.1038/onc.2008.310
Chatterjee, S., and Mukherjee, T. K. (2014). Spectroscopic investigation of interaction between bovine serum albumin and amine-functionalized silicon quantum dots. Phys. Chem. Chem. Phys. 16, 8400–8408. doi: 10.1039/c4cp00372a
Chen, Y., Guan, R., Zhang, C., Huang, J., Ji, L., and Chao, H. (2016). Two-photon luminescent metal complexes for bioimaging and cancer phototherapy. Coord. Chem. Rev. 310, 16–40. doi: 10.1016/j.ccr.2015.09.010
Corberán, R., Sanaú, M., and Peris, E. (2006). Aliphatic and aromatic intramolecular C-H activation on Cp*Ir(NHC) complexes. Organometallics 25, 4002–4008. doi: 10.1021/om060343r
Daum, S., Reshetniko, M. S. V., Sisa, M., Dumych, T., Lootsik, M. D., Bilyy, R., et al. (2017). Lysosome-targeting amplifiers of reactive oxygen species as anticancer prodrugs. Angew. Chem. Int. Ed. 56, 15545–15549. doi: 10.1002/anie.201706585
Deubel, D. V., and Lau, J. K.-C. (2006). In silico evolution of substrate selectivity: comparison of organometallic ruthenium complexes with the anticancer drug cisplatin. Chem. Commun. 23, 2451–2453. doi: 10.1039/b601590e
Esteghamat-Panah, R., Hadadzadeh, H., Farrokhpour, H., Mortazavi, M., and Amirghofran, Z. (2017). A mononuclear Ru(II) complex with meloxicam: DNA and BSA-binding, molecular modeling and anticancer activity against human carcinoma cell lines. Inorg. Chim. Acta 454, 184–196. doi: 10.1016/j.ica.2016.04.037
Ge, X., Liu, X., Tian, Z., Chen, S., Liu, X., Guo, L., et al. (2019). Half-sandwich Ruthenium(II) complexes with triphenylamine modified dipyridine skeleton and application in biology/luminescence imaging. Appl. Organomet. Chem. 33:e5171. doi: 10.1002/aoc.5171
Gómez-Sintes, R., Ledesma, M. D., and Boya, P. (2016). Lysosomal cell death mechanisms in aging. Ageing Res. Rev. 32, 150–168. doi: 10.1016/j.arr.2016.02.009
Grossi, M., Morgunova, M., Cheung, S., Scholz, D., Conroy, E., Terrile, M., et al. (2016). Lysosome triggered near-infrared fluorescence imaging of cellular trafficking processes in real time. Nat. Commun. 10855, 1–13. doi: 10.1038/ncomms10855
Han, Y., Tian, Z., Zhang, S., Liu, X., Li, J., Li, Y., et al. (2018). Half-sandwich Iridium(III) N-heterocyclic carbene antitumor complexes and biological applications. J. Inorg. Biochem. 189, 163–171. doi: 10.1016/j.jinorgbio.2018.09.009
Hao, H., Liu, X., Ge, X., Zhao, Y., Tian, X., Ren, T., et al. (2019). Half-sandwich iridium(III) complexes with α-picolinic acid frameworks and antitumor applications. J. Inorg. Biochem. 192, 52–61. doi: 10.1016/j.jinorgbio.2018.12.012
He, X., Liu, X., Tang, Y., Du, J., Tian, M., Xu, Z., et al. (2019). Half-sandwich Iridium(III) complexes with triphenylamine-substituted dipyridine frameworks and bioactivity applications. Dyes Pigm. 160, 217–226. doi: 10.1016/j.dyepig.2018.08.006
Jayabharathi, J., Thanikachalam, V., and Venkatesh Perumal, M. (2011). Mechanistic investigation on binding interaction of bioactive imidazole with protein bovine serum albumin-A biophysical study. Spectrochim. Acta A Mol. Biomol. Spectrosc. 79, 502–507. doi: 10.1016/j.saa.2011.03.020
Jemal, A., Bray, F., Center, M. M., Ferlay, J., Ward, E., and Forman, D. (2011). Global cancer statistics. CA Cancer J Clin. 61, 69–90. doi: 10.3322/caac.20107
Li, C., Yu, M., Sun, Y., Wu, Y., Huang, C., and Li, F. (2011). A nonemissive iridium(III) complex that specifically lights-up the nuclei of living cells. J. Am. Chem. Soc. 133, 11231–11239. doi: 10.1021/ja202344c
Li, J., Liu, X., Zhang, H., Ge, X., Tang, Y., Xu, Z., et al. (2019). Ferrocenyl-triphenyltin complexes as lysosome-targeted Imaging and anticancer agents. Inorg. Chem. 58, 1710–1718. doi: 10.1021/acs.inorgchem.8b03305
Liu, L.-J., Lu, L., Zhong, H.-J., He, B., Kwong, D. W. J., Ma, D.-L., et al. (2015). An iridium(III) complex inhibits JMJD2 activities and acts as a potential epigenetic modulator. J. Med. Chem. 58, 6697–6703. doi: 10.1021/acs.jmedchem.5b00375
Liu, Z., Habtemariam, A., Pizarro, A. M., Fletcher, S. A., Kisova, A., Vrana, O., et al. (2011a). Organometallic half-sandwich iridium anticancer complexes. J. Med. Chem. 54, 3011–3026. doi: 10.1021/jm2000932
Liu, Z., and Sadler, P. J. (2014). Organoiridium complexes: anticancer agents and catalysts. Acc. Chem. Res. 47, 1174–1185. doi: 10.1021/ar400266c
Liu, Z., Salassa, L., Habtemariam, A., Pizarro, A. M., Clarkson, G. J., and Sadler, P. J. (2011b). Contrasting reactivity and cancer cell cytotoxicity of isoelectronic organometallic iridium(III) complexes. Inorg. Chem. 50, 5777–5783. doi: 10.1021/ic200607j
Lu, L., Wang, M., Liu, L.-J., Leung, C.-H., and Ma, D.-L. (2015). Label-free luminescent switch-on probe for ochratoxin a detection using a G-quadruplex-selective iridium(III) complex. ACS Appl. Mater. Interfaces 7, 8313–8318. doi: 10.1021/acsami.5b01702
Ma, D.-L., Chan, D. S.-H., and Leung, C.-H. (2014). Group 9 organometallic compounds for therapeutic and bioanalytical applications. Acc. Chem. Res. 47, 3614–3631. doi: 10.1021/ar500310z
Muhammad, N., and Guo, Z. (2014). Metal-based anticancer chemotherapeutic agents. Curr. Opin. Chem. Biol. 19, 144–153. doi: 10.1016/j.cbpa.2014.02.003
Naz, H., Khan, P., Tarique, M., Rahman, S., Meena, A., Ahamad, S., et al. (2017). Binding studies and biological evaluation of β-carotene as a potential inhibitor of human calcium/calmodulin-dependent protein kinase IV. Int. J. Biol. Macromol. 96, 161–170. doi: 10.1016/j.ijbiomac.2016.12.024
Nichi, M., Rijsselaere, T., Losano, J. D. A., Angrimani, D. S. R., Kawai, G. K. V., Goovaerts, I. G. F., et al. (2016). Evaluation of epididymis storage temperature and cryopreservation conditions for improved mitochondrial membrane potential, membrane integrity, sperm motility and in vitro fertilization in bovine epididymal sperm. Reprod. Domest. Anim. 52, 257–263. doi: 10.1111/rda.12888
Nobre, S. M., and Monteiro, A. L. (2004). Synthesis of diarylmethane derivatives from Pd-catalyzed cross-coupling reactions of benzylic halides with arylboronic acids. Tetrahedron Lett. 45, 8225–8228. doi: 10.1016/j.tetlet.2004.09.020
Pacheco, M. E., and Bruzzone, L. (2013). Synchronous fluorescence spectrometry: conformational investigation or inner filter effect? J. Lumin. 137, 138–142. doi: 10.1016/j.jlumin.2012.12.056
Pellei, M., Gandin, V., Marinelli, M., Orsetti, A., Del Bello, F., Santini, C., et al. (2015). Novel triazolium based 11th group NHCs: synthesis, characterization and cellular response mechanisms. Dalton Trans. 44, 21041–21052. doi: 10.1039/C5DT02934A
Powell, A. B., Bielawski, C. W., and Cowley, A. H. (2010). Design, synthesis, and study of main chain poly(N-Heterocyclic carbene) complexes: applications in electrochromic devices. J. Am. Chem. Soc. 132, 10184–10194. doi: 10.1021/ja104051x
Richens, D. T. (2005). Ligand substitution reactions at inorganic centers. Chem. Rev. 105, 1961–2002. doi: 10.1021/cr030705u
Romero-Canelón, I., and Sadler, P. J. (2013). Next-generation metal anticancer complexes: multitargeting via redox modulation. Inorg. Chem. 52, 12276–12291. doi: 10.1021/ic400835n
Rubbiani, R., Can, S., Kitanovic, I., Alborzinia, H., Stefanopoulou, M., Kokoschka, M., et al. (2011). Comparative in vitro evaluation of N-heterocyclic carbene gold(I) complexes of the benzimidazolylidene type. J. Med. Chem. 54, 8646–8657. doi: 10.1021/jm201220n
Sabharwal, S. S., and Schumacker, P. T. (2014). Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles heel? Nat. Rev. Cancer 14, 709–721. doi: 10.1038/nrc3803
Saftig, P., and Klumperman, J. (2009). Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–631. doi: 10.1038/nrm2745
Schuh, E., Pflüger, C., Citta, A., Folda, A., Rigobello, M. P., Bindoli, A., et al. (2012). Gold(I) carbene complexes causing thioredoxin 1 and thioredoxin 2 oxidation as potential anticancer agents. J. Med. Chem. 55, 5518–5528. doi: 10.1021/jm300428v
Senese, S., Lo, Y. C., Huang, D., Zangle, T. A., Gholkar, A. A., Robert, L., et al. (2014). Chemical dissection of the cell cycle: probes for cell biology and anti-cancer drug development. Cell Death Dis. 5:e1462. doi: 10.1038/cddis.2014.420
Singh, A. K., Yadav, M., Pandey, R., Kumar, P., and Pandey, D. S. (2010). Half-sandwich ruthenium, rhodium and iridium complexes containing dipyridyl amine based ligands. J. Organomet. Chem. 695, 1932–1939. doi: 10.1016/j.jorganchem.2010.04.022
Sun, R. W.-Y., and Che, C.-M. (2009). The anti-cancer properties of gold(III) compounds with dianionic porphyrin and tetradentate ligands. Coord. Chem. Rev. 253, 1682–1691. doi: 10.1016/j.ccr.2009.02.017
Thota, S., Rodrigues, D. A., Crans, D. C., and Barreiro, E. J. (2018). Ru(II) compounds: next-generation anticancer metallotherapeutics? J. Med. Chem. 61, 5805–5821. doi: 10.1021/acs.jmedchem.7b01689
Trachootham, D., Alexandre, J., and Huang, P. (2009). Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 8, 579–591. doi: 10.1038/nrd2803
Valastyan, S., and Weinberg Robert, A. (2011). Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292. doi: 10.1016/j.cell.2011.09.024
Wang, C., Liu, J., Tian, Z., Tian, M., Tian, L., Zhao, W., et al. (2017). Half-sandwich iridium N-heterocyclic carbene anticancer complexes. Dalton Trans. 46, 6870–6883. doi: 10.1039/C7DT00575J
Weaver, J., Gaillard, S., Toye, C., Macpherson, S., Nolan, S. P., and Riches, A. (2011). Cytotoxicity of gold(I) N-heterocyclic carbene complexes assessed by using human tumor cell lines. Chem.-Eur. J. 17, 6620–6624. doi: 10.1002/chem.201100321
Yamabe, M., Kumagai, T., Shimogawara, R., Blay, E. A., Hino, A., Ichimura, K., et al. (2016). Novel synthetic compounds with endoperoxide structure damage juvenile stage of Schistosoma mansoni by targeting lysosome-like organelles. Parasitol. Int. 66, 917–924. doi: 10.1016/j.parint.2016.10.013
Ye, R.-R, Cao, J.-J., Tan, C.-P., Ji, L.-N., and Mao, Z.-W. (2017). Valproic acid-functionalized cyclometalated iridium(III) complexes as mitochondria-targeting anticancer agents. Chem.-Eur. J. 23, 15166–15176. doi: 10.1002/chem.201703157
Yellol, G. S., Donaire, A., Yellol, J. G., Vasylyeva, V., Janiak, C., and Ruiz, J. (2013). On the antitumor properties of novel cyclometalated benzimidazole Ru(II), Ir(III) and Rh(III) complexes. Chem. Commun. 49, 11533–11535. doi: 10.1039/c3cc46239k
Zamora, A., Pérez, S. A., Rodríguez, V., Janiak, C., Yellol, G. S., and Ruiz, J. (2015). Dual antitumor and antiangiogenic activity of organoplatinum(II) complexes. J. Med. Chem. 58,1320–1336. doi: 10.1021/jm501662b
Zeng, L., Gupta, P., Chen, Y., Wang, E., Ji, L., Chao, H., et al. (2017). The development of anticancer ruthenium(ii) complexes: From single molecule compounds to nanomaterials. Chem. Soc. Rev. 46, 5771–5804. doi: 10.1039/C7CS00195A
Zhang, H., Guo, L., Tian, Z., Tian, M., Zhang, S., Xu, Z., et al. (2018). Significant effects of counteranions on the anticancer activity of iridium(III) complexes. Chem. Commun. 54, 4421–4424. doi: 10.1039/C8CC01326H
Zhang, P., Zhuo, S., Sun, L., Zhang, P., and Zhu, C. (2015). Determination of gamma-globulin at nanogram levels by its quenching effect on the fluorescence of a red emitting conjugated polymer. New J. Chem. 39, 4551–4555. doi: 10.1039/C5NJ00286A
Zhitomirsky, B., and Assaraf, Y. G. (2017). Lysosomal accumulation of anticancer drugs triggers lysosomal exocytosis. Oncotarget 8, 45117–45132. doi: 10.18632/oncotarget.15155
Keywords: iridium(III) compounds, anticancer, N-heterocyclic carbene, organelle targeting, imidazole
Citation: Liu X, Han Y, Ge X and Liu Z (2020) Imidazole and Benzimidazole Modified Half-Sandwich IridiumIII N-Heterocyclic Carbene Complexes: Synthesis, Anticancer Application, and Organelle Targeting. Front. Chem. 8:182. doi: 10.3389/fchem.2020.00182
Received: 09 January 2020; Accepted: 27 February 2020;
Published: 17 March 2020.
Edited by:
Xiaoda Yang, Peking University, ChinaReviewed by:
Zongwan Mao, Sun Yat-sen University, ChinaAnnaluisa Mariconda, Università degli Studi della Basilicata, Italy
Yangzhong Liu, University of Science and Technology of China, China
Copyright © 2020 Liu, Han, Ge and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhe Liu, bGl1emhlcWRAMTYzLmNvbQ==
 Xicheng Liu
Xicheng Liu Yali Han
Yali Han