
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Chem. , 21 August 2019
Sec. Theoretical and Computational Chemistry
Volume 7 - 2019 | https://doi.org/10.3389/fchem.2019.00536
This article is part of the Research Topic Proton Transfer Processes in Biological Reactions: A Computational Approach View all 7 articles
Hydrogen bonds play a critical role in nucleobase studies as they encode genes, map protein structures, provide stability to the base pairs, and are involved in spontaneous and induced mutations. Proton transfer mechanism is a critical phenomenon that is related to the acid–base characteristics of the nucleobases in Watson–Crick base pairs. The energetic and dynamical behavior of the proton can be depicted from these characteristics and their adjustment to the water molecules or the surrounding ions. Further, new pathways open up in which protonated nucleobases are generated by proton transfer from the ionized water molecules and elimination of a hydroxyl radical in this review, the analysis will be focused on understanding the mechanism of untargeted mutations in canonical, wobble, Hoogsteen pairs, and mutagenic tautomers through the non-covalent interactions. Further, rare tautomer formation through the single proton transfer (SPT) and the double proton transfer (DPT), quantum tunneling in nucleobases, radiation-induced bystander effects, role of water in proton transfer (PT) reactions, PT in anticancer drugs–DNA interaction, displacement and oriental polarization, possible models for mutations in DNA, genome instability, and role of proton transfer using kinetic parameters for RNA will be discussed.
The cellular machinery, which has been managed by nature for millions of years, still shows errors during DNA replication. These errors generate mutations that affect health disorders. Two types of mutations occur during DNA replica. One is the induced mutation, which is caused by the external agents as free radicals or radiations, but can be reduced by prevention of high-risk factors. The other is the spontaneous mutations that are caused by the action of any external element. The origin of these mutations is still unclear. The protons change their positions during the interchanging of interbase hydrogen bonding in DNA bases. The process occurs due to unwinding of DNA strands during proton transfer (PT) reactions with the formation of “rare tautomers.” Rare tautomers promote the spontaneous mutations and are not detected easily. These mutations are not referred to the random mutations in Watson–Crick (WC) pairs [the genetic code being located in the guanine–cytosine (GC) bases]. The position of SPT or DPT differs in these tautomers. As the detection of these tautomers is difficult, computational approaches are applied to study the PT reactions and its biological consequences in DNA and RNA nucleobases. PT mechanism is defined by the movement of a proton (H+) from one molecule to another. These reactions play an important role in acid/base chemistry, heterogeneous and homogeneous catalysis, corrosion, proton exchange membrane fuel cells, and biochemical processes. The DNA consists of adenine (A), guanine (G), cytosine (C), and thymine (T), while in RNA, thymine is replaced by uracil (U), with the other bases remaining the same. The schematic representation of the structures is given in Figure 1. Thus, RNA gives the corresponding base pairs as AU, and GC. Theoretical studies predict that AU is energetically more stable than AT. Sixty-four possible triplet codons are translated into defined sequence of 20 amino acids linked via peptide bonds.
During tautomeric equilibrium, the interbase hydrogen bonds are transferred as positively charged (proton) carriers. Proton transfer in hydrogen bonding also involves many protons, and these multiproton transfers occur in a planned or stepwise manner between the acid–base sites. These bonds act as the stabilizing factor for the DNA duplexes, even when there are stacking interactions between the bases. The hydrogen bonds join the “stair-step” base pairs in DNA duplexes. The strength of hydrogen bond is important to control the replication and transcription process during cell division along with the other controlling factors as solvent, counter ions, hydrophobicity, local dielectric of surrounding water, pH, pKa, and temperature.
In natural and synthetic catalysis, the transfer of a single proton is difficult due to the size of the catalysts and the complex multi-step or multi component nature of the reactions. The proton transfer reaction involves three basic steps: (a) hydrogen bonding of the acid (A-H+) site to the base (B) forming intermediate A-H+…B complex, (b) movement of proton from A to B, and, finally, (c) dissociation of A and B-H+. Proton transfer reactions occur rapidly, with diffusion being the slowest with a lifetime of 1 × 10−10 s (see Figure 2).
The most interesting feature of these PT reactions is the unexpected results of the experiments and computational calculations given for the studied structures. The stabilization of intermediates also has a vital role in mutagenic studies of these nucleobases apart from the other factors. The critical steps of mutations involve intra- and intermolecular tautomerizations (Löwdin, 1963; Sevilla et al., 1995). The tautomers are important in medicinal chemistry, drug design, and chemical information systems mostly for the organic and bioorganic reactions (Miller et al., 1994; Armitage, 1998; Burrows and Muller, 1998; Boudaïffa et al., 2000) (see Figure 3). DPT is the possible path for mutations with ions, radicals, or surrounding molecules as the supporting factors. The error introduced in formation of rare tautomers during PT reactions affects the important biochemistry processes as diseases.
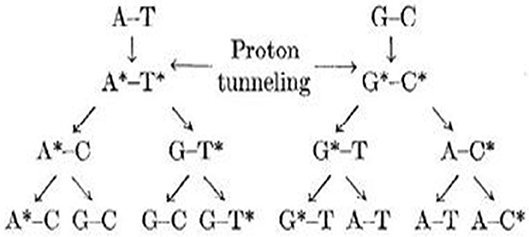
Figure 3. Schematic representation of various possible rare tautomeric states of Watson–Crick (WC) base pairs.
In this section, we will discuss the occurrence of proton tunneling, the effect of radiation on the frequency of proton tunneling, and its result on DNA replication integrity. DNA is used as an information storage biological species as the encoded double-stranded helix structure of nucleobases contain the necessary genetic information with the mirror copy of the other strand (Lodish et al., 2000). This recorded information is just like the basic unit of information (bits) on the magnetic tape, i.e., the data storage system. The encoded messages are transferred to the ribosome during protein synthesis, which transforms it into the receipts of protein assembling. During the replication process, the unzipped DNA base pairs are processed by one of DNA-polymerase (DNA-pol) enzymes. All DNA-pols have intricate geometrical structures. The second strand is assembled by the required information and a new floating deoxynucleotide triphosphates (dNTP) molecule is constructed. The performance of DNA and RNA during the transfer of genetic information is described by the computational algorithms. The dNTP selections that use the quantum tunneling of protons as the main dynamical mechanism require the multistep quantum process. The quantum tunneling effect of quantized wave packet is shown in Figure 4.
Hydrogen bonding occurs by the sharing of a single proton between two lone electrons (electronegative atoms N, O) in nucleobases. The two separate atoms contain an extra unpaired electron (outer orbital shell) to possess the single proton. In tautomeric forms, a proton is moved from its original lone pair to another position. The genetic code is affected when the changes occur from normal to the tautomeric forms, generating errors in DNA replication, and causing mutations. The amplified mutations further lead to severe mutations. In DNA, huge competition occurs to catch the protons in the surrounding environment by the multiple electron lone pairs of several molecules. The single proton–electron system is modeled as a single-well potential. The double-well potential is formed with bumps or potential barrier, when two lone pair electrons compete for a single proton in the hydrogen bonding, which are represented as equilibrium shifts in the following form.
The protons opt tunneling (quantum jump) from one equilibrium state to another through the potential barrier. Protons move in the classically forbidden regions to produce the oscillations with a non-stationary state, extended to symmetric and an asymmetric double-well potential (Parker and Van Everv, 1971). In DNA structures, a minimum of two hydrogen bonds are always involved in the process. Hence, it can be considered as a quantum mechanical two-body problem. For maximum stability, it was assumed that the double-well-potential is highly asymmetric in DNA duplexes. A large potential barrier is required to reduce the frequency of tautomeric formation and to ensure high purity of DNA replication. The proton tunneling time depends on the form and height of the barrier. Tunneling occurs in both directions through the potential barrier. Reverse tunneling also occurs in two ways either with radiation or through normal proton tunneling. In the ground state of the AT base pair, the lowest tunneling level has a proton lifetime of 0.165 × 10−2 s with a corresponding tunneling time of τ1, 2 = 0.346 × 10−4 s, and the highest tunneling level has a proton lifetime of 0.366 × 10−2 s and a tunneling time of τ1, 2 = 0.247 × 10−11 s.
The influence of quantum tunneling, which has significant contribution on proton transfer and the reaction pathways (Godbeer et al., 2015) was studied for the canonical and tautomeric charge-neutral forms of the adenine–thymine base pair (A–T and A*-T*, respectively) with the density functional theory (DFT). The conformational and tautomeric composition of monomers of (tetrazol-5-yl)-acetic acid (TAA) (Araujo-Andrade et al., 2014) were also studied with B3LYP/6-311++G (d,p) level using DFT. 2D potential energy was generated at MP2/6-311++G (d,p) level and a proposal was given to study the tautomerization process during sublimation.
In a latest study, the hydrogen bonds and its use in molecular recognition were studied by comparing the tunneling-assisted quantum entanglement shared in the ground states of covalent and hydrogen bonds (Pusuluk et al., 2018). It was seen that there is significant amounts of quantum entanglement for the thermal state of hydrogen bond. The density-functional theory calculations with hybrid functionals and van der Waals corrections, and optimized path-integral ring-polymer methods (Litman et al., 2019) were used to calculate the quantum vibrational spectra and reaction rates for the nuclear quantum dynamics of intramolecular DPT in porphycene. The results indicated that below 100 K, the concerted double hydrogen transfer (DHT) tunneling pathway dominates, while including the nuclear quantum effects for 100 and 300 K, the concerted and stepwise pathways are selected. Further, the researchers provided architecture for the physical understanding of hydrogen transfer dynamics in the complex systems.
The mutations (transitions) are reversible in which the DNA replicates continuously. Repeated transitions revert back to their original structures. In these mutations, the DNA strand turns either into junk DNA and thus fails to pass on its encoded information and directions, or turns into malfunctioned DNA to give destructive information. In the twentieth century, quantum-mechanical studies were carried out to predict the tautomeric equilibria of heterocyclic compounds. Few parameters such as ultraviolet spectra, dipole moments, and ionization potentials were calculated using semi-empirical or non-empirical quantum-mechanical computational method as per the requirements. These calculations were found to be useful to predict the relative stability of the tautomers in vapor phase in solution and the influence of substituents (Kwiatkowski et al., 1986). The change in tautomeric equilibria of DNA bases that occurs due to the changes from an inert to a polar environment was discussed in a review with the extensive work of the past 14 years (Person et al., 1989). The tautomeric equilibria of 2(4)-mono oxopyrimidines in the gas phase and solution were studied at low-temperature matrices (Nowak et al., 1980). Energies, heats of vaporization, and UV spectra were calculated and compared to the known experimental data. A study on potential energy surface (PES) of guanine performed by Prof. Leszczynski's group showed that the PES of guanine is not flat. The DFT studies were carried out at MP2/6-31G(d,p) reference geometries with MP4(SDTQ)/6-31G(d,p), MP4(SDQ)/6-311G(d,p), and MP2/6-311++G- (df,pd) levels (Leszczynski, 1992, 1998). Further photo-induced proton transfer for 2-(2′-hydroxyphenyl)benzoxazole (HBO) was studied (Ogawa et al., 2000). The basic feature of this model was the study of tautomerization in the biological environment of duplex DNA. Further, it was seen that the biochemical knowledge on biosynthesis of nucleotides, DNA (and RNA) sequence information and transmission, interaction of nucleic acids with proteins, and DNA sequence rearrangements and alterations are the most important things (Blackburn and Gait, 1990). Computational studies about the tautomerism and protonation of guanine and cytosine in the gas phase and in aqueous solution were carried out for the most stable tautomeric forms of the neutral and protonated nucleic acid bases (Colominas et al., 1996). The nature of 10 nucleic acid base stacking was studied at that time by non-empirical ab initio and empirical potential (EP) characterization with planar MP2/6-31G* optimized geometries and many-body correction at the HF/MINI-1 level. Base stacking was investigated in six B-DNA and two Z-DNA base pair steps. The sequence-dependent variations of the total base pair step stacking energies range from −9.9 to −14.7 kcal/mol with the adjustment of the standard combination of a Lennard–Jones and atomic point charge terms (Sponer et al., 1996). DFT studies were carried out for 28 H-bonded DNA (formed by A, T, G, C) base pairs at HF/MINI-1 DFT level of theory (Sponer and Hobza, 1994). The results indicated the H-bonded structure of the cytosine dimer to be more stable than the stacked structures of these pairs. Similarly, the double-proton transfer in adenine–thymine (AT) and GC base pairs in gas phase (Gorb et al., 2004) within room temperature was studied with the inclusion of environmental effects using DFT with B3LYP/6-31G(d) and MP2/6-31G(d) level. The results revealed that the hydrogen-bonded bases possess non-planar geometries. The calculations predicted greater stability for canonic or rare forms of the DNA bases occurring in water molecules and metal cations. Several other computational studies were also carried out for the nucleobases (Hobza, 1986; Stewart et al., 1994; Gould et al., 1995; Jarzynski, 1997).
The intramolecular proton transfer in mono- and dihydrated tautomers of guanine (Gorb and Leszczynski, 1998; Gorb et al., 2004) was studied by HF and the MP2 levels of theory with MP4(SDQ)/6-31G(d)//MP2/6-31G(d) and the MP2/6-311++G(d,p)//MP2/6-31G(d) approximations. It was found that the two-fold water influences the NH2-non-planarity phenomena and decreases the non-planarity for the oxo-tautomers. The two-fold water is the source of non-planarity for the hydroxo tautomers and it decreases the non-planarity for the oxo tautomers.
Another study was carried out for infrared laser-driven double proton transfer (DPT) by a 2D model (Abdel-Latif and Kühn, 2010) using the stepwise and concerted transfer pathways. It was concluded that the driven wave packet is dependent on the parameters of the model Hamiltonian and the propagation time. The stepwise mechanism was dominated in most of the cases, while concerted transfer via tunneling occurs for high barrier systems. The field of proton transfer in hydrogen-bonded networks was also explained in the review by Marx (2006). The review gave insights into Grotthuss diffusion in water, excited-state proton transfer in solution, phase transitions in ice, and protonated water networks in the membrane protein bacteriorhodopsin with the ab initio simulation techniques.
Computational calculations on the hydrogen atom transfer in the cytosine–guanine base pair (Villani, 2010) and its coupling with electronic rearrangement were studied. It was observed that a different behavior occurred when the hydrogen transfer begins with a H of the guanine or of the cytosine and concerted (synchronic in the N–N and asynchronic in the N–O) double-hydrogen transfer can be activated only when the first H atom of guanine is moved. The concerted double-hydrogen process begins with the hydrogen atom of a purinic base. Ab initio constrained molecular dynamics and metadynamics were used to study the mechanism of proton transfer in DNA base pairs (AT, GC) in the gas phase at room temperature (Xiao et al., 2012). Interestingly, the results reveal the DPT in the GC base pair to be a concerted and asynchronous mechanism and in AT to be a stepwise and an asynchronous mechanism. The double-proton transfer reactions in WC GC base pairs were studied after the addition of hydrogen atom (Lin et al., 2012) and the structural changes and energy differences were compared to explore the DPT mechanism. The results revealed that the concerted double-proton transfer mechanism is favorable in the gas phase and the stepwise mechanism is favorable in water with the PT products being energetically less favored.
Oxidized nucleobases (removal of one valence electron) also exhibit enhanced acidity, which leads to PT between the strands of DNA (Ghosh and Schuster, 2006) and competes with the migration of electron hole along the strands (Charkaborty, 2007). The experimental and electronic structure calculations (Khistyaev et al., 2013) showed that ionization-induced PT between nucleobases is endothermic in AT (neutral state) while exothermic in ionized species. The barrierless H-bonded pairs in ionization-induced species predict higher efficiency for the process. PT is slightly endothermic in π-stacked systems as no hydrogen bonding exists in the complexes. PT reactions occur via transition state (TS) (Hratchian and Schlegel, 2005) with one imaginary frequency connecting the reagent and product at the PES. Then, the reaction proceeds over or under the barrier via the tunneling (Löwdin, 1963; Bell, 1980; Koch et al., 2017; Piatkowski et al., 2018; Smedarchina et al., 2018). At this stage, the energy level of proton is equal for the reactants and products (Li et al., 2011; Ceriotti et al., 2016). The path integral molecular dynamics (PIMD) reveal that the nuclear quantum effects (NQEs) can also change the relative stability of tautomeric forms of DNA base pairs (Pérez et al., 2010). Car–Parrinello-based PIMD has been used to see the effect of NQEs during the DPT process in similar DNA structures.
Water is considered as a very important complex in the PT biological systems (Ball, 2013). For example, water filled ion channels through interfaces and membranes and in aerosols (Voth, 2006). A relay-type transport of protons is provided by water wires, which is important for all processes (Agmon, 1995; Chandler et al., 2012). Water wires are involved in the proton-coupled electron transfer in DNA. Experimental studies have been carried out for the NO+(H2O)n clusters with proton-coupled water activation in the ionosphere (Relph et al., 2010). The excited state photo acid structures (Mohammed et al., 2005), proton vs. hydrogen transfer pathways, and the effect of water solvation on catalytic action on tautomers equilibria via PT (Sukhodub, 1987; Kim et al., 1996, 2007; Mizuse et al., 2011) were also studied. The experimental and energetic calculations on dimethyluracil dimers predict superficial proton transfer in the absence of hydrogen bonds (Golan et al., 2012). Experiments were carried out to investigate the nature and dynamics of proton transfer in stacked systems and molecular dynamics calculations were carried out to visualize the actual proton transfer mechanism.
The charge transport in DNA also has potential applications in molecular electronic devices, DNA damage, and repair (Seeman, 2001; Boon et al., 2003; Schuster, 2004; Holman et al., 2007; Jacquemin et al., 2014; Zhang et al., 2015). We have carried out studies on neutral silver cluster interaction to DNA bases/WC base pairs (Srivastava, 2018) using DFT with hybrid density functional B3LYP (Lee et al., 1988; Becke, 1993) potential and LANL2DZ (Hay, 2002)/6–31+G** basis set. See Figure 5. The results reveal strong silver–WC pair's interaction and absorption in the visible region. Similarly, theoretical studies on the electronic and optoelectronic properties of [A.2AP(w)/A*0.2AP(WC)/C.2AP(w)/C*0.2AP(WC)/C.A(w)/C*.A(WC)]–Au8 mismatch nucleobase complexes (Srivastava, 2017) were carried out at the B3LYP/6-311++G(d,p) level. After analyzing the different parameters, the results indicated the applicability of these complexes in fluorescent bioimaging. The smaller HOMO–LUMO (HL) band gap suggested strong chances of electron transfer through DNA duplexes (see Figure 6).
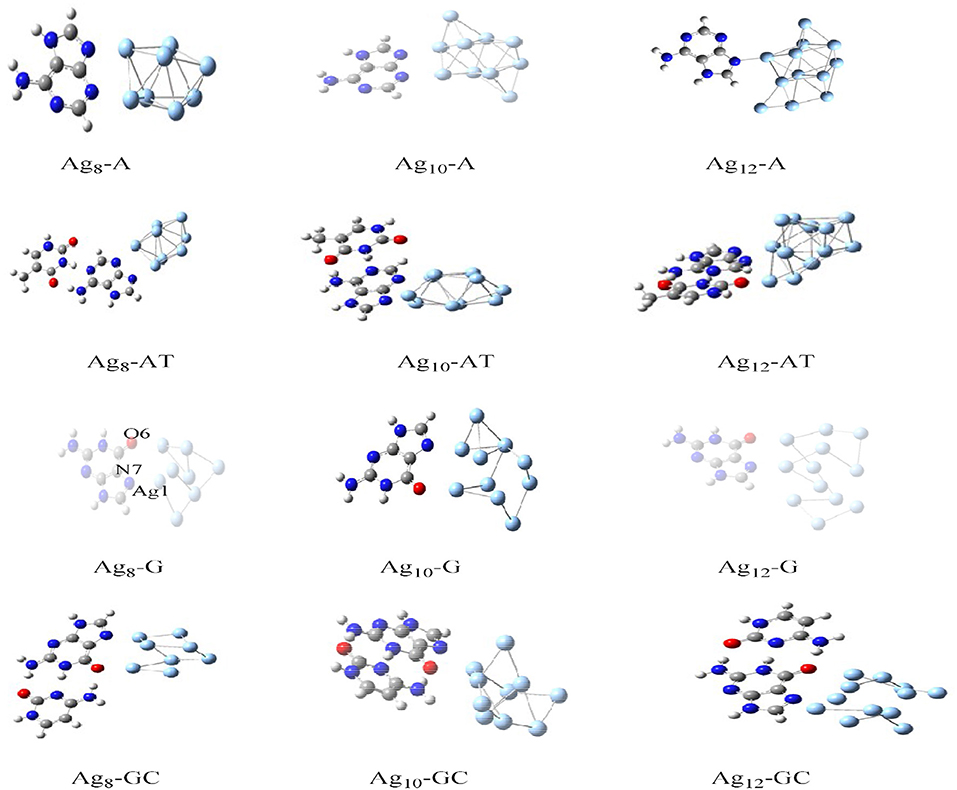
Figure 5. Optimized structures of Ag8-A, AT, G, and GC base pairs (Adapted with permission from Taylor and Francis).
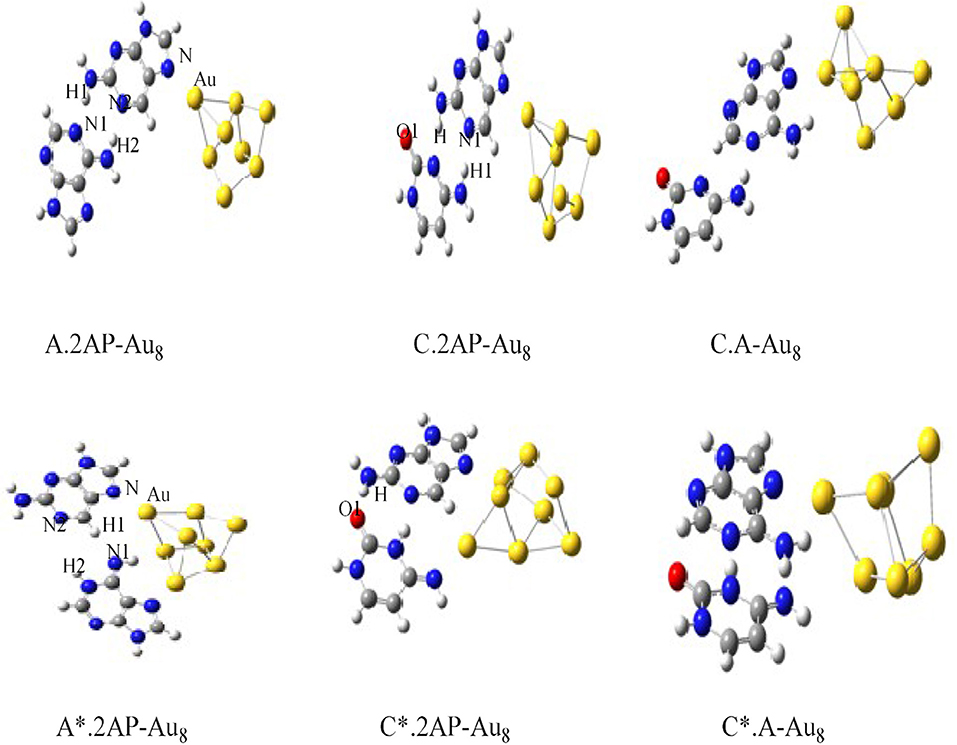
Figure 6. Optimized structures of [A.2AP(w)/A*.2AP(WC)/C.2AP(w)/C*.2AP(WC)/C.A(w)/C*.A(WC)]–Au8 complexes (Adapted with permission from Taylor and Francis).
Studies have been carried out to find the relationship between charge transport and proton transfer. The effect of water molecules led to proton transfer for the cationic stack of AT while a pKa experiment does not predict proton transfer in AT base pairs (Steenken, 1989; Colson et al., 1992). A recent study reported that proton transfer occurs at high temperature as well. Here, two different mechanisms (displacement and oriental polarization) were proposed for the CT and PT at different degrees of temperatures (Zengtao et al., 2019). It was observed that multiple proton transfer occurred at low temperatures and the water-assisted PT occurred by displacement polarization and oriented polarization mechanisms. The rate of PT decreased at higher temperature with water shifting the polarization mode to enhance the PT rate. Further, it was concluded that higher temperature lowers the probability of proton transfer and PT is favored at low temperatures. The DPT mechanism for hydrated AT and GC pairs is given in Figure 7.
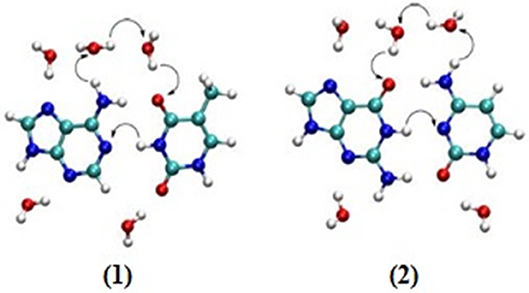
Figure 7. Suggested mechanisms (by arrows) for double proton transfer in hydrated (1) AT and (2) GC base pairs.
The results for the series of nucleobase complexes with organic or inorganic proton donors indicated that electron attachment to these complexes in the gas phase induces PT, which leads to the strong stability for the valence anions (Dabkowska et al., 2004; Harańczyk et al., 2004a,b; Haranczyk et al., 2005; Radisic et al., 2005; Mazurkiewicz et al., 2007b). The probability of stable anion formation in the surrounding of DNA complexes was also studied. The intermolecular PT to excess electron attachment of adenine–formic acid hydrogen-bonded complexes indicated more involvement of adenine bases in proton-donor and -acceptor centers and hydrogen-bonding interactions (Mazurkiewicz et al., 2007a). Further, it was predicted that the stability of the valence ion will increase with the involvement of more species with the nucleobases. As the large affinities of adenine complexes are counter balanced by many physiological environmental factors, they play an important role in the induced mutations by low energy electrons.
Studies have been conducted for the selectively modified DNA structures, which show promising applications for ultrashort electric pulses in medicine. QM/MM (Schrödinger, 1945; Watson and Crick, 1953) became the perfect choice to study the condensed phase biomolecular reactions (Kryachko, 2002; Bertran et al., 2006). QM/MM approach is a reliable and sensitive computational approach for the larger systems. The effect of QM size, method, and schemes on the computed proton transfer energetics in a DNA double helix (Floriań and Leszczyński, 1996), the role of external agents on the PT tautomeric equilibria, and the mutations in electric fields were studied by QM/MM approaches. The influence of embedding and coupling schemes by the QM/MM approach (Roßbach and Ochsenfeld, 2017) was investigated for the transfer of a proton in a DNA base pair. The results indicated that the embedding scheme (mechanical or electrostatic) has much greater impact on the convergence behavior than the coupling scheme (additive QM/MM or subtractive ONIOM).
An analytical review on two-dimensional (2D) potential-energy surface based on two equal hydrogen bonds coupled by a correlation term was given to describe the dynamics of the DPT mechanism (Smedarchina et al., 2008). The quantum-mechanical tunneling and its role in chemical transformations were explained in a recent review (Meisner and Kästner, 2016). The “Tunnel effect” affects the reaction paths and branching ratios and influences the biochemical processes. In another review, a new discipline called “quantum biology” was introduced (McFadden and Al-Khalili, 2018), which included not only the parameters of quantum mechanics such as coherence, tunneling, and entanglement but also photosynthesis, enzyme catalysis, avian navigation, or olfaction. The work has been carried out for the dominant role of tunneling in condensed phases and at high temperatures for DHT in porphycenes by experimental means. These experimental results have provided deep experimental insight into the hydrogen transfer phenomena (Ciaćka et al., 2016).
The SPT and DPT mechanisms in DNA were carried out by many refined models (Kryachko, 2002; Bertran et al., 2006; Kumar and Sevilla, 2010) using second-order Møller–Plesset (MP2) energies on Hartree–Fock (HF) geometries in WC base pairs (Floriań et al., 1994; Floriań and Leszczyński, 1996). In these studies, the mutations were energetically favorable for GC pairs. The tautomeric equilibria of isolated WC base pairs in gas phase and solvent phase (Gorb et al., 2004) were studied using the CASSCF/CASPT2 method to map the 2D PESs for the GC pair by three different mechanisms, which is stepwise double proton transfer (SDPT), DHT, and the concerted double proton transfer (CDPT). The accurate theoretical predictions for biological activities were given with the proposed chemical models. See Figure 8 for SPT and DPT in canonical WC pairs.
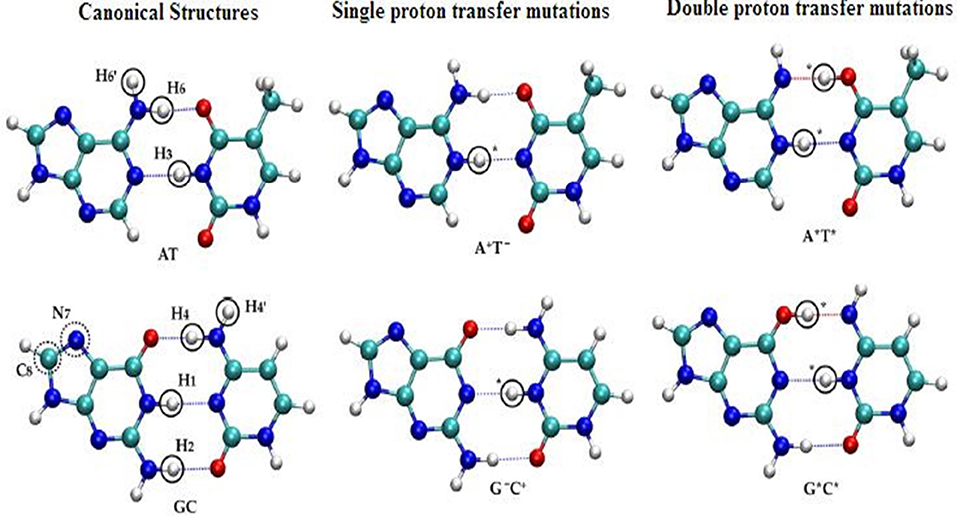
Figure 8. Structural representation of the single and double proton transfer mechanisms of canonical (AT, GC) pairs.
The effect of the surroundings on the spontaneous tautomeric mutation in DNA (Cerón-Carrasco et al., 2009a,b) showed that water molecules are responsible for the double-helix DNA architecture (Kabelác and Hobza, 2007). Catalyzing the spontaneous mutation increases the proton acceptance and donation with DNA. The SPT zwitterionic product acts as a transient species, and the shift of equilibrium to the canonical form occurred for the direct and water-assisted mechanism in AT base pair. Mutations can induce in GC only during the DPT mechanism. The spontaneous mutation for DPT was observed in a propeller-like conformation for the studies of single GC base pairs (Floriań and Leszczyński, 1996; Gorb et al., 2004). The PT reaction for GC radical anion (Chen et al., 2009) with hydrated model and stacking effects was carried out with a different level of DFT. Liang and co-workers used the molecular dynamics (MD) method to study the motions of proton (Xiao et al., 2012).
Prof. Hovorun's group has carried out studies for the canonical A·T(WC) and G·C(WC) WC complexes (Löwdin, 1963; Gorb et al., 2004; Brovarets' et al., 2012; Brovarets' and Hovorun, 2014a,b, 2015e; Roßbach and Ochsenfeld, 2017), wobble pairs Padermshoke et al., 2008; Brovarets' et al., 2015, model protein–DNA complexes (Brovarets' et al., 2012), water-assisted proton transfer in nucleosides (Markova et al., 2017), and mutagenic tautomers (Kondratyuk et al., 2000; Samijlenko et al., 2004; Platonov et al., 2005) to search the reason for the spontaneous point mutations, and other important biological functions such as heredity, aging, and diseases (Löwdin, 1963). These mutations can be considered as “quantum jump” between the undamaged chromosomes and its mutated counterpart states. PT reactions in DNA can be altered by metals and free radicals or the high-energy radiation and electric field.
Though the genotoxic agents create a damaging role in spontaneous mutations, its influence in cancer cells can be positively controlled (Xiao et al., 2012). Chemical reagents as Pt–DNA adduct support SPT with near-planar structures (Korolev et al., 2009). PT reactions also support the biological activity of cisplatin, carboplatin, and oxalaplatin for sequence-specific mutations by the effects of metal ions (Na+, K+, Ca2+, and Mg2+) (Rozsnyai and Ladik, 1970). A novel approach for the internal treatment of cancer with the action of an external physical agent with chemotherapeutic drug was suggested, which controls the exposure region and time duration. Different computational methods have been opted on small DNA models (Jissy and Datta, 2010; Arabi and Matta, 2011) and GC base pairs (Cerón-Carrasco and Jacquemin, 2013) to study the PT reactions by applying intense external electric fields.
The intramolecular PT reactions were studied for both isolated and hydrated DNA bases, while the intermolecular SP and DP transfer reactions are seen in the nucleic acid bases (dimers) at both ground and excited electronic states (Sekiya and Sakota, 2008). Small activation barriers were seen for GC (anionic and cationic) pairs. The tautomeric equilibria and kinetic parameters (lifetime and rate constants) were defined by the activation barrier (Atkins, 1998).
Further, it was studied that the PT reaction is more favorable for the anion as compared to the cation. In the uracil and alanine interactions, the results demonstrated the possibility of electron-induced mutations in DNA–protein complexes. Neutral radicals of hydrogenated pyrimidine nucleobases also lead to mutations in DNA and RNA by low-energy electrons (Beierlein et al., 2011). The effect of polarity was also studied for the dipole-bound anionic states.
DNA replication errors that are caused by genomic instability (Liu et al., 2014; Tomasetti et al., 2017) occurred by the mutagenic tautomerization of WC pairs (Watson and Crick, 1953). The high-energy tautomerized states (Brovarets' and Hovorun, 2010, 2015f) and wobble pairs (Brovarets' and Hovorun, 2015b) for the mutagenic tautomerization of canonical base pairs were studied by Prof. Brovarets et al. The reverse barrier is absent in the tautomeric A·T(WC) pair of DNA bases and have a small kT value for the G·C(WC) DNA base pair (Gorb et al., 2004; Bertran et al., 2006; Brovarets' et al., 2012; Brovarets' and Hovorun, 2014a, 2015a), indicating the non-applicability of Löwdin theory of DPT. The tautomerization occurred via the sequential intrapair proton transfer and shifted to the related pairs (Brovarets' and Hovorun, 2015b). The studies were followed by other similar studies (Brovarets' and Hovorun, 2009, 2015b,c,d, 2016) by the same group. Results indicated that the non-dissociative transitions occurred via specific intermolecular interaction along the intrinsic reaction coordinate (IRC) (Brovarets' and Pérez-Sánchez, 2017). It showed that the tautomeric transitions for A·T DNA base pairs are non-planar and high-energetic intrapair bases (Brovarets' and Hovorun, 2014a, 2015b,d; Brovarets' et al., 2015, 2018a,b,c,d).
Cytosine plays a crucial role in DNA/RNA base pairing as several hydrogen-bonding patterns due to involvement in the genetic codon of 17 amino acids. Interestingly, the proton transfer reactions for cytosine-specific DNA-binding proteins for cytosine and 5-fluorocytosine with 5-nitrouracil, CytNit, and 5FcytNit were also studied (Portalone, 2011).
The strength and the directional properties of non-covalent binding interactions (hydrogen bonding, ionic interactions, van der Waals, and π-π stacking) were used to design the self-assembled or self-associated compounds (Watson and Crick, 1953; Dahm, 2008). The hydrogen-bonding association results in complete proton transfer for an ionic compound with the reinforcement of non-covalent interactions (Merchán and Serrano-Andrés, 2003). The rare tautomeric forms were produced by the intermolecular single/double proton transfer (SPT/DPT) reactions in the hydrogen-bonding DNA pairs. For the unusual combination of NABs pair, normal hydrogen-bonding pattern in DNA is altered, which leads to spontaneous mutations. The DPT reaction is more favorable than the SPT for the neutral systems. The SPT products for base pairs are largely stabilized due to the transfer of a positive charge. The output products are stable and involved in the mutagenic processes. SPT occurs in the formation of the ion-pair complex, while the DPT process retains the electroneutrality. In DPT, the energy barrier is high with the thermodynamically unstable double tautomers. DPT reaction does not show any mutagenic effects. The structural mechanisms for spontaneous point mutations (Turaeva and Brown-Kennerly, 2015) in DNA occur through the transitions and transversions, and various enzymatic complexes for DNA lesions are involved in the repair process, such as DNA glycolases, polymerases, and photolyases (Fromme et al., 2004; Braithwaite et al., 2005). The stable canonical NAB pairs were studied to understand the mechanism of genetic code (Blancafort et al., 2005; Chen and Li, 2005; Marian, 2005; Perun et al., 2005; Tomić et al., 2005). The protonation of NABs also contributes to the stabilization of triple helix, which is stabilized at acidic pH. The PT reaction is also seen in the molecular identification of cytosine–acidic nucleobase derivatives (Park et al., 2011). The DPT in prototropic tautomerisms for many amidine systems and porphyrins was studied experimentally, and the rates and the kinetic isotope effects for both types of DPT were reported. The interaction of amidines with carboxylic acids is useful as the guanidine moiety serves as the binding site for carboxylic acids.
Twenty-two biologically important nucleotide base pairs were studied (Brovarets' and Hovorun, 2010, 2015f) to understand the microstructural mechanisms via DPT and intermolecular hydrogen bonding. It was predicted that the short-lived, low-populated mispairs provide long-lived A·C* (Brovarets' and Hovorun, 2015a), G*·T (Brovarets' and Hovorun, 2015b,c), and H·T* base pairs. The DPT tautomerizations are dipole active and depend on the orientation and change in dipole moment. These complexes can be used for molecular generators under external electric fields (Cerón-Carrasco and Jacquemin, 2013; Mandal et al., 2015; Zhang et al., 2015; Ruiz-Blanco et al., 2017). Further reactive forces via IRC were studied for the reactant, transition states, and product. The cooperativity of the specific intermolecular interactions, i.e., non-classical bonds, loosened covalent bridges, and attractive van der Waals contacts were also analyzed.
New pathways for the mutagenic tautomerization of the classical A.T DNA base pairs in free state, canonical A.T Watson-Crick (WC), reverse A.T Watson-Crick (rWC), A.T Hoogsteen (H), and reverse A.T Hoogsteen (rH) pairs were studied via a sequential PT method. The WC base pairs consist of GC and AT base pairs, while the Hoogsteen base pair is a variation of base-pairing in nucleic acids, such as A•T base pair. The two nucleobases, one on each strand, can be held together by hydrogen bonds in the major groove. A Hoogsteen base pair applies the N7 position of the purine base and C6 amino group to bind the WC (N3–C4) face of the pyrimidine base. The results revealed significant changes in the mutual orientation of the base pairs. Various factors were studied for the process, such as transition states, symmetry, tight ion pairs, Gibbs free energies of activation, and the tautomeric transitions. It was observed that the product of the tautomerization of these pairs was transformed into the energetically favorable [A.T*(rwWC), A.T*(rwH), and A.T*O2(wH)] wobble mispairs through DPT. In these PT processes, the DNA bases shift laterally relative to each other, leading to the wobble pairs of mutagenic tautomers (Brovarets' et al., 2015). This mechanism was applied for the mutagenic tautomerization of various other mispairs (Brovarets' and Hovorun, 2009, 2015b,c,d; Brovarets' et al., 2018e). The mechanism does not provide sufficient long-lived mutagenic DNA tautomers and escape from the replicative DNA-pol, transforming into the canonical tautomeric forms. The optimized structures and the transition states of AT and GC base pairs are given in Figure 9.
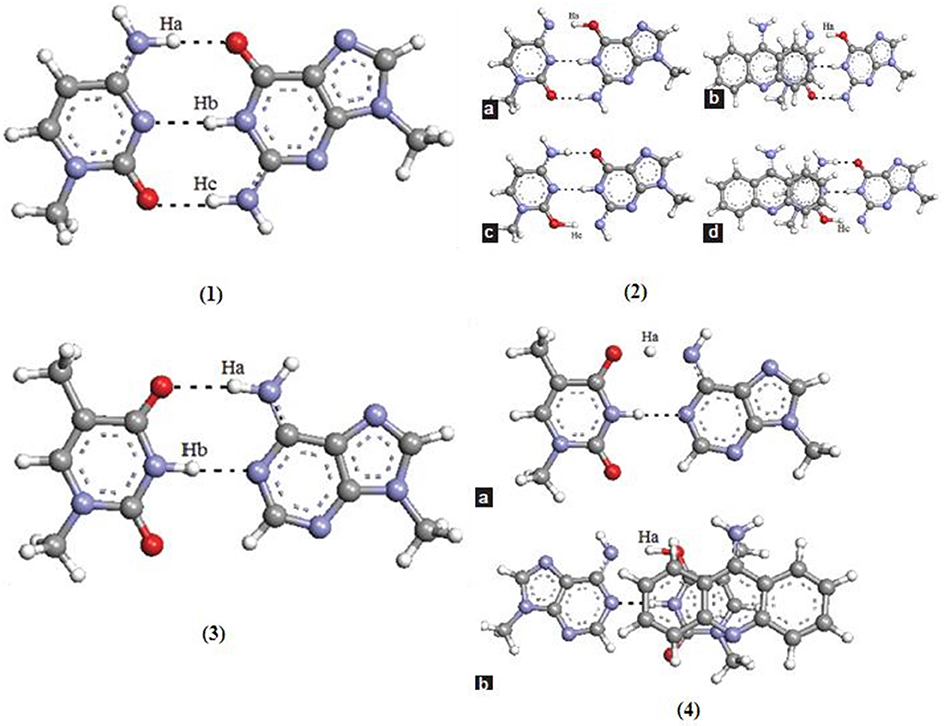
Figure 9. Optimized structure of (1) guanine–cytosine base pair; transition states of (2a) unstacked GC base pair for proton (Ha) transfer type PT1, (2b) stacked GC base pair for proton (Ha) transfer type PT1, (2c) unstacked GC base pair for proton (Hc) transfer type PT2, and (2d) stacked GC base pair for proton (Hc) transfer type PT2; (3) optimized structure of (3) adenine–thymine base pair; transition states of (4a) unstacked GC base pair for proton (Ha) transfer type PT1, (4b) stacked GC base pair for proton (Ha) transfer type PT1 (Adapted from Bezbaruah and Medhi, 2016).
GC radical cation base pair was studied theoretically by the use of DFT, B3LYP/6-31+G** calculations (Kumar and Sevilla, 2009) to analyze the role of hydration on PT mechanism. In GC pairs, holes and electrons were created due to direct interaction of ionizing radiation. Holes were initially captured by guanine (lowest oxidation potential), while thymine and cytosine have captured the electron due to higher electron affinities. Recent studies indicated that hydration has a pronounced effect on the experimental results, and it favors the proton transfer reaction within the base pairs (Adhikary et al., 2006; Witwicki et al., 2009). The effect of bulk solvent is negligible in fully hydrated equilibrium and the stability of the deprotonated states depend on the interactions of water molecules (Lill and Helms, 2001). The energy surface of intermolecular proton transfers can also be reproduced when the assisted water molecules are replaced by point charges. The charges need to be enhanced at close distances due to the induced polarization.
Latest reports included the bystander effects (damage created by irradiated cells in the environment) to study the untargeted mutations (Whiteside et al., 2011). Bystander effects are basically the combination of untargeted and delayed mutations and can be studied experimentally. Untargeted base substitution mutations include some nucleotides inserted to the undamaged sites of DNA. Five rare tautomeric forms of AT pairs within the neighborhood from cyclobutane dimers were studied during DNA synthesis (Niwa, 2006). It was predicted that radiation itself induces genomic instability in cells, which cause genetic changes and increase the rate of mutations. Genomic instability occurs due to the delayed mutations and untargeted mutations that lead to cancer. Six mechanistic explanations were provided to explain the bystander effects. The most probable explanation is the occurrence of non-specific molecular damage due to an irreversible regulatory change in the dynamic interaction network of the cellular gene products (Averbeck, 2010; Campa et al., 2015). Though the conventional reason for mutations depends on sporadic errors of DNA polymerases (Taylor, 2002; Wright, 2010; Watanabe, 2015), one more explanation is given by A-rule, in which the non-complimentary bases are inserted to the DNA polymerases against the damages (Wright, 2010; Watanabe, 2015). Still, there remain many unanswered questions related to the untargeted mutations. For example, why do mutations occur on some undamaged sites of DNA? What is the difference between the undamaged and untargeted mutation sites? Why does genomic instability increase in an unnatural manner with the number of untargeted and delayed mutations? Which external agent is responsible to selectively damage DNA, leaving the non-target sites?
The explanation was given by the polymerase-tautomeric model, which is used to explain the formation of targeted (base substitution, insertions, and deletions) mutations and UV-induced mutagenesis. Also, the polymerase-tautomeric model for bystander effects explains the mechanism for the formation of delayed targeted and untargeted base substitution mutations.
The interactions of anticancer drugs to the DNA nucleobases were studied by the proton transfer mechanism (Bezbaruah and Medhi, 2016). The interaction was carried out for normal base pairs and drug–base pair stacked models. The stacking interactions of drugs–DNA bases results in change in PT energies. PT may also change the acid–base characteristics during the process. The changes in barrier of proton transfer and shifting of equilibrium were also seen in these drugs–DNA bases interactions.
DNA and RNA are both essential for the living organism. DNA keeps the hereditary information of the cell whereas RNA contains the necessary codes to convert that information into functional products. Both RNA and DNA duplexes have large sugar–phosphate chains. The polymerization of amino acids forms the messenger RNA (mRNA) (Sprinzl, 2006). Translational adaptor recognizes the position of the mRNA and the particular place of amino acid is fulfilled by the transfer RNA (tRNA). Each of the other 40 different tRNAs has two specific functions: (i) recognition of its anticodon triplet, a three-letter code of the mRNA, and (ii) the tRNA accepts the incoming amino acid on its chemical site of 3′-terminal adenosine and carries it to the A-site of peptidyl transferees for the formation of peptide bond at the ribosomal center. The post-transcriptional modification of nucleotides in RNA occurs as tRNA, rRNA, mRNA, snRNA, snoRNA, and tmRNA. In this way, 96 different types of nucleotides are identified till now (Rozenski et al., 1999). Other suggested modifications were methylation, pseudouridylation, and thiouridylation, which occurred earlier (Sloan et al., 2017). The role of modified transitions from RNA to DNA was suggested by Martínez Giménez et al. (1998). Thirty-five modifications were found in the peptidyl transferase center, and the A, P, and E sites of tRNA, mRNA, the polypeptide tunnel, and subunit interface. Though these modifications are not essential for ribosomal functioning, they improve the efficiency for ribosomes at a global level (Decatur and Fournier, 2002). The folding and unfolding of hairpin ribozyme have multiple conformations of the RNA with distinct kinetics, and it was studied that the mechanical stability of RNA pseudoknots has strong correlation with their frame shifting efficiency during translation. RNA species hairpin (Li and Tinoco, 2009), pseudoknot (Chen et al., 2007), ribozyme (Onoa et al., 2003), kissing complex (Li and Tinoco, 2009), and riboswitch (Greenleaf et al., 2008) were also studied. The kinetics of reactions was measured by the lifetimes of each species in the reaction. The change in Gibbs was obtained from the mechanical work done (reversible). The reversibility and the unreversibility of the unfolding/folding reaction were measured by the occurrence of the same force in the forward process and no hysteresis, respectively. However, the reversible work for non-reversible reaction was obtained from the distribution of non-reversible work values.
RNA is a four-helix-junction structure (self-cleaving), which contains two internal loops (Wilson et al., 2005). RNA helicases use nucleotide triphosphates (NTPs) to unwind RNA duplexes and are involved in many RNA metabolism processes (Jankowsky and Fairman, 2007; Pyle, 2008). The hairpin ribozyme was derived from tobacco ring spot virus satellite. Investigations were carried out for the role of proton transfer using kinetic parameters in the catalytic mechanism of polyadenylate polymerase (PAP) for the forward (adenylyl transfer) and reverse (pyrophosphorolysis) reactions. Forward reaction suggested the involvement of two protonic species. The multi-conformational continuum electrostatic (MCCE) method was performed to calculate the kinetic parameters and pH variation. Results indicated that PAP contains three globular domains and adopts a closed, active conformation (Balbo and Bohm, 2007; Balbo et al., 2007) and (Mg2+) ion mechanism (Bard et al., 2000; Martin et al., 2000).
The involvement of proton transfer in the ribozyme mechanism is observed in the past few years. Proton transfer is known as general/specific acid–base catalysis (Jencks, 1969; Silverman, 2000) when it occurs to/from biopolymer side chains, water, or OH and hydronium ions, respectively. Acid–base chemistry is observed in every enzyme-catalyzed chemical reaction, except the radical reactions. In ribozyme reactions, the PT prevents the accumulation of unfavorable intermediates that bear charge on bridging oxygen atoms. In general acid–base catalysis, the charge does not accumulate on the 2′- or 5′-oxygens. To optimize general acid–base chemistry, RNA uses the pKa shifting mechanism. In class 1 method, the loaded proton is sequestered in hydrogen bonding, for example, RNA and DNA helices (Wang et al., 1997; Allawi and Santa Lucia, 1998; Pan et al., 1999), cleavage site of the hairpin ribozyme (Cai and Tinoco, 1996; Butcher et al., 1999), lead-dependent ribozyme (Legault and Pardi, 1994), protonated adenine in a Hoogsteen interaction with guanine in a DNA duplex (Gao and Patel, 1988; Leonard et al., 1990; Carbonnaux et al., 1991), protonated cytosine forming a wobble base pair with guanine in the tetrahymena ribozyme (Knitt et al., 1994), protonated cytosine–cytosine base pair (Borah and Wood, 1976), protonated cytosine forming a Hoogsteen pair, and DNA–antibiotic (Quigley et al, 1986) complexes. In class 2, proton is exposed, for example, a model for the transition state of the human hepatitis D virus (HDV) ribozyme self-cleavage reaction (Nakano et al., 2000), Hoogsteen–Hoogsteen interactions for two protonated adenines (Rich et al., 1961), adenine and protonated adenine in a DNA duplex (Chou et al., 1992; Maskos et al., 1993), and guanine and protonated adenine in a Hoogsteen pairing. In RNA folding, co-operativity acts as a driving force in pKa shifting (Moody et al., 2005). Larger interactions lead to bigger pKa shift. Further proton transfer in five different small ribozymes, Hepatitis Delta Virus Ribozyme, Hairpin Ribozyme, Hammerhead Ribozyme, VS Ribozyme, and glmS Ribozyme, were studied with novel biochemical experiments and theoretical means to understand the mechanism. Computational studies have been carried out for the charged nucleobases in RNA, which indicated the thermodynamically unstable ionized nucleobases at pH ~7.4 (Halder et al., 2019). Water affects the systems through hydrogen bonding, Coulomb interactions, and the mediation of proton transfer as an intrinsic component in the secondary structure of proteins.
The catalytic role for a conserved adenosine residue in 23S RNA of the large subunit of ribosome was proposed from the experimental crystallographic and chemical modification methods (Ban et al., 2000; Muth et al., 2000; Nissen et al., 2000). The RNA side chains were not suited for acid–base catalysis as the pKa values of these side chains have significantly higher or lower values than neutral pH. Thus, a significant shift in pKa will be required.
Interestingly, the nucleic acid bases (NABs) with sulfur atom were also studied in natural tRNAs (Yekeler, 2000). These pairs are useful in many biological activities such as thyroid-regulating activities and antitumor agents and in numerous metabolic processes. The effect of sulfur atom on the conformation of the RNA helix was studied as these thiouracil derivatives are useful for the treatment of anti-HIV (Al-Masoudi et al., 2011), cardiovascular diseases (Ribeiro Da Silva et al., 2013), and others (Wen et al., 2006). Various mechanisms were suggested to study these structures as the effect on 2-thiouridine on the RNA structure remains unclear (Sochacka et al., 2015). The DFT studies show lower stability of these base pairs with sulfur and the retard/deform shape of the strand with the RNA viral/tumoral, used for the pharmaceutical reagents. It also gives predictive insight into the structural and energetic effects of these nucleobase modifications, such as post-transcriptional changes, ionization, tautomerization, and metal ion coordination to the nucleobases.
Proton transfer reactions are a very important catalytic activity to control virtually all chemical reactions. Proton shuttles are used in the transfer mechanism of ribosomes and large ribozymes. Even after 50 years of Löwdin's hypothesis, the mechanism behind the rare tautomeric forms in DNA remains unclear. However, quantum computational calculation has accredited several research groups to traverse proton transfer reactions in DNA. Recently, investigations were carried out for the possible modifications of natural tautomeric equilibria, which are induced by external agents. In DNA base pairs, GC base pair emerged as the foundation of DNA replication. It clearly bound the double strands and promotes mutations through induced PT reactions either by chemical or by physical agents. There is a least but finite probability for protons to change place within the hydrogen bond due to quantum tunneling, which will alter the genetic code and cause mutations. These mutations could be the cause of several diseases such as cancer. As the PT reactions occurring in DNA base pairs are explored now, the progress in the control of the spontaneous mutation mechanism is anticipated in the future, which will manage to control the suicide of malignant cells. Water-mediated interactions, altering of energy levels in solvated species, and modified PES along reaction coordinates facilitate effective proton transport through the interfaces and ion channels, which need to be explored.
The regulation of proton tunneling has opened a new area of research in bioelectronics and biocomputing as well. This effect is applied as a “control gate in biocomputing” and realized by a large number of enzymatic reactions. As the tunneling in semiconductors is sensitive to their environment, the devices can be used as sensors. The enzyme tunneling of protons is sensitive to the temperature, external pressure, and others, so focus should be toward the studies of external parameters such as the magnetic field, electromagnetic wave frequency and intensity, etc., which will open new prospects in the external control and information transfer in bioelectronics.
The author confirms being the sole contributor of this work and has approved it for publication.
RS acknowledges the financial assistance by the DST WOS-A (SR/WOS-A/CS-69/2018) scheme.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
RS is also thankful to her mentor Dr. Shrish Tiwari, Bioinformatics, CSIR-Centre for Cellular and Molecular Biology, Hyderabad, for the support.
Abdel-Latif, M., and Kühn, O. (2010). Infrared laser driven double proton transfer. An optimal control theory study. Chem. Phys. 368, 76–82. doi: 10.1016/j.chemphys.2009.12.021
Adhikary, A., Kumar, A., Becker, D., and Sevilla, M. D. (2006). The guanine cation radical: investigation of deprotonation states by ESR and DFT. J. PhysChem. B 110, 24171–24180. doi: 10.1021/jp064361y
Agmon, N. (1995). The Grotthuss mechanism. Chem. Phys. Lett. 244, 456–462. doi: 10.1016/0009-2614(95)00905-J
Allawi, H. T., and Santa Lucia, J. Jr. (1998). Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: sequence dependence and pH effects. Biochemistry 37, 9435–9444. doi: 10.1021/bi9803729
Al-Masoudi, N. A., Saleh, B. A., Abdul Karim, N., Issa, A. Y., and Pannecouque, C. (2011). Synthesis and anti-HIV activity of new 2-thiolumazine and 2-thiouracil metal complexes. Heteroatom. Chem. 22, 44–50. doi: 10.1002/hc.20654
Arabi, A. A., and Matta, C. F. (2011). Effects of external electric fields on double proton transfer kinetics in the formic acid dimer. Phys. Chem. Chem. Phys. 13, 13738–13748. doi: 10.1039/c1cp20175a
Araujo-Andrade, C., Reva, I., and Fausto, R. (2014). Tetrazole acetic acid: tautomers, conformers, and isomerisation. J. Chem. Phys. 140:064306. doi: 10.1063/1.4864119
Armitage, B. (1998). Photocleavage of nucleic acids. Chem. Rev. 98, 1171–1200. doi: 10.1021/cr960428+
Averbeck, D. (2010). Non-targeted effects as a paradigm breaking evidence. Mutat. Res. 687, 7–12. doi: 10.1016/j.mrfmmm.2010.01.004
Balbo, P. B., and Bohm, A. (2007). Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure 15, 1117–1131. doi: 10.1016/j.str.2007.07.010
Balbo, P. B., Toth, J., and Bohm, A. (2007). X-ray crystallographic and steady state fluorescence characterization of the protein dynamics of yeast polyadenylate polymerase. J. Mol. Biol. 366, 1401–1415. doi: 10.1016/j.jmb.2006.12.030
Ball, P. (2013). “The importance of water,” in Astrochemistry and Astrobiology, Physical Chemistry in Action, eds I. W. M. Smith, C. S. Cockell, and S. Leach (Berlin: Springer), 169–210.
Ban, N., Nissen, P., Hansen, J., Moore, P. B., and Steitz, T. A. (2000). The complete atomic structure of the large ribosomal subunit at 2.4? Resol. Sci. 289, 905–920. doi: 10.1126/science.289.5481.905
Bard, J., Zhelkovsky, A. M., Helmling, S., Earnest, T. N., Moore, C. L., and Bohm, A. (2000). Structure of yeast poly(A) polymerase alone and in complex with 3'-dATP. Science 289, 1346–1349. doi: 10.1126/science.289.5483.1346
Becke, A. D. (1993). Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652. doi: 10.1063/1.464913
Beierlein, F. R., Kneale, G. G., and Clark, T. (2011). Predicting the effects of basepair mutations in DNA-protein complexes by thermodynamic integration. Biophys. J. 101, 1130–1138. doi: 10.1016/j.bpj.2011.07.003
Bertran, J., Blancafort, L., Noguera, M., and Sodupe, M. (2006).“Proton transfer in DNA base pairs,”in Computational Studies of RNA and DNA, eds J. Šponer and F. Lankaš (Dordrecht: Springer), 411–432.
Bezbaruah, B., and Medhi, B. C. (2016). Quantum mechanical study on the proton transfer mechanism within adenine-thymine and guanine-cytosine base pairs of DNA nucleobase. Ind. J. Adv. Chem. Sci. 4, 314–320.
Blackburn, G. M., and Gait, M. J. (1990). Nucleic Acids in Chemistry and Biology. New York, NY: IRL Press at Oxford University Press, 467:446.
Blancafort, L., Cohen, B., Hare, P. M., Kohler, B., and Robb, M. A. (2005). Singlet excited-state dynamics of 5-fluorocytosine and cytosine: an experimental and computational study. J. Phys. Chem. A 109, 4431–4436. doi: 10.1021/jp045614v
Boon, E. M., Livingston, A. L., Chmiel, N. H., David, S. S., and Barton, J. K. (2003). DNA mediated charge transport for DNA repair. Proc. Natl. Acad. Sci. U.S.A. 100, 12543–12547. doi: 10.1073/pnas.2035257100
Borah, B., and Wood, J. L. (1976). The cytidinium—cytidine complex: infrared and Raman spectroscopic studies. J. Mol. Struct. 30, 13–30. doi: 10.1016/0022-2860(76)85042-9
Boudaïffa, B., Cloutier, P., Hunting, D., Huels, M. A., and Sanche, L. (2000). Resonant formation of DNA strand breaks by low-energy (3 to 20 eV) electrons. Science. 287, 1658–1660. doi: 10.1126/science.287.5458.1658
Braithwaite, E. K., Prasad, R., Shock, D. D., Hou, E. W., Beard, W. A., and Wilson, S. H. (2005). DNA polymerase λ mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J. Biol. Chem. 280, 18469–18475. doi: 10.1074/jbc.M411864200
Brovarets', O. O., and Hovorun, D. M. (2009). Physicochemical mechanism of the wobble DNA base pairs GuaThy and AdeCyt transition into the mismatched base pairs Gua*Thy and AdeCyt*formed by the mutagenic tautomers. UkrBioorgActa. 8, 12–18.
Brovarets', O. O., and Hovorun, D. M. (2010). IR vibrational spectra of Hbonded complexes of adenine, 2-aminopurine and 2-aminopurine+ with cytosine and thymine: quantum-chemical study. Optics Spectr. 111, 750–757. doi: 10.1134/S0030400X11120058
Brovarets', O. O., and Hovorun, D. M. (2014a). Can tautomerisation of the A·T Watson-Crick base pair via double proton transfer provoke point mutations during DNA replication? A comprehensive QM and QTAIM analysis. J. Biomol. Struct. Dyn. 32, 127–154. doi: 10.1080/07391102.2012.755795
Brovarets', O. O., and Hovorun, D. M. (2014b). Why the tautomerization of the G·C Watson-Crick base pair via the DPT does not cause point mutations during DNA replication? QM and QTAIM comprehensive analysis. J. Biomol. Struct. Dyn. 32, 1474–1499. doi: 10.1080/07391102.2013.822829
Brovarets', O. O., and Hovorun, D. M. (2015a). New structural hypostases of the AT and GC Watson-Crick DNA base pairs caused by their mutagenic tautomerisation in a wobble manner: a QM/QTAIM prediction. RSC Adv. 5, 99594–99605. doi: 10.1039/C5RA19971A
Brovarets', O. O., and Hovorun, D. M. (2015b). Wobble↔Watson-Crick tautomeric transitions in the homo-purine DNA mismatches: a key to the intimate mechanisms of the spontaneous transversions. J. Biomol. Struct. Dynam. 33, 2710–2715. doi: 10.1080/07391102.2015.1077737
Brovarets', O. O., and Hovorun, D. M. (2015c). Novel physico-chemical mechanism of the mutagenic tautomerisation of the Watson–Crick-like AG and CT DNA base mispairs: a quantum-chemical picture. RSC Adv. 5, 66318–66333. doi: 10.1039/C5RA11773A
Brovarets', O. O., and Hovorun, D. M. (2015d). A novel conception for spontaneous transversions caused by homopyrimidine DNA mismatches: a QM/QTAIM highlight. Phys. Chem. Chem. Phys. 17, 21381–21388. doi: 10.1039/C5CP03211C
Brovarets', O. O., and Hovorun, D. M. (2015e). Proton tunneling in the A·T Watson-Crick DNA base pair: Myth or reality? J. Biomol. Struct. Dyn. 33, 2716–2720. doi: 10.1080/07391102.2015.1092886
Brovarets', O. O., and Hovorun, D. M. (2015f). Tautomeric transition between wobble AC DNA base mispair and Watson-Crick-like AC*mismatch: miscrostructural mechanism and biological significance. Phys. Chem. Chem. Phys. 17, 15103–15110. doi: 10.1039/C5CP01568E
Brovarets', O. O., and Hovorun, D. M. (2016). By how many tautomerisation routes the Watson-Crick-like A·C*DNA base mispair is linked with the wobble mismatches? A QM/QTAIM vision from a biological point of view. Struct. Chem. 27, 119–131. doi: 10.1007/s11224-015-0687-4
Brovarets', O. O., and Pérez-Sánchez, H. (2017). Whether 2-aminopurine induces incorporation errors at the DNA replication? A quantum-mechanical answer on the actual biological issue. J. Biomol. Struct. Dyn. 35, 3398–3411. doi: 10.1080/07391102.2016.1253504
Brovarets', O. O., Tsiupa, K. S., and Hovorun, D. M. (2018a). Surprising conformers of the biologically important A·T DNA base pairs: QM/QTAIM proofs. Front. Chem. 6:8. doi: 10.3389/fchem.2018.00008
Brovarets', O. O., Tsiupa, K. S., and Hovorun, D. M. (2018b). Non-dissociative structural transitions of the Watson-Crick and reverse Watson-Crick A·T DNA basepairs into the Hoogsteen and reverse Hoogsteen forms. Sci. Rep. 8:10371. doi: 10.1038/s41598-018-28636-y
Brovarets', O. O., Tsiupa, K. S., and Hovorun, D. M. (2018c). Novel pathway for mutagenic tautomerization of classical A·T DNA base pairs via sequential proton transfer through quasi-orthogonal transition states: a QM/QTAIM investigation. PLoS ONE 13:e0199044. doi: 10.1371/journal.pone.0199044
Brovarets', O. O., Tsiupa, K. S., and Hovorun, D. M. (2018d). The A·T(rWC)/A·T(H)/A·T(rH)↔A·T*(rwWC)/A·T*(wH)/A·T*(rwH) mutagenic tautomerization via sequential proton transfer: a QM/QTAIM study. RSC Adv. 8, 13433–13445. doi: 10.1039/C8RA01446A
Brovarets', O. O., Tsiupa, K. S., and Hovorun, D. M. (2018e). Unexpected A·T(WC)↔A·T(rWC)/A·T(rH) and A·T(H)↔A·T(rH)/A·T(rWC) conformational transitions between the classical A·T DNA base pairs: a QM/QTAIM comprehensive study. Int. J. Quantum. Chem. 118:e25674. doi: 10.1002/qua.25692
Brovarets', O. O., Yurenko, Y. P., Dubey, I. Y., and Hovorun, D. M. (2012). Can DNA-binding proteins of replisometautomerize nucleotide bases? Ab initio model study. J. Biomol. Struct. Dyn. 29, 1101–1109. doi: 10.1080/07391102.2011.672624
Brovarets', O. O., Zhurakivsky, R. O., and Hovorun, D. M. (2015). DPT tautomerisation of the wobble guanine·thymine DNA base mispair is not mutagenic: QM and QTAIM arguments. J. Biomol. Struct. Dyn. 33, 674–689. doi: 10.1080/07391102.2014.897259
Burrows, C. J., and Muller, J. G. (1998). Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 98, 1109–1152. doi: 10.1021/cr960421s
Butcher, S. E., Allain, F. H., and Feigon, J. (1999). Solution structure of the loop B domain from the hairpin ribozyme. Nat. Struct. Biol. 6, 212–216. doi: 10.1038/6651
Cai, Z., and Tinoco, I. Jr. (1996). Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry 35, 6026–6036. doi: 10.1021/bi952985g
Campa, A., Balduzzi, M., Dini, V., Esposito, G., and Tabocchini, M. A. (2015). The complex interactions between radiation induced non-targeted effects and cancer. Cancer Lett. 356, 126–136. doi: 10.1016/j.canlet.2013.09.030
Carbonnaux, C., van der Marel, G. A., van Boom, J. H., Guschlbauer, W., and Fazakerley, G. V. (1991). Solution structure of an oncogenic DNA duplex containing a G.A mismatch. Biochemistry 30, 5449–5458. doi: 10.1021/bi00236a018
Ceriotti, M., Fang, W., Kusalik, P. G., McKenzie, R. H., Michaelides, A., Morales, M. A. et al. (2016). Nuclear quantum effects in water and aqueous systems: experiment, theory, and current challenges. Chem. Rev. 116, 7529–7550. doi: 10.1021/acs.chemrev.5b00674
Cerón-Carrasco, J. P., and Jacquemin, D. (2013). Electric-field induced mutation of DNA: a theoretical investigation of the GC base pair. Phys. Chem. Chem. Phys. 15, 4548–4553. doi: 10.1039/c2cp44066k
Cerón-Carrasco, J. P., Requena, A., Michaux, C., Perpète, E. A., and Jacquemin, D. (2009a). Effects of hydration on the proton transfer mechanism in the adenine–thymine base pair. J. Phys. Chem. A 113, 7892–7898. doi: 10.1021/jp900782h
Cerón-Carrasco, J. P., Requena, A., Zún iga, J., Michaux, C., Perpète, E., and Jacquemin, D. (2009b). Intermolecular proton transfer in microhydrated guanine–cytosine base pairs: a new mechanism for spontaneous mutation in DNA. J. Phys. Chem. A, 113, 10549–10556. doi: 10.1021/jp906551f
Chandler, D., Dellago, C., and Geissler, P. (2012) Wired-upwater. Nat. Chem. 4, 245–247. doi: 10.1038/nchem.1300
Charkaborty, T. (ed.). (2007). Charge Migration in DNA. Berlin: Springer, 1–204. doi: 10.1007/978-3-540-72494-0
Chen, G., Wen, J. D., and Tinoco, I., Jr. (2007). Single-molecule mechanical unfold-ing and folding of a pseudoknot in human telomerase RNA. RNA 13, 2175–2188. doi: 10.1261/rna.676707
Chen, H., and Li, S. (2005). Theoretical study toward understanding ultrafast internal conversion of excited 9H-adenine. J. Phys. Chem. A 109, 8443–8446. doi: 10.1021/jp0537207
Chen, H.-Y., Kao, C.-L., and Hsu, S. C. (2009). Proton transfer in guanine-cytosine radical anion embedded in B-form DNA. J. Am. Chem. Soc. 131, 15930–15938. doi: 10.1021/ja906899p
Chou, S.-H., Cheng, J.-W., Fedoroff, O. Y., Chuprina, V. P., and Reid, B. R. (1992). Novel cross-strand three-purine stack of the highly conserved 5′-GA/AAG-5′ internal loop at the 3′-end termini of Parvovirus Genomes. J. Am. Chem. Soc. 114, 3114–3115. doi: 10.1021/ja00034a055
Ciaćka, P., Fita, P., Listkowski, A., Radzewicz, C., and Waluk, J. (2016). Evidence for dominant role of tunneling in condensed phases and at high temperatures: double hydrogen transfer in porphycenes. J. Phys. Chem. Lett. 7, 283–288. doi: 10.1021/acs.jpclett.5b02482
Colominas, C., Luque, F. J., and Orozco, M. (1996). Tautomerism and protonation of guanine and cytosine. Implications in the formation of hydrogen-bonded complexes. J. Am. Chem. Soc. 118, 6811–6821. doi: 10.1021/ja954293l
Colson, A. O., Besler, B., Close, D. M., and Sevilla, M. D. (1992). Ab initio molecular orbital calculations of DNA bases and their radical ions in various protonation states: evidence for proton transfer in GC base pair radical anions. J. Phys. Chem. 96, 661–668. doi: 10.1021/j100181a028
Dabkowska, I., Rak, J., Gutowski, M., Nilles, J. M., Stokes, S. T., and Bowen, K. H., Jr. (2004). Barrier-free intermolecular proton transfer induced by excess electron attachment to the complex of alanine with uracil. J. Chem. Phys. 120, 6064–6071. doi: 10.1063/1.1666042
Dahm, R. (2008). Discovering DNA: Friedrich Miescher and the early years of nucleic acid research. Hum. Gen. 122, 565–581. doi: 10.1007/s00439-007-0433-0
Decatur, W. A., and Fournier, M. J. (2002). rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351. doi: 10.1016/S0968-0004(02)02109-6
Floriań, J., Hrouda, V., and Hobza, P. (1994). Proton transfer in the adenine–thymine base pair. J. Am. Chem. Soc. 116, 1457–1460. doi: 10.1021/ja00083a034
Floriań, J., and Leszczyński, J. (1996). Spontaneous DNA mutations induced by proton transfer in the guanine cytosine base pairs: an energetic perspective. J. Am. Chem. Soc. 118, 3010–3017. doi: 10.1021/ja951983g
Fromme, J. C., Banerjee, A., and Verdine, G. L. (2004). DNA glycosylase recognition and catalysis. Curr. Opin. Struct. Biol. 14, 43–49. doi: 10.1016/j.sbi.2004.01.003
Gao, X., and Patel, D. J. (1988). G(syn).cntdot.A(anti) mismatch formation in DNA dodecamers at acidic pH: pH-dependent conformational transition of G.cntdot.Amispairs detected by proton NMR. J. Am. Chem. Soc. 110, 5178–5182. doi: 10.1021/ja00223a045
Ghosh, A. K., and Schuster, G. B. (2006). Role of the guanine N1 imino proton in the migration and reaction of radical cations in DNA oligomers. J. Am. Chem. Soc. 128, 4172–4175. doi: 10.1021/ja0573763
Godbeer, A. D., Al-Khalili, J. S., and Stevenson, P. D. (2015). Modelling proton tunnelling in the adenine-thymine base pair. Phys. Chem. Chem. Phys. 17, 13034–13044. doi: 10.1039/C5CP00472A
Golan, A., Bravaya, K. B., Kudirka, R., Kostko, O., Leone, S. R., Krylov, A. I., et al. (2012). Ionization of dimethyluracil dimers leads to facile proton transfer in the absence of hydrogen bonds. Nat. Chem. 4, 323–329. doi: 10.1038/nchem.1298
Gorb, L., and Leszczynski, J. (1998). Intramolecular proton transfer in mono- and dihydrated tautomers of guanine: an ab initio post hartree-fock study. J. Am. Chem. Soc. 120, 5024–5032. doi: 10.1021/ja972017w
Gorb, L., Podolyan, Y., Dziekonski, P., Sokalski, W. A., and Leszczyński, J. (2004). Double-proton transfer adenine-thymine and gunine-cytosine base pairs. A post-hartree-fockab initio study. J. Am. Chem. Soc. 126, 10119–10129. doi: 10.1021/ja049155n
Gould, I. R., Burton, N. A., Hall, R. J., and Hillier, I. H. (1995). Tautomerism in uracil, cytosine and guanine: a comparison of electron correlation predicted by ab initio and density functional theory methods. J. Mol. Struct. 331, 147–154. doi: 10.1016/0166-1280(94)03887-Q
Greenleaf, W. J., Frieda, K. L., Foster, D. A., Woodside, M. T., and Block, S. M. (2008). Di-rect observation of hierarchical folding in single riboswitch aptamers. Science 319, 630–633. doi: 10.1126/science.1151298
Halder, A., Vemuri, S., Roy, R., Katuri, J., Bhattacharyya, D., and Mitra, A. (2019). Evidence for hidden involvement of N3-protonated guanine in RNA structure and function. ACS Omega 4, 699–709. doi: 10.1021/acsomega.8b02908
Harańczyk, M., Dabkowska, I., Rak, J., Gutowski, M., Nilles, J. M., Stokes, S.T., et al. (2004b). Excess electron attachment induces barrier-free proton transfer in anionic complexes of thymine and uracil with formic acid. J. Phys. Chem. B 108, 6919–6921. doi: 10.1021/jp0379143
Haranczyk, M., Rak, J., Gutowski, M., Radisic, D., Stokes, S. T., and Bowen, K. H. (2005). Intermolecular proton transfer in anionic complexes of uracil with alcohols. J. Phys. Chem. B 109, 13383–13391. doi: 10.1021/jp050246w
Harańczyk, M., Rak, J., Gutowski, M., Radisic, D., Stokes, S. T., Nilles, J. M., et al. (2004a) Effect of hydrogen bonding on barrier-free proton transfer in anionic complexes of uracil with weak acids: (U…HCN)− versus (U…H2S)−. Israel J. Chem. 44, 157–170. doi: 10.1560/PBM0-CM9L-7YBH-W719.
Hay, P. J. (2002). Theoretical studies of the ground and excited electronic states in cyclometalated phenylpyridine Ir(III) complexes using density functional theory. J. Phys. Chem. A. 106, 1634–1641. doi: 10.1021/jp013949w
Hobza, P. (1986). Nonempirical ab initio calculations on DNA base pairs. Chem. Phys. 204, 365–372. doi: 10.1016/0301-0104(95)00311-8
Holman, M. R., Ito, R. T., and Rokita, S. E. (2007). Self-repair of thymine dimer in duplex DNA. J. Am. Chem. Soc. 129, 6–7. doi: 10.1021/ja0668365
Hratchian, H. P., and Schlegel, H. B. (2005). “Finding minima, transition states, and following reaction pathways on ab initio potential energy surfaces,” in Theory and Applications of Computational Chemistry, eds C. E. Dykstra, G. Frenking. Amsterdam: Elsevier, 195–249.
Jacquemin, D., Zúñiga, J., Requena, A., and Céron-Carrasco, J. P. (2014). Assessing the importance of proton transfer reactions in DNA. Acc. Chem. Res. 47, 2467–2474. doi: 10.1021/ar500148c
Jankowsky, E., and Fairman, M. E. (2007) RNA helicases-one fold for many functions. Curr. Opin. Struct. Biol. 17, 316–324. doi: 10.1016/j.sbi.2007.05.007
Jarzynski, C. (1997). Nonequilibrium equality for free energy differences. Phys. Rev. Lett. 78, 2690–2693. doi: 10.1103/PhysRevLett.78.2690
Jissy, A. K., and Datta, A. (2010). Designing molecular switches based on DNA-base mispairing. J. Phys. Chem. B 114, 15311–15318. doi: 10.1021/jp106732u
Kabelác, M., and Hobza, P. (2007). Hydration and stability of nucleic acid bases and base pairs. Phys. Chem. Chem. Phys. 9, 903–917. doi: 10.1039/B614420A
Khistyaev, K., Golan, A., Bravaya, K. B., Orms, N., Krylov, A. I., and Ahmed, M. (2013). Proton transfer in nucleobases is mediated by water. J. Phys. Chem. A 117, 6789–6797. doi: 10.1021/jp406029p
Kim, N. J., Kim, H. M., and Kim, S. K. (2007). Ionization-induced proton transfer in thymine ammonia van der Waals clusters. Int. J. Mass. Spectr. 261, 32–37. doi: 10.1016/j.ijms.2006.07.017
Kim, S. K., Lee, W., and Herschbach, D. R. (1996). Cluster beam chemistry: hydration of nucleic acid bases; Ionization potentials of hydrated adenine and thymine. J. Phys. Chem. 100, 7933–7937. doi: 10.1021/jp960635d
Knitt, D. S., Narlikar, G. J., and Herschlag, D. (1994). Dissection of the role of the conserved G.U pair in group I RNA self-splicing. Biochemistry 33, 13864–13879. doi: 10.1021/bi00250a041
Koch, M., Pagan, M., Persson, M., Gawinkowski, S., Waluk, J., and Kumagai, T. (2017). Direct observation of double hydrogen transfer via quantum tunneling in a single porphycene molecule on a Ag (110) surface. J. Am. Chem. Soc. 139, 12681–12687. doi: 10.1021/jacs.7b06905
Kondratyuk, I. V., Samijlenko, S. P., Kolomiets, I. M., and Hovorun, D. M. (2000). Prototropic molecular-zwitterionic tautomerism of xanthine and hypoxanthine. J. Mol. Struct. 523, 109–118. doi: 10.1016/S0022-2860(99)00385-3
Korolev, N., Lyubartsev, A. P., Rupprecht, A., and Nordenskiöld, L. (2009). Competitive binding of Mg2+, Ca2+, Na+, and K+ ions to DNA in oriented DNA fibers: experimental and Monte Carlo simulation results. Biophys. J. 77, 2736–2749. doi: 10.1016/S0006-3495(99)77107-9
Kryachko, E. S. (2002). The origin of spontaneous point mutations in DNA via Löwdin mechanism of proton tunneling in DNA base pairs: cure with covalent base pairing. Int. J. Quantum Chem. 90, 910–923. doi: 10.1002/qua.975
Kumar, A., and Sevilla, M. D. (2009). Influence of hydration on proton transfer in the guanine-cytosine radical cation (G•+-C) base pair: a density functional theory study. J. Phys. Chem. B 113, 11359–11361. doi: 10.1021/jp903403d
Kumar, A., and Sevilla, M. D. (2010). Proton-coupled electron transfer in DNA on formation of radiation-produced ion radicals. Chem. Rev. 110, 7002–7023. doi: 10.1021/cr100023g
Kwiatkowski, J. S., Julia, T., and Rein, Z. R. (1986). Quantum-mechanical prediction of tautomeric equilibria. Adv. Quantum Chem. 18, 85–130. doi: 10.1016/S0065-3276(08)60048-9
Lee, C., Yang, W., and Parr, R. G. (1988). Development of the Colle-Salvetti correlation energy formula into a functional of the electron density. Phys. Rev. B 37, 785–789. doi: 10.1103/PhysRevB.37.785
Legault, P., and Pardi, A. (1994). In situ probing of adenine protonation in RNA by 13C NMR. J. Am. Chem. Soc. 116, 8390–8391. doi: 10.1021/ja00097a066
Leonard, G. A., Booth, E. D., and Brown, T. (1990). Structural and thermodynamic studies on the adenine guanine mismatch in B-DNA. Nucleic Acids Res. 18, 5617–5623. doi: 10.1093/nar/18.19.5617
Leszczynski, J. (1992). Structure and properties of uracil and its sulfur analogs: a systematic study of basis set effects in Ab InitioSCF calculations. Int. J. Quantum Chem. 19, 43–55.
Leszczynski, J. (1998). The potential energy surface of guanine is not flat: an ab initio study with large basis sets and higher order electron correlation contributions. J. Phys. Chem. A 102, 2357–2362. doi: 10.1021/jp972950l
Li, P. T., and Tinoco, I. Jr. (2009). Mechanical unfolding of two DIS RNA kissing complexes from HIV-1. J. Mol. Biol. 386, 1343–1356. doi: 10.1016/j.jmb.2009.01.023
Li, X. Z., Walker, B., and Michaelides, A. (2011). Quantum nature of the hydrogen bond. Proc. Natl. Acad. Sci. U.S.A. 108, 6369–6373. doi: 10.1073/pnas.1016653108
Lill, M. A., and Helms, V. (2001). Molecular dynamics simulation of proton transport with quantum mechanically derived proton hopping rates (Q-HOP MD). J. Chem. Phys. 115, 7993–8005. doi: 10.1063/1.1407293
Lin, Y., Wang, H., Gao, S., Li, R., and Schaefer III, H. F. (2012). Hydrogen-bonded double-proton transfer in five guanine–cytosine base pairs after hydrogen atom addition. J. Phys. Chem. B 116, 8908–8915. doi: 10.1021/jp3048746
Litman, Y., Richardson, J. O., Kumagai, T., and Rossi, M. (2019). Elucidating the nuclear quantum dynamics of intramolecular double hydrogen transfer in porphycene. J. Am. Chem. Soc. 14, 162526–162534. doi: 10.1021/jacs.8b12471
Liu, T., Wang, N., Cao, J., Sofiadis, A., Dinets, A., Zedenius, J., et al. (2014). The age- and shorter telomere-dependent TERT promoter mutation in follicular thyroid cell-derived. Oncogene 33, 4978–4984. doi: 10.1038/onc.2013.446
Lodish, H., Berk, A., Zipursky, S. L., and Matsudaira, P. (2000). Molecular Cell Biology. 4th Edn. New York, NY: Freeman.
Löwdin, P. O. (1963). Proton tunneling in DNA and its biological implications. Rev. Mod. Phys. 35, 724–732. doi: 10.1103/RevModPhys.35.724
Mandal, D., Ramanan, R., Usharani, D., Janardanan, D., Wang, B., and Shaik, S. (2015). How does tunneling contribute to counterintuitive H-abstraction reactivity of nonheme Fe (IV) O oxidants with alkanes? J. Am. Chem. Soc. 137, 722–733. doi: 10.1021/ja509465w
Marian, C. M. (2005). A new pathway for the rapid decay of electronically excited adenine. Chem. Phys. 122, 104314–104327. doi: 10.1063/1.1861452
Markova, N., Pejov, L., Stoyanova, N., and Enchev, V. (2017). Hybrid MC/QC simulations of water-assisted proton transfer in nucleosides. Guanosine and its analog acyclovir. J. Biomol. Struct. Dyn. 35, 1168–1188. doi: 10.1080/07391102.2016.1179594
Martin, G., Keller, W., and Doublie, S. (2000). Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 19, 4193–4203. doi: 10.1093/emboj/19.16.4193
Martínez Giménez, J. A., Sáez, G. T., and Seisdedos, R. T. (1998). On the function of modified nucleosides in the RNA world. J. Theor. Biol. 194, 485–490. doi: 10.1006/jtbi.1998.0770
Marx, D. (2006). Proton transfer 200 years after von grotthuss: insights from Ab initio simulations. ChemPhysChem 7, 1848–1870. doi: 10.1002/cphc.200600128
Maskos, K., Gunn, B. M., LeBlanc, D. A., and Morden, K. M. (1993). NMR study of G.cntdot.A and A.cntdot.A pairing in (dGCGAATAAGCG)2. Biochemistry 32, 3583–3595. doi: 10.1021/bi00065a009
Mazurkiewicz, K., Haranczyk, M., Gutowski, M., Rak, J., Radisic, D., Eustis, S.N., et al. (2007a). Valence anions in complexes of adenine and 9-methyladenine with formic acid: stabilization by intermolecular proton transfer. J. Am. Chem. Soc. 129, 1216–1224. doi: 10.1021/ja066229h
Mazurkiewicz, K., Haranczyk, M., Storoniak, P., Gutowski, M., Rak, J., Radisic, D., et al. (2007b). Intermolecular proton transfer induced by excess electron attachment to adenine (formic acid)n (n = 2, 3) hydrogen-bonded complexes. Chem. Phys. 342, 215–222. doi: 10.1016/j.chemphys.2007.10.005
McFadden, J., and Al-Khalili, J. (2018). The origins of quantum biology. Proc. R. Soc. A 474:20180674. doi: 10.1098/rspa.2018.0674
Meisner, J., and Kästner, J. (2016). Atom tunnelling in chemistry. Angew. Chem. Int. Ed. 55, 5400–5413. doi: 10.1002/anie.201511028
Merchán, M., and Serrano-Andrés, L. (2003). Ultrafast internal conversion of excited cytosine via the lowest ππ* electronic singlet state. J. Am. Chem. Soc. 125, 8108–8109. doi: 10.1021/ja0351600
Miller, J. H., Wilson, W. W., and Ritchie, R. H. (1994). Computational Approaches in Molecular Radiation Biology. eds M. N. Varma and J. Chatterjee (New York, NY: Plenum), 65–76.
Mizuse, K., Kuo, J.-L., and Fujii, A. (2011). Structural trends of ionized water networks: infrared spectroscopy of water cluster radical cations (H2O)n+ (n = 3-11). Chem. Sci. 2, 868–876. doi: 10.1039/C0SC00604A
Mohammed, O. F., Pines, D., Dreyer, J., Pines, E., and Nibbering, E. T. (2005). Sequential proton transfer through water bridges in acid-base reactions. Science 310, 83–86. doi: 10.1126/science.1117756
Moody, E. M., Lecomte, J. T., and Bevilacqua, P. C. (2005). Linkage between proton binding and folding in RNA: a thermodynamic framework and its experimental application for investigating pKa shifting. RNA 11, 157–172. doi: 10.1261/rna.7177505
Muth, G. W., Ortoleva-Donnelly, L., and Strobel, S. A. (2000). A single adenosine with a neutral pKa in the ribosomal peptidyltransferasecenter. Science 289, 947–950. doi: 10.1126/science.289.5481.947
Nakano, S., Chadalavada, D. M., and Bevilacqua, P. C. (2000). General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science 287, 1493–1497. doi: 10.1126/science.287.5457.1493
Nissen, P., Hansen, J., Ban, N., and Moore, P. B., and Steitz, T. A. (2000). The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930. doi: 10.1126/science.289.5481.920
Niwa, O. (2006). Radiation induced dynamic mutations and transgenerational effects. J. Radiat. Res. 47, B25–B30. doi: 10.1269/jrr.47.B25
Nowak, M. J., Szczepaniak, A., and Shugar, B. D. (1980). Tautomeric equilibria of 2(4)-monooxopyrimidines in the gas phase, in low-temperature matrices and in solution. J. Mol. Struct. 62, 47–69. doi: 10.1016/0022-2860(80)85222-7
Ogawa, A. K., Abou-Zied, O. K., Tsui, V., Jimenez, R., Case, D. A., and Romesberg, F. E. (2000). A phototautomerizable model DNA base pair. J. Am. Chem. Soc. 122, 9917–9920. doi: 10.1021/ja001778n
Onoa, B., Dumont, S., Liphardt, J., Smith, S. B., Tinoco, I., and Bustamante, C. (2003). Identifying kinetic barriers to mechanical unfolding of the T thermophile ribozyme. Science 299, 1892–1895. doi: 10.1126/science.1081338
Padermshoke, A., Katsumoto, Y., Masaki, R., and Aida, M. (2008). Thermally induced double proton transfer in GG and wobble GT base pairs: a possible origin of the mutagenic guanine. Chem. Phys. Lett. 457, 232–236. doi: 10.1016/j.cplett.2008.04.029
Pan, B., Mitra, S. N., and Sundaralingam, M. (1999). Crystal structure of an RNA 16-mer duplex R(GCAGAGUUAAAUCUGC)2 with nonadjacent G(syn).A+(anti) mispairs. Biochemistry 38, 2826–2831. doi: 10.1021/bi982122y
Park, K., Kim, Y., Kim, K. H., and Kim, Y. (2011). Quantum mechanical studies for structures and energetic of double proton transfer in biologically important hydrogen-bonded complexes. Bull. Korean Chem. Soc. 32, 3634–3641. doi: 10.5012/bkcs.2011.32.10.3634
Parker, B. R., and Van Everv, J. (1971). Quantum tunnelling in{DNA}. Chem. Phys. Lett. 8, 94–99. doi: 10.1016/0009-2614(71)80586-9
Pérez, A., Tuckerman, M. E., Hjalmarson, H. P., and von Lilienfeld, O. A. (2010). enol tautomers of watson–crick base pair models are metastable because of nuclear quantum effects. J. Am. Chem. Soc. 132, 11510–11515. doi: 10.1021/ja102004b
Person, W. B., Szczepaniak, K., Szczesniak, M., Kwiatkowski, J. S., and Czerminski, L. R. (1989). Tautomerism of nucleic acid bases and the effect of molecular interactions on tautomeric equilibria. J. Mol. Struct. 194, 239–258. doi: 10.1016/0022-2860(89)80084-5
Perun, S., Sobolewski, A. L., and Domcke, W. (2005). Ab initio studies on the radiationless decay mechanisms of the lowest excited singlet states of 9H-adenine. J. Am. Chem. Soc. 127, 6257–6265. doi: 10.1021/ja044321c
Piatkowski, L., Schanbacher, C., Wackenhut, F., Jamrozik, A., Meixner, A. J., and Waluk, J. (2018). Nature of large temporal fluctuations of hydrogen transfer rates in single molecules. J. Phys. Chem. Lett. 9, 1211–1215. doi: 10.1021/acs.jpclett.8b00299
Platonov, M. O., Samijlenko, S. P., Sudakov, O. O., Kondratyuk, I. V., and Hovorun, D. M. (2005). To what extent can methyl derivatives be regarded as stabilized tautomers of xanthine? Spectrochim. Acta A 62, 112–114. doi: 10.1016/j.saa.2004.12.012
Portalone, G. (2011). Solid-phase molecular recognition of cytosine based on proton-transfer reaction. Part, I. supramolecular architecture in the cocrystals of cytosine and its 5-Fluoroderivative with 5-Nitrouracil. Portalone Chem. Cent. J. 5:51. doi: 10.1186/1752-153X-5-51
Pusuluk, O., Torun, G., and Deliduman, C. (2018). Quantum entanglement shared in hydrogen bonds and its usage as a resource in molecular recognition. Modern Phys. Lett. B 32:1850308. doi: 10.1142/S0217984918503086
Pyle, A. M. (2008). Translocation and unwinding mechanisms of RNA and DNA helicases. Ann. Rev. Biophys. 37, 317–336. doi: 10.1146/annurev.biophys.37.032807.125908
Quigley, G. J., et al. (1986). Non-Watson-Crick G. C and A. T base pairs in a DNA-antibiotic complex. Science 232, 1255–1258. doi: 10.1126/science.3704650
Radisic, D., Bowen, K. H., Dabkowska, I., Storoniak, P., Rak, J., and Gutowski, M. (2005). AT base pair anions versus (9-methyl-A)(1-methyl-T) base pair anions. J. Am. Chem. Soc. 127, 6443–6450. doi: 10.1021/ja050724g
Relph, R. A., Guasco, T. L., Elliott, B. M., Kamrath, M. Z., McCoy, A. B., Steele, R. P., et al. (2010). How the shape of an h-bonded network controls proton-coupled water activation in HONO formation. Science 327, 308–312. doi: 10.1126/science.1177118
Ribeiro Da Silva, M. A. V., Amaral, L. M. P. F., and Szterner, P. (2013). Szterner Piotr: Experimental study on the thermochemistry of 2-thiouracil, 5-methyl-2-thiouracil and 6-methyl-2-thiouracil. J. Chem. Thermodyn. 57, 380–386. doi: 10.1016/j.jct.2012.08.004
Rich, A., Davies, D. R., Crick, F. H., and Watson, J. D. (1961). The molecular structure of polyadenylic acid. J. Mol. Biol. 3, 71–86. doi: 10.1016/S0022-2836(61)80009-0
Roßbach, S., and Ochsenfeld, C. (2017). Influence of coupling and embedding schemes on QM size convergence in QM/MM approaches for the example of a proton transfer in DNA. J. Chem. Theory Comput. 13, 1102–1107. doi: 10.1021/acs.jctc.6b00727
Rozenski, J., Crain, P. F., and McCloskey, J. A. (1999). The RNA modification database: 1999 update. Nucleic Acids Res. 27, 196–197. doi: 10.1093/nar/27.1.196
Rozsnyai, B. F., and Ladik, J. (1970). Calculation of the effects of divalent metal ions on nucleotide base pairs. II. Single G–C and A–T and Poly(G–C) and (A–T). J. Chem. Phys. 53, 4325–4333. doi: 10.1063/1.1673941
Ruiz-Blanco, Y. B., Almeida, Y., Sotomayor-Torres, C. M., and García, Y. (2017). Unveiled electric profiles within hydrogen bonds suggest DNA base pairs with similar bond strengths. PLoS ONE 12:e0185638. doi: 10.1371/journal.pone.0185638
Samijlenko, S. P., Krechkivska, O. M., Kosach, D. A., and Hovorun, D. M. (2004). Transitions to high tautomeric states can be induced in adenine by interactions with carboxylate and sodium ions: DFT calculation data. J. Mol. Struct. 708, 97–104. doi: 10.1016/j.molstruc.2004.05.034
Schrödinger, E. (1945). What is Life? The Physical Aspect of the Living Cell. Cambridge: Cambridge University Press.
Schuster, G. B. (ed.). (2004). “Long-Range Charge Transfer in DNA I-II,” in Topics in Current Chemstry. Vol. 236–237. Heidelberg: Springer. doi: 10.1007/b14032
Seeman, N. C. (2001). DNA nicks and nodes and nanotechnology. Nano Lett. 1, 22–26. doi: 10.1021/nl000182v
Sekiya, H., and Sakota, K. (2008). Excited-state double-proton transfer in a model DNA base pair: resolution for stepwise and concerted mechanism controversy in the 7-azaindole dimer revealed by frequency- and time-resolved spectroscopy. J. Photochem. Photobiol. C Photochem. Rev. 9, 81–91. doi: 10.1016/j.jphotochemrev.2008.04.001
Sevilla, M. D., Besler, B., and Colson, A. O. (1995). Ab initio molecular orbital calculations of dna radical ions. 5. Scaling of calculated electron affinities and ionization potentials to experimental values. J. Phys. Chem. 99, 1060–1995. doi: 10.1021/j100003a032
Silverman, R. B. (2000). The Organic Chemistry of Enzyme-Catalyzed Reactions. San Diego, CA: Academic Press.
Sloan, K. E., Warda, A. S., Sharma, S., Entian, K. D., Lafontaine, D. L. J., and Bohnsack, M. T. (2017). Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152. doi: 10.1080/15476286.2016.1259781
Smedarchina, Z., Siebrand, W., and Fernández-Ramos, A. (2018). Entanglement and co-tunneling of two equivalent protons in hydrogen bond pairs. J. Chem. Phys. 148, 102307-1–102307-15. doi: 10.1063/1.5000681
Smedarchina, Z., Siebrand, W., Fernández-Ramos, A., and Meana-Pañeda, R. (2008). Mechanisms of double proton transfer theory and applications. Z. Phys. Chem. 222, 1291–1309. doi: 10.1524/zpch.2008.5389
Sochacka, E., Szczepanowski, R. H., Cypryk, M., Sobczak, M., Janicka, M., Kraszewska, K., et al. (2015). 2-Thiouracil deprived of thiocarbonyl function preferentially base pairs with guanine rather than adenine in RNA and DNA duplexes. Nucleic Acids Res. 43, 2499–25121. doi: 10.1093/nar/gkv109
Sponer, J., and Hobza, P. (1994). Nonplanar geometries of DNA bases. Ab initio second-order Moeller-Plesset study. J. Phys. Chem. 98, 3161–3164. doi: 10.1021/j100063a019
Sponer, J., Leszczynski, J., and Hobza, P. (1996). Hydrogen bonding and stacking of DNA bases: a review of quantum-chemical ab initio studies. Biol. Struct. Dynam. 14, 117–135. doi: 10.1080/07391102.1996.10508935
Sprinzl, M. (2006). Chemistry of aminoacylation and peptide bond formation on the 3'terminus of Trna. J. Biosci. 31, 489–496. doi: 10.1007/BF02705188
Srivastava, R. (2017). Theoretical studies on the electronic and optoelectronic properties of [A.2AP(w)/A*0.2AP(WC)/C.2AP(w)/C*0.2AP(WC)/C.A(w)/C*.A(WC)]–Au8 mismatch nucleobase complexes. Mol. Phys. 116, 263–272. doi: 10.1080/00268976.2017.1382737
Srivastava, R. (2018). Complexes of DNA bases and Watson–Crick base pairs interaction with neutral silver Agn (n = 8, 10, 12) clusters: a DFT and TDDFT study. J. Biomol. Struct. Dyn. 36, 1050–1062. doi: 10.1080/07391102.2017.1310059
Steenken, S. (1989). Purine bases, nucleosides, and nucleotides: aqueous solution redox chemistry and transformation reactions of their radical cations and e– and OH adducts. Chem. Rev. 89, 503–520. doi: 10.1021/cr00093a003
Stewart, E. L., Foley, C. K., Allinger, N. L., and Bowen, J. P. (1994). Ab-initio calculations with electronic correlation (MP2) on the nucleic-acid bases and their methyl derivatives. J. Am. Chem. Soc. 116, 7282–7286. doi: 10.1021/ja00095a035
Sukhodub, L. F. (1987). Interactions and hydration of nucleic acid bases in a vacuum. Experimental study. Chem. Rev. 87, 589–606. doi: 10.1021/cr00079a006
Taylor, J. S. (2002). New structural and mechanistic insight into the A-rule and the instructional and non-instructional behavior of DNA photoproducts and other lesions. Mutat. Res. 510, 55–70. doi: 10.1016/S0027-5107(02)00252-X
Tomasetti, C. R., Li, L., and Vogelstein, B. (2017). Stem cell divisions, somatic mutations, cancer etiology, and cancer preventions. Science 355, 1330–1334. doi: 10.1126/science.aaf9011
Tomić, K., Tatchen, J., and Marian, C. M. (2005). quantum chemical investigation of the electronic spectra of the keto, enol, and keto–imine tautomers of cytosine. J. Phys. Chem. A 109, 8410–8418. doi: 10.1021/jp051510o
Turaeva, N., and Brown-Kennerly, V. (2015). Marcus model of spontaneous point mutation in DNA. Chem. Phys. 461, 106–110. doi: 10.1016/j.chemphys.2015.09.005
Villani, G. (2010). Theoretical investigation of hydrogen atom transfer in the cytosine-guanine base pair and its coupling with electronic rearrangement. Concerted vs Stepwise Mechanism. J. Phys. Chem. B 114, 9653–9662. doi: 10.1021/jp102457s
Voth, G. A. (2006). Computer simulation of proton solvation and transport in aqueous and biomolecular systems. Acc. Chem. Res. 39, 143–150. doi: 10.1021/ar0402098
Wang, C., Gao, H., Gaffney, B. L., and Jones, R. A. (1997). Nitrogen-15-labeled oligodeoxynucleotides. 3. Protonation of the adenine N1 in the A.cntdot.C and A.cntdot.Gmispairs of the duplexes {d[CG(15N1)AGAATTCCCG]}2 and {d[CGGGAATTC(15N1)ACG]}2. J. Am. Chem. Soc. 113, 5486–5488. doi: 10.1021/ja00014a068
Watanabe, M. (2015). The first target of radiation carcinogenesis is not DNA. Int. Congress Series. 42, 2409–2413. doi: 10.1016/j.ics.2006.10.009
Watson, J. D., and Crick, F. H. C. (1953). Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171, 737–738. doi: 10.1038/171737a0
Wen, D. X., Zhu, F., Zhao, L., Huang, B., and Zeng, J. (2006). Influence of cationic gemini surfactants on the electrochemical behavior of 2-thiouracil at silver electrodes. Solid State Electrochem. 10, 69–77. doi: 10.1007/s10008-005-0657-7
Whiteside, J. R., Allinson, S. L., Trevor, T. J., et al. (2011). Timeframes of UVA-induced bystander effects in human keratinocytes. Photochem. Photobiol. 87, 435–440. doi: 10.1111/j.1751-1097.2010.00881.x
Wilson, T. J., Nahas, M., Ha, T., and Lilley, D. M. (2005). Folding and catalysis of the hairpin ribozyme. Biochem. Soc. Trans. 33, 461–465. doi: 10.1042/BST0330461
Witwicki, M., Jezierska, J., and Ozarowski, A. (2009). Solvent effect on EPR, molecular and electronic properties of semiquinone radical derived from 3,4-dihydroxybenzoic acid as model for humic acid transient radicals: high-field EPR and DFT studies. Chem. Phys. Lett. 473, 160–166. doi: 10.1016/j.cplett.2009.03.035
Wright, E. G. (2010). Manifestations and mechanisms of non-targeted effects of ionizing radiation. Mutat. Res. 687, 28–33. doi: 10.1016/j.mrfmmm.2010.01.014
Xiao, S., Wang, L., Liu, Y., Lin, X., and Liang, H. (2012). Theoretical investigation of the proton transfer mechanism in guanine-cytosine and adenine-thymine base pairs. J. Chem. Phys. 137:195101. doi: 10.1063/1.4766319
Yekeler, H. (2000). Ab initio study on tautomerism of 2-thiouracil in the gas phase and in solution. J. Comp. Aided Mol. Design 14, 243–250. doi: 10.1023/A:1008132202838
Zengtao, L., Cui, S., Guo, F., and Zhang, G. (2019). Two possible mechanisms of water-assisted proton transfer in the stack of adenine-thymine: displacement polarization and oriented polarization. AIP Adv. 9:015015. doi: 10.1063/1.5064505
Keywords: proton transfer, mutations, tautomerism, quantum tunneling, oriental polarization
Citation: Srivastava R (2019) The Role of Proton Transfer on Mutations. Front. Chem. 7:536. doi: 10.3389/fchem.2019.00536
Received: 05 April 2019; Accepted: 15 July 2019;
Published: 21 August 2019.
Edited by:
Dmytro Hovorun, National Academy of Sciences of Ukraine, UkraineReviewed by:
Xugeng Guo, Henan University, ChinaCopyright © 2019 Srivastava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruby Srivastava, YW1pdHJ1YnkxQGdtYWlsLmNvbQ==; cnVieUBjY21iLnJlcy5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.