- 1Graduate School of Environmental Studies, Tohoku University, Sendai, Japan
- 2Graduate School of Engineering, Tohoku University, Sendai, Japan
- 3Frontier Research Institute for Interdisciplinary Sciences, Tohoku University, Sendai, Japan
Microfluidic devices are widely used for cell analysis, including applications for single-cell analysis, healthcare, environmental monitoring, and organs-on-a-chip that mimic organs in microfluidics. Moreover, to enable high-throughput cell analysis, real-time monitoring, and non-invasive cell assays, electric and electrochemical systems have been incorporated into microfluidic devices. In this mini-review, we summarize recent advances in these systems, with applications from single cells to three-dimensional cultured cells and organs-on-a-chip. First, we summarize microfluidic devices combined with dielectrophoresis, electrophoresis, and electrowetting-on-a-dielectric for cell manipulation. Next, we review electric and electrochemical assays of cells to determine chemical section activity, and oxygen and glucose consumption activity, among other applications. In addition, we discuss recent devices designed for the electric and electrochemical collection of cell components from cells. Finally, we highlight the future directions of research in this field and their application prospects.
Introduction
Cell analysis is essential for healthcare and environmental monitoring, and has recently benefited from the organs-on-a-chip technique that can effectively mimic organs and their microfluidics (Bhatia and Ingber, 2014; Rogal et al., 2017). Recently, a wide diversity of microfluidic devices have been adapted for cell analysis, which allows for many samples to be assessed simultaneously and for the construction of functional tissue models. Microfluidic techniques can compartmentalize, monitor, sort, collect, lyse, and culture individual cells and cell aggregates; in addition, cell-derived signals can be amplified. In cell culture, microfluidic platforms can enable cell stimulation and mimic cellular environments. Therefore, the techniques are considered superior to conventional methods such as microscopic imaging, flow cytometry, and cell culture. Although optical approaches are commonly used for evaluating cells, novel electric and electrochemical approaches are also increasingly reported. This mini-review summarizes recent advances in electric and electrochemical microfluidic devices developed for cell analysis with a range of applications, from single cells to three-dimensional (3D) cultured cells and organs-on-a-chip (Figure 1). In the first section, we summarize the microfluidic devices currently available for cell manipulation. In the second section, we discuss the common electric and electrochemical-based cell assays, followed by a description of approaches for collecting cell components. Finally, we provide a summary and discussion of future directions of research and prospects in this field.
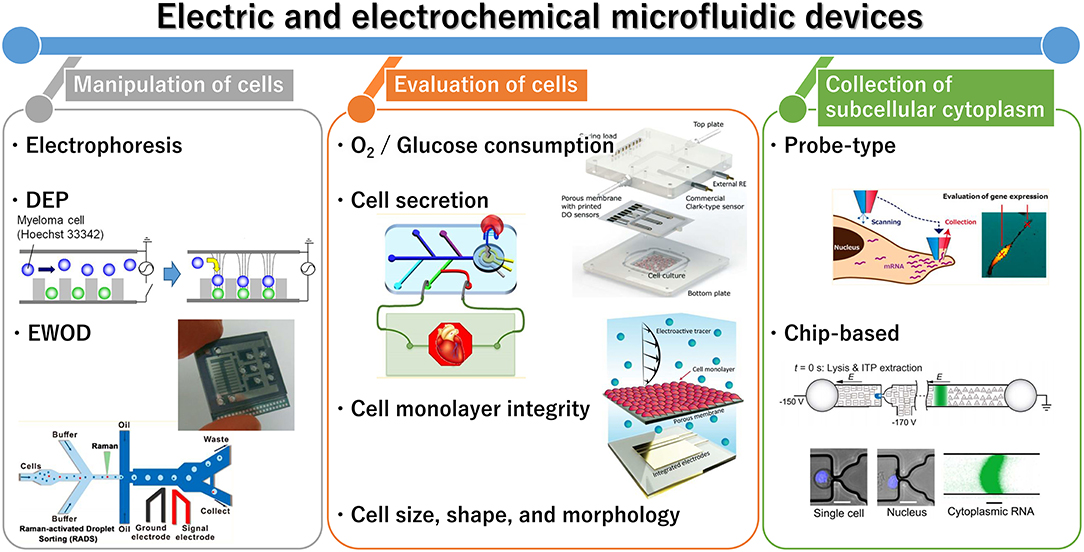
Figure 1. General outline of electric and electrochemical microfluidic devices for cell analysis. Reprinted with permission from Yoshimura et al. (2014); Nashimoto et al. (2016); Shin et al. (2016); Wang et al. (2017). Copyright (2014, 2016, and 2017) American Chemical Society. Reprinted with permission from Rival et al. (2014); Moya et al. (2018); Wong and Simmons (2019). Reproduced from Abdelmoez et al. (2018).
Manipulation of Cells and Droplets Containing Cells
In some cases, the cells are introduced from an inlet to be trapped and assayed sequentially in a microfluidic chip. In addition, droplets containing cells can be manipulated for cell lysis and polymerase chain reaction (PCR)-based assays. Electric and electrochemical approaches are widely used for all of these applications, including electrophoresis, dielectrophoresis (DEP), and electrowetting-on-dielectric (EWOD).
Electrophoresis
Driven by the negative charges of cell membranes, cells can be moved from a microfluidic channel to an electrode applied at a positive potential. For example, yeast cells were trapped using electrophoresis, and a reporter protein was electrochemically evaluated using the electrode (Yasukawa et al., 2008; Ino et al., 2009). After the evaluation, the cells are pushed to the microfluidic channel. This allowed for sequential assays of the cells using only a single electrochemical sensor.
In particular, Capillary electrophoresis (CE) devices are widely used for cell manipulation (Mellors et al., 2010). In this case, the cells migrate via electro-osmotic flow toward the lysis intersection where the electric field is increased. Ultimately, these cells can be evaluated with several methods, including mass spectrometry (Mellors et al., 2010).
Thus, the manipulation of molecules and particles is based on their charges. The following section describes DEP, which is used for the manipulation of non-charged particles.
DEP
DEP is a specific form of electrophoresis that is used for manipulating biosamples that contain non-charged particles (Pohl, 1951). In general, DEP involves the application of AC voltage, and the cells are pushed toward high or low intensity of electric fields by inducing positive and negative DEP, respectively, depending on the AC frequency. DEP manipulation can be used for assays of single cells (Thomas et al., 2009) to 3D cultured cells (Kanno et al., 2015c). Recently, the DEP technique was combined with a bipolar electrode system, and the cells were manipulated in a microfluidic device via electrode arrays in the absence of ohmic contacts (Anand et al., 2015). In addition, a DEP device has been used for cell paring by trapping single cells sequentially (Sen et al., 2013b; Yoshimura et al., 2014; Wu et al., 2017), and a microfluidic DEP device was applied for separation of mesenchymal stem cells (MSCs) from a cell mixture according to their dielectric properties (Yoshioka et al., 2018) and controlled cell differentiation (Yoshioka et al., 2016). Moreover, enrichment and detection of target cancer cells were performed by combining DEP manipulation and impedance measurements (Ngoc-Viet and Jen, 2018).
DEP devices have also been proposed for high-throughput droplet sorting, and single cells compartmentalized in droplets are monitored using a fluorescence method (Baret et al., 2009) and Raman spectroscopy (Wang et al., 2017).
DEP is widely used for manipulation of cells and droplets in a solution. For manipulating droplets in air, other techniques, such as EWOD, are utilized.
EWOD
The EWOD technique can modulate the surface tension between a liquid droplet and solid substrate through application of voltage to an electrode. In this technique, individual droplets are moved along an array of electrodes. As an application of the technique for cell analysis, droplets containing cells and chemicals were manipulated in a microfluidic device for the isolation of a single cell for subsequent RNA purification and gene expression analysis (Rival et al., 2014). More recently, Sinha et al. developed a droplet-based digital microfluidic platform for gene editing (Sinha et al., 2018). The system enabled all of the necessary steps for gene editing, including cell culture, delivery, and analysis. Specifically, lung cells were cultured on the device for up to 6 days, and gene transfection and knockout procedures were performed.
In this section, we focuses on microfluidic electric and electrochemical devises for manipulating cells and droplets. These devices evaluate cells by collecting cellular components, stimulating cells, and so on. These methods are summarized in the following section.
Evaluation of Cells
Cells in microfluidic devices are mainly measured or observed using optical techniques. For example, cells are immuno-stained in the devices, or the cells and resulting culture medium are collected from the devices, and their components are evaluated with immunostaining, enzyme-linked immunosorbent assay (ELISA), and PCR. However, these traditional techniques are time-consuming, require manual sample collection from the microfluidic system, and necessitate large working volumes. Therefore, there is strong demand for the integration of online sensor capabilities.
Recently, electric and electrochemical systems have been used for evaluating cells in microfluidic devices, and some of these key applications are highlighted below.
Oxygen and Glucose Consumption
Since oxygen is essential for the energy metabolism of cells, it is important to determine the respiratory activity of cells. In general, platinum (Pt) electrodes are used for measuring the amount of dissolved oxygen near the cells, because O2 is reduced to H2O to obtain oxygen reduction currents at the electrodes. Some electrochemical sensors have been developed with gas-permeable membranes, known as Clark-type sensors. Moya et al. reported a liver chip integrated with multiple sensors and a porous, delicate membrane (Moya et al., 2018). The sensor electrodes were inkjet-printed along the microfluidic channel allowing for local online monitoring of oxygen concentrations. In addition to liver cells, single bovine embryos were evaluated using a microfluidic device (Wu et al., 2007). In this study, single embryos were manipulated to the sensing area with a pressure driving force, and the same force was used to collect the embryos from the microfluidic device after measurements.
Along with oxygen, glucose is a major cell energy source; thus, glucose consumption is a key parameter of cell activity. In general, enzymes such as glucose oxidase and glucose dehydrogenase can be modified with electrodes, and the resulting products of the reaction with glucose, such as H2O2 or redox mediators, are measured. Misun et al. reported a droplet array device with enzyme-modified electrodes for body-on-a-chip applications (Misun et al., 2016). This microfluidic platform is based on hanging-drop networks for the cultivation and analysis of 3D cultured cells.
In this section, we summarize the detection of consumption of target analytes. In the next section, we discuss the detection of cell secretion products.
Cell Secretion
Many types of cell secretion products are monitored using electrochemical techniques. For example, a microfluidic chip-based perfusion system was designed to maintain viable and functioning heart tissue samples and to measure reactive oxygen species (ROS) using electrochemical methods (Cheah et al., 2010). Since ROS play a major role in cell interactions, and are related to the development and progression of many diseases, measurements of ROS levels have important clinical applications. H2O2 is a relatively stable metabolite among ROS. Therefore, it is widely utilized as a key marker of immune and inflammation processes in living cells and tissues. Inoue et al. developed an electrochemical-sensing device for the continuous monitoring of H2O2 secreted from human promyelocytic leukemia cells (Inoue et al., 2010). The electrode chip was coated with an osmium polyvinylpyridine gel polymer conjugated to horseradish peroxidase (Os-HRP), which allowed for the selective detection of extracellular H2O2 of the leukocytes. In a more recent study, Prasad et al. integrated a bare Pt electrode and an Os-HRP-modified silver electrode to monitor both the oxygen consumption and H2O2 generation of cells in a single chip, which enabled evaluation of the respiratory burst of immune cells (Prasad et al., 2016).
Lactate ions are the end product of the anaerobic pathway. Therefore, it is important to measure their accumulation in the culture medium to monitor cell metabolic activity. Bavli et al. reported a liver-on-chip device that could maintain human tissue properties for over 1 month in vitro under physiological conditions, and lactate was measured with enzyme-modified electrodes (Bavli et al., 2016).
Shin et al. reported a microfluidic aptamer-based electrochemical sensor for monitoring damage to cardiac organoids (Shin et al., 2016, 2017). They integrated a microfluidic bioreactor and an electrochemical biosensor in a single platform, which enabled the in situ detection of creatine kinase (CK)-MB as a biomarker secreted from a damaged cardiac spheroid. Electrochemical impedance spectroscopy (EIS) was adopted to the sensor system comprising a microelectrode functionalized with CK-MB-specific aptamers.
Exosomes are small (50–150 nm in diameter) vesicles secreted from various cells, and are recognized as important mediators of intracellular communication or transporters of pathogenic proteins. Moreover, exosomes have recently attracted attention as candidate biomarkers of various diseases such as cancers and metabolic disorders. Exosomes have been monitored using aptamer-based electrochemical sensors (Zhou et al., 2016). Since redox mediator-labeled probes are removed from the capture DNAs when capturing exosomes, the redox currents are decreased. In this study, exosomes were introduced from the inlets of the devices. In the future, exosomes from cells on chips will also be evaluated.
Microcapillary electrophoresis (microCE) is another approach used to analyze exosomes and extracellular vesicles. Akagi et al. developed a microCE chip and applied it to an on-chip immunoelectrophoresis assay for extracellular vesicles (EVs) of human breast cancer cells (Akagi et al., 2015). Since EVs from living bodies are heterogeneous in size, individual EVs could not be characterized by conventional methods. The microCE chip characterizes EVs according to variations in their zeta potential, which is expected to become a robust system for the sensitive profiling of EVs.
Thus, for detection of some of targets, it is important to modify electrodes. Enzymes, such as glucose oxidase, HRP, and lactate dehydrogenase are widely used to transfer electrons from target analyte to redox mediators or electrodes. In addition, several types of aptamers and antigens are modified at electrodes to capture target analytes, and the capture is electrochemically detected. These modifications are summarized in Table 1.
In the first half of this section, we summarize the detection of consumption and secretion of analytes. In contrast, it is important to measure cell shape and morphology for cell evaluation. In the second half, we discuss their detection.
Cell Monolayer Integrity
Trans-epithelial electrical resistance (TEER) is a widely used parameter for evaluating in vitro barrier tissue integrity (Elbrecht et al., 2016). TEER measurements are performed by applying an AC voltage at electrodes set on both sides of a cell monolayer, and the voltage and current are measured to calculate the electrical resistance of the barrier. Takayama's group evaluated epithelial and endothelial barriers in a microfluidic chip using TEER measurements (Douville et al., 2010). In addition, a blood–brain barrier (BBB) model was evaluated with this approach (Wang et al., 2016). Ingber's group also described a microfluidic device containing electrodes for assessing lung chips (Henry et al., 2017). In addition to enabling the real-time, non-invasive monitoring of barrier functions, multi-electrode arrays (MEAs) were combined with TEER measurements for heart-on-a-chip (Maoz et al., 2017).
Similar to TEER measurements, an electrochemical permeability assay was reported for evaluating cell monolayer permeability (Wong and Simmons, 2019). In this case, the ubiquitous fluorescent tracer was replaced with an electroactive tracer, and the barrier function of endothelial cells was assessed by monitoring the diffusion of the electroactive tracer across a cell monolayer.
Cell Size, Shape, and Morphology
Impedance detection has also been applied for evaluating the allergic response in a microfluidic device. RBL-2H3 mast cells and ANA-1 macrophages were co-cultured and their allergic response to a stimulus was observed (Jiang et al., 2016). Moreover, Schmid et al. combined EIS with a microfluidic hanging-drop platform for monitoring spheroid sizes and contractions of human cardiac spheroids (Schmid et al., 2016).
Ion currents via nano- or micropores are measured for the electrical discrimination of various biomolecules, cells, bacteria, and viruses. Yasaki et al. reported a rational methodology that can detect samples within a particle volume of 0.01% of the pore volume by measuring the transient current generated in a microfluidic bridge circuit (Yasaki et al., 2017). The device was subsequently applied for the size detection of bacterial cells (Yasaki et al., 2018).
Thus, we discuss cell evaluation techniques in this section. In contrast, it is important to obtain intracellular information. In the following section, we summarize the techniques used for collection of subcellular cytoplasm.
Collection of Subcellular Cytoplasm
We previously reviewed the progress in intracellular electrochemical sensing techniques (Ino et al., 2018b). Here, we focus on two main types of electric and electrochemical microfluidic devices for lysing cells and collecting components of cells by applying pulse voltage.
Probe-Type Microfluidic Devices
A probe-type microfluidic device with a Pt-ring electrode at its tip was used to collect the cytosol of a single adherent cell on an ITO electrode, and cellular mRNA was analyzed at the single-cell level (Nashimoto et al., 2007). The bias was applied between the Pt and ITO electrodes. To collect the mRNA of single cells in 3D cultured cells, a double-barrel carbon probe was developed (Ito et al., 2016). The device was used to measure local gene expression levels during sprouting angiogenesis in embryonic stem cell-derived embryoid bodies.
Laforge et al. reported an electrochemical syringe containing organic and liquid solutions for manipulating aL–fL-scale volumes (Laforge et al., 2007). The interface between the organic and liquid solution in a pipet was moved by applying a potential. Previously, we combined a syringe with scanning ion conductance microscopy (SICM) for measuring cellular morphology and collecting the cytosol from living cells (Nashimoto et al., 2016).
Chip-Based Devices
Shintaku's group reported a microfluidic device containing a hydrodynamic trap for electrical lysis (Subramanian Parimalam et al., 2018). All on-chip processes were completed in <5 min. In addition, a microfluidic system was designed to physically separate nuclear and cytoplasmic RNAs from a single cell, allowing for separate analyses of these RNAs (Abdelmoez et al., 2018).
Since electrical techniques can provide precise control of applied potentials, they enable the collection of subcellular cytoplasm at a single cell level. By combining these techniques with microfluidic devices, the throughput of the collection will be improved.
Conclusion and Prospects
In this mini-review, we have presented an overview of recent research progress in electric and electrochemical microfluidic devices for cell analysis. These applications and devices are summarized in Table 1.
In typical microfluidic devices, only a few electrodes or electrochemical sensors are incorporated. However, the future goal of microfluidic device design is to provide multiplex assays and electrochemical imaging of cell activity. Electrochemical imaging enables chemical mapping using electrode arrays. For these purposes, it is important to incorporate several individual and addressable electrodes. Although numerous reports have described such devices in which the electrodes are simply arranged, with useful applications in cell analysis, including neurons (Kasai et al., 2005), the areas of the leading electrodes and connector pads are large, which limits the density and number of electrodes that can be incorporated. Recently, several kinds of integrated electrode arrays have been developed to overcome this limitation (Ino et al., 2012, 2014; Kätelhön et al., 2014; Kanno et al., 2015c; Zhang et al., 2017). These systems can provide electrochemical images consisting of 2D current values. Compared to assays using a single electrode, spatial information can be obtained and/or multiplex sensing can be performed with an integrated system.
Moreover, in the future, electrode array devices will be combined with microfluidic systems. In particular, large-scale integration (LSI) technology will be used for incorporating many addressable electrodes into microfluidic devices. Indeed, LSI devices have already been used for electrochemical imaging (Inoue et al., 2012, 2015; Sen et al., 2013a, 2014; Abe et al., 2015, 2016; Kanno et al., 2015a,b, 2016, 2017; Ino et al., 2017, 2018b,c,d, 2019a,b). Since signal amplifiers and switching elements are incorporated into the sensing area, highly sensitive sensors can be incorporated with high density. In addition, measurement modes such as amperometry and potentiometry can be selected separately for each sensor, allowing for multiplex analytes to be simultaneously measured. Further, electrochemical imaging in microfluidic devices will be combined with optical imaging analysis using mathematical approaches (Kato et al., 2016; Nagasaka et al., 2017) to improve their analytical features.
Since electrodes are inevitably fouled during the measurement, long-term assays remain a challenge of this technology. For example, measurement of oxygen consumption using non-modified electrodes can only be continuously performed for 2 days. For long-term monitoring, the electrodes should be modified with certain materials, or the electrodes will need to be changed every 2 days. For this purpose, the attachment and connectors should be improved. In addition, enzymes might be unsuitable for long-term measurement because their activity gradually decreases. To solve these problems, non-enzyme electrochemical sensors using metal nanoparticles and porous surfaces have been reported (Niu et al., 2013). The sensors are expected to be widely used for cell analysis in microfluidic devices.
For multiplex assays, individual sensors should be modified with different materials such as glucose oxidase and HRP. For this purpose, electrodeposition is a useful technique for addressable and individual electrode arrays. Since electrodeposition is also applied for hydrogel formation (Ozawa et al., 2013a,b, 2016, 2017; Ino et al., 2018a,d; Taira et al., 2018, 2019; Li et al., 2019), biomaterials including cells are easily patterned only on the target electrodes.
In general device fabrication, photolithography, and metal-sputtering techniques are typically used. In contrast, it is expected that printing techniques will become more widely used in future applications, including inkjet printing (Bachmann et al., 2017) and printed organic transistor-based enzyme sensors (Mano et al., 2018). Lind et al. reported cardiac physiological devices designed via multilateral 3D printing (Lind et al., 2017). Curto et al. reported an organic transistor platform with integrated microfluidics for in-line multi-parametric in vitro cell monitoring (Curto et al., 2017). Moreover, printed carbon electrodes have been reported (Shitanda et al., 2018) for reducing the sensor cost. Although the majority of probe and chip devices are currently made from glass, silicon, and plastic materials, paper devices (Yang et al., 2017; Akyazi et al., 2018; Jin et al., 2018) are also widely used for low-cost assays. In the future, paper-based devices are expected to be more commonly used for on-chip cell analysis.
Nanodevices are also increasingly used for cell analysis in microfluidic chips. For example, nanopores were used for measuring oligonucleotides and microvesicles via detection of ion currents when passing through the pores (Anderson et al., 2015; Kawano, 2018). For highly sensitive and selective assays, a redox cycling system was used in nanofluidic and nanocavity devices including two sets of electrodes in close proximity to one another (Lemay et al., 2013; Kätelhön et al., 2014; Kanno et al., 2015c). A redox species generated at one electrode diffuses to the other electrode, which generates an electrochemical reaction to regenerates the original species, resulting in signal amplification. In addition to these nanosystems, a pyrolyzed carbon mesh was integrated into a microchannel for a good redox cycling efficiency (Lee et al., 2018). These systems are expected to be incorporated into microfluidic devices for improved cell analysis.
In this review, we described the electric and electrochemical devices for heart-on-a-chip, liver-on-a-chip, carcinoma microtissue models, and so on. Since lung-on-chip (Huh et al., 2010) and gut-on-a-chip (Kim et al., 2012) devices have attracted considerable attention, electric and electrochemical systems will be incorporated into these organ-on-a-chip devices.
In this review, we discuss the cell manipulation for cell analysis in microfluidic devices. In future, these techniques will also be used for fabrication of functional tissue models. Some studies have reported the fabrication of tissue models for manipulating cells and scaffold materials using DEP (Ramón-Azcón et al., 2012, 2013); these approaches will be combined with microfluidic techniques.
We believe, the presented electric and electrochemical techniques have high expectations to broaden the possibilities of microfluidic devices and cell analysis.
Author Contributions
All authors wrote the manuscript. KoI and HS supervised preparation of the paper.
Funding
This work was supported by a Grant-in-Aid for Scientific Research (A) (No. 16H02280), a Grant-in-Aid for Scientific Research (B) (Nos. 18H01840, and 18H01999), and a Grant-in-Aid for Young Scientists (B) (No. 16K16386) from the Japan Society for the Promotion of Science (JSPS). This work was also supported by Program for Creation of Interdisciplinary Research from Frontier Research Institute for Interdisciplinary Sciences, Tohoku University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdelmoez, M. N., Iida, K., Oguchi, Y., Nishikii, H., Yokokawa, R., Kotera, H., et al. (2018). SINC-seq: correlation of transient gene expressions between nucleus and cytoplasm reflects single-cell physiology. Genome Biol. 19:66. doi: 10.1186/s13059-018-1446-9
Abe, H., Ino, K., Li, C. Z., Kanno, Y., Inoue, K. Y., Suda, A., et al. (2015). Electrochemical imaging of dopamine release from three-dimensional-cultured PC12 cells using large-scale integration-based amperometric sensors. Anal. Chem. 87, 6364–6370. doi: 10.1021/acs.analchem.5b01307
Abe, H., Kanno, Y., Ino, K., Inoue, K. Y., Suda, A., Kunikata, R., et al. (2016). Electrochemical imaging for single-cell analysis of cell adhesion using a collagen-coated large-scale integration (LSI)-based amperometric device. Electrochemistry 84, 364–367. doi: 10.5796/electrochemistry.84.364
Akagi, T., Kato, K., Kobayashi, M., Kosaka, N., Ochiya, T., and Ichiki, T. (2015). On-chip immunoelectrophoresis of extracellular vesicles released from human breast cancer cells. PLoS ONE 10:e0123603. doi: 10.1371/journal.pone.0123603
Akyazi, T., Basabe-Desmonts, L., and Benito-Lopez, F. (2018). Review on microfluidic paper-based analytical devices towards commercialisation. Anal. Chim. Acta 1001, 1–17. doi: 10.1016/j.aca.2017.11.010
Anand, R. K., Johnson, E. S., and Chiu, D. T. (2015). Negative dielectrophoretic capture and repulsion of single cells at a bipolar electrode: the impact of faradaic ion enrichment and depletion. J. Am. Chem. Soc. 137, 776–783. doi: 10.1021/ja5102689
Anderson, W., Lane, R., Korbie, D., and Trau, M. (2015). Observations of tunable resistive pulse sensing for exosome analysis: improving system sensitivity and stability. Langmuir 31, 6577–6587. doi: 10.1021/acs.langmuir.5b01402
Bachmann, B., Adly, N., Schnitker, J., Yakushenko, A., Rinklin, P., Offenhaeusser, A., et al. (2017). All-inkjet-printed gold microelectrode arrays for extracellular recording of action potentials. Flex. Print. Electr. 2:035003. doi: 10.1088/2058-8585/aa7928
Baret, J. C., Miller, O. J., Taly, V., Ryckelynck, M., El-Harrak, A., Frenz, L., et al. (2009). Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip 9, 1850–1858. doi: 10.1039/b902504a
Bavli, D., Prill, S., Ezra, E., Levy, G., Cohen, M., Vinken, M., et al. (2016). Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl. Acad. Sci. U.S.A. 113, E2231–E2240. doi: 10.1073/pnas.1522556113
Bhatia, S. N., and Ingber, D. E. (2014). Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772. doi: 10.1038/nbt.2989
Cheah, L. T., Dou, Y. H., Seymour, A. M., Dyer, C. E., Haswell, S. J., Wadhawan, J. D., et al. (2010). Microfluidic perfusion system for maintaining viable heart tissue with real-time electrochemical monitoring of reactive oxygen species. Lab Chip 10, 2720–2726. doi: 10.1039/c004910g
Curto, V. F., Marchiori, B., Hama, A., Pappa, A. M., Ferro, M. P., Braendlein, M., et al. (2017). Organic transistor platform with integrated microfluidics for in-line multi-parametric in vitro cell monitoring. Microsyst. Nanoeng. 3:17028. doi: 10.1038/micronano.2017.28
Douville, N. J., Tung, Y. C., Li, R., Wang, J. D., El-Sayed, M. E., and Takayama, S. (2010). Fabrication of two-layered channel system with embedded electrodes to measure resistance across epithelial and endothelial barriers. Anal. Chem. 82, 2505–2511. doi: 10.1021/ac9029345
Elbrecht, D. H., Long, C. J., and Hickman, J. J. (2016). Transepithelial/endothelial electrical resistance (TEER) theory and applications for microfluidic body-on-a-chip devices. J. Rare Dis. Res. Treat. 1, 46–52. doi: 10.29245/2572-9411/2016/3.1026
Henry, O. Y. F., Villenave, R., Cronce, M. J., Leineweber, W. D., Benz, M. A., and Ingber, D. E. (2017). Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip 17, 2264–2271. doi: 10.1039/C7LC00155J
Huh, D., Matthews, B. D., Mammoto, A., Montoya-Zavala, M., Hsin, H. Y., and Ingber, D. E. (2010). Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668. doi: 10.1126/science.1188302
Ino, K., Kanno, Y., Inoue, K. Y., Suda, A., Kunikata, R., Matsudaira, M., et al. (2017). Electrochemical motion tracking of microorganisms using a large-scale-integration-based amperometric device. Angew. Chem. Int. Edn. 56, 6818–6822. doi: 10.1002/anie.201701541
Ino, K., Kanno, Y., Nishijo, T., Komaki, H., Yamada, Y., Yoshida, S., et al. (2014). Densified electrochemical sensors based on local redox cycling between vertically separated electrodes in substrate generation/chip collection and extended feedback modes. Anal. Chem. 86, 4016–4023. doi: 10.1021/ac500435d
Ino, K., Kitagawa, Y., Watanabe, T., Shiku, H., Koide, M., Itayama, T., et al. (2009). Detection of hormone active chemicals using genetically engineered yeast cells and microfluidic devices with interdigitated array electrodes. Electrophoresis 30, 3406–3412. doi: 10.1002/elps.200900244
Ino, K., Matsumoto, T., Taira, N., Kumagai, T., Nashimoto, Y., and Shiku, H. (2018a). Hydrogel electrodeposition based on bipolar electrochemistry. Lab Chip 18, 2425–2432. doi: 10.1039/C8LC00465J
Ino, K., Nashimoto, Y., Taira, N., Ramon Azcon, J., and Shiku, H. (2018b). Intracellular electrochemical sensing. Electroanalysis 30, 2195–2209. doi: 10.1002/elan.201800410
Ino, K., Nishijo, T., Arai, T., Kanno, Y., Takahashi, Y., Shiku, H., et al. (2012). Local redox-cycling-based electrochemical chip device with deep microwells for evaluation of embryoid bodies. Angew. Chem. Int. Edn. 51, 6648–6652. doi: 10.1002/anie.201201602
Ino, K., Onodera, T., Kanno, Y., Suda, A., Kunikata, R., Matsue, T., et al. (2018c). Electrochemicolor imaging of endogenous alkaline phosphatase and respiratory activities of mesenchymal stem cell aggregates in early-stage osteodifferentiation. Electrochim. Acta 268, 554–561. doi: 10.1016/j.electacta.2018.02.094
Ino, K., Onodera, T., Nashimoto, Y., and Shiku, H. (2019a). Differential electrochemicolor imaging using LSI-based device for simultaneous detection of multiple analytes. Sens. Mater. 31, 13–22. doi: 10.18494/SAM.2019.2035
Ino, K., Terauchi, M., Gakumasawa, M., Taira, N., Suda, A., Kunikata, R., et al. (2018d). Local hydrogel fabrication based on electrodeposition with a large-scale integration (LSI)-based amperometric device. Sens. Actuators B Chem. 277, 95–101. doi: 10.1016/j.snb.2018.08.135
Ino, K., Yokokawa, Y., Taira, N., Suda, A., Kunikata, R., Nashimoto, Y., et al. (2019b). Electrochemical imaging of cell activity in hydrogels embedded in grid-shaped polycaprolactone scaffolds using a large-scale integration-based amperometric device. Anal. Sci. 35, 39–43. doi: 10.2116/analsci.18SDP01
Inoue, K. Y., Ino, K., Shiku, H., Kasai, S., Yasukawa, T., Mizutani, F., et al. (2010). Electrochemical monitoring of hydrogen peroxide released from leucocytes on horseradish peroxidase redox polymer coated electrode chip. Biosens. Bioelectr. 25, 1723–1728. doi: 10.1016/j.bios.2009.12.014
Inoue, K. Y., Matsudaira, M., Kubo, R., Nakano, M., Yoshida, S., Matsuzaki, S., et al. (2012). LSI-based amperometric sensor for bio-imaging and multi-point biosensing. Lab Chip 12, 3481–3490. doi: 10.1039/c2lc40323d
Inoue, K. Y., Matsudaira, M., Nakano, M., Ino, K., Sakamoto, C., Kanno, Y., et al. (2015). Advanced LSI-based amperometric sensor array with light-shielding structure for effective removal of photocurrent and mode selectable function for individual operation of 400 electrodes. Lab Chip 15, 848–856. doi: 10.1039/C4LC01099J
Ito, H., Nashimoto, Y., Zhou, Y., Takahashi, Y., Ino, K., Shiku, H., et al. (2016). Localized gene expression analysis during sprouting angiogenesis in mouse embryoid bodies using a double barrel carbon probe. Anal. Chem. 88, 610–613. doi: 10.1021/acs.analchem.5b04338
Jiang, H., Jiang, D., Zhu, P., Pi, F., Ji, J., Sun, C., et al. (2016). A novel mast cell co-culture microfluidic chip for the electrochemical evaluation of food allergen. Biosens. Bioelectr. 83, 126–133. doi: 10.1016/j.bios.2016.04.028
Jin, J. H., Kim, J. H., Lee, S. K., Choi, S. J., Park, C. W., and Min, N. K. (2018). A fully integrated paper-microfluidic electrochemical device for simultaneous analysis of physiologic blood ions. Sensors 18:104. doi: 10.3390/s18010104
Kanno, Y., Ino, K., Abe, H., Sakamoto, C., Onodera, T., Inoue, K. Y., et al. (2017). Electrochemicolor imaging using an LSI-based device for multiplexed cell assays. Anal. Chem. 89, 12778–12786. doi: 10.1021/acs.analchem.7b03042
Kanno, Y., Ino, K., Inoue, K. Y., Sen, M., Suda, A., Kunikata, R., et al. (2015a). Feedback mode-based electrochemical imaging of conductivity and topography for large substrate surfaces using an LSI-based amperometric chip device with 400 sensors. J. Electroanal. Chem. 741, 109–113. doi: 10.1016/j.jelechem.2015.01.020
Kanno, Y., Ino, K., Inoue, K. Y., Suda, A., Kunikata, R., Matsudaira, M., et al. (2015b). Simulation analysis of positional relationship between embryoid bodies and sensors on an LSI-based amperometric device for electrochemical imaging of alkaline phosphatase activity. Anal. Sci. 31, 715–719. doi: 10.2116/analsci.31.715
Kanno, Y., Ino, K., Sakamoto, C., Inoue, K. Y., Matsudaira, M., Suda, A., et al. (2016). Potentiometric bioimaging with a large-scale integration (LSI)-based electrochemical device for detection of enzyme activity. Biosens. Bioelectr. 77, 709–714. doi: 10.1016/j.bios.2015.10.021
Kanno, Y., Ino, K., Shiku, H., and Matsue, T. (2015c). A local redox cycling-based electrochemical chip device with nanocavities for multi-electrochemical evaluation of embryoid bodies. Lab Chip 15, 4404–4414. doi: 10.1039/C5LC01016K
Kasai, N., Han, C. X., and Torimitsu, K. (2005). Hydrogen peroxide distribution and neuronal cell death in a rat hippocampal slice. Sens. Actuators B Chem. 108, 746–750. doi: 10.1016/j.snb.2004.11.096
Kätelhön, E., Mayer, D., Banzet, M., Offenhäusser, A., and Wolfrum, B. (2014). Nanocavity crossbar arrays for parallel electrochemical sensing on a chip. Beilstein J. Nanotechnol. 5, 1137–1143. doi: 10.3762/bjnano.5.124
Kato, R., Matsumoto, M., Sasaki, H., Joto, R., Okada, M., Ikeda, Y., et al. (2016). Parametric analysis of colony morphology of non-labelled live human pluripotent stem cells for cell quality control. Sci. Rep. 6:34009. doi: 10.1038/srep34009
Kawano, R. (2018). Nanopore decoding of oligonucleotides in DNA computing. Biotechnol. J. 13:e1800091. doi: 10.1002/biot.201800091
Kim, H. J., Huh, D., Hamilton, G., and Ingber, D. E. (2012). Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174. doi: 10.1039/c2lc40074j
Laforge, F. O., Carpino, J., Rotenberg, S. A., and Mirkin, M. V. (2007). Electrochemical attosyringe. Proc. Natl. Acad. Sci. U.S.A. 104, 11895–11900. doi: 10.1073/pnas.0705102104
Lee, J., Sharma, D., Lim, Y., and Shin, H. (2018). Redox cycling effect at microchannel-integrated sandwich electrodes consisting of a suspended mesh and a substrate-bound planar electrode. Sens. Actuators B Chem. 267, 467–475. doi: 10.1016/j.snb.2018.04.003
Lemay, S. G., Kang, S., Mathwig, K., and Singh, P. S. (2013). Single-molecule electrochemistry: Present status and outlook. Acc. Chem. Res. 46, 369–377. doi: 10.1021/ar300169d
Li, J., Wu, S., Kim, E., Yan, K., Liu, H., Liu, C., et al. (2019). Electrobiofabrication: electrically-based fabrication with biologically-derived materials. Biofabrication 11:032002. doi: 10.1088/1758-5090/ab06ea
Lind, J. U., Busbee, T. A., Valentine, A. D., Pasqualini, F. S., Yuan, H., Yadid, M., et al. (2017). Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater. 16, 303–308. doi: 10.1038/nmat4782
Mano, T., Nagamine, K., Ichimura, Y., Shiwaku, R., Furusawa, H., Matsui, H., et al. (2018). Printed organic transistor-based enzyme sensor for continuous glucose monitoring in wearable healthcare applications. ChemElectroChem 5, 3881–3886. doi: 10.1002/celc.201801129
Maoz, B. M., Herland, A., Henry, O. Y. F., Leineweber, W. D., Yadid, M., Doyle, J., et al. (2017). Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 17, 2294–2302. doi: 10.1039/C7LC00412E
Mellors, J. S., Jorabchi, K., Smith, L. M., and Ramsey, J. M. (2010). Integrated microfluidic device for automated single cell analysis using electrophoretic separation and electrospray ionization mass spectrometry. Anal. Chem. 82, 967–973. doi: 10.1021/ac902218y
Misun, P. M., Rothe, J., Schmid, Y. R. F., Hierlemann, A., and Frey, O. (2016). Multi-analyte biosensor interface for real-time monitoring of 3D microtissue spheroids in hanging-drop networks. Microsyst. Nanoeng. 2:16022. doi: 10.1038/micronano.2016.22
Moya, A., Ortega-Ribera, M., Guimerà, X., Sowade, E., Zea, M., Illa, X., et al. (2018). Online oxygen monitoring using integrated inkjet-printed sensors in a liver-on-a-chip system. Lab Chip 18, 2023–2035. doi: 10.1039/C8LC00456K
Nagasaka, R., Gotou, Y., Yoshida, K., Kanie, K., Shimizu, K., Honda, H., et al. (2017). Image-based cell quality evaluation to detect irregularities under same culture process of human induced pluripotent stem cells. J. Biosci. Bioeng. 123, 642–650. doi: 10.1016/j.jbiosc.2016.12.015
Nashimoto, Y., Takahashi, Y., Yamakawa, T., Torisawa, Y. S., Yasukawa, T., Ito-Sasaki, T., et al. (2007). Measurement of gene expression from single adherent cells and spheroids collected using fast electrical lysis. Anal. Chem. 79, 6823–6830. doi: 10.1021/ac071050q
Nashimoto, Y., Takahashi, Y., Zhou, Y., Ito, H., Ida, H., Ino, K., et al. (2016). Evaluation of mRNA localization using double barrel scanning ion conductance microscopy. ACS Nano 10, 6915–6922. doi: 10.1021/acsnano.6b02753
Ngoc-Viet, N., and Jen, C. P. (2018). Impedance detection integrated with dielectrophoresis enrichment platform for lung circulating tumor cells in a microfluidic channel. Biosens. Bioelectr. 121, 10–18. doi: 10.1016/j.bios.2018.08.059
Niu, X., Lan, M., Zhao, H., and Chen, C. (2013). Highly sensitive and selective nonenzymatic detection of glucose using three-dimensional porous nickel nanostructures. Anal. Chem. 85, 3561–3569. doi: 10.1021/ac3030976
Ozawa, F., Ino, K., Arai, T., Ramón-Azcón, J., Takahashi, Y., Shiku, H., et al. (2013a). Alginate gel microwell arrays using electrodeposition for three-dimensional cell culture. Lab Chip 13, 3128–3135. doi: 10.1039/c3lc50455g
Ozawa, F., Ino, K., Shiku, H., and Matsue, T. (2016). Electrochemical hydrogel lithography of calcium-alginate hydrogels for cell culture. Materials 9:744. doi: 10.3390/ma9090744
Ozawa, F., Ino, K., Shiku, H., and Matsue, T. (2017). Cell sheet fabrication using RGD peptide-coupled alginate hydrogels fabricated by an electrodeposition method. Chem. Lett. 46, 605–608. doi: 10.1246/cl.170003
Ozawa, F., Ino, K., Takahashi, Y., Shiku, H., and Matsue, T. (2013b). Electrodeposition of alginate gels for construction of vascular-like structures. J. Biosci. Bioeng. 115, 459–461. doi: 10.1016/j.jbiosc.2012.10.014
Pohl, H. A. (1951). The motion and precipitation of suspensoids in divergent electric fields. J. Appl. Phys. 22, 869–871. doi: 10.1063/1.1700065
Prasad, A., Kikuchi, H., Inoue, K. Y., Suzuki, M., Sugiura, Y., Sugai, T., et al. (2016). Simultaneous real-time monitoring of oxygen consumption and hydrogen peroxide production in cells using our newly developed chip-type biosensor device. Front. Physiol. 7:109. doi: 10.3389/fphys.2016.00109
Ramón-Azcón, J., Ahadian, S., Estili, M., Liang, X., Ostrovidov, S., Kaji, H., et al. (2013). Dielectrophoretically aligned carbon nanotubes to control electrical and mechanical properties of hydrogels to fabricate contractile muscle myofibers. Adv. Mater. 25, 4028–4034. doi: 10.1002/adma.201301300
Ramón-Azcón, J., Ahadian, S., Obregón, R., Camci-Unal, G., Ostrovidov, S., Hosseini, V., et al. (2012). Gelatin methacrylate as a promising hydrogel for 3D microscale organization and proliferation of dielectrophoretically patterned cells. Lab Chip 12, 2959–2969. doi: 10.1039/c2lc40213k
Rival, A., Jary, D., Delattre, C., Fouillet, Y., Castellan, G., Bellemin-Comte, A., et al. (2014). An EWOD-based microfluidic chip for single-cell isolation, mRNA purification and subsequent multiplex qPCR. Lab Chip 14, 3739–3749. doi: 10.1039/C4LC00592A
Rogal, J., Probst, C., and Loskill, P. (2017). Integration concepts for multi-organ chips: how to maintain flexibility? Future Sci OA 3:FSO180. doi: 10.4155/fsoa-2016-0092
Schmid, Y. R. F., Burgel, S. C., Misun, P. M., Hierlemann, A., and Frey, O. (2016). Electrical impedance spectroscopy for microtissue spheroid analysis in hanging-drop networks. ACS Sens. 1, 1028–1035. doi: 10.1021/acssensors.6b00272
Sen, M., Ino, K., Inoue, K. Y., Arai, T., Nishijo, T., Suda, A., et al. (2013a). LSI-based amperometric sensor for real-time monitoring of embryoid bodies. Biosens. Bioelectr. 48, 12–18. doi: 10.1016/j.bios.2013.03.069
Sen, M., Ino, K., Inoue, K. Y., Suda, A., Kunikata, R., Matsudaira, M., et al. (2014). Electrochemical evaluation of sarcomeric alpha-actinin in embryoid bodies after gene silencing using an LSI-based amperometric sensor array. Anal. Methods 6, 6337–6342. doi: 10.1039/C4AY00791C
Sen, M., Ino, K., Ramón-Azcón, J., Shiku, H., and Matsue, T. (2013b). Cell pairing using a dielectrophoresis-based device with interdigitated array electrodes. Lab Chip 13, 3650–3652. doi: 10.1039/c3lc50561h
Shin, S. R., Kilic, T., Zhang, Y. S., Avci, H., Hu, N., Kim, D., et al. (2017). Label-free and regenerative electrochemical microfluidic biosensors for continual monitoring of cell secretomes. Adv. Sci. 4:1600522. doi: 10.1002/advs.201600522
Shin, S. R., Zhang, Y. S., Kim, D. J., Manbohi, A., Avci, H., Silvestri, A., et al. (2016). Aptamer-based microfluidic electrochemical biosensor for monitoring cell-secreted trace cardiac biomarkers. Anal. Chem. 88, 10019–10027. doi: 10.1021/acs.analchem.6b02028
Shitanda, I., Komoda, M., Hoshi, Y., and Itagaki, M. (2018). Screen-printed paper-based three-electrode system with long-term stable and instantaneously usable reference electrode. Chem. Lett. 47, 1502–1504. doi: 10.1246/cl.180809
Sinha, H., Quach, A. B. V., Vo, P. Q. N., and Shih, S. C. C. (2018). An automated microfluidic gene-editing platform for deciphering cancer genes. Lab Chip 18, 2300–2312. doi: 10.1039/C8LC00470F
Subramanian Parimalam, S., Oguchi, Y., Abdelmoez, M. N., Tsuchida, A., Ozaki, Y., Yokokawa, R., et al. (2018). Electrical lysis and RNA extraction from single cells fixed by dithiobis(succinimidyl propionate). Anal. Chem. 90, 12512–12518. doi: 10.1021/acs.analchem.8b02338
Taira, N., Ino, K., Ida, H., Nashimoto, Y., and Shiku, H. (2019). Electrodeposition-based rapid bioprinting of 3D-desinged hydrogels with a pin art device. Biofabrication 11:035018. doi: 10.1088/1758-5090/ab166e
Taira, N., Ino, K., Robert, J., and Shiku, H. (2018). Electrochemical printing of calcium alginate/gelatin hydrogel. Electrochim. Acta 281, 429–436. doi: 10.1016/j.electacta.2018.05.124
Thomas, R. S., Morgan, H., and Green, N. G. (2009). Negative DEP traps for single cell immobilisation. Lab Chip 9, 1534–1540. doi: 10.1039/b819267g
Wang, J. D., Khafagy El, S., Khanafer, K., Takayama, S., and Elsayed, M. E. (2016). Organization of endothelial cells, pericytes, and astrocytes into a 3D microfluidic in vitro model of the blood-brain barrier. Mol. Pharm. 13, 895–906. doi: 10.1021/acs.molpharmaceut.5b00805
Wang, X., Ren, L., Su, Y., Ji, Y., Liu, Y., Li, C., et al. (2017). Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells. Anal. Chem. 89, 12569–12577. doi: 10.1021/acs.analchem.7b03884
Wong, J. F., and Simmons, C. A. (2019). Microfluidic assay for the on-chip electrochemical measurement of cell monolayer permeability. Lab Chip 19, 1060–1070. doi: 10.1039/c8lc01321g
Wu, C., Chen, R., Liu, Y., Yu, Z., Jiang, Y., and Cheng, X. (2017). A planar dielectrophoresis-based chip for high-throughput cell pairing. Lab Chip 17, 4008–4014. doi: 10.1039/C7LC01082F
Wu, C.-C., Saito, T., Yasukawa, T., Shiku, H., Abe, H., Hoshi, H., et al. (2007). Microfluidic chip integrated with amperometric detector array for in situ estimating oxygen consumption characteristics of single bovine embryos. Sens. Actuators B Chem. 125, 680–687. doi: 10.1016/j.snb.2007.03.017
Yang, Y., Noviana, E., Nguyen, M. P., Geiss, B. J., Dandy, D. S., and Henry, C. S. (2017). Paper-based microfluidic devices: emerging themes and applications. Anal. Chem. 89, 71–91. doi: 10.1021/acs.analchem.6b04581
Yasaki, H., Shimada, T., Yasui, T., Yanagida, T., Kaji, N., Kanai, M., et al. (2018). Robust Ionic current sensor for bacterial cell size detection. ACS Sens. 3, 574–579. doi: 10.1021/acssensors.8b00045
Yasaki, H., Yasui, T., Yanagida, T., Kaji, N., Kanai, M., Nagashima, K., et al. (2017). Substantial expansion of detectable size range in ionic current sensing through pores by using a microfluidic bridge circuit. J. Am. Chem. Soc. 139, 14137–14142. doi: 10.1021/jacs.7b06440
Yasukawa, T., Nagamine, K., Horiguchi, Y., Shiku, H., Koide, M., Itayama, T., et al. (2008). Electrophoretic cell manipulation and electrochemical gene-function analysis based on a yeast two-hybrid system in a microfluidic device. Anal. Chem. 80, 3722–3727. doi: 10.1021/ac800143t
Yoshimura, Y., Tomita, M., Mizutani, F., and Yasukawa, T. (2014). Cell pairing using microwell array electrodes based on dielectrophoresis. Anal. Chem. 86, 6818–6822. doi: 10.1021/ac5015996
Yoshioka, J., Ohsugi, Y., Yoshitomi, T., Yasukawa, T., Sasaki, N., and Yoshimoto, K. (2018). Label-free rapid separation and enrichment of bone marrow-derived mesenchymal stem cells from a heterogeneous cell mixture using a dielectrophoresis device. Sensors 18:3007. doi: 10.3390/s18093007
Yoshioka, J., Yoshitomi, T., Yasukawa, T., and Yoshimoto, K. (2016). Alternation of gene expression levels in mesenchymal stem cells by applying positive dielectrophoresis. Anal. Sci. 32, 1213–1216. doi: 10.2116/analsci.32.1213
Zhang, H., Oellers, T., Feng, W., Abdulazim, T., Saw, E. N., Ludwig, A., et al. (2017). High-density droplet microarray of individually addressable electrochemical cells. Anal. Chem. 89, 5832–5839. doi: 10.1021/acs.analchem.7b00008
Keywords: electric devices, electrochemical devices, microfluidic devices, cell manipulation, cell analysis, organs-on-a-chip
Citation: Hiramoto K, Ino K, Nashimoto Y, Ito K and Shiku H (2019) Electric and Electrochemical Microfluidic Devices for Cell Analysis. Front. Chem. 7:396. doi: 10.3389/fchem.2019.00396
Received: 04 April 2019; Accepted: 16 May 2019;
Published: 04 June 2019.
Edited by:
Huan-Tsung Chang, National Taiwan University, TaiwanReviewed by:
Aihua Liu, Qingdao University, ChinaWei-Lung Tseng, National Sun Yat-sen University, Taiwan
Copyright © 2019 Hiramoto, Ino, Nashimoto, Ito and Shiku. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kosuke Ino, a29zdWtlLmlub0B0b2hva3UuYWMuanA=; Hitoshi Shiku, aGl0b3NoaS5zaGlrdS5jM0B0b2hva3UuYWMuanA=
 Kaoru Hiramoto1
Kaoru Hiramoto1 Kosuke Ino
Kosuke Ino Yuji Nashimoto
Yuji Nashimoto