- 1Department of Chemistry, University of Milan, Milan, Italy
- 2Department of Pharmaceutical Sciences, University of Milan, Milan, Italy
- 3Department of Food, Environmental and Nutritional Sciences, University of Milan, Milan, Italy
Unnatural amino acids have tremendously expanded the folding possibilities of peptides and peptide mimics. While α,α-disubstituted and β-amino acids are widely studied, γ-derivatives have been less exploited. Here we report the conformational study on the bicyclic unnatural γ amino acid, 4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-3-carboxylic acid 1. In model peptides, the (+)-(3aR6aS)-enantiomer is able to stabilize α-turn conformation when associated to glycine, as showed by 1H-NMR, FT-IR, and circular dichroism experiments, and molecular modeling studies. α-turn is a structural motif occurring in many biologically active protein sites, although its stabilization on isolated peptides is quite uncommon. Our results make the unnatural γ-amino acid 1 of particular interest for the development of bioactive peptidomimetics.
Introduction
Amino acids are the key building blocks of proteins and biomolecules and are widely exploited in different applications, from pharmaceutical chemistry and biomedicine to material science, optoelectronics and catalysis (Zhang et al., 2012; Mikhalevich et al., 2017; Solomon et al., 2017; López-Andarias et al., 2018; Raymond and Nilsson, 2018). The insertion of unnatural amino acids (UAAs) in peptides and peptide mimics could add specific features to these molecules, such as proteolytic stability, active functional groups and new reactivity (Clerici et al., 2016; Pellegrino et al., 2016, 2017; Bucci et al., 2017). α,α disubstituted and β- homologs of natural amino acids have been particularly studied during the years for their ability to introduce conformational constrains in peptides and to stabilize specific secondary structures (Bonetti et al., 2015; Pellegrino et al., 2015; Fanelli et al., 2017; Kobayashi et al., 2017). On the other hand, γ UAAs provide a further opportunity to engineer the available backbone through the incorporation of an additional methylene group (Vasudev et al., 2011; Sohora et al., 2018). Recent studies report that γ-peptides are able to form helices, sheets and turns, whose conformational stability is increased by introducing substituents on the backbone chain (Bouillère et al., 2011). Moreover, cyclic γ UAAs, as gabapentin (Gpn) (Chatterjee et al., 2009; Konda et al., 2018) and γ-cyclohexyl amino acid (Guo et al., 2012) are able to stabilize both turn and helix conformation in oligomers and in mixed α-γ and β-γ sequences. On the other hand, the use of bicyclic γ amino acids for the stabilization of the peptide structure is much less investigated (Machetti et al., 2000).
In this work, we investigated the conformational behavior of both the enantiomers of the bicyclic unnatural γ amino acid 4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-3-carboxylic acid 1, obtained starting from the corresponding ethyl esters recently described by us (Tamborini et al., 2015). Compound 1 is a conformationally constrained dipeptide analog and, in principle, it could substitute two amino acids in a peptide chain. The presence of the constrained bicyclic system could induce specific secondary structure allowing a proper orientation of the peptide arms at C- and N- termini. Furthermore, the presence of the isoxazoline ring, a core often found in biologically active compounds, could be particularly useful for future applications in the pharmaceutical field. In fact, isoxazoline derivatives are important scaffolds found in many naturally occurring and biologically active compounds possessing a wide range of bioactivities, such as antibacterial, antifungal, antiparasitic (Conti et al., 2011; Bruno et al., 2014; Pinto et al., 2016a), anticancer (Castellano et al., 2011; Kaur et al., 2014), anti-inflammatory and anticonvulsant activity (Sperry and Wright, 2005; Pinto et al., 2011, 2016b). Isoxazolines are also considered to be important precursors for the synthesis of β-hydroxyketones (Kozikowski and Park, 1990; Tsantali et al., 2007), β-aminoalcohols (Fuller et al., 2005), isoxazolidines (Itoh et al., 2002), and many other valuable compounds. Recently, peptidomimetics containing the isoxazoline ring have been reported as β-turn mimics (Bucci et al., 2018; Memeo et al., 2018).
Starting from the (–)-(3aS6aR)-1 and (+)-(3aR6aS)-1 enantiomers, model peptides containing Leu-Val dipeptide at C-terminus and variable sequences at N-terminus were prepared. Their conformational behavior was investigated by NMR spectroscopy, FT-IR, circular dichroism, and molecular modeling. Our results indicated that (+)-(3aR6aS)-1 enantiomer, in combination with glycine, is effective in stabilizing the α-turn conformation in peptides (Figure 1). This structural motif occurs quite often in many key sites of proteins, such as enzyme active site, and metal binding domains (Wintjens et al., 1996), although few molecules are known to mimic or stabilize it on isolated peptides (Kelso et al., 2004; Hoang et al., 2011; Krishna et al., 2014; Wang et al., 2018.). Our results make thus the unnatural γ-amino acid 1 of particular interest for future development of bioactive peptidomimetics.
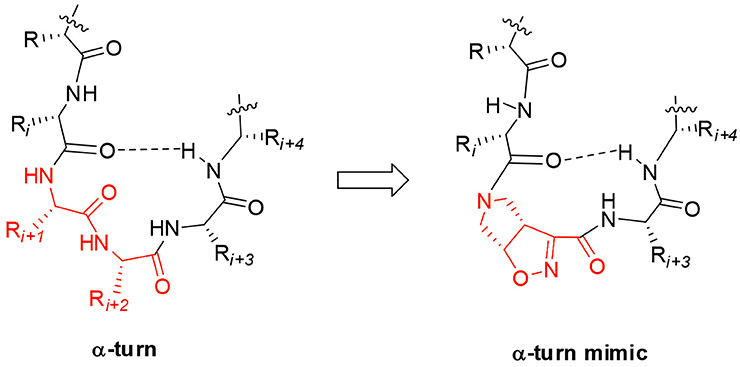
Figure 1. Natural α-turn and the proposed α-turn mimic containing the bicyclic pyrrolidine-isoxazoline γ amino acid.
Materials and Methods
Materials
Chemicals were obtained from Zentek (Italy) and used without further purification. Melting points were determined in a Stuart Scientific melting point apparatus in open capillary tubes and are certified. ESI mass spectra were recorded on an LCQESI. MS was recorded on a LCQ Advantage spectrometer from Thermo Finningan and a LCQ Fleet spectrometer from Thermo Scientific. CD experiments were carried out on a Jasco J-810 instrument. Spectra were obtained from 195 to 250 nm with a 0.1 nm step and 1 s collection time per step, taking three averages. The CD spectra were plotted as mean residue ellipticity θ (degree × cm2 × dmol−1) vs. wave length λ (nm). Noise-reduction was obtained using a Fourier-transform filter program from Jasco. The NMR spectroscopic experiments were carried out on a Varian OXFORD 300 MHz (300 and 75 MHz for 1H and 13C, respectively). Chemical shifts, δ, are given in ppm relative to the CHCl3 internal standard, and the coupling constants, J, are reported in hertz (Hz).
Methods
Synthesis of (–)-(3aS,6aR)-1 and (+)-(3aR,6aS)-1
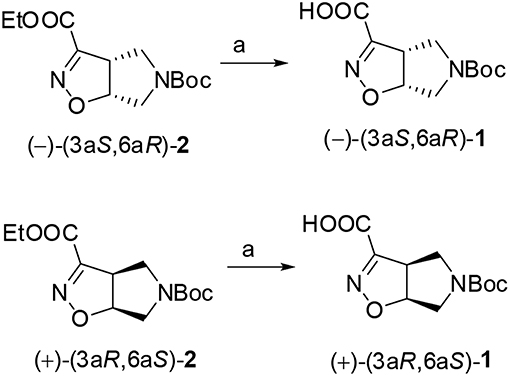
Compound (–)-2 or (+)-2 (200 mg, 0.70 mmol) was dissolved in MeOH (3.0 mL) and treated with 1N NaOH aqueous solution (1.4 mL). The mixture was stirred at room temperature for 1 h and the disappearance of the starting material was monitored by TLC (Cyclohexane/EtOAc 7:3). The mixture was diluted in water (20 mL), made acidic with 2N aqueous HCl and extracted with EtOAc. The organic phase was dried over anhydrous Na2SO4 and after evaporation of the solvent, the acid derivate (–)-(3aS,6aR)-1 or (+)-(3aR,6aS)-1 (170 mg, 95%) was obtained as a white solid.
(–)-(3aS,6aR)-1: Rf (CH2Cl2/MeOH 9:1 + 1% AcOH): 0.40; [α]: (–) 192.2 (c: 0.55 in MeOH); mp = dec. T > 137°C; 1H NMR (300 MHz, CD3OD): δ 5.35 (dd, J = 4.8, 9.2, 1H); 4.10 (ddd, J = 2.2, 9.2, 9.2, 1H); 3.86–3.96 (m, 2H); 3.38–3.50 (m, 2H); 1.44 (s, 9H) ppm; 13C NMR (75 MHz, CD3OD): δ 161.28, 154.77, 152.95, 87.62, 80.33, 53.06, 50.93, 49.10, 27.12 (3C) ppm.
HRMS (ESI) [M + Na]+ calculated for C11H16N2O5Na: 279.0957, found: 279.0950.
(+)-(3aR,6aS)-1: [α]: (+) 192.5 (c: 0.57 in MeOH).
General Procedure for Coupling Reaction
Free carboxylic acid (1 equiv.) was dissolved in CH2Cl2 (0.01M solution) and the mixture was cooled to 0°C. HOBt (1.1 equiv.) and EDC (1.1 equiv.) were added. After 1 h, free amino compound (1 equiv.) was added, followed by the addition of DIEA (2 equiv.). The reaction mixture was stirred at room temperature for 24 overnight (TLC analysis). The organic layer was washed with a 5% solution of KHSO4 (3 times), with a saturated solution of NaHCO3 (3 times) and with brine (1 time). The organic phase was dried over Na2SO4 and the solvent was removed under vacuum. The products were purified by column flash chromatography on silica gel (hexane/ethyl acetate gradient) followed by a crystallization from a mixture of ethyl acetate/hexane.
N-Boc-(–)-Δ2-isox-Leu-Val-CONH2 (4)
1H NMR (300 MHz, CDCl3): δ 7.11 (bs, 1H), 6.88 (d, J = 8.8 Hz, 1H), 6.19 (bs, 1H), 5.72 (bs, 1H), 5.34–5.29 (m, 1H), 4.56–4.48 (m, 1H), 4.31 (dd, J = 8.2; 6.9 Hz, 1H), 4.17–3.83 (m, 3H), 3.56 – 3.42 (m, 2H), 2.23–2.07 (m, 1H), 1.85–1.61(m, 3H), 1.43 (s, 9H), 0.99 – 0.92 (m, 12H) ppm.
13C NMR (75 MHz, CDCl3): δ 173.04, 171.32, 159.44, 154.28, 153.93, 87.40, 80.43, 58.24, 53.26, 52.30, 50.53, 49.32, 40.87, 30.53, 28.31, 24.82, 22.91, 21.92, 19.27, 17.93 ppm.
HRMS (ESI) [M + Na]+ calculated for C22H37N5O6Na: 490.2642, found: 490.2635.
N-Boc- (+)-Δ2-isox-Leu-Val-CONH2 (5)
1H NMR (300 MHz, CDCl3): δ 7.05 (bs, 1H), 6.73 (d, J = 7.9 Hz, 1H), 6.13 (bs, 1H), 5.69 (bs, 1H), 5.37–5.28 (m, 1H), 4.56–4.47 (m, 1H), 4.36–4.27 (m, 1H), 4.18–4.07 (m, 1H), 3.99–3.77 (m, 2H), 3.61–3.41 (m, 2H), 2.28–2.10 (m, 1H), 1.78–1.57(m, 3H), 1.49 (s, 9H), 1.01 – 0.90 (m, 12H) ppm.
13C NMR (75 MHz, CDCl3): δ 173.16, 171.54, 159.33, 154.16, 153.96, 80.34, 60.38, 58.16, 53.27, 52.25, 49.41, 41.02, 30.67, 28.32, 24.83, 22.87, 21.99, 19.20, 18.02 ppm.
HRMS (ESI) [M + Na]+ calculated for C22H37N5O6Na: 490.2642, found: 490.2635.
NH-Boc-Phe-(-)-Δ2-isox-Leu-Val-CONH2 (8) –Mixture of two conformers-
1H NMR (300 MHz, CDCl3): δ 7.36–7.16 (m, 5H), 7.15–7.08 (m, 1H), 7.05–6.97 (m, 1H), 6.88–6.77 (m, 1H), 6.05 (bs, 1H), 5.71 (bs, 1H), 5.43 (d, J = 8.4 Hz 1H), 5.33–5.22 (m, 3H), 5.09–4.9 (m, 1H), 4.64–4.44 (m, 3H), 4.33–4.23 (m, 2H), 4.17–4.03 (m, 2H), 3.99–3.91 (m, 1H), 3.87–3.75(m, 2H), 3.70–3.50 (m, 2H), 3.42–3.30 (m, 1H), 3.13–3.03 (m, 1H), 2.97–2.74 (m, 3H), 2.51(dd, J = 12.2; 4.8 Hz, 1H), 2.21–2.05(m, 1H), 1.51–1.33 (m, 9H), 1.06 – 0.84 (m, 12H) ppm.
13C NMR (75 MHz, CDCl3): δ 172.91, 171.38, 170.09, 159.11, 157.88, 157.30, 153.83, 152.33, 136.34, 129.46, 129.30, 128.56, 128.44, 127.12, 126.85, 96.33, 87.21, 85.98, 58.09, 53.31, 52.77, 52.36, 50.95, 49.64, 49.00, 48.59, 41.05, 40.91, 30.83, 30.62, 29.69, 28.35, 24.84, 22.81, 22.71, 19.18, 17.94 ppm.
HRMS (ESI) [M + Na]+ calculated for C31H46N6O7Na: 637.3326, found: 637.3332.
NH-Boc-Phe-(+)-Δ2-isox-Leu-Val-CONH2 (9) –Mixture of two conformers-
1H NMR (300 MHz, CDCl3): δ 7.33–7.05 (m, 6H) 7.04–6.91 (m, 1H), 6.70 (d, J = 8.5 Hz, 1H), 6.13–5.91 (m, 2H), 5.46 (d, J = 8.5 Hz, 1H), 5.36–5.10 (m, 2H), 4.87–4.71 (m, 1H), 4.67–4.43 (m, 1H), 4.38–4.23 (m, 2H), 4.23–3.69 (m, 4H), 3.69–3.41 (m, 1H), 3.21 (dd, J = 14.3, J = 5.0 Hz, 1H), 3.10–2.79 (m, 2H), 2.54 (dd, J = 14.3, J = 5.0 Hz, 1H), 2.30–1.99 (m, 1H), 1.89–1.55 (m, 3H), 1.49 (m, 9H), 1.31 (m, 12 H) ppm.
13C NMR (75 MHz, CDCl3): δ 173.14, 172.90, 171.92, 171.23, 170.66, 170.49, 159.68, 158.86, 155.92, 155.18, 153.77. 153.59, 136.00, 129.63, 129.43, 129.15, 128.60, 128.41, 127.14, 127.05, 86.96, 85.53, 79.99, 79.88, 58.23, 57.68, 54.13, 53.47, 52.97, 52.19, 51.05, 48.82, 41.04, 40.85, 39.32, 30.88, 30.66, 28.45, 28.30, 25.05, 24.81, 22.91, 22.01, 21.51, 19.28, 17.98, 17.84 ppm.
HRMS (ESI) [M + Na]+ calculated for C31H46N6O7Na: 637.3326, found: 637.3332.
NH-Boc-Phe-Gly-(+)-Δ2-isox-Leu-Val-CONH2 (10)
1H NMR (300 MHz, CD3CN): δ 7.47 (d, J = 6.9 Hz, 1H), 7.30–7.26 (m, 6H), 6.95 (d, J = 8.5 Hz, 1H), 6.72 (bs, 1H), 6.42 (bs, 1H), 5.63 (bs, 1H), 5.47–5.31 (m, 1H), 4.52–4.47 (m, 1H), 4.40–3.39 (m, 8H), 3.23–3.07 (m, 1H), 2.70–2.93 (m, 1H), 2.14–2.08 (m, 1H), 1.78–1.55 (m, 3H), 1.35 (s, 9H), 1.02–0.77 (m, 12H) ppm.
13C NMR (75 MHz, CD3CN): δ 173.43, 171.63, 167.09, 159.52, 154.34, 137.64, 129.27, 128.31, 126.53, 109.99, 87.41, 86.02, 79.09, 58.06, 56.82, 55.76, 53.67, 52.65, 51.39, 49.79, 48.71, 41.62, 40.43, 40.10, 37.60, 31.51, 30.55, 29.34, 27.49, 24.77, 22.27, 20.81, 18.86, 17.21, 16.63 ppm.
HRMS (ESI) [M + Na]+ calculated for C33H49N7O8Na: 694.3540, found: 694.3539.
General Procedure for N-Boc-Deprotection
N-Boc protected compound (1 equiv.) was dissolved in CH2Cl2 (0.01M) and the solution cooled at 0°C. TFA (50% v/v) was added dropwise, the solution was warmed up at room temperature and was stirred for 2 h. The solvent was removed under vacuum with the obtainment of the trifluoroacetate salt that was directly used in the next coupling step.
NH-(–)-Δ2-isox-Leu-Val-CONH2 (6)
1H NMR (300 MHz, CD3OD): δ 7.90 (d, J = 8.7 Hz, 1H), 5.53 (dd, J = 9.5, 4.9 Hz, 1H), 4.54 (dd, J = 9.8, 4.9 Hz, 1H), 4.42 (t, J = 8.8 Hz, 1H), 4.26 – 4.17 (m, 1H), 3.90 – 3.84 (m, 1H), 3.89 – 3.75 (m, 2H), 3.58 – 3.49 (m, 2H), 3.22 (dd, J = 5.9, 4.1 Hz, 1H), 2.05 (dq, J = 13.6, 6.8 Hz, 1H), 1.80 – 1.59 (m, 3H), 1.05 – 0.82 (m, 12H) ppm.
13C NMR (75 MHz, CD3OD): δ 147.47, 172.82, 159.52, 152.51, 85.48, 58.22, 52.96, 52.21, 50.55, 49.27, 40.31, 30.68, 24.54, 21.98, 20.47, 18.32, 17.08 ppm.
HRMS (ESI) [M + Na]+ calculated for C17H29N5O4Na: 390.2117, found: 390.2111.
NH-(+)-Δ2-isox-Leu-Val-CONH2 (7)
1H NMR (300 MHz, CD3OD); δ 7.97 (d, J = 8.6 Hz, 1H), 5.53 (dd, J = 9.5, 4.9 Hz, 1H), 4.55 (m, 1H), 4.39 (m, 1H), 4.26 – 4.16 (m, 1H), 3.90 – 3.72 (m, 2H), 3.58 – 3.43 (m, 2H), 3.24 – 3.16 (m, 1H), 2.05 (m, 1H), 1.75 – 1.58 (m, 3H), 1.02 – 0.81 (m, 12H) ppm.
13C NMR (75 MHz, CD3OD): δ 147.47, 172.99, 159.40, 152.40, 128.72, 85.21, 58.05, 53.12, 52.17, 50.66, 49.14, 40.21, 30.91, 29.36, 24.84, 21.99, 20.25, 18.26, 16.90 ppm.
HRMS (ESI) [M + Na]+ calculated for C17H29N5O4Na: 390.2117, found: 390.2111.
Molecular Modeling
Compound 9 and 10 were modeled in explicit solvent with periodic boundary conditions using metadynamics and restrained molecular dynamics simulations. For each of the compounds two isomers (cis and trans with respect to the amide bond) were considered, resulting in a set of four peptides. Such molecules were solvated in a cubic box of 4 nm with chloroform (compound 8) and acetonitrile (compound 10) to reproduce the NMR experiment conditions. Every system has been submitted to geometry optimization with the steepest descent algorithm with a convergence of 100 kJ mol−1 nm−1. Then we performed a 1 ns equilibration in NVT conditions at 300 K, followed by a 1 ns NPT equilibration at the 1 bar and at the same temperature. After the equilibration phase, we ran a 50 ns Well Tempered Metadynamics (WTMD) with a 8.0 kJ mol−1 bias-factor, at a reference temperature of 300 K. To study the open and closed state of both peptides, we selected as collective variable (CV) the distance between CβVAL and the N-tert-butoxy carbonyl (N-Boc) quaternary carbon. A Gaussian hill with σ = 0.1 Å and an initial height of 1 kJ mol−1 was added once every 100 steps. The simulation was sped up by saving the Gaussian hills on a grid that ranged from 0 to 16 Å and spaced 0.02 Å. Compound 10 was also simulated using a 50 ns restrained molecular dynamics. Special potentials are used for imposing restraints on the motion of the system, to include knowledge from experimental data such as the NMR derived interatomic distances. Distance restraints add a penalty to the potential when the distance between specified pairs of atoms exceeds a threshold value (Abraham et al., 2014). The threshold values used in our simulation are reported in the Supplementary Materials.
The restraints are applied in a progressive way to give time to the peptide to relax, using a τ equal to 500 ps. Since we are using multiple distance restraints, they are not necessary satisfied at each simulation step but as a time average. The atoms restrained and the relative distances are shown in Table S1. Compound 9 and 10 have been described using the Amber99SB-ILDN Force Field (Hornak et al., 2006). 4,5,6,6a-Tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-3-carboxylic acid residues were parameterized according to standard procedures as explained in reference (Gandini et al., 2018). During molecular dynamics (MD) and metadynamics simulations, temperature was held constant with the v-rescale algorithm (Bussi et al., 2007), while pressure was kept constant through the Berendsen barostat (Berendsen et al., 1984). MD simulations were performed using the leap-frog algorithm with 2 fs time-step, with holonomic constraints on every bond enforced using the LINCS algorithm. Simulations and subsequent analysis were performed with the GROMACS 5.0.4 (Van Der Spoel et al., 2005) program suite, while metadynamics was run along with PLUMED version 2.2.2 (Tribello et al., 2014).
Results and Discussion
The two enantiomers of compound 1 were obtained starting from the corresponding ethyl esters recently described by us (Tamborini et al., 2015). Racemate (±)-2 (Conti et al., 2006) was synthesized in a flow chemistry reactor exploiting the 1,3-dipolar cycloaddition reaction of N-Boc-3-pyrroline with ethoxycarbonyl formonitrile oxide. An excellent enantiomeric separation (e.e. >99%) of the racemate was achieved by semi-preparative chiral HPLC. Alkaline hydrolysis of (–)-(3aS6aR)-2 and (+)-(3aR6aS)-2 gave the desired (–)-(3aS6aR)-1 and (+)-(3aR6aS)-1, respectively (Scheme 1).
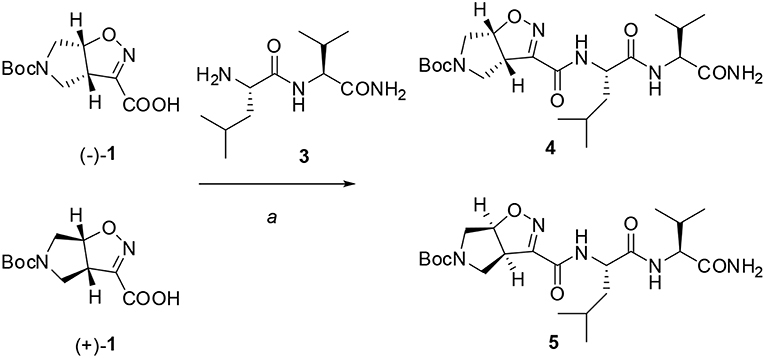
The ability of compound 1 to stabilize secondary structures was evaluated in model peptides containing (L)-Phe at N-terminus and (L)-Leu-(L)-Val sequence at C-terminus and (Schemes 2, 3). This last dipeptide was chosen as it normally adopts extended conformation in solution. Compound 1 was used in both the 3aS6aR and 3aR6aS stereochemistries [(–)-1 and (+)-1, respectively], as a different effect on peptide conformations could be expected depending on the stereochemistry of the unnatural amino acids (Pellegrino et al., 2012).
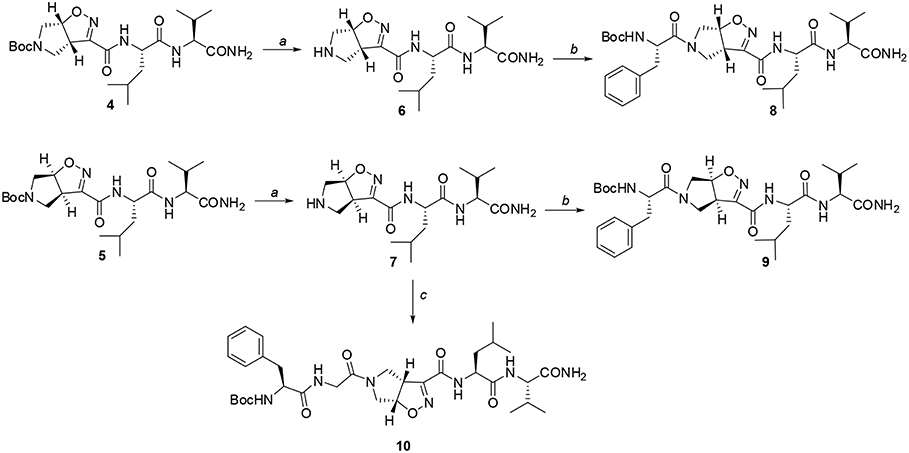
Scheme 3. Reagents and conditions: a) TFA, CH2Cl2 from 0°C to r.t, 2 h; b) EDC/HOBt/DIEA, N-Boc-(L)-Phe, CH2Cl2 from 0°C to r.t, 24 h; c) EDC/HOBt/DIEA, N-Boc-(L)-Phe-Gly-OH, CH2Cl2 from 0°C to r.t, 24 h.
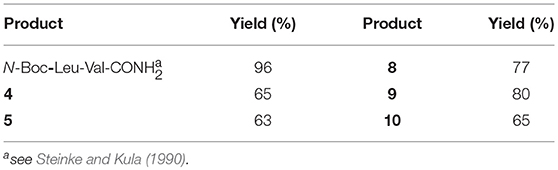
Firstly, Leu-Val dipeptide 3 was prepared starting from (L)-Valinamide and NH-Boc-(L)-Leu, according to the general coupling procedure, followed by N-terminus deprotection in TFA (91% overall yield). Diastereoisomeric dipeptides 4 and 5 were then achieved in good yields (65% and 63%, respectively) by a coupling reaction of (–)-1 or (+)-1 with dipeptide 3 (Scheme 2).
Both 4 and 5 compounds were completely characterized by NMR spectroscopy (CDCl3, see SI for complete proton assignment). In both tripeptides, no significant Noesy effects between Leu-Val dipeptide and the scaffold were detected. Furthermore, in variable temperature experiments, the obtained Δδ/ΔT is higher than 5 ppb/K for all the amide protons, indicating the absence of H-bonds stabilizing a particular conformation. The JNH−CHα value for NH-Val is of 8.8 Hz and 7.9 Hz, for 4 and 5, respectively (NH-Leu appears as a broad signal for each compound). From these findings, we hypothesized that both 4 and 5 adopt an extended conformation and that the isoxazoline scaffold is not interacting with Leu-Val dipeptide.
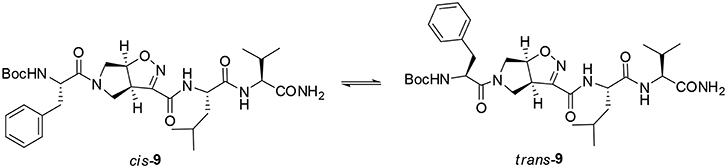
The peptide chain was then elongated at N-terminus, through Boc-deprotection and coupling with (L)-Phe. Diastereoisomeric dipeptides 8 and 9 were achieved in good overall yields (73% and 76%, respectively) (Scheme 3). From NMR studies, it was found that both model peptides 8 and 9 are present as a mixture of conformers in 2:1 ratio in CDCl3 solution. The presence of these two conformers is probably due to the low-energy barrier cis/trans isomerization of the tertiary amide on the pyrrolidine as it is frequently observed on the acylated proline and in the case of tertiary cyclic amides (Laursen et al., 2013; Pellegrino et al., 2014). Regarding compound 8, this rotation led to a splitting of the (L)-Phe proton signals only. Furthermore, no significative Noesy effects were detected, and, in variable temperature experiments, the obtained Δδ/ΔT coefficient is high for all the amide protons. Taking all these data together, we can assume that Phe and Leu-Val dipeptide are oriented in opposite directions and that the 3aS6aR stereochemistry of the scaffold is not effective in inducing specific secondary structures when used in combination with (L)-α-amino acids.
A different scenario was observed for compound 9. Its two rotamers are indeed characterized by different chemical shifts, suggesting that the isomerization of the tertiary amide leads to two different structures conformations influencing the entire molecule. Unfortunately, due to several overlapping signals, it was not possible to assessing significative Noesy proximities. Variable temperature experiments were thus done, in order to investigate if the NH were involved in hydrogen bonds and, as a consequence, if the two isomers of 9 were characterized by a stable conformation in solution. In the case of the major isomer, the obtained Δδ/ΔT is around 4 ppb/K for NH-Phe, indicating that this proton could be involved in a weak/medium hydrogen bond. All the other amide protons had higher Δδ/ΔT. On the other hand, in the minor conformer the obtained Δδ/ΔT is of around 2 ppb/K for NH-Val, indicating that this proton is involved in a strong hydrogen bond. Metadynamic studies were then performed on the cis/trans tertiary amide conformers of compound 9 (Figure 2).
In particular, we performed two 50 ns long metadynamics simulations, using the distance between Boc quaternary carbon and CβVal as collective variable (CV). This geometric parameter was selected as a suitable CV because it discriminates well between closed and extended states of the peptide. In this way, it could be possible to evaluate if the two conformers had an intrinsic tendency toward turn conformation. The cis isomer showed a broad free energy minimum corresponding to CV values between 5 and 8 Å, while the trans isomer showed much higher free energy values in this region, exhibiting a shallow minimum around 10–15 Å (Figure 3).
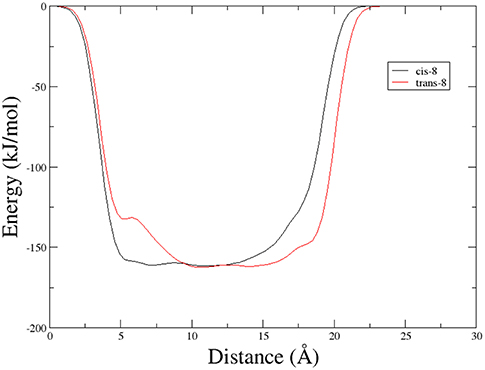
Figure 3. Potential energy as a function of distance between Boc quaternary carbon and CβVal obtained from Metadynamics for the two isomers of compound 9 in chloroform.
This different behavior could be indicative of a preference of cis-9 toward a more closed conformation, although from these data it was not possible to make any further assumptions. For this reason, we envisaged that the introduction of a Gly residue between Phe and isoxazoline scaffold could increase the conformational flexibility of the N-terminus peptide arm favoring its interaction with the C-teminus part of the molecule. Compound 7 was thus elongated at N-terminus, through coupling with N-Boc-(L)-Phe-Gly-OH dipeptide using general coupling reaction conditions to obtain peptide 10 (65%, Scheme 3). The NMR study on 10 (CD3CN, see SI for complete assignment), showed that it is present in solution as a single stable conformer (only trace amounts of a second isomer were detected). A complete set of NHi-NHi+1 Noesy proximities, whose calculated distances ranged from 2.68 to 2.96 Å, were observed (Figure 4 on the top right). Furthermore, long range Noesy effects involving HαGly and NHVal (3.03 Å) and HβLeu (2.73 Å) were found (Figure 4 on the bottom).
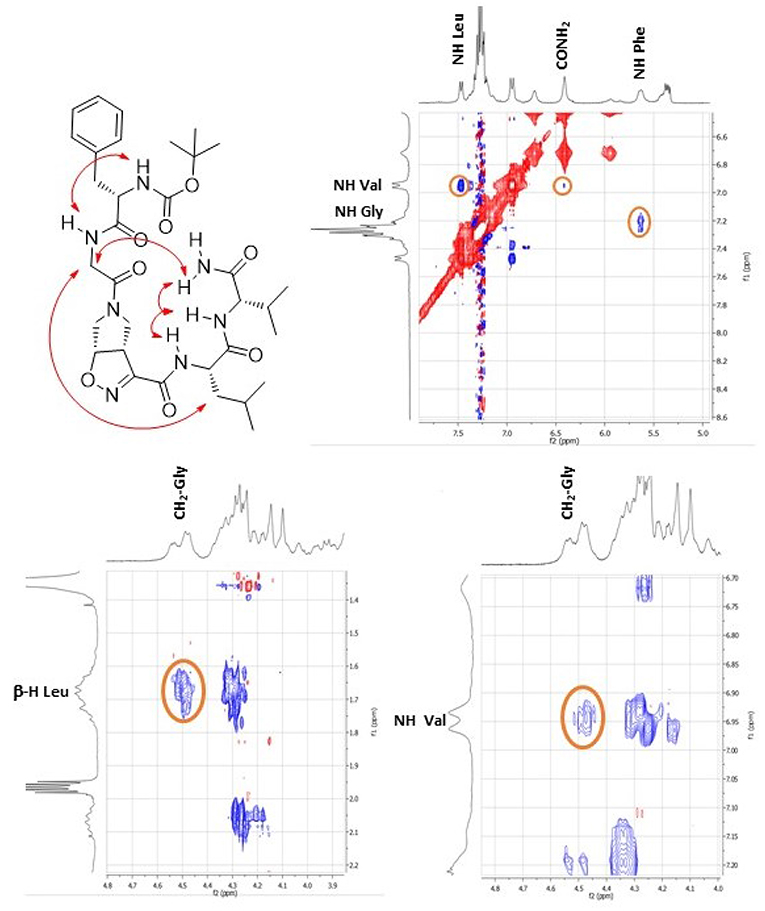
Figure 4. Noesy proximity (red) observed for compound 10 (CD3CN). On the top right: zoom of the amide region. On the bottom left: Zoom of the aliphatic region. On the bottom right: zoom of the NH-aliphatic region.
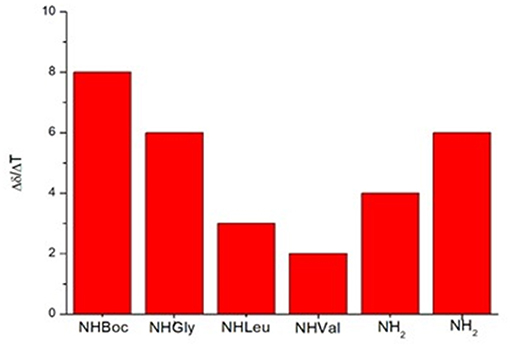
In variable temperature experiments (Figure 5), the obtained Δδ/ΔT is of around 2 ppb/K for NH-Val, while NH-Leu and one of the C-terminus amide protons possess Δδ/ΔT of around 3 and 4 ppb/K, respectively. The presence of hydrogen bonds was also confirmed by FT-IR analysis. N-H stretching bands A and B (3,500–3,000 cm−1region, Figure S20) are indeed downshifted as frequently observed in intramolecular hydrogen bonded conformations (Tonan and Ikawa, 1996; Barth, 2007). Furthermore, the deconvolution of the amide I band (1,700–1,600 cm−1) showed the presence of a band at 1,655 cm−1 (Figure S20), typical of α structures (Kong and Yu, 2007).
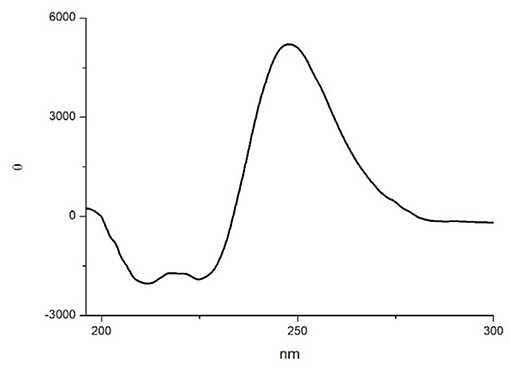
In order to gain more information on the conformation of compound 10, far-UV circular dichroism (CD) analysis in CH3CN (0.1 mM solution) was then performed. In Figure 6 the obtained CD spectrum is reported.
It showed a maximum at around 250 nm due to the strong absorption of the isoxazoline ring, as observed by Memeo et al. (2018) on similar compounds (see also the UV spectrum in the SI). In the amide bond region, two negative minima at 225 and 208 nm, with the same intensity, and a slightly positive band at 200 nm were also observed. The presence of the exciton splitting of the π → π* transition band suggested that a α-turn conformation was present (Wang et al., 2018), although in short peptides it is difficult to correctly evaluate the intensity of Cotton effects with respect to secondary structure motifs (Chin et al., 2002; Wang et al., 2018).
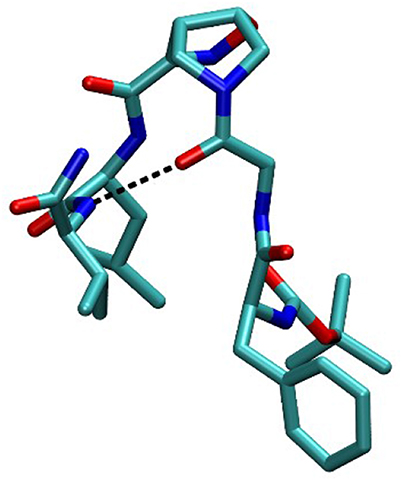
In order to completely elucidate the conformation of peptide 10, a restrained molecular dynamic was finally performed. The restraints were based on the obtained NOESY values. When the hydrogen was not uniquely defined, like in HαGly and HβLeu, we used the closest carbon, e.g., CαGly and CβLeu to implement distance restrains (Figure S3). The analysis showed that CαGly-CβLeu (A) and CαGly-HNHVal (B) are mutually exclusive in the trans isomer but not in the cis one as shown in Figure S1. Furthermore, H-bond analysis resulted in the observation that a H-bond is between CO-Gly and NH-Val. In Figure 7 the most representative structure is reported.
Taking together both experimental and molecular modeling data, we can assume that, in solution, compound 10 effectively adopts a α-turn conformation. This motif is normally formed by 5 α-amino acids and is characterized by a H-bond between i and i+4 residues. In our case, the isoxazoline scaffold replaces two of the three core amino acids (Figure 1) and the H-bond is formed by the CO-Gly and NH-Val, leading to the formation of a 12 member pseudo-cycle. Furthermore, the overall structure of the peptidomimetic is reinforced by medium-weak H-bonds involving NH-Leu and one C-terminus-NH2, as evicted from NMR data. The presence of a α-turn conformation is also confirmed by the computational analysis of the dihedral angles of the residue i+3 and of the distances between residues i and i+4 (Table S9), as their values rely within the α-turn parameters (Pavone et al., 1996).
In conclusion, as α-turns are often found on biologically active sites and few molecules have been reported to mimic or stabilize them, the ability of compound (+)-1 to stabilize α-turn conformation on isolated peptide is particularly important in view of future biological applications.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files.
Author Contributions
FO and RB equally contributed to this work. The project was conceived by SaP, LT, and AP. SaP designed and coordinated the research. LT and RB chemically synthesized and analyzed the materials, RB performed NMR, FT-IR and CD experiments, FO and StP performed molecular modeling. SaP, StP, LT, and AP analyzed and compiled the data and co-wrote the manuscript. The final manuscript was read and approved by all the authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Donatella Nava for the NMR analysis and Michele Consonni and Sabrina Giofrè for practical help in the synthesis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2019.00133/full#supplementary-material
Abbreviations
ACN, Acetonitrile; Boc, tert-butyloxycarbonyl; DIEA, N, N-Diisopropylethylamine; DMF, Dimethylformamide; EDC, 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide; Gly, Glycine; HOBt, Hydroxybenzotriazole; Leu, Leucine; Phe, Phenylalanine; TFA, Trifluoroacetic acid; Val, Valine.
References
Abraham, M., Van Der Spoel, D., Lindahl, E., and Hess, B. (2014). GROMACS User Manual Version 5.0.4. Available online at: https://ftp.gromacs.org/pub/manual/manual-5.0.4.pdf
Barth, A. (2007). Infrared spectroscopy of proteins. BBA Bioenerg. 1767, 1073–1101. doi: 10.1016/j.bbabio.2007.06.004
Berendsen, H. J. C., Postma, J. P. M., van Gusteren, W. F., and DiNola, A. (1984). Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690. doi: 10.1063/1.448118
Bonetti, A., Pellegrino, S., Das, P., Yuran, S., Bucci, R. Ferri, N., et al. (2015). Dipeptide Nanotubes Containing Unnatural Fluorine-Substituted β2,3-Diarylamino Acid and l-Alanine as candidates for biomedical applications. Org. Lett. 17, 4468–4471. doi: 10.1021/acs.orglett.5b02132
Bouillère, F., Thétiot-Laurent, S., Kouklovsky, C., and Alezra, V. (2011). Foldamers containing γ-amino acid residues or their analogues: structural features and applications. Amino Acids 41, 687–707. doi: 10.1007/s00726-011-0893-3
Bruno, S., Pinto, A., Paredi, G., Tamborini, L., De Micheli, C., La Pietra, V., et al. (2014). Discovery of covalent inhibitors of glyceraldehyde-3-phosphate dehydrogenase, a target for the treatment of malaria. J. Med. Chem. 57, 7465–7471. doi: 10.1021/jm500747h
Bucci, R., Das, P., Iannuzzi, F., Feligioni, M., Gandolfi, R., Gelmi, M. L., et al. (2017). Self-assembly of an amphipathic ααβ-tripeptide into cationic spherical particles for intracellular delivery. Org. Biomol, Chem. 15, 6773–6779. doi: 10.1039/C7OB01693J
Bucci, R., Giofré, S., Clerici, F., Contini, A., Pinto, A., Erba, E., et al. (2018). Tetrahydro-4H-(pyrrolo[3,4-d]isoxazol-3-yl)methanamine: a bicyclic diamino scaffold stabilizing parallel turn conformations. J. Org. Chem. 83, 11493–11501. doi: 10.1021/acs.joc.8b01299
Bussi, G., Donadio, D., and Parrinello, M. (2007). Canonical sampling through velocity rescaling. J. Phys Chem. 126:014101. doi: 10.1063/1.2408420
Castellano, S., Kuck, D., Viviano, M., Yoo, J., López-Vallejo, F., Conti, P., et al. (2011). Synthesis and biochemical evaluation of δ 2-isoxazoline derivatives as DNA methyltransferase 1 inhibitors. J. Med. Chem. 54, 7663–7677. doi: 10.1021/jm2010404
Chatterjee, S., Vasudev, P. G., Raghothama, S., Ramakrishnan, C., Shamala, N., and Balaram, P. (2009). Expanding the peptide β-turn in αγ hybrid sequences: 12 atom hydrogen bonded helical and hairpin turns. J. Am. Chem. Soc. 131, 5956–5965. doi: 10.1021/ja900618h
Chin, D.-H., Woody, R. W., Rohl, C. A., and Baldwin, R. L. (2002). Circular dichroism spectra of short, fixed-nucleus alanine helices. Proc. Natl. Acad. Sci. U.S.A. 99, 15416–15421. doi: 10.1073/pnas.232591399
Clerici, F., Erba, E., Gelmi, M. L., and Pellegrino, S. (2016). Non-standard amino acids and peptides: from self-assembly to nanomaterials Tetrahedron Lett. 57, 5540–5550. doi: 10.1016/j.tetlet.2016.11.022
Conti, P., De Amici, M., Pinto, A., Tamborini, L., Grazioso, G., Frolund, B., et al. (2006). Synthesis of 3-hydroxy- and 3-carboxy-Δ2-isoxazoline amino acids and evaluation of their interaction with GABA receptors and transporters. Eur. J. Org. Chem. 24, 5533–5542. doi: 10.1002/ejoc.200600628
Conti, P., Pinto, A., Wong, P. E., Major, L. L., Tamborini, L., Iannuzzi, M. C., et al. (2011). Synthesis and in vitro/in vivo evaluation of the antitrypanosomal activity of 3-bromoacivicin, a potent CTP synthetase inhibitor. ChemMedChem 6, 329–333. doi: 10.1002/cmdc.201000417
Fanelli, R., Berthomieu, D., Didierjean, C., Doudouh, A., Lebrun, A., Martinez, J., et al. (2017). Hydrophobic α,α-disubstituted disilylated TESDpg induces incipient 310-Helix in short tripeptide sequence. Org. Lett. 19, 2937–2940. doi: 10.1021/acs.orglett.7b01172
Fuller, A., Chen, B., Minter, A. R., and Mapp, A. K. (2005). Succinct synthesis of β-amino acids via chiral isoxazolines. J. Am. Chem. Soc. 127, 5376–5383. doi: 10.1021/ja0431713
Gandini, E., Dapiaggi, F., Oliva, F., Pieraccini, S., and Sironi, M. (2018) Well-Tempered MetaDynamics based method to evaluate universal peptidomimetics. Chem. Phys. Lett. 706, 729–735. doi: 10.1016/j.cplett.2018.07.029
Guo, L., Zhang, W., Guzei, I. A., Spencer, L. C., and Gellman, S. H. (2012). New preorganized γ-amino acids as foldamer building blocks. Org. Lett. 14, 2582–2585. doi: 10.1021/ol3008815
Hoang, H. N., Driver, R. W., Beyer, R. L., Malde, A. K., Le, G. T., Abbenante, G, et al (2011). Protein α-turns recreated in structurally stable small molecules. Angew Chem. Int. Ed. 50, 11107–11111. doi: 10.1002/anie.201105119
Hornak, V., Abel, R., Okur, A., Strockbine, B., Roitberg, A., and Simmerling, C. (2006). Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725. doi: 10.1002/prot.21123
Itoh, T., Watanabe, M., and Fukuyama, T. (2002). Synthetic approach to tetrodotoxin. Synlett 8, 1323–1325. doi: 10.1055/s-2002-32984
Kaur, K., Kumar, V., Sharma, A. K., and Gupta, G. K. (2014). Isoxazoline containing natural products as anticancer agents: a review. Eur. J. Med. Chem. 77, 121–133. doi: 10.1016/j.ejmech.2014.02.063
Kelso, M. J., Beyer, R. L., Hoang, H. N., Lakdawala, A. S., Snyder, J. P., Oliver, W. V., et al. (2004). Alpha-turn mimetics: short peptide alpha-helices composed of cyclic metallopentapeptide modules. J Am. Chem. Soc. 126, 4828–4842. doi: 10.1021/ja037980i
Kobayashi, H., Misawa, T. Matsuno, K., and Demizu, Y. (2017). Preorganized cyclic α,α-disubstituted α-amino acids bearing functionalized side chains that act as peptide-helix inducers. J. Org. Chem. 82, 10722–10726. doi: 10.1021/acs.joc.7b01946
Konda, M., Jadhav, R. G., Maiti, S., Mobin, S. M., Kauffmannb, B., and Das, A. K. (2018). Understanding the conformational analysis of gababutin based hybrid peptides. Org. Biomol. Chem. 16, 1728–1735. doi: 10.1039/C8OB00035B
Kong, J., and Yu, S. (2007). Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sinica. 39, 549–559. doi: 10.1111/j.1745-7270.2007.00320.x
Kozikowski, A. P., and Park, P. U. (1990). Synthesis of streptazolin: use of the aza-Ferrier reaction in conjunction with the INOC process to deliver a unique but sensitive natural product J. Org. Chem. 55, 4668–4682. doi: 10.1021/jo00302a036
Krishna, Y., Sharma, S., Ampapathi, R. S., and Koley, D. (2014). Furan-based locked Z-vinylogous γ-amino acid stabilizing protein α-turn in water-soluble cyclic α3γ tetrapeptides. Org. Lett. 16, 2084–2087. doi: 10.1021/ol5002126
Laursen, J. S., Engel-Andreasen, J., Fristrup, P., Harris, P., and Olsen, C. A. (2013). Cis–trans amide bond rotamers in β-peptoids and peptoids: evaluation of stereoelectronic effects in backbone and side chains. J. Am. Chem. Soc. 135, 2835–2844. doi: 10.1021/ja312532x
López-Andarias, A., López-Andarias, J., Atienza, C., Chichón, F. J., Carrascosa, J. L., and Martín, N. (2018). Tuning optoelectronic and chiroptic properties of peptide-based materials by controlling the pathway complexity. Chem. Eur. J. 24, 7755–7760. doi: 10.1002/chem.201801238
Machetti, F., Ferrali, A., Menchi, G., Occhiato, E. G., and Guarna, A. (2000). Oligomers of enantiopure bicyclic γ/δ-amino acids (BTAa). 1. Synthesis and conformational analysis of 3-aza-6,8-dioxabicyclo[3.2.1]octane-7-carboxylic acid oligomers (PolyBTG). Org. Lett. 2, 3987–3990. doi: 10.1021/ol006548s
Memeo, M. G., Bruschi, M., Bergonzi, L., Desimoni, G., Faita, G., and Quadrelli, P. (2018). Cyclopenta[d]isoxazoline β-turn mimics: synthetic approach, turn driving force, scope, and limitations. ACS Omega 3, 13551–13558. doi: 10.1021/acsomega.8b01670
Mikhalevich, V., Craciun, I., Kyropoulou, M., Palivan, C. G., and Meier, W. (2017). Amphiphilic peptide self-assembly: expansion to hybrid materials. Biomacromolecules 18, 3471–3480. doi: 10.1021/acs.biomac.7b00764
Pavone, V., Gaeta, G., Lombardi, A., Nastri, F., Maglio, O., Isernia, C., et al. (1996). Discovering protein secondary structures: classification and description of isolated α-turns. Biopolymers 38, 705–721. doi: 10.1002/(SICI)1097-0282(199606)38:6<705::AID-BIP3>3.0.CO;2-V
Pellegrino, S., Bonetti, A., Clerici, F., Contini, A., Moretto, A., Soave, R., et al. (2015). 1H-azepine-2-oxo-5-amino-5-carboxylic acid: a 310 helix inducer and an effective tool for functionalized gold-nanoparticles. J. Org. Chem, 80, 5507–5516. doi: 10.1021/acs.joc.5b00396
Pellegrino, S., Contini, A., Clerici, F., Gori, A., Nava, D., and Gelmi, M. L. (2012). 1H-Azepine-4-amino-4-carboxylic acid: a new α,α disubstituted ornithine analogue capable of inducing helix conformations in short Ala-Aib pentapeptides. Chem. Eur. J. 18, 8705–8715. doi: 10.1002/chem.201104023
Pellegrino, S., Contini, A., Gelmi, M. L., Lo Presti, L., Soave, R., and Erba, E. (2014). Asymmetric modular synthesis of a semirigid dipeptide mimetic by cascade cycloaddition/ring rearrangement and borohydride reduction. J. Org. Chem, 79, 3094–3102. doi: 10.1021/jo500237j
Pellegrino, S., Facchetti, G., Contini, A., Gelmi, M. L., Erba, E., Gandolfi, R., et al. (2016). Ctr-1 Mets7 motif inspiring new peptide ligands for Cu(I)-catalyzed asymmetric Henry reaction under green conditions. RSC Adv. 6, 71529–71533. doi: 10.1039/C6RA16255J
Pellegrino, S., Tonali, N., Erba, E., Kaffy, J., Taverna, M., Contini, A., et al. (2017). ß-Hairpin mimics containing a piperidine-pyrrolidine scaffold modulate the ß-amyloid aggregation process preserving the monomer species. Chem. Sci. 8, 1295–1302. doi: 10.1039/C6SC03176E
Pinto, A., Conti, P., Grazioso, G., Tamborini, L., Madsen, U., Nielsen, B., et al. (2011). Synthesis of new isoxazoline-based acidic amino acids and investigation of their affinity and selectivity profile at ionotropic glutamate receptors. Eur. J. Med.Chem. 46, 787–793. doi: 10.1016/j.ejmech.2010.12.020
Pinto, A., Tamborini, L., Cullia, G., Conti, P., and De Micheli, C. (2016a). Inspired by nature: the 3-halo-4,5-dihydroisoxazole moiety as a novel molecular warhead for the design of covalent inhibitors. ChemMedChem 1, 10–14. doi: 10.1002/cmdc.201500496
Pinto, A., Tamborini, L., Pennacchietti, E., Coluccia, A., Silvestri, R., Cullia, G., et al. (2016b). Bicyclic γ-amino acids as inhibitors of γ-aminobutyrate aminotransferase. J. Enzyme Inhib. Med. Chem. 2, 295–301. doi: 10.3109/14756366.2015.1021251
Raymond, D. M., and Nilsson, B. L. (2018). Multicomponent peptide assemblies. Chem. Soc. Rev. 47, 3659–3720. doi: 10.1039/C8CS00115D
Smith, E. L., and Spackman, D. H. (1955). Leucine aminopeptidase. V. activation, specificity, and mechanism of action. J. Biol. Chem. 122, 271–299.
Sohora, M., Vazdar, M., Sović, I., Mlinarić-Majerski, K., and Basarić, N. (2018). Photocyclization of tetra- and pentapeptides containing adamantylphthalimide and phenylalanines: reaction efficiency and diastereoselectivity. J. Org. Chem. 83, 14905–14922. doi: 10.1021/acs.joc.8b01785
Solomon, L. A., Kronenberg, J. B., and Fry, H. C. (2017). Control of heme coordination and catalytic activity by conformational changes in peptide–amphiphile assemblies. J. Am. Chem. Soc. 139, 8497–8507. doi: 10.1021/jacs.7b01588
Sperry, J. B., and Wright, D. L. (2005). Furans, thiophenes and related heterocycles in drug discovery. Curr. Opin. Drug Discov. Dev. 8, 723–740. doi: 10.1002/chin.200615242
Steinke, D., and Kula, M. R. (1990). Selektive desamidierung von peptidamiden. Angew Chemie 102, 1204–1206. doi: 10.1002/ange.19901021035
Tamborini, L., Mastronardi, F., Dall'Oglio, F., De Micheli, C., Nielsen, B., Lo Presti, L., et al. (2015). Synthesis of unusual isoxazoline containing β and γ-dipeptides as potential glutamate receptor ligands MedChemComm 6, 1260–1266. doi: 10.1039/C5MD00159E
Tonan, K., and Ikawa, S. (1996). Intramolecular hydrogen bonding and conformation of small peptides: variable-temperature FTIR study on N-Acetyl-l-Pro-l-Leu-Gly-NH2 and related compounds. J. Am. Chem Soc. 118, 6960–6965. doi: 10.1021/ja953380a
Tribello, G. A., Bonomi, M., Branduardi, D., Camilloni, C., and Bussi, G. (2014). PLUMED 2: new feathers for an old bird. Comp. Phys. Comm. 185, 604–613. doi: 10.1016/j.cpc.2013.09.018
Tsantali, G. G., Dimtsas, J., Tsoleridis, C. A., and Takakis, I. M. (2007). Preparation of sixteen 3-hydroxy-4- and 7-hydroxy-1-hydrindanones and 3-hydroxy-4- and 8-hydroxy-1-hydroazulenones. Eur. J. Org. Chem. 2007, 258–265. doi: 10.1002/ejoc.200600639
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E,, and Berendsen, H. J. (2005). GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718. doi: 10.1002/jcc.20291
Vasudev, P. G., Chatterjee, S., Shamala, N., and Balaram, P. (2011). Structural chemistry of peptides containing backbone expanded amino acid residues: conformational features of β, γ, and hybrid peptides. Chem. Rev. 111, 657–687. doi: 10.1021/cr100100x
Wang, L., Coric, P., Zhu, K., Liu, W.-Q., Vidal, M., Bouaziz, S., et al. (2018). Synthesis and characterization of water-soluble macrocyclic peptides stabilizing protein α-turn. Org. Biomol. Chem. 16, 459–471. doi: 10.1039/C7OB02852K
Wintjens, R. T., Rooman, M. J., and Wodak, S. J. (1996). Automatic classification and analysis of alpha alpha-turn motifs in proteins. J. Mol. Biol. 255, 235–253. doi: 10.1006/jmbi.1996.0020
Keywords: unnatural γ-amino acids, peptidomimetic, isoxazoline, α-turn, metadynamic studies, conformational analysis
Citation: Oliva F, Bucci R, Tamborini L, Pieraccini S, Pinto A and Pellegrino S (2019) Bicyclic Pyrrolidine-Isoxazoline γ Amino Acid: A Constrained Scaffold for Stabilizing α-Turn Conformation in Isolated Peptides. Front. Chem. 7:133. doi: 10.3389/fchem.2019.00133
Received: 29 November 2018; Accepted: 20 February 2019;
Published: 18 March 2019.
Edited by:
Jutta Eichler, Friedrich-Alexander-Universität Erlangen-Nürnberg, GermanyCopyright © 2019 Oliva, Bucci, Tamborini, Pieraccini, Pinto and Pellegrino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefano Pieraccini, c3RlZmFuby5waWVyYWNjaW5pQHVudW5pbWkuaXQ=
Andrea Pinto, YW5kcmVhLnBpbnRvQHVuaW1pLml0
Sara Pellegrino, c2FyYS5wZWxsZWdyaW5vQHVuaW1pLml0
 Francesco Oliva
Francesco Oliva Raffaella Bucci2
Raffaella Bucci2 Andrea Pinto
Andrea Pinto Sara Pellegrino
Sara Pellegrino