- 1Pharmacognosy Department, Faculty of Pharmacy, Cairo University, Cairo, Egypt
- 2Pharmacognosy Department, Faculty of Pharmacy, Beni-Suef University, Beni-Suef, Egypt
- 3Pharmacognosy Department, Faculty of Pharmacy, Nahda University, Beni-Suef, Egypt
- 4Faculty of Pharmacy, The University of Jordan, Amman, Jordan
- 5College of Pharmacy, Alfaisal University, Riyadh, Saudi Arabia
- 6School of Computing, Engineering and Physical Sciences, University of the West of Scotland, Paisley, United Kingdom
- 7Marine Biodiscovery Centre, University of Aberdeen, Aberdeen, United Kingdom
- 8Marine Invertebrates, National Institute of Oceanography and Fisheries, Red Sea Branch, Hurghada, Egypt
- 9Department of Biochemistry, Faculty of Science, Center for Science and Medical Research, University of Jeddah, Jeddah, Saudi Arabia
- 10Department of Chemistry and Center for Integrated Protein Science Munich (CIPSM), Biosystems Chemistry, Technical University of Munich, Garching, Germany
- 11Chair of Technical Biochemistry, Technische Universität Dresden, Dresden, Germany
- 12Department of Pharmacognosy, Faculty of Pharmacy, Minia University, Minia, Egypt
Saccharomonospora sp. UR22 and Dietzia sp. UR66, two actinomycetes derived from the Red Sea sponge Callyspongia siphonella, were co-cultured and the induced metabolites were monitored by HPLC-DAD and TLC. Saccharomonosporine A (1), a novel brominated oxo-indole alkaloid, convolutamydine F (2) along with other three known induced metabolites (3-5) were isolated from the EtOAc extract of Saccharomonospora sp. UR22 and Dietzia sp. UR66 co-culture. Additionally, axenic culture of Saccharomonospora sp. UR22 led to isolation of six known microbial metabolites (6-11). A kinase inhibition assay results showed that compounds 1 and 3 were potent Pim-1 kinase inhibitors with an IC50 value of 0.3 ± 0.02 and 0.95 ± 0.01 μM, respectively. Docking studies revealed the binding mode of compounds 1 and 3 in the ATP pocket of Pim-1 kinase. Testing of compounds 1 and 3 displayed significant antiproliferative activity against the human colon adenocarcinoma HT-29, (IC50 3.6 and 3.7 μM, respectively) and the human promyelocytic leukemia HL-60, (IC50 2.8 and 4.2 μM, respectively). These results suggested that compounds 1 and 3 act as potential Pim-1 kinase inhibitors that mediate the tumor cell growth inhibitory effect. This study highlighted the co-cultivation approach as an effective strategy to increase the chemical diversity of the secondary metabolites hidden in the genomes of the marine actinomycetes.
Introduction
Marine sponge-associated microorganisms have been proved an essential source of biologically active natural products (Thomas et al., 2010; Roue et al., 2012; Abdelmohsen et al., 2014a). Large numbers of secondary metabolites with novel molecular scaffolds and diverse biological activities including antimicrobial (Hentschel et al., 2001; Eltamany et al., 2014), anti-parasitic (Abdelmohsen et al., 2014b; Viegelmann et al., 2014), immunomodulatory (Tabares et al., 2011), and anticancer (Simmons et al., 2011; Yi-Lei et al., 2014) effects have been isolated from sponge-associated actinomycetes. For example, salinosporamide A, a potent inhibitor of the 20S proteasome that has been isolated from a Salinospora species (Feling et al., 2003; Gulder and Moore, 2010), entered clinical trials for multiple myeloma treatment, only three years after its discovery (Fenical et al., 2009). Due to the continuous discovery of bioactive natural products from marine microbes, re-isolation of known microbial secondary metabolites has become a real challenge (Hong et al., 2009). However, microbial genome sequencing has confirmed the presence of a large number of silent biosynthetic gene clusters that encode for secondary metabolites which are not produced under normal laboratory conditions (Dashti et al., 2014). Microbial competition for nutrition and other resources is considered one of the most important factors for induction of novel bioactive secondary metabolites (Oh et al., 2005). Crosstalk between microbes inhabiting the same environment induces the unexpressed biosynthetic pathways leading to production of unusual secondary metabolites (Pettit, 2009; Schroeckh et al., 2009; Zuck et al., 2011). Co-cultivation of two different microbial strains together in one culture allows direct interaction between them, which may lead to the induction of new cryptic secondary metabolites not previously detected in the axenic cultures (Rateb et al., 2013). Examples of the production of induced new natural products by co-fermentation of marine derived microorganisms include a rare class of pseurotins, 11-O-methylpseurotin A2 derived from mixed fermentation of Streptomyces bullii and the fungus Aspergillus fumigatus MBC-F1-10 (Rateb et al., 2013), the cyclic depsipeptides emericellamides A and B isolated from a co-culture of marine-derived fungus Emericella sp. (CNL-878) and the marine bacterium Salinispora arenicola (Oh et al., 2007), the diterpenoids libertellenones A–D isolated from mixed fermentation of the marine α-proteobacterium strain CNJ-328 with the fungus and Libertella sp. CNL-52 (Oh et al., 2005) and a chlorinated benzophenone pestalone sourced from a co-culture of the same bacterial strain CNJ-328 with Pestalotia sp. strain (Cueto et al., 2001). Recently, co-culture has also proved that both strains affect each other and induce new fungal and bacterial metabolites which were not detected in axenic cultures (Wakefield et al., 2017). In this study, we report on the induction of new bioactive secondary metabolites (Figure 1) saccharomonosporine A (1) and convolutamydine F (2) along with other three known metabolites 3-5 in response to microbial co-cultivation of two marine actinomycetes, Saccharomonospora sp. UR22 and Dietzia sp. UR66, derived from the Red Sea sponge Callyspongia siphonella. The HPLC and TLC chromatograms of the axenic cultures together with the co-culture derived extracts showed that compounds 1-5 were produced only during co-fermentation of both microbes. On the other hand, fermentation of Saccharomonospora sp. UR22 alone led to the isolation of a set of known microbial metabolites (6-11). Performing in vitro assay and docking studies on the isolated compounds revealed the potential of Pim-1 kinase as a promising target for only compound 1 and 3. Cytotoxicity evaluation of the isolated compounds showed that compound 1 and 3 have significant antiproliferative activities against HT-29 and HL-60 cell lines. These results coincide with the enzyme inhibition assay ones.
Material and Methods
General Apparatus and Chemicals
Ultra violet (UV) spectra were acquired on a ultra-violet visible (UV-vis) spectrometer (Shimadzu UV 1800 spectro, Japan). Optical rotation values were acquired at Bellingham + Stanley ADP600 Series Polarimeter at the sodium D line (589 nm) and 25°C. IR spectra were recorded as KBr disks on a IR spectrophotometer (Shimadzu S8400, Japan) High performance liquid chromatography (HPLC) analysis was performed by Thermofisher dionex ultimate 3000 with PDA detector and Xterra (Waters) C18 RP analytical HPLC column (5 μm, 4.6 × 250 mm). High resolution mass spectrometric data were obtained using a Thermo Instruments MS system (LTQ XL/LTQ Orbitrap Discovery) coupled to a Thermo Instruments HPLC system (Accela PDA detector, Accela PDA autosampler, and Accela pump). 1D and 2D NMR spectra were recorded on Bruker Avance III 400 MHz (Bruker AG, Switzerland) with BBFO Smart Probe and Bruker 400 AEON Nitrogen-Free Magnet. Data were analyzed using Topspin 3.1 Software. Each sample was dissolved in suitable deuterated solvent. Chemical shifts were recorded and expressed in ppm related to the TMS signal at 0.00 ppm as internal reference. All solvent used for preparing extracts were of technical grade (ADWIC; El-Nasr Pharmaceutical Chemicals Co., Egypt); and reagents used for preparing samples were of analytical grade (E-Merck, Darmstadt, Germany). Silica gel (60–120 mesh, 50 g, Fluka®) was used for chromatographic isolation and purification. TLC analysis was performed using Merck 9385 pre-coated aluminum plate silica gel (Kieselgel 60) with F254 indicator thin layer plates.
Isolation and Cultivation of the Actinomycetes
Callyspongia siphonella was collected at a depth of 10 m in the Red Sea (Hurghada, Egypt) in November 2015. A voucher specimen was reserved at the National Institute of Oceanography and Fisheries, Red Sea Branch, Invertebrates Department. Sponge biomass was transferred to plastic bag containing seawater and transported to the laboratory. Sponge specimens were rinsed in sterile seawater, cut into pieces of ca. 1 cm3, and then thoroughly homogenized in a sterile mortar with 10 volumes of sterile seawater. The supernatant was diluted in ten-fold series (10−1, 10−2, 10−3) and subsequently plated out on agar plates. Five different media M1 (Mincer et al., 2002), ISP2 medium (Shirling and Gottlieb, 1966), oligotrophic medium (OLIGO) (Olson et al., 2000), actinomycete isolation agar (AIA) (Lechevalier and Lechevalier, 1975) and marine agar (MA) (Weiner et al., 1985) were used for the isolation of actinomycetes. All media were supplemented with 0.2 μm pore size filtered cycloheximide (100 μg/mL), nystatin (25 μg/mL), and nalidixic acid (25 μg/mL) to facilitate the isolation of slow-growing actinomycetes. Cycloheximide and nystatin inhibit fungal growth, while nalidixic acid inhibits many fast-growing Gram-negative bacteria. All media contained DifcoBacto agar (18 g/L) and were prepared in 1 L artificial sea water (NaCl 234.7 g, MgCl2.6 H2O 106.4 g, Na2SO4 39.2 g, CaCl2 11.0 g, NaHCO3 1.92 g, KCl 6.64 g, KBr 0.96 g, H3BO3 0.26 g, SrCl2 0,24 g, NaF 0.03 g, and ddH2O to 10.0 L) (Lyman and Fleming, 1940). The inoculated plates were incubated at 30°C for 6–8 weeks. Distinct colony morphotypes were picked and re-streaked until visually free of contaminants. Saccharomonospora sp. UR22 and Dietzia sp. UR66 were cultivated on ISP2 medium. The isolates were maintained on plates for short-term storage and long-term strain collections were set up in medium supplemented with 30% glycerol at −80°C.
Molecular Identification
16S rRNA gene amplification, cloning and sequencing were performed according to Hentschel et al. using the universal primers 27F and 1492R (Lane, 1991). Chimeric sequences were identified by using the Pintail program (Ashelford et al., 2005). The genus-level affiliation of the sequence was validated using the Ribosomal Database Project Classifier. The genus-level identification of all the sequences was done with RDP Classifier (-g 16srrna, -f allrank) and validated with the SILVA Incremental Aligner (SINA) (search and classify option) (Pruesse et al., 2012). An alignment was calculated again using the SINA web aligner (variability profile: bacteria). Gap-only position were removed with trimAL (-noallgaps). For phylogenetic tree construction, the best fitting model was estimated initially with Model Generator. RAxML (-f a -m GTRGAMMA –x 12345 –p 12345 –# 1000) and the estimated model was used with 1,000 bootstrap resamples to generate the maximum-likelihood tree. Visualization was done with Interactive Tree of Life (ITOL).
Fermentation, Extraction, and Isolation
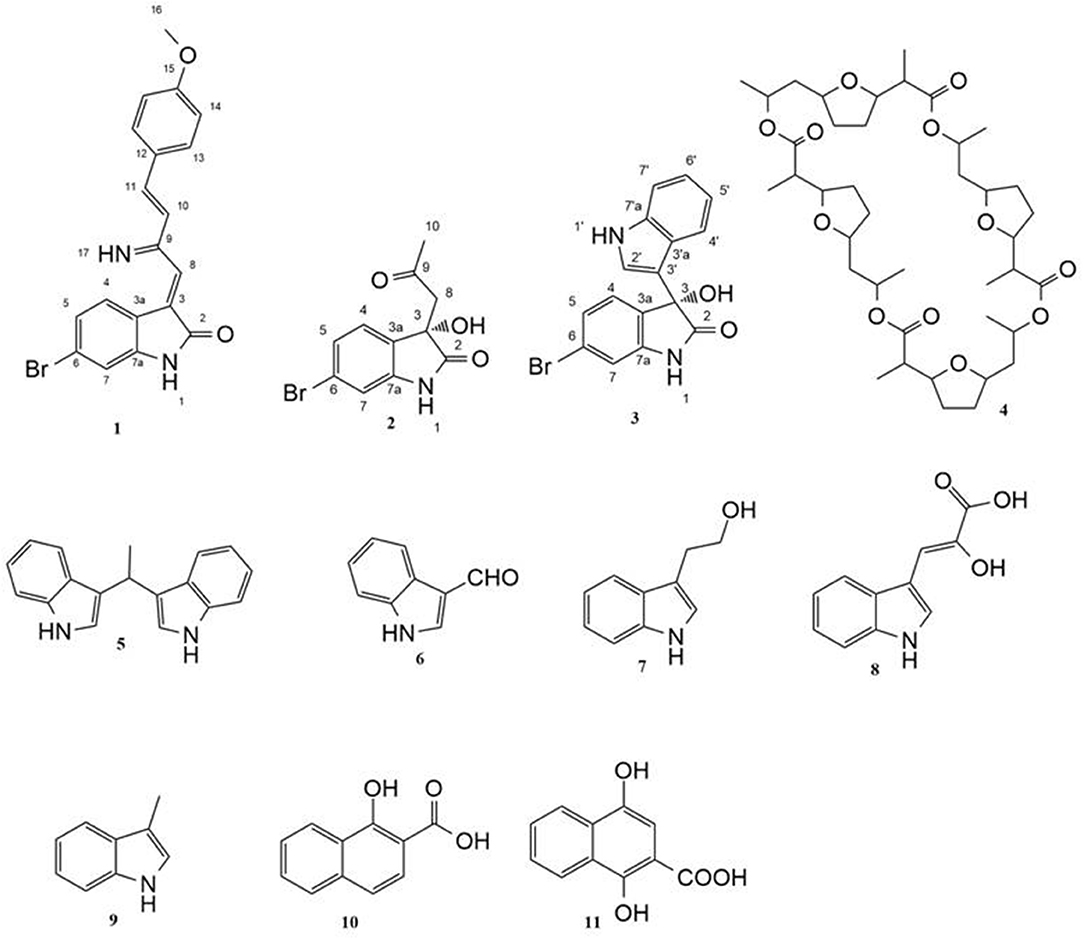
Each strain was fermented in 10 Erlenmeyer flasks (2 L), each containing 1 L of ISP 2 (International Streptomyces Project) medium in artificial sea water and incubated at 30°C for 14 days with shaking at 150 rpm. For co-cultivation experiment, 10 mL of 5-day-old culture of Saccharomonospora sp. UR22 was inoculated into 15 Erlenmeyer flasks (2 L), each containing 1 L of ISP 2 medium inoculated with 10 mL of 5-day-old culture of Dietziasp. UR66. After fermentation of single cultures and co-culture, filtration was done, and the supernatant was extracted with ethyl acetate (3 × 500 mL) to give the ethyl acetate extracts. These extracts (400 mg for Saccharomonospora sp. UR22, 950 mg for co-culture) were fractionated on a Sephadex LH20 (32-64 μm, 100 x 25 mm, Fluka®) column eluting with MeOH/H2O (90:10%), to yield six fractions. Fraction Nr. 2 (from co-culture) was subjected to C18 reversed-phase column chromatography (0.04–0.063 mm, 50 × 10 mm, Merck®) with an isocratic elution using acetonitrile/water (40:60) to give five crude compounds 1-5. Further purification was performed by preparative TLC on a (20 cm × 20 cm) silica gel plates that developed in CH2Cl2/MeOH/NH3 (7:4:0.5, v/v) solvent system. Fraction Nr. 4 (from Saccharomonospora sp. UR22) was treated in the same manner to obtain six known compounds 6-11 (Figure 1).
HPLC Analysis
All actinomycete-derived extracts were analyzed by HPLC-DAD. Fifty micro liters (1 mg/mL in acetonitrile) of bacterial crude extracts was injected. Then, isocratic elution was performed with 50% aqueous acetonitrile containing 0.1% trifluoroacetic as amobile phase over 15 min at a flow rate of 1 mL/min and UV detection at different wavelengths (210, 254, 270, and 300 nm).
Pim-1 Kinase Assay
The kinase activity was measured following the manufacturer's instructions (HTScan Kinase Assays from Cell Signaling Technology) using staurosporine as a positive control. Different concentrations of test compounds were incubated with 50 ng of Pim-1 kinase enzyme in the reaction buffer [25 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.1 mM Na3VO4, 5 mM–glycerophosphate, 2 mM dithiothreitol (DTT)] and the ATP/substrate cocktail [200 μM ATP + 1.5 μM biotinylated BAD Ser112 peptide] at room temperature for 30 min. The reaction was stopped by the addition of stop buffer (50 mM EDTA, pH 8) and transferred to a streptavidincoated 96-well plate. After incubation for 1 h at room temperature, the wells were washed three times with phosphate-buffered saline Tween-20 (PBST) buffer. After incubation with anti BADSer112 antibody for 2 h, the wells were washed three times with 1 × PBST and incubated with the peroxidase-conjugated secondary antibody for 30 min. Following three washes with 1 × PBST, the substrate (3,3,5,5-tetramethylbenzidine) was added and the samples were incubated at room temperature for 15 min. The reaction was stopped by the addition of 2 N HCl and the absorbance was measured with a spectrophotometer at 450 nm. The assay was performed in triplicate.
Docking Studies
The crystal structure of Pim-1 of PDB code 3umw was used. Docking experiments were conducted employing LigandFit docking engine. This docking software considers the ligand as flexible and the receptor as rigid structure. The binding site was generated from the “Find sites as volume of selected ligands” option in Discovery Studio 2.5. The number of trials of Monte Carlo search parameters = 30,000; and search step for torsions with polar hydrogens = 30.0°. The root mean square threshold for ligand-to-binding-site shape matching was set to 2.0 Å, employing a maximum of 1.0 binding-site partitions. The interaction energies were assessed employing the CFF force field (v.1.02) with a non-bonded cutoff distance of 10.0 Å and distance-dependent dielectric. An energy grid extending 5.0 Å from the binding site was implemented. The interaction energy was estimated with a trilinear interpolation value using soft potential energy approximations. Rigid body ligand minimization parameters: 40 steepest descent iterations followed by the 80 Broyden–Fletcher–Goldfarb–Shannon minimization iterations were applied to every orientation of the docked ligand. The proposed inhibitors were further energy minimized within the binding site by implementing the “Smart Minimization” option for a maximum of 1,000 iterations (Khanfar and Taha, 2013).
MTT Assay
Cell proliferation was evaluated in cell lines by the MTT assay in triplicates. 104 cells were plated in a 96-well microtiter plate in a final volume of 100 μl of culture medium. Cells were treated for 24 h with test compound at 37 °C with 5% CO2. After treatment, the cells were immediately incubated with 10 μl MTT (5.0 mg/mL) for 4 h at 37°C. The cells were then lysed in 100 μl of lysis buffer (isopropanol, conc. HCl and Triton X-100) for 10 min at room temperature and 300 rpm/min shaking. The enzymatic reduction of MTT to formazan crystals that dissolved in DMSO was quantified by photometry at 570 nm. Dose-response curves were generated and the IC50 values were define d as the concentration of compound required to inhibit cell proliferation by 50%. 5-Flurouracil was used as a positive control.
Results and Discussion
Mono and Co-culture HPLC Profiles
HPLC and TLC analysis of EtOAc extracts obtained from axenic fermentation of each actinomycete compared to that from mixed fermentation indicated a very different chemical profiles (Figure 2). The co-culture derived extract showed a higher metabolic diversity than extracts from mono cultures. Exclusive detection of metabolites 1-5 in the co-culture illustrated that they are produced only upon co-fermentation of the two microbes.
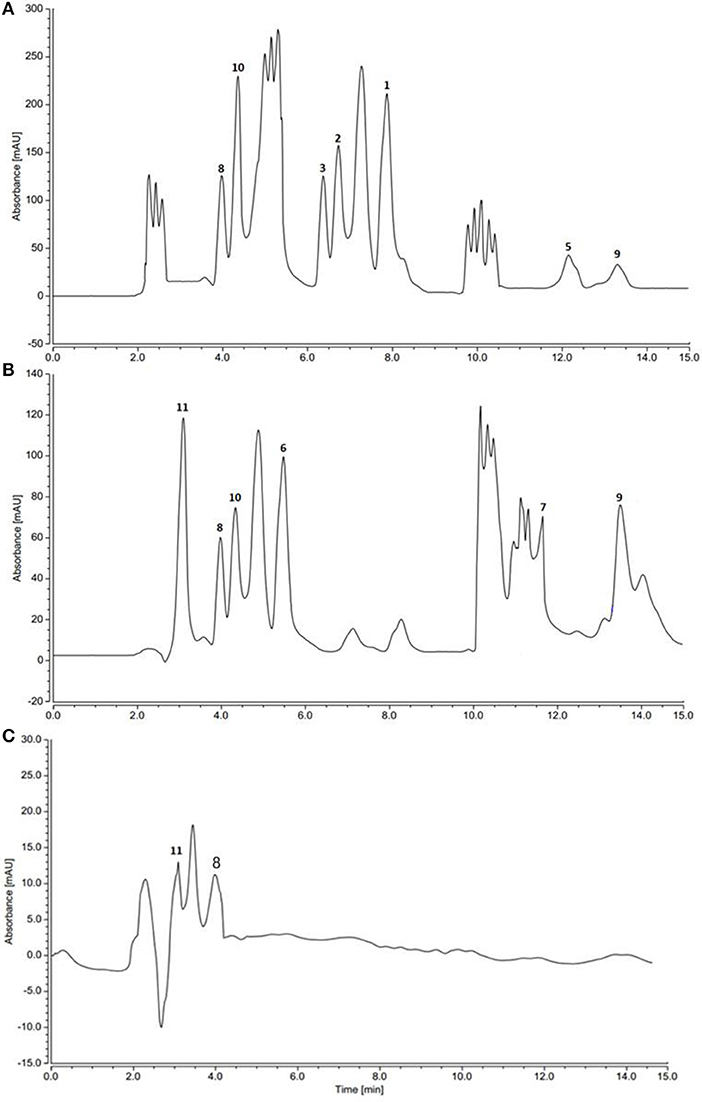
Figure 2. HPLC profiles of actinomycetes extracts. (A) Saccharomonospora sp. UR22 and Dietzia sp. UR66 co-culture (B) Saccharomonospora sp. UR22 mono-culture (C) Dietzia sp. UR66 mono-culture.
Isolation and Structural Characterization
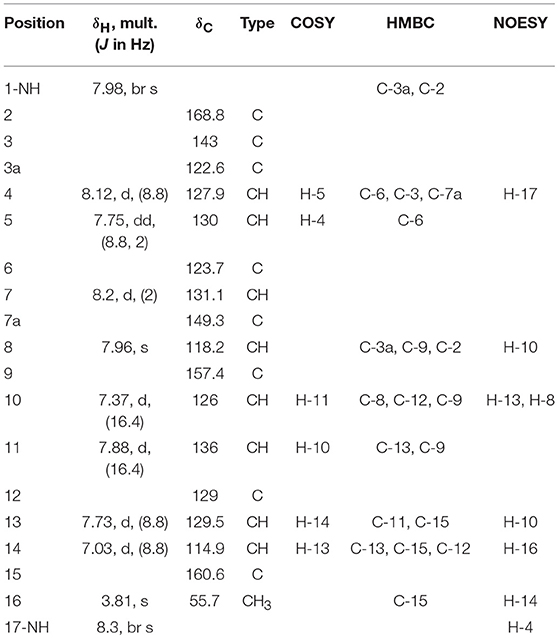
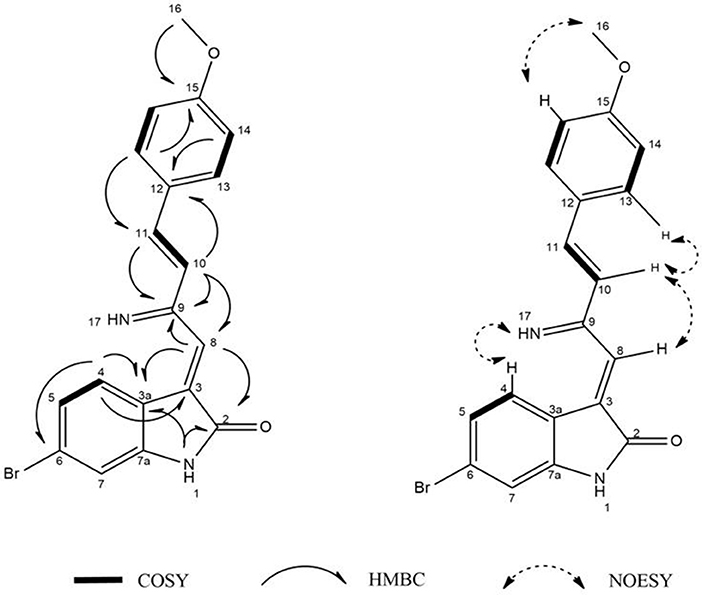
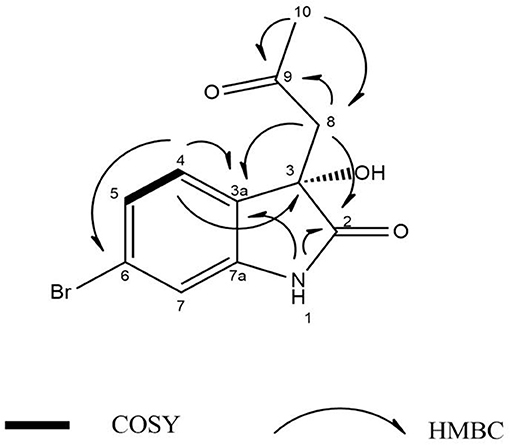
Guided by HPLC-DAD, the EtOAc extracts from the liquid cultures of the axenic fermentations and mixed fermentation were subjected to Sephadex LH-20 column chromatography followed by C18 reversed-phase column chromatography and final preparative TLC purification to target the isolation of the new metabolites 1 and 2 together with the other nine known metabolites 3-11. Compound 1 was isolated as yellow amorphous powder. The molecular formula C19H15O2N2Br was suggested on the basis of positive HRESIMS ion at m/z 383.0392 [M+H]+, indicating 13 degrees of unsaturation. Furthermore, the presence of a bromine atom was confirmed by its characteristic isotope cluster (Figure S1). 1H NMR spectral data of 1 (Figure S2) (Table 1) in DMSO-d6 suggested the presence of a methyl singlet at δH 3.81 (3H, s, H-16) and two exchangeable protons (δH 8.32, 7.98) due to NH. The splitting pattern of resonances at δH 8.20 (H-7), 8.12 (H-4), and 7.75 (H-5) together with the COSY correlation (Figure S5) between (H-4) and (H-5) suggested the presence of a 1,3,4-trisubstituted benzene ring. On the other hand, the signals at δH 7.73 (H-13), 7.03 (H-14) and the 1H-1H COSY correlation between them indicated the presence of 1,4 disubstituted benzene ring. The DEPTQ spectrum (Figure S3) (Table 1) displayed seventeen signals, with eight sp2 aromatic carbons including eight CH groups and six quaternary carbons, and one sp3 aliphatic carbon (CH3 group). The spectra also revealed an aminocarbonyl at δC168.8 (C-2), an imino carbon at δC 157.4 (C-9) and one oxygenated carbon at δC 55.7 (C-16). The assignment of protonated carbons was achieved by the HSQC data (Figure S4). The key HMBC correlations (Figure S6) from H-4 (δH 8.12) to C-3 (δC 143), and from the exchangeable proton H-1(δH 7.98) to the carbonyl carbons C-2 (δC 168.8) and C-3a (δC 122.6) together with the previous reported data (Guo et al., 2010) suggested the presence of brominated indolin-2-one skeleton at C-6 (δC 123.7). The sub-structure of a methoxyphenyl group was deduced from the HMBC correlation between the methyl protons H-16 (δH 3.81) and C-15 (δC 160.6). 1H-1H COSY correlation between H-11 (δH 7.88) and H-10 (δH 7.37) together with HMBC correlations from H-11 to C-10 (δC 126) and C-9 (δC 157.4), from H-10 to C-9 and C-8 (δC 118.2) indicated the presence of an imino butenylidene moiety. The E configuration of H-11 (δH 7.88, d) and H-10 (δH 7.37, d) was deduced from the J-values extracted from the 1H NMR spectrum (J = 16.4 Hz). The connection of the imino butenylidene moiety to the methoxyphenyl group was established through the HMBC correlation from H-13 (δH 7.73) to C-11 (δC 136). Moreover, the HMBC correlations (Figure 3) from H-8 (δH 7.96) to C-2 (δC 168.8) and C-3a (δC 122.6) connected this moiety to the indolin-2-one skeleton at C-3. The chemical shifts of H-8 at δH 7.96 and H-4 at δH 8.12 are very characteristic for E configuration at the double bond between C-3 and C-9 (Faita et al., 1994). Furthermore, NOESY correlation (Figures S7, S8) between the NH proton H-17 and H-4, and absence of NOESY cross-peak between H-8 and H-4, also pointed to the E configuration at the C-3/C-8 double bond. On that basis, the structure of 1 was established as (E)-6-bromo-3-((E)-2-imino-4-(4-methoxyphenyl)but-3-en-1-ylidene)indolin-2-one, a new secondary metabolite to which were named as Saccharomonosporine A.
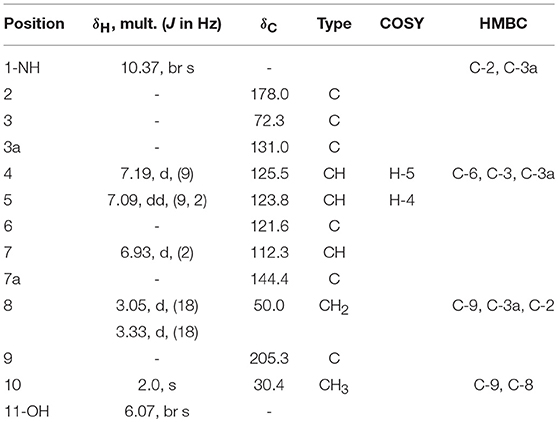
Compound 2 was isolated as white crystals, the molecular formula C11H10O3NBr was suggested on the basis of positive HRESIMS ion at m/z 283.9899 [M+H]+, indicating seven degrees of unsaturation. Furthermore, the presence of a bromine atom was confirmed by its characteristic isotope cluster (Figure S9). Similar to compound 1, the 1H and 13C NMR spectra (Figures S10, S11) of 2 (Table 2) also revealed an aminocarbonyl at δC178.0 and one oxygenated carbon at δC 72.3 (C-3). The key HMBC correlations (Figure 4 and Figure S14) from H-4 (δH 7.19) to C-3 (δC 72.3), and from the exchangeable proton H-1(δH 10.37) to the carbonyl carbons C-2 (δC 178) and C-3a (δC 131) together with the previous reported data (Guo et al., 2010) suggested the presence of brominated indolin-2-one skeleton. A 2-oxopropyl unit was deduced from the HMBC cross-peaks of H-8 (δH 3.05) to C-9 (δC 205.3), H-10 (δH 2.0) to C-8 (δC 50), and H-10 to C-9 and was indicated to be attached at C-3 (δC 72.3) through the HMBC correlation of H-8 to C-3. These spectroscopic data together with the optical rotation value –13.8 (c 0.85, MeOH), revealed that this compound is the previously unreported (S) enantiomer of (R)-6-Bromo-3-hydroxy-3-(2 oxopropyl)indolin-2-one, a synthetic 3-hydroxyoxindole derivative (Guo et al., 2010). Several 3-hydroxyoxindole derivatives have been reported from natural marine sources (Kamano et al., 1995; Zhang et al., 1995). For example, convolutamydine A, a brominated (R) enantiomer of 2 (Kamano et al., 1995), so the name convolutamydine F was suggested to compound 2.
Compound 3 was isolated as white crystals. HRESIMS, 1D and 2D NMR spectral data (Table S1, Figures S15–S20) revealed that compound 3 has a planer structure identical to compound 3f, a previously reported synthetic intermediate (Wang and Ji, 2006), which obtained as a racemic mixture. Compound 3 had a negative optical rotation of −8.7 (c 0.3, MeOH), in contrast to previous reports on similar optically active compounds that showed a positive optical rotations for the (R) enantiomers (Deng et al., 2010; Guo et al., 2010). On this basis compound 3 was identified as (S) 6-bromo-3-hydroxy-3-(1H-indol-3-yl) indolin-2-one. To the best of our knowledge, compounds 2 and 3 are considered the first (S) 3-hydroxyoxindole derivatives reported from natural sources. Nonactin (4) (Wu and Sun, 2006) and vibrindole (5) (Ronit et al., 1994) were also isolated from the co-culture of Saccharomonospora sp. UR22 and Dietzia sp. UR66. In addition, indole-3-carbaldehyde (6) (Ashour et al., 2007), tryptophol (7) (Gore et al., 2012), indole-3-pyruvic acid (8) (Wishart et al., 2007), skatole (9) (Chen et al., 2015), 1-hydroxy-2-naphthoic acid (10) (Kauko and Lajunen, 1978) and 1,4 dihydroxy-2-naphthoic acid (11) (Isawa et al., 2002) were isolated from the axenic culture of Saccharomonospora sp. UR22. All isolated known metabolites were identified based on their accurate mass analyses and comparison of their NMR spectroscopic data (Figures S21–S28) with those reported in the literature.
Pim-1 Kinase Assay
Pim-1 kinase is a well-established oncoprotein in several tumor entities, e.g., myeloid leukemia, prostate cancer, colorectal cancer, or pancreatic cancer (Weirauch et al., 2013). In addition, over expression of Pim-1 kinase has been described in both human colon adenocarcinoma HT-29 cell (Weirauch et al., 2013) and human promyelocytic leukemia HL-60 (Fan et al., 2016) cell lines. Treatment of the aforementioned cell lines with Pim-1 kinase inhibitors resulted in a potent growth inhibitory activity (Weirauch et al., 2013; Fan et al., 2016). Furthermore, previous compounds structurally similar to compounds 1 and 3 showed potent and selective Pim-1 kinase inhibitory activity (Nakano et al., 2012; Sun et al., 2015). In order to investigate Pim-1 kinase as a potential target that mediates tumor cell growth inhibitory effect, all isolated compounds were evaluated for their efficacy to inhibit the in vitro enzymatic activity of Pim-1 kinase (Table 3). Compounds 1 and 3 exhibited significant inhibitory effects on Pim-1 enzyme activity with an IC50 value of 0.3 ± 0.02 and 0.95 ± 0.01 μM, respectively.
Docking of the Active Compounds
The potential binding mode of compounds 1 and 3 with Pim-1 kinase was analyzed by docking in the ATP-binding site of Pim-1. The protein data bank (PDB) contains several Pim-1 crystallographic proteins, however, Pim-1 of PDB code (3umw) was selected for docking experiments since it has optimum resolution (2.08 Å) and is co-crystalized with Pim-1 inhibitor that is structurally similar to compound 1 and 3 (Nakano et al., 2012). The sphere surrounding the co-crystallized inhibitor was selected as an active site for docking. Both compounds 1 and 3 showed similar docking poses (Figure 5) and comparable to the co-crystalized inhibitor (Nakano et al., 2012). The bromobenzene moiety of compound 1 is impeded within a hydrophobic pocket of ALA65, LEU44, VAL126, and LEU175. The oxindole proton of amidic group is interacted with the hinge region through hydrogen bonding to the main chain carbonyl of GLU121. Hydrogen bonding of this nature was reported with several Pim-1 inhibitors (Nakano et al., 2012). Moreover, the oxindole carbonyl oxygen is hydrogen bonded to the peptidic proton of ASP186 bridged through the conserved water molecule (HOH30). The imine nitrogen of compound 1 is hydrogen bonded to the terminal amino moiety of LYS67. The phenoxy ring is sandwiched between two hydrophobic surfaces of PHE49 and VAL52 (Figure 5A). Alternatively, compound 3 share similar interactions as in compound 1 within the hydrophobic pocket of ALA65, LEU44, VAL126, and LEU175, and hydrogen bonding interaction with GLU121 and ASP186. Moreover, the indole nitrogen is hydrogen bonded with peptidic oxygen of ILE185. However, the interaction with LYS67 is a π-cation interaction with the indole ring of compound 3 (Figure 5C). The binding interactions of compounds 1 and 3 showed high resemblance with the binding mode of the co-crystalized ligand (PDB code: 3umw) within the ATP-binding site of Pim-1 (Nakano et al., 2012). The co-crystalized structure forms similar hydrogen bonding interactions (with GLU121 and LYS67), hydrophobic interactions (with LEU174, VAL126, LEU44, VAL52, and ALA65), and π-stacking (with PHE49) as with compounds 1 and 3 (Figure 5E). However, the co-crystalized ligand forms additional electrostatically-enforced hydrogen bonding interaction with ASP128. Such interaction can explain the superior activity of the co-crystalized ligand (IC50 = 3 nM) over compounds 1 and 3. Therefore, the electrostatic interaction with ASP128 should be taking into consideration for future designing of more active analogs of compounds 1 and 3.
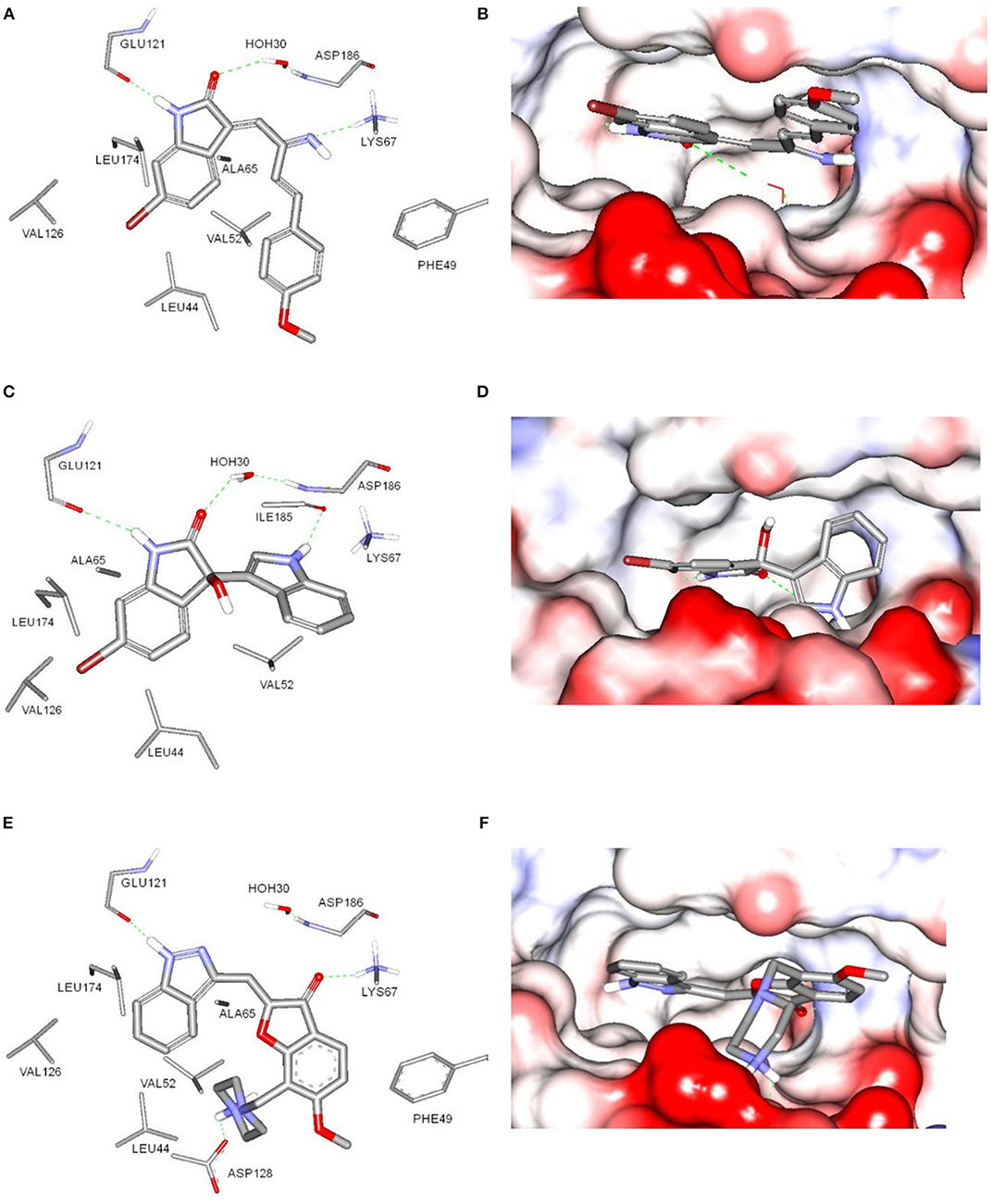
Figure 5. Docking of compounds 1 (A,B) and 3 (C,D) within the ATP-binding site of Pim-1 kinase (PDB code 3umw). (E,F) The key binding interactions of Pim-1 co-crystallized ligand. The amino acid side chains were depicted in (A,C,E) for clarification.
Antiproliferative Activity
The antiproliferative properties of the nine isolated compounds were evaluated against lung adenocarcinoma H1650, the human promyelocytic leukemia HL-60 and the human colon adenocarcinoma HT-29 cell lines (Table 4). Compounds 1 and 3 displayed potent antiproliferative activity against HL-60 and HT-29 cells (IC50 2.8, 3.6 and 4.2, 3.7 μM, respectively). The other compounds did not exhibit any cytotoxicity at the tested concentrations. These results came in great accordance with the Pim-1 kinase assay results.
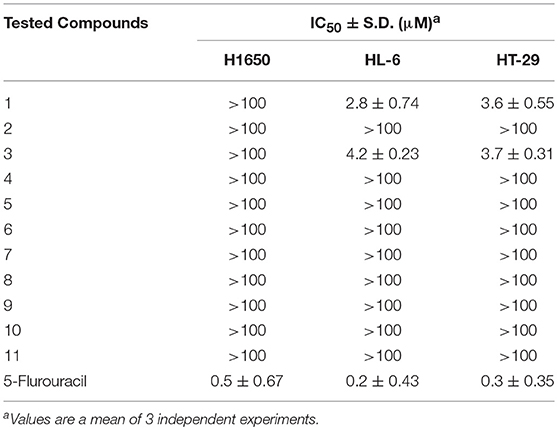
Table 4. Antiproliferative activity of isolated metabolites against H1650, HL-60, and HT-29 cancer cells.
Data for Saccharomonosporine A (1)
Yellow powder (18 mg); UV (MeOH) λmax (log ε) 343 (4.22), 425 (3.16); IR (KBr) νmax 3165, 1716, 1605, 1570, cm−1; 1H-NMR (DMSO-d6, 400 MHz); and 13C-NMR (DMSO-d6, 100 MHz) data, see Table 1; HRESIMS m/z 383.0392 [M + H]+ (calcd for C19H15O2N2Br).
Data for Convolutamydine F (2)
White crystals (10 mg); UV (MeOH) λmax (log ε) 323 (3.18), 408 (2.83); –13.8 (c 0.85, MeOH), IR (KBr) νmax 3331, 1742, 1714 cm−1; 1H-NMR (DMSO-d6, 400 MHz) and 13C-NMR (DMSO-d6, 100 MHz) data, see Table 2; HRESIMS m/z 283.9899 [M + H]+ (calcd for C11H10O3NBr).
Conclusion
In this study, two sponge-derived actinomycetes, Saccharomonospora sp. UR22 and Dietzia sp. UR66, were co-fermented in liquid media. The presence of induced metabolites was studied by comparison of the HPLC-DAD and TLC chromatograms of the crude extracts of the two axenic cultures and the co-culture. Co-cultivation of Saccharomonospora sp. UR22 and Dietzia sp. UR66 induced the biosynthesis of novel oxindole alkaloid saccharomonosporine A (1), convolutamydine F (2) along with other three induced metabolites (3-5) which were not detected in either microorganism in a single culture. Axenic culture of Saccharomonospora sp. UR22 led to isolation of common known microbial metabolites 6-11. Compounds 1 and 3 exhibited potent antiproliferative activities toward HL-60 and HT-29. Based on previous reports on similar compounds, Pim-1 inhibitory assay results and docking studies in the ATP-binding site of Pim-1 kinase, we suggested that both compounds 1 and 3 mediated their cytotoxicity by inhibiting the well-known oncoprotein Pim-1 kinase. These findings highlighted the co-cultivation approach as an effective strategy to enhance the chemical diversity of the secondary metabolites hidden in the genomes of the marine actinomycetes.
Author Contributions
SE, RM, and HH designed the experiments. TM collected and identified the marine sponge. AS, HH, and UA performed the experiments and isolated the compounds. AS and MR performed data acquisition and structure elucidation. DH and TG performed the biological assays. MK performed the docking study. UA, RM, MR, TG, AS and MK drafted and revised the manuscript.
Funding
Research in the TAMG lab is generously funded by the DFB (GU 1233/1-1, CIPSM). This work was supported by the German Research Foundation (DFG) and the Technical University of Munich (TUM) in the framework of the Open Access Publishing Program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would also like to thank Dr. Mohammed E. Deraz for his support in the HPLC analysis work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2018.00538/full#supplementary-material
References
Abdelmohsen, U. R., Bayer, K., and Hentschel, U. (2014a). Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 31, 381–399. doi: 10.1039/c3np70111e
Abdelmohsen, U. R., Cheng, C., Viegelmann, C., Zhang, T., Grkovic, T., Ahmed, S., et al. (2014b). Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora sp. EG49. Mar. Drugs 12, 1220–1244. doi: 10.3390/md12031220
Ashelford, K. E., Chuzhanova, N. A., Fry, J. C., Jones, A. J., and Weightman, A. J. (2005). At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71, 7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005
Ashour, M. A., Elkhayat, E. S., Ebel, R., Edrada, R., and Proksch, P. (2007). Indole alkaloid from the Red Sea sponge Hyrtios erectus. ARKIVOC 2007, 225–231. doi: 10.3998/ark.5550190.0008.f22
Chen, S., Lu, G. P., and Cai, C. (2015). Iridium-catalyzed methylation of indoles and pyrroles using methanol as feedstock. RSC Adv. 86, 70329–70332. doi: 10.1039/C5RA15822B
Cueto, M., Jensen, P. R., Kauffman, C., Fenical, W., Lobkovsky, E., and Clardy, J. (2001). Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod. 64, 1444–1446. doi: 10.1021/np0102713
Dashti, Y., Grkovic, T., Abdelmohsen, U. R., Hentschel, U., and Quinn, R. J. (2014). Production of induced secondary metabolites by a co-culture of sponge-associated actinomycetes, Actinokineospora sp. EG49 and Nocardiopsis sp. RV163. Mar. Drugs 12, 3046–3059. doi: 10.3390/md12053046
Deng, J., Zhang, S., Ding, P., Jiang, H., Wang, W., and Li, J. (2010). Facile creation of 3-indolyl-3-hydroxy-2-oxindoles by an organocatalytic enantioselective Friedel–Crafts reaction of indoles with isatins. Adv. Synth. Catal. 352, 833–838. doi: 10.1002/adsc.200900851
Eltamany, E. E., Abdelmohsen, U. R., Ibrahim, A. K., Hassanean, H. A., Hentschel, U., and Ahmed, S. A. (2014). New antibacterial xanthone from the marine sponge-derived Micrococcus sp. EG45. Bioorg. Med. Chem. Lett. 24, 4939–4942. doi: 10.1016/j.bmcl.2014.09.040
Faita, G., Mella, M., Righetti, P. P., and Tacconi, G. (1994). An easy lewis acid-mediated isomerization from (E)-to (Z)-oxoindolin-3-ylidene ketones. Tetrahedron 37, 10955–10962. doi: 10.1016/S0040-4020(01)85706-9
Fan, Y. B., Huang, M., Cao, Y., Gong, P., Liu, W. B., Jin, S. Y., et al. (2016). Usnic acid is a novel Pim-1 inhibitor with the abilities of inhibiting growth and inducing apoptosis in human myeloid leukemia cells. RSC Adv. 6, 24091–24096. doi: 10.1039/C6RA01159D
Feling, R. H., Buchanan, G. O., Mincer, T. J., Kauffman, C. A., Jensen, P. R., and Fenical, W. (2003). Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. 42, 355–357. doi: 10.1002/anie.200390115
Fenical, W., Jensen, P. R., Palladino, M. A., Lam, K. S., Lloyd, G. K., and Potts, B. C. (2009). Discovery and development of the anticancer agent salinosporamide A (NPI-0052). Bioorg. Med. Chem. 17, 2175–2180. doi: 10.1016/j.bmc.2008.10.075
Gore, S., Baskaran, S., and König, B. (2012). Fischer indole synthesis in low melting mixtures. Org. Lett. 17, 4568–4571. doi: 10.1021/ol302034r
Gulder, T. A. M., and Moore, B. S. (2010). The salinosporamide natural product family: potent 20S proteasome inhibitors as potential cancer chemotherapeutics. Angew. Chem. Int. Ed. 49, 9346–9367. doi: 10.1002/anie.201000728
Guo, Q., Bhanushali, M., and Zhao, C.G. (2010). Quinidine thiourea-catalyzed aldol reaction of unactivated ketones: highly enantioselective synthesis of 3-alkyl-3-hydroxyindolin-2-ones. Angew. Chem. Int. Ed. 49, 9460–9464. doi: 10.1002/anie.201004161
Hentschel, U., Schmid, M., Wagner, M., Fieseler, L., Gernert, C., and Hacker, J. (2001). Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 35, 305–312. doi: 10.1111/j.1574-6941.2001.tb00816.x
Hong, K., Gao, A. H., Xie, Q. Y., Gao, H. G., Zhuang, L., Lin, H. P., et al. (2009). Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 7, 24–44. doi: 10.3390/md7010024
Isawa, K., Hojo, K., Yoda, N., Kamiyama, T., Makino, S, Saito, M., et al. (2002). Isolation and identification of a new bifidogenic growth stimulator produced by Propionibacterium freudenreichii ET-3. Biosci. Biotech. Biochem. 3, 679–681. doi: 10.1271/bbb.66.679
Kamano, Y., Zhang, H. P., Ichihara, Y., Kizu, H., Komiyama, K., and Pettit, G.R. (1995). Convolutamydine A, a novel bioactive hydroxyoxindole alkaloid from marine bryozoan Amathia convoluta. Tetrahedron Lett. 36, 2783–2784. doi: 10.1016/0040-4039(95)00395-S
Kauko, R., and Lajunen, L. H. J. (1978). NMR Studies on aromatic compounds I-13C NMR spectra of some mono- and disulphonato-substituted hydroxynaphthoic acids in DMSO-&water solution. Mag. Res. Chem. 11, 12–15. doi: 10.1002/mrc.1270110104
Khanfar, M., and Taha, M. (2013). Elaborate ligand-based modeling coupled with multiple linear regression and K nearest neighbor QSAR analyses unveiled new nanomolar mTOR inhibitors. J. Chem. Inform. Model. 53, 2587–2612. doi: 10.1021/ci4003798
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds. E. Stackebrandt, and M. Goodfellow (Chichester: John Wiley and Sons), 115–175.
Lechevalier, M. P., and Lechevalier, H.A. (1975). Actinoplanete with cylindrical sporangia, actinoplanes-Rectilineatus sp. nov. Int. J. Syst. Bacteriol. 25, 371–376. doi: 10.1099/00207713-25-4-371
Mincer, T. J., Jensen, P. R., Kauffman, C. A., and Fenical, W. (2002). Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 68, 5005–5011. doi: 10.1128/AEM.68.10.5005-5011.2002
Nakano, H., Saito, N., Parker, L., Tada, Y., Abe, M., Tsuganezawa, K., et al. (2012). Rational evolution of a novel type of potent and selective proviral integration site in moloney murine leukemia virus kinase 1 (PIM1) inhibitor from a screening-hit compound. J. Med. Chem. 55, 5151–5164. doi: 10.1021/jm3001289
Oh, D. C., Jensen, P. R., Kauffman, C. A., and Fenical, W. (2005). Libertellenones A–D: induction of cytotoxic diterpenoid biosynthesis by marine microbial competition. Bioorg. Med. Chem. 13, 5267–5273. doi: 10.1016/j.bmc.2005.05.068
Oh, D. C., Kauffman, C. A., Jensen, P. R., and Fenical, W. (2007). Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 70, 515–520. doi: 10.1021/np060381f
Olson, J. B., Lord, C. C., and McCarthy, P. J. (2000). Improved recoverability of microbial colonies from marine sponge samples. Microb. Ecol. 40, 139–147. doi: 10.1007/s002480000058
Pettit, R.K. (2009). Mixed fermentation for natural product drug discovery. Appl. Microbiol. Biotechnol. 83, 19–25. doi: 10.1007/s00253-009-1916-9
Pruesse, E., Peplies, J., and Glockner, F.O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829. doi: 10.1093/bioinformatics/bts252
Rateb, M. E., Hallyburton, I., Houssen, W. E., Bull, A. T., Goodfellow, M., Santhanam, R., et al. (2013). Induction of diverse secondary metabolites in Aspergillus fumigatus by microbial co-culture. RSC Adv. 3, 14444–14450. doi: 10.1039/c3ra42378f
Ronit, B., Carmeli, S., and Sar, N. (1994). Vibrindole A, a metabolite of the marine bacterium, Vibrio parahaemolyticus, isolated from the toxic mucus of the boxfish Ostracion cubicus. J. Nat. Prod. 11, 1587–1590.
Roue, M., Quevrain, E., Domart-Coulon, I., and Bourguet-Kondracki, M. L. (2012). Assessing calcareous sponges and their associated bacteria for the discovery of new bioactive natural products. Nat. Prod. Rep. 29, 739–751. doi: 10.1039/c2np20040f
Schroeckh, V., Scherlach, K., Nutzmann, H. W., Shelest, E., Schmidt-Heck, W., Schuemann, J., et al. (2009). Intimate bacterial–fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. 106, 14558–14563. doi: 10.1073/pnas.0901870106
Shirling, E., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 317–327. doi: 10.1099/00207713-16-3-313
Simmons, L., Kaufmann, K., Garcia, R., Schwar, G., Huch, V., and Muller, R. (2011). Bendigoles D–F, bioactive sterols from the marine sponge-derived Actinomadura sp. SBMs009. Bioorg. Med. Chem. 19, 6570–6575. doi: 10.1016/j.bmc.2011.05.044
Sun, H. B., Wang, X. Y., Li, G. B., Zhang, L. D., Liu, J., and Zhao, L. F. (2015). Design, synthesis and biological evaluation of novel C3-functionalized oxindoles as potential Pim-1 kinase inhibitors. RSC Adv. 5, 29456–29466. doi: 10.1039/C5RA00177C
Tabares, P., Pimentel-Elardo, S. M., Schirmeister, T., Hunig, T., and Hentschel, U. (2011). Anti-protease and immunomodulatory activities of bacteria associated with Caribbean sponges. Mar. Biotechnol. 13, 883–892. doi: 10.1007/s10126-010-9349-0
Thomas, T. R., Kavlekar, D. P., and LokaBharathi, P. A. (2010). Marine drugs from sponge-microbe association—A review. Mar. Drugs 8, 1417–1468. doi: 10.3390/md8041417
Viegelmann, C., Parker, J., Ooi, T., Clements, C., Abbott, G., Young, L., et al. (2014). Isolation and identification of antitrypanosomal and antimycobacterial active steroids from the sponge Haliclona simulans. Mar. Drugs 12, 2937–2952. doi: 10.3390/md12052937
Wakefield, J., Hassan, H. M., Jaspars, M., Ebel, R., and Rateb, M. E. (2017). Dual induction of new microbial secondary metabolites by fungal bacterial co-cultivation. Front. Microbiol. 8:1284. doi: 10.3389/fmicb.2017.01284
Wang, S. Y., and Ji, S. J. (2006). Facile synthesis of 3,3-di (heteroaryl) indolin-2-one derivatives catalyzed by ceric ammonium nitrate (CAN) under ultrasound irradiation. Tetrahedron 62, 1527–1535. doi: 10.1016/j.tet.2005.11.011
Weiner, R. M., Segall, A. M., and Colwell, R. R. (1985). Characterization of a marine bacterium associated with Crassostrea Virginica (the Eastern Oyster). Appl. Environ. Microbiol. 49, 83–90.
Weirauch, U., Beckmann, N., Thomas, M., Grünweller, A., Huber, K., Bracher, F., et al. (2013). Functional role and therapeutic potential of the pim-1 kinase in colon carcinoma. Neoplasia 15, 783–794. doi: 10.1593/neo.13172
Wishart, D. S., Tzur, D., Knox, C., et al. (2007). HMDB: the human metabolome database. Nucleic Acids Res. 35, D521–D526. doi: 10.1093/nar/gkl923
Wu, Y., and Sun, Y.P. (2006). Synthesis of nonactin and the proposed structure of trilactone. Org. Lett. 13, 2831–2834. doi: 10.1021/ol0609661
Yi-Lei, N., Yun-Dan, W., Chuan-Xi, W., Ru, L., Yang, X., Dong-Sheng, F., et al. (2014). Compounds from marine-derived Verrucosispora sp. FIM06054 and their potential antitumour activities Nat. Prod. Res. 28, 2134–2139. doi: 10.1080/14786419.2014.926350
Zhang, H. P., Kamano, Y., Ichihara, Y., Kizu, H., Komiyama, K., Itokawa, H., et al. (1995). Isolation and structure of convolutamydines B~D from marine bryozoan Amathia convoluta. Tetrahedron 51, 5523–5528. doi: 10.1016/0040-4020(95)00241-Y
Keywords: Saccharomonospora sp., Dietzia sp., actinomycetes, Saccharomonosporine A, convolutamydine F, docking, Pim-1 kinase, co-cultivation
Citation: El-Hawary SS, Sayed AM, Mohammed R, Khanfar MA, Rateb ME, Mohammed TA, Hajjar D, Hassan HM, Gulder TAM and Abdelmohsen UR (2018) New Pim-1 Kinase Inhibitor From the Co-culture of Two Sponge-Associated Actinomycetes. Front. Chem. 6:538. doi: 10.3389/fchem.2018.00538
Received: 04 June 2018; Accepted: 16 October 2018;
Published: 15 November 2018.
Edited by:
Peter Proksch, Heinrich Heine Universität Düsseldorf, GermanyReviewed by:
Margherita Brindisi, Università degli Studi di Siena, ItalyDiego Brancaccio, Università degli Studi di Napoli Federico II, Italy
Copyright © 2018 El-Hawary, Sayed, Mohammed, Khanfar, Rateb, Mohammed, Hajjar, Hassan, Gulder and Abdelmohsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tobias A. M. Gulder, dG9iaWFzLmd1bGRlckBjaC50dW0uZGU=
Usama Ramadan Abdelmohsen, dXNhbWEucmFtYWRhbkBtdS5lZHUuZWc=
 Seham S. El-Hawary1
Seham S. El-Hawary1 Ahmed M. Sayed
Ahmed M. Sayed Mohammad A. Khanfar
Mohammad A. Khanfar Mostafa E. Rateb
Mostafa E. Rateb Hossam M. Hassan
Hossam M. Hassan Tobias A. M. Gulder
Tobias A. M. Gulder Usama Ramadan Abdelmohsen
Usama Ramadan Abdelmohsen