
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 26 June 2018
Sec. Chemical Biology
Volume 6 - 2018 | https://doi.org/10.3389/fchem.2018.00254
 Lin Jiang1
Lin Jiang1 Hong Pu1
Hong Pu1 Jingxi Xiang1
Jingxi Xiang1 Meng Su1
Meng Su1 Xiaohui Yan2
Xiaohui Yan2 Dong Yang2
Dong Yang2 Xiangcheng Zhu1,3
Xiangcheng Zhu1,3 Ben Shen2,4,5
Ben Shen2,4,5 Yanwen Duan1,3,6*
Yanwen Duan1,3,6* Yong Huang1,6*
Yong Huang1,6*Three natural products of nonaketide biosynthetic origin, probably biosynthesized from nine molecules of malonyl-CoA, have been isolated. Herein we described the isolation and structure elucidation of huanglongmycin (HLM) A-C and identification of the putative hlm biosynthetic gene cluster from Streptomyces sp. CB09001, isolated from a karstic cave in Xiangxi, China. Albeit previously isolated, HLM A was reported for the first time to exhibit moderate cytotoxicity against A549 lung cancer cell line (IC50 = 13.8 ± 1.5 μM) and weak antibacterial activity against gram-negative clinical isolates. A putative biosynthetic pathway for HLM A, featuring a nonaketide-specific type II polyketide synthase, was proposed. It would be consistent with the isolation of HLM B and C, which are two new natural products and likely shunt metabolites during HLM A biosynthesis.
Aromatic polyketides of microbial origin are a large family of natural products with important biological activities, including anticancer agents doxorubicin and mithramycin, and antibiotics tetracyclines. Aromatic polyketides are typically biosynthesized by type II polyketide synthases (PKSs), minimally consisting of two ketosynthases KSα and KSβ (also named “chain length factor”) and an acyl carrier protein (ACP), that are associated with additional ketoreductases and cylases/aromatases (Hopwood, 1997; Shen, 2000; Hertweck et al., 2007; Das and Khosla, 2009; Zhou et al., 2010; Zhang et al., 2017). The KSα, KSβ, and ACP are often clustered together and called minimal PKSs. Extensive biosynthetic studies of these polyketides, such as actinorhodin (1), frenolicin (2), and tetracenomycin C (3), have helped establish certain “design rules” to make designer analogs, such as DMAC (5) and SEK26 (6), by a biotechnology platform now often termed “combinatorial biosynthesis” (Figure 1) (McDaniel et al., 1993, 1995; Kramer et al., 1997; Yu et al., 1998). However, only a few natural nonaketide-derived aromatic polyketides, including the biaryl compounds julichromes (11), setomimycin and spectomycins, have been proposed to be biosynthesized by the Claisen-like condensation of nine malonyl-CoA (Marti et al., 2000; Zhang et al., 2008; Präg et al., 2014). Interestingly, the nonaketide precursors could form homo- or heterodimers through oxidative phenol coupling (Präg et al., 2014).
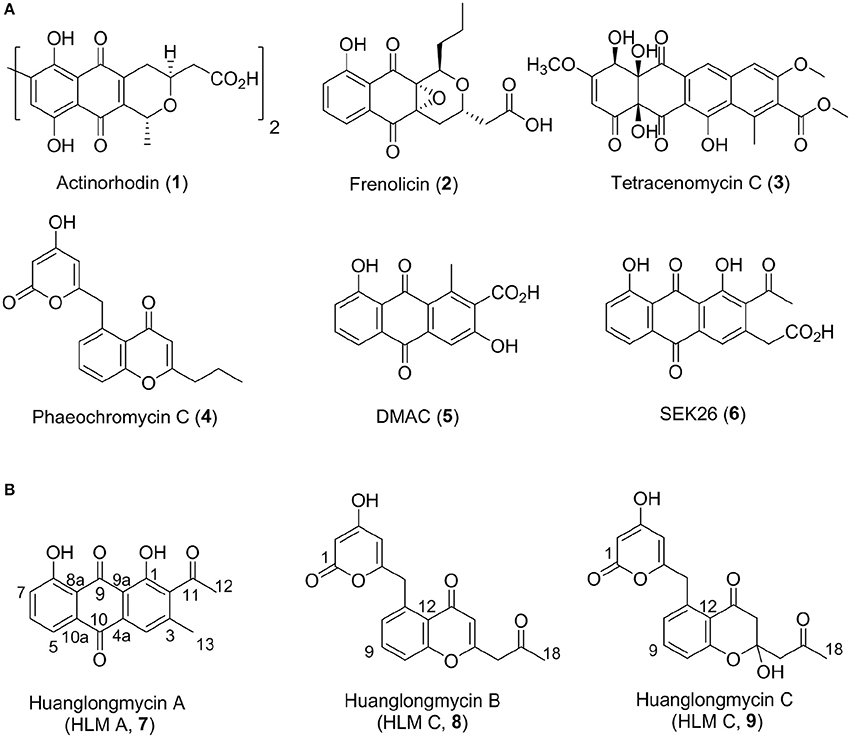
Figure 1. Structures of selected aromatic polyketides and their biosynthetic shunt products resulting from combinatorial biosynthesis (1–6) (A) and the three huanglongmycin A–C (7–9) isolated from S. sp. CB09001 (B).
We have been interested in collecting diverse microbial strains for natural product discovery and recently isolated several new natural products with antibacterial and anticancer activities (Luo et al., 2014; Ma et al., 2015, 2017; Yan et al., 2016; Pan et al., 2017). The western part of Hunan province (also named “Xiangxi”) in China is on the east of Yunnan-Kweichow plateau, which was geologically formed through epeirogeny uplifts in the late Permian period about 200-million years ago (Luo et al., 2014). Xiangxi is well-known for its characterized karstic topography and biodiversity (Chen et al., 2015). In this report, we describe the isolation, structure elucidation, biosynthesis, and biological activities of three aromatic polyketides from strain S. sp. CB09001, isolated from a karstic cave in Xiangxi (Figure 1).
IR spectra were recorded on Nicolet iS50 FT-IR (Thermo Scientific). CD spectra were recorded on J-815 from JASCO. HRMS spectra were recorded on a LTQ-ORBITRAP-ETD instrument. NMR spectra were acquired using a Brucker 400 or 500 MHz spectrometer. Chemical shifts were reported in ppm relative to CDCl3 (δ = 7.26 ppm) or DMSO-d6 (δ = 2.50 ppm) for 1H NMR and CDCl3 (δ = 77.23 ppm) or DMSO-d6 (δ = 39.60 ppm) for 13C NMR spectroscopy. Column chromatography (CC) was carried out on silica gel (100–200 mesh and 300–400 mesh, Yantai Jiangyou Silica Gel Development Co., Ltd., Yantai, China) and polyamide (100–200 mesh, Sinopharm Chemical Reagent Co., Ltd., Shanghai, China). Semipreparative reversed phase-high-performance liquid chromatography (RP-HPLC) was performed using a Waters 1525 Binary HPLC Pump equipped with a Waters 2489 UV/Visible Detector and using a Welch Ultimate AQ-C18 column (250 × 10 mm, 5 μm).
The S. sp. CB09001 strain was grown on G1 agar (2% soluble starch, 0.1% KNO3, 0.05% K2HPO4, 0.05% MgSO4·7H2O, 0.05% NaCl, 0.001% FeSO4·7H2O, 1.5% agar) plates and incubated at 28°C to obtain spores. For the genomic DNA isolation, S. sp. CB09001 was cultured in 250-mL baffled flasks containing 50 mL TSB liquid medium with 0.5% glycine and cultivated at 28°C for 36 h. Genomic DNA was isolated using standard protocols (Kieser et al., 2000). The 16S rRNA of S. sp. CB09001 was PCR-amplified and sequenced. The genome of S. sp. CB09001 was sequenced using the combination of Illumina Miseq/Hiseq and Pacific RSII platforms (Shanghai Majorbio Co., Ltd., Shanghai, China).
The strain S. sp. CB09001 was grown in 250 mL Erlenmeyer flasks containing 50 mL of G1 medium and were incubated at 30°C on rotary shakers (230 rpm) for 72 h. Then 3% macroporous resin DA201-H (Jiangsu Su Qing Water Treatment Engineering Group Co., Ltd., Jiangyin, China) or different concentration triclosan (0–20 μM) were added, and fermented for additional 4 days. S. sp. CB09001 was also fermented in G1 fermentation medium with different fermentation volume in 250-mL flat-bottom Erlenmeyer flasks from 25 to 150 mL per flask. In scale-up fermentation, each of the seed cultures in G1 medium (50 mL) was aseptically transferred to 2 L Erlenmeyer flasks containing 1 L of G1 medium. All of these flasks were then incubated at 30°C on rotary shakers (200 rpm) for 7 days.
After fermentation, the culture (6 L) was centrifuged (4,000 rpm, 10 min) to yield the supernatant. After its pH was adjusted to 0.9 using concentrated HCl, the supernatant was extracted with EtOAc (3 × 6 L), which was concentrated under reduced pressure and subjected to silica gel CC using petroleum ether (PE)/EtOAc and EtOAc/MeOH containing 1% formic acid. Fractions A2-A3 (PE/EtOAc 90/10-80/20) was further purified by silica gel CC eluting with PE/EtOAc (90/10) to give three fractions. The first fraction was applied to silica gel CC eluted with CH2Cl2/MeOH (95/5) to obtain compound 7 (5.5 mg), which was further purified by recrystallization. Fractions A12-A15 (EtOAc/MeOH 90/10-60/40) were combined and subjected to silica gel CC using gradient elution with CH2Cl2/MeOH. The fractions (CH2Cl2/MeOH 92/8-90/10) were combined and subjected to polyamide CC using gradient elution with CH2Cl2/MeOH containing 1% formic acid (100/0, 94/6, 92/8, 90/10, 88/12, 86/14, 84/16, 80/20, 70/30, 60/40, 50/50, 40/60, 20/80, 0/100). The fraction C6 (86/14) was further purified by semipreparative RP-C18 HPLC (Welch Ultimate AQ 5 μm, 250 × 10 mm) with a flow rate of 3 mL/min and a gradient elution of CH3CN/H2O (containing 0.2% formic acid) in 20 min (5 to 95% for 15 min, followed by 95% for 2 min, and 95 to 5% for 0.5 min, followed by 5% for 2.5 min), to afford compounds 8 (tR = 12.0 min, 5.3 mg) and 9 (tR = 11.8 min, 33.3 mg), respectively.
Huanglongmycin A (7): Yellowish-brown solid; UV (MeOH) λmax (Abs) 192.8 (0.268), 225.8 (0.388), 255.4 (0.300), 428.7 (0.144); 1H, 13C, and 2D NMR spectroscopic data, see Table 1; (−)-HRESIMS m/z 295.0619 [M - H]− (calcd. for C17H11O5, 295.0612).
Huanglongmycin B (8): Light yellow needle crystal; UV (MeOH) λmax (Abs) 200.4 (0.473), 229.8 (0.297), 298.6 (0.200); IR(νmax) 3,110, 2,960, 1,730, 1,710, 1,640, 1,590, 1,570, 1,480, 1,400, 1,320, 1,290, 1,160 cm−1; 1H, 13C, and 2D NMR spectroscopic data, see Table 1; (+)-HRESIMS m/z 327.0869 [M + H]+ (calcd. for C18H15O6, 327.0863), m/z 349.0687 [M + Na]+, m/z 653.1659 [2M + H]+, m/z 675.1480 [2M + Na]+.
Huanglongmycin C (9): Light yellow solid; UV (MeOH) λmax (Abs) 199.2 (0.488), 257.0 (0.170), 287.9 (0.142); IR(νmax) 3,440, 2,950, 1,650, 1,440, 1,310, 1,030 cm−1; 1H, 13C, and 2D NMR spectroscopic data, see Table 1; (+)-HRESIMS m/z 367.0794 [M + Na]+ (calcd. for C18H16O7Na, 367.0788), m/z 327.0870 [M - H2O + H]+, m/z 689.1872 [2M + H]+.
Cytotoxicity assay: Compounds 7–9 were evaluated for their cytotoxicity against the human cancer cell lines A549, SKOV3, Hela, and Caco-2 using the standard MTT method (Tai et al., 2014).
Antibacterial assay: Compounds 7–9 were tested for their antibacterial activities against Staphyloccocus aureus ATCC 29213, methicillin-sensitive Staphylococcus aureus, methicillin-resistant Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa using a previously described method (Wiegand et al., 2008).
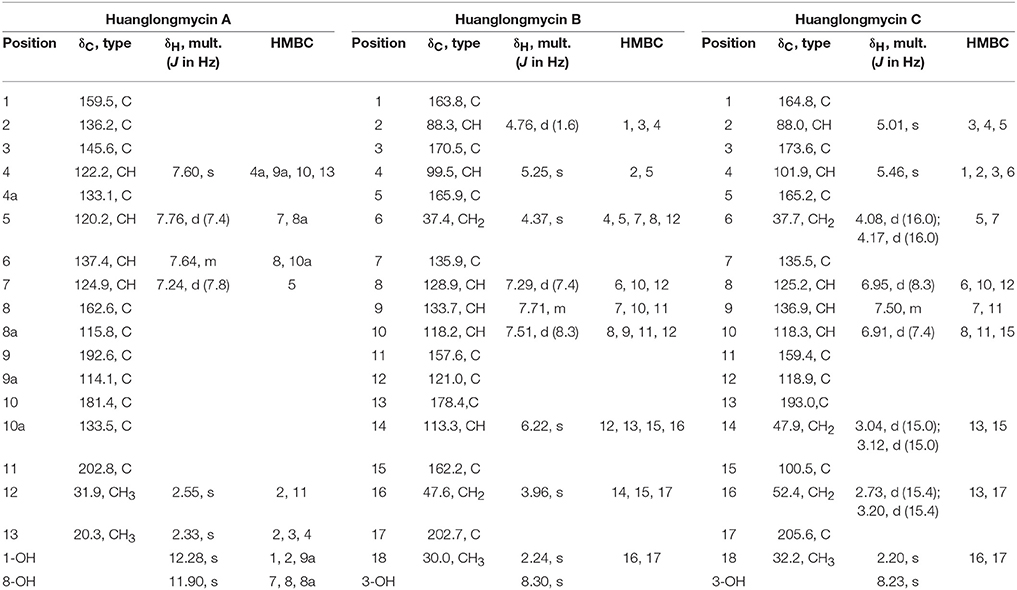
Table 1. NMR Spectroscopic Data for huanglongmycin A (HLM A, 7), huanglongmycin B (HLM B, 8) and huanglongmycin C (HLM C, 9).
The 16S rRNA sequence and genome sequence of S. sp. CB09001 have been submitted to GenBank with accession number MG890330 and CP026730, respectively.
We collected several soil samples from Huanglong cave in Xiangxi, from which new actinomycetes strains were isolated. One of the strains, CB09001, was classified as a Streptomyces species based on its morphological characteristics and the 16S rRNA sequence (NCBI accession no. MG890330). Encouraged by its unique metabolite profiles, S. sp CB09001 was fermented under various fermentation conditions, such as the addition of elicitor triclosan, and the variation of the fermentation volume and the addition of macroporous resin (Figures S1–S3) (Bode et al., 2002; Yoon and Nodwell, 2014; Tanaka et al., 2017). For large scale fermentation, S. sp. CB9001 was fermented in six 2-L Erlenmeyer flasks, containing 1 L of G1 medium, and isolation of natural products from the fermentation culture afforded three compounds, we named huanglongmycin (HLM) A–C (7–9). Their structures were established on the basis of HR-ESI-MS and extensive 1D and 2D NMR analysis (Figure 2, Figures S4–S25, Table 1).
HLM A (7) was obtained as a yellowish-brown solid. The molecular formula of 7 was determined as C17H12O5 based on HR-ESI-MS analysis, affording an [M - H]− ion at m/z 295.0619 (calculated for [M - H]− ion at m/z 295.0606), which indicated twelve degrees of unsaturation (Figure S8). Analysis of the 1H NMR, 13C NMR, HSQC, HMBC, and 1H-1H COZY spectra suggested that the structure of 7 was similar to several known anthraquinones DMAC (5) and SEK26 (6) from rationally designed type II PKSs (Figure 1, Figures S11–S15) (McDaniel et al., 1993, 1995). The presence of an acetyl group was based on the HMBC from H-12 (δH 2.55, -CH3) to C-11 (δC 202.8). The observation of HMBC from H-4 (δH 7.60) to C-4a (δC 133.1), C-9a (δC 114.1), C-3 (δC 145.6), and C-13 (δC 20.3), established the methyl group at C-3, hence the unambiguous assignment of the structure of 7 (Figure S14). Compound 7 was firstly obtained in the process to elucidate the structure of an anthraquinone protetrone through chemical degration, and later it was obtained in a medicinal chemistry program and also isolated from a Streptomyces strain, which was discovered from a soil sample in Mansoura, Egypt, but no detailed biological characterization was reported (McCormick and Jensen, 1968; Tietze et al., 2007; Abdelfattah, 2009). Compound 7 isolated from S. sp. CB9001 in this study showed consistent MS and NMR data with the previous reports.
HLM B (8) was obtained as a light yellow needle crystal. HR-ESI-MS data established the molecular formula of 8 as C18H14O6 (m/z 349.0687 [M + Na]+, calculated for [M + Na]+ ion at m/z 349.0687), indicating twelve degrees of unsaturation (Figure S9). The 1H NMR spectroscopic data of 8 revealed three aromatic protons at H-8 (δH 7.29, 1H, d, J = 7.4 Hz), H-9 (δH 7.71 dd, J = 7.4, 8.3 Hz), H-10 (δH 7.5, d, J = 8.3 Hz), which can be grouped into one ABX spin system with the aid of the COZY correlations of H-8/H-9/H-10 (Figure 2, Figures S16, S20). Further analysis of the NMR spectra suggested that the structure of 8 was similar to the known polyketide phaeochromycin C (4), except the presence of one additional keto carbonyl group C-17 (δC 202.7) (Figure 1, Figure S17) (Graziani et al., 2005). Presence of the pyrone moiety in 8 was confirmed by analyzing its proton and carbon NMR spectroscopic data, combined with the correlations of H-2 (δH 4.76) to C-1 (δC 163.8), C-3 (δC 170.5), and C-4 (δC 99.5), and H-4 (δH 5.25) to C-2 (δC 88.3), C-3 (δC 170.5), C-5 (δC 165.9), and C-6 (δC 37.4) in the HMBC spectrum (Figures S16–S19).
HLM C (9) was isolated as a light yellow solid. The molecular formula C18H16O7, was established upon analysis of the HR-ESI-MS peak at m/z 327.0870 [M - H2O + H]+ and 689.1872 [2M + H]+ (calculated for [M - H2O + H]+ ion at m/z 327.0869, and [2M + H]+ ion at m/z 689.1870), which afforded eleven degrees of unsaturation (Figure S10). The 1H, 13C and 2D NMR data obtained for HLM C (9) were similar to those obtained for 8, except the presence of a methylene C-14 (δC 37.4), and a quaternary carbon C-15 (δC 100.5) connected to a hydroxy group (Table 1, Figures S21–S25). Its structure was further supported by the HMBC of H-14 (δH 47.9) to C-12 (δC 118.9), C-13 (δC 193.0), C-15 (δC 100.5), and C-16 (δC 52.4) (Figure S24).
Inspired by the previous combinatorial biosynthesis approaches to generate the “unnatural” natural products by type II PKSs, such as 5 and 6, and the fact that only a few biosynthetic gene clusters of nonaketides have been reported, we decided to identify the huanglongmycin (hlm) gene cluster in S. sp. CB09001 to shed light on the biosynthesis of HLM A–C (Figure 3, Table 2). The genome of S. sp. CB09001 was sequenced and bioinformatic analysis using antiSMASH revealed that there are three putative type II PKS gene clusters (GenBank accession number CP026730) (Blin et al., 2017). One gene cluster is likely to be involved in the biosynthesis of HLMs, and the other two share very high sequence homology to gray pigment or angucycline biosynthesis (Tables S1, S2). The putative KSβ gene hlmF is located in the KSβ group for nonaketide biosynthesis, based on the phylogenetic analysis of known 42 type II PKS gene clusters whose secondary metabolite products have been structurally characterized (Figure 4) (Hillenmeyer et al., 2015). Pairwise comparison of the annotated proteins between the hlm cluster with the julichrome biosynthetic gene cluster revealed high amino acid identity, strongly supporting that the hlm gene cluster most likely encodes the biosynthesis of HLMs in S. sp. CB09001 (Table 2).
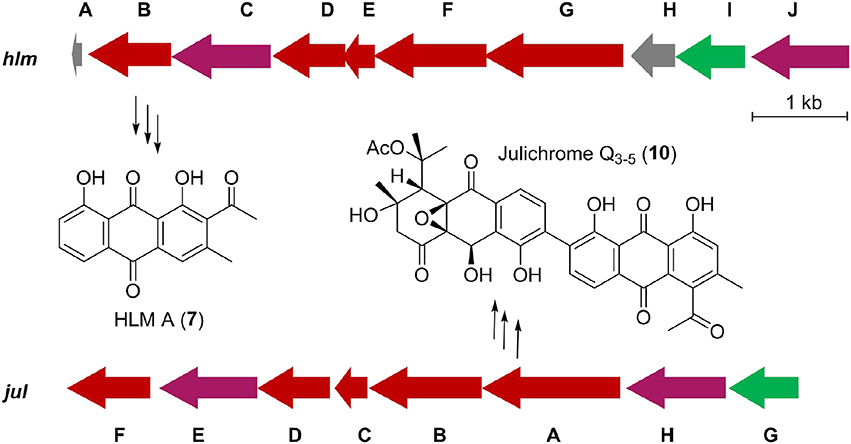
Figure 3. Predicted gene organization of huanglongmycin (hlm) cluster and julichrome (jul) cluster. Genes are color-coded according to their proposed functions. Red, purple, green, and gray represent core biosynthetic genes, additional biosynthetic genes, regulatory genes and other genes, respectively.
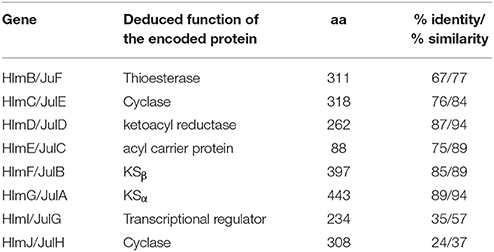
Table 2. Alignment of the homologous proteins from the huanglongmycin (hlm) and the julichrome (jul) biosynthetic gene cluster.
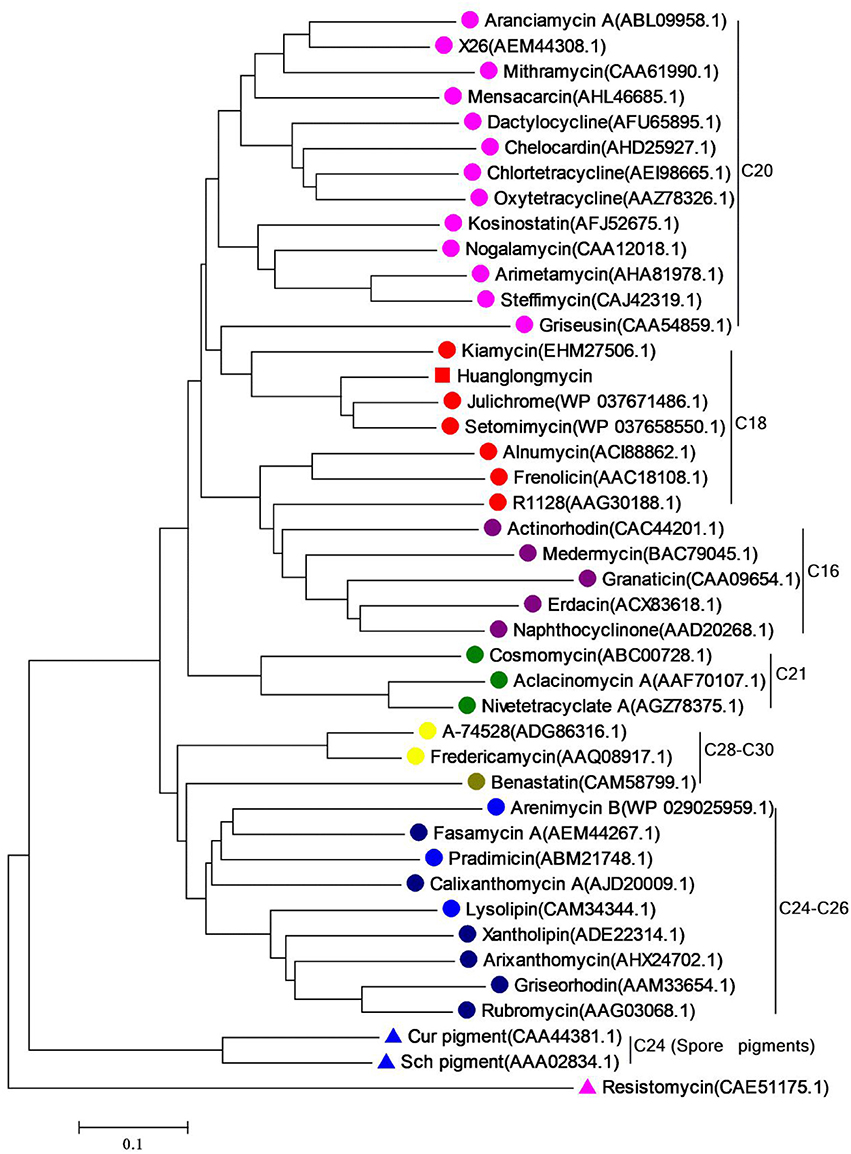
Figure 4. Phylogenetic analysis of the KSβ gene of huanglongmycin gene cluster. The same color circles (or triangles) represent the same polyketide chain length (noted on the right).
Thus, we now propose a pathway for HLM A–C biosynthesis, featuring a nonaketide intermediate by the HLM minimal PKS, containing KSα (HlmG), KSβ (HlmF), and ACP (HlmE), which catalyzes the decarboxylative condensation of nine molecules of malonyl-CoA (Figure 5). Reduction of the poly-β-keto intermediate by the C-9 specific KR (HlmD), followed by the di-domain BexL-type reducing cyclase HlmC might afford the initial C7-C12 cyclization (Caldara-Festin et al., 2015). Interestingly, the isolation of the shunt metabolites HLM B and C suggested the leakage of the HLM polyketide intermediate during its transfer to the cyclase HlmJ under specific fermentation conditions, which were often observed in the engineered mutants or in vitro enzymatic assays (Shen and Hutchinson, 1993; Kharel et al., 2010). The formation of HLM A might be from the previous SEK26 (6) polyketide through decarboxylation, the MS of which was also observed in the fermentation extract from an E. coli strain containing a fungi PKS, one KR and two cyclases (Zhang et al., 2008). It was similar to the formation of anthraquione aloesaponarin II from DMAC (McDaniel et al., 1993). The above proposal was in agreement with the previous engineered biosynthesis of aromatic polyketide RM 18 (11) and nonaSEK14 (12) from the same nonaketide intermediate, albeit from a wild-type Streptomyces strain (Figure 5) (McDaniel et al., 1993; Hopwood, 1997; Zhang et al., 2008). The similar biosynthetic pathway for huanglongmycin A might be also operational in the Streptomyces strain isolated from Egypt, which suggested that natural nonaketides might be underappreciated (Abdelfattah, 2009).
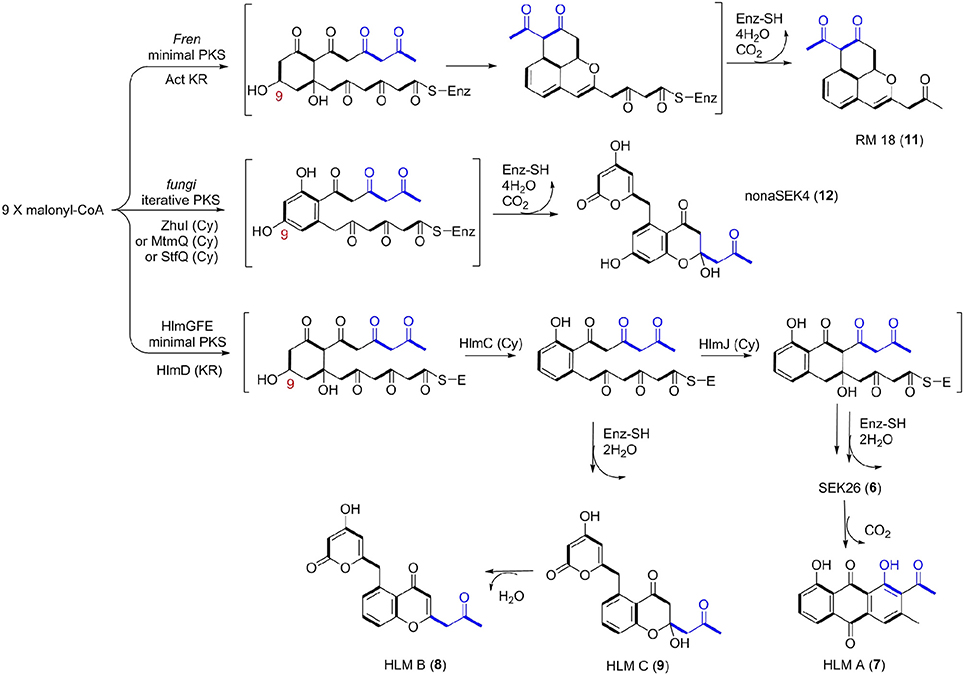
Figure 5. Biosynthetic Pathways for 18-Carbon Aromatic Polyketides. Biosynthesis between the known aromatic polyketides (i.e., nonaSEK4 and RM 18) and the putative HLM gene cluster were compared. The putative hlm minimal PKS contains HlmG, HlmF, and HlmE, the Fren PKS is the minimal PKS of frenolicin, while act KR is the ketoacyl reductase from actinorhodin PKS. The fungal iterative PKS is the megasynthase PKS4 from Gibberella fujikuroi. ZhuI, MtmQ, and StfQ are the cyclases of aromatic polyketide R1128, mithramycin, and steffimycin, respectively, which catalyzed the region-specific cyclization at C7–C12.
HLM A–C (7–9) were first evaluated for their antibacterial activities against Staphylococcus aureus ATCC 29213, and several clinical isolates from local hospitals, including methicillin-sensitive S. aureus (MSSA) strain and methicillin-resistant S. aureus (MRSA) strains, as well as strains of Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa, using chloramphenicol and linezolid as controls (Table 3). HLM A showed weak antibacterial activities against the gram-negative pathogens P. aeruginosa and E. coli, with minimum inhibitory concentrations (MICs) of 64 μg/mL. HLM B showed weak antibacterial activities against the tested S. aureus strains, with a MIC of 64 μg/mL, while HLM C showed no antibacterial activities against the five indicator strains (Table 3).
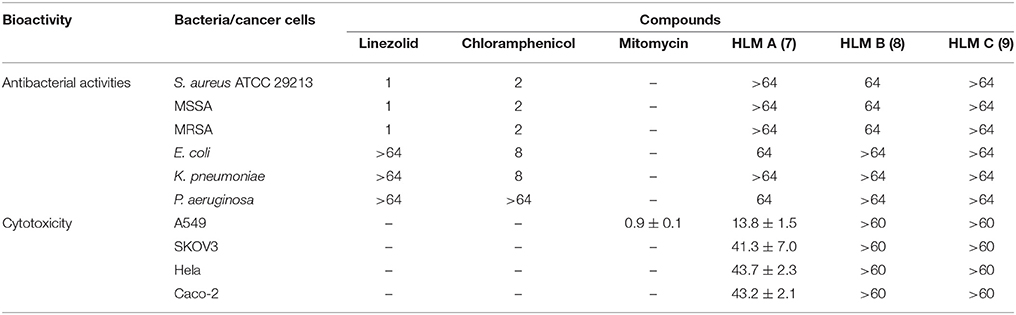
Table 3. Antibacterial activities (MICs, μg/mL) and cytotoxic activities (IC50, μM) of huanglongmycin A–C (7–9). S. aureus 29213, Staphylococcus aureus ATCC 29213; MSSA, Methicillin-sensitive Staphylo-coccus aureus; MRSA, Methicillin-resistant Staphylococcus aureus; –, not tested.
The in vitro cytotoxicity of compounds 7–9 were evaluated against four human cancer cell lines, including non-small cell lung cancer cell line A549, epithelial cancer cell line SKOV3, Hela and human epithelial colorectal adenocarcinoma (Caco-2) using MTT assay. HLM A showed moderate cytotoxicity against A549 (IC50 = 13.8 ± 1.5 μM) and weak cytotoxicities against SKOV3, Hela and Caco-2 (IC50 = 41.3 ± 7.0, 43.7 ± 2.3, and 43.2 ± 2.1 μM), while HLM B and C showed no cytotoxicities at the tested condition (IC50 > 60 μM).
In summary, this work describes the isolation and characterization of three natural products HLM A-C, and the identification of a putative gene cluster, featuring a type II PKS specific for nonaketide biosynthesis from S. sp. CB09001. The proposal for HLMs biosynthesis agreed well with the previous engineered biosynthesis for bacteria aromatic polyketides in the past few decades (McDaniel et al., 1993, 1995; Hopwood, 1997; Kramer et al., 1997; Yu et al., 1998; Shen, 2000; Hertweck et al., 2007; Zhang et al., 2008, 2017; Das and Khosla, 2009; Zhou et al., 2010). Albeit previously isolated, HLM A was reported for the first time, to exhibit moderate cytotoxicity against A549 cancer cell line (IC50 = 13.8 ± 1.5 μM) and weak antibacterial activity against P. aeruginosa (MICs = 64 μg/mL). This study showcased once again Nature is the ultimate combinatorial biosynthetic chemist, and should inspire continued efforts for natural product discovery from unexplored and underexplored ecological niches.
YH, YD, and BS: conceived the project; LJ and HP: performed experiments; JX and MS: carried out the cytotoxicity assays; XY, DY, and XZ: contributed to bioinformatics analysis; YH and LJ: wrote the manuscript with help from the other authors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported in parts by NSFC grants 81473124 (to YH). We thank the Center for Advanced Research in Central South University for the HRMS and NMR experiments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2018.00254/full#supplementary-material
Abdelfattah, M. S. (2009). Mansoquinone: isolation and structure elucidation of new antibacterial aromatic polyketides from terrestrial. Nat. Prod. Res. 23, 212–218. doi: 10.1080/14786410801961550
Blin, K., Wolf, T., Chevrette, M. G., Lu, X., Schwalen, C. J., Kautsar, S. A., et al. (2017). antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucl. Acids Res. 45, W36–W41. doi: 10.1093/nar/gkx319
Bode, H. B., Bethe, B., Höfs, R., and Zeeck, A. (2002). Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem 3, 619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9
Caldara-Festin, G., Jackson, D. R., Barajas, J. F., Valentic, T. R., Patel, A. B., Aguilar, S., et al. (2015). Structural and functional analysis of two di-domain aromatase/cyclases from type II polyketide synthases. Proc. Natl. Acad. Sci. U.S.A. 112, e6844–e6851. doi: 10.1073/pnas.1512976112
Chen, G. X., Liao, W. B., Xiong, L. Z., and Zhang, Y. K. (2015). Xiangxi Medicinal Plant Resources Development and Sustainable Utilization. Chengdou: Southwest Jiaotong University Press.
Das, A., and Khosla, C. (2009). Biosynthesis of aromatic polyketides in bacteria. Acc. Chem. Res. 42, 631–639. doi: 10.1021/ar8002249
Graziani, E. I., Ritacco, F. V., Bernan, V. S., and Telliez, J. B. (2005). Phaeochromycins A-E, anti-inflammatory polyketides isolated from the soil actinomycete Streptomyces phaeochromogenes LL-P018. J. Nat. Prod. 68, 1262–1265. doi: 10.1021/np0500629
Hertweck, C., Luzhetskyy, A., Rebets, Y., and Bechthold, A. (2007). Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 24, 162–190. doi: 10.1039/B507395M
Hillenmeyer, M. E., Vandova, G. A., Berlew, E. E., and Charkoudian, L. K. (2015). Evolution of chemical diversity by coordinated gene swaps in type II polyketide gene clusters. Proc. Natl. Acad. Sci. U.S.A. 112, 13952–13957. doi: 10.1073/pnas.1511688112
Hopwood, D. A. (1997). Genetic contributions to understanding polyketide synthases. Chem. Rev. 97, 2465–2497. doi: 10.1021/cr960034i
Kharel, M. K., Pahari, P., Lian, H., and Rohr, J. (2010). Enzymatic total synthesis of rabelomycin, an angucycline group antibiotic. Org. Lett. 12, 2814–2817. doi: 10.1021/ol1009009
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation.
Kramer, P. J., Zawada, R. J. X., McDaniel, R., Hutchinson, C. R., Hopwood, D. A., and Khosla, C. (1997). Rational design and engineered biosynthesis of a novel 18-carbon aromatic polyketide. J. Am. Chem. Soc. 119, 635–639. doi: 10.1021/ja962888q
Luo, J. X., He, Y. B., Wang, R., Liu, M., and Hu, X. F. (2014). Lithofacies palaeogeography of the Late Permian Wujiaping Age in the Middle and Upper Yangtze Region, China. J. Palaeogeogr. 3, 384–409. doi: 10.3724/SP.J.1261.2014.00063
Ma, M., Rateb, M. E., Teng, Q., Yang, D., Rudolf, J. D., Zhu, X., et al. (2015). Angucyclines and Angucyclinones from Streptomyces sp. CB01913 Featuring C-Ring Cleavage and Expansion. J. Nat. Prod. 78, 2471–2480. doi: 10.1021/acs.jnatprod.5b00601
Ma, M., Rateb, M. E., Yang, D., Rudolf, J. D., Zhu, X., Huang, Y., et al. (2017). Germicidins H-J from Streptomyces sp. CB00361. J. Antibiot. (Tokyo) 70, 200–203. doi: 10.1038/ja.2016.100
Marti, T., Hu, Z., Pohl, N. L., Shah, A. N., and Khosla, C. (2000). Cloning, nucleotide sequence, and heterologous expression of the biosynthetic gene cluster for R1128, a non-steroidal estrogen receptor antagonist. Insights into an unusual priming mechanism. J. Biol. Chem. 275, 33443–33448. doi: 10.1074/jbc.M006766200
McCormick, J. R. D., and Jensen, E. R. (1968). Biosynthesis of tetracyclines. X. protetrone1. J. Am. Chem. Soc. 90, 7126–7127. doi: 10.1021/ja01027a045
McDaniel, R., Ebert-Khosla, S., Hopwood, D. A., and Khosla, C. (1993). Engineered biosynthesis of novel polyketides: manipulation and analysis of an aromatic polyketide synthase with unproven catalytic specificities. J. Am. Chem. Soc. 115, 11671–11675. doi: 10.1021/ja00078a002
McDaniel, R., Ebert-Khosla, S., Hopwood, D. A., and Khosla, C. (1995). Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature 375, 549–554. doi: 10.1038/375549a0
Pan, G., Xu, Z., Guo, Z., Hindra Ma, M., Yang, D., et al. (2017). Discovery of the leinamycin family of natural products by mining actinobacterial genomes. Proc. Natl. Acad. Sci. U.S.A. 114, e11131–e11140. doi: 10.1073/pnas.1716245115
Präg, A., Grüning, B. A., Häckh, M., Lüdeke, S., Wilde, M., Luzhetskyy, A., et al. (2014). Regio- and stereoselective intermolecular oxidative phenol coupling in Streptomyces. J. Am. Chem. Soc. 136, 6195–6198. doi: 10.1021/ja501630w
Shen, B., and Hutchinson, C. R. (1993). Enzymatic synthesis of a bacterial polyketide from acetyl and malonyl coenzyme A. Science 262, 1535–1540. doi: 10.1126/science.8248801
Tai, W., Chen, Z., Barve, A., Peng, Z., and Cheng, K. (2014). A novel rapamycin-polymer conjugate based on a new poly(ethylene glycol) multiblock copolymer. Pharm. Res. 31, 706–719. doi: 10.1007/s11095-013-1192-3
Tanaka, Y., Izawa, M., Hiraga, Y., Misaki, Y., Watanabe, T., and Ochi, K. (2017). Metabolic perturbation to enhance polyketide and nonribosomal peptide antibiotic production using triclosan and ribosome-targeting drugs. Appl. Microbiol. Biotechnol. 101, 4417–4431. doi: 10.1007/s00253-017-8216-6
Tietze, L. F., Gericke, K. M., and Schuberth, I. (2007). Synthesis of highly functionalized anthraquinones and evaluation of their antitumor activity. Eur. J. Org. Chem. 27, 4563–4577. doi: 10.1002/ejoc.200700418
Wiegand, I., Hilpert, K., and Hancock, R. E. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Yan, X., Ge, H., Huang, T., Hindra Yang, D., Teng, Q., et al. (2016). Strain prioritization and genome mining for enediyne natural products. mBio 7, e02104–e02116. doi: 10.1128/mBio.02104-16
Yoon, V., and Nodwell, J. R. (2014). Activating secondary metabolism with stress and chemicals. J. Ind. Microbiol. Biotechnol. 41, 415–424. doi: 10.1007/s10295-013-1387-y
Yu, T. W., Shen, Y., McDaniel, R., Floss, H. G., Khosla, C., Hopwood, D. A., et al. (1998). Engineered biosynthesis of novel polyketides from Streptomyces spore pigment polyketide synthases. J. Am. Chem. Soc. 120, 7749–7759. doi: 10.1021/ja9803658
Zhang, W., Li, Y., and Tang, Y. (2008). Engineered biosynthesis of bacterial aromatic polyketides in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 20683–20688. doi: 10.1073/pnas.0809084105
Zhang, Z., Pan, H. X., and Tang, G. L. (2017). New insights into bacterial type II polyketide biosynthesis. F1000Res. 6, 172–183. doi: 10.12688/f1000research.10466.1
Keywords: nonaketide, type II polyketide synthase, karstic cave, huanglongmycin, cytotoxicity
Citation: Jiang L, Pu H, Xiang J, Su M, Yan X, Yang D, Zhu X, Shen B, Duan Y and Huang Y (2018) Huanglongmycin A-C, Cytotoxic Polyketides Biosynthesized by a Putative Type II Polyketide Synthase From Streptomyces sp. CB09001. Front. Chem. 6:254. doi: 10.3389/fchem.2018.00254
Received: 01 April 2018; Accepted: 08 June 2018;
Published: 26 June 2018.
Edited by:
Maria Luisa Mangoni, Sapienza Università di Roma, ItalyReviewed by:
Max Julian Cryle, Monash University, AustraliaCopyright © 2018 Jiang, Pu, Xiang, Su, Yan, Yang, Zhu, Shen, Duan and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanwen Duan, eXdkdWFuNjZAc2luYS5jb20=
Yong Huang, am9uZ2h1YW5nQGNzdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.