
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Chem. Eng., 30 July 2024
Sec. Biochemical Engineering
Volume 6 - 2024 | https://doi.org/10.3389/fceng.2024.1446454
 Xiang Li1†
Xiang Li1† Rongshuai Jiang1†
Rongshuai Jiang1† Shenghou Wang2
Shenghou Wang2 Chenyang Li1
Chenyang Li1 Yongping Xu3,4
Yongping Xu3,4 Shuying Li3
Shuying Li3 Qian Li1,3*
Qian Li1,3* Liang Wang3,5*
Liang Wang3,5*Cordycepin, an adenosine analog, exhibits diverse bioactivities and holds significant potential for applications in healthcare and agriculture. Fungi of the genus Cordyceps, such as Cordyceps militaris, can naturally produce cordycepin. Current sources of cordycepin primarily involve extraction from fruiting bodies or isolation from liquid fermentation using C. militaris, presenting challenges such as low production intensity, complex separation and purification systems, and high production costs, limiting industrial feasibility. Recent advancements have witnessed the utilization of various fungal chassis cells to successfully engineer heterologous biosynthetic platforms for cordycepin, such as Saccharomyces cerevisiae and unconventional yeasts, offering advantages of high yield, short fermentation cycles, and a broad substrate spectrum. This mini review summarizes the biosynthetic pathways of cordycepin and focused on the comparison of the characteristics, advantages, current performance and prospects for the microbial cell factories, analyzing potential targets for metabolic pathway modification and giving strategies in both genetic engineering and process engineering to enhance production intensity. The mini review particularly emphasizes the crucial role of chassis cell stress tolerance to the toxic product in determining cordycepin yield and highlights the urgent need for high-throughput screening methods for high-yield strains.
Cordycepin (3′-deoxyadenosine) is a unique adenosine analog first identified by Cunningham et al. (1950) in the fermentation broth of the medicinal fungus Cordyceps militaris. As a valuable natural compound, cordycepin exhibits a range of biological functions (Tuli et al., 2014; Radhi et al., 2021), including antitumor (Nakamura et al., 2015; Khan and Tania, 2023), anti-inflammatory (Tan et al., 2020), antiviral (Rabie, 2022), immunomodulatory activities (Lee et al., 2020). It also helps alleviate symptoms of diabetes (Shin et al., 2009) and hyperlipidemia (Guo et al., 2010), showing broad prospects for drug development and health products (Kunhorm et al., 2019; Yang et al., 2020). To relieve the rapid deamination of cordycepin in vivo, cordycepin derivatives with N6 protection (Schwenzer et al., 2021; Cui et al., 2024) have been developed, and the ProTide NUC-7738, featuring a protective phosphoramidate cap, has overcoming cellular resistance (Schwenzer et al., 2021) and is currently in Phase II clinical trials. Cordycepin directly inhibits cancer cell proliferation and disrupts the tumor microenvironment (Khan and Tania, 2020; Chen et al., 2024), making it suitable for combination therapies (Liao et al., 2020). Additionally, cordycepin exhibits antibacterial (Jiang et al., 2019), insecticidal (Woolley et al., 2020), and allelopathic effects (Quy et al., 2019) comparable to glyphosate in dicotyledonous plants, indicating potential agricultural applications. However, research on the mechanisms underlying cordycepin’s activity has predominantly focused on mammalian cells, involving pathways such as PI3K/Akt/mTOR, MAPK and AMPK signaling (Radhi et al., 2021), with limited data on microbial or plant systems. Phosphorylation of cordycepin (CoTP) (Hawley et al., 2020) plays a central role in its activity.
Cordycepin can be obtained via chemical synthesis and biological methods (Wang et al., 2022). Chemical synthesis faces challenges of efficiency, cost, precursor availability, and environmental concerns (Hansske and Robins, 1985; Aman et al., 2000), making it unsuitable for industrial applications. Biological methods mainly involve using naturally cordycepin-synthesizing microorganisms or artificially constructed microbial cell factories through solid or liquid fermentation (Duan X. et al., 2023). Fungi with natural cordycepin biosynthesis pathways include C. militaris (Xia et al., 2017), Ophiocordyceps sinensis (Xiang et al., 2014), Cordyceps kyushuensis (Zhao et al., 2019), Cordyceps cicadae (Liu et al., 2018), Aspergillus nidulans (Xia et al., 2017), and Irpex lacteus (Liu et al., 2022). These fungi employ various fermentation techniques, such as solid-state fermentation, static fermentation, and submerged fermentation. Previous review articles have analyzed the evolutionary relationships among these fungi and compared their yields (Wang et al., 2022; Duan X. et al., 2023). Currently, extraction from C. militaris fruiting bodies or fermentation broth remains the primary source of cordycepin.
Although C. militaris exhibits the highest fermentation yield among known natural cordycepin hosts (Duan X. et al., 2023), its long fermentation cycle results in low production intensity. Solid and surface fermentation typically require 45–60 days, and submerged liquid fermentation takes at least 15–20 days (Yang et al., 2020). Additionally, the strains are highly unstable, facing issues such as strain degeneration (e.g., abnormal color change, failure to form fruiting bodies) (Lou et al., 2019) during fruiting body cultivation and complex environmental responses during liquid fermentation. Factors like light, temperature, oxygen supply, redox state, and metal ions significantly affect yield, leading to substantial batch-to-batch variation (Zhao et al., 2020; Wang et al., 2022). Consequently, with the development of synthetic biology, alternative microbial cell factories such as Saccharomyces cerevisiae (Huo et al., 2021; Wang et al., 2024), unconventional yeasts Komagataella phaffii (Tan et al., 2023) and Yarrowia lipolytica (Duan et al., 2022; Song et al., 2023), and Aspergillus oryzae (Jeennor et al., 2023) have been developed in recent years for cordycepin production (Table 1). Some of these engineered strains surpass natural producers in terms of production intensity and give simpler downstream purification due to their defined culture media. Given the toxicity of cordycepin to host cells, improving cell tolerance to cordycepin stress and elucidating its underlying mechanisms are crucial for constructing efficient cell factories.
This review will compare and analyze the characteristics and advantages of different chassis cells cordycepin factories, discuss key targets affecting cordycepin yield based on elucidated biosynthetic pathways, and propose feasible tools and strategies for simultaneously enhancing stress tolerance to cordycepin and the yield.
The genome of C. militaris was sequenced in 2011 (Zheng et al., 2011). Subsequently, Xia et al. (2017) identified the key gene cluster responsible for cordycepin biosynthesis through comparative genomic analysis with A. nidulans. In their work, this gene cluster, designated as Cns1-4, proved pivotal in the biosynthetic pathway of cordycepin (Figure 1), whereas 3′-AMP serves as the substrate of dephosphorylation by Cns2, a metal-dependent phosphohydrolase, to form 2′-carbonyl-3′-deoxyadenosine (2′-C-3′-dA). This intermediate is then converted to cordycepin through a redox reaction catalyzed by Cns1, an oxidoreductase, and 3′-AMP can be produced from adenosine via the nucleotide kinase domain (NK) at the N-terminus of Cns3 or from 2′,3′-cAMP, a product of mRNA degradation that is ubiquitously present in various cells (Wongsa et al., 2020). Therefore, theoretically, the overexpression of Cns1 and Cns2 in a host cell can utilize endogenous 3′-AMP to achieve heterologous cordycepin production. The C-terminal HisG domain of Cns3 catalyzes the conversion of adenosine to pentostatin (PTN), an adenosine deaminase inhibitor that prevents the deamination and subsequent inactivation of cordycepin. Cns4, an ABC transporter, exports PTN extracellularly. This mechanism ensures that when intracellular concentrations of cordycepin become excessively high, cordycepin can be converted to the non-toxic 3′-deoxyinosine (3′-dI) form, thereby protecting the cells. This system is referred to as the Protector-Protégé feedback mechanism. Additionally, Xia et al. (2017) also found that Cns1 and Cns2 could form a functional protein complex localized to lipid droplets. Later, similar gene clusters with analogous functions to Cns1-4 were also identified in C. kyushuensis (designated as ck1-4) (Zhao et al., 2019) and C. cicadae (Liu et al., 2018).
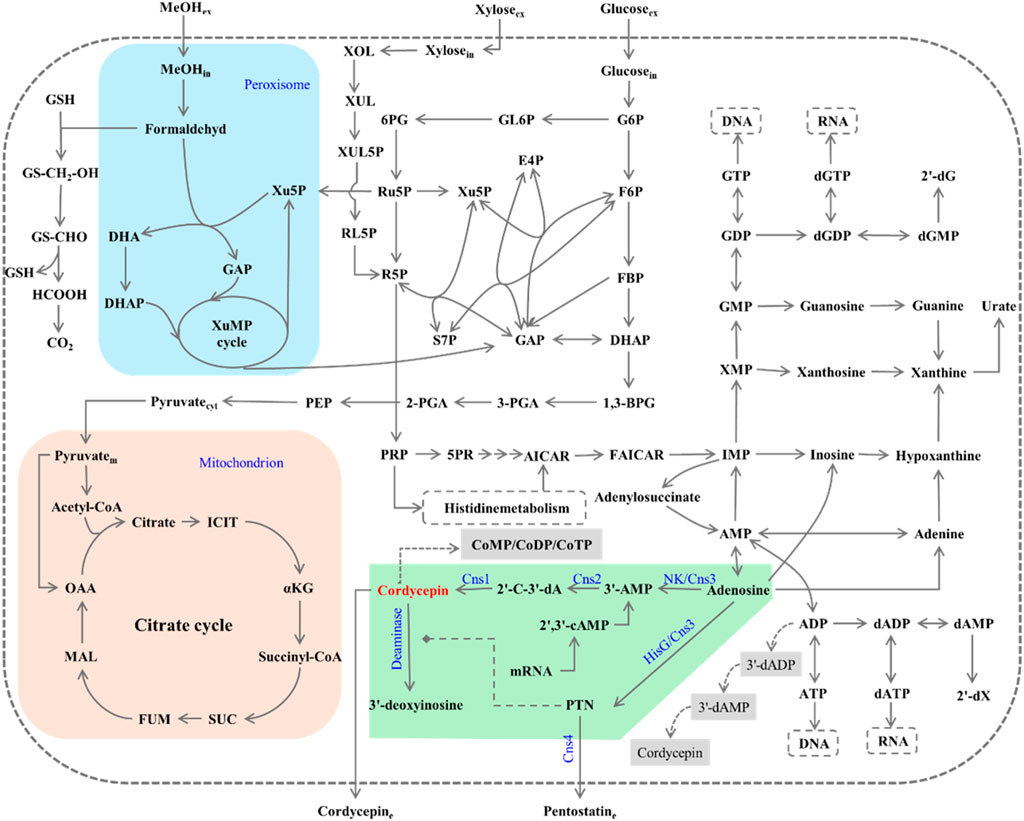
Figure 1. The metabolic pathways related to the biosynthesis of cordycepin in Cordyceps militaris and engineered microorganisms. GSH, glutathione; GS-CH2-OH, S-(Hydroxymethyl)glutathione; GS-CHO, S-Formylglutathione; HCOOH, Formate; CO2, Carbon dioxide; DHA, Dihydroxyacetone; DHAP, Dihydroxyacetone phosphate; GAP, Glyceraldehyde 3-phosphate; Xu5P, xylulose-5-phosphate; Ru5P, Ribulose 5-phosphate; R5P, Ribose 5-phosphate; XOL, Xylitol; XUL, D-Xylulose; XUL5P, D-Xylulose 5-phosphate; RL5P, D-Ribulose 5-phosphate; 6PG, 6-Phospho gluconate; GL6P, Glucono-1,5-lactone 6-phosphate; G6P, Glucose 6-phosphate; F6P, Fructose 6-phosphate; FBP, Fructose-1,6-bisphosphatase I; E4P, Erythrose 4-phosphate; S7P, Sedoheptulose 7-phosphate; 1,3-BPG, Glycerate 1,3-diphosphate; 3-PGA, 3-phosphoglycerate; 2-PGA, 2-phosphoglycerate; PEP, Phosphoenolpyruvate; ICIT, Isocitrate; αKG, 2-Ketoglutaric acid; SUC, Succinate; FUM, Fumarate; MAL, Malate; OAA, Oxaloacetate; PRPP, 5-Phosphoribosyl diphosphate; 5PR, 5-Phosphoribosylamine; AICAR, 5′-Phospho-ribosyl-5-amino-4-imidazole carboxamide; FAICAR, 5′-Phosphoribosyl-5-formamido-4-imidazolecarboxamide; IMP, Inosine 5′-phosphate; XMP, Xanthosine 5′-phosphate; GMP, Guanosine 5′-phosphate; GDP, Guanosine 5′-diphosphate; GTP, Guanosine 5′-triphosphate; dGDP, 2′-Deoxyguanosine 5′-diphosphate; dGTP, 2′-Deoxyguanosine 5′-triphosphate; dGMP, 2′-Deoxyguanosine 5′-monophosphate; 2′-dG, 2′-Deoxyguanosine; AMP, Adenosine 5′-monophosphate; ADP, Adenosine 5′-diphosphate; ATP, Adenosine 5′-triphosphate; dADP, 2′-Deoxyadenosine 5′-diphosphate; dATP, 2′-Deoxyadenosine 5′-triphosphate; dAMP, 2′-Deoxyadenosine 5′-monophosphate; 2′-dX, 2′-Deoxyadenosine; 3′-dADP, 3′-Deoxyadenosine 5′-diphosphate; 3′-dAMP, 3′-Deoxyadenosine 5′-monophosphate; 3′-AMP, 3′-Adenylic acid; 2′-C-3′ -dA, 2′-carbonyl-3′ -deoxyadenosine; CNS1, Oxidordeuctase involved of cordycepin biosynthesis; CNS2, Metal dependent phosphohydrolase involved of cordycepin biosynthesis; 2′-3′-cAMP, 2′,3′-cyclic adenosine monophosphate; NK/Cns3, Cns3 contains an N-terminal nucleoside/nucleotide kinase; HisG/Cns3, Cns3 contains a C-terminal HisG family of ATP phosphoribosyltransferases; Cns4, Transporter proteins of Pentostatin; CoMP, Cordycepin monophosphate; CoDP, Cordycepin diphosphate; CoTP, Cordycepin triphosphate.
In contrast, no similar gene clusters have been identified in O. sinensis (Xia et al., 2017). Transcriptomic analysis suggests that O. sinensis may rely on an alternative reductive metabolic pathway for cordycepin biosynthesis (Xiang et al., 2014) (lower right corner in Figure 1): AMP is converted to ADP by adenylate kinase (ADEK), then reduced to 3′-dADP by ribonucleotide reductase (NRDJ), followed by dephosphorylation to 3′-dAMP by ADEK, and finally converted to cordycepin through a dephosphorylation reaction catalyzed by 5′-nucleotidase (NT5E). However, conclusive experimental evidence is still lacking.
Despite the development of various microbial chassis cells as cordycepin-producing cell factories, some already achieving high production levels, C. militaris remains irreplaceable for cordycepin production for several reasons. Firstly, C. militaris demonstrates excellent biosafety, can be artificially cultivated (Kontogiannatos et al., 2021), and has a broad substrate spectrum, utilizing not only silkworm pupae, but also rice, barley, and xylose (Wongsa et al., 2020) as carbon sources. In countries like China, it has a long history of use as both food and medicine (Cui, 2015), leading to high social acceptance. Notably, as early as 2009 C. militaris was announced as a kind of safe food by Chinese government, providing regulatory support for the application and promotion of cordycepin products from C. militaris, a benefit that cordycepin products from yeast cell factories currently lack. Additionally, in 2016, the CRISPR-edited, browning-resistant Agaricus bisporus was approved by the US Department of Agriculture (USDA) (Waltz, 2016), suggesting that other mushrooms, including C. militaris, could potentially receive authorization for genetic editing in the future.
Secondly, C. militaris contains various bioactive molecules such as cordyceps polysaccharides, D-mannitol, N6-(2-hydroxyethyl)-adenosine (HEA), and carotenoids, etc. (Wu et al., 2021). Cordycepin, its signature active substance, can synergize with these compounds to target multiple sites, enhancing health and therapeutic effects. Cordycepin is also a key indicator for grading the quality of C. militaris fruiting bodies in China. Therefore, breeding C. militaris strains to increase cordycepin yield is of importance.
However, due to the lack of genetic editing tools, previous research mainly focused on optimizing fermentation strategies (solid, static, and submerged fermentation) for C. militaris, as reviewed previously by Wang et al. (2022). Recently, with the complete elucidation of the C. militaris genome and advances in molecular biology techniques, breeding methods for C. militaris have diversified. These methods now include the development of gene editing toolkit (Chen et al., 2018; Chen et al., 2022; Meng et al., 2022) and metabolic pathway optimization (Zhang et al., 2022), genome shuffling (Wang et al., 2017), and multi-omics approaches to identify key metabolic nodes.
Chen et al. (2022) successfully developed the CRISPR-Cas9-TRAMA system, achieving precise multiple gene editing and large DNA fragment deletions. Unlike exogenously synthesized gRNA, this system synthesizes gRNA endogenously using tRNA and PtrpC promoters. The polyethylene glycol-mediated transformation (PMT) was found superior to Agrobacterium tumefaciens-mediated transformation (ATMT) for genetic transformation. The plasmid includes the AMA1 sequence (Meng et al., 2022) from A. nidulans Glasgow strains, making it an episomal plasmid that can be eliminated by selective removal in the culture medium. The study also found that C. militaris can repair DNA through non-homologous end joining (NHEJ) or homology-directed repair (HDR). Future efforts to enhance genetic editing and metabolic modification in C. militaris should focus on elucidating key components influencing NHEJ and HDR, identifying neutral sites for gene (cluster) integration and expression, discovering efficient constitutive or inducible promoters (Lyu et al., 2022) (including AI-predicted promoters), and developing abundant selection markers (Lou et al., 2021) with more efficient transformation/screening methods.
Genome shuffling (Biot-Pelletier and Martin, 2014), combined with traditional physical (such as plasma, proton beam irradiation or UV) or chemical (such as nitrosoguanidine) mutagenesis (Masuda et al., 2011; Zhang et al., 2020), and adaptive laboratory evolution (ALE) (Mavrommati et al., 2022) remain effective breeding methods, though they often yield uncertain results and require substantial labor. Current methods for detecting cordycepin primarily rely on HPLC or thin-layer chromatography. There is a lack of colorimetric or fluorescent labeling methods for high-throughput screening. Breakthroughs in this area could significantly enhance the efficiency of both rational and non-rational genetic engineering. One solution is to identify endogenous promoters able to linearly respond to cordycepin concentration as biosensors, converting different cordycepin yields into phenotypes such as growth rate variations which could easily be recognized. Besides, Omics research plays a crucial role in exploring cordycepin biosynthesis pathways, identifying key enzyme genes and metabolic nodes (Chen et al., 2020; Zhang et al., 2023; Chai et al., 2024).
Cordycepin was first biosynthesized in the model organism S. cerevisiae, establishing the initial yeast cell factory for heterologous cordycepin expression (Xia et al., 2017; Huo et al., 2021). It was demonstrated that expressing only Cns1 and Cns2 is sufficient for cordycepin heterologous expression, while the yield was merely 137.27 mg/L even after fermentation optimization (Huo et al., 2021). By overexpressing the cpdB gene (3′-cyclic-nucleotide 2′-phosphodiesterase/3′-nucleotidase) from Escherichia coli, which converts 2′, 3′-cAMP to 3′-AMP, cordycepin concentration was significantly increased (Wang et al., 2024). This indicates that 3′-AMP in S. cerevisiae is similarly to that in Cordyceps fungi, derived via RNA degradation pathways, and that Cns3 is not necessary for heterologous cordycepin production. This finding has also been validated in other yeast cell factories.
Subsequently, Wang et al. (2024) improved cordycepin yield to 725.16 mg/L through rational metabolic engineering. Their study specifically compared the catalytic efficiency of Cns1/2 homologs from the marine fugus Emericellopsis atlantica and found that codon-optimized EA1/2 from the marine fungus increased cordycepin titer by 18.6% compared to Cns1/2, which underscores the importance of enzyme activity in the yeast host for cordycepin yield and the potential for mining potential sources of cordycepin synthesis genes from microorganisms in unique environments or through enzyme evolution and screening.
In metabolic engineering, enhancing the pentose phosphate pathway and purine synthesis pathway genes, including ZWF1 (glucose-6-phosphate dehydrogenase), PRS4 (ribose phosphate diphosphokinase), and ADE4 (amidophosphoribosyl transferase), significantly increased cordycepin yield (Wang et al., 2024). Overexpressing Cns1-2 in S. cerevisiae also upregulated these genes at the transcriptional level (Huo et al., 2021), indicating that synthesis of PRPP (5′-phosphoribosyl diphosphate) and the diversion of carbon flux into purine metabolism are critical nodes for improving cordycepin synthesis, partially consistent with previous findings in C. militaris (Chen et al., 2020).
Notably, deleting the ADO1 gene significantly increased cordycepin yield (Wang et al., 2024). One possible explanation is that this deletion enhances host tolerance to cordycepin stress, providing a novel approach to increasing cell factory yields, suggesting the tolerance of the chassis cells to toxic products may determine yield limits. Cordycepin can be phosphorylated to cordycepin triphosphate (Hawley et al., 2020), which strongly inhibits RNA synthesis and causes cellular toxicity (Holbein et al., 2009). Deleting ADO1 may disrupt adenosine phosphorylation to AMP, potentially affecting phosphorylation levels and ratios of cordycepin (Wang et al., 2024). Further evidence, such as measuring cordycepin phosphorylation levels in yeast or identifying endogenous enzymes involved in cordycepin phosphorylation, could validate this hypothesis.
Non-conventional yeasts have recently emerged as effective cell factories for the production of various natural products (Rebello et al., 2018). Cordycepin-producing cell factories have been successfully developed in the non-conventional yeasts K. phaffii (Tan et al., 2023) and Y. lipolytica (Duan et al., 2022; Song et al., 2023). These systems utilize codon-optimized Cns1 and Cns2 genes derived from C. militaris, achieving yields of 2.68 g/L in shake flasks and 4,362.54 mg/L in fermenters, respectively.
In the construction and optimization of the Y. lipolytica cordycepin cell factory, various molecular biology strategies were employed (Duan et al., 2022; Duan X. Y. et al., 2023; Song et al., 2023), including multi-copy integration of the key synthesis enzymes Cns1-2 at the rDNA locus, promoter optimization, and different expression methods for key enzymes (linker/2A peptide/RIAD-RIDD protein scaffold). Typically, exogenous gene expression in engineered strains using free plasmids results in unstable yields due to uneven plasmid distribution during cell division, not mentioning the need for antibiotics to maintain plasmid presence (Friehs, 2004). In contrast, integration at loci such as rDNA (Le Dall et al., 1994), delta sequences (Sakai et al., 1990), or IS elements (Lee et al., 2016) could ensure both the stability and multi-copy integration.
To optimize metabolic pathways, several genes in Y. lipolytica were overexpressed including GLK1 (Glucokinase) and phosphoglucomutases (PGM1) to enhance the pentose phosphate pathway (PPP) and PRPP production (Duan X. Y. et al., 2023). Overexpression of adenylate succinate synthase (ADE12) and adenylosuccinate lyase (ADE13) upregulated the purine synthesis pathway. Furthermore, the overexpression of 3′-phosphoglycerate kinase (PGK) and pyruvate kinase (PK) effectively increased intracellular ATP levels, significantly boosting cordycepin production (Song et al., 2023). This indicates that PPP, purine synthesis pathways (affecting precursor supply for cordycepin synthesis), and ATP supply and consumption are crucial metabolic targets for cordycepin biosynthesis.
In K. phaffii, a two-stage fermentation process using glycerol/methanol was employed, with the inducible promoters AOX1p and FLD1p expressing Cns1 and Cns2 from C. militaris, respectively (Tan et al., 2023). This approach, achieving inducible expression of cordycepin in a cell factory for the first time, spatially separated the cell growth phase (glycerol) from the cordycepin production phase (methanol), avoiding too early accumulation of cordycepin that might hinder cell growth. Transcriptome analysis revealed downregulation of genes related to methanol assimilation and dissimilation, peroxisome biogenesis, and the pentose phosphate pathway, providing targets for further strain improvement. The broad substrate utilization spectrum of K. phaffii, especially its ability to utilize methanol as a sole carbon and energy source (Ata et al., 2021; Zha et al., 2023), positions it competitively in controlling cordycepin fermentation costs and downstream product purification.
As in Cordyceps militaris, the yield of cordycepin in yeast cell factories can also be increased through process engineering optimization. For example, the addition of adenine has been shown to significantly enhance cordycepin production in all reported yeast cell factories, whereas adding adenosine can only partially increase the yield in the engineered Y. lipolytica (Song et al., 2023). Furthermore, optimizing the type and concentration of carbon and nitrogen sources, as well as the C/N ratio, has been crucial for the engineered Y. lipolytica (Duan X. Y. et al., 2023). In the case of engineered K. phaffii, the initial pH and methanol concentration in flask fermentation have significant effects on the final productivity (Tan et al., 2023).
Based on a comparative analysis of three yeast cell factories, the following optimization strategies are proposed.
The superior cordycepin synthesis capabilities in Y. lipolytica and K. phaffii compared to S. cerevisiae are largely attributed to their greater tolerance to cordycepin stress. In Y. lipolytica, cordycepin concentrations up to 3.0 g/L (Duan et al., 2022) did not affect final biomass, revealing the importance of considering host stress tolerance when generating toxic products. Techniques of genome evolution (Huang C. et al., 2022) like Synthetic Chromosome Rearrangement and Modification by LoxP-mediated Evolution (SCRaMbLE) (Cheng et al., 2024) can potentially enhance both cordycepin stress tolerance and production yield. NHEJ-mediated genomic library construction in Y. lipolytica can also introduce numerous LoxP sites (Bai et al., 2021). Additionally, adaptive laboratory evolution (Fernandes et al., 2023) remains effective; however, mutants with improved cordycepin production ability lack visible phenotypes or growth-coupled traits, making them difficult to screen. Therefore, efficient screening methods based on high-throughput platforms are critical. Traditional methods relying on HPLC are too laborious and costly to efficiently identify targets from thousands of (or much more) mutants.
One widely implemented strategy in cell factories is the use of transcription factor (TF)-based biosensors. For instance, the fatty acid and phospholipid regulator FapR and its operator fapO from Bacillus subtilis have been extensively studied and applied in both prokaryotic and eukaryotic systems as a malonyl-CoA sensor (Zhang and Shi, 2021). The allosteric conformational changes in transcription factors (TFs) when binding a specific metabolite leads to their binding to or dissociation from the operator region, thus regulating transcription. Endogenous transcription factors that respond to cordycepin concentration can be identified through transcriptome data mining of the host cell. Additionally, artificial transcription factors can be developed using reported cordycepin-binding receptors (Nakamura et al., 2015). These artificial TFs can be fused with reporter gene modules (such as fluorescent proteins or antibiotic resistance genes) to establish intracellular cordycepin biosensors.
Adenosine cannot freely permeate biological membranes and its transport relies on selected protein carriers (Pastor-Anglada and Perez-Torras, 2018), whereas its structural analogue cordycepin has been reported to be abundantly present extracellularly in various existing cell factories, indicating the presence of endogenous cordycepin transport proteins. Future work should assess intracellular versus extracellular cordycepin distribution to identify and enhance expression of potential cordycepin export proteins.
Deamination is a detoxification mechanism used by cells to neutralize cordycepin (Chen et al., 2023), but in cell factory fermentation for cordycepin production, deamination should be avoided. The deamination issue observed in K. phaffii (Tan et al., 2023) suggests the presence of endogenous deaminases that recognize cordycepin, necessitating study on the mechanism and stringent fermentation control.
Given cordycepin’s toxicity, timely removal from the fermentation broth can alleviate product feedback inhibition and maintain cell activity. Coupled fermentation-separation systems, established in C. militaris (Guan et al., 2019), could be adapted for yeast, especially using self-flocculating yeasts to enhance efficiency. S. cerevisiae can achieve flocculation through the function of FLO genes (Verstrepen et al., 2003), and expressing the FLO1 gene from S. cerevisiae in K. phaffii has shown some degree of flocculation (Sae-Tang et al., 2023). Flocculation can also increase cell density, reinforcing high-density fermentation for yield improvement.
In C. militaris, Cns1/2 form complexes localized on lipid droplets (Xia et al., 2017), a phenomenon also observed in heterologous expression in S. cerevisiae (Wang et al., 2024). Although no such information was available for non-conventional yeasts, compartmentalization can mitigate toxicity of the product. Y. lipolytica’s natural advantage in lipid production (Lazar et al., 2018) might partially explain its high yield if Cns1-2 localizes on lipid droplets. K. phaffii, under methanol induction, efficiently expands peroxisomes, suggesting a potential for organelle-targeted enzyme localization via signal peptide fusion (Ye et al., 2024).
For substrate conversion, similar metabolic modules and key pathways are followed across host cells, necessitating PPP and purine synthesis pathway enhancement to improve 3′-AMP supply. Complex feedback inhibition of enzymes like PRPP synthetase could be addressed by enzyme mutation or introducing high-activity enzymes from other species (Huang Z. et al., 2022). ATP supply and consumption also require consideration. Inducible promoters can temporally separate growth and production phases. Tailored metabolic pathway optimization strategies should be designed for different substrates, like enhancing methanol metabolism in K. phaffii or incorporating synthetic, more efficient methanol utilization pathways to supplement with the current Xu-5-P cycle (Antoniewicz, 2019).
Synthetic pathways for cordycepin production have been successfully engineered in A. oryzae (Jeennor et al., 2023), where the integration of Cns1-2 from C. militaris under constitutive promoters resulted in a productivity of 564.64 ± 9.59 mg/L/day and 98% cordycepin secretion. Similar to C. militaris, supplementation with adenine, adenosine, or glycine could enhance production, with adenine being the most effective precursor. This research provides a food-grade platform for heterologous expression of cordycepin and accommodates a broader range of carbon sources.
It is noteworthy that the aforementioned hosts predominantly consist of eukaryotic cells, with limited progress observed in prokaryotic platforms. Despite advantages in growth rates and heterologous protein expression at a high titer, the complex structure of Cns1 hampers its soluble expression in prokaryotic microbes like E. coli (Xia et al., 2017) and B. subtilis (Duan et al., 2022).
In conclusion, the advancement of synthetic biology promises the development of more diverse chassis cells for cordycepin production, expanding its application scenarios. Non-conventional yeast cell factories hold great potential for achieving high yields and offer advantages in separation and purification processes following fermentation. Investigating the stress mechanisms of cordycepin in host cells and enhancing stress tolerance through genetic and process engineering is crucial for yield improvement. The task of establishing high-throughput screening platforms using biosensing systems for cordycepin production is urgent. Further research is needed to explore genetic elements responsible for cordycepin synthesis from broader sources, with mutation and screening facilitating compatibility with the chosen chassis cell.
XL: Writing–original draft, Visualization. RJ: Writing–original draft. SW: Conceptualization, Writing–original draft. CL: Writing–original draft. YX: Conceptualization, Funding acquisition, Writing–review and editing. SL: Conceptualization, Funding acquisition, Writing–review and editing. QL: Writing–original draft, Writing–review and editing, Conceptualization, Funding acquisition. LW: Conceptualization, Funding acquisition, Project administration, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Open Funding Project of Guangdong Provincial Key Laboratory of Silviculture, Protection and Utilization (SPU2023-07), Research Project of Applied Basic Research Program of Department of Science and Technology of Liaoning Province (2022JH2/101300137), and Dalian University Interdisciplinary Project (DLUXK-2-23-QN-006).
Authors YX and SL were employed by Dalian SEM Bio-Engineering Technology Co., Ltd., while QL and LW are conducting postdoctoral research at the postdoctoral workstation of Dalian SEM Bio-Engineering Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aman, S., Anderson, D. J., Connolly, T. J., Crittall, A. J., and Ji, G. (2000). From adenosine to 3’-deoxyadenosine: development and scale up. Org. Process. Res. Dev. 4 (6), 601–605. doi:10.1021/op000209x
Antoniewicz, M. R. (2019). Synthetic methylotrophy: strategies to assimilate methanol for growth and chemicals production. Curr. Opin. Biotechnol. 59, 165–174. doi:10.1016/j.copbio.2019.07.001
Ata, Ö., Ergün, B. G., Fickers, P., Heistinger, L., Mattanovich, D., Rebnegger, C., et al. (2021). What makes Komagataella phaffii non-conventional? FEMS Yeast Res. 21 (8), foab059. doi:10.1093/femsyr/foab059
Bai, Q., Cheng, S., Zhang, J., Li, M., Cao, Y., and Yuan, Y. (2021). Establishment of genomic library technology mediated by non-homologous end joining mechanism in Yarrowia lipolytica. Sci. China Life Sci. 64 (12), 2114–2128. doi:10.1007/s11427-020-1885-x
Biot-Pelletier, D., and Martin, V. J. (2014). Evolutionary engineering by genome shuffling. Appl. Microbiol. Biotechnol. 98 (9), 3877–3887. doi:10.1007/s00253-014-5616-8
Chai, L., Li, J., Guo, L., Zhang, S., Chen, F., Zhu, W., et al. (2024). Genomic and transcriptome analysis reveals the biosynthesis network of cordycepin in Cordyceps militaris. Genes 15 (5), 626. doi:10.3390/genes15050626
Chen, B. X., Wei, T., Xue, L. N., Zheng, Q. W., Ye, Z. W., Zou, Y., et al. (2020). Transcriptome analysis reveals the flexibility of cordycepin network in Cordyceps militaris activated by L-alanine addition. Front. Microbiol. 11, 577. doi:10.3389/fmicb.2020.00577
Chen, B. X., Wei, T., Ye, Z. W., Yun, F., Kang, L. Z., Tang, H. B., et al. (2018). Efficient CRISPR-Cas9 gene disruption system in edible-medicinal mushroom Cordyceps militaris. Front. Microbiol. 9, 1157. doi:10.3389/fmicb.2018.01157
Chen, B. X., Xue, L. N., Wei, T., Wang, N., Zhong, J. R., Ye, Z. W., et al. (2022). Multiplex gene precise editing and large DNA fragment deletion by the CRISPR-Cas9-TRAMA system in edible mushroom Cordyceps militaris. Microb. Biotechnol. 15 (12), 2982–2991. doi:10.1111/1751-7915.14147
Chen, M., Luo, J., Jiang, W., Chen, L., Miao, L., and Han, C. (2023). Cordycepin: a review of strategies to improve the bioavailability and efficacy. Phytother. Res. 37 (9), 3839–3858. doi:10.1002/ptr.7921
Chen, R., Feng, C., Chen, L., Zheng, X., Fang, W., Wu, S., et al. (2024). Single-cell RNA sequencing indicates cordycepin remodels the tumor immune microenvironment to enhance TIGIT blockade’s anti-tumor effect in colon cancer. Int. Immunopharmacol. 126, 111268. doi:10.1016/j.intimp.2023.111268
Cheng, L., Zhao, S., Li, T., Hou, S., Luo, Z., Xu, J., et al. (2024). Large-scale genomic rearrangements boost SCRaMbLE in Saccharomyces cerevisiae. Nat. Commun. 15 (1), 770. doi:10.1038/s41467-023-44511-5
Cui, J. (2015). Biotechnological production and applications of Cordyceps militaris, a valued traditional Chinese medicine. Crit. Rev. Biotechnol. 35 (4), 475–484. doi:10.3109/07388551.2014.900604
Cui, L., Zhao, L., Shen, G., Yu, D., Yuan, T., Zhang, Y., et al. (2024). Antitumor mechanism and therapeutic potential of cordycepin derivatives. Molecules 29 (2), 483. doi:10.3390/molecules29020483
Cunningham, K. G., Manson, W., Spring, F. S., and Hutchinson, S. A. (1950). Cordycepin, a metabolic product isolated from cultures of Cordyceps militaris (Linn.) Link. Nature 166 (4231), 949. doi:10.1038/166949a0
Duan, X., Yang, H., Wang, C., Liu, H., Lu, X., and Tian, Y. (2023a). Microbial synthesis of cordycepin, current systems and future perspectives. Trends Food Sci. Technol. 132, 162–170. doi:10.1016/j.tifs.2023.01.006
Duan, X. Y., Liu, H. H., Song, L. P., Wang, C., Yang, H., Lu, X. Y., et al. (2023b). Efficient production of cordycepin by engineered Yarrowia lipolytica from agro-industrial residues. Bioresour. Technol. 377, 128964. doi:10.1016/j.biortech.2023.128964
Duan, X. Y., Tian, Y., Song, Z. Q., Song, L. P., Lin, W. B., Wang, C., et al. (2022). High-level de novo biosynthesis of cordycepin by systems metabolic engineering in Yarrowia lipolytica. Bioresour. Technol. 363, 127862. doi:10.1016/j.biortech.2022.127862
Fernandes, T., Osório, C., Sousa, M. J., and Franco-Duarte, R. (2023). Contributions of adaptive laboratory evolution towards the enhancement of the biotechnological potential of non-conventional yeast species. J. Fungi 9 (2), 186. doi:10.3390/jof9020186
Friehs, K. (2004). Plasmid copy number and plasmid stability. Adv. Biochem. Eng. Biotechnol. 86, 47–82. doi:10.1007/b12440
Guan, H., Li, H., Li, Q., Liu, B., Wang, J., and Wang, L. J. C. J. (2019). Efficient production of cordycepin during submerged liquid fermentation by Cordyceps militaris coupled with macroporous resin adsorption. CIESC J. 70 (7), 2675–2683. doi:10.11949/0438-1157.20190076
Guo, P., Kai, Q., Gao, J., Lian, Z. Q., Wu, C. M., Wu, C. A., et al. (2010). Cordycepin prevents hyperlipidemia in hamsters fed a high-fat diet via activation of AMP-activated protein kinase. J. Pharmacol. Sci. 113 (4), 395–403. doi:10.1254/jphs.10041fp
Hansske, F., and Robins, M. J. J. T. l. (1985). Regiospecific and stereoselective conversion of ribonucleosides to 3’-deoxynucleosides. A high yield three-stage synthesis of cordycepin from adenosine. Tetrahedron Lett. 26 (36), 4295–4298. doi:10.1016/s0040-4039(00)98716-1
Hawley, S. A., Ross, F. A., Russell, F. M., Atrih, A., Lamont, D. J., and Hardie, D. G. (2020). Mechanism of activation of AMPK by cordycepin. Cell. Chem. Biol. 27 (2), 214–222.e4. doi:10.1016/j.chembiol.2020.01.004
Holbein, S., Wengi, A., Decourty, L., Freimoser, F. M., Jacquier, A., and Dichtl, B. (2009). Cordycepin interferes with 3' end formation in yeast independently of its potential to terminate RNA chain elongation. RNA 15 (5), 837–849. doi:10.1261/rna.1458909
Huang, C., Wang, C., and Luo, Y. (2022a). Research progress of pathway and genome evolution in microbes. Synth. Syst. Biotechnol. 7 (1), 648–656. doi:10.1016/j.synbio.2022.01.004
Huang, Z., Li, N., Yu, S., Zhang, W., Zhang, T., and Zhou, J. (2022b). Systematic engineering of Escherichia coli for efficient production of nicotinamide mononucleotide from nicotinamide. ACS Synth. Biol. 11 (9), 2979–2988. doi:10.1021/acssynbio.2c00100
Huo, C., Li, H., Li, Q., Wang, J., Li, C., and Wang, L. (2021). Construction and optimization of cordycepin-producing Saccharomyces cerevisiae. Chin. J. Biotechnol. 37 (9), 3334–3347. doi:10.13345/j.cjb.200738
Jeennor, S., Anantayanon, J., Panchanawaporn, S., Chutrakul, C., Vongsangnak, W., and Laoteng, K. (2023). Efficient de novo production of bioactive cordycepin by Aspergillus oryzae using a food-grade expression platform. Microb. Cell. Fact. 22 (1), 253. doi:10.1186/s12934-023-02261-5
Jiang, Q., Lou, Z., Wang, H., and Chen, C. (2019). Antimicrobial effect and proposed action mechanism of cordycepin against Escherichia coli and Bacillus subtilis. J. Microbiol. 57 (4), 288–297. doi:10.1007/s12275-019-8113-z
Khan, M. A., and Tania, M. (2020). Cordycepin in Anticancer research: molecular mechanism of therapeutic effects. Curr. Med. Chem. 27 (6), 983–996. doi:10.2174/0929867325666181001105749
Khan, M. A., and Tania, M. (2023). Cordycepin and kinase inhibition in cancer. Drug Discov. Today 28 (3), 103481. doi:10.1016/j.drudis.2022.103481
Kontogiannatos, D., Koutrotsios, G., Xekalaki, S., and Zervakis, G. I. (2021). Biomass and cordycepin production by the medicinal mushroom Cordyceps militaris-a review of various aspects and recent trends towards the exploitation of a valuable fungus. J. Fungi 7 (11), 986. doi:10.3390/jof7110986
Kunhorm, P., Chaicharoenaudomrung, N., and Noisa, P. (2019). Enrichment of cordycepin for cosmeceutical applications: culture systems and strategies. Appl. Microbiol. Biotechnol. 103 (4), 1681–1691. doi:10.1007/s00253-019-09623-3
Lazar, Z., Liu, N., and Stephanopoulos, G. (2018). Holistic approaches in lipid production by Yarrowia lipolytica. Trends Biotechnol. 36 (11), 1157–1170. doi:10.1016/j.tibtech.2018.06.007
Le Dall, M. T., Nicaud, J. M., and Gaillardin, C. (1994). Multiple-copy integration in the yeast Yarrowia lipolytica. Curr. Genet. 26 (1), 38–44. doi:10.1007/bf00326302
Lee, C. T., Huang, K. S., Shaw, J. F., Chen, J. R., Kuo, W. S., Shen, G., et al. (2020). Trends in the immunomodulatory effects of Cordyceps militaris: total extracts, polysaccharides and cordycepin. Front. Pharmacol. 11, 575704. doi:10.3389/fphar.2020.575704
Lee, H., Doak, T. G., Popodi, E., Foster, P. L., and Tang, H. (2016). Insertion sequence-caused large-scale rearrangements in the genome of Escherichia coli. Nucleic Acids Res. Nucleic Acids Res. 44 (15), 7109–7119. doi:10.1093/nar/gkw647
Liao, X., Tao, L., Guo, W., Wu, Z. X., Du, H., Wang, J., et al. (2020). Combination of cordycepin and apatinib synergistically inhibits nsclc cells by down-regulating VEGF/PI3K/Akt signaling pathway. Front. Oncol. 10, 1732. doi:10.3389/fonc.2020.01732
Liu, T., Liu, Z., Yao, X., Huang, Y., Qu, Q., Shi, X., et al. (2018). Identification of cordycepin biosynthesis-related genes through de novo transcriptome assembly and analysis in Cordyceps cicadae. R. Soc. Open Sci. 5 (12), 181247. doi:10.1098/rsos.181247
Liu, Z., Leng, G., Wen, J., Deng, G., and Jiang, J. (2022). Cordycepin production by a novel endophytic fungus Irpex lacteus CHG05 isolated from Cordyceps hawkesii Gray. Folia Microbiol. 67 (6), 851–860. doi:10.1007/s12223-022-00981-6
Lou, H., Lin, J., Guo, L., Wang, X., Tian, S., Liu, C., et al. (2019). Advances in research on Cordyceps militaris degeneration. Appl. Microbiol. Biotechnol. 103 (19), 7835–7841. doi:10.1007/s00253-019-10074-z
Lou, H., Zhao, Y., Zhao, R., Ye, Z., Lin, J., and Guo, L. (2021). Screening and functional verification of selectable marker genes for Cordyceps militaris. J. Food Qual. 2021, 1–8. doi:10.1155/2021/6687768
Lyu, M., Zeng, J., Zhou, Y., Zhang, T., Wang, A., Ma, J., et al. (2022). Overlapping promoter library designed for rational heterogenous expression in Cordyceps militaris. Microb. Cell. Fact. 21 (1), 107. doi:10.1186/s12934-022-01826-0
Masuda, M., Das, S. K., Fujihara, S., Hatashita, M., and Sakurai, A. (2011). Production of cordycepin by a repeated batch culture of a Cordyceps militaris mutant obtained by proton beam irradiation. J. Biosci. Bioeng. 111 (1), 55–60. doi:10.1016/j.jbiosc.2010.08.018
Mavrommati, M., Daskalaki, A., Papanikolaou, S., and Aggelis, G. (2022). Adaptive laboratory evolution principles and applications in industrial biotechnology. Biotechnol. Adv. 54, 107795. doi:10.1016/j.biotechadv.2021.107795
Meng, G., Wang, X., Liu, M., Wang, F., Liu, Q., and Dong, C. (2022). Efficient CRISPR/Cas9 system based on autonomously replicating plasmid with an AMA1 sequence and precisely targeted gene deletion in the edible fungus, Cordyceps militaris. Microb. Biotechnol. 15 (10), 2594–2606. doi:10.1111/1751-7915.14107
Nakamura, K., Shinozuka, K., and Yoshikawa, N. (2015). Anticancer and antimetastatic effects of cordycepin, an active component of Cordyceps sinensis. J. Pharmacol. Sci. 127 (1), 53–56. doi:10.1016/j.jphs.2014.09.001
Pastor-Anglada, M., and Pérez-Torras, S. (2018). Who is who in adenosine transport. Front. Pharmacol. 9, 627. doi:10.3389/fphar.2018.00627
Quy, T. N., Xuan, T. D., Andriana, Y., Tran, H. D., Khanh, T. D., and Teschke, R. (2019). Cordycepin isolated from Cordyceps militaris: its newly discovered herbicidal property and potential plant-based novel alternative to glyphosate. Molecules 24 (16), 2901. doi:10.3390/molecules24162901
Rabie, A. M. (2022). Potent inhibitory activities of the adenosine analogue cordycepin on SARS-CoV-2 replication. ACS Omega 7 (3), 2960–2969. doi:10.1021/acsomega.1c05998
Radhi, M., Ashraf, S., Lawrence, S., Tranholm, A. A., Wellham, P. A. D., Hafeez, A., et al. (2021). A systematic review of the biological effects of cordycepin. Molecules 26 (19), 5886. doi:10.3390/molecules26195886
Rebello, S., Abraham, A., Madhavan, A., Sindhu, R., Binod, P., Karthika Bahuleyan, A., et al. (2018). Non-conventional yeast cell factories for sustainable bioprocesses. FEMS Microbiol. Lett. 365 (21). doi:10.1093/femsle/fny222
Sae-Tang, K., Bumrungtham, P., Mhuantong, W., Champreda, V., Tanapongpipat, S., Zhao, X. Q., et al. (2023). Engineering flocculation for improved tolerance and production of D-lactic acid in Pichia pastoris. J. Fungi 9 (4), 409. doi:10.3390/jof9040409
Sakai, A., Shimizu, Y., and Hishinuma, F. (1990). Integration of heterologous genes into the chromosome of Saccharomyces cerevisiae using a delta sequence of yeast retrotransposon Ty. Appl. Microbiol. Biotechnol. 33 (3), 302–306. doi:10.1007/bf00164526
Schwenzer, H., De Zan, E., Elshani, M., van Stiphout, R., Kudsy, M., Morris, J., et al. (2021). The novel nucleoside analogue ProTide NUC-7738 overcomes cancer resistance mechanisms in vitro and in a first-in-human phase I clinical trial. Clin. Cancer Res. 27 (23), 6500–6513. doi:10.1158/1078-0432.Ccr-21-1652
Shin, S., Lee, S., Kwon, J., Moon, S., Lee, S., Lee, C. K., et al. (2009). Cordycepin suppresses expression of diabetes regulating genes by inhibition of lipopolysaccharide-induced inflammation in macrophages. Immune Netw. 9 (3), 98–105. doi:10.4110/in.2009.9.3.98
Song, Z., Lin, W., Duan, X., Song, L., Wang, C., Yang, H., et al. (2023). Increased cordycepin production in Yarrowia lipolytica using combinatorial metabolic engineering strategies. ACS Synth. Biol. 12 (3), 780–787. doi:10.1021/acssynbio.2c00570
Tan, H., Wang, L., Wang, H., Cheng, Y., Li, X., Wan, H., et al. (2023). Engineering Komagataella phaffii to biosynthesize cordycepin from methanol which drives global metabolic alterations at the transcription level. Synth. Syst. Biotechnol. 8 (2), 242–252. doi:10.1016/j.synbio.2023.03.003
Tan, L., Song, X., Ren, Y., Wang, M., Guo, C., Guo, D., et al. (2020). Anti-inflammatory effects of cordycepin: a review. Phytother. Res. 35, 1284–1297. doi:10.1002/ptr.6890
Tuli, H. S., Sandhu, S. S., and Sharma, A. K. (2014). Pharmacological and therapeutic potential of Cordyceps with special reference to cordycepin. 3 Biotech. 4 (1), 1–12. doi:10.1007/s13205-013-0121-9
Verstrepen, K. J., Derdelinckx, G., Verachtert, H., and Delvaux, F. R. (2003). Yeast flocculation: what brewers should know. Appl. Microbiol. Biotechnol. 61 (3), 197–205. doi:10.1007/s00253-002-1200-8
Waltz, E. (2016). Gene-edited CRISPR mushroom escapes US regulation. Nature 532 (7599), 293. doi:10.1038/nature.2016.19754
Wang, H. B., Fu, X. M., Zuo, X. R., Zhang, C. B., and Lu, W. Y. (2024). Overproduction of cordycepin in Saccharomyces cerevisiae by cordycepin synthase screening and metabolic engineering. AIChE J. 318361 (10), 3103–3113. doi:10.1002/aic.18361
Wang, L., Yan, H., Zeng, B., and Hu, Z. (2022). Research progress on cordycepin synthesis and methods for enhancement of cordycepin production in Cordyceps militaris. Bioengineering 9 (2), 69. doi:10.3390/bioengineering9020069
Wang, Y., Zhang, G., Zhao, X., and Ling, J. (2017). Genome shuffling improved the nucleosides production in Cordyceps kyushuensis. J. Biotechnol. 260, 42–47. doi:10.1016/j.jbiotec.2017.08.021
Wongsa, B., Raethong, N., Chumnanpuen, P., Wong-Ekkabut, J., Laoteng, K., and Vongsangnak, W. (2020). Alternative metabolic routes in channeling xylose to cordycepin production of Cordyceps militaris identified by comparative transcriptome analysis. Genomics 112 (1), 629–636. doi:10.1016/j.ygeno.2019.04.015
Woolley, V. C., Teakle, G. R., Prince, G., de Moor, C. H., and Chandler, D. (2020). Cordycepin, a metabolite of Cordyceps militaris, reduces immune-related gene expression in insects. J. Invertebr. Pathol. 177, 107480. doi:10.1016/j.jip.2020.107480
Wu, X., Wu, T., Huang, A., Shen, Y., Zhang, X., Song, W., et al. (2021). New insights into the biosynthesis of typical bioactive components in the traditional Chinese medicinal fungus Cordyceps militaris. Front. Bioeng. Biotechnol. 9, 801721. doi:10.3389/fbioe.2021.801721
Xia, Y., Luo, F., Shang, Y., Chen, P., Lu, Y., and Wang, C. (2017). Fungal cordycepin biosynthesis is coupled with the production of the safeguard molecule pentostatin. Cell. Chem. Biol. 24 (12), 1479–1489.e4. doi:10.1016/j.chembiol.2017.09.001
Xiang, L., Li, Y., Zhu, Y., Luo, H., Li, C., Xu, X., et al. (2014). Transcriptome analysis of the Ophiocordyceps sinensis fruiting body reveals putative genes involved in fruiting body development and cordycepin biosynthesis. Genomics 103 (1), 154–159. doi:10.1016/j.ygeno.2014.01.002
Yang, L., Li, G., Chai, Z., Gong, Q., and Guo, J. (2020). Synthesis of cordycepin: current scenario and future perspectives. Fungal Genet. Biol. 143, 103431. doi:10.1016/j.fgb.2020.103431
Ye, C., Hong, H., Gao, J., Li, M., Gou, Y., Gao, D., et al. (2024). Characterization and engineering of peroxisome targeting sequences for compartmentalization engineering in Pichia pastoris. Biotechnol. Bioeng. 121 (7), 2091–2105. doi:10.1002/bit.28706
Zha, J., Liu, D., Ren, J., Liu, Z., and Wu, X. (2023). Advances in metabolic engineering of Pichia pastoris strains as powerful cell factories. J. Fungi 9 (10), 1027. doi:10.3390/jof9101027
Zhang, H., Chen, P., Xu, L., Xu, D., Hu, W., Cheng, Y., et al. (2022). Construction of cordycepin high-production strain and optimization of culture conditions. Curr. Microbiol. 80 (1), 12. doi:10.1007/s00284-022-03110-1
Zhang, H., Deng, L., Zhang, Z., Guan, Y., Li, B., Yang, J., et al. (2020). Enhanced cordycepin production in caterpillar medicinal mushroom, Cordyceps militaris (Ascomycetes), mutated by a multifunctional plasma mutagenesis system. Int. J. Med. Mushrooms 22 (12), 1147–1159. doi:10.1615/IntJMedMushrooms.2020037153
Zhang, H., Yang, J., Luo, S., Liu, L., Yang, G., Gao, B., et al. (2023). A novel complementary pathway of cordycepin biosynthesis in Cordyceps militaris. Int. Microbiol. doi:10.1007/s10123-023-00448-9
Zhang, Y., and Shi, S. (2021). Transcription factor-based biosensor for dynamic control in yeast for natural product synthesis. Front. Bioeng. Biotechnol. 9, 635265. doi:10.3389/fbioe.2021.635265
Zhao, X., Li, Q., Liu, W., Guan, H., Li, C., Wang, J., et al. (2020). Advances in biosynthesis of cordycepin from Cordyceps militaris. Chin. J. Biotechnol. 36 (7), 1293–1304. doi:10.13345/j.cjb.190500
Zhao, X., Zhang, G., Li, C., and Ling, J. (2019). Cordycepin and pentostatin biosynthesis gene identified through transcriptome and proteomics analysis of Cordyceps kyushuensis Kob. Microbiol. Res. 218, 12–21. doi:10.1016/j.micres.2018.09.005
Keywords: cordycepin, Cordyceps militaris, biosynthesis, non-conventional yeast, microbial cell factory
Citation: Li X, Jiang R, Wang S, Li C, Xu Y, Li S, Li Q and Wang L (2024) Prospects for cordycepin biosynthesis in microbial cell factories. Front. Chem. Eng. 6:1446454. doi: 10.3389/fceng.2024.1446454
Received: 09 June 2024; Accepted: 12 July 2024;
Published: 30 July 2024.
Edited by:
Zhuangrong Huang, Bristol Myers Squibb, United StatesReviewed by:
Yuxin Liu, Sanofi Genzyme, United StatesCopyright © 2024 Li, Jiang, Wang, Li, Xu, Li, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Li, bGlxaWFuQGRsdS5lZHUuY24=; Liang Wang, d2FuZ2xpYW5nQGRscHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.